Abstract
Chemical proteomics has taken shape as a powerful technique that seeks to understand small molecule and protein function in the physiological system. Chemical proteomics that integrates activity-based protein profiling (ABPP) technology with mass spectrometry has been introduced to evaluate small molecule and protein interaction and expand the druggable proteome.
DNA sequencing technologies have facilitated the discovery of many genes that play fundamental roles in human disease1. The protein products encoded by some of these genes have served as targets for ground-breaking new medicines2. Other genes, however, code for proteins, such as transcription factors and adaptor proteins, that lack small-molecule ligands and are often considered “undruggable”. In these cases, our knowledge of human genetics has not yet been effectively translated into new therapies. A critical challenge has thus emerged in biomedical research – how can the massive gains in understanding of the genetics of human disease be translated into new therapies? Over the past decade, chemical proteomics has taken shape as a powerful technique that seeks to understand small molecule and protein function in the physiological system and plays an irreplaceable role in drug discovery3. The development of the activity-based protein profiling (ABPP) technology, pioneered by Benjamin Cravatt, uses broad-spectrum chemical probes to functionally characterize large numbers of proteins in native biological systems4. Chemical proteomics that integrates ABPP with mass spectrometry (MS) has been introduced for simultaneously evaluating the reactivity and small-molecule interactions of thousands of sites on proteins from diverse structural and mechanistic classes (Figure 1). This relationship extends far beyond enzyme active sites to include more cryptic sites of functionality/druggability at, for instance, protein-DNA/protein-protein interfaces in transcription factors, scaffolding/adaptor proteins, and E3 ligases. Accordingly, an outstanding proteome coverage has now been achieved, with a still growing inventory of 30,000+ sites on 10,000+ human proteins5-7.
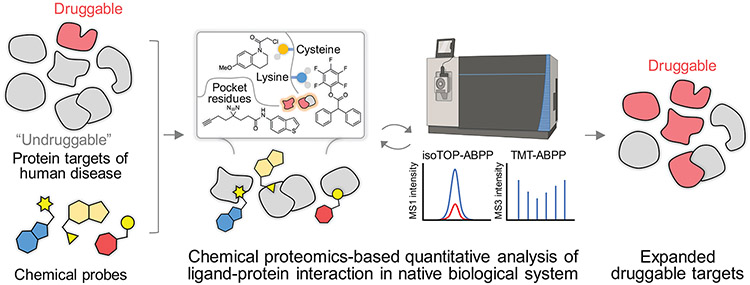
Figure 1.
Schematic depicting the chemical proteomic platform for expanding druggability of human proteome. The structures shown are cysteine reactive5, lysine reactive6 and fully functionalized7 probes. The ligand-protein interaction can be quantitatively analyzed using isoTOP-ABPP (isotopic Tandem Orthogonal Proteolysis – ABPP)9 or TMT-ABPP (Tandem Mass Tag – ABPP)10 technology.
One example that leverages chemical proteomic platform to expand druggability is from a recent study by Bar-Peled et al. that identifies NR0B1 as a co-dependent vulnerable target in KEAP1-mutant non-small-cell lung cancers (NSCLCs)8. NR0B1 is a poorly understood orphan nuclear receptor with a very shallow pocket in the typical ligand-binding domain and considered as a ligandless adaptor. The authors used their ligand-discovery platform that integrates ABPP and chemical proteomics to identify druggable sites on proteins that are preferentially expressed in KEAP1-mutant NSCLCs. NR0B1 was found to engage in a multimeric protein complex to regulate the transcriptional output of KEAP1-mutant NSCLC cells, and, more crucially, possess a ligandable cysteine in its protein interaction domain. The authors then developed small molecules that covalently target the ligandable cysteine, thereby disrupting NR0B1 complexes and suppressing the growth of KEPA1-mutant NSCLCs. This study underscores the value of emerging chemical proteomic methods in discovering druggable vulnerabilities in generically defined cancers, in this case disclosing a cysteine in the protein-protein interaction domain of a transcriptional regulator that is amenable to inactivation by small molecules.
The systematic advancements that have been made by chemical proteomics and ABPP over the past decade have furnished the first technology platform for realizing the complete druggability of the human proteome. Now a much larger fraction of the human proteome can be targeted by small molecules than estimated by past predictions of protein druggability. Accordingly, by leveraging and continuing to advance the chemical proteomic and ABPP platforms, many more chemical probes can be identified for biologically credentialed, but, as-of-yet, undrugged disease targets.
Acknowledgement
I would like to thank Prof. Benjamin Cravatt and Prof. Hening Lin for their support and advice in my early scientific career. My research is supported by the National Cancer Institute of the National Institutes of Health under Award Number K99CA248715.
Biography

Xiaoyu Zhang was born in China and earned his B.S. degree in Pharmaceutical Science in 2008 and M.S. degree in Pharmaceutical Science in 2011 from Zhejiang University. He obtained his Ph.D. degree in Biochemistry and Chemical Biology in 2017 from Cornell University under the guidance of Dr. Hening Lin. His doctoral thesis work focused on understanding the biological roles of NAD+-dependent deacylases in human disease. He is currently a postdoctoral fellow in Dr. Benjamin Cravatt’s laboratory at The Scripps Research Institute and interested in combining chemical tools, chemoproteomic platforms and molecular and cell biology approaches to broadly interrogate and discover E3 ubiquitin ligases that support small molecule-induced protein degradation. He has been recognized with a number of scientific honors, including The NIH Pathway to Independence Award (K99/R00), Damon Runyon Postdoctoral Fellowship Award, Eli Lilly Asia Outstanding Graduate Thesis Award, Keystone Symposia Future of Science Fund Scholarship, and Chu Kochen Scholarship.
References
- 1.Gonzaga-Jauregui C, Lupski JR & Gibbs RA Human genome sequencing in health and disease. Annu Rev Med 63, 35–61, doi: 10.1146/annurev-med-051010-162644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berndt N, Karim RM & Schonbrunn E Advances of small molecule targeting of kinases. Curr Opin Chem Biol 39, 126–132, doi: 10.1016/j.cbpa.2017.06.015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drewes G & Knapp S Chemoproteomics and Chemical Probes for Target Discovery. Trends Biotechnol 36, 1275–1286, doi: 10.1016/j.tibtech.2018.06.008 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Cravatt BF, Wright AT & Kozarich JW Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem 77, 383–414, doi: 10.1146/annurev.biochem.75.101304.124125 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Backus KM et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574, doi: 10.1038/nature18002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hacker SM et al. Global profiling of lysine reactivity and ligandability in the human proteome. Nat Chem 9, 1181–1190, doi: 10.1038/nchem.2826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker CG et al. Ligand and Target Discovery by Fragment-Based Screening in Human Cells. Cell 168, 527–541 e529, doi: 10.1016/j.cell.2016.12.029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bar-Peled L et al. Chemical Proteomics Identifies Druggable Vulnerabilities in a Genetically Defined Cancer. Cell 171, 696–709 e623, doi: 10.1016/j.cell.2017.08.051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weerapana E et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795, doi: 10.1038/nature09472 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinogradova EV et al. An Activity-Guided Map of Electrophile-Cysteine Interactions in Primary Human T Cells. Cell, doi: 10.1016/j.cell.2020.07.001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]