Abstract
Background/Aim:
Hydrogen sulfide (H2S) and the enzymes that synthesize it, cystathionine-b-synthase, cystathionine γ-lyase, and 3-mercaptopyruvate, are increased in different human malignancies. Due to its short half-life, H2S concentrations have not been directly measured in a human malignancy. Here we directly measured in vivo H2S levels within oral squamous cell carcinoma (OSCC).
Patients and Methods:
Punch biopsies of OSCC and benign mucosae from 15 patients were analyzed by HPLC, western blotting, and tissue microarray analyses.
Results:
H2S concentrations were significantly higher in OSCC compared to adjacent benign oral mucosae. Western blot and tissue microarray studies revealed significantly increased cystathionine-b-synthase, cystathionine γ-lyase, and 3-mercaptopyruvate, phopho-Stat3, mitoNEET, hTERT, and MAPK protein levels in OSCC.
Conclusion:
H2S concentrations and the enzymes that synthesize it are significantly increased in OSCC. Here, for the first time H2S concentrations within a living human malignancy were measured and compared to adjacent counterpart benign tissue.
Keywords: Hydrogen sulfide, H2S, mitoNEET, cystathionine-β-synthase, cystathionine γ-lyase, 3-mercaptopyruvate
Worldwide head and neck cancers are the sixth most common cancer type and are frequently epithelial aerodigestive tract malignancies (1). Approximately 75% are oral cancers, with 90% of these being oral squamous cell carcinomas (OSCC, 1–3). The remaining 10% consist of salivary gland tumors, lymphomas, and rare sarcomas (3). The five- and ten-year survival rates for these cancers are 59% and 48%, respectively, with these survival rates not having significantly improved for thirty years (1–3). The tongue is commonly effected, particularly the lateral-ventral surface, with other common locations being the oropharynx, floor of the mouth, and soft palate (4).
The risk factors for OSCC include tobacco, alcohol, areca nut, ultraviolet radiation exposure, and immunosuppression (5). OSCC typically arises from precursor lesions, such as submucosal fibrosis, erythro- and leukoplakias, actinic cheilosis, and oral lichen planus (2, 4, 5). Compared to benign oral squamous epithelium, OSCC precursors accumulate complex molecular changes that evolve over time as the lesions progress from precursor lesions, to oral OSCC, and eventually to metastatic disease (5–8). These changes include regional chromosomal gains and losses, tumor suppressor gene loss/inactivation, activation of cell growth promoting signal transduction pathways, and complex alterations in miRNA expression and epigenetic gene regulatory patterns (5–7). One study found 2,891 genes differentially expressed in OSCC compared to benign oral squamous epithelium, demonstrating the high complexity of OSCC malignant transformation (8). Altered signal transduction pathways important in OSCC growth include the changes/increases in the cyclin D1, epidermal growth factor/transforming growth factor-α, vascular endothelial growth factor, extracellular-signal regulated kinase, and AKT kinase-initiated pathways (5).
Hydrogen sulfide (H2S) is a recently discovered gasotransmitter that exerts extensive regulatory effects in health and disease (9). Three enzymes synthesize H2S: cystathionine-β-synthase (CBS), cystathionine γ-lyase (CSE), and 3-mercaptopyruvate (3-MST, 9, 10). Intracellular H2S concentrations, and one or more of the H2S-synthesizing enzymes, show increased expression in cells derived from several malignant human tumors, including melanoma, and colon, breast, bladder, and ovarian carcinomas, when compared to adjacent non-cancerous tissue or non-transformed cells (11–16). Increased intracellular CBS protein expression stimulates tumor xenograft growth, angiogenesis, and peritumoral vascular tone, indicating that H2S promotes tumor growth (11–16). H2S concentrations, and CBS, CSE, and 3-MST protein expressions are also often increased at higher tumor grades and stages (12–16).
The half-life of H2S is short, with one study done under aerobic conditions revealing H2S half-lives of 2.0, 2.8, and 10.0 min in murine hepatic, renal, and brain homogenates, respectively (17). For this reason, increased tumor H2S levels have been inferred from studies correlating the effects of H2S donors/inhibitors and/or increases or decreases in CBS, CSE, and 3-MST enzyme levels with parameters such as tumor growth and invasion, clinical stage, tumor grade, and angiogenesis (11–16). One study showed that buffered bladder tumor homogenates provided with exogenous L-cysteine and phosphatepyridoxine aldehyde produced nearly four times more H2S than comparable benign bladder tissue homogenates (15).
Exogenous H2S increases the proliferation and cell cycle progression of human OSCC cell lines via activation of the COX2/AKT/ERK1/2 axis (18, 19). Additionally, compared to benign oral squamous epithelium, CBS protein levels are increased in OSCC and with higher expression seen at higher tumor grades (14). This data suggests that H2S and the enzymatic pathways involved in its synthesis are increased in OSCC. Since OSCC and adjacent benign oral mucosae could easily and quickly be biopsied by punch biopsy technique and preserved in liquid N2 for later H2S analysis, we performed these studies to directly measure OSCC H2S concentrations and compare these measurements to adjacent benign oral mucosae. Additionally, tissue microarray was used to examine CSE and 3-MST levels in OSCC and western blotting to examine CBS, CSE, 3-MST, nicotinamide phosphoribosyltransferase (Nampt), mitoNEET, phospho-Stat3, hTERT, mitogen-activated protein kinase (MAPK), sulfide: quinone oxidoreductase (SQOR) and ataxia-telangiectasia mutated (ATM) protein levels in OSCC and adjacent benign oral mucosae.
Materials and Methods
Materials.
Monobromobimane (MBB), Tris (2-carboxyethyl) phosphine hydrochloride (TCEP), sulfosalicylic acid (SSA), 1-fluoro-2,4-dinitrobenzene (DNFB), diethylenetriaminepentaacetic acid (DTPA), sulfosalicylic acid (SSA), and N-ethylmaleimide (NEM) were purchased from Sigma (St. Louis, MO, USA). Polyvinylidene difluoride membranes were the source of the western blot membrane (Bio-Rad, Hercules, CA, USA, catalog number 170–4270). Antibodies used were anti-CBS (Santa Cruz Biotechnology, Santa Cruz, CA, USA, catalog number sc-67154), anti-CSE (Santa Cruz Biotechnology, catalog number sc-101924), anti-3-MST (Santa Cruz Biotechnology, catalog number sc-135993), anti-Visfatin (Nampt, Bethyl Laboratories, Montgomery TX, catalog number A300–779A), anti-phospho-Stat3 (ser727, Cell Signaling, Danvers, MA, USA, catalog number 9134S), anti-Stat3 (Cell Signaling, catalog number 12640S), anti-mitoNEET (Proteintech, Danvers, MA, USA, catalog number 16006–1-AP), anti-hTERT, (Millipore, Burlington, MA, USA, catalog number MABD55), anti-mitogen-activated protein kinase (MAPK, Cell Signaling, catalog number 4370), anti-SQOR (Boster Biological Technology, Pleasanton, CA, USA, catalog number A11155X), and anti-ATM (abcam, Cambridge, MA, catalog number ab199726). Secondary antibodies were goat anti-rabbit IgG (abcam, catalog numbers ab6721 and ab2040) and goat anti-mouse IgG (abcam catalog number ab205719), and goat anti-mouse IgG (Santa Cruz Biotechnology, catalog number sc358920).
