Graphical abstract
Keywords: Heavy metal(loid) resistant microbiota; Distribution pattern; Metal resistome; Biotic selection; Copper and silver; Zinc, lead and cadmium; Arsenic; Mercury
Abstract
Heavy metal(loid)s exert selective pressure on microbial communities and evolution of metal resistance determinants. Despite increasing knowledge concerning the impact of metal pollution on microbial community and ecological function, it is still a challenge to identify a consistent pattern of microbial community composition along gradients of elevated metal(loid)s in natural environments. Further, our current knowledge of the microbial metal resistome at the community level has been lagging behind compared to the state-of-the-art genetic profiling of bacterial metal resistance mechanisms in a pure culture system. This review provides an overview of the core metal resistant microbiome, development of metal resistance strategies, and potential factors driving the diversity and distribution of metal resistance determinants in natural environments. The impacts of biotic factors regulating the bacterial metal resistome are highlighted. We finally discuss the advances in multiple technologies, research challenges, and future directions to better understand the interface of the environmental microbiome with the metal resistome. This review aims to highlight the diversity and wide distribution of heavy metal(loid)s and their corresponding resistance determinants, helping to better understand the resistance strategy at the community level.
1. Introduction
Heavy metal(loid)s are natural constituents of the earth’s crust and persistent in the environment. Release of heavy metal(loid)s naturally occurs through weathering, geothermal activities, forest fires, and microbial activities. However, this process has been accelerated rapidly by anthropogenic activities, causing the increased global dispersion of heavy metal(loid)s thereby constituting a great threat for human and ecosystem health [1], [2]. The widespread heavy metal(loid) pollution is highly coincident with increasingly advanced civilizations in human history [2]. For instance, changes of lead in dated deposits were coincident with the prosperity and eclipse of the Roman Empire (over 2000 years ago), the Industrial Revolution (over 200 years ago), as well as the promotion and phase-out of leaded gasoline (over 50 years ago) [3], [4]. Similarly, elevated copper deposition in Greenland ice could be correlated to ancient mining and smelting activities during Roman and medieval times in Europe and China [5]. Nowadays, anthropogenic heavy metal(loid) emissions into the environment has exceeded the tolerable baseline of natural cycling in many ecosystems. Not surprisingly, urbanized areas with intensive mining and industrial activities are typical hot spots of heavy metal(loid) contamination. The antimicrobial properties of heavy metal(loid)s, such as silver, mercury, copper, zinc and arsenic, have long been appreciated before the discovery and widespread use of antibiotics [6], [7]. Some useful products, such as copper alloy, exhibit a high biocidal efficacy against a range of nosocomial pathogens, including methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococci, Escherichia coli O157: H7, Pseudomonas aeruginosa and SARS-CoV-2 [8], [9], [10], [11], [12]. Due to the high antimicrobial efficacy, heavy metal(loid)s-based antimicrobial agents or commercial products are broadly applied in public health, medical treatments, agriculture and livestock industry. Additionally, metal(loid)s are supplemented in feed for growth promotion and disease prevention. Over the last decades, elevated copper and zinc contents have been reported in livestock manures and sewage sludge worldwide [13], [14], [15], [16]. With manure/sewage-sludge application and irrigation with wastewater, thousands tons of heavy metal(loid)s enter and accumulate in soils, sediments and aquatic ecosystems, posing a global threat to agriculture products, ecosystems and human health [6], [17], [18].
Enriched heavy metal(loid)s in the environment, either released from natural sources or anthropogenic activities, constitute a great selective pressure for the development and spread of heavy metal(loid) resistance determinants. As a side effect, increased abundance and selection for heavy metal(loid) resistance also boosts the spread of antibiotic resistance through co-selection [19]. Antimicrobial heavy metal(loid)s target multiple cellular processes by inducing oxidative stress, destabilizing protein function, impairing the integrity of membranes, interfering with nutrient assimilation, and causing DNA damage [6]. The mechanisms leading to toxicity might differ and depend on the intrinsic properties of the individual metal. In some circumstances, different mixtures of metals can have joint toxic effects. To defend themselves from metal toxicity, microbes have evolved sophisticated mechanisms of adaption and resistance. To date, the genetic basis of microbial heavy metal(loid) resistance has been elucidated in detail from different perspectives [20], [21], [22], [23], [24]. Essentially, the diverse determinants maintaining heavy metal(loid) homeostasis can be divided into four categories: energy-dependent efflux (ATPase, RND, CDF family), enzymatic detoxification (redox and (de)methylation), intracellular sequestration, and reduction of uptake. Other unspecific mechanisms enhancing bacterial resistance to heavy metal(loid)s are also well documented. For example, biofilm formation protects microorganisms from toxic heavy metal(loid)s [25], [26]. Bacteria secret extracellular polymers or siderophores to trap heavy metal(loid)s, reduce the bioavailability, and alleviate stress [25], [27]. Today, studies of metal resistance mechanisms have facilitated the development of many fields such as bioremediation of metal-contaminated environments [28], [29], [30], and bio-mining of minerals from ores or effluents from industrial operations [31]. Additionally, it is beneficial to elucidate the increasing development of microbial resistance to heavy metal(loid)s and antibiotics, and to prevent failure when using antimicrobial products [32], [33].
Metal(loid)s are of importance for many biological functions such as respiration, photosynthesis, carbon and nitrogen cycling [34], [35], [36]; however, most microorganisms and ecological processes in the environment are vulnerable to heavy metal(loid) pollution [37]. The resistance and resilience of environmental microorganisms to heavy metal(loid) perturbation rely on many factors. For example, microbial intrinsic detoxification systems largely determine their survival in heavy metal(loid) polluted environment. Other (a)biotic factors influencing heavy metal(loid) toxicity and microbial fitness may also play a vital role in complex environments. Despite increasing concerns on the ecological effect of heavy metal(loid) exposure on the microbial community and function, several crucial questions on the core metal resistant microbiome are still not solved. The term “heavy metal(loid) resistant microbiome” contains the resistant microbiota, metal resistome, and their interactions with surrounding biotic and abiotic environments. The core heavy metal(loid) resistant microbiota is a suite of members shared among microbial consortia from an individual or a multiple-metal polluted environment. They are not necessarily found in every environment and resistant to all heavy metal(loid)s, but are fairly constant regardless of geographical region. Heavy metal(loid) resistome is termed as “the collection of all genes conferring metal resistance in a given environment, including both intrinsic genes and acquired resistance genes via horizontal gene transfer (HGT). This review chronicles the discovery and development of major microbial heavy metal(loid) resistance determinants, and discuss the prevailing knowledge gap between resistome studies based on individual isolates and the microbial community. By investigating previously published sequencing data, we summarized the profile of metal-resistant microbiota and resistome distributed in natural/managed ecosystems, and further interpreted the major biotic and abiotic factors driving the structure of the core metal resistant microbiome. The influence of heavy metal(loid)s on the animal-related microbiome is not included. Finally, we discussed methodological challenges and the future direction of heavy metal(loid) resistome.
2. Bacterial heavy metal(loid) resistome: From pure culture to microbiome
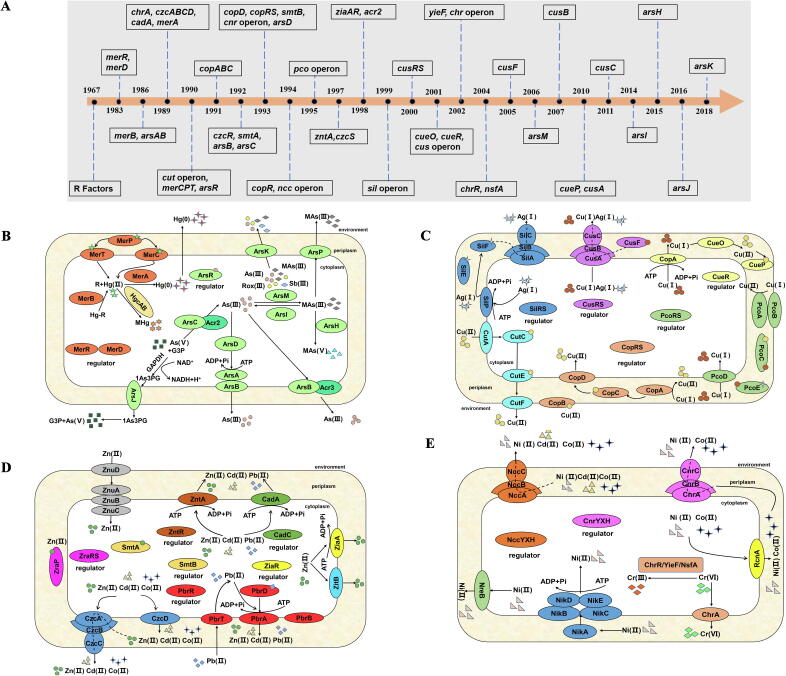
Investigation of bacterial heavy metal(loid) resistance mechanisms, which began using several prominent isolates, has been carried out for decades. To date, substantial efforts have been devoted to identify the genetic basis of heavy metal(loid) resistance, elaborate relevant biochemical pathways, and characterize the structural basis of critical components in heavy metal(loid) resistance determinants. Another related goal has been the elucidation of the evolution, distribution and biological function of these heavy metal(loid) resistance determinants across prokaryotes. These studies laid a solid foundation for the systemic understanding on how bacteria deal with toxic heavy metal(loid)s and maintain homeostasis. Here, we summarize the discovery of several crucial heavy metal(loid) resistance systems, including mercury, arsenic, copper, silver, zinc, cobalt, nickel and chromate (Fig. 1).
Fig. 1.
Summary of heavy metal(loid) resistance mechanisms. A. A timeline for highlighting the discovery of several important heavy metal(loid) resistance determinants. Related references are cited when elaborating resistance mechanism of each heavy metal(loid) below. B-E. Schematic diagram showing heavy metal(loid) resistance mechanisms in Gram-negative bacteria. All proteins are colored by operon. B. Mercury and arsenic resistance mechanism. The mer operon, which is regulated by MerR or MerD, confers bacterial mercury resistance [40], [48], [49], [50], [52]. Hg(II) enters the cell through transporters MerP, MerT and MerC [45], [46], [47], and is reduced by a mercuric reductase MerA [41], [42]. Methylation and demethylation of Hg(II) are mediated by HgcAB [53] and MerB [43], [198], respectively. As (III) combined with ArsR (repressor protein) triggers expression of the ars operon, which contributes to bacterial arsenic resistance [59], [61], [62], [63], [64], [65], [199], [200]. As(V) is reduced to As(III) by arsenate reductase ArsC/Acr2 [62]. As(III) is either pumped out directly via ArsB/Acr3/ArsK, or bound to the ArsD chaperone and delivered to the ArsAB ATP-dependent efflux pump. ArsM and ArsI are responsible for As(III) methylation and demethylation, respectively [69], [70]. Detoxification of MAs(III) includes oxidation of MAs(III) to less toxic MAs(V) by ArsH [71], or efflux via ArsP and ArsK [66], [67], [72]. ArsJ is responsible for organoarsenical efflux [68]. C. Copper and silver resistance mechanisms. The cut operon involves in copper uptake (CutA), delivery (CutC), intracellular storage (CutE), and efflux (CutF) [201], [202], [203]. Several mechanisms handle periplasmic copper detoxification, including the CusCFBA efflux system [83], [86], [87], [166], [204], [205], [206], a multicopper oxidase CueO, as well as two homologous CopABCDRS [73], [74], [75], [207], [208], [209], [210] and the plasmid-borne PcoABCDERS systems [77], [79], [80]. The P-type ATPase CopA is responsible for cytoplasmic Cu(I) efflux. The sil operon, which is regulated by a two-component regulator SilRS, mediates bacterial silver resistance [91]. D. Zinc, lead, cadmium and cobalt resistance mechanisms. Three types of transporters involves in Zn(II)/Cd(II)/Pb(II)/Co(II) efflux, including the RND transporter CzcCBA [92], [93], cation-translocating P-type ATPases ZntA, ZiaA and CadA [95], [96], [98], and CDF transporters ZitB or CzcD. SmtA and ZraP serve as metallothioneins involving in cytoplasmic and periplasmic Zn(II) binding, respectively [99], [211], [212]. The Znu system is a high-affinity Zn(II) uptake system. The pbr operon confers bacterial lead resistance [213], [214]. E. Nickel, cobalt, cadmium and chromium resistance mechanisms. The cnr operon and ncc operon export Ni(II)/Co(II)/Cd(II) out of the cell [94], [215]. A permease NreB and efflux protein RcnA mediate Ni(II) or Co(II) export [216], [217]. The nik operon is responsible for Ni(II) uptake [218], [219]. Cr detoxification includes Cr(VI) efflux via the ChrA transporter [103], [104], [105], and Cr(VI) reduction to less toxic Cr(III) through several reductases such as ChrR, YieF and NsfA [101], [102].
Mercury resistance was the first thoroughly studied bacterial heavy metal(loid) resistance system, which started from discovering mercurial resistance R-factors in clinical isolates and followed with the generation of mercury hypersensitivity mutants in the late 1960s-1970s [38], [39], [40]. Later, genes involved in mercury resistance were gradually elucidated, including mercuric reduction (merA) [41], [42], organomercurical cleavage (merB) [43], [44], mercuric ion transport (merP, merT, merC) [45], [46], [47], regulator (merR and merD) [40], [48], [49], [50], [51], [52], and mercury methylation (hgcA and hgcB) [53]. Fig. 1B shows the schematic representation of mercury resistance mechanism. Detailed information on mercury resistance was discussed in other excellent reviews [54], [55].
