Abstract
Understanding the SARS-CoV-2 virus’ pathways of infection, virus–host–protein interactions, and mechanisms of virus-induced cytopathic effects will greatly aid in the discovery and design of new therapeutics to treat COVID-19. Chloroquine and hydroxychloroquine, extensively explored as clinical agents for COVID-19, have multiple cellular effects including alkalizing lysosomes and blocking autophagy as well as exhibiting dose-limiting toxicities in patients. Therefore, we evaluated additional lysosomotropic compounds to identify an alternative lysosome-based drug repurposing opportunity. We found that six of these compounds blocked the cytopathic effect of SARS-CoV-2 in Vero E6 cells with half-maximal effective concentration (EC50) values ranging from 2.0 to 13 μM and selectivity indices (SIs; SI = CC50/EC50) ranging from 1.5- to >10-fold. The compounds (1) blocked lysosome functioning and autophagy, (2) prevented pseudotyped particle entry, (3) increased lysosomal pH, and (4) reduced (ROC-325) viral titers in the EpiAirway 3D tissue model. Consistent with these findings, the siRNA knockdown of ATP6V0D1 blocked the HCoV-NL63 cytopathic effect in LLC-MK2 cells. Moreover, an analysis of SARS-CoV-2 infected Vero E6 cell lysate revealed significant dysregulation of autophagy and lysosomal function, suggesting a contribution of the lysosome to the life cycle of SARS-CoV-2. Our findings suggest the lysosome as a potential host cell target to combat SARS-CoV-2 infections and inhibitors of lysosomal function could become an important component of drug combination therapies aimed at improving treatment and outcomes for COVID-19.
Keywords: SARS-CoV-2, cytopathic effect, autophagy, small molecule inhibitors, coronavirus
The COVID-19 global viral pandemic caused by SARS-CoV-2 began in late 2019 and originated from Wuhan, Hubei Province, China.1 The betacoronavirus SARS-CoV-2 is a member of the Coronaviridae family of positive single-stranded RNA viruses. As of November 19, 2020, there have been over 55,000,000 infections worldwide and over 1,300,000 deaths.2 While not the deadliest virus in the past century, it is highly infectious (estimated R0 = 5.7).3 The absolute number of infections and mortality will not be known for several years, and after only seven months, it is already in the top five most deadliest pandemic outbreaks of this century.4
The SARS-CoV-2 infection in humans produces a disease called coronavirus disease of 2019, COVID-195.6 It is related to the 2003 coronavirus outbreak of SARS-CoV, the original SARS. For the current COVID-19, symptoms range from mild fever, tiredness, and dry cough to acute respiratory distress syndrome, stroke due to blood clots, cardiac and renal damage, and death.7 While some clinical symptoms are common among patients with severe disease, its epidemiology and the mechanisms of disease pathology are still unclear and need to be further studied. Evidence has emerged that, in addition to the airway epithelium, endothelial cells in the vascular system and cardiomyocytes in the heart are also major sites of SARS-CoV-2 infection.8,9
The research and clinical responses have been unprecedented, and much of the effort is focused on identifying therapeutics, including drug repurposing efforts with the experimental anti-Ebola virus drug remdesivir,10,11 and developing vaccines. Chloroquine (CQ), an older FDA-approved antimalarial drug, along with its better tolerated analog hydroxychloroquine (HCQ) have been reported to inhibit SARS-CoV-2 infection in vitro and show some promise in patients.12−14 In mice, CQ and HCQ display antiviral effects against human coronavirus strain OC43,15 human enterovirus EV71,16 Zika virus,17 and human influenza virus H5N1.18 CQ was not effective in reducing viral titers in the lungs of mice infected with SARS-CoV, although it did induce a reduction in markers of inflammation.19 CQ and HCQ have been reported to elicit antiviral activity via a number of mechanisms of action including its alkalizing effects on acidic compartments such as the late endosomes and lysosomes. However, HCQ has been reported to be ineffective in reducing viral replication/shedding in animal models of SARS-CoV-2 and clinical disease symptoms.20 Indeed, most clinical trials on CQ and HCQ have shown no positive effect on morbidity and mortality in either prophylaxis or treatment.21 It is clear that other repurposing and improved molecular entities are needed to reduce clinical symptoms of COVID-19 and death due to the viral pandemic.
CQ, in addition to its inhibitory effects on the lysosome and autophagy, has been reported to have broad antiviral effects through several mechanisms of action. One in particular is the disruption of the early steps in the viral life cycle including the release of the virus from the endosome when endocytosis is used for viral entry.22,23 The basic amine property of CQ and similar molecules leads to their accumulation in cellular acidic compartments and raises their pH.24 Viruses such as SARS-CoV that depend on low acidic pH for entry and uncoating can no longer execute functions required for viral entry into host cells after CQ treatment.25 While these compounds exert multiple cellular effects, their characterized inhibition of autophagic flux and elevation of vesicular pH are consistent with the antiviral efficacy in vitro.26 Accordingly, a SARS-CoV-2 study by Liu and co-workers has proposed that these drugs may act by preventing the progression of the virions through the endocytic pathway after binding to the angiotensin coverting enzyme 2 (ACE2) on the cell surface, thereby inhibiting release of the viral genome.14 Additionally, cathepsin proteases active at low pH in acidic organelles such as endosomes and lysosomes have been reported to play a role in viral entry where endocytosis is the main entry mechanism.27
In this study, we have identified 6 annotated molecules including CQ and HCQ that reduce the cytopathic effect (CPE) of SARS-CoV-2 in Vero E6 cells. We have investigated the activities of these molecules on endocytosis, lysosome, and autophagy using LC3B immunostaining as well as LysoTracker dye staining.28 To explore the compounds’ mechanism of action, we used SARS-CoV-2 pseudotyped particles (PPs) that transduce cells with luciferase through a viral spike-mediated endocytosis.29 We also evaluated the compounds for endocytosis of dextran, changes in vesicular pH, p62 accumulation, and a model virion system using quantum dots conjugated to the SARS-CoV-2 receptor binding domain (QD-RBD). We obtained QD-RBD immunofluorescence colocalization data that reveals the subcellular localization of the nanoparticle after endocytosis. We explored the host proteins involved in coronavirus infection by knocking down genes related to the maintenance of acidic pH and autophagosome formation. We further evaluated SARS-CoV-2 infected Vero E6 cell lysate for changes in lysosomal and autophagy pathways to shed light on the cellular pathologies that follow infection. Lastly, we demonstrated that ROC-325 reduces the viral titer of a 3D lung tissue model infected with SARS-CoV-2. Altogether, our work suggests that an alkalization of the acidic compartments in host cells is an effective strategy to reduce viral infection and that the lysosome is a viable target organelle for COVID-19 drug discovery.
Results
Lysosomotropic Compounds Block the SARS-CoV-2 CPE
We employed a cell-based assay using Vero E6 host cells that measures the CPE of SARS-CoV-2 (Figure S1).
The CPE reduction assay is a widely employed assay format to screen for antiviral agents, and it can be scaled for high-throughput screening.30,31 In this assay, host cell death is a consequence of the viral infection and cell viability is a surrogate readout for viral infection that can be measured with a range of cell viability assays. The CPE assay was optimized to result in 5% viability at the 72 h time point. All compounds were tested in dose–response assays, and “hit” antiviral compounds were those that protected the host cells from the viral CPE. To increase infectivity of SARS-CoV-2 in the CPE assay, we used a clone of Vero E6 that had previously been selected for high ACE2 expression.30 The cell viability measurements were normalized to cells not infected with the virus (100% activity) and untreated cells infected with the virus (0% activity; virus completely kills the cells). As a counter assay, all compounds were tested against cells not exposed to the virus in order to identify compounds that exerted cytotoxicity against Vero E6 cells.
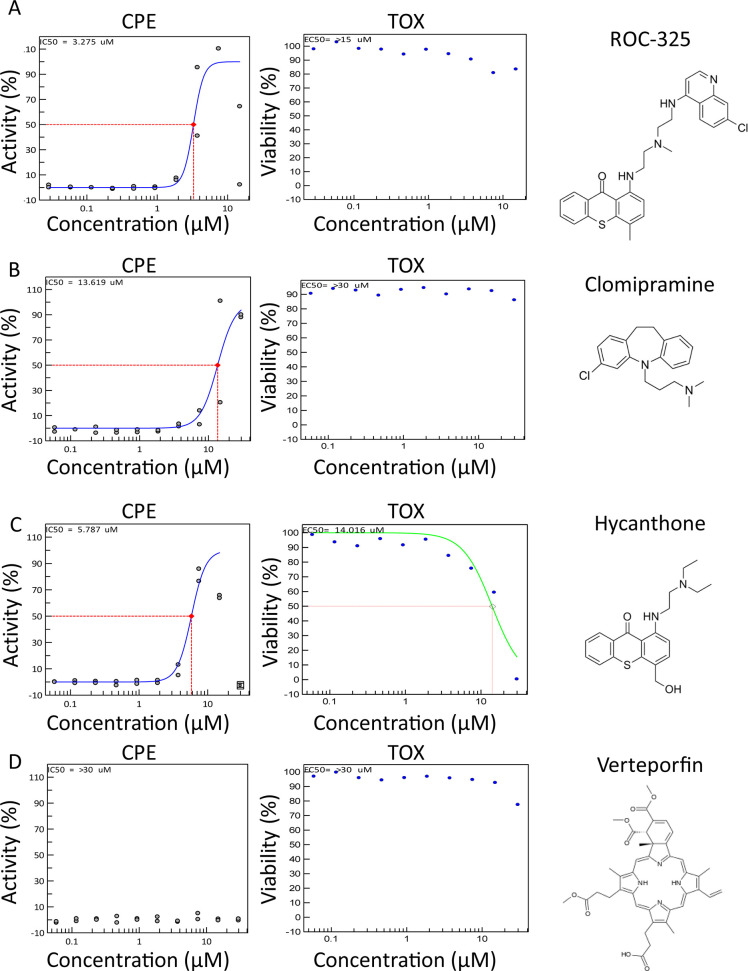
Given that lysosomotropic compounds and autophagy inhibitors including HCQ have shown efficacy against many different types of viruses22 including SARS-CoV-2 in CPE assays,14 we assessed the protective effect of a group of inhibitors including ROC-325, clomipramine, hycanthone, verteporfin, CQ, HCQ, and mefloquine in 384-well plates (Figure 1).
Figure 1.
CPE activity and toxicity for ROC-325, clomipramine, hycanthone, and verteporfin. (A) ROC-325, (B) clomipramine, (C) hycanthone, and (D) verteporfin CPE activity (blue curve, left graph) and toxicity (green curve, right graph) in 10 point, 1:2 dilution concentration–response curves starting at 30.0 μM down to 2.29 nM, along with their structures. ROC-325 started at 15 μM down to 1.14 nM. The red dashed line indicates EC50 or CC50 for CPE and toxicity assays, respectively. Duplicate values are shown for each concentration. Curves were generated using nonlinear regression.
While CQ was the most potent compound (discussed below), ROC-325 was the second most potent with a half-maximal effective concentration (EC50) of 3.28 μM and less than 20% cytotoxicity at 30.0 μM (Figure 1A), indicating a greater than 10-fold selectivity index (SI) between antiviral and cytotoxic concentrations. Clomipramine exhibited an EC50 of 13.6 μM while inducing less than 20% cytotoxicity at 30.0 μM (Figure 1B). Hycanthone demonstrated an EC50 of 5.79 μM and a half-maximal cytotoxic concentration (CC50) of 14.0 μM (Figure 1C). Hycanthone’s concentration–response was bell-shaped due to the reduction of cell viability by almost 100% at 30 μM. Verteporfin was inactive in the screen against SARS-CoV-2 CPE and reduced cell viability by approximately 22% at 30.0 μM (Figure 1D).
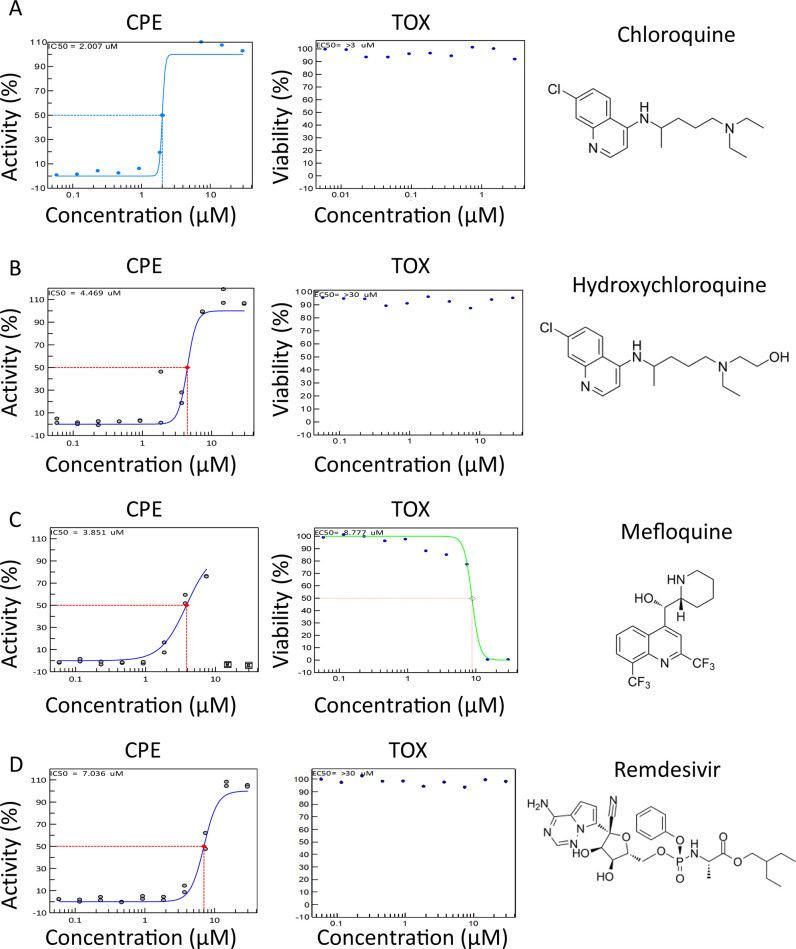
The antimalarial drugs CQ and HCQ inhibited viral CPE with an EC50 of 2.01 and 4.47 μM, respectively, with no associated cell toxicity (Figure 2A,B). HCQ was the third most potent compound tested in the CPE. Mefloquine, a related antimalarial compound, exhibited an EC50 of 3.85 μM with an associated CC50 of 8.78 μM and 100% cytotoxicity at 15.0 to 30.0 μM (Figure 2C). For comparison, remdesivir, the nucleotide analog inhibitor of RNA-dependent RNA polymerase for a number of viruses and FDA approved for hospitalized COVID-19 patients,10,11,32 exhibited an EC50 of 7.04 μM with no apparent cytotoxicity (Figure 2D). The EC50 values for all of the compounds are summarized in Table 1.