Tissue Microarray (TMA).
Upon Institutional Review Board (IRB) approval by LSU Health Shreveport, two TMAs catalog number OR208 were purchased from US Biomax, Inc. (Rockville, MD, USA). Two TMAs were purchased, so that they could be interrogated with both anti-CSE and anti-3-MST antibodies. Together the TMAs contained 28 benign oral cavity squamous epithelial samples and 186 oral SCC samples: 152 OSSC grade I, 22 OSSC grade II, and six OSCC grade III tumor samples. All tissue samples in the TMAs were in 1.0 mm diameter.
CSE and 3-MST immunohistochemistry (IHC).
The concentration of primary CSE and 3-MST antibodies were optimized to normal kidney as control tissue. The staining of the TMAs was performed in the Tissue Core Histology Lab Facility at the Moffitt Cancer Center. The microarray slides were stained using a Ventana Discovery XT automated system (Ventana Medical Systems, Tucson, AZ, USA) as per the manufacturer’s protocol with proprietary reagents. Briefly, the slides were deparaffinized on the automated system with EZ Prep solution (#950–100; Ventana Medical Systems). The heat-induced antigen retrieval method was used in Cell Conditioning 1 (#950–124; Ventana Medical Systems). Mouse monoclonal antibody to human CSE (abcam, catalog number ab54573), was used at a 1:1,000 concentration in Dako antibody diluent (Dako, Carpentaria, CA, USA, catalog number S0809) and incubated for 60 min. The Santa Cruz mouse monoclonal antibody to human 3-MST (Santa Cruz Biotechnology, catalog number sc-135993), was used under the same conditions. The Ventana anti-mouse secondary antibodies were used for 16 min. The detection system used was the Ventana OmniMap kit. Slides were then dehydrated and cover-slipped as per standard laboratory protocol.
Evaluation of CSE and 3-MST staining.
Relative CSE and 3-MST protein expression was determined as immunostain intensity scored on a zero to three scale as follows: no staining as zero, light staining as one, moderate staining as two, and heavy staining as three. The percentage of cells stained was measured, with no detectable staining as zero, 1–33% as one, 34–66% as two, and 67–100% as three. The final IHC score was the product of the percentage of cells stained, multiplied by the intensity score, allowing for a minimal score of zero and a maximal score of nine. Nuclear and cytoplasmic Nampt and CBS staining were seen in all tissue samples examined, although at low levels in benign oral epithelia. We therefore measured and quantified CSE and 3-MST staining in the nuclear and cytoplasmic compartments.
TMA Statistical analysis.
The standard error of the mean (SEM) IHC score was calculated by taking the mean of each data set, subtracting the mean from each number in the set and squaring the result, then calculating the mean of the squared differences, and taking the square root of this number to find the standard deviation. The standard deviation was divided by the square root of the number of tissue samples in the sample set to find the SEM. This statistical analysis was the same as performed on a previous TMA study of CBS levels in OSCC (14).
Patients.
After obtaining IRB approval, patients were selected based on the presence of a T2 or greater OSCC. From February 2017 to December 2017, 116 patients presented to one of the oncology clinics within the department of Oral and Maxillofacial Surgery at our institution for evaluation of an oral cavity lesion. Either the lesions had been previously biopsied by an outside physician and were likely OSCC, or they were biopsied by one of our physicians and returned as OSCC. If surgical resection was chosen as the primary treatment modality, patients with T2 or greater OSCCs were asked if they would approve of being enrolled in our study. If they opted to participate, IRB-approved informed consent was obtained. After inclusion criteria had been met, twenty patients were enrolled in the study. They ranged in age from 47 to 83 years, with various oral cavity tumor locations (Table I). On the morning of each surgery, the tumor was identified and a 1.5 cm margin was marked circumferentially around the tumor in the standard oncologic fashion. Once the surgical margin was demarcated, three separate 3.0 mm punch biopsies were obtained from the periphery of the resection margin in what was deemed to be benign tissue, as well as three separate 3 mm punch biopsies from the center of the malignancy. A separate punch biopsy was used for the benign specimens as well as the malignant specimens to prevent cross-contamination. Any excess blood on the specimens was blotted away with a surgical lap sponge to prevent any blood contamination. These specimens were individually placed in Eppendorf tubes labeled as “benign” numbers one to three and “malignant” numbers one to three. They were immediately sealed, placed into a bath of liquid nitrogen, and passed on to the pathologist for further testing. Less than twenty sec elapsed between obtaining each specimen and final placement in the liquid nitrogen bath. All tissue samples were examined and staged by experienced, board-certified pathologists, to confirm that the tissues used were either benign oral mucosae or OSCC. Of the twenty samples, three were lost due to excess necrosis or being too bloody, and two were non-OSCC tumors; thus, leaving fifteen OSCCs and benign mucosal-paired samples analyzed (Table I).
Table I.
Profiles of the fifteen patients enrolled in this study.
| Age | Tumor size | Tumor type | Tumor grade | Tumor location |
|---|---|---|---|---|
| 62 | T2 | OSCC | II | Right tongue |
| 49 | T4a | OSCC | II | Right mandible |
| 54 | T4a | OSCC | II | Mandible/Floor of mouth |
| 69 | T2 | OSCC | II | Floor of mouth |
| 51 | T2 | OSCC | I | Left retromolar trigone |
| 56 | T2 | OSCC | I-II | Left tongue |
| 73 | T4a | OSCC | II | Anterior Mandible |
| 77 | T2 | OSCC | III | Left floor of mouth |
| 55 | T2 | OSCC | II | Right mandibular alveolar ridge |
| 47 | T3 | OSCC | II | Left tongue |
| 77 | T4a | OSCC | II | Left mandible |
| 49 | T2 | OSCC | II | Left tongue |
| 55 | T3 | OSCC | II | Right tongue |
| 52 | T2 | OSCC | II | Left buccal mucosa |
| 83 | T4 | OSCC | II | Right mandible |
Western blots.