Arsenic biotransformation and transport is another well-studied resistance system, which was mainly conducted by Rosen and colleagues since 1981/1982 [56], [57]. The ars operon with many diverse variants is widely distributed in nearly all Prokaryotes to defend against ubiquitous arsenic in the environment [58]. Arsenic resistance mechanisms can be divided into inorganic and organic arsenic detoxification systems (Fig. 1B). The molecular mechanism of inorganic arsenic detoxification, including the typical arsRDABC operon [59], [60], [61], [62], [63], [64] and acr3 [65], was initially studied in several pioneer R plasmids. With the massive application of organoarsenic-containing herbicides and pesticides worldwide [1], organoarsenical detoxification pathways were soon identified in bacteria isolated from organoarsenic contaminated environments. The detoxification system is composed of ArsP responsible for Roxasone (Rox) and Monomethylarsonous acid (MMA(III)) efflux [66], [67], ArsJ conferring 1-arseno-3-phosphoglycerate (1As3PG) efflux [68], and enzymes responsible for biotransformations of organic arsenicals encoded by arsM, arsH and arsI [69], [70], [71]. The newly reported efflux transporter ArsK is responsible for As(III), Methylarsenite (MAs(III)) and Rox(III) resistance [72].
Copper is one of the essential but also toxic metal(loid)s for microorganisms. The intracellular copper homeostasis is strictly controlled via delicate mechanisms (Fig. 1C). The plasmid-born cop operon was initially identified in Pseudomonas syringae isolated from Californian tomato fields using copper as an antifungal agent. CopABCDRS in P. syringae is the first identified plasmid-encoded system involving the sequestration of copper in the periplasm and outer membrane [73], [74], [75]. Chromosomal homologs of copABCD were also identified in P. syringae, conferring an enhanced copper resistance [76]. Meanwhile, another plasmid-encoded pco operon was identified in E. coli strain isolated from the piggery, where copper was used as a feed additive in Australia [77]. The pco operon is co-located with the sil operon, constituting a larger operon with a high copper resistance [78]. The basic pco operon pcoABCDRS in E. coli is homologous to cop operon in P. syringae [77], [79], [80]. Additional cop genes, assembled as copVTMKNSRABCDIJGFLQHE, were later identified in Cupravidus metallidurans CH34 megaplasmid pMoL30, handling both periplasmic and cytoplasmic copper detoxification [81]. In addition to plasmid-encoded pco systems, there are three major chromosomal determinants responsible for copper resistance in E. coli. Copper-translocating P-type ATPase, named as CopA, is essential transporter for cytoplasmic Cu(I) efflux in E. coli [82]. Other independent copper resistant systems, including cueO and cus operon, are responsible for periplasmic copper detoxification. The multicopper oxidase CueO contains similar methionine-rich regions as present in PcoA in E. coli and CopA in P. syringae, and could oxidate the toxic Cu(I) into less toxic Cu(II) in the periplasm [83], [84], [85]. CusCFBARS is another important system for periplasmic copper detoxification under both aerobic and anaerobic conditions [83], [86], [87], [88]. Microorganisms lacking CusCFBA homologs, such as many strains of Salmonella and Yersinia, possess alternative CueP as functional substitutes [89], [90]. In the case of silver, the molecular basis of silver resistance, the sil operon, was well studied in Salmonella [91]. The sil operon is regulated by a two-component membrane sensor and regulator SilRS. It consists of two parallel efflux pumps: a specific Ag(I)-translocating P-type ATPase (SilP) and a cation/proton antiporter (SilCBA) with SilE serving as silver-specific periplasmic binding protein (Fig. 1C).
As is the case with copper, zinc, cobalt and nickel are also essential micronutrients required for microorganisms, but are toxic when in excess. CzcCBA (for cadmium, zinc and cobalt resistance) and CnrCBA (Co and Ni resistance) are two homologous divalent cation efflux systems, which were initially reported in Alcaligenes eutrophus (today Cupriavidus metallidurans CH34 plasmid pMOL30 [92], [93]. Ncc is the third divergent divalent cation resistance system for nickel, cadmium, and cobalt resistance [94]. Additionally, ZntA from E. coli and CadA from Staphylococcus aureus are two important cation-translocating P-type ATPases catalyzing the efflux of zinc, lead and cadmium [95], [96], [97]. Other systems, such as zinc exporter ZiaA in Synechocystis [98] and metallothionein SmtA [99], also contribute to zinc homeostasis (Fig. 1D).
Chromate is a highly toxic metal widespread in the environment. Bacterial chromate reduction could convert the soluble and toxic Cr(VI) to insoluble and less toxic Cr(III), which is vital for environmental bioremediation [100]. Chromate reduction is conducted by several reductases, such as ChrR, YieF, NfsA, and lipoyl dehydrogenase (LpDH) [101], [102] (Fig. 1E). ChrA is a chromate efflux transporter found in plasmids of P. aeruginosa [103] and C. metallidurans (formerly known as Ralstonia metallidurans, or A. eutrophus) [104], [105]. Additional chr determinants, including chrIBACEF on plasmid pMOL28 and chrBAF on the chromosome, were further identified in C. metallidurans [106]. These determinants are widespread on both plasmids and chromosomes.
The natural environment is composed of an enormous abundance and diversity of microorganisms with various physiological properties and differing tolerance to pollutants. When encountering heavy metal(loid) stress, the discrepancy in species adaption to heavy metal(loid)s may lead to a dramatic shift within natural populations and their ecological function. The interference and re-construction of the population will not only be accompanied with a succession of the microbiome, but will also affect the distribution and longevity of specific resistance genes in the environment. Elaborating the response and development of core resistant microbiomes and resistome under heavy metal(loid) selective pressure is essential to understand the ecological impact of heavy metal(loid)s on microbial succession and their potential environmental risks. However, compared to molecular mechanisms investigated in pure culture isolates, our knowledge on the microbial resistome to heavy metal(loid)s at the community level has lagged far behind. The fundamental mechanisms by which microbial populations adapted to heavy metal(loid) stress and their preference for resistance strategy in the natural environment remain virtually unknown. The knowledge gap could be partly due to the diverse microbes and methodological challenges, which will be discussed in the sections below.
3. The core heavy metal(loid) resistant microbiota
Microorganisms drive vital biogeochemical processes, including carbon turnover, nutrient cycling and pollutant transformation. However, they are highly sensitive to heavy metal(loid)s compared to animals and plants in the same environment [107]. In particular, prokaryotic populations, in terms of biomass, activity and diversity, are more vulnerable to heavy metal(loid) pollution than eukaryotes [108], [109]. Thus, microorganisms are frequently considered as potential ecological indicators for heavy metal(loid) pollution [110]. With advances in Next-Generation Sequencing (NGS), a growing number of studies have illustrated the impact of heavy metal(loid) pollution on microbial communities in many ecosystems [111], [112], [113], [114], [115]. However, our knowledge regarding community changes under heavy metal(loid) pollution is mainly from studies at a local scale; little is known about the core resistant microbiome at a scale across different ecosystems.
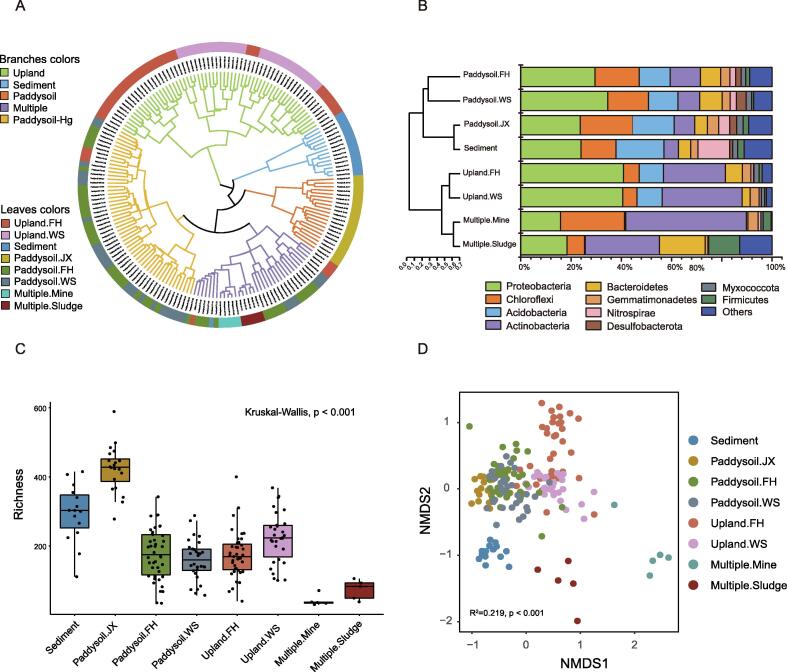
Here, we retrived metadata from published studies in the Web of Science database to give an overview on the profile of core heavy metal(loid) resistant microbiota and their distribution pattern. We only focus on the influence of heavy metal pollution on microbiota and resistome in natural environments. Our search terms included “microbial community” and “heavy metal”. Research articles were checked manually to remove duplicates, irrelevant articles, and articles without accession number. We use a bacterial dataset with V3–V4 hypervariable regions of the 16S rRNA gene to illustrate the preliminary profile of the core microbiota under heavy metal(loid) polluted environments (Fig. 2). Eventually, 186 samples from 5 independent research articles met the criteria and were selected for analysis (Table S1). Proteobacteria, Chloroflexi, Acidobacteria, Actinobacteria, Bacteroidetes, Gemmatimonadetes, Nitrospirae, Desulfobacterota, Myxococcota and Firmicutes were identified as the dominant phyla persistent in a variety of heavy metal(loid) contaminated environments. Proteobacteria was the most widespread and ubiquitous bacterial group in the environment. It contains both slow-growing oligotrophic and fast-growing copiotrophic taxa with extensive metabolic properties and wide niche breadth, enabling Proteobacteria to occupy a wide range of habitats [116]. Furthermore, bacteria belonging to Proteobacteria possess a variety of heavy metal(loid) resistance determinants. Metagenome analysis of river sediments with chronic heavy metal pollution identified the most variety and abundance of heavy metal(loid) resistance genes in Proteobacteria, followed by Actinobacteria and Bacteroidetes [117], [118]. With the capability of strong adaption and tolerance, Proteobacteria has been reported to be a predominant heavy metal(loid) resistant phylum in many polluted environments [119], [120], [121]. The importance of Proteobacteria as a core heavy metal(loid) resistant phylum is also confirmed in studies using isolation-based methods. By compiling literature related to heavy metal(loid) resistant isolates from various environments, we found over 66% of studied isolates belong to Proteobacteria, most of which were Gammaproteobacteria (52.84%) (Fig. 3). Acidobacteria is another prevalent population present in heavy metal(loid) polluted areas. As k-strategists, Acidobacteria species grow slowly, but are well adapted to acidic and oligotrophic niches due to their extensive metabolic versatility and acidic tolerance [122]. Therefore, acidification and reduced availability of nutrients in heavy metal(loid) polluted areas favor the survival of Acidobacteria. Other abundant phyla, such as Chloroflexi and Nitrospirae, are reportedly involved in nitrogen cycling in heavy metal(loid) polluted sites [123].
Fig. 2.
Profile of the core heavy metal(loid) resistant microbiome. A. Hierarchical cluster of samples retrieved from studies on heavy metal(loid) polluted environments. Cluster was analyzed based on the Bray-Curtis dissimilarity of relative abundance of amplicon sequence variants (ASVs) at the phylum level. Branches of the tree are colored according to habitat. Colors in the outer ring represent detailed information including both habitat and location. B. The mean relative abundance of top 10 phyla across samples from different environments. The remaining phyla are classified into “Others”. Community composition is clustered according to the Bray-Curtis dissimilarity of the mean relative abundance. Silva 138.1 was used for taxonomy classification. C. Boxplot showing a difference in richness among different groups (P < 0.001, Kruskal-Wallis test). D. Non-metric multidimensional scaling (NMDS) analysis based on the unweighted-UniFrac distance of ASV matrix showing changes in community structure in different groups (PERMANOVA, P < 0.05).
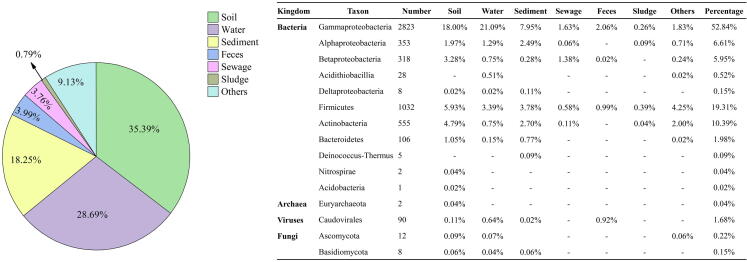
Fig. 3.
Habitat and taxonomic distribution of heavy metal(loid) resistant bacteria isolated from polluted environments. A total of 370 research articles, which are correlated to the isolation of heavy metal(loid) resistant bacteria from different environments, were collected from the Web of Science using keywords “(metal resistant bacteria) AND (isolation)”. Information on the isolation environment, taxonomic and number of isolates were retrieved. Most heavy metal(loid) resistant strains were isolated from soil, water, and sediments (82.33% of collected articles). The majority of isolates belong to Proteobacteria (66.07%). Others represent habitats including the root nodules, mosses, copper alloy coins, fly ash, slag, mine tailing and uranium ore.