Figure 2.
CPE activity and toxicity for CQ, HCQ, mefloquine, and remdesivir. (A) CQ, (B) HCQ, (C) mefloquine, and (D) remdesivir CPE activity (blue curve, left graph) and toxicity (green curve, right graph) in 10 point, 1:2 dilution concentration–response curves starting at 30.0 μM down to 2.29 nM, along with their structures. The dashed line indicates EC50 or CC50 for CPE and toxicity assays, respectively. Duplicate values are shown for each concentration. Curves were generated using nonlinear regression.
Table 1. CPE Assay in Vero E6 and Average LC3B-Based Autophagy Assay Parameters from Four Cell Linesa.
| compound | reported MoA | CPE EC50 (μM)b | CPE CC50 (μM) | CPE SI | autophagy EC50 (μM)c | autophagy CC50 (μM)c | autophagy SI | MoA ref. |
|---|---|---|---|---|---|---|---|---|
| chloroquine | ↓ lysosome fusion | 2.01 | >30 | >10 | 3.29 ± 1.86, 1.82 | >50 | >10 | (80) |
| ROC-325 | ↓ lysosome fusion | 3.28 ± 0.57 | >30 | >10 | 5.2 ± 1.71, 1.68 | >25 | >10 | (38) |
| mefloquine | ↓ autophagic flux | 3.85 ± 0.24 | 8.78 | 2.3 | 7.3d | 18.4 ± 2.08, 2.04 | 2.6 | (45) |
| hydroxychloroquine | ↓ lysosome fusion | 4.47 | >30 | >10 | 6.55 ± 6.67, 6.53 | >50 | >10 | (14) |
| hycanthone | ↑ lysosomal membrane permeabilization | 5.79 ± 0.26 | 14.2 | 2.5 | 7.35 ± 4.7, 4.61 | 11.3 ± 2.73,e 2.68 | 1.5 | (44) |
| clomipramine | ↓ autophagic flux | 13.6 ± 2.96 | >30 | >10 | 13.2 ± 5.4, 5.28 | >50 | >10 | (41) |
| verteporfin | ↓ autophagosome formation | ND | >30 | ND | ND | ND | ND | (81) |
ND: not determined. SI > 10 used when no CC50 was calculated.
Data shown as mean ± confidence interval (CI).
Data shown as mean ± SD, CI.
Could only be calculated from Huh-7.5.
Max inhibition of cell viability: ∼60%.
To demonstrate the day to day reproducibility of the CPE assay, compound concentration–response curves and EC50 values from multiple experiments are shown for CQ, HCQ, and remdesivir (Figure S2). Compound EC50 values in the CPE assay were consistent across multiple experiments with geometric mean EC50’s of 1.96 μM for CQ, 4.75 μM for HCQ, and 4.75 μM for remdesivir.
Lysosomotropic Compounds Increase LC3B and LysoTracker Dye Staining
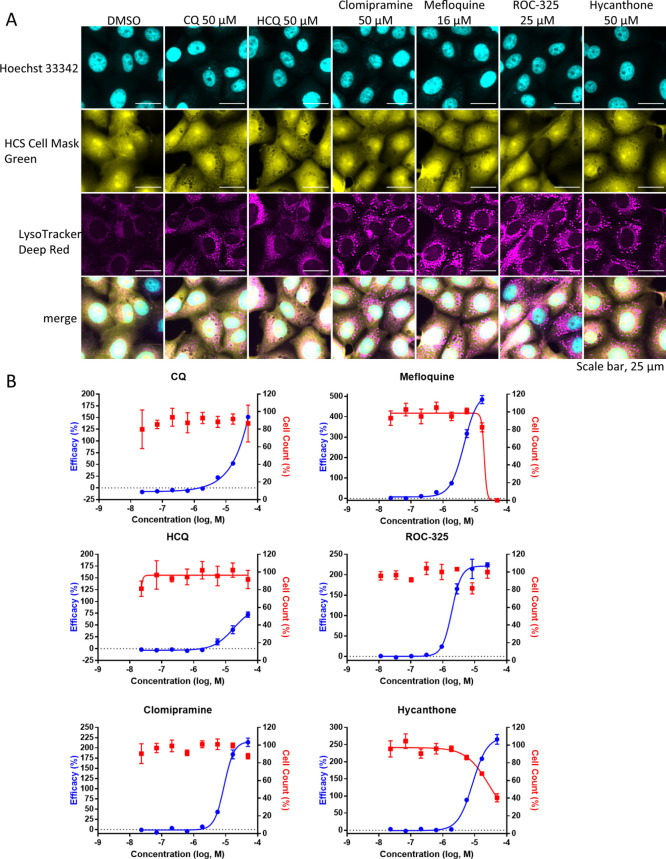
Because 6 out of the 7 autophagy inhibitors (ROC-325, clomipramine, hycanthone, CQ, HCQ, and mefloquine) showed activity in the CPE assay, we sought to confirm their effect on autophagy in Vero E6, HeLa, HEK293T, and Huh-7.5 cells using immunostaining for autophagy marker LC3B as well as LysoTracker dye staining. LC3B immunostaining directly visualizes autophagosomes, while LysoTracker Dye accumulates preferentially in acidic organelles. These assays allow for the visualization of autophagosome accumulation and acidic organelles such as endosomes and lysosomes, respectively. Compounds that block autophagic flux by way of lysosomal disruption are expected to increase LC3B and LysoTracker staining measurements.28
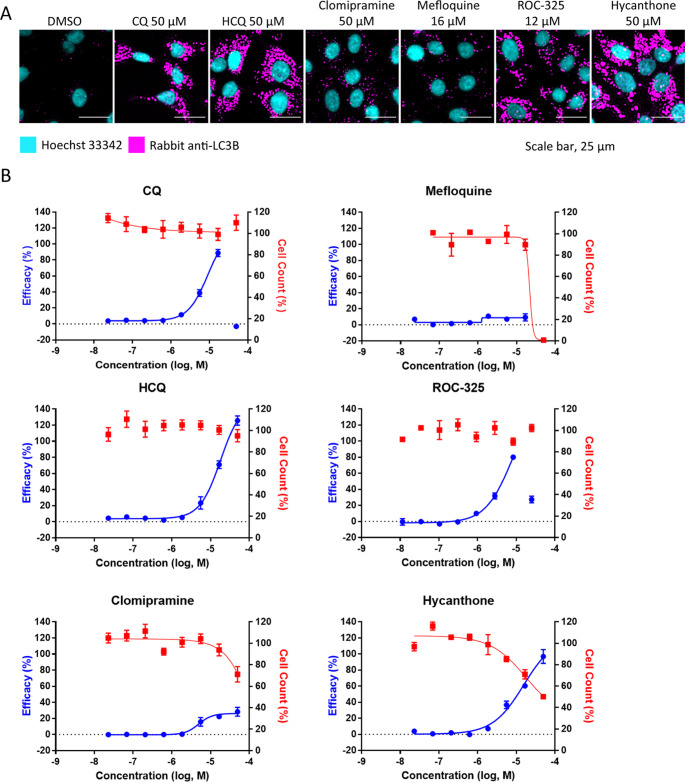
To carry out this assay, cells were allowed to adhere overnight and were then treated with compounds at concentrations ranging from 50 to 0.02 μM for approximately 16 h. In Vero E6 cells, increases in the intracellular LC3B spot, also called spots, were concentration dependent for all of the compounds except for mefloquine (Figure 3A,B). CQ, HCQ, and hycanthone treatment produced maximal spot counts, while ROC-325 and clomipramine produced a submaximal increase of 80% and 40%, respectively. Mefloquine was ineffective at inducing LC3B spot accumulation. Increases in LC3B spots indicate an accumulation of LC3B that is localized to the autophagosome when autophagic flux is blocked. The potent effect of CQ and HCQ on LC3B spot counts was apparent in all cell lines tested (Figures 3 and S3–S5). On the basis of nuclei counts, CQ, HCQ, clomipramine, and ROC-325 were not cytotoxic at the highest concentrations (50 μM for all except for ROC-325 at 25 μM). In line with the drug toxicity data from the CPE assay, mefloquine was completely toxic at 50 μM, while hycanthone killed approximately 60% of the cells at 50 μM. The compound CC50 data was consistent between the two assays.
Figure 3.
Autophagy inhibition assay using LC3B immunostaining in Vero E6 cells. (A) Image montage of DMSO, CQ, HCQ, clomipramine, mefloquine, ROC-325, and hycanthone stained with Hoechst 33342 (cyan) and LC3B (magenta). CQ and HCQ images were taken from wells in positive control column 2. Scale bar, 25 μm. (B) 8 point, 1:3 dilution concentration–response curves starting at 50 μM down to 0.023 μM for compounds in (A). The blue curve indicates the efficacy in LC3B accumulation (increase in spot count), and the red curve indicates cell counts. The efficacy data was normalized to DMSO (0%) and CQ (100%). Cell count data was normalized to DMSO (100%) and 0 (no cells 0%). Error bars indicate SD. N = 3 intraplate replicates. Curves were generated using nonlinear regression.
In Vero E6 cells, we observed drug-induced increases in LysoTracker relative spot intensity measurements that were concentration dependent (Figure 4A,B). With the exception of HCQ, the maximum efficacy was higher than the CQ positive control (100%) that was used to normalize the responses. Interestingly, clomipramine and mefloquine, which did not induce large increases in Vero E6 LC3B spot counts, produced dramatic elevations in LysoTracker relative spot intensity similar to ROC-325 and hycanthone (Figure 4B). In further support of the CPE assay data, mefloquine was toxic at the highest concentration.
Figure 4.
LysoTracker Deep Red staining in Vero E6 cells. (A) Image montage of DMSO, CQ, HCQ, clomipramine, mefloquine, ROC-325, and hycanthone stained with Hoechst 33342 (cyan), HCS Cell Mask Green (yellow), and LysoTracker Deep Red (magenta). CQ and HCQ images were taken from wells in positive control column 2. Scale bar, 25 μm. (B) 8 point, 1:3 dilution concentration–response curves starting at 50 μM down to 0.023 μM for the compounds in (A). The blue curve indicates efficacy, and the red curve indicates cell counts. Efficacy data are normalized to DMSO (0%) and CQ (100%). Cell count data are normalized to DMSO (100%) and 0 (no cells 0%). Error bars indicate SD. N = 3 intraplate replicates. Curves were generated using nonlinear regression.
In addition to Vero E6 monkey epithelial kidney cells, we also examined the effects of these compounds in three human cell lines and observed some differences between them (Figures S3–S8). For example, in Huh-7.5, mefloquine increased LC3B spot counts even at low concentrations (Figure S4), whereas in other cell lines, it was not a potent inducer of autophagosome accumulation. Clomipramine was effective in increasing LC3B in all cell lines except for Vero E6 (Figures 3 and S3–S5). In contrast, hycanthone and mefloquine produced the strongest effect on LysoTracker measurements in Vero E6 compared to the other three cell lines (Figures 4 and S6–S8). Although there were some interesting variations in compound effects among the cell lines tested, the average EC50 and CC50 values from the LC3B spot count measurements in all four cell lines corresponded well with the data from the CPE assay, indicating that the effects of the compounds related to lysosomal dysfunction and protection from viral-induced cell death were well-correlated (Table 1).