Whole cell lysates where prepared from the liquid N2 frozen surgical biopsies. Following weighing and quick thawing, the tissue samples were lysed in chilled RIPA buffer (20 mM Tris-HCl, 150 mM NaCl, 1% Triton-X100, 0.1% SDS, 1 mM EDTA, 1% sodium deoxycholate, 1 mM PMSF, 5 μg/ml Leupeptin) at 1 mg tissue/10 μl RIPA buffer. The tissue was next homogenized by tekmar tissumizer, centrifuged at 12,000 g at 4°C for 10 min, and the supernatant transferred to a new tube. The protein concentration was measured by Bio-rad DC protein assay. Ten-20 μg protein supernatant extract was mixed with 2X SDS loading buffer according to the protein concentration. The protein sample was then loaded onto an SDS gel or placed at −20°C for storage. Lysates separated by SDS-PAGE were transferred to polyvinylidene difluoride membranes, and membranes were blocked in 5% nonfat dry milk in 1X PBS before the addition of the primary antibodies. GAPDH was used as a western blot loading control, except for the phospho-Stat3 ser727 blots, where an anti-Stat3 antibody was employed. Some of the antibodies to specific proteins were added to the study at later time points than were CBS, CSE, 3-MST, and Nampt, and hence there were fewer OSCC/benign mucosal pairs interrogated.
H2S measurements.
Bioavailable H2S levels were measured as previously reported (20, 21). Levels of free sulfide in the OSCC/benign mucosal-paired samples were measured by high performance liquid chromatography (HPLC) after derivatization with excess MBB as stable products sulfide-dibimane (SDB). Briefly, oral tissue samples were homogenized in Tris·HCl buffer [100 mM Tris·HCl (pH 9.5) and 0.1 mM diethylenetriaminepentaacetic acid (DTPA)]. Cell lysates were derivatized with MBB and then measured by Shimadzu Prominence 20A equipment with RF-10AXL (excitation wavelength: 390 mm and emission wavelength: 475 mm) and an Eclipse XDB-C18 column (4.6×250 mm, 5 μm). Typical retention times of SDB were around 16.5 min. Free H2S levels were calculated according to standard SDB (20, 21).
H2S and western blot statistical analysis.
Data for the western blots and H2S measurements were normalized with image J densitometry software and significance was calculated by using prism software version 5.02 (GraphPad Inc., San Diego, CA, USA).
Results
Tissue microarrays.
Following processing, four benign oral cavity squamous epithelial samples were lost in each TMA. The remaining tissue samples were not lost. As shown in Figure 1 and Table II, both CSE and 3-MST were increased in OSCC compared to benign oral epithelia by TMA analysis. CSE and 3-MST expression increased with higher OSCC tumor grades, with CSE minimally expressed in benign squamous epithelia and increased in OSCC (Table II). This data, combined with a previous study, demonstrates that CBS, CSE, and 3-MST show higher protein expression in OSCC compared to benign oral squamous epithelia, with CBS more highly induced as measured by previous TMA analysis (14).
Figure 1.
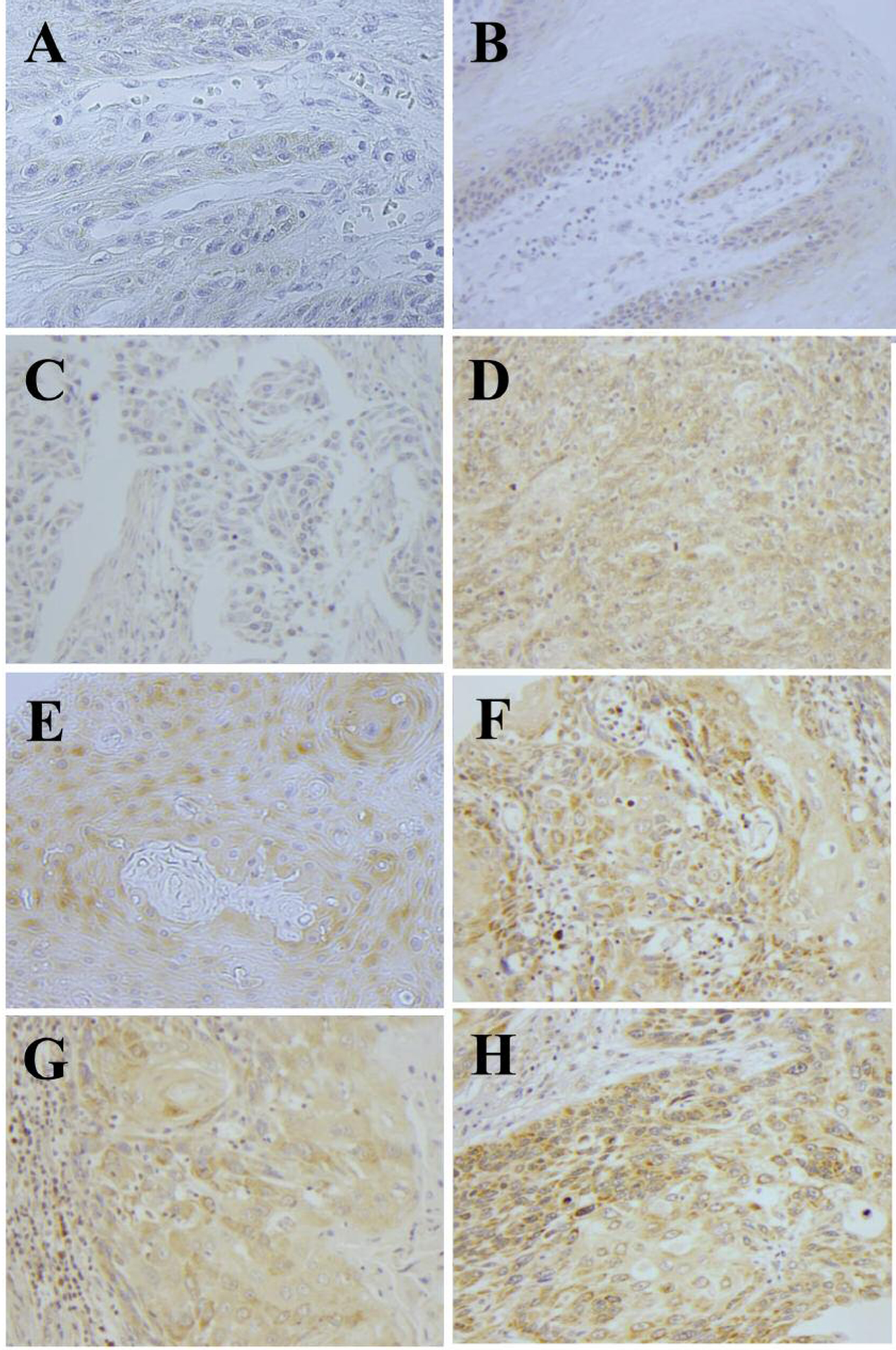
Representative immunostaining of benign oral squamous mucosae with CSE and 3-MST (A) and (B), respectively. Grade I (C and D), grade II (E and F), and grade III (G and H) OSCC were also stained for CSE and 3-MST, respectively. High-power views (×400).
Table II.