The distribution and relative abundance of dominant phyla vary greatly with environmental habitats, as reflected in the clustering of all samples (Fig. 2A) and in isolation-based results (Fig. 3). Samples from heavy metal(loid) contaminated paddy soil and sediment show a comparable bacterial composition at the phylum level. In dryland, we observed a higher relative abundance of Proteobacteria and Actinobacteria, but less Chloroflexi than those in paddy soil and sediment. Communities from the mining area and sludge are distinct. In these habitats, Proteobacteria are less abundant than in other environments, while other phyla such as Actinobacteria, Chloroflexi, or Bacteroidetes become dominant (Fig. 2B). The taxonomic diversity of core heavy metal(loid) resistant microbiomes varies significantly with environmental habitats (P < 0.001), where the mining area and sludge show the lowest diversity (Fig. 2C). The non-metric multidimensional scaling (NMDS) ordination revealed that samples from sediment, sludge and mining sites possess distinct bacterial communities compared to other habitats. Samples from paddy soil overlap extensively regardless of the geographical location, but the microbiome is separate from dryland samples (Fig. 2D). These results suggest that environmental habitat has a strong impact on heavy metal(loid) resistant microbiome, which exceeds the effects from heavy metal(loid)s (types and concentration) and geographical locations.
4. Microbial responses to heavy metal(loid) pollution
4.1. Contrasting taxonomic response
Although the overall shift in bacterial composition under heavy metal(loid) pollution has been consistently reported [124], [125], [126], [127], [128], [129], [130], [131], contrasting responses are observed when comparing specific phyla across studies. For instance, the relative abundance of Actinobacteria increased in metal contaminated soil from Hunan, China [132], and in nickel polluted rhizosphere soil in Greece [133], but decreased in metal-contaminated paddy soils from Zhejiang, China [134]. Similar conflicting results were also found for Proteobacteria, where a steady decline in the relative abundance of Proteobacteria was observed along a copper gradient in grassland soil, Denmark [135], but a consistent or increased abundance was reported in other field studies [115], [136]. These inconsistent responses not only reflect the complex interaction between heavy metal(loid)s and diverse bacterial communities, but also indicate heavy metal(loid) stress could not be the only key factor shaping the core resistant microbiome.
Many variables could contribute to bacterial community shifts under heavy metal(loid) pollution, including profiles of heavy metal(loid)s (e.g., types, toxicity, pollution level, exposure history and synergistic effects), environmental factors, and the legacy effect of the local microbiome. For instance, chronic or short-term exposure to the same metal pollutant led to an opposite trend in Actinobacteria abundance [134]. In addition to heavy metal(loid)s, other abiotic factors, such as pH, organic matter and nutrient content, directly or indirectly affect microbial responses through influencing the fate and toxicity of heavy metal(loid)s entering the environment [108], [137], or affecting the physiological status of bacterial cells. Some studies indicate that even in highly contaminated environments, environmental factors, especially nutrient availability, could mitigate the selective pressure of heavy metal(loid)s on the microbial community, especially in the nutrient-limited marine environment [122], [138], [139]. Additionally, physiological traits and intrinsic resistance of the initial microbial community are of importance for taxonomic responses under heavy metal(loid) stress. Microbial traits such as life-history strategy, stress tolerance, physiological plasticity, dormancy potential and detoxification systems are directly related to microbial community resistance to heavy metal(loid)s [140]. For example, microbial species with the ability to produce spores/myxospores or generate a robust biofilm are prevalent in soil with high copper pollution [141]. Further, horizontal gene transfer of heavy metal(loid) resistance genes could enhance community tolerance without phylogenic changes. It should be noted that taxonomic responses to heavy metal(loid) perturbation is usually quantified by the relative abundance of taxa within a community. It better reflects the rank order of taxa rather than changes in real numbers. Certain taxa with the same absolute abundance in different communities could display different trends in relative abundance [142]. Therefore, after an investigation of overall community composition changes through amplicon sequencing, absolute quantification of specific taxa is highly recommended to access their responses to heavy metal(loid)s.
4.2. Microbial diversity patterns under heavy metal(loid) stress
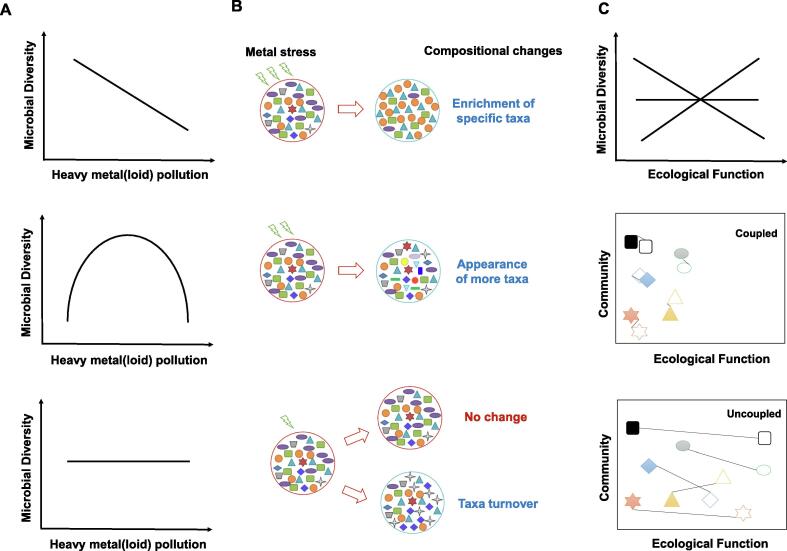
Higher microbial diversity is insurance for resistance and resilience of microbial community and function (the insurance hypothesis) [143]. Therefore, microbial diversity is a commonly used index for heavy metal(loid) polluted environments. However, contrasting observations always appear when assessing the impact of heavy metal(loid)s on microbial diversity and functionality in both terrestrial and aquatic ecosystems. Microbial diversity could either decrease, increase, or remain constant along with heavy metal(loid) gradients (Fig. 4A). In general, reduced diversity is frequently observed in studies with severe heavy metal(loid) contamination, or short-term exposure experiments in the laboratory. Strong selection occurs by heavy metal(loid)s purging sensitive taxa, leading to the proliferation of a few specific resistant groups and resulting in a subsequent decrease in diversity. In other cases, exposure to a low concentration of essential metals such as copper and zinc will promote growth of a wide range of bacteria, which is accompanied by an increase in microbial diversity. Previous studies have shown that the diversity patterns of soil and benthic prokaryotic community in metal polluted environments followed the intermediate disturbance hypothesis [123], [144]. Alternatively, microbial diversity may not be affected, even under chronic and high levels of pollution [145]. The constant diversity could be due to the strong resistance and resilience of the initial community, the capacity to acquire resistance through horizontal gene transfer, or turnover of dominant taxa without influencing microbial diversity [133], [146], [147]. Further, different responses of the rare biosphere to heavy metal(loid) pressure could result in changes in microbial diversity in different ways. The rare biosphere possesses huge genetic diversity and contributes greatly to microbial diversity, functionality, and ecosystem stability [148]. When encountering a disturbance, rare species susceptible to heavy metal(loid) toxicity will be eliminated. Extinction of highly diverse rare species leads to a severe decline of microbial diversity [37], [149]. For example, a previous study showed heavy metal pollution reduced more than 99.9% of the bacterial diversity via eliminating rare taxa in pristine soil [37]. However, the rare biosphere may contain taxa that are tolerant to heavy metal(loid) pollution. These rare taxa could remain rare under adaptive condition, or become dominant when the conditions are favorable [150], [151], leading to different responses of microbial diversity.
Fig. 4.
Potential responses of microbial diversity, taxonomic composition, and function to heavy metal(loid) contamination. A. Shifts of microbial diversity along with heavy metal(loid) gradients. Microbial diversity could either decrease (upper), increase (middle), or remain constant (bottom) along with heavy metal(loid) gradients. B. Potential impacts of heavy metal(loid)s on community composition. Heavy metal(loid) exposure selects specific metal resistant taxa, leading to a decrease in diversity. Moderate-heavy metal(loid) loading or low concentration of essential metals benefit the growth of various taxa, which is accompanied by an increase in microbial diversity. Either unaffected microbial composition or turnover of dominant taxa under heavy metal(loid) exposure will lead to a constant microbial diversity. C. Relationship between microbial diversity/taxonomic community and function in heavy metal(loid) polluted environment. Increased microbial diversity may lead to increase, decrease, or constant in functional profiling (upper). Coupled and uncoupled patterns between microbial community and function could be due to horizontal gene transfer, functional redundancy, and changes in rare taxa.
Links between microbial composition, diversity and functionality are complex (Fig. 4B and C). Liu et al. reported consistent responses of microbial taxonomic and functional traits with increased mercury content at the regional scale [123]. However, uncoupled patterns between microbial community and function are also observed under heavy metal(loid) pollution. The underlying mechanisms for the discrepancy in microbial community structure and function after disturbance have been interpreted in another review [152]. Essentially, the microbial community with a high functional redundancy is relatively stable. In this case, the lost function due to a taxonomic shift will be offset by other taxa with similar functional traits. The opposite pattern (small compositional change but large functional change) often occurs when the microbial community can acquire mobile genetic elements conferring function. Alternatively, metal stress could inhibit rare but keystone species responsible for important functional processes, leading to great changes in function rather than in composition.
5. Evolution and distribution of heavy metal(loid) resistance genes in the environment
Due to their ability of rapid adaption, microbes are considered as living fossils recording contemporary environmental changes such as heavy metal(loid) pollution [153]. Heavy metal(loid)s were high in primordial anoxic oceans and hydrothermal vents where life likely first arose, presenting one of the first challenges for early microorganisms. Therefore, microbial heavy metal(loid) resistance determinants are believed to be as ancient as the evolution of life on Earth. With the rise of oxygen during the Great Oxidation Event (GOE, 2.4 Bya), more diverse heavy metal(loid) species appeared and became bioavailable [154], [155]. Bacteria, in turn, evolved novel enzymatic systems to accommodate dramatic changes in heavy metal(loid) toxicity. Therefore, the current heavy metal(loid) resistance determinants are thought to be the consequence of billions of years of selection by geological heavy metal(loid)s and later by anthropogenic influence. This is verified by a recent study showing the expansion of arsenic resistance systems from reduced arsenic detoxification to diverse ars operon conferring arsenic resistance to both reduced and oxidized arsenic species during/after the GOE [153].
Vertical inheritance, horizontal gene transfer, gene duplication, and mutation are major mechanisms involved in the evolution of heavy metal(loid) resistance genes in contaminated environments. As we have discussed above, heavy metal(loid)s provide a selective advantage for resistant populations, leading to the thriving of specific populations. The inherent resistance determinants possessed by surviving populations could be passed down to subsequent generations, and thereby become abundant in the polluted environment. For instance, enriched Rhodanobacter contributes to the overabundance of multiple genes encoding heavy metal(loid) resistance determinants in uranium-contaminated groundwater [156]. A Geobacter population with the gene hgcA encoding enzymes responsible for mercury methylation was shown to dominate mercury methylating communities in metal-affected paddy soil and lake sediments [157], [158]. Horizontal gene transfer is also of importance for the development of heavy metal(loid) resistance. It has been suggested that heavy metal(loid)s modulate conjugation-mediated horizontal gene transfer in a type and dose-dependent manner [159]. In natural and anthropogenic environments, re-assembly of gene clusters or genomic islands conferring heavy metal(loid) resistance is commonly identified in isolates from heavy metal(loid) enriched environments such as mining sites, livestock farms, hospitals and sewage [160], [161], [162]. These reassembled clusters contain multiple resistance operons flanked with mobile genetic elements, conferring a robust resistance to multiple heavy metal(loid)s. For instance, copper homeostasis and silver resistance island (CHASRI), is widely present in Enterobacteriaceae members. CHASRI arose from the assembly of several pre-existing cus and pco clusters in E. cloacae, Pseudomonas spp. and Shewanella spp., but has a distinct evolution path from inherited clusters. Recombination of multiple types of genes encoding functions conferring copper resistance enables a robust copper resistance across (an)aerobic environments. Interestingly, CHASRI diversification highly coincided with historically anthropogenic copper production during the Roman Empire, the Song Dynasty and the post-Industrial Revolution, indicating anthropogenic impacts on the generation of novel genetic clusters [161]. In addition, the generation of genetic variation by a point mutation in existing resistance genes is another way to accelerate heavy metal(loid) detoxification. For example, previous studies observed a 10-fold decline in abundance of the merA gene, but a constant abundance of housekeeping genes, from the old to newly deposited sediments with higher mercury content. Instead of increasing gene copy numbers, bacteria living in the newly deposited sediment evolved more effective mercury reductases (MerA) by replacing one amino acid. Not surprisingly, the rapid evolution of merA again coincided with changes in anthropogenic mercury emission during the Industrial Revolution [163], [164].
Genes encoding heavy metal(loid) resistance determinants are prevalent across different taxonomic groups. However, an uneven distribution and a great divergence of these heavy metal(loid) resistance determinants are present in a host-dependent way. Metagenome analysis of environmental and human-related samples displayed a strong correlation between metal resistance genes richness and bacterial genus richness [165]. This result is also supported by investigating the distribution of heavy metal(loid) resistance genes on bacterial genomes. For instance, genes encoding CusB and CusF-like proteins are only prevalent in Proteobacteria groups, particularly in alpha-, beta-, delta- and gamma- Proteobacteria [166]. Even for highly homologous genes (e.g., mer and ars operon), variations still exist across bacterial hosts. For example, the merB gene encoding an organomercurial lysase was identified in about 50% of Pseudomonas mer operons [167], but rarely found in E. coli [39]. Uneven distribution of other homologous operons is observed in different bacterial groups, such as the copper resistance operon cop in Pseudomonas and Xanthomonas, and pco operon in E. coli [22]. Further, horizontal gene transfer results in a random distribution of genes encoding heavy metal(loid) resistance determinants across phylogenetic boundaries, generating a more complicated distribution of heavy metal(loid) resistance genes in natural environments.