Exploring the Mechanism for CPE Inhibition
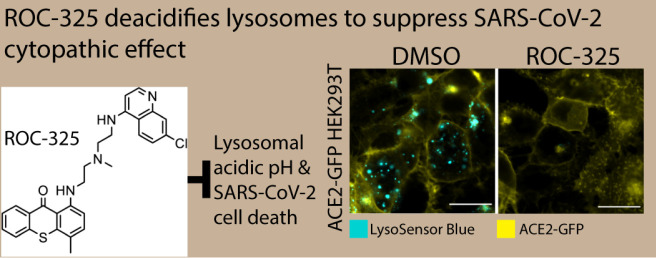
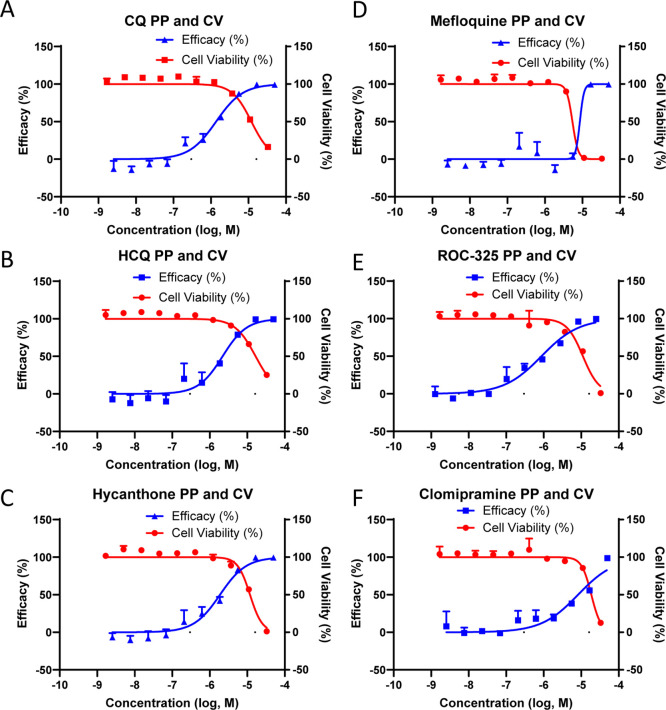
The inhibition of autophagic flux in cells treated with CQ is a secondary effect to the increase of lysosomal pH that prevents autophagosome/lysosome fusion to form autophagolysosomes. Autophagosomes are nascent organelles formed to degrade cellular waste and as a first line of defense against pathogens. To probe the mechanism of SARS-CoV-2 infection inhibition, we utilized the SARS-CoV-2 spike pseudotyped particle (PP) assay in HEK239T cells stably transfected with ACE2 fused to green fluorescent protein (ACE2-GFP) that are permissive to spike-mediated PP entry and transduction of luciferase.33 All compounds inhibited PP entry, and while some toxicity was observed as measured by ATP content, the CC50/EC50 ratio, or SI, indicated an efficacy independent of compound toxicity (Figure 5, Table 2). The data suggested these compounds blocked the cellular entry of SARS-CoV-2 PPs that rely on spike-mediated endocytosis and fusion in ACE2-expressing cells.
Figure 5.
Compounds inhibit SARS-CoV-2 PP transduction of luciferase in ACE2-GFP HEK293T cells. Activity and cytotoxicity curves for (A) CQ, (B) HCQ, (C) hycanthone, (D) mefloquine, (E) ROC-325, and (F) clomipramine treated cells for 24 h followed by 48 h of transduction with PP. Curves were generated using nonlinear regression. Data are normalized to delEnv particles lacking the SARS-CoV-2 envelope protein (no transduction, 100%) and DMSO treated cells (0%). Error bars indicate SD. N = 3 triplicate wells in a 384-well plate.
Table 2. PP EC50 and CC50 Average Values with the SI (CC50/EC50 Ratio)a.
| compound | PP EC50 (μM) | CC50 (μM) | SI |
|---|---|---|---|
| bafilomycin A1 | ND | ND | ND |
| CQ in ddH2O | 1.37 ± 0.10 | 12.0 ± 0.51 | 8.7 |
| HCQ in ddH2O | 2.19 ± 0.07 | 16.7 ± 20.3 | 7.6 |
| hycanthone | 1.88 ± 0.08 | 11.7 ± 0.49 | 6.2 |
| mefloquine | 8.84 ± 0.64 | 5.40 ± 0.01 | 0.6 |
| ROC-325 | 0.87 ± 0.12 | 11.0 ± 0.71 | 12.7 |
| clomipramine | 8.28 ± 0.46 | 18.6 ± 0.29 | 2.2 |
Data shown as mean ± SD. ND: not determined.
We tested the inhibition of endocytosis by these compounds directly using a pHrodo Red Dextran endocytosis assay whose fluorescence readout depends on both endocytosis and the acidic pH of the endosomes and lysosomes. All compounds blocked pHrodo Red Dextran uptake and fluorescence, suggesting an alkalizing effect on the acidic compartments of the endolysosomal system that controls endocytic uptake of extracellular material (Figure S9).
pHrodo Red Dextran readouts are affected by both endocytosis and pH levels in endolysosomes. To isolate the effect of the compounds on endocytosis and acidic organelle pH, a LysoSensor Blue dye was used to determine the pH of acidic compartments after cells were treated with the compounds. This pH dye has a pKa of approximately 5.1 that fluoresces brightly at acidic pH and dims at more alkaline pH. The compounds potently reduced the fluorescence of LysoSensor Blue, indicating that the compounds have an alkalizing effect on normally acidic organelles (Figure S10). We also confirmed that the alkalizing effect of these compounds on acidic compartments is correlated with the inhibition of autophagy in Expi293F cells stably expressing ACE2 and transduced with GFP-p62. A shorter compound treatment of 16 h elevated p62 puncta counts after the compound treatment, confirming a blockade of autophagic flux (Figure S11). We applied the same method to the tandem GFP-RFP LC3B autophagy sensor and found a similar increase in both GFP- and RFP-positive spot counts, indicating blocked autophagic flux.
Monitoring SARS-CoV-2 Endocytosis and Localization Using a Quantum Dot Nanoparticles
We previously developed a quantum dot-conjugated SARS-CoV-2 receptor binding domain nanoparticle (QD-RBD) that tracks viral spike binding to ACE2 and subsequent endocytosis into the cell.34 Upon treating cells with QD-RBD, the nanoparticle rapidly binds ACE2 on the cell surface and induces endocytosis. In ACE2-GFP expressing cells, both the QD-RBD and ACE2 can be visualized using fluorescence microscopy. In compound treated cells, the QD-RBD was still able to enter the cells as many puncta similar to the DMSO treated cells were observed (Figure S12). However, we noticed an enlargement of the puncta size, suggesting that, while the QD-RBD was endocytosed, there was an accumulation of the endocytic vesicles compared to the DMSO treated cells.
The QD-RBD nanoparticle treated cells were fixed and immunostained for markers of endosomes, lysosomes, and autophagosomes (Figure S13). Interestingly, the QD-RBD probe most strongly colocalized with Rab7, LAMP1, and GABARAPL1. ATP6V0D1, a subunit of the V-ATPase, GABARAP, and cathepsin B were next in order of the most to least colocalization as measured by the Pearson’s R value. The least amount of colocalization was observed with ATG12, EEA1, and ATG5. While the entry and progression through the endocytic pathway is time dependent and these cells were treated with QD-RBD for 3 h, the strong colocalization of the late endosomal and lysosomal markers indicated the virus could reach these organelles rapidly.
We relied on the availability of screening data using the OpenData Portal for SARS-CoV-235,36 to understand more about the relationship of endosomal trafficking inhibitors and pH modulators. One such molecule is VPS34-IN-1, an inhibitor of class III phosphotidylinositide 3-kinase vacuolar protein sorting 34 (VPS34) originally identified in yeast, which blocks the maturation of endosomes. VPS34-IN-1 potently inhibited the SARS-CoV-2 CPE at concentrations that were not cytotoxic, although cytotoxicity was observed at higher concentrations (Figure S14). Bafilomycin A1, the classic inhibitor of V-ATPase that prevents acidification of the lysosome, is highly cytotoxic but nonetheless was able to block SARS-CoV-2 CPE at nontoxic nanomolar concentrations (Figure S14). Combined, the data suggested that endocytosis and the maturation of endosomes toward the lysosome are critical routes for viral infection in cells expressing ACE2.
Interrogating the Host Factors Necessary for Coronavirus Infection
To directly test whether acidic organelles or autophagosomes are involved in coronavirus infection, we used siRNA-mediated knockdown of ATP6 V0D1, a subunit of the V-ATPase, or ATG5 and ATG7, respectively (Figure S15). The ATG5 and ATG7 proteins are important components of the phagophore initiation and elongation stages of autophagosome formation. We were able to substantially reduce the expression of ATP6V0D1 but were less successful in knocking down ATG5 and ATG7. siRNA treated LLC-MK2 cells, a monkey epithelial cell line, were then infected with human coronavirus-NL63 (HCoV-NL63), which caused a significant CPE. The knockdown of ATP6V0D1 rescued the CPE by 60%. While autophagosomal involvement could not be ruled out given the insufficient knockdown, the reduction in expression of one V-ATPase subunit reduced the coronaviral CPE. The siRNA knockdown of ATP6V0D1 in ACE2-GFP HEK239T cells also appeared to prevent the PP entry, although the data was unclear because the scrambled siRNA control interfered with PP transduction (Figure S16).
Assessing the Impact of SARS-CoV-2 Infection on Cellular Pathways
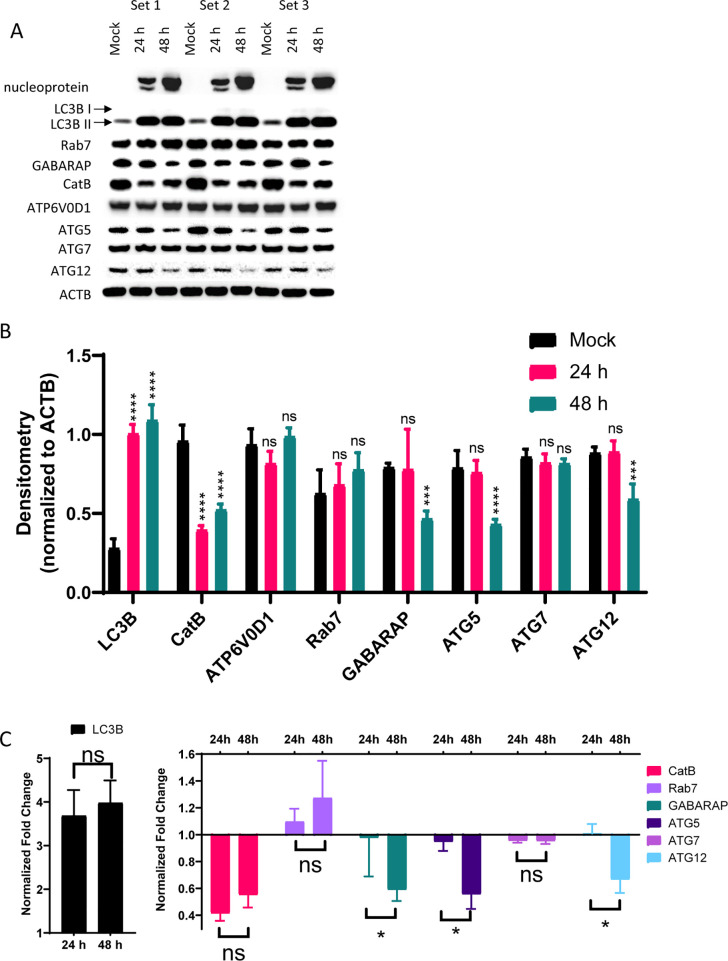
SARS-CoV-2 replication starts after its entry into the cells and release of its viral genome into the cytosol. The virus hijacks the cellular machinery for all stages of viral replication that are crucial for its infectivity. We analyzed SARS-CoV-2 infected Vero E6 cell lysates for protein expression of endolysosomal- and autophagy-related systems. Protein concentrations in mock, 24, and 48 h infected lysates were first measured to normalize protein loading. Lysates were immunoblotted for SARS-CoV-2 nucleoprotein, LC3B-II, cathepsin B (CatB), ATP6V0D1, GABARAP, ATG5, ATG7, and ATG12 with ACTB as a loading control (Figure 6A,B). A significant increase in LC3B-II was observed after 24 and 48 h of infection (Figure 6B,C). CatB was significantly downregulated at both 24 and 48 h. We did not observe a significant change in ATP6V0D1, Rab7, or ATG7. GABARAP, ATG5, and ATG12 were decreased significantly after 48 h. The change in expression between 24 and 48 h for ATG5, ATG7, and GABARAP was significant. The changes we observed in these infected lysates suggested that autophagic flux was blocked and lysosomal function was decreased after SARS-CoV-2 infection.
Figure 6.
SARS-CoV-2 blocks autophagic flux and reduces lysosomal protein expression. (A) Western blot densitometry for blots normalized to ACTB. (B) Densitometry normalized to the ACTB loading control for each blot (single blot shown). (C) Fold-change of samples normalized to mock infected samples. N = triplicate samples from separate wells per condition. ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.005; ****, p < 0.0001 according to two-way ANOVA with Dunnett’s (for B) or Tukey’s (for C) multiple comparison tests.