Relative CSE and 3-MST staining in the two tissue microarrays comparing benign squamous epithelium, and OSCC, grades I-III.
| Antibody | CSE number | 3-MST number | CSE IHC score | 3-MST IHC score | CSE SEM | 3-MST SEM |
|---|---|---|---|---|---|---|
| Benign squamous epithelium | 24 | 24 | 0.42 | 0.79 | 0.27 | 0.60 |
| SCC grade I | 152 | 152 | 0.71 | 1.41 | 0.40 | 0.19 |
| SCC grade II | 22 | 22 | 2.13 | 2.68 | 0.20 | 0.23 |
| SCC grade III | 6 | 6 | 3.33 | 4.00 | 0.61 | 0.18 |
IHC: Averaged immunohistochemical score; SEM: standard error of the mean.
Western blotting.
TMA analyses, while useful, can lack robustness and controls (22). To confirm and extend the TMA results, western blot analyses were performed on the fifteen surgical OSCC/benign mucosal samples (Table I). Western blotting was performed for CBS (15 patients), CSE (15 patients), 3-MST (15 patients), Nampt (15 patients), phospho-Stat3 (10 patients), mitoNEET (14 patients), MARPK (10 patients), hTERT (9 patients), SQOR (10 patients), and ATM (11 patients, Table I). As shown in Figure 2A and D and 2F–2H, CBS, CSE, 3-MST, Nampt mitoNEET, MARPK, and hTERT protein expression were increased in OSCC compared to benign oral mucosae. For Figure 2E, Stat3 ser727 phosphorylation was measured and was increased in the OSCC samples compared to benign oral mucosae. The significance value for these eight benign/OSCC comparisons was p<0.001. Mito NEET showed both the highest expression increases and the most reliable increases in OSCC compared to benign oral epithelium (Figure 2F, Table III). The SQOR and ATM proteins (Figures 2I and J), were not induced and the p-values for these western blots were not significant. The proteins examined, the number of cases, and their statistical significances in OCSS cases compared to counterpart benign tissue are shown in Table III.