Considering the discrepancy in microbial phylogenetic community structure in different habitats, we propose that the distribution of genes encoding heavy metal(loid) resistance determinants may vary between environmental habitats, while horizontal gene transfer could lessen the correlation between heavy metal(loid)s resistome and phylogenetic signature [168]. This hypothesis is supported by several studies investigating resistance strategies to heavy metal(loid)s. For instance, metagenome comparison across various environmental habitats showed a niche differentiation in both taxonomic composition and heavy metal(loid) resistome [165]. In this study, samples from external environments (e.g., soil, water, sediment, and smog, etc.) contain more abundance, higher richness, but lower beta diversity of biocide/metal resistance genes than human and animal-related samples. This result indicated the natural environment represents a large reservoir harboring heavy metal(loid) resistance genes. However, the heavy metal(loid) resistome is not always coincident with taxonomic profiles. For example, biocide and metal resistome in different sites of the human body (an exception being oral samples) displayed overlap, although the different sites have distinct taxonomic profiles [165]. Niches differentiation of the heavy metal(loid) resistome was also observed when comparing terrestrial, mangroves and ocean ecosystems [169], even in soil with different physicochemical properties [170]. As the main reservoir of various anthropogenic pollutants, estuarine and marine sediment could represent specific niches for the resistant microbiome [131]. For instance, previous studies identified the presence of divergent mer genes in estuarine and marine environment compared to the well-characterized mer operons from terrestrial, clinical and freshwater environments [171], [172].
Environmental physicochemical properties (e.g., pH, organic matter, (an)aerobic status, Fe-Mn oxides, etc.) were shown to affect heavy metal(loid) speciation, bioavailability, and transport in the environment, leading to discrepancies in heavy metal(loid) resistomes. A previous study showed that soil with low pH favors genes encoding functions involved in metal acquisition and multiple heavy metal(loid) and antibiotic efflux systems [170]. The uneven distribution of heavy metal(loid) resistance genes could be due to the enhanced impact of toxicants under low pH conditions. Additionally, heavy metal(loid) efflux by the cation diffusion facilitator (CDF) is pH-dependent and driven by a proton motive force. Increased H+ at a low pH environment could affect the efflux of CDF transporters (e.g., ZitB and CzcD) [173]. The impact of environmental properties was observed in the mercury transporter MerC, where the uneven distribution of merC in brackish water and freshwater could be related to the effects of Na+ on Hg2+ transport [174]. Together, discrepancies in environmental properties and initial microbial community in different habitats may result in niches-preference of the core heavy metal(loid) resistome. However, the distribution pattern, driving factor and mechanism behind the ecological division of heavy metal(loid) resistance genes remain virtually unknown. Further studies are needed to investigate the development of resistance strategies in different niches and underlying reasons.
6. Overlooked biotic factors shaping bacterial heavy metal(loid) resistome
There is no doubt that geographic and anthropogenic activities contribute greatly to the emergence, persistence, and evolution of bacterial heavy metal(loid) resistance in the environment. However, compared to the increased knowledge on the impact of abiotic and anthropogenic factors, the role of microbial interactions in maintaining metal resistance is largely ignored. The evolutionary history of microbial interaction could be tracked back to 3.5 billion years ago [175]. In the figuratively speaking microbial jungle, heavy metal(loid)s are used as an offensive weapon against competitors or predators. Thus, the intense interaction among bacteria or even across microbial kingdoms could represent a long-term and robust driver for the development of the heavy metal(loid) resistome. In addition, heavy metal(loid) pollution altered microbial interactions, which could further change host-associated microbiota and resistome. As reported in previous studies, heavy metal pollution affected the interaction between heterotrophic bacteria and cyanobacteria from estuary [176], and archaea-bacteria interaction in long-term contaminated soil [132]. In this section, we will provide insight into heavy metal(loid)-mediated interactions and selection for heavy metal(loid) resistant bacteria (Fig. 5).
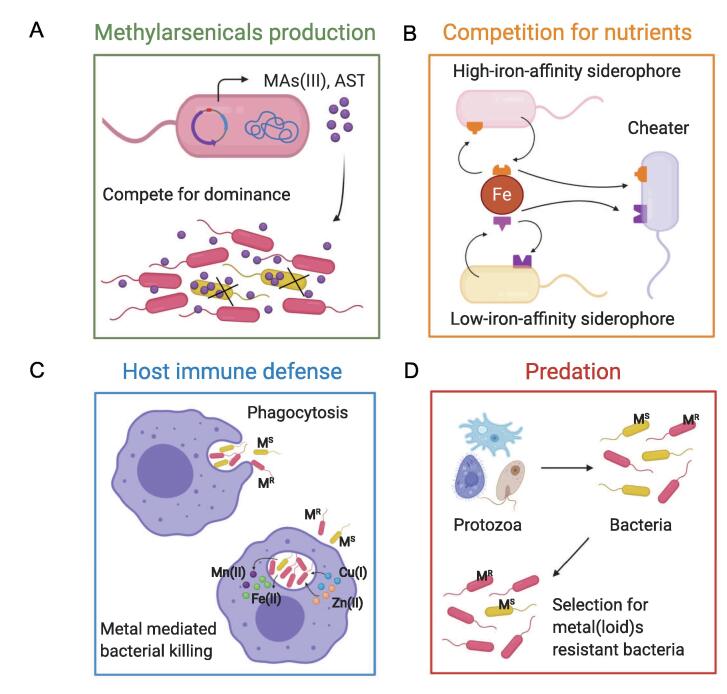
Fig. 5.
Selection of microbial interactions in heavy metal(loid) resistant bacteria. A. Bacteria secret toxic methylarsenicals (MAs(III) and AST) to kill off their competitors without detoxification system. MAs(III): methylarsenite; AST: arsinothricin. B. Siderophore-mediated interspecies cooperation and competition. C. Heavy metal(loid)-mediate host innate immune defense against pathogens. D. Selection of protozoan predation for bacterial heavy metal(loid) resistance.
The secretion of antimicrobial compounds, such as antibiotics, is one of the critical competitive strategies employed by microorganisms to kill or suppress the growth of competitors. Microorganisms were shown to produce secondary metabolites with antimicrobial activity to maintain a competitive advantage under nutrient-limited conditions [177]. A recent study proposes an antibiotic property of methylarsenicals, including MAs(III) (methylarsenite) and AST (arsinothricin), in bacterial warfare for dominance. Some microbial groups which are capable of reducing MAs(V) (methylarsenate) to MAs(III), utilize the produced toxic MAs(III) to inhibit or kill off competitors without corresponding detoxification system [178]. To survive in a MAs(III)-mediated battle, microbial species have evolved several MAs(III) detoxification mechanisms including re-oxidation of MAs(III) to less toxic MAs(V) by ArsH, degradation of MAs(III) to less toxic As(III) by ArsI, or efflux MAs(III) out of the cell through ArsP and ArsK. AST is another broad-spectrum methylarsenical antibiotic synthesized by some soil bacteria. Bacteria with the arsN gene could detoxify AST, thereby having a competitive advantage over sensitive bacteria [179].
Competition for micronutrients such as iron is another metal-mediated strategy in microbial interaction. Bacteria secrete siderophores to scavenge iron in iron-limited environments for growth and function. To date, siderophore-mediated interspecies cooperation and competition have been shown as critical drivers for bacterial co-evolutionary dynamics in nature and infectious settings [180], [181], [182], [183]. Bacteria producing high-iron-affinity siderophores are more competitive than those producing low-iron-affinity siderophores under iron-limiting conditions. A recent study reports the suppression of pathogens by the rhizosphere microbiome through the depletion of iron from pathogens [184]. Simultaneously, the use of heterologous siderophore also provokes competition between siderophore producers and cheaters taking up heterologous siderophore without pay. Further, the evolutionary arms race for iron is also of importance in host-pathogen interactions. The iron restriction is one of the main mechanisms of the host immune response against invading pathogens. To inhibit pathogen growth and repair, host cells evolve a series of iron-withdrawal mechanisms counteracting iron acquisition pathways in pathogens [185].
The other typical example of an interaction involving heavy metal(loid)s is the host innate immune defense against infecting pathogens utilizing heavy metal(loid) toxicity. Copper and zinc have been reported as important weapons to kill invading pathogens. In contrast to simultaneously occurring iron depletion, copper and zinc are accumulated in the phagosome of macrophages to poison invading bacteria during infection. Simultaneously, iron and manganese, which are required for Fe-S cluster repair and oxidative defense, are withdrawn by NRAMP1 (natural resistance-associated macrophage protein 1) and other iron and manganese transporters. The overload of toxic copper and zinc, together with the depletion of iron and manganese in the phagosome, increases the susceptibility of pathogens. In turn, copper and zinc efflux and detoxification determinants protect engulfed bacteria against metal toxicity and are proposed as essential virulence factors [20], [33]. A similar heavy metal(loid)-mediated bacterial killing mechanism is also observed during protozoan predation. Recent studies document upregulations of several genes encoding Cu(I) and Zn(II) transporters, as well as NRAMP-type transporters in amoeba during predation. Meanwhile, the deletion of copper, zinc, or arsenic resistance determinants reduced the fitness of intracellular bacteria against protozoa digestion [186], [187]. Protozoa are highly diverse and ubiquitous in natural environments, and they are thought to regulate microbial diversity, composition and functionality through trophic regulation [188], [189]. Considering the long evolutionary history between protozoa and bacteria, protozoan predation may play an essential role in maintaining bacterial heavy metal(loid) resistance. However, this continual selection has long been overlooked.
7. Summary and outlook
Heavy metal(loid)s pose an intense selective pressure for the development of resistant populations and diverse detoxification systems. Previous studies in individual isolates provide a state-of-art molecular mechanism responsible for bacterial heavy metal(loid) resistance, including both ubiquitous operons and specific determinants with narrow-spectrum hosts. However, the isolated bacteria only represent a small and partly biased fraction of the microbiota in the environment. We know little about the distribution patterns of genes encoding heavy metal(loid) resistance determinants and the microbial community’s strategies to avoid heavy metal(loid) toxicity. Filling this knowledge gap would enable to better understand the results obtained pure culture and at the community level.
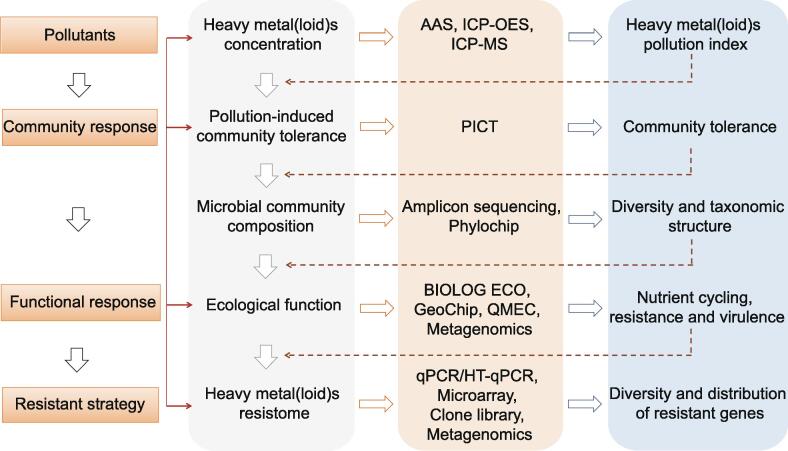
Fig. 6 summarizes the framework to investigate microbial resistance profiles from different perspectives. For instance, pollution-induced community tolerance (PICT) is a commonly used approach to determine the community tolerance to heavy metal(loid)s, and BIOLOG ECO microplates are widely employed to interpret metabolic activities of the microbial community of interest via determining their capacities to utilize different carbon sources [190]. Amplicon sequencing is mainly employed to illuminate taxonomic changes of the microbial community. Only a few studies have used this method to monitor the diversity and structure of genes conferring heavy metal(loid) resistance [157], [163]. The less frequent use of amplicon sequencing in heavy metal(loid) resistome analysis could partly be due to the high diversity and low conservation of heavy metal(loid) resistance genes, and the lack of reliable primers and specialized databases for analysis of the bacterial heavy metal(loid) resistome [191]. Metagenomics allows deep mining of the heavy metal(loid) resistome and identification of novel genes. It allowed resolving several technical limitations presented in other approaches, such as PCR-related biases and low-resolution views. However, the increased cost for high-resolution analysis hinders the large-scale application of the metagenomics approach. Therefore, metagenomic sequencing is often combined with amplicon sequencing, where amplicon sequencing analyzes a broad set of samples and provides an overall survey of the community composition, while metagenomics is used to further interpret functional profiling in subsamples [192]. The lack of comprehensive primer or probe sets is another challenge for detecting a comprehensive heavy metal(loid) resistome. Currently, there is no independent microarray-based chip available for genes conferring heavy metal(loid) resistance. Only a metal resistance module in Geochip is available, which consists of 44 metal(loid)s resistance genes involved in the detoxification of 13 metals [193]. However, most of the probes in the metal resistance module detect genes functioning as transporters, and only a few genes are present as arsenic and mercuric reduction and organomercural lyase [194]. For qPCR-based quantification approach, currently available degenerate primers for heavy metal(loid) resistance genes are limited, which cannot represent the diverse detoxification systems in the environment. As shown in Table 1, the majority of available primers target genes encoding heavy metal(loid) transporters. Additionally, some degenerate primers were initially designed for only specific groups of bacteria. The coverage and specificity of these primers should be of concern when used to quantify environmental samples. A recent developed As-Chip is a high throughput detection tool using high-throughput-qPCR (HT-qPCR) to evaluate arsenic biogeochemical cycling [195]. This chip contains 81 primer sets targeting 19 genes involved in different arsenic metabolic and resistance pathways. It has been successfully applied to investigate the arsenic resistome in soil and the earthworm intestine [195], [196].