Viral Titers Were Suppressed by ROC-325 in a 3D Lung Tissue Model of SARS-CoV-2 Infection
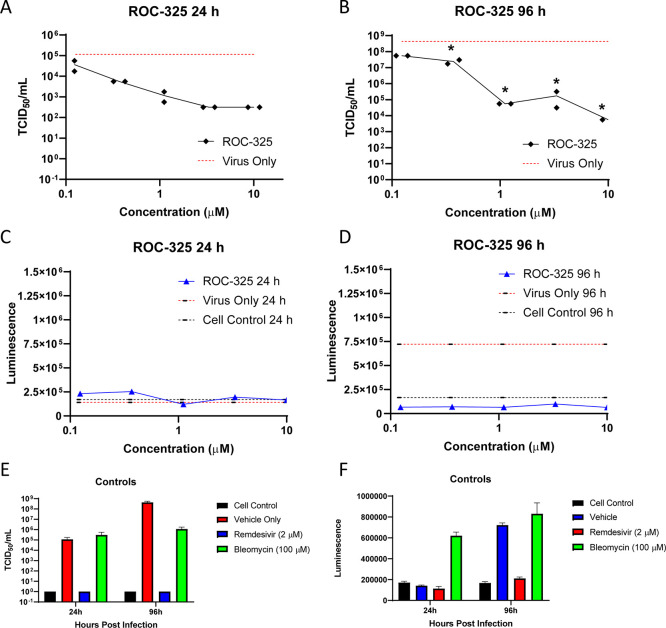
The compound ROC-325 was previously shown to be effective in certain cancer models and superior to other autophagy inhibitors.37 We tested ROC-325 in the EpiAirway 3D tissue model that uses human bronchial epithelial cells. ROC-325 reduced viral titers in a viral titer reduction assay as measured by the median tissue culture infectious dose (TCID50) by more than two log units after 24 h (Figure 7A) and more than three log units after 96 h of infection (Figure 7B). ROC-325 did not exhibit any cytotoxicity in this model (Figure 7C,D). Remdesivir as a positive control was highly effective in reducing viral titers without any cytotoxicity. Altogether, lysosomotropic ROC-325 reduced the CPE in Vero E6 cells and decreased viral titer in a human bronchial epithelium 3D tissue model after SARS-CoV-2 infection.
Figure 7.
ROC-325 reduces viral load after 24 or 96 h of SARS-CoV-2 infection in the EpiAirway 3D lung tissue model. TCID50/mL measurements in a viral titer reduction assay for ROC-325 after (A) 24 h or (B) 96 h of SARS-CoV-2 infection. LDH luminescence values as a measure of cell viability for ROC-325 at (C) 24 h or (D) 96 h of SARS-CoV-2 infection. (E) TCID50/mL measurements in a viral titer reduction assay for the cell control (no virus), vehicle only, remdesivir, or bleomycin. (F) LDH luminescence values as a measure of cell viability for the cell control, vehicle, remdesivir, and bleomycin. ns, not significant; *, p < 0.05 according to a two-way ANOVA with Sidak’s multiple comparison test.
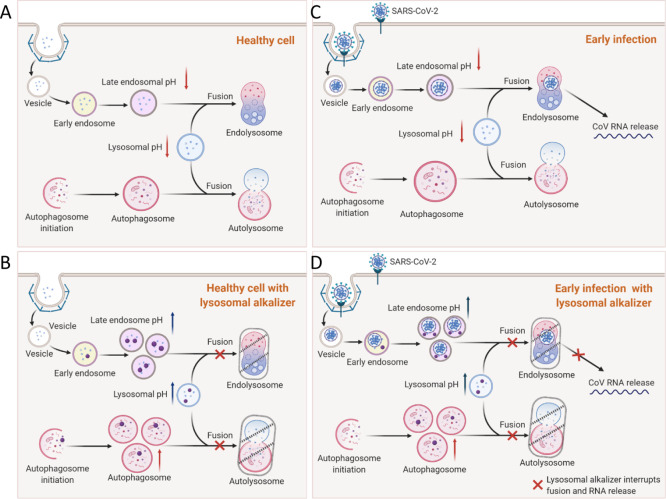
We have illustrated our working hypothesis in Figure 8 as one potential mechanism for the reduction of viral infection and subsequent CPE by lysosomotropic compounds during early infection. First, in a healthy cell, there is normal endocytosis of extracellular material and cellular components at the plasma membrane (Figure 8A). Autophagic flux proceeds normally with autophagosome and lysosome fusion to form autolysosomes. When healthy cells are treated with lysosomotropic compounds, the process of endolysosome fusion is disrupted and resident proteases are unable to function, leading to an increase in the number of late endosomes and autophagosomes (Figure 8B). In the case of an infected cell, endocytosis of SARS-CoV-2 leads to the release of viral RNA into the cell (Figure 8C). We hypothesize that, when lysosomotropic compounds are present during early viral infection, the interference of lysosomal function (red X) leads to containment of the virus and a reduction in viral replication (Figure 8D).
Figure 8.
Illustration of lysosomotropic compounds and their blockade of endocytosis-dependent viral infection. (A) Healthy cells have normal autophagic flux, and the endocytic pathway is functional. (B) Lysosomal alkalizer treatments in healthy cells increase vesicular pH, causing a blockade of normal vesicle fusion and a buildup of endosomes and autophagosomes. (C) In the early stages of viral infection, endocytosis leads to the release of viral RNA after endosome lysosome fusion. (D) Lysosomal alkalizer compounds can block (red Xs) the fusion of endosomes with the lysosome to prevent the release of viral RNA and subsequent cell death.
Discussion
New antiviral drug repurposing opportunities are necessary for helping to treat COVID-19. In this work, we have identified several lysosomotropic compounds that protected Vero E6 cells against the CPE of SARS-CoV-2. Through a series of assays to evaluate potential mechanisms of action, we confirmed these compounds inhibited normal lysosomal function through an alkalization of acidic cellular compartments. This led to a suppression of autophagic flux as well as a blockade of endosomal maturation to the lysosome, a critical part of SARS-Cov-2 endocytosis and viral replication.
The compounds tested included the preclinical drug ROC-32537−39 and FDA-approved clomipramine,40,41 which suppressed the SARS-CoV-2 CPE in Vero E6. Hycanthone, an FDA-approved schistosomicide and oxidative metabolite of lucanthone,42−44 and mefloquine45−47 both showed moderate levels of activity against SARS-CoV-2 CPE and exhibited cytotoxicity at high drug concentrations. To evaluate how CPE protective compounds interrupted lysosomal function, we examined their effects on the autophagy marker LC3B48 as well as the endosomes and lysosomes with the LysoTracker Deep Red dye. We found that the inhibition of lysosomal function as measured by LC3B spot counts correlated well with the inhibition of SARS-CoV-2 measured in the CPE assay for ROC-325, clomipramine, hycanthone, and mefloquine. Interestingly, ROC-325 elevated LysoTracker Deep Red staining only in Vero E6. This may be due a significant increase in the pH of the acidic organelles normally stained by LysoTracker Deep Red that interferes with its activity and movement into the organelles. The effect of ROC-325 may be more pronounced in Vero E6 cells. To our knowledge, this is the first report showing that ROC-325 and hycanthone are efficacious against SARS-CoV-2. Using a SARS-CoV-2 pseudotyped particle, we demonstrated potent inhibition of viral entry in cells that expressed ACE2. We further investigated the compounds’ lysosomotropic effects through a series of assays and found a specific effect of increasing lysosomal pH leading to accumulations of SARS-CoV-2 nanoparticles after endocytosis.
The 72 h SARS-CoV-2 CPE assay measures the phenotypic consequence of viral infection and replication in cells.49−51 SARS-CoV-2 can induce cell death14,52−54 after 48 to 72 h of infection, and thus, cell viability is a surrogate measure of viral replication in vitro. However, there are limitations to the CPE assay including its dependence on the host response and the fact that it is an indirect measurement of SARS-CoV-2 infection and replication. The phenotypic outcome can also vary depending on culture conditions and the viral multiplicity of infection (MOI), the number of virions that are added per cell during the infection.55 The potencies of drug protection against virally induced cell death can be lower than in other assays that directly measure viral load. Nevertheless, this study confirms that SARS-CoV-2 infection in Vero E6 cells results in cell death similar to other reports and that CPE can be suppressed by increasing lysosomal pH with small molecule compounds to the same extent as the positive control remdesivir.56,57 Recently, a drug-repurposing screen of FDA-approved compounds, using a similar CPE assay with SARS-CoV-2 in Vero E6 cells, found clomipramine (EC50 = 5.93 μM; CC50 > 30 μM) and mefloquine (EC50 = 7.11 μM; CC50 > 18.5 μM) to be active with low toxicity.58 The same study found HCQ to be more active than CQ with an EC50 of 9.21 and 42.03 μM, respectively. Mefloquine was also found to be active in another SARS-CoV-2 CPE screen using Caco-2 cells with an EC50 of 14.1 μM.59 In our study, the SI was calculated using the ratio of the EC50, the half-maximal effective concentration, and the CC50, the half-maximal cytotoxic concentration. Between the CPE and the autophagy assays, there was good correspondence in the cytotoxicity measurements by CellTiter-Glo and nuclei counts, respectively. The SI is an important measure for future preclinical development, as it provides insights into the potential clinical safety of a compound at a cellular level.
Evolution has endowed many viruses with the ability to escape autophagic degradation by using the autophagosome membrane for the formation of viral double membrane vesicles (DMVs), although the precise mechanism is still unclear. It has also been reported that some coronavirus proteins such as open reading frame protein 8b (ORF-8b) directly contribute to cell death following viral infection.60 Interestingly, ORF-8b causes the induction of autophagosome formation accompanied by damaging effects on lysosomal function and autophagy flux. ORF-8b also forms aggregates in cells that caused ER stress and lysosome malfunction, which could be responsible for reduced clearance of viral particles by autophagic flux.60 The nonstructural protein 6 (NSP-6) of the infectious bronchitis virus (IBV), an avian coronavirus, significantly increased the number of autophagosomes in host cells.61 The SARS-CoV accessory protein ORF-3a has three transmembrane domains that insert into the lysosomal membrane causing lysosome function dysregulation and necrotic cell death.62 It appears paradoxical that viral infection inhibits autophagic clearance while autophagy inhibitors, also known to block autophagosome to lysosome fusion, suppress viral infection. Our data, combined with the reported mechanism of action for CQ as an antiviral, suggest that these autophagy inhibitors may also interrupt the early steps in the viral life cycle, namely, the fusion of the virus containing endosomes with the lysosome, thereby reducing viral replication and protecting cells from viral-induced cell death. The effect of altering endosomal and lysosomal pH among other mechanisms appears to make acidotropic compounds like HCQ and CQ highly effective against SARS-CoV-2 and other viruses at least in vitro.63 Other host targets for viral inhibition include the point of entry with clathrin-mediated endocytosis of the virus,64 p38 MAPK involved in viral replication,65 post-translational processing of viral proteins in the Golgi apparatus,66 and budding of the virus from the infected cell.67,68 We refer the reader to two excellent reviews by Bello-Perez and co-workers69 and Yang and Shen26 on the role of autophagy and endocytosis in coronavirus infection.
It is critically important to note that host proteases such as the transmembrane serine protease 2 (TMPRSS2)70 at the cell surface along with endosomal cysteine proteases such as cathepsins activated at low pH inside of the cell27 facilitate membrane fusion and release of the viral contents. It is yet unclear how these viral entry events are coordinated, but infection of different cell types may depend on cell surface spike priming for membrane fusion by TMPRSS2 such as that reported for lung epithelial cells,70 endocytosis after ACE2 binding34 followed by membrane fusion, or both. While cell surface membrane fusion mediated by TMPRSS2 cleavage of SARS-CoV-2 is important for infection in airway epithelium, a recent report utilizing genome-wide CRISPR screening in lung epithelial cells identified vacuolar ATPase, Arp2/3, and Rab7A as important targets mediating viral infection, suggesting endocytic mechanisms and trafficking may still play an important role in these cell types.71 Indeed, in our study, the knockdown of ATP6V0D1, a subunit of the V-ATPase responsible for acidification of cellular compartments like the lysosome and late endosome, prevented a HCoV-NL63 CPE in LLC-MK2 cells. Further, we also show QD-RBD nanoparticle colocalization with Rab7, ATP6V0D1, and LAMP1. Lastly, ROC-325 reduced viral titers in the EpiAirway human bronchial epithelial cell 3D tissue model. EpiAirway uses normal human bronchial epithelial cells that have been shown to moderately express TMPRSS2 at the mRNA and protein level.70,72,73 The inhibition of SARS-CoV-2 viral titers in a lung model that expresses TMPRSS2 with a lysosome alkalizing molecule is an intriguing result and warrants further study into the interplay of viral infection mechanisms and host targets such as the lysosome.
If a lysosomotropic compound’s activity was only dependent on its autophagy inhibition, such compounds would be expected to show a significantly greater efficacy in lung models as in other tissue compartments. Our study utilized cell lines with an endocytic entry, limiting the majority of our conclusions to be applied to cells without TMPRSS2 cell surface priming of the spike protein to mediate fusion at the plasma membrane. The utilization of primary cells derived from various human tissues or differentiated iPSCs will be critical to understanding the interplay between viral entry mechanisms, viral effects on the lysosome and cellular pathology, and the impact that lysosomotropic compounds may have on viral infection in both prophylaxis and treatment drug regimens.
ROC-325 was originally developed as an orally available inhibitor of autophagy designed to incorporate the chemical motifs of HCQ and lucanthone, with the goal of both improved autophagic inhibition and consequent single-agent anticancer activity.39,44 ROC-325 is a preclinical candidate with low in vitro and in vivo toxicity and strong anticancer properties.37,74 Our study shows that it may also be a candidate for repositioning as a treatment for COVID-19. Clomipramine, a centrally acting, FDA-approved, tricyclic antidepressant used for the treatment of obsessive-compulsive disorder, panic disorder, major depressive disorder, and chronic pain35,75 may also be an interesting preclinical candidate with its existing regulatory status easing a path toward use in the clinic, although the human Cmax does not cover the CPE EC50 and its use has been replaced with next-generation medications. Because most of these compound EC50 values were higher than their human plasma concentrations at the clinically efficacious doses, they likely will not be efficacious as single agents for the treatment of COVID-19 (Table 3). Indeed, caution must be taken with clinical application of CQ and HCQ because of potential cardiotoxicity.76 Furthermore, a large observational trial did not find a reduction in the death of patients taking HCQ.77 Nonetheless, the sum of this work indicates that targeting steps of the viral life cycle in cells with lysosomotropic compounds could be a valid drug discovery strategy for combating SARS-CoV-2. In support of our conclusions, advanced methods to mine high-throughput screening databases have identified autophagy and AP-1 (clathrin adaptor) signaling as prime targets for antiviral activity.78 The compounds described here also have value as research tools to better understand the interplay between the lysosome, endocytic pathways, and viral live cycle. Interestingly, the lysosome has recently been discovered to be a platform for coronaviral egress as opposed to the classic biosecretory pathway used by other viruses. Recent work by Ghosh et al. found a strong inactivation of lysosomal function by way of increased lysosomal pH, effectively preventing lysosomal substrate degradation.79 Our analysis of SARS-CoV-2 infected lysates suggested the same, wherein cathepsin B expression was downregulated and LC3B II was upregulated, pointing toward lysosomal and dysfunction and a disruption in autophagic flux in the context of viral infection.