Figure 2.
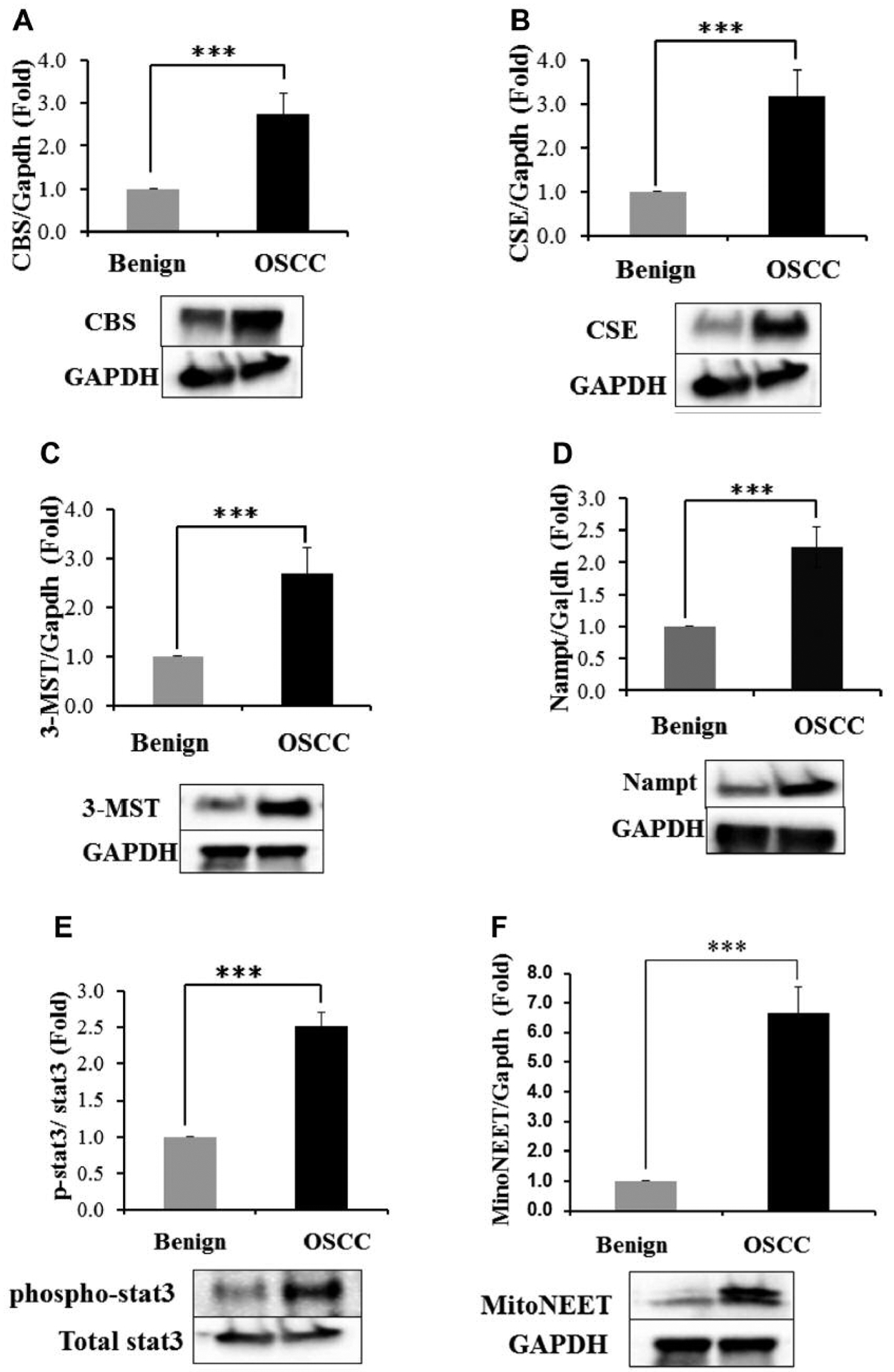
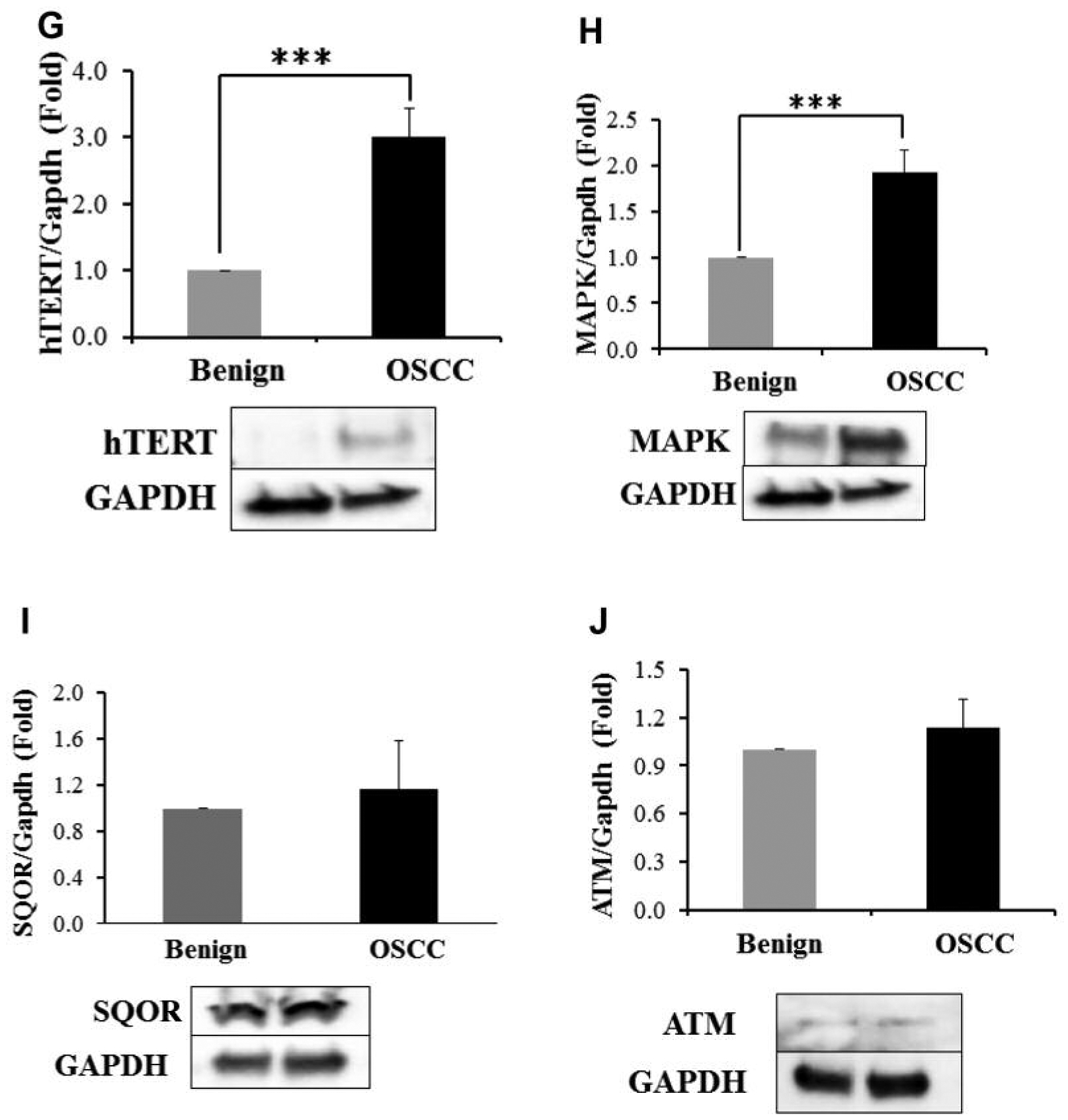
Western blot analyses of benign oral mucosae and OSCC for the CBS (A), CSE (B), 3-MST (C), Nampt (D), mitoNEET (E), phospho-Stat3 (F), hTERT (G), MAPK (H), SQOR (I) and ATM (J) proteins. ***p<0.001. SQOR and ATM (2I and 2J) did not have statistically significant differences between benign oral epithelium and OSCC.
Table III.
The average increases in each protein seen when specific protein levels in OCSS were compared to benign oral mucosae. Numbers were rounded off to two decimal places. For all OSCC/benign tissue pairs where the p-value was significant, the protein examined by western blotting was increased in every OSCC/benign tissue pair.
| Protein | Average induction in OSCC/Benign oral mucosae | Number of cases examined | p-Value for 15 pairs analyzed |
|---|---|---|---|
| CBS | 2.76 | 15 | Significant |
| CSE | 2.96 | 15 | Significant |
| 3-MST | 2.73 | 15 | Significant |
| Nampt | 2.25 | 15 | Significant |
| p-Stat3 | 2.39 | 11 | Significant |
| mitoNEET | 6.76 | 14 | Significant |
| hTERT | 2.92 | 11 | Significant |
| MAPK | 1.81 | 12 | Significant |
| SQOR | 1.04 | 10 | Not Significant |
| ATM | 1.17 | 11 | Not Significant |
Benign oral mucosal and OSCC H2S measurements.
When fifteen benign tissue/OSCC samples were compared, free H2S was significantly increased in the OCSS compared to benign oral muscae (Figure 3).
Figure 3.
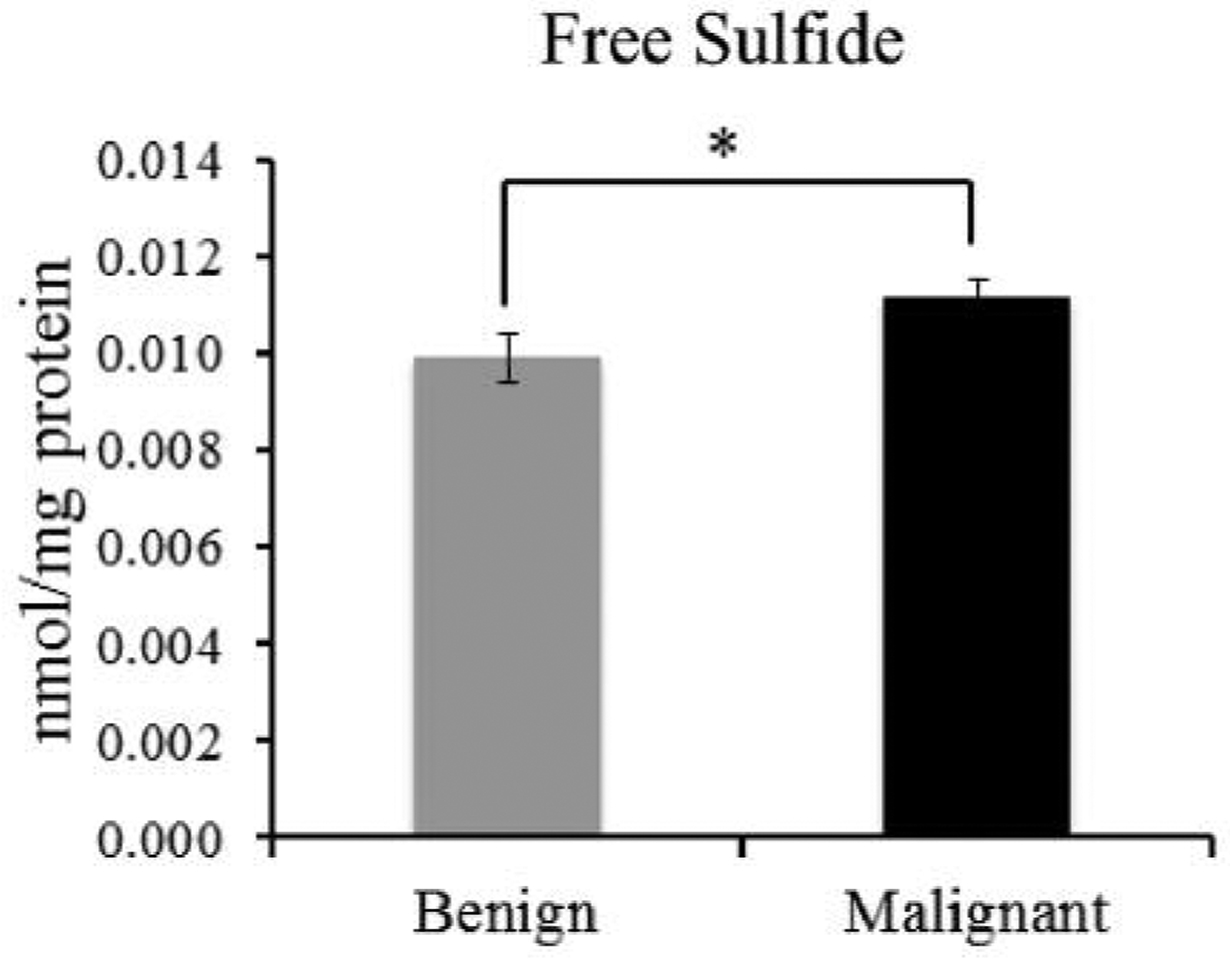
Comparison of the cellular free sulfide (H2S) pools of benign oral mucosae and OSCC in fifteen patients. Free sulfide (H2S) is pmol/mg protein. *p<0.05.