Fig. 6.
Schematic diagram of approaches accessing pollution level, microbial community structure, functional response, and resistance strategy in heavy metal(loid) polluted environments. AAS: Atomic absorption spectrometry; ICP-OES/MS: Inductively coupled plasma-optical emission spectroscopy/mass spectroscopy; PICT: Pollution-induced community tolerance; QMEC: Quantitative microbial element cycling; HT-qPCR: High-throughput-qPCR.
Table 1.
Primers published previously for quantification of heavy metal(loid) resistance genes by qPCR.
| Heavy metal(loid)s | Genes | Function | Sequence(5′-3′) | Sample type | Reference |
|---|---|---|---|---|---|
| Cu | copA | Efflux | F: GGTGCTGATCATCGCCTG | Sediments from mining‐waste discharge canal and marine, intertidal samples, freshwater | [220] |
| R: GGGCGTCGTTGATACCGT | |||||
| cusA | Efflux | F: ATGCSACVGGYGTTGGCTGG | Marine sediments, sewage sludge, swine manure | [221] | |
| R: CCRTTCAGYTCGGCRATRCC | |||||
| pcoA | Redox | F: GCTGCAGATGGCCAGTATGTAAA | Swine manure | [222] | |
| R: CCCTCGAGCGTAACCGGTCC | |||||
| pcoD | Binding | F: ATAACTTCAAGCCGGGGACCCAG | Swine manure | [222] | |
| R: AATGCACAGAGCGTCATTGT | |||||
| tcrB | Efflux | F: CATCACGGTAGCTTTAAGGAGATTTTC | Swine manure | [223] | |
| R: ATAGAGGACTCCGCCACCATTG | |||||
| Zn | zntA | Efflux | F: GGTCGGGTCTGGCATTGAAG | Swine manure | [222] |
| R: TTGCAGCATCGGCGCGCAGGGTA | |||||
| Ni | nreB | Efflux | F: CCTTCACGCCGACTTTCCAG | Rhizosphere | [224] |
| R: CGGATAGGTAATCAGCCAGCA | |||||
| cnrA | Efflux | F: AACAAGCAGGTSCAGATCAAC | Rhizosphere | [224] | |
| R: TGATCAGGCCGAAGTCSAGCG | |||||
| Co/Zn/Cd | czcA | Efflux | F: GGSGCGMTSGAYTTCGGC | Sediment, seawater | [225] |
| R: GCCATYGGNYGGAACAT | |||||
| czcC | Efflux | F: AGCCGYCAGTATCCGGATCTGAC | Sediment, water, biofilm, soil | [226] | |
| R: GTGGTCGCCGCCTGATAGGT | |||||
| czcD | Efflux | F: TCATCGCCGGTGCGATCATCAT | Sediment, water, biofilm, soil | [226] | |
| R: TGTCATTCACGACATGAACC | |||||
| Ni/Co | nccA | Efflux | F: TTYAGCCAGGTVACSGTSATYTT | Sediment, water, biofilm, soil | [226] |
| R: GCYGCRTCSGCRCGCACCAGRTA | |||||
| Pb | pbrT | Uptake | F: AGCGCGCCCAGGAGCGCAGCGTCTT | Sediment, water, biofilm, soil | [226] |
| R: GGCTCGAAGCCGTCGAGRTA | |||||
| Hg | hgcA | Redox | F: GGNRTYAAYRTCTGGTGYGC | Paddy Soils, forest soils, lakes, wastewater, compost | [227] |
| R: CGCATYTCCTTYTYBACNCC | |||||
| merA | Redox | F: CCTGCGTCAACGTCGGCTG | Sediment, seawater | [164] | |
| R: GCGATCAGGCAGCGGTCGAA | |||||
| merB | C-Hg lyase | F: TCGCCCCATATATTTTAGAAC | Fecal, soil, sediment | [228] | |
| R: GTCGGGACAGATGCA AAGAAA | |||||
| merC | Uptake | F: CATCGGGCTGGGCTTCTTGAG | Fecal, soil, sediment | [228] | |
| R: CATCGTTCCTTATTCGTGTGG | |||||
| merD | Regulator | F CCAGGCGGCTACGGCTTGTT | Fecal, soil, sediment | [228] | |
| R: GGTGGCCAACTGCACTTCCAG | |||||
| merR | Regulator | F: GCCGGGGTCAATGTGGAGAC | Wastewater treatment plant | [229] | |
| R: TAGTCACCCCGTGACTCCCCC | |||||
| merP | Binding | F: CCGCYTGYCCGATCACWGTC | Sediment, seawater | [164] | |
| R: CGGATAGCCSGCGTCYKCGG | |||||
| merT | Uptake | F: RGTGGCGYTGTTYTTCGCCT | Sediment, seawater | [164] | |
| R: CCAGCRCGGCCACGAYCCAG |
Taken together, approaches used for metal resistant microbiome study have different merits and drawbacks (Table 2). Therefore, integrating multi-scaled methods will provide better insights into heavy metal(loid) resistant microbiome and resistome, especially for samples from the natural environment with a high level of spatial and temporal heterogeneity. For instance, a recent study assessed a mercury-methylating bacterial community via multiple approaches [197]. 16S rRNA gene pyrosequencing along with a mercury contamination gradient did not have sufficient resolution to identify mercury-methylating microbes; while amplicon sequencing of hgcAB and metagenomic analysis provided comparable results. A qPCR-based approach is recommended to quantify the abundance of specific mercury-methylating clades. Additionally, a combination of a culture-dependent method with high-throughput sequencing technologies is of importance for future studies, since findings of novel resistance determinants in environmental isolates will expand our knowledge on detoxification systems, and benefit functional annotation or PCR primer design. In turn, investigation of the core resistant microbiome and resistome through high-throughput sequencing technologies will promote a better understanding of the microbial response and adaption under heavy metal(loid) exposure, which may guide isolation and application of specific isolates for remediation.
Table 2.
Advantages and limitations of methods for heavy metal(loid) resistome study.
| Method | Advantages | Limitations |
|---|---|---|
| qPCR |
|
|
| HT-qPCR |
|
|
| Clone library |
|
|
| Microarray |
|
|
| Amplicon sequencing |
|
|
| Metagenomics and Metatranscriptomics |
|
|
Progress in understanding the core heavy metal(loid) resistant microbiome and resistance strategy from local to large-scale or across ecosystems is still a challenge. Rapid advances in high-throughput sequencing technologies enable us to interpret heavy metal(loid) resistant microbiomes in an integrative way. With an increase in high-throughput sequencing data in hand, it is possible to integrate individual local studies from various environments, and gain clues in feature and ecological function of the core heavy metal(loid) resistant microbiota, adaption of a resistance strategy and environmental driving factors. However, the discrepancy in both sample and data processing in different studies is still problematic for integrative analysis. Here, we propose the following five key questions which should be addressed in future studies. 1. What are the features of core heavy metal(loid) resistant microbiomes, and what are the driving factors shaping assemblages of core heavy metal(loid) resistant microbiomes. 2. What is the diversity, abundance, and geographic distribution of heavy metal(loid) resistance genes across different ecosystems? Is there a preference for a specific resistance strategy in different habitats, and what are the underlying mechanisms? 3. To what extend do biotic and abiotic factors affect the core heavy metal(loid) resistant microbiome and resistome? 4. Does the core heavy metal(loid) resistant microbiome also contribute to microbial antibiotic resistance? 5. How can we integrate multiple and novel technologies to interpret the development of the heavy metal(loid) resistant microbiome and resistome?
CRediT authorship contribution statement
Xiuli Hao: Conceptualization, Writing - original draft, Visualization. Jiaojiao Zhu: Writing - original draft, Visualization. Christopher Rensing: Conceptualization, Writing - review & editing. Ying Liu: Data curation, Formal analysis. Shenghan Gao: Data curation, Formal analysis. Wenli Chen: Resources, Writing - review & editing. Qiaoyun Huang: Resources, Writing - review & editing. Yu-Rong Liu: Conceptualization, Resources, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (41877330, 41503076 and 41877120). We also thank the Fundamental Research Funds for the Central Universities, China (Program no. 2662018QD065 and 2662019PY010).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.12.006.
Contributor Information
Xiuli Hao, Email: xlhao@mail.hzau.edu.cn.
Yu-Rong Liu, Email: yrliu@mail.hzau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhu Y.-G., Yoshinaga M., Zhao F.-J., Rosen B.P. Earth abides arsenic biotransformations. Annu Rev Earth Planet Sci. 2014;42(1):443–467. doi: 10.1146/annurev-earth-060313-054942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nriagu J.O. A history of global metal pollution. Science. 1996;272(5259):223–230. doi: 10.1126/science:272.5259.223. [DOI] [Google Scholar]
- 3.Hong S.M. Greenland ice evidence of hemispheric lead pollution 2-millennia ago by Greek and Roman civilizations. Science. 1994;265(5180):1841–1843. doi: 10.1126/science.265.5180.1841. [DOI] [PubMed] [Google Scholar]
- 4.Renberg I., Persson M.W., Emteryd O. Pre-industrial atmospheric lead contamination detected in Swedish lake sediments. Nature. 1994;368(6469):323–326. doi: 10.1038/368323a0. [DOI] [Google Scholar]
- 5.Hong S., Candelone J.-P., Patterson C.C., Boutron C.F. History of ancient copper smelting pollution during Roman and medieval times recorded in Greenland ice. Science. 1996;272(5259):246–249. doi: 10.1126/science:272.5259.246. [DOI] [Google Scholar]
- 6.Lemire J.A., Harrison J.J., Turner R.J. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol. 2013;11(6):371–384. doi: 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- 7.Oremland R.S., Stolz J.F. The ecology of arsenic. Science. 2003;300(5621):939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- 8.Michels H.T., Keevil C.W., Salgado C.D., Schmidt M.G. From laboratory research to a clinical trial: copper alloy surfaces kill bacteria and reduce hospital-acquired infections. HERD. 2015;9(1):64–79. doi: 10.1177/1937586715592650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould S.W.J., Fielder M.D., Kelly A.F., Morgan M., Kenny J., Naughton D.P. The antimicrobial properties of copper surfaces against a range of important nosocomial pathogens. Ann Microbiol. 2009;59(1):151–156. doi: 10.1007/BF03175613. [DOI] [Google Scholar]
- 10.Noyce J.O., Michels H., Keevil C.W. Potential use of copper surfaces to reduce survival of epidemic meticillin-resistant Staphylococcus aureus in the healthcare environment. J Hosp Infect. 2006;63(3):289–297. doi: 10.1016/j.jhin.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Elguindi J., Wagner J., Rensing C. Genes involved in copper resistance influence survival of Pseudomonas aeruginosa on copper surfaces. J Appl Microbiol. 2009;106(5):1448–1455. doi: 10.1111/j.1365-2672.2009.04148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debski B. Supplementation of pigs diet with zinc and copper as alternative to conventional antimicrobials. Pol J Vet Sci. 2016;19(4):917–924. doi: 10.1515/pjvs-2016-0113. [DOI] [PubMed] [Google Scholar]
- 14.Bolan N., Khan M., Donaldson J., Adriano D., Matthew C. Distribution and bioavailability of copper in farm effluent. Sci Total Environ. 2003;309(1-3):225–236. doi: 10.1016/S0048-9697(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 15.Zhang F.S. Content of heavy metals in animal feeds and manures from farms of different scales in Northeast China. Int J Environ Res Public Health. 2012;9(8):2658–2668. doi: 10.3390/ijerph9082658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ihnat M., Fernandes L. Trace elemental characterization of composted poultry manure. Bioresour Technol. 1996;57(2):143–156. [Google Scholar]
- 17.Arjoon A., Olaniran A.O., Pillay B. Co-contamination of water with chlorinated hydrocarbons and heavy metals: challenges and current bioremediation strategies. Int J Environ Sci Technol. 2013;10(2):395–412. doi: 10.1007/s13762-012-0122-y. [DOI] [Google Scholar]
- 18.Hou D., O’Connor D., Igalavithana A.D., Alessi D.S., Luo J., Tsang D.C.W., Sparks D.L., Yamauchi Y., Rinklebe J., Ok Y.S. Metal contamination and bioremediation of agricultural soils for food safety and sustainability. Nat Rev Earth Environ. 2020;1(7):366–381. doi: 10.1038/s43017-020-0061-y. [DOI] [Google Scholar]
- 19.Baker-Austin C., Wright M.S., Stepanauskas R., McArthur J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14(4):176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Chandrangsu P., Rensing C., Helmann J.D. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol. 2017;15(6):338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruins M.R., Kapil S., Oehme F.W. Microbial resistance to metals in the environment. Ecotoxicol Environ Saf. 2000;45(3):198–207. doi: 10.1006/eesa:1999.1860. [DOI] [PubMed] [Google Scholar]
- 22.Silver S., Phung L.T. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50(1):753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 23.Nies D.H. Microbial heavy-metal resistance. Appl Microbiol Biotechnol. 1999;51(6):730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 24.Nies D.H., Silver S. Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol. 1995;14(2):186–199. doi: 10.1007/BF01569902. [DOI] [PubMed] [Google Scholar]
- 25.Harrison J.J., Ceri H., Turner R.J. Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microbiol. 2007;5(12):928–938. doi: 10.1038/nrmicro1774. [DOI] [PubMed] [Google Scholar]
- 26.Xu S., Xing Y., Liu S., Luo X., Chen W., Huang Q. Co-effect of minerals and Cd(II) promoted the formation of bacterial biofilm and consequently enhanced the sorption of Cd(II) Environ Pollut. 2020;258:113774. doi: 10.1016/j.envpol.2019.113774. [DOI] [PubMed] [Google Scholar]
- 27.Chaturvedi K.S., Hung C.S., Crowley J.R., Stapleton A.E., Henderson J.P. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat Chem Biol. 2012;8(8):731–736. doi: 10.1038/nchembio.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao X., Xie P., Zhu Y.-G., Taghavi S., Wei G., Rensing C. Copper tolerance mechanisms of Mesorhizobium amorphae and its role in aiding phytostabilization by Robinia pseudoacacia in copper contaminated soil. Environ Sci Technol. 2015;49(4):2328–2340. doi: 10.1021/es504956a. [DOI] [PubMed] [Google Scholar]
- 29.Das S., Dash H.R., Chakraborty J. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl Microbiol Biotechnol. 2016;100(7):2967–2984. doi: 10.1007/s00253-016-7364-4. [DOI] [PubMed] [Google Scholar]
- 30.Sher S., Rehman A. Use of heavy metals resistant bacteria—a strategy for arsenic bioremediation. Appl Microbiol Biotechnol. 2019;103(15):6007–6021. doi: 10.1007/s00253-019-09933-6. [DOI] [PubMed] [Google Scholar]