Table 3. Clinical Features of Autophagy Inhibitor Compounds.
| compound | primary indication | regulatory status | Cmax (μM) | pharmacological characteristics | reference |
|---|---|---|---|---|---|
| ROC-325 | autophagy inhibitor for cancer | preclinical development | ND | highly bioavailable in vivo, effective against renal cell carcinoma, well tolerated in vivo | (37, 39) |
| clomipramine | serotonin reuptake inhibitor for OCD | FDA approved | 0.29 | well-absorbed, metabolized to desmethylclomipramine, high Vd, T1/2 = 24 h | (82) |
| hycanthone | antihelminthic | FDA approved | ND | active metabolite of lucanthone, T1/2 = 3–5 h, well-absorbed in monkey, human data lacking | (83) |
| chloroquine | antimalarial | FDA approved | 0.17–0.32 | slow elimination, T1/2 = 30–60 days, 75% bioavailable, Vd = 100 L/kg, 50–70% plasma bound, accumulates in blood cells | (84, 85) |
| hydroxychloroquine | antimalarial | FDA approved | 1.5 | similar to CQ, rapid and complete absorption, T1/2 around 40 days | (86) |
| mefloquine | antimalarial | FDA approved | 4.5 | high Cmax can cause toxicity, slow elimination like CQ and HCQ | (87) |
Because such compounds target host cells to suppress SARS-CoV-2 CPE, they have the potential to be combined with other drugs that directly target viral proteins for treatment. This type of combination therapy has certain advantages including synergistic activity from different mechanisms of action and the reduction of the development of viral drug resistance due to the involvement of a host cell target. Furthermore, individual drug concentrations can be lowered in combination therapies to prevent the toxicity seen at higher doses when treating with a single drug. Further tests of drug combinations using advanced 3D tissue models and SARS-CoV-2 animal models will be needed to confirm the therapeutic usage of these compounds.
Materials and Methods
Reagents
The following items were purchased from Gibco: OptiMEM I (11058021), MEM (11095), DMEM (11965092), HI FBS (14000), Pen/Strep (15140), TrypLE (12604013), PBS −/– (w/o Ca2+ or Mg2+) (10010049), and Trypsin-EDTA (25300-054). Hyclone FBS (SH30071.03) was purchased from GE Healthcare. The following items were purchased from ATCC: EMEM (30-2003), Vero E6 (CRL-1586, RRID:CVCL_0574), HeLa (CCL-2, RRID:CVCL_0030), and HEK293T (CRL-3216, RRID:CVCL_0063). ACE2-GFP and ACE2-Expi293F cells were from Codex Biosolutions. The following reagent was obtained through BEI Resources, NIAID, NIH: Human Coronavirus, NL63, NR-470. LLC-MK2 and HCoV-NL63 were a gift from Dr. Emily Lee at NCATS. Huh-7.5 cells were a gift from the Tang Lab at FSU. The following items were purchased from Invitrogen: Lipofectamine RNAiMax (13778030), Live Cell Imaging Buffer (A14291DJ), LysoTracker Deep Red (L12492), goat-antimouse AlexaFluor-647 (A-21242, RRID:AB_2535811), goat-antirabbit AlexaFluor-647 (A32733), HCS Cell Mask Green (H32714), Hoechst 33342 (H3570), LysoSensor Blue (L753), pHrodo Red Dextran (P10361), Premo Autophagy Sensor GFP-p62 (P63240), and Premo Autophagy Tandem Sensor RFP-GFP-LC3B (P36239). siRNAs against ATP6V0D1 (assay ID s17396), ATG5 (assay ID s18158), and ATG7 (assay ID s41770) were purchased from ThermoFisher. LC3B (3868, RRID:AB_2137707), cathepsin B (31718, RRID:AB_2687580), GABARAPL1 (26332S), GABARAP (13733, RRID:AB_9091), LAMP1 (9091, RRID:AB_2687579), Rab7 (9367, RRID:AB_1904103), EEA1 (2411, RRID:AB_2096814), and ATG12 (2010, RRID:AB_2059086) were purchased from Cell Signaling Technologies. Cell Staining Buffer (420201) was purchased from BioLegend. The following items were purchased from Corning: 384-well plates (3764 BC), BioCoat 384-well poly-d-lysine coated plates (354663 BC), and amphotericin B (30-003-CF). 100% methanol (34860) was purchased from Sigma-Aldrich. Calpain Inhibitor IV (208724) was purchased from CalbioChem.
Cell Culture
Vero E6 cells previously selected for high ACE2 expression88 were cultured in MEM/10% HI FBS supplemented with 0.5 μg/mL amphotericin B and passaged twice per week at 1:5 dilutions using trypsin. Briefly, cell culture media were aspirated, and cells were washed twice with PBS. Two milliliters of trypsin is added for 1–2 min at room temperature, and 10 mL of EMEM is added to wash the flask and create a single cell suspension. Cells are spun at 800 rpm for 5 min. The supernatant was aspirated, and the cells were resuspended in fresh media for seeding into flasks or multiwell plates.
Vero E6 (grown in EMEM, 10% FBS, and 1% penicillin/streptomycin), HeLa CCL-2, HEK293T, and Huh-7.5 (grown in DMEM, 10% FBS, and 1% penicillin/streptomycin) cells were cultured in T175 flasks and passaged at 95% confluency. Briefly, cells were washed once with PBS and dissociated from the flask using TrypLE. Cells were counted prior to seeding.
Preparation of Assay Ready Plates for the CPE Assay
An 80 μL aliquot of each compound stock solution (10 mM in 100% DMSO) is transferred into empty wells in columns 3 and 13 of an Echo Qualified 384-Well Polypropylene Source Microplate (Labcyte P-05525). Compounds are diluted 2-fold by transferring 40 μL of each sample into 40 μL of DMSO in the adjacent well (columns 4 and 14) and mixing. This process is repeated to create 8 more 2-fold serially diluted samples in the wells of columns 5–12 and 6–22. Using a Labcyte ECHO 550 (San Jose, CA) acoustic liquid handling system, a 90 nL aliquot of each diluted sample is dispensed into the corresponding wells of a Corning 3764BC plate. An equal volume of DMSO is added to the control wells to maintain a 0.3% DMSO final assay concentration in all the wells. These are referred to as assay ready plates (ARPs) and are stored at −20 °C.
Method for Measuring the Anti-CPE Effects of Compounds
A CPE assay previously used to measure antiviral effects against SARS-CoV30 was adapted for the performance in 384-well plates to measure the CPE of SARS CoV-2 with the following modifications. Cells harvested and suspended at 160 000 cells/mL in MEM/1% PSG/1% HEPES supplemented with 2% HI FBS were batch inoculated with SARS CoV-2 (USA_WA1/2020) at an MOI of approximately 0.002, which resulted in approximately 5% cell viability 72 h postinfection. ARPs were brought to room temperature, and 5 μL of assay media was dispensed to all the wells. The plates were transported into the BSL-3 facility, and a 25 μL aliquot of virus inoculated cells (4000 Vero E6 cells/well) was added to each well in columns 3–24. The wells in columns 23–24 contained virus infected cells only (no compound treatment). A 25 μL aliquot of uninfected cells was added to columns 1–2 of each plate for the cell only (no virus) controls. After incubating plates at 37 °C with 5% CO2 and 90% humidity for 72 h, 30 μL of Cell Titer-Glo (Promega, Madison, WI) was added to each well. Following incubation at room temperature for 10 min, the plates were sealed with a clear cover and surface decontaminated, and luminescence was read using a PerkinElmer Envision (Waltham, MA) plate reader to measure cell viability. Raw data from each test well was normalized to the average signal of the noninfected cells (avg. cells; 100% inhibition) and virus infected cells only (avg. virus; 0% inhibition) to calculate the % inhibition of the CPE using the following formula: % inhibition CPE = 100 × (test cmpd – avg. virus)/(avg. cells – avg. virus).
Method for Measuring the Cytotoxic Effect of Compounds
Compound cytotoxicity was assessed in a BSL-2 counter screen as follows: Host cells in media were added in 25 μL aliquots (4000 cells/well) to each well of the assay ready plates prepared with test compounds as above. Cells only (100% viability) and cells treated with hyamine at 100 μM final concentration (0% viability) serve as the high and low signal controls, respectively, for the cytotoxic effect in the assay. DMSO was maintained at a constant concentration for all wells (0.3%) as dictated by the dilution factor of stock test compound concentrations. After incubating plates at 37 °C/5% CO2 and 90% humidity for 72 h, plates were brought to room temperature and 30 μL of Cell Titer-Glo (Promega) was added to each well. Luminescence was read using a BMG PHERAstar plate reader following incubation at room temperature for 10 min to measure cell viability.
Autophagy Assays
Twenty microliters of cells were seeded into 384-well, black, clear-bottom, poly-d-lysine coated plates to achieve 60% confluent wells. Plates were covered with metal lids and placed in a 37 °C incubator with 95% humidity and 5% CO2 overnight before compound treatment. 100 nL of compound per well was dispensed using the Labcyte Echo 655. The compounds were added at 8 concentrations with 1:3 dilutions starting at 50 μM down to 0.02 μM. ROC-325 was dispensed at the highest working concentration of 25 μM due to a maximum solubility of 5 mM in DMSO.
For LysoTracker staining, 5 μL of a 5× 250 nM LysoTracker Deep Red (Invitrogen, Carlsbad, CA) diluted in Live Cell Imaging Buffer (Invitrogen) was added to the plates mentioned above and incubated for 1 h at 37 °C with 5% CO2 after which the cells were fixed using 4% PFA (Electron Microscopy Sciences, Hatfield, PA) and incubated at room temperature for 30 min. Media in the wells were then evacuated, and the cells were washed three times with PBS using the automated Bluewasher plate washing system from Blue Cat Bio (Concord, MA). Plates were then sealed and imaged on the IN Cell 2500 HS (GE Healthcare, Chicago, IL) automated high-content imaging system. Images were uploaded to the Columbus Analyzer and processed for high-content analysis.
For LC3B immunostaining, media were evacuated on the Bluewasher and 100% ice-cold methanol was added to the wells for 10 min at −30 °C. Plates were washed three times with PBS and blocked with Cell Staining Buffer (BioLegend, San Diego, CA). Plates were then incubated with rabbit-anti-LC3B (Cell Signaling Technologies, Danvers, MA) antibodies in Cell Staining Buffer for 2 h at room temperature. Plates were washed three times with PBS, and secondary antibody goat-antirabbit AlexaFluor-647 (Invitrogen) was added in Cell Staining Buffer for 1 h. The plates were washed three times in PBS before adding 1:5000 Hoechst 33342 (Invitrogen). After a final three washes in PBS, the plates were sealed and imaged on the IN Cell 2500 HS automated high-content imaging system. Images were uploaded to the Columbus Analyzer and processed for high-content analysis.
For LC3B imaging in the 647 nm fluorescence channel, we measured the average number of puncta, or spots, in each cell. Efficacy data was normalized to DMSO (0%) and CQ (100%). Cell count data was normalized to DMSO (100%) and 0 (no cells 0%). An increase in the number of puncta in each image would increase the efficacy (%).
For LysoTracker imaging in the 647 nm fluorescence channel, we measured the average intensity of the puncta, or spots, in each cell. Efficacy data was normalized to DMSO (0%) and CQ (100%). Cell count data was normalized to DMSO (100%) and 0 (no cells 0%). An increase in the average intensity of the puncta in each cell would increase the efficacy (%). Image montages were prepared using Fiji (ImageJ, NIH).
SARS-CoV-2-S PP Assay
SARS-CoV2-S PP and delEnv (bald) PP were custom produced by Codex Biosolutions (Gaithersburg, MD) using previously reported methods using a murine leukemia virus (MLV) pseudotyping system.89,90 The SARS-CoV2-S construct with the Wuhan-Hu-1 sequence (BEI #NR-52420) was C-terminally truncated by 19 amino acids for pseudotyping. ACE2-GFP HEK293T cells were treated with SARS-CoV-2 PPs and evaluated as previously described.29 Briefly, 3500 ACE2-GFP cells were seeded in 15 μL of media in 384-well plates and incubated at 37 °C and 5% CO2 overnight. Cells were treated with compound using acoustic dispensing. Fifteen microliters of PPs was added, and plates were spin-inoculated at 1500 rpm (453g) for 45 min and incubated for 48 h at 37 °C and 5% CO2. The supernatant was removed, and 20 μL/well of Bright-Glo (Promega) was added; the mixture was incubated for 5 min at room temperature. The luminescence signal was measured using a PHERAStar plate reader (BMG Labtech). All data were normalized to DMSO and SARS-CoV-2 PP treated wells as 0% efficacy and DMSO and delEnv PP treated wells as 100% efficacy. The cytotoxicity of the compounds was measured in mock PP treated plates using ATPlite reagent (PerkinElmer). Data was normalized to DMSO treated cells as 100% cell viability and DMSO treated media as 0% cell viability.