Discussion
Previously TMA analysis revealed that CBS is increased in OSCC compared to benign oral squamous epithelium (14). In this study, a similar microarray analysis was used to establish that CSE and 3-MST are also increased in OSCC compared to benign oral epithelium (Table II, Figure 1). Since TMA analyses can lack adequate controls and can show low assay robustness, western blotting was used on our OCSS/benign oral mucosae pairs to look for changes in ten proteins (22). Each of these proteins was examined for specific reasons; 1) to examine the levels of the H2S-synthesizing enzymes (CBS, CSE, and 3-MST), 2) to identify new proteins increased in OCSS which may be related to H2S metabolism (mitoNEET, phospho-Stat3, and SQOR), and 3) validify our results by examining proteins previously found to be either increased or not increased in OSCC (Nampt, MARPK, hTERT, and ATM). Due to the very short half-life of H2S, a time-consuming microdissection of the benign punch biopsy to include only benign squamous epithelium was not possible, hence here the term “benign oral mucosae” was use for these samples (17).
To initiate these studies CBS, CSE, and 3-MST protein expression were analyzed in the 15 OCSS/benign tissue sample pairs. As shown in Figure 2A and C all three enzymes were increased roughly 2.7-fold in OSCC compared to adjacent benign tissue. Each enzyme was consistently increased in OSCC compared to benign tissue pair for all 15 pairs analyzed (Table III). Nampt levels were next examined as it is commonly increased in malignant tumors (23, 24). Nampt is regulated by CBS and CSE, with inhibition of either enzyme suppressing Nampt expression and increased cellular H2S increasing Nampt expression (23, 24). Like the three H2S-synthesizing enzymes, Nampt was reliably increased on average 2.5-fold in every tumor-benign tissue sample comparison (Figure 2D, Table III). Our finding that Nampt is co-induced with CBS and CSE in OSCC lends some support to this circuit playing a role in OSCC (23).
Phospho-Stat3 ser727 plays an important role in oncogenic transformation, the regulation of Stat3 activity, and is highly expressed in cervical squamous cell carcinoma (25–28). Stat3 phosphorylation is also increased following H2S exposure, promotes cell survival and cyclin D1 expression, and increases CSE expression in breast cancer (29–31). Therefore, its expression in OSCC was examined as it is likely H2S regulated and in turn may regulate CSE. As shown in Figure 2E, phospho-Stat3 ser727 was increased in OSCC compared to benign oral mucosae about 3.4-fold. To our knowledge, this is the first time this Stat3 phosphorylation has been observed in OSCC. We are currently examining its role in CSE regulation in OSCC.
MitoNEET is an outer mitochondrial iron-sulfur protein that shows increased expression in several human malignancies (32, 33). High tumor mitoNEET expression suppresses apoptosis, autophagy, and lowers intramitochondrial iron concentrations, likely allowing tumors cells to tolerate higher reactive oxygen species, while avoiding ferroptosis (33). The oxidation state of mitoNEET is regulated by glutathione reductase, a cellular redox regulator enzyme that catalyses the conversion of glutathione disulfide to glutathione, an enzymatic reaction requiring nicotinamide adenine dinucleotide phosphate (NADPH) as a substrate (33, 34). Since CBS, CSE, 3-MST, and glutathione reductase are highly expressed in OSCC and H2S is a powerful activator of glucose-6-phosphate dehydrogenase, which leads to increased pentose phosphate shunt activation and subsequent increased intracellular NADPH levels, we hypothesized that mitoNEET would be increased in OSCC (35, 36). As shown in Figure 2F and Table III, mitoNEET is strongly and significantly increased in OSCC compared to benign mucosae. MitoNEET was, on average, induced over six-fold in all fourteen OSCC and benign mucosal samples interrogated, higher than any other protein here examined. To our knowledge, this is the first time that mitoNEET has been demonstrated to be increased in OCSS. The reason for this induction in unknown, however higher mitoNEET levels might promote tumor survival via ferroptosis, apoptosis, and autophagy suppression (32).
Next, we looked at TERT and MAPK expression in ten each benign mucosal and OSCC samples. TERT and MARK were induced roughly 2.0- and 2.5-fold, in OSCC compared with the benign mucosal samples, respectively (Figures 2G and H, and Table III). hTERT is increased in many tumors, including OSCC, where it promotes OSCC progression and the epithelial-mesenchymal transition (37). Similarly, MAPK promotes OSCC progression though multiple pathways, including the induction of cyclin D1 and activation of the PI3K/Akt pathways (38, 39).
SQOR protein levels were next examined in ten OSCC and benign mucosal samples. SQOR mediates the first step in H2S mitochondrial metabolism where it catalyzes a two-electron oxidation of H2S to sulfane sulfur, while using coenzyme Q as an electron acceptor (40, 41). We hypothesized that SQOR protein expression may be increased in OSCC and hence, use H2S as an electron source for the mitochondrial electron transport chain. SQOR however, was not induced in OSCC compared to benign oral mucosae (Figure 2I, Table III). Last, ATM protein levels were examined in eleven patient samples (Figure 2J, Table III). The ATM protein is not induced in OSCC (42). Similar to previous studies, the ATM protein was not induced in OSCC in our study. Based on this, and our finding that hTERT and MAPK were induced in OSCC as previously found, we conclude that our western blot protocol yielded valid results (37–39, 42).