- 31.Orell A., Navarro C.A., Arancibia R., Mobarec J.C., Jerez C.A. Life in blue: copper resistance mechanisms of bacteria and Archaea used in industrial biomining of minerals. Biotechnol Adv. 2010;28(6):839–848. doi: 10.1016/j.biotechadv.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Pal C., Bengtsson-Palme J., Kristiansson E., Larsson D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015;16(1) doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodgkinson V., Petris M.J. Copper homeostasis at the host-pathogen interface. J Biol Chem. 2012;287(17):13549–13555. doi: 10.1074/jbc.R111.316406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyes C., Hodgskiss L.H., Kerou M., Pribasnig T., Abby S.S., Bayer B., Kraemer S.M., Schleper C. Genome wide transcriptomic analysis of the soil ammonia oxidizing archaeon Nitrososphaera viennensis upon exposure to copper limitation. ISME J. 2020;14(11):2659–2674. doi: 10.1038/s41396-020-0715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher O.S., Kenney G.E., Ross M.O., Ro S.Y., Lemma B.E., Batelu S., Thomas P.M., Sosnowski V.C., DeHart C.J., Kelleher N.L., Stemmler T.L., Hoffman B.M., Rosenzweig A.C. Characterization of a long overlooked copper protein from methane- and ammonia-oxidizing bacteria. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-06681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gwak J.-H., Jung M.-Y., Hong H., Kim J.-G., Quan Z.-X., Reinfelder J.R., Spasov E., Neufeld J.D., Wagner M., Rhee S.-K. Archaeal nitrification is constrained by copper complexation with organic matter in municipal wastewater treatment plants. ISME J. 2020;14(2):335–346. doi: 10.1038/s41396-019-0538-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gans J., Wolinsky M., Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309(5739):1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 38.Smith D.H. R factors mediate resistance to mercury, nickel, and cobalt. Science. 1967;156(3778):1114–1116. doi: 10.1126/science:156.3778.1114. [DOI] [PubMed] [Google Scholar]
- 39.Schottel Janet, Mandal Amalendu, Clark Dan, Silver Simon, Hedges R.W. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature. 1974;251(5473):335–337. doi: 10.1038/251335a0. [DOI] [PubMed] [Google Scholar]
- 40.Foster T.J. Transposon A-generated mutations in the mercuric resistance genes of plasmid R100–1. J Bacteriol. 1979;140(1):167–181. doi: 10.1128/jb.140.1.167-181.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore M.J. Purification, crystallization, and preliminary x-ray diffraction studies of the flavoenzyme mercuric ion reductase from Bacillus sp. strain RC607. J Biol Chem. 1989;264(24):14386–14388. [PubMed] [Google Scholar]
- 42.Schiering N., Kabsch W., Moore M.J., Distefano M.D., Walsh C.T., Pai E.F. Structure of the detoxification catalyst mercuric ion reductase from Bacillus sp. strain RC607. Nature. 1991;352(6331):168–172. doi: 10.1038/352168a0. [DOI] [PubMed] [Google Scholar]
- 43.Begley T.P., Walts A.E., Walsh C.T. Mechanistic studies of a protonolytic organomercurial cleaving enzyme: bacterial organomercurial lyase. Biochemistry. 1986;25(22):7192–7200. doi: 10.1021/bi00370a064. [DOI] [PubMed] [Google Scholar]
- 44.Walts A.E., Walsh C.T. Bacterial organomercurial lyase: novel enzymatic protonolysis of organostannanes. J Am Chem Soc. 1988;110(6):1950–1953. [Google Scholar]
- 45.Lund P.A., Brown N.L. Role of the merT and merP gene products of transposon Tn501 in the induction and expression of resistance to mercuric ions. Gene. 1987;52(2-3):207–214. doi: 10.1016/0378-1119(87)90047-3. [DOI] [PubMed] [Google Scholar]
- 46.Kusano T. Constitutive synthesis of a transport function encoded by the Thiobacillus ferrooxidans merC gene cloned in Escherichia coli. J Bacteriol. 1990;172(5):2688–2692. doi: 10.1128/jb.172.5.2688-2692.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamlett N.V. Roles of the Tn21 merT, merP, and merC gene products in mercury resistance and mercury binding. J Bacteriol. 1992;174(20):6377–6385. doi: 10.1128/jb.174.20.6377-6385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foster T.J., Brown N.L. Identification of the merR gene of R100 by using mer-lac gene and operon fusions. J Bacteriol. 1985;163(3):1153–1157. doi: 10.1128/jb.163.3.1153-1157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nucifora G. Mercury operon regulation by the merR gene of the organomercurial resistance system of plasmid pDU1358. J Bacteriol. 1989;171(8):4241–4247. doi: 10.1128/jb.171.8.4241-4247.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ni'Bhriain N.N., Silver S., Foster T.J. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J Bacteriol. 1983;155(2):690–703. doi: 10.1128/jb.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukhopadhyay D. Purification and functional characterization of MerD: a coregulator of the mercury resistance operon in Gram-negative bacteria. J Biol Chem. 1991;266(28):18538–18542. [PubMed] [Google Scholar]
- 52.Misra T.K. Bacterial resistances to inorganic mercury salts and organomercurials. Plasmid. 1992;27(1):4–16. doi: 10.1016/0147-619X(92)90002-R. [DOI] [PubMed] [Google Scholar]
- 53.Parks J.M. The genetic basis for bacterial mercury methylation. Science. 2013;339(6125):1332. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 54.Foster T.J. The genetics and biochemistry of mercury resistance. CRC Crit Rev Microbiol. 1987;15(2):117–140. doi: 10.3109/10408418709104455. [DOI] [PubMed] [Google Scholar]
- 55.Silver S., Misra T.K. Plasmid-mediated heavy metal resistances. Annu Rev Microbiol. 1988;42(1):717–743. doi: 10.1146/annurev.mi.42.100188.003441. [DOI] [PubMed] [Google Scholar]
- 56.Silver S. Inducible plasmid-determined resistance to arsenate, arsenite, and antimony(III) in Escherichia coli and Staphylococcus aureus. J Bacteriol. 1981;146(3):983–996. doi: 10.1128/jb.146.3.983-996.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mobley H.L., Rosen B.P. Energetics of plasmid-mediated arsenate resistance in Escherichia coli. Proc Natl Acad Sci. 1982;79(20):6119–6122. doi: 10.1073/pnas.79.20.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ben Fekih I. Distribution of arsenic resistance genes in prokaryotes. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen C.M. Nucleotide sequence of the structural genes for an anion pump. The plasmid-encoded arsenical resistance operon. J Biol Chem. 1986;261(32):15030–15038. [PubMed] [Google Scholar]
- 60.Rosen B.P. Molecular characterization of an anion pump - the arsA gene-product is an arsenite (antimonate)-stimulated ATPase. J Biol Chem. 1988;263(7):3067–3070. [PubMed] [Google Scholar]
- 61.Wu J., Tisa L.S., Rosen B.P. Membrane topology of the ArsB protein, the membrane subunit of an anion-translocating ATPase. J Biol Chem. 1992;267(18):12570–12576. [PubMed] [Google Scholar]
- 62.Ji G., Silver S. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci. 1992;89(20):9474–9478. doi: 10.1073/pnas.89.20.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J., Rosen B.P. The arsD gene encodes a second trans-acting regulatory protein of the plasmid-encoded arsenical resistance operon. Mol Microbiol. 1993;8(3):615–623. doi: 10.1111/j.1365-2958.1993.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 64.Wu J., Rosen B.P. The ArsR protein is a trans-acting regulatory protein. Mol Microbiol. 1991;5(6):1331–1336. doi: 10.1111/j.1365-2958.1991.tb00779.x. [DOI] [PubMed] [Google Scholar]
- 65.Ghosh M., Shen J., Rosen B.P. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1999;96(9):5001–5006. doi: 10.1073/pnas.96.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen Z., Luangtongkum T., Qiang Z., Jeon B., Wang L., Zhang Q. Identification of a novel membrane transporter mediating resistance to organic arsenic in Campylobacter jejuni. Antimicrob Agents Chemother. 2014;58(4):2021–2029. doi: 10.1128/AAC.02137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen J., Madegowda M., Bhattacharjee H., Rosen B.P. ArsP: a methylarsenite efflux permease: a trivalent organoarsenical efflux permease. Mol Microbiol. 2015;98(4):625–635. doi: 10.1111/mmi.13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J., Yoshinaga M., Garbinski L.D., Rosen B.P. Synergistic interaction of glyceraldehydes-3-phosphate dehydrogenase and ArsJ, a novel organoarsenical efflux permease, confers arsenate resistance: a new pathway of arsenate resistance. Mol Microbiol. 2016;100(6):945–953. doi: 10.1111/mmi.13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qin J., Rosen B.P., Zhang Y., Wang G., Franke S., Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S -adenosylmethionine methyltransferase. PNAS. 2006;103(7):2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshinaga M., Rosen B.P. A C*As lyase for degradation of environmental organoarsenical herbicides and animal husbandry growth promoters. Proc Natl Acad Sci. 2014;111(21):7701–7706. doi: 10.1073/pnas.1403057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen J., Bhattacharjee H., Rosen B.P. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone: a trivalent organoarsenical oxidase. Mol Microbiol. 2015;96(5):1042–1052. doi: 10.1111/mmi.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi K. Efflux transporter ArsK is responsible for bacterial resistance to arsenite, antimonite, trivalent roxarsone, and methylarsenite. Appl Environ Microbiol. 2018;84(24):e01842–e01848. doi: 10.1128/AEM.01842-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mellano M.A., Cooksey D.A. Induction of the copper resistance operon from Pseudomonas syringae. J Bacteriol. 1988;170(9):4399–4401. doi: 10.1128/jb.170.9.4399-4401.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bender C.L., Cooksey D.A. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J Bacteriol. 1986;165(2):534–541. doi: 10.1128/jb.165.2.534-541.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bender C.L., Cooksey D.A. Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1987;169(2):470–474. doi: 10.1128/jb.169.2.470-474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lim C.K., Cooksey D.A. Characterization of chromosomal homologs of the plasmid-borne copper resistance operon of Pseudomonas Syringae. J Bacteriol. 1993;175(14):4492–4498. doi: 10.1128/jb.175.14.4492-4498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tetaz T.J., Luke R.K. Plasmid-controlled resistance to copper in Escherichia coli. J Bacteriol. 1983;154(3):1263–1268. doi: 10.1128/jb.154.3.1263-1268.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hao X., Lüthje F.L., Qin Y., McDevitt S.F., Lutay N., Hobman J.L., Asiani K., Soncini F.C., German N., Zhang S., Zhu Y.-G., Rensing C. Survival in amoeba—a major selection pressure on the presence of bacterial copper and zinc resistance determinants? Identification of a “copper pathogenicity island”. Appl Microbiol Biotechnol. 2015;99(14):5817–5824. doi: 10.1007/s00253-015-6749-0. [DOI] [PubMed] [Google Scholar]
- 79.Rouch D. Inducible plasmid-mediated copper resistance in Escherichia coli. J Gen Microbiol. 1985;131(4):939–943. doi: 10.1099/00221287-131-4-939. [DOI] [PubMed] [Google Scholar]
- 80.Brown N.L., Barrett S.R., Camakaris J., Lee B.T.O., Rouch D.A. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol Microbiol. 1995;17(6):1153–1166. doi: 10.1111/j.1365-2958.1995.mmi_17061153.x. [DOI] [PubMed] [Google Scholar]
- 81.Monchy S. Transcriptomic and proteomic analyses of the pMOL30-encoded copper resistance in Cupriavidus metallidurans strain CH34. Microbiology. 2006;152:1765–1776. doi: 10.1099/mic.0.28593-0. [DOI] [PubMed] [Google Scholar]
- 82.Rensing C., Fan B., Sharma R., Mitra B., Rosen B.P. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci. 2000;97(2):652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Outten F.W., Huffman D.L., Hale J.A., O'Halloran T.V. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem. 2001;276(33):30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 84.Roberts S.A., Weichsel A., Grass G., Thakali K., Hazzard J.T., Tollin G., Rensing C., Montfort W.R. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc Natl Acad Sci. 2002;99(5):2766–2771. doi: 10.1073/pnas.052710499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rensing C., Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev. 2003;27(2-3):197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 86.Munson G.P., Lam D.L., Outten F.W., O'Halloran T.V. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol. 2000;182(20):5864–5871. doi: 10.1128/JB.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Franke S., Grass G., Rensing C., Nies D.H. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185(13):3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grass G., Rensing C. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol. 2001;183(6):2145–2147. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Osman D. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J Biol Chem. 2010;285(33):25259–25268. doi: 10.1074/jbc.M110.145953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pontel L.B., Soncini F.C. Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol Microbiol. 2009;73(2):212–225. doi: 10.1111/j.1365-2958.2009.06763.x. [DOI] [PubMed] [Google Scholar]
- 91.Gupta A., Matsui K., Lo J.-F., Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat Med. 1999;5(2):183–188. doi: 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 92.Nies D.H., Nies A., Chu L., Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci. 1989;86(19):7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nies D. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol. 1987;169(10):4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt T., Schlegel H.G. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J Bacteriol. 1994;176(22):7045–7054. doi: 10.1128/jb.176.22.7045-7054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nucifora G., Chu L., Misra T.K., Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci. 1989;86(10):3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beard S.J., Hashim R., Membrillo‐Hernández J., Hughes M.N., Poole R.K. Zinc(II) tolerance in Escherichia coli K‐12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol. 1997;25(5):883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 97.Rensing C., Mitra B., Rosen B.P. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci. 1997;94(26):14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thelwell C., Robinson N.J., Turner-Cavet J.S. An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc Natl Acad Sci. 1998;95(18):10728–10733. doi: 10.1073/pnas.95.18.10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gupta A. Amplification and rearrangement of a prokaryotic metallothionein locus smt in Synechococcus PCC 6301 selected for tolerance to cadmium. Proc Biol Sci. 1992;248(1323):273–281. doi: 10.1098/rspb.1992.0072. [DOI] [PubMed] [Google Scholar]