LysoSensor Blue and pHrodo Red Dextran Assay
ACE2-GFP cells grown in a 384-well black clear bottom poly-d-lysine coated plate were treated with 75 nL of compound using acoustic dispensing into 15 μL of DMEM and incubated for 48 h. On the day of the assay, media were aspirated and replaced with Live Cell Imaging Solution (LCIS) containing 2 μg/mL LysoSensor Blue or 50 μg/mL pHrodo Red Dextran. Cells were incubated for 30 min at 37 °C and 5% CO2 followed by a final aspiration, and LCIS was added before live cell imaging on the Opera Phenix high content imaging system using 405 nm excitation for LysoSensor Blue or 532 nm excitation for pHrodo Red Dextran. Corrected spot intensity (background subtracted) was used as the parameter for measuring LysoSensor Blue positive puncta. Spot area (μm2) was used as the parameter for measuring pHrodo Red Dextran positive puncta.
Premo Autophagy Sensor GFP-p62 and Tandem GFP-RFP-LC3B Assay
ACE2-Expi293F cells grown in a 96-well black clear bottom poly-d-lysine coated plate were transduced with 6 μL of Premo Autophagy Sensor GFP-p62 or Tandem Sensor GFP-RFP-LC3B. After confirming transduction by visual inspection under fluorescence, the cells were treated with 1 to 3 dilutions of compounds for 16 h. Cells were incubated for 30 min at 37 °C and 5% CO2 before live cell imaging on the Opera Phenix high content imaging system using digital phase contrast to visualize cell bodies and 488 nm for GFP-p62 and the GFP of GFP-RFP-LC3B or 568 nm for the RFP of GFP-RFP-LC3B.
QD-RBD Endocytosis Assay
ACE2-GFP cells grown in a 96-well black clear bottom poly-d-lysine coated plate were treated directly with 10 nM QD-RBD in Optimem I reduced serum media as previously described.34 After 3 h of incubation at 37 °C, the cells were imaged on the Opera Phenix using 405 nm excitation.
QD-RBD Colocalization Immunostaining
Following QD-RBD treatment, the cells were washed once with PBS and fixed with either 4% PFA for 30 min at room temperature or 100% ice-cold methanol for 15 min depending on the primary antibody. Cells were blocked with Cell Staining Buffer and permeabilized with 0.1% Triton X-100 for 30 min. Primary antibodies were added at various dilutions: 1 to 1000 for CatB, GABARAPL1, GABARAP, LAMP1, Rab7, EEA1, ATG12, ATG7, and ATG5. ATP6V0D1. One to 2000 was used for ATP6V0D1. Primary antibodies were washed 3× with PBS, and secondary antibodies goat-antimouse or antirabbit 647 were added depending on the species of the primary antibody. Cells were washed 3× with PBS and incubated with Hoechst 33342 to visualize the nuclei. Cells were imaged on the Opera Phenix confocal imaging system.
Preparation of HCoV-NL63 Viral Inoculate
LLC-MK2 cells (ATCC) were grown in EMEM with 2% FBS for 1 day at 90% confluence. Virus was added to cells and incubated for several days. Cells were monitored for CPE, and supernatant was collected for future use.
siRNA Knockdown Experiments and HCoV-NL63 Infection
LLC-MK2 rhesus macaque kidney epithelial cells were grown in 96-well white solid bottom plates (for CPE assay) or 6-well dishes for lysate collection without pen/strep. siRNA treatment was performed using 1 pmol (96-well) or 25 pmol (6-well) and Lipofectamine RNAiMax (ThermoFisher) according to the manufacturer’s suggestion.
For viral infection, the supernatant of cells treated with siRNA for 48 h in a 96-well plate was aspirated and viral inoculate was added at an MOI of 1.0 for 72 h. ATPlite reagent was used to determine the CPE. Data was normalized to cells not infected with virus (100% viability) and cells infected with the virus and mock siRNA (0% viability).
Lysate was collected from siRNA treated wells using a standard RIPA lysis buffer procedure. Briefly, cells were rinsed once with PBS and trypsinized. Media were added to the cell suspension and spun down at 300g for 5 min. The cell pellet was resuspended in RIPA Lysis Buffer 2 (Enzo) containing cOmplete Mini, EDTA-free, protease inhibitor cocktail (Roche), and PhosSTOP phosphatase inhibitor cocktail (Sigma-Aldrich). Lysates were vortexed for 10 s followed by 10 min on ice 3 times. Lysates were spun down at 4 °C for 30 min, and the supernatant was collected for Western Blot analysis.
Western Blot Analysis
The Pierce BCA Assay (ThermoFisher) was used to quantify the protein concentration in lysates. 4× LDS buffer (ThermoFisher) and 10× reducing agent (ThermoFisher) were added to the lysate, and PBS was added to equalize volumes for each sample. Samples were heated at 70 °C for 10 min. Samples were loaded in precast SDS-PAGE gels (ThermoFisher) and run in denaturing conditions with MOPS buffer. Gels were transferred to PVDF using the ThermoFisher iBlot 2 semidry transfer system. Blots were blocked with StartBlock (ThermoFisher) for 1 h before the addition of primary antibodies incubated at 4 °C overnight. Blots were rinsed 3× for 5 min each in TBS-Tween 20 followed by 1 h of incubation with secondary HRP antibody. Blots were imaged on a G Box (Syngene) gel imaging system using chemiluminscence detection. When densitometry was used, the images of the blots were analyzed using Fiji (ImageJ). Densitometry was normalized to the loading control (actin or GAPDH).
SARS-CoV-2 Infection and Collection of Viral Lysates
Vero E6 cells were seeded at 45 000 per well in 12-well plates in 1.25 mL of growth media and incubated for 24 h at 37 °C. 250 μL of SARS-CoV-2 (USA_WA1/2020) at an MOI of 0.05 was incubated for 45 min with cells at 37 °C. The supernatant was removed; fresh media were added to the wells and further incubated for 24 and 48 h. For lysate collection, PBS + 0.5% Triton X-100 with 1× protease inhibitor cocktail was added to the cells. Lysates were collected and stored at −20 °C until needed.
SARS-CoV-2 Infection of EpiAirway
Step 1: In the first part of the experiment, the test compound was diluted in assay medium (AIR-100-ASY). The test compound dilutions were added to each insert on the apical layer (0.15 mL) and basolateral layer (0.85 mL). Following the 1 h treatment, the apical medium was removed, and the basolateral medium was replaced with fresh compound. Virus (0.15 mL) was then added to each insert on the apical layer, removed after 1 h, and washed with 0.4 mL of TEER buffer. The basal side medium/compound was replaced with 1 mL of assay medium. Every 24 hours, the basolateral medium was replaced with 1 mL of fresh medium containing fresh compound. At 24 and 96 h postinfection (p.i.), the apical layer of the tissues was washed with 0.4 mL of TEER buffer and aliquoted to separate microfuge tubes. At 24 and 96 h p.i., the basolateral medium (1.0 mL) was collected from each well, aliquoted into separate microfuge tubes, and stored at −80 °C.
Step 2: The apical layer supernatants from all treatments were titered by TCID50 to determine the amount of virus present in each sample. Medium from the basolateral layer of the tissue culture inserts will be assayed using a lactate dehydrogenase (LDH) release assay as a measure of cell viability.
Collection of Samples from the Apical Surface for 50% Tissue Culture Infectious Dose (TCID50) Assay
At 24 and 96 h p.i., the apical layer of the tissues was washed with 0.4 mL of TEER buffer, aliquoted to separate microfuge tubes (1.5 mL), and stored at −80 °C.
Procedure for TCID50 Assay
Vero E6 cells were plated in a 96-well plate (20 000/well) and incubated overnight at 37 °C in a CO2 incubator. Virus assay medium was serially diluted 1:10 and added to the Vero E6 cells after decanting the existing supernatant. After 3 days, crystal violet was added to the cells and viral titers were calculated using the Reed and Muench method.91
LDH Release Assay
Five microliters of basolateral medium was removed from all the wells and added to 95 μL of LDH storage buffer. Samples were stored at −80 °C. On the day of the assay, samples were thawed and further diluted in LDH storage buffer. A no cell control was included as a negative control to determine the culture medium background. 50 μL of diluted sample was added into a 96-well opaque-walled, nontransparent assay plate (with clear bottom), and 50 μL of LDH detection reagent was added to each well. Samples were incubated for 60 min at room temperature. Luminescence was recorded.
Image and Statistical Analysis
CPE assay raw data from each test well was normalized to the average signal of noninfected cells (avg. cells; 100% inhibition) and virus infected cells only (avg. virus; 0% inhibition) to calculate the % inhibition of CPE using the following formula: % inhibition CPE = 100 × (test cmpd – avg. virus)/(avg. cells – avg. virus). EC50 values were obtained using nonlinear regression.
High-content image analysis data was downloaded as a Microsoft Excel spreadsheet. DMSO negative control (0% activity) (col. 1 and 24 for acoustic dispensing) and CQ positive control (100% activity) (col. 2 for acoustic dispensing, 8 wells) were used to normalize each compound concentrations’ response. The other 8 wells of column 2 contained 10 mM HCQ. EC50 values were obtained using nonlinear regression in Graphpad Prism 7.04. In some cases, the highest concentration point was not included in the curve fit due to technical issues during experimental execution, although the measured value was shown. When cell viability was below 20%, the efficacy point was excluded altogether (i.e., mefloquine at 24 or 8 μM). Six fields per well were imaged on the IN Cell 2500HS. LC3B and LysoTracker data was obtained using a single well with hundreds of cells for each compound concentration from three intraplate replicate wells that were imaged when acoustic dispensing was used for compound treatment. Cell counts were also reported using nuclear object segmentation. GraphPad Prism 7.04v and 8.4.3v were used for visualizing autophagy data. EC50 and CC50 values from high-content imaging were obtained using nonlinear regression. Images were uploaded to the Columbus Analyzer and processed for high-content analysis. Image montages were prepared using Fiji (ImageJ, NIH). The illustration in Figure 8 was generated using Biorender.com.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Center for Advancing Translational Sciences, NIH. The Naval Research Laboratory provided funding via its internal Nanoscience Institute. Reagent preparation was supported via the NRL COVID-19 base fund. We thank Richard Eastman (NCATS) and Sara McKellip (SRI) for assistance with acoustic dispensing support and compound management. We thank the laboratory of Dr. Hengli Tang for providing the Huh-7.5 cells. We thank Dr. Emily Lee for providing the HCoV-NL63 and LLC-MK2 cells. We thank the researchers at the University of Louisville, Drs. Severson, Gabbard, and Palmer, for their help in the EpiAirway infection research. The plasmids and protocols for PP generation were kind gifts from Dr. Gary Whittaker at Cornell University.
Glossary
Abbreviations
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- COVID-19
coronavirus disease that was discovered in 2019
- CQ
chloroquine
- HCQ
hydroxychloroquine
- LC3B
microtubule-associated proteins 1B light chains 3B
- CPE
cytopathic effect
- SI
selectivity index
- TOX
toxicity measurement for CC50 calculation
- MOI
multiplicity of infection
- PP
pseudotyped particles
- ATG
autophagy related
- CatB
cathepsin B
- QD-RBD
quantum dot-conjugated SARS-CoV-2 RBD
- siRNA
silencer RNA
- HCoV-NL63
Human Coronavirus NL63
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00349.
Study design; Figure S1, CPE assay workflow; Figure S2, CPE assay reproducibility; Figure S3, autophagy inhibition assay using LC3B immunostaining in HeLa cells; Figure S4, autophagy inhibition assay using LC3B immunostaining in HEK293T cells; Figure S5, autophagy inhibition assay using LC3B immunostaining in Huh-7.5 cells; Figure S6; LysoTracker Deep Red staining in HeLa cells; Figure S7, LysoTracker Deep Red staining in HEK293T cells; Figure S8, LysoTracker Deep Red staining in Huh-7.5 cells; Figure S9, compounds inhibit pHrodo dextran uptake in ACE2-GFP HEK293T cells; Figure S10, compounds inhibit LysoSensor Blue staining in ACE2-GFP HEK293T cells; Figure S11, compounds induce accumulation of p62 LC3B; Figure S12, compounds increase puncta size of QD-RBD; Figure S13, endosomal trafficking inhibitor VPS34-IN-1 and Bafilomycin A1 prevent the SARS-CoV-2 cytopathic effect in Vero E6 cells; Figure S14, QD-RBD colocalizes with endosomal and lysosomal markers; Figure S15, LLC-MK2 cells are protected from HCoV-NL63 CPE after siRNA knockdown of ATP6 V0D1; Figure S16, ATP6V0D1 knockdown partially blocks SARS-CoV-2 PP transduction in ACE2-GFP HEK293T cells (PDF)
Author Contributions
Experimental: K.G., C.Z.C., R.B., B.N.T., Y.-S.C, L.R., M.X., M.P., M.H., E.O., K.S., M.W., and J.C.T. Data analysis: K.G., B.N.T., X.H., and M.S. Compound management and selection: K.G., X.H., P.S., and Z.I. Manuscript conception: K.G., C.Z.C, and W. Zheng. Manuscript writing: K.G., C.Z.C., K.S., W.H., and W. Zheng. Preparation of figures: K.G., C.Z.C., and W. Zhu. Critical manuscript editing and discussion: K.G., M.P., C.Z.C., W. Zheng, A.S., M.D.H., M.S., and D.C.L. Laboratory automation: C.K.-T. and S.G.M.