A large body of data indicates that H2S, CBS, CSE, and 3-MST are increased in many cancer types, where increased H2S and enzyme levels promote tumor growth and invasion, angiogenesis, and metastasis (11–19). Here, all three H2S-synthesizing enzymes were increased in OCSS compared to benign oral mucosae, comparable with previous results for other malignancies (Figure 2A and C (11–16). Additionally, for the first time total cellular free H2S was directly measured in a human tumor. H2S was significantly increased in OSCC compared to benign oral mucase (Figure 3). Interestingly, although the increase in H2S was statistically significant, the increase was modest and less than the 2.7-fold increases in the protein expression of CBS, CSE, and 3-MST seen here. Similarly, it is also less than the nearly fourfold increase in cellular H2S concentrations seen in buffered bladder tumor homogenates compared to benign bladder tissue homogenates, and in cancer-derived human cell lines compared to counterpart benign tissues (11, 13, 15). Although our study and many previous studies did not examine H2S-synthesizing enzyme activities, the modest increase in OCSS H2S levels seen here suggest that the OSCC is rapidly metabolizing H2S, possibly to support malignant cell growth. Support for this comes from previous studies showing that suppression of H2S-synthesizing enzyme activities or suppression of their protein expression slows tumor cell growth, while exogenous H2S can increase it (11–15, 18, 19).
Our findings here are preliminary and more work needs to be done. For example, OSCC relative CBS, CSE, 3-MST, and SQOR enzymatic activities should be measured. The activities of CBS, CSE, and 3-MST may be increased, increasing H2S synthesis and availability in OSCC, however the work here does not address this. Possible differences in H2S and protein expression between different tumor grades and stages should be examined, which is not possible with the small sample size used herein. Similarly, while SQOR levels were not increased in OSCC, its activity might be increased allowing H2S electrons to be shunted into the mitochondrial electron transport chain to serve as an energy source. Additionally, the H2S cellular half-life in benign oral mucosae and OSCC should be examined, as well as the differences in the specific sulfane species found in benign and malignant tissues. Last, the possible roles of mitoNEET and phospho-Stat3 ser727 in OSCC as related to H2S and tumor growth should be examined. This work is currently underway in our laboratory.
Acknowledgements
The Authors thank the Histology Section of the Tissue Core at the Moffitt Cancer Center and Research Institute for the support in performing the IHC stains. This work was supported in part by HL113303 to C.G.K.
Footnotes
Conflicts of Interest
The Authors have no conflicts of interest regarding this work.
References
- 1.Johnson NW, Warnakulasuriya S, Gupta PC, Dimba E, Chindia M, Otoh EC, Sankaranarayanan R, Califano J and Kowalski L: Global oral health inequalities in incidence and outcomes for oral cancer: causes and solutions. Adv Dent Res 23: 237–246, 2011. [DOI] [PubMed] [Google Scholar]
- 2.Warnakulasuriya S: Global epidemiology of oral and oropharyngeal cancer. Oral Oncol 45: 309–316, 2009. [DOI] [PubMed] [Google Scholar]
- 3.Shiboski CH, Shiboski SC and Silverman S Jr.: Trends in oral cancer rates in the United States, 1973–1996. Community Dent Oral Epidemiol 28: 249–256, 2000. [DOI] [PubMed] [Google Scholar]
- 4.Natarajan E and Eisenberg E: Contemporary concepts in the diagnosis of oral cancer and precancer. Dent Clin North Am 55: 63–88, 2011. [DOI] [PubMed] [Google Scholar]
- 5.Huber MA and Tantiwongkosi B: Oral and oropharyngeal cancer. Med Clin North Am 98: 1299–1321, 2014. [DOI] [PubMed] [Google Scholar]
- 6.Salahshourifar I, Vincent-Chong VK, Kallarakkal TG and Zain RB: Genomic DNA copy number alterations from precursor oral lesions to oral squamous cell carcinoma. Oral Oncol 50: 404–412, 2014. [DOI] [PubMed] [Google Scholar]
- 7.Rivera C: Essentials of oral cancer. Int J Clin Exp Pathol 8: 1884–11894, 2015. [PMC free article] [PubMed] [Google Scholar]
- 8.Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL and Gaffney PM: Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res 64: 55–63, 2004. [DOI] [PubMed] [Google Scholar]
- 9.Wang R: Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev 92: 791–896, 2012. [DOI] [PubMed] [Google Scholar]
- 10.Kabil O and Banerjee R: Redox biochemistry of hydrogen sulfide. J Biol Chem 285: 21903–21907, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szabo C, Coletta C, Chao C, Módis K, Szczesny B, Papapetropoulos A and Hellmich MR: Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Natl Acad Sci USA 110: 12474–12479, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharyya S, Saha S, Giri K, Lanza IR, Nair KS, Jennings NB, Rodriguez-Aguayo C, Lopez-Berestein G, Basal E, Weaver AL, Visscher DW, Cliby W, Sood AK, Bhattacharya R and Mukherjee P: Cystathionine beta-synthase (CBS) contributes to advanced ovarian cancer progression and drug resistance. PLoS One 8: e79167, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen S, Kawahara B, Gupta D, Tsai R, Khachatryan M, Roy-Chowdhuri S, Bose S, Yoon A, Faull K, Farias-Eisner R and Chaudhuri G: Role of cystathionine β-synthase in human breast Cancer. Free Radic Biol Med 86: 228–238, 2015. [DOI] [PubMed] [Google Scholar]
- 14.Patel S, Ansari J, Meram A, Abdulsattar J, Cotelingam J, Coppola D, Ghali G and Shackelford R: Increased nicotinamide phosphoribosyltransferase and Cystathionine-Beta-Synthase in oral cavity squamous cell carcinomas. Int J Clin Exp Pathol 10: 702–707, 2017. [Google Scholar]
- 15.Gai JW, Qin W, Liu M, Wang HF, Zhang M, Li M, Zhou WH, Ma QT, Liu GM, Song WH, Jin J and Ma HS: Expression profile of hydrogen sulfide and its synthases correlates with tumor stage and grade in urothelial cell carcinoma of bladder. Urol Oncol 34: 166.e15–20, 2016. [DOI] [PubMed] [Google Scholar]
- 16.Jurkowska H, Placha W, Nagahara N and Wrobel M: The expression and activity of cystathionine-γ-lyase and 3-mercaptopyruvate sulfurtransferase in human neoplastic cell lines. Amino Acids 41: 151e158, 2011. [DOI] [PubMed] [Google Scholar]