- 100.Thatoi H., Das S., Mishra J., Rath B.P., Das N. Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review. J Environ Manage. 2014;146:383–399. doi: 10.1016/j.jenvman.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 101.Ackerley D.F. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl Environ Microbiol. 2004;70(2):873–882. doi: 10.1128/AEM.70.2.873-882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ackerley D.F. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ Microbiol. 2004;6(8):851–860. doi: 10.1111/j.1462-2920.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 103.Cervantes C. Cloning, nucleotide sequence, and expression of the chromate resistance determinant of Pseudomonas aeruginosa plasmid pUM505. J Bacteriol. 1990;172(1):287–291. doi: 10.1128/jb.172.1.287-291.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nies A., Nies D.H., Silver S. Cloning and expression of plasmid genes encoding resistances to chromate and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989;171(9):5065–5070. doi: 10.1128/jb.171.9.5065-5070.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nies A., Nies D.H., Silver S. Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J Biol Chem. 1990;265(10):5648–5653. [PubMed] [Google Scholar]
- 106.Juhnke S. New genes involved in chromate resistance in Ralstonia metallidurans strain CH34. Arch Microbiol. 2002;179(1):15–25. doi: 10.1007/s00203-002-0492-5. [DOI] [PubMed] [Google Scholar]
- 107.Zhou D.N. Effects of heavy metal pollution on microbial communities and activities of mining soils in Central Tibet, China. J Food Agric Environ. 2013;11(1):676–681. [Google Scholar]
- 108.Frossard A., Hartmann M., Frey B. Tolerance of the forest soil microbiome to increasing mercury concentrations. Soil Biol Biochem. 2017;105:162–176. [Google Scholar]
- 109.Rajapaksha R.M., Tobor-Kaplon M.A., Baath E. Metal toxicity affects fungal and bacterial activities in soil differently. Appl Environ Microbiol. 2004;70(5):2966–2973. doi: 10.1128/AEM.70.5.2966-2973.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li C. Effects of heavy metals on microbial communities in sediments and establishment of bioindicators based on microbial taxa and function for environmental monitoring and management. Sci Total Environ. 2020;749:141555. doi: 10.1016/j.scitotenv.2020.141555. [DOI] [PubMed] [Google Scholar]
- 111.Stefanowicz A.M. Pine forest and grassland differently influence the response of soil microbial communities to metal contamination. Sci Total Environ. 2010;408(24):6134–6141. doi: 10.1016/j.scitotenv.2010.08.056. [DOI] [PubMed] [Google Scholar]
- 112.Roane T.M., Pepper I.L. Microbial responses to environmentally toxic cadmium. Microb Ecol. 1999;38(4):358–364. doi: 10.1007/s002489901001. [DOI] [PubMed] [Google Scholar]
- 113.Bååth E. Measurement of heavy metal tolerance of soil bacteria using thymidine incorporation into bacteria extracted after homogenization-centrifugation. Soil Biol Biochem. 1992;24(11):1167–1172. [Google Scholar]
- 114.Fließbach A., Martens R., Reber H.H. Soil microbial biomass and microbial activity in soils treated with heavy metal contaminated sewage sludge. Soil Biol Biochem. 1994;26(9):1201–1205. [Google Scholar]
- 115.Fagnano M. Copper accumulation in agricultural soils: risks for the food chain and soil microbial populations. Sci Total Environ. 2020;734:139434. doi: 10.1016/j.scitotenv.2020.139434. [DOI] [PubMed] [Google Scholar]
- 116.Zhang C. Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol Biochem. 2016;97:40–49. [Google Scholar]
- 117.Chen Y. Long-term and high-concentration heavy-metal contamination strongly influences the microbiome and functional genes in Yellow River sediments. Sci Total Environ. 2018;637:1400–1412. doi: 10.1016/j.scitotenv.2018.05.109. [DOI] [PubMed] [Google Scholar]
- 118.Ellis R.J. Cultivation-dependent and -independent approaches for determining bacterial diversity in fleavy-metal-contaminated soil. Appl Environ Microbiol. 2003;69(6):3223–3230. doi: 10.1128/AEM.69.6.3223-3230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao X.Q. Study on the influence of soil microbial community on the long-term heavy metal pollution of different land use types and depth layers in mine. Ecotox Environ Saf. 2019;170:218–226. doi: 10.1016/j.ecoenv.2018.11.136. [DOI] [PubMed] [Google Scholar]
- 120.Li S. A comprehensive survey on the horizontal and vertical distribution of heavy metals and microorganisms in soils of a Pb/Zn smelter. J Hazard Mater. 2020;400:123255. doi: 10.1016/j.jhazmat.2020.123255. [DOI] [PubMed] [Google Scholar]
- 121.Oyetibo G.O. Comparative geochemical evaluation of toxic metals pollution and bacterial communities of industrial effluent tributary and a receiving estuary in Nigeria. Chemosphere. 2019;227:638–646. doi: 10.1016/j.chemosphere.2019.04.048. [DOI] [PubMed] [Google Scholar]
- 122.Naether A. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl Environ Microbiol. 2012;78(20):7398–7406. doi: 10.1128/AEM.01325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu Y.R. Consistent responses of soil microbial taxonomic and functional attributes to mercury pollution across China. Microbiome. 2018;6(183) doi: 10.1186/s40168-018-0572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Z. Application of 16S rDNA-PCR amplification and DGGE fingerprinting for detection of shift in microbial community diversity in Cu-, Zn-, and Cd-contaminated paddy soils. Chemosphere. 2006;62(8):1374–1380. doi: 10.1016/j.chemosphere.2005.07.050. [DOI] [PubMed] [Google Scholar]
- 125.Turpeinen R., Kairesalo T., Häggblom M.M. Microbial community structure and activity in arsenic-, chromium- and copper-contaminated soils. FEMS Microbiol Ecol. 2004;47(1):39–50. doi: 10.1016/S0168-6496(03)00232-0. [DOI] [PubMed] [Google Scholar]
- 126.Hemme C.L. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 2010;4(5):660–672. doi: 10.1038/ismej.2009.154. [DOI] [PubMed] [Google Scholar]
- 127.Wyszkowska J. Effect of cadmium, copper and zinc on plants, soil microorganisms and soil enzymes. J Elem. 2013;18(4):769–796. [Google Scholar]
- 128.Müller A.K. The effect of long-term mercury pollution on the soil microbial community. FEMS Microbiol Ecol. 2001;36(1):11–19. doi: 10.1111/j.1574-6941.2001.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 129.Joynt J. Microbial community analysis of soils contaminated with lead, chromium and petroleum hydrocarbons. Microb Ecol. 2006;51(2):209–219. doi: 10.1007/s00248-005-0205-0. [DOI] [PubMed] [Google Scholar]
- 130.Gołębiewski M. 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb Ecol. 2014;67(3):635–647. doi: 10.1007/s00248-013-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen J.R. A pollution gradient contributes to the taxonomic, functional, and resistome diversity of microbial communities in marine sediments. Microbiome. 2019;7 doi: 10.1186/s40168-019-0714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li X.Q. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ Pollut. 2017;231:908–917. doi: 10.1016/j.envpol.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 133.Lopez S. Nickel drives bacterial community diversity in the rhizosphere of the hyperaccumulator Alyssum murale. Soil Biol Biochem. 2017;114:121–130. [Google Scholar]
- 134.Song J.W. Effects of Cd, Cu, Zn and their combined action on microbial biomass and bacterial community structure. Environ Pollut. 2018;243:510–518. doi: 10.1016/j.envpol.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 135.Nunes I. Coping with copper: legacy effect of copper on potential activity of soil bacteria following a century of exposure. FEMS Microbiol Ecol. 2016;92(11) doi: 10.1093/femsec/fiw175. [DOI] [PubMed] [Google Scholar]
- 136.Kou S.M. The response of a 16S ribosomal RNA gene fragment amplified community to lead, zinc, and copper pollution in a Shanghai field trial. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Griffiths B.S., Philippot L. Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol Rev. 2013;37(2):112–129. doi: 10.1111/j.1574-6976.2012.00343.x. [DOI] [PubMed] [Google Scholar]
- 138.Chodak M. Diversity of microorganisms from forest soils differently polluted with heavy metals. Appl Soil Ecol. 2013;64:7–14. [Google Scholar]
- 139.Di Cesare A. The role of metal contamination in shaping microbial communities in heavily polluted marine sediments. Environ Pollut. 2020;265(Pt B):114823. doi: 10.1016/j.envpol.2020.114823. [DOI] [PubMed] [Google Scholar]
- 140.Bardgett R.D., Caruso T. Soil microbial community responses to climate extremes: resistance, resilience and transitions to alternative states. Philos T R Soc B. 2020;375(1794) doi: 10.1098/rstb.2019.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shaw J.L.A. Long-term effects of copper exposure to agricultural soil function and microbial community structure at a controlled and experimental field site. Environ Pollut. 2020;263 doi: 10.1016/j.envpol.2020.114411. [DOI] [PubMed] [Google Scholar]
- 142.Isobe K. Phylogenetic conservation of bacterial responses to soil nitrogen addition across continents. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yachi S., Loreau M. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci U S A. 1999;96(4):1463–1468. doi: 10.1073/pnas.96.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Misson B. Chemical multi-contamination drives benthic prokaryotic diversity in the anthropized Toulon Bay. Sci Total Environ. 2016;556:319–329. doi: 10.1016/j.scitotenv.2016.02.038. [DOI] [PubMed] [Google Scholar]
- 145.Li J. Long-term nickel exposure altered the bacterial community composition but not diversity in two contrasting agricultural soils. Environ Sci Pollut Res Int. 2015;22(14):10496–10505. doi: 10.1007/s11356-015-4232-1. [DOI] [PubMed] [Google Scholar]
- 146.Hong C. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ Sci Pollut Res. 2015;22(14):10788–10799. doi: 10.1007/s11356-015-4186-3. [DOI] [PubMed] [Google Scholar]
- 147.Giller K.E., Witter E., McGrath S.P. Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol Biochem. 1998;30(10–11):1389–1414. [Google Scholar]
- 148.Lynch M.D.J., Neufeld J.D. Ecology and exploration of the rare biosphere. Nat Rev Microbiol. 2015;13(4):217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 149.Jiao S. Dominant role of abundant rather than rare bacterial taxa in maintaining agro-soil microbiomes under environmental disturbances. Chemosphere. 2019;235:248–259. doi: 10.1016/j.chemosphere.2019.06.174. [DOI] [PubMed] [Google Scholar]
- 150.Shade A. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio. 2014;5(4) doi: 10.1128/mBio.01371-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kurm V., Geisen S., Hol W.H.G. A low proportion of rare bacterial taxa responds to abiotic changes compared with dominant taxa. Environ Microbiol. 2019;21(2):750–758. doi: 10.1111/1462-2920.14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bissett A. Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecol Lett. 2013;16:128–139. doi: 10.1111/ele.12109. [DOI] [PubMed] [Google Scholar]
- 153.Chen S.C. The Great Oxidation Event expanded the genetic repertoire of arsenic metabolism and cycling. Proc Natl Acad Sci U S A. 2020;117(19):10414–10421. doi: 10.1073/pnas.2001063117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fru E.C. Cu isotopes in marine black shales record the Great Oxidation Event. Proc Natl Acad Sci U S A. 2016;113(18):4941–4946. doi: 10.1073/pnas.1523544113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dupont C.L., Grass G., Rensing C. Copper toxicity and the origin of bacterial resistance-new insights and applications. Metallomics. 2011;3(11):1109–1118. doi: 10.1039/c1mt00107h. [DOI] [PubMed] [Google Scholar]
- 156.Hemme C.L. Lateral gene transfer in a heavy metal-contaminated-groundwater microbial community. mBio. 2016;7(2) doi: 10.1128/mBio.02234-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu Y.R. Unraveling microbial communities associated with methylmercury production in paddy soils. Environ Sci Technol. 2018;52(22):13110–13118. doi: 10.1021/acs.est.8b03052. [DOI] [PubMed] [Google Scholar]
- 158.Bravo A.G. Geobacteraceae are important members of mercury-methylating microbial communities of sediments impacted by waste water releases. ISME J. 2018;12(3):802–812. doi: 10.1038/s41396-017-0007-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Klumper U. Metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner. ISME J. 2017;11(1):152–165. doi: 10.1038/ismej.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li Y.P. Genomic islands confer heavy metal resistance in Mucilaginibacter kameinonensis and Mucilaginibacter rubeus isolated from a gold/copper mine. Genes. 2018;9(12):E573. doi: 10.3390/genes9120573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Staehlin B.M. Evolution of a heavy metal homeostasis/resistance island reflects increasing copper stress in Enterobacteria. Genome Biol Evol. 2016;8(3):811–826. doi: 10.1093/gbe/evw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Luthje F.L. Genome sequences of two copper-resistant Escherichia coli strains isolated from copper-fed pigs. Genome Announc. 2014;2(6) doi: 10.1128/genomeA.01341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ruuskanen M.O., Aris-Brosou S., Poulain A.J. Swift evolutionary response of microbes to a rise in anthropogenic mercury in the Northern Hemisphere. ISME J. 2020;14(3):788–800. doi: 10.1038/s41396-019-0563-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Poulain A.J. Microbial DNA records historical delivery of anthropogenic mercury. ISME J. 2015;9(12):2541–2550. doi: 10.1038/ismej.2015.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pal C. The structure and diversity of human, animal and environmental resistomes. Microbiome. 2016;4(54) doi: 10.1186/s40168-016-0199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kim E.H., Rensing C., McEvoy M.M. Chaperone-mediated copper handling in the periplasm. Nat Prod Rep. 2010;27(5):711–719. doi: 10.1039/b906681k. [DOI] [PubMed] [Google Scholar]