The authors declare no competing financial interest.
Supplementary Material
References
- Yang X.; Yu Y.; Xu J.; Shu H.; Liu H.; Wu Y.; Zhang L.; Yu Z.; Fang M.; Yu T. (2020) Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 8, 475. 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E.; Du H.; Gardner L. (2020) An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533. 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanche S.; Lin Y. T.; Xu C.; Romero-Severson E.; Hengartner N.; Ke R. (2020) High Contagiousness and Rapid Spread of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 26, 1470. 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E.; Cyranoski D.; Mallapaty S.; Stoye E.; Tollefson J. (2020) The coronavirus pandemic in five powerful charts. Nature 579 (7800), 482–483. 10.1038/d41586-020-00758-2. [DOI] [PubMed] [Google Scholar]
- Rothan H. A.; Byrareddy S. N. (2020) The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 109, 102433. 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Naming the coronavirus disease (COVID-19) and the virus that causes it, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it.
- Xu Z.; Shi L.; Wang Y.; Zhang J.; Huang L.; Zhang C.; Liu S.; Zhao P.; Liu H.; Zhu L.; et al. (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet respiratory medicine 8 (4), 420–422. 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby P.; Lüscher T. (2020) COVID-19 is, in the end, an endothelial disease. Eur. Heart J. 41 (32), 3038–3044. 10.1093/eurheartj/ehaa623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z.; Flammer A. J.; Steiger P.; Haberecker M.; Andermatt R.; Zinkernagel A. S.; Mehra M. R.; Schuepbach R. A.; Ruschitzka F.; Moch H. (2020) Endothelial cell infection and endotheliitis in COVID-19. Lancet 395 (10234), 1417–1418. 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko W.-C.; Rolain J.-M.; Lee N.-Y.; Chen P.-L.; Huang C.-T.; Lee P.-I.; Hsueh P.-R. (2020) Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int. J. Antimicrob. Agents 55 (4), 105933–105933. 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J.; Ohmagari N.; Shin D.; Diaz G.; Asperges E.; Castagna A.; Feldt T.; Green G.; Green M. L.; Lescure F.-X.; Nicastri E.; Oda R.; Yo K.; Quiros-Roldan E.; Studemeister A.; Redinski J.; Ahmed S.; Bernett J.; Chelliah D.; Chen D.; Chihara S.; Cohen S. H.; Cunningham J.; D’Arminio Monforte A.; Ismail S.; Kato H.; Lapadula G.; L’Her E.; Maeno T.; Majumder S.; Massari M.; Mora-Rillo M.; Mutoh Y.; Nguyen D.; Verweij E.; Zoufaly A.; Osinusi A. O.; DeZure A.; Zhao Y.; Zhong L.; Chokkalingam A.; Elboudwarej E.; Telep L.; Timbs L.; Henne I.; Sellers S.; Cao H.; Tan S. K.; Winterbourne L.; Desai P.; Mera R.; Gaggar A.; Myers R. P.; Brainard D. M.; Childs R.; Flanigan T. (2020) Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 382, 2327. 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P.; Lagier J. C.; Parola P.; Hoang V. T.; Meddeb L.; Mailhe M.; Doudier B.; Courjon J.; Giordanengo V.; Vieira V. E.; Dupont H. T.; Honore S.; Colson P.; Chabriere E.; La Scola B.; Rolain J. M.; Brouqui P.; Raoult D. (2020) Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 56, 105949. 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yao X.; Ye F.; Zhang M.; Cui C.; Huang B.; Niu P.; Liu X.; Zhao L.; Dong E.; Song C.; Zhan S.; Lu R.; Li H.; Tan W.; Liu D. (2020) In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 71, 732. 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z.; Diao B.; Wang R.; Wang G.; Wang C.; Tan Y.; Liu L.; Wang C.; Liu Y.; Liu Y.; Yuan Z.; Ren L.; Wu Y.; Chen Y. (2020) The Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Directly Decimates Human Spleens and Lymph Nodes. medRxiv 10.1101/2020.03.27.20045427. [DOI] [Google Scholar]
- Keyaerts E.; Li S.; Vijgen L.; Rysman E.; Verbeeck J.; Van Ranst M.; Maes P. (2009) Antiviral activity of chloroquine against human coronavirus OC43 infection in newborn mice. Antimicrob. Agents Chemother. 53 (8), 3416–3421. 10.1128/AAC.01509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y. W.; Yam W. K.; Sun J.; Chu J. J. H. (2018) An evaluation of Chloroquine as a broad-acting antiviral against Hand, Foot and Mouth Disease. Antiviral Res. 149, 143–149. 10.1016/j.antiviral.2017.11.017. [DOI] [PubMed] [Google Scholar]
- Shiryaev S. A.; Mesci P.; Pinto A.; Fernandes I.; Sheets N.; Shresta S.; Farhy C.; Huang C. T.; Strongin A. Y.; Muotri A. R.; Terskikh A. V. (2017) Repurposing of the anti-malaria drug chloroquine for Zika Virus treatment and prophylaxis. Sci. Rep. 7 (1), 15771. 10.1038/s41598-017-15467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.; Zou Z.; Sun Y.; Li X.; Xu K.-F.; Wei Y.; Jin N.; Jiang C. (2013) Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 23 (2), 300–302. 10.1038/cr.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D. L.; Day C. W.; Bailey K.; Heiner M.; Montgomery R.; Lauridsen L.; Chan P. K. S.; Sidwell R. W. (2006) Evaluation of Immunomodulators, Interferons and Known in Vitro SARS-CoV Inhibitors for Inhibition of SARS-Cov Replication in BALB/c Mice. Antiviral Chem. Chemother. 17 (5), 275–284. 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- Rosenke K.; Jarvis M. A.; Feldmann F.; Schwarz B.; Okumura A.; Lovaglio J.; Saturday G.; Hanley P. W.; Meade-White K.; Williamson B. N.; Hansen F.; Perez-Perez L.; Leventhal S.; Tang-Huau T.-L.; Nason M.; Callison J.; Haddock E.; Scott D.; Sewell G.; Bosio C. M.; Hawman D.; de Wit E.; Feldmann H. (2020) Hydroxychloroquine Proves Ineffective in Hamsters and Macaques Infected with SARS-CoV-2. bioRxiv 10.1101/2020.06.10.145144. [DOI] [Google Scholar]
- Hernandez A. V.; Roman Y. M.; Pasupuleti V.; Barboza J. J.; White C. M. (2020) Hydroxychloroquine or Chloroquine for Treatment or Prophylaxis of COVID-19: A Living Systematic Review. Ann. Intern. Med. 173 (4), 287–296. 10.7326/M20-2496. [DOI] [PubMed] [Google Scholar]
- Savarino A.; Di Trani L.; Donatelli I.; Cauda R.; Cassone A. (2006) New insights into the antiviral effects of chloroquine. Lancet Infect. Dis. 6 (2), 67–69. 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino A.; Boelaert J. R.; Cassone A.; Majori G.; Cauda R. (2003) Effects of chloroquine on viral infections: an old drug against today’s diseases?. Lancet Infect. Dis. 3 (11), 722–727. 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogstad D. J.; Schlesinger P. H. (1987) The basis of antimalarial action: non-weak base effects of chloroquine on acid vesicle pH. Am. J. Trop. Med. Hyg. 36 (2), 213–220. 10.4269/ajtmh.1987.36.213. [DOI] [PubMed] [Google Scholar]
- Yang Z.-Y.; Huang Y.; Ganesh L.; Leung K.; Kong W.-P.; Schwartz O.; Subbarao K.; Nabel G. J. (2004) pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78 (11), 5642–5650. 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N.; Shen H.-M. (2020) Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int. J. Biol. Sci. 16 (10), 1724–1731. 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Wan Y.; Luo C.; Ye G.; Geng Q.; Auerbach A.; Li F. (2020) Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 117 (21), 11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N.; Yoshimori T.; Levine B. (2010) Methods in mammalian autophagy research. Cell 140 (3), 313–326. 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Chen C. Z.; Swaroop M.; Xu M.; Wang L.; Lee J.; Wang A. Q.; Pradhan M.; Hagen N.; Chen L.; Shen M.; Luo Z.; Xu X.; Xu Y.; Huang W.; Zheng W.; Ye Y. (2020) Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov 6 (1), 80. 10.1038/s41421-020-00222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson W. E.; Shindo N.; Sosa M.; Fletcher T. 3rd; White E. L.; Ananthan S.; Jonsson C. B. (2007) Development and validation of a high-throughput screen for inhibitors of SARS CoV and its application in screening of a 100,000-compound library. J. Biomol. Screening 12 (1), 33–40. 10.1177/1087057106296688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox C. B.; Rasmussen L.; White E. L. (2008) Adapting Cell-Based Assays to the High Throughput Screening Platform: Problems Encountered and Lessons Learned. JALA (1998-2010) 13 (3), 168–173. 10.1016/j.jala.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Cao R.; Zhang L.; Yang X.; Liu J.; Xu M.; Shi Z.; Hu Z.; Zhong W.; Xiao G. (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30 (3), 269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Z.; Xu M.; Pradhan M.; Gorshkov K.; Petersen J. D.; Straus M. R.; Zhu W.; Shinn P.; Guo H.; Shen M.; Klumpp-Thomas C.; Michael S. G.; Zimmerberg J.; Zheng W.; Whittaker G. R. (2020) Identifying SARS-CoV-2 Entry Inhibitors through Drug Repurposing Screens of SARS-S and MERS-S Pseudotyped Particles. ACS Pharmacol. Transl. Sci. 3, 1165–1175. 10.1021/acsptsci.0c00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorshkov K.; Susumu K.; Chen J.; Xu M.; Pradhan M.; Zhu W.; Hu X.; Breger J. C.; Wolak M.; Oh E. (2020) Quantum Dot-Conjugated SARS-CoV-2 Spike Pseudo-Virions Enable Tracking of Angiotensin Converting Enzyme 2 Binding and Endocytosis. ACS Nano 14 (9), 12234–12247. 10.1021/acsnano.0c05975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe K. R.; Zhao T.; Eastman R. T.; Hu X.; Wang K.; Backus M.; Baljinnyam B.; Chen C. Z.; Chen L.; Eicher T.; Ferrer M.; Fu Y.; Gorshkov K.; Guo H.; Hanson Q. M.; Itkin Z.; Kales S. C.; Klumpp-Thomas C.; Lee E. M.; Michael S.; Mierzwa T.; Patt A.; Pradhan M.; Renn A.; Shinn P.; Shrimp J. H.; Viraktamath A.; Wilson K. M.; Xu M.; Zakharov A. V.; Zhu W.; Zheng W.; Simeonov A.; Mathé E. A.; Lo D. C.; Hall M. D.; Shen M. (2020) An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv 10.1101/2020.06.04.135046. [DOI] [Google Scholar]
- Chen C. Z.; Shinn P.; Itkin Z.; Eastman R. T.; Bostwick R.; Rasmussen L.; Huang R.; Shen M.; Hu X.; Wilson K. M.; Brooks B.; Guo H.; Zhao T.; Klump-Thomas C.; Simeonov A.; Michael S. G.; Lo D. C.; Hall M. D.; Zheng W. (2020) Drug Repurposing Screen for Compounds Inhibiting the Cytopathic Effect of SARS-CoV-2. bioRxiv 10.1101/2020.08.18.255877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew J. S.; Espitia C. M.; Zhao W.; Han Y.; Visconte V.; Phillips J.; Nawrocki S. T. (2017) Disruption of Autophagic Degradation with ROC-325 Antagonizes Renal Cell Carcinoma Pathogenesis. Clin. Cancer Res. 23 (11), 2869–2879. 10.1158/1078-0432.CCR-16-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew J. S.; Nawrocki S. T. (2017) Drain the lysosome: Development of the novel orally available autophagy inhibitor ROC-325. Autophagy 13 (4), 765–766. 10.1080/15548627.2017.1280222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. M.; Espitia C.; Wang W.; Nawrocki S. T.; Carew J. S. (2019) Moving beyond hydroxychloroquine: the novel lysosomal autophagy inhibitor ROC-325 shows significant potential in preclinical studies. Cancer Communications 39 (1), 1–3. 10.1186/s40880-019-0418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. J.; Chee C. E.; Huang S.; Sinicrope F. (2011) Autophagy modulation for cancer therapy. Cancer Biol. Ther. 11 (2), 169–176. 10.4161/cbt.11.2.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M.; Munarriz E. R.; Bartesaghi S.; Milanese M.; Dinsdale D.; Guerra-Martin M. A.; Bampton E. T.; Glynn P.; Bonanno G.; Knight R. A.; et al. (2009) Desmethylclomipramine induces the accumulation of autophagy markers by blocking autophagic flux. J. Cell Sci. 122 (18), 3330–3339. 10.1242/jcs.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi D.; Peruzzotti G.; Dennis E. W.; Berberian D. A.; Freele H.; Tullar B. F.; Archer S. (1967) Hycanthone, a new active metabolite of lucanthone. J. Med. Chem. 10 (5), 867–876. 10.1021/jm00317a025. [DOI] [PubMed] [Google Scholar]