- 17.Vitvitsky V, Kabil O and Banerjee R: High turnover rates for hydrogen sulfide allow for rapid regulation of its tissue concentrations. Antioxid Redox Signal 17: 22–31, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Bian H, Li X, Wu H, Bi Q, Yan Y and Wang Y: Hydrogen sulfide promotes cell proliferation of oral cancer through activation of the COX2/AKT/ERK1/2 axis. Oncol Rep 35: 2825–2832, 2016. [DOI] [PubMed] [Google Scholar]
- 19.Ma Z, Bi Q and Wang Y: Hydrogen sulfide accelerates cell cycle progression in oral squamous cell carcinoma cell lines. Oral Dis 21: 156–162, 2015. [DOI] [PubMed] [Google Scholar]
- 20.Leskova A, Pardue S, Glawe JD, Kevil CG and Shen X: Role of thiosulfate in hydrogen sulfide-dependent redox signaling in endothelial cells. Am J Physiol Heart Circ Physiol 313: H256–H264, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen X, Peter EA, Bir S, Wang R and Kevil CG: Analytical measurement of discrete hydrogen sulfide pools in biological specimens. Free Radic Biol Med 15: 2276–2283, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joos T and Bachmann J: Protein microarrays: potentials and limitations. Front Biosci 14: 4376–4385, 2009. [DOI] [PubMed] [Google Scholar]
- 23.Sanokawa-Akakura R, Ostrakhovitch EA, Akakura S, Goodwin S and Tabibzadeh S: A H2S-Nampt dependent energetic circuit is critical to survival and cytoprotection from damage in cancer cells. PLoS One 23: e108537, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shackelford RE, Mayhall K, Maxwell NM, Kandil E and Coppola D: Nicotinamide phosphoribosyltransferase in malignancy: a review. Genes Cancer 4: 447–456, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Wu X, Zhang F, Mo S, Lu Y, Wei W, Chen X, Lan L, Lu B and Liu Y: Paclitaxel induces apoptosis of esophageal squamous cell carcinoma cells by downregulating STAT3 phosphorylation at Ser727. Oncol Rep 37: 2237–2244, 2017. [DOI] [PubMed] [Google Scholar]
- 26.Plaza-Menacho I, van der Sluis T, Hollema H, Gimm O, Buys CH, Magee AI, Isacke CM, Hofstra RM and Eggen BJ: Ras/ERK1/2-mediated STAT3 Ser727 phosphorylation by familial medullary thyroid carcinoma-associated RET mutants induces full activation of STAT3 and is required for c-fos promoter activation, cell mitogenicity, and transformation. J Biol Chem 282: 6415–6424, 2007. [DOI] [PubMed] [Google Scholar]
- 27.Wakahara R, Kunimoto H, Tanino K, Kojima H, Inoue A, Shintaku H and Nakajima K: Phospho-Ser727 of STAT3 regulates STAT3 activity by enhancing dephosphorylation of phospho-Tyr705 largely through TC45. Genes Cells 17: 132–145, 2012. [DOI] [PubMed] [Google Scholar]
- 28.Yang SF, Yuan SS, Yeh YT, Wu MT, Su JH, Hung SC and Chai CY: The role of p-STAT3 (ser727) revealed by its association with Ki-67 in cervical intraepithelial neoplasia. Gynecol Oncol 98: 446–452, 2005. [DOI] [PubMed] [Google Scholar]
- 29.Calvert JW, Jha S, Gundewar S, Elrod JW, Ramachandran A, Pattillo CB, Kevil CG and Lefer DJ: Hydrogen sulfide mediates cardioprotection through Nrf2 signaling. Circ Res 105: 365–374, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deepak Roshan VG, Sinto MS, Thomas S and Kannan S: Cyclin D1 overexpression associated with activation of STAT3 in oral carcinoma patients from South India. J Cancer Res Ther 14: 403–408, 2018. [DOI] [PubMed] [Google Scholar]
- 31.You J, Shi X, Liang H, Ye J, Wang L, Han H, Fang H, Kang W and Wang T: Cystathionine-γ-lyase promotes process of breast cancer in association with STAT3 signaling pathway. Oncotarget 8: 65677–65686, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohn YS, Tamir S, Song L, Michaeli D, Matouk I, Conlan AR, Harir Y, Holt SH, Shulaev V, Paddock ML, Hochberg A, Cabanchick IZ, Onuchic JN, Jennings PA, Nechushtai R and Mittler R: NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc Natl Acad Sci USA 110: 14676–14681, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittler R, Darash-Yahana M, Sohn YS, Bai F, Song L, Cabantchik IZ, Jennings PA, Onuchic JN and Nechushtai R: NEET proteins: A new link between iron metabolism, reactive oxygen species, and cancer. Antioxid Redox Signal, 2018. 10.1089/ars.2018.7502. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landry AP, Cheng Z and Ding H: Reduction of mitochondrial protein mitoNEET [2Fe-2S] clusters by human glutathione reductase. Free Radic Biol Med 81: 19–127, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rawal RM, Patel DD, Patel BP, Patel MM, Wadhwa MK, Patel PS and Bhatavdekar JM: Assessment of glutathione-S-transferase and glutathione reductase in patients with squamous-cell carcinoma of buccal mucosa. Int J Cancer 83: 727–731, 1999. [DOI] [PubMed] [Google Scholar]
- 36.Chhabra A, Mishra S, Kumar G, Gupta A, Keshri GK, Bharti B, Meena RN, Prabhakar AK, Singh DK, Bhargava K and Sharma M: Glucose-6-phosphate dehydrogenase is critical for suppression of cardiac hypertrophy by H2S. Cell Death Discov 4: 6, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao T, Hu F, Qiao B, Chen Z and Tao Q: Telomerase reverse transcriptase potentially promotes the progression of oral squamous cell carcinoma through induction of epithelial-mesenchymal transition. Int J Oncol 46: 2205–2215, 2015. [DOI] [PubMed] [Google Scholar]
- 38.Fu X and Feng Y: QKI-5 suppresses cyclin D1 expression and proliferation of oral squamous cell carcinoma cells via MAPK signalling pathway. Int J Oral Maxillofac Surg 44: 562–567, 2015. [DOI] [PubMed] [Google Scholar]
- 39.Murugan AK, Munirajan AK and Tsuchida N: Ras oncogenes in oral cancer: the past 20 years. Oral Oncol 48: 383–392, 2012. [DOI] [PubMed] [Google Scholar]
- 40.Jackson MR, Melideo SL and Jorns MS: Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry 51: 6804–6815, 2012. [DOI] [PubMed] [Google Scholar]
- 41.Jackson MR, Melideo SL and Jorns MS: Role of human sulfide: quinone oxidoreductase in H2S metabolism. Methods Enzymol 554: 255–270, 2015. [DOI] [PubMed] [Google Scholar]
- 42.He Y, Chen Q and Li B: ATM in oral carcinogenesis: association with clinicopathological features. J Cancer Res Clin Oncol 134: 1013–1020, 2008. [DOI] [PubMed] [Google Scholar]