- 167.Clark D.L., Weiss A.A., Silver S. Mercury and organomercurial resistances determined by plasmids in Pseudomonas. J Bacteriol. 1977;132(1):186–196. doi: 10.1128/jb.132.1.186-196.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Forsberg K.J. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509(7502):612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Imchen M. Comparative mangrove metagenome reveals global prevalence of heavy metals and antibiotic resistome across different ecosystems. Sci Rep. 2018;8 doi: 10.1038/s41598-018-29521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Malik A.A. Bacterial physiological adaptations to contrasting edaphic conditions identified using landscape scale metagenomics. mBio. 2017;8(4) doi: 10.1128/mBio.00799-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Reyes N.S., Frischer M.E., Sobecky P.A. Characterization of mercury resistance mechanisms in marine sediment microbial communities. FEMS Microbiol Ecol. 1999;30(3):273–284. doi: 10.1111/j.1574-6941.1999.tb00655.x. [DOI] [PubMed] [Google Scholar]
- 172.Barkay T., Liebert C., Gillman M. Environmental significance of the potential for Mer(Tn21)-mediated reduction of Hg2+ to Hg0 in natural waters. Appl Environ Microbiol. 1989;55(5):1196–1202. doi: 10.1128/aem.55.5.1196-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chao Y., Fu D. Kinetic study of the antiport mechanism of an Escherichia coli zinc transporter, ZitB. J Biol Chem. 2004;279(13):12043–12050. doi: 10.1074/jbc.M313510200. [DOI] [PubMed] [Google Scholar]
- 174.Sensfuss C., Reh M., Schlegel H.G. No correlation exists between the conjugative transfer of the autotrophic character and that of plasmids in Nocardia opaca strains. J Gen Microbiol. 1986;132:997–1007. doi: 10.1099/00221287-132-4-997. [DOI] [PubMed] [Google Scholar]
- 175.Hassani M.A., Duran P., Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6(58) doi: 10.1186/s40168-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jasmin C. Aberrations in the microbiome of cyanobacteria from a tropical estuary polluted by heavy metals. Mar Pollut Bull. 2020;160 doi: 10.1016/j.marpolbul.2020.111575. [DOI] [PubMed] [Google Scholar]
- 177.Leisner J.J., Jorgensen N.O., Middelboe M. Predation and selection for antibiotic resistance in natural environments. Evol Appl. 2016;9(3):427–434. doi: 10.1111/eva.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chen J., Yoshinaga M., Rosen B.P. The antibiotic action of methylarsenite is an emergent property of microbial communities. Mol Microbiol. 2019;111(2):487–494. doi: 10.1111/mmi.14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chen J., Rosen B.P. The arsenic methylation cycle: how microbial communities adapted methylarsenicals for use as weapons in the continuing war for dominance. Front Environ Sci. 2020;8 [Google Scholar]
- 180.Butaite E. Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ford S.A. Microbe-mediated host defence drives the evolution of reduced pathogen virulence. Nat Commun. 2016;7 doi: 10.1038/ncomms13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Andersen S.B. Long-term social dynamics drive loss of function in pathogenic bacteria. P Natl Acad Sci USA. 2015;112(34):10756–10761. doi: 10.1073/pnas.1508324112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hibbing M.E. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8(1):15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gu S.H. Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat Microbiol. 2020;5(8):p. 1002-+. doi: 10.1038/s41564-020-0719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nairz M. The struggle for iron - a metal at the host-pathogen interface. Cell Microbiol. 2010;12(12):1691–1702. doi: 10.1111/j.1462-5822.2010.01529.x. [DOI] [PubMed] [Google Scholar]
- 186.Hao X. A role for copper in protozoan grazing - two billion years selecting for bacterial copper resistance. Mol Microbiol. 2016;102(4):628–641. doi: 10.1111/mmi.13483. [DOI] [PubMed] [Google Scholar]
- 187.Hao X. Bacterial resistance to arsenic protects against protist killing. Biometals. 2017;30(2):307–311. doi: 10.1007/s10534-017-0003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Oliverio A.M. The global-scale distributions of soil protists and their contributions to belowground systems. Sci Adv. 2020;6(4) doi: 10.1126/sciadv.aax8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gao Z. Protists: puppet masters of the rhizosphere microbiome. Trends Plant Sci. 2019;24(2):165–176. doi: 10.1016/j.tplants.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 190.Garland J.L., Mills A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol. 1991;57(8):2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pal C. BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014;42(D1):D737–D743. doi: 10.1093/nar/gkt1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Handley K.M. Determining microbial roles in ecosystem function: redefining microbial food webs and transcending kingdom barriers. mSystems. 2019;4(3) doi: 10.1128/mSystems.00153-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tu Q.C. GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour. 2014;14(5):914–928. doi: 10.1111/1755-0998.12239. [DOI] [PubMed] [Google Scholar]
- 194.He Z.L. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010;4(9):1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- 195.Zhao Y. AsChip: a high-throughput qPCR chip for comprehensive profiling of genes linked to microbial cycling of arsenic. Environ Sci Technol. 2019;53(2):798–807. doi: 10.1021/acs.est.8b03798. [DOI] [PubMed] [Google Scholar]
- 196.Wang H.T. Effects of arsenic on gut microbiota and its biotransformation genes in earthworm Metaphire sieboldi. Environ Sci Technol. 2019;53(7):3841–3849. doi: 10.1021/acs.est.8b06695. [DOI] [PubMed] [Google Scholar]
- 197.Christensen G.A. Determining the reliability of measuring mercury cycling gene abundance with correlations with mercury and methylmercury concentrations. Environ Sci Technol. 2019;53(15):8649–8663. doi: 10.1021/acs.est.8b06389. [DOI] [PubMed] [Google Scholar]
- 198.Walts A.E., Walsh C.T. Bacterial organomercurial lyase: novel enzymic protonolysis of organostannanes. J Am Chem Soc. 1988;110(6):1950–1953. [Google Scholar]
- 199.San Francisco M.J. Identification of the metalloregulatory element of the plasmid-encoded arsenical resistance operon. Nucleic Acids Res. 1990;18(3):619–624. doi: 10.1093/nar/18.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yang J. Arsenic binding and transfer by the ArsD As(III) metallochaperone. Biochemistry. 2010;49(17):3658–3666. doi: 10.1021/bi100026a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gupta S.D. Identification of cutC and cutF (nlpE) genes involved in copper tolerance in Escherichia coli. J Bacteriol. 1995;177(15):4207–4215. doi: 10.1128/jb.177.15.4207-4215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Snyder W.B. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol. 1995;177(15):4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rogers S.D. Cloning and characterization of CutE, a gene involved in copper transport in Escherichia coli. J Bacteriol. 1991;173(21):6742–6748. doi: 10.1128/jb.173.21.6742-6748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Grass G., Rensing C. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol. 2001;183(6):2145. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chacón K.N. Tracking metal ions through a Cu/Ag efflux pump assigns the functional roles of the periplasmic proteins. Proc Natl Acad Sci U S A. 2014;111(43):15373–15378. doi: 10.1073/pnas.1411475111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kim E.-H. Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol. 2011;193(10):2381–2387. doi: 10.1128/JB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mellano M.A., Cooksey D.A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1988;170(6):2879–2883. doi: 10.1128/jb.170.6.2879-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cervantes C., Gutierrez-Corona F. Copper resistance mechanisms in bacteria and fungi. FEMS Microbiol Rev. 1994;14(2):121–137. doi: 10.1111/j.1574-6976.1994.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 209.Cha J.-S., Cooksey D.A. Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon. Appl Environ Microbiol. 1993;59(5):1671. doi: 10.1128/aem.59.5.1671-1674.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Arnesano F. A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc Natl Acad Sci U S A. 2003;100(7):3814–3819. doi: 10.1073/pnas.0636904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Morby A.P. SmtB is a metal-dependent repressor of the cyanobacterial metallothionein gene smtA: identification of a Zn inhibited DNA-protein complex. Nucleic Acids Res. 1993;21(4):921–925. doi: 10.1093/nar/21.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cavet J. Construction of Zn2+/Cd2+ hypersensitive cyanobacterial mutants lacking a functional metallothionein locus. J Appl Biol Chem. 1993;268:4494–4498. [PubMed] [Google Scholar]
- 213.Borremans B. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol. 2001;183(19):5651–5658. doi: 10.1128/JB.183.19.5651-5658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hynninen A. An efflux transporter PbrA and a phosphatase PbrB cooperate in a lead-resistance mechanism in bacteria. Mol Microbiol. 2009;74(2):384–394. doi: 10.1111/j.1365-2958.2009.06868.x. [DOI] [PubMed] [Google Scholar]
- 215.Liesegang H. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J Bacteriol. 1993;175(3):767–778. doi: 10.1128/jb.175.3.767-778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Grass G. NreB from Achromobacter xylosoxidans 31A Is a nickel-induced transporter conferring nickel resistance. J Bacteriol. 2001;183(9):2803–2807. doi: 10.1128/JB.183.9.2803-2807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rodrigue A., Effantin G., Mandrand-Berthelot M.A. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J Bacteriol. 2005;187(8):2912–2916. doi: 10.1128/JB.187.8.2912-2916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Navarro C., Wu L.F., Mandrand-Berthelot M.A. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993;9(6):1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- 219.Contreras M. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol Microbiol. 2003;49(4):947–963. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- 220.De la Iglesia R. Novel polymerase chain reaction primers for the specific detection of bacterial copper P-type ATPases gene sequences in environmental isolates and metagenomic DNA. Lett Appl Microbiol. 2010;50(6):552–562. doi: 10.1111/j.1472-765X.2010.02832.x. [DOI] [PubMed] [Google Scholar]
- 221.Besaury L. Abundance and diversity of copper resistance genes cusA and copA in microbial communities in relation to the impact of copper on Chilean marine sediments. Mar Pollut Bull. 2013;67(1):16–25. doi: 10.1016/j.marpolbul.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 222.Xiong W. Fate of metal resistance genes in arable soil after manure application in a microcosm study. Ecotoxicol Environ Saf. 2015;113:59–63. doi: 10.1016/j.ecoenv.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 223.Hasman H. Copper resistance in Enterococcus faecium, mediated by the tcrB gene, is selected by supplementation of pig feed with copper sulfate. Appl Environ Microbiol. 2006;72(9):5784–5789. doi: 10.1128/AEM.02979-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Chaintreuil C. Nickel resistance determinants in bradyrhizobium strains from nodules of the endemic New Caledonia legume Serianthes calycina. Appl Environ Microbiol. 2007;73(24):8018–8022. doi: 10.1128/AEM.01431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kaci A. Distinct diversity of the czcA gene in two sedimentary horizons from a contaminated estuarine core. Environ Sci Pollut Res Int. 2014;21(18):10787–10802. doi: 10.1007/s11356-014-3029-y. [DOI] [PubMed] [Google Scholar]
- 226.Roosa S. Bacterial metal resistance genes and metal bioavailability in contaminated sediments. Environ Pollut. 2014;189:143–151. doi: 10.1016/j.envpol.2014.02.031. [DOI] [PubMed] [Google Scholar]
- 227.Liu Y.R. Analysis of the microbial community structure by monitoring an Hg methylation gene (hgcA) in paddy soils along an Hg gradient. Appl Environ Microbiol. 2014;80(9):2874–2879. doi: 10.1128/AEM.04225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liebert C.A. Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl Environ Microbiol. 1997;63(3):1066–1076. doi: 10.1128/aem.63.3.1066-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yuan L. Mercury/silver resistance genes and their association with antibiotic resistance genes and microbial community in a municipal wastewater treatment plant. Sci Total Environ. 2019;657:1014–1022. doi: 10.1016/j.scitotenv.2018.12.088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.