- Petcherski A.; Chandrachud U.; Butz E. S.; Klein M. C.; Zhao W.-N.; Reis S. A.; Haggarty S. J.; Ruonala M. O.; Cotman S. L. (2019) An Autophagy Modifier Screen Identifies Small Molecules Capable of Reducing Autophagosome Accumulation in a Model of CLN3-Mediated Neurodegeneration. Cells 8 (12), 1531. 10.3390/cells8121531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew J. S.; Espitia C. M.; Esquivel J. A.; Mahalingam D.; Kelly K. R.; Reddy G.; Giles F. J.; Nawrocki S. T. (2011) Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J. Biol. Chem. 286 (8), 6602–6613. 10.1074/jbc.M110.151324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N.; Thomas S.; Golden E. B.; Hofman F. M.; Chen T. C.; Petasis N. A.; Schönthal A. H.; Louie S. G. (2012) Inhibition of autophagy and induction of breast cancer cell death by mefloquine, an antimalarial agent. Cancer Lett. 326 (2), 143–154. 10.1016/j.canlet.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Sharma N. (2013) Inhibition of Tumor Cell Growth by Mefloquine via Multimechanistic Effects Involving Increased Cellular Stress, Inhibition of Autophagy, and Impairment of Cellular Energy Metabolism, Ph.D. Dissertation, University of Southern California. [Google Scholar]
- Shin J. H.; Park S. J.; Jo Y. K.; Kim E. S.; Kang H.; Park J.-H.; Lee E. H.; Cho D.-H. (2012) Suppression of autophagy exacerbates Mefloquine-mediated cell death. Neurosci. Lett. 515 (2), 162–167. 10.1016/j.neulet.2012.03.040. [DOI] [PubMed] [Google Scholar]
- Tanida I.; Waguri S. (2010) Measurement of autophagy in cells and tissues. Methods Mol. Biol. (N. Y., NY, U. S.) 648, 193–214. 10.1007/978-1-60761-756-3_13. [DOI] [PubMed] [Google Scholar]
- Familletti P. C.; Rubinstein S.; Pestka S. (1981) A convenient and rapid cytopathic effect inhibition assay for interferon. Methods in Enzymology 78, 387–394. 10.1016/0076-6879(81)78146-1. [DOI] [PubMed] [Google Scholar]
- Darling A. J.; Boose J. A.; Spaltro J. (1998) Virus assay methods: accuracy and validation. Biologicals 26 (2), 105–110. 10.1006/biol.1998.0134. [DOI] [PubMed] [Google Scholar]
- Noah J. W.; Severson W.; Noah D. L.; Rasmussen L.; White E. L.; Jonsson C. B. (2007) A cell-based luminescence assay is effective for high-throughput screening of potential influenza antivirals. Antiviral Res. 73 (1), 50–59. 10.1016/j.antiviral.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Guzzi P. H.; Mercatelli D.; Ceraolo C.; Giorgi F. M. (2020) Master Regulator Analysis of the SARS-CoV-2/Human Interactome. J. Clin. Med. 9 (4), 982. 10.3390/jcm9040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y.; Liu Y.; Cao L.; Wang D.; Guo M.; Jiang A.; Guo D.; Hu W.; Yang J.; Tang Z.; Wu H.; Lin Y.; Zhang M.; Zhang Q.; Shi M.; Liu Y.; Zhou Y.; Lan K.; Chen Y. (2020) Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerging Microbes Infect. 9 (1), 761–770. 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K. L.; Yuen K. S.; Castaño-Rodriguez C.; Ye Z. W.; Yeung M. L.; Fung S. Y.; Yuan S.; Chan C. P.; Yuen K. Y.; Enjuanes L.; Jin D. Y. (2019) Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 33 (8), 8865–8877. 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabram P.; Aguilar-Cordova E. (2000) Multiplicity of Infection/Multiplicity of Confusion. Mol. Ther. 2 (5), 420–421. 10.1006/mthe.2000.0212. [DOI] [PubMed] [Google Scholar]
- Jeon S.; Ko M.; Lee J.; Choi I.; Byun S. Y.; Park S.; Shum D.; Kim S. (2020) Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 64, e00819-20. 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touret F.; Gilles M.; Barral K.; Nougairède A.; Decroly E.; de Lamballerie X.; Coutard B. (2020) In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 10, 13093. 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S.; Coleman C. M.; Haupt R.; Logue J.; Matthews K.; Frieman M. (2020) Broad anti-coronaviral activity of FDA approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. J. Virol. 94, e01218-20. 10.1128/JVI.01218-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger B.; Bojkva D.; Zaliani A.; Cinatl J.; Claussen C.; Westhaus S.; Reinshagen J.; Kuzikov M.; Wolf M.; Geisslinger G.; Gribbon P.; Ciesek S. (2020) Identification of inhibitors of SARS-CoV-2 in-vitro cellular toxicity in human (Caco-2) cells using a large scale drug repurposing collection. Research Square 10.21203/rs.3.rs-23951/v1. [DOI] [Google Scholar]
- Shi C.-S.; Nabar N. R.; Huang N.-N.; Kehrl J. H. (2019) SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov 5 (1), 101. 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E. M.; Whelband M. C.; Wileman T. (2014) Coronavirus NSP6 restricts autophagosome expansion. Autophagy 10 (8), 1426–1441. 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y.; Nabar N. R.; Shi C.-S.; Kamenyeva O.; Xiao X.; Hwang I.-Y.; Wang M.; Kehrl J. H. (2018) SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 9 (9), 904. 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bari M. A. A. (2017) Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 5 (1), e00293–e00293. 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T. Y.; Frieman M.; Wolfram J. (2020) Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat. Nanotechnol. 15 (4), 247–249. 10.1038/s41565-020-0674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono M.; Tatsumi K.; Imai A. M.; Saito K.; Kuriyama T.; Shirasawa H. (2008) Inhibition of human coronavirus 229E infection in human epithelial lung cells (L132) by chloroquine: involvement of p38 MAPK and ERK. Antiviral Res. 77 (2), 150–152. 10.1016/j.antiviral.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C. A.; Rolain J.-M.; Colson P.; Raoult D. (2020) New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19?. Int. J. Antimicrob. Agents 55, 105938–105938. 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.-L.; Tan S.-H.; See E.-E.; Ooi E.-E.; Ling A.-E. (2003) Proliferative growth of SARS coronavirus in Vero E6 cells. J. Gen. Virol. 84 (12), 3291–3303. 10.1099/vir.0.19505-0. [DOI] [PubMed] [Google Scholar]
- Ferreira D.; Santo M.; Rebello M.; Rebello M. (2000) Weak bases affect late stages of Mayaro virus replication cycle in vertebrate cells. J. Med. Microbiol. 49 (4), 313–318. 10.1099/0022-1317-49-4-313. [DOI] [PubMed] [Google Scholar]
- Bello-Perez M.; Sola I.; Novoa B.; Klionsky D. J.; Falco A. (2020) Canonical and Noncanonical Autophagy as Potential Targets for COVID-19. Cells 9 (7), 1619. 10.3390/cells9071619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S.; Chua R. L.; Trefzer T.; Kahn N. C.; Schneider M. A.; Muley T.; Winter H.; Meister M.; Veith C.; Boots A. W.; Hennig B. P.; Kreuter M.; Conrad C.; Eils R. (2020) SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 39 (10), e105114–e105114. 10.15252/embj.2020105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z.; Jordan T. X.; Wessels H.-H.; Hoagland D. A.; Kasela S.; Legut M.; Maniatis S.; Mimitou E. P.; Lu L.; Geller E.; Danziger O.; Rosenberg B. R.; Phatnani H.; Smibert P.; Lappalainen T.; tenOever B. R.; Sanjana N. E. (2020) Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzig K. E.; Canepa-Escaro F.; Schiliro M.; Berdnikovs S.; Prakash Y. S.; Chiarella S. E. (2020) Estrogen regulates the expression of SARS-CoV-2 receptor ACE2 in differentiated airway epithelial cells. Am. J. Physiol Lung Cell Mol. Physiol 318 (6), L1280–L1281. 10.1152/ajplung.00153.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Kovalchuk A.; Li D.; Rodriguez-Juarez R.; Ilnytskyy Y.; Kovalchuk I.; Kovalchuk O. (2020) In search of preventive strategies: novel high-CBD Cannabis sativa extracts modulate ACE2 expression in COVID-19 gateway tissues. Aging 12 (22), 22425–22444. 10.18632/aging.202225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki S. T.; Han Y.; Visconte V.; Phillips J. G.; Przychodzen B. P.; Maciejewski J. P.; Kelly K. R.; Carew J. S. (2016) Development of ROC-325: A Novel Small Molecule Inhibitor of Autophagy with Promising Anti-Leukemic Activity. Blood 128 (22), 525–525. 10.1182/blood.V128.22.525.525. [DOI] [Google Scholar]
- Kelly M. W.; Myers C. W. (1990) Clomipramine: A Tricyclic Antidepressant Effective in Obsessive Compulsive Disorder. DICP, Ann. Pharmacother. 24 (7–8), 739–744. 10.1177/106002809002400718. [DOI] [PubMed] [Google Scholar]
- Bauman J. L.; Tisdale J. E. (2020) Chloroquine and Hydroxychloroquine in the Era of SARS – CoV2: Caution on Their Cardiac Toxicity. Pharmacotherapy 40, 387. 10.1002/phar.2387. [DOI] [PubMed] [Google Scholar]
- Geleris J.; Sun Y.; Platt J.; Zucker J.; Baldwin M.; Hripcsak G.; Labella A.; Manson D.; Kubin C.; Barr R. G.; Sobieszczyk M. E.; Schluger N. W. (2020) Observational Study of Hydroxychloroquine in Hospitalized Patients with Covid-19. N. Engl. J. Med. 382, 2411. 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Chen C. Z., Sakamuru S., Simeonov A., Hall M. D., Xia M., Zheng W., and Huang R. (2020) Mining of high throughput screening database reveals AP-1 and autophagy pathways as potential targets for COVID-19 therapeutics. arXiv 2007.12242, https://arxiv.org/abs/2007.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.; Dellibovi-Ragheb T. A.; Kerviel A.; Pak E.; Qiu Q.; Fisher M.; Takvorian P. M.; Bleck C.; Hsu V. W.; Fehr A. R.; Perlman S.; Achar S. R.; Straus M. R.; Whittaker G. R.; de Haan C. A. M.; Kehrl J.; Altan-Bonnet G.; Altan-Bonnet N. (2020) β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 183, 1520. 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthe M.; Orhon I.; Rocchi C.; Zhou X.; Luhr M.; Hijlkema K. J.; Coppes R. P.; Engedal N.; Mari M.; Reggiori F. (2018) Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14 (8), 1435–1455. 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue E.; Tovey A.; Vogl A. W.; Arns S.; Sternberg E.; Young R. N.; Roberge M. (2011) Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J. Biol. Chem. 286 (9), 7290–7300. 10.1074/jbc.M110.139915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balant-Gorgia A. E.; Gex-Fabry M.; Balant L. P. (1991) Clinical pharmacokinetics of clomipramine. Clin. Pharmacokinet. 20 (6), 447–462. 10.2165/00003088-199120060-00002. [DOI] [PubMed] [Google Scholar]
- Edwards G.; Breckenridge A. M. (1988) Clinical pharmacokinetics of anthelmintic drugs. Clin. Pharmacokinet. 15 (2), 67–93. 10.2165/00003088-198815020-00001. [DOI] [PubMed] [Google Scholar]
- Krishna S.; White N. J. (1996) Pharmacokinetics of quinine, chloroquine and amodiaquine. Clinical implications. Clin. Pharmacokinet. 30 (4), 263–299. 10.2165/00003088-199630040-00002. [DOI] [PubMed] [Google Scholar]
- Ducharme J.; Farinotti R. (1996) Clinical pharmacokinetics and metabolism of chloroquine. Focus on recent advancements. Clin. Pharmacokinet. 31 (4), 257–274. 10.2165/00003088-199631040-00003. [DOI] [PubMed] [Google Scholar]
- Laaksonen A.-L.; Koskiahde V.; Juva K. (1974) Dosage of antimalarial drugs for children with juvenile rheumatoid arthritis and systemic lupus erythematosus: a clinical study with determination of serum concentrations of chloroquine and hydroxychloroquine. Scand. J. Rheumatol. 3 (2), 103–108. 10.3109/03009747409115809. [DOI] [PubMed] [Google Scholar]
- Kollaritsch H.; Karbwang J.; Wiedermann G.; Mikolasek A.; Na-Bangchang K.; Wernsdorfer W. H. (2000) Mefloquine concentration profiles during prophylactic dose regimens. Wien. Klin. Wochenschr. 112 (10), 441–447. [PubMed] [Google Scholar]
- Li W.; Moore M. J.; Vasilieva N.; Sui J.; Wong S. K.; Berne M. A.; Somasundaran M.; Sullivan J. L.; Luzuriaga K.; Greenough T. C.; Choe H.; Farzan M. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426 (6965), 450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J. K.; Tang T.; Nathan L.; Jaimes J. A.; Hsu H. L.; Daniel S.; Whittaker G. R. (2019) Production of Pseudotyped Particles to Study Highly Pathogenic Coronaviruses in a Biosafety Level 2 Setting. J. Visualized Exp. 145, e59010. 10.3791/59010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J. K.; Whittaker G. R. (2016) Murine Leukemia Virus (MLV)-based Coronavirus Spike-pseudotyped Particle Production and Infection. Bio-protocol 6 (23), e2035. 10.21769/BioProtoc.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. J.; Muench H. (1938) A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS12. Am. J. Epidemiol. 27 (3), 493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.