Summary:
Few data exist on the prognostic and predictive impact of erb-b2 receptor tyrosine kinase 4 (ERBB4) in ovarian cancer. Thus, we evaluated ERBB4 expression by immunohistochemistry in a tumor microarray consisting of 100 ovarian serous carcinoma specimens (50 complete responses [CRs] and 50 incomplete responses [IRs] to platinum-based therapy), 51 normal tissue controls, and 16 ovarian cancer cell lines. H scores were used to evaluate expression and were semiquantitatively classified into low, intermediate, and high categories. Category frequencies were compared between tumor specimens vs controls using an unpaired t test. Among tumors, category frequencies were compared between CR and IR to chemotherapy. Overall survival (OS) was stratified by category. In total, 74 ovarian serous carcinoma samples (32 CRs and 42 IRs), 28 normal controls, and 16 ovarian cancer cell lines were evaluable. High-level ERBB4 expression was observed at a significantly higher frequency in ovarian serous carcinoma compared with normal control tissue. Among tumor specimens, ERBB4 expression was significantly higher for those with an IR to chemotherapy compared with CR (P = .033). OS was inversely correlated with ERBB4 expression levels. Median rates of OS were 18, 22, and 58 months among high-, intermediate-, and low-expression tumors, respectively. Our results indicate that ERBB4 expression by immunohistochemistry may correlate with chemotherapy-resistant ovarian serous carcinoma and shortened OS.
Introduction
Ovarian cancer is the fifth most common cause of cancer-related death in American women.1 Approximately 220,000 women develop epithelial ovarian cancer worldwide every year.2 In 2016, an estimated 22,280 women living in the United States were diagnosed with the disease, and 14,240 of US women were expected to die of the disease that same year.1 More than 90% of ovarian malignancies are of epithelial origin, and ovarian serous carcinoma is the most common subtype, representing nearly 40% of all epithelial ovarian cancers.3,4 Approximately 65% of patients with ovarian cancer present with an advanced stage of disease at diagnosis.5 Standard first-line treatment of peritoneal disease is surgical cytoreduction combined with platinum-based chemotherapy. Although several new and promising agents have been investigated in recent years, platinum-based cytotoxic chemotherapy (systemic or intraperitoneal) remains the standard initial approach to advanced and recurrent ovarian cancer.6 Individuals with a poor prognosis include those whose disease progresses on platinum-based treatment (platinum refractory) or disease that persists or recurs within 6 months of treatment (platinum-resistant disease).
In the last decade, several studies have explored possible molecular pathways involved in platinum resistance, including the epidermal growth factor receptor (EGFR) family, which has 4 members7: ERBB1 (formerly known as HER1/EGFR), ERBB2 (formerly known as HER2/neu), ERBB3 (formerly known as HER3), and ERBB4 (formerly known as HER4). Although ERBB1 and ERBB2 have been studied in various cancers, less is known about the cancer biology of ERBB3 and ERBB4.8 All EGFR family members, including ERBB4, have a heavily glycosylated ecto domain that contains a ligand-binding site, a single transmembrane domain, an intracellular protein-tyrosine kinase catalytic domain, and a tyrosine-containing cytoplasmic tail.9 ERRB4 forms homodimers or heterodimers with the other ERBB members to activate signaling via multiple second messengers, which mediate its final biological effects.10 Four different ERBB4 isoforms derived from alternative splicing of ERBB4 messenger RNA — JM-a/CYT-1, JM-a-CYT-2, JM-b/CYT-1, and JM-b/CYT-2 — have been reported in the literature.11,12
Unlike other members of the EGFR family, activated erb-b2 receptor tyrosine kinase 4 (ERBB4) undergoes proteolytic cleavage at the cell surface to release a soluble intracellular domain (4ICD). Once released from the plasma membrane, 4ICD can localize to cytosol or nucleus.13 ERBB4 isoforms activate or inhibit downstream molecular pathways, which can have different or opposing roles in developing resistance to chemotherapeutic agents. ERBB4 mediates proapoptotic signaling to trigger cell death and also promotes the up- or down-regulation of diverse genes required for cells to survive under stress.14,15 In certain tissues, it appears to exert a suppressive role on tumor development by activating genes that promote cellular differentiation and inhibit proliferation. By contrast, mutations of ERBB4 have also been associated with multiple cancer types.16,17
Therefore, to identify a possible role of ERBB4 in the development of platinum resistance, we sought to study ERBB4 expression by immunohistochemistry in clinical samples obtained from patients with complete response (CR) and incomplete response (IR) to platinum-based chemotherapy and ovarian cancer cell lines.
Materials and Methods
Study Design
After approval from the Institutional Review Board at the H. Lee Moffitt Cancer Center & Research Institute (Tampa, FL), ERBB4 expression levels by immunohistochemistry were evaluated in a tissue microarray consisting of samples from 100 ovarian serous carcinoma specimens (50 cases of CR and 50 cases of IR to platinum-based chemotherapy), 51 tissue samples from normal, non-neoplastic ovarian and fallopian tube control specimens, and 16 ovarian cancer cell lines.
Evaluation of ERBB4 expression was performed by light microscopy and digital imaging. The paired results from each sample were compared by Pearson correlation tests. Clinical data were extracted from medical records, including patient age at diagnosis, response to chemotherapy, disease-specific survival rate, and pathological tumor, node, and metastasis stage.
CR to primary therapy was defined as the normalization of cancer antigen 125 level following treatment, the absence of disease on computed tomography, or the absence of disease on second-look surgery. An IR to primary therapy was defined as disease demonstrating partial response to therapy, stable disease, disease progression during treatment, or positive findings on second-look surgery.
Cell Lines
Ovarian cancer cell lines were obtained from various sources, including the American Type Culture Collection (Manassas, VA), the European Collection of Cell Cultures (Salisbury, England), Kyoto University (Kyoto, Japan), the University of South Florida (Tampa, FL), or Duke University (Durham, NC). Cell lines were maintained in RPMI-1640 medium (Thermo Fisher Scientific, Carlsbad, CA) supplemented with 10% fetal bovine serum, 1% sodium pyruvate, 1% penicillin/streptomycin, and 1% nonessential amino acids. Cells were genotyped by short-tandem repeat profiling to confirm the tissue of origin and mycoplasma testing was performed every 6 months, in accordance with manufacturer protocol.
Cell Viability Assays
Drug activity was evaluated using high-throughput cell viability assays. A total of 2.5 × 103 cells/well were plated in 384 well plates using complete media with 10% fetal bovine serum and allowed to adhere overnight. After cell adherence, an increasing concentration of cisplatin or carboplatin was added to appropriate wells using an automated pipetting station. Four replicate wells were used for each drug concentration and vehicle controls. Initially, drug dilutions consisted of 1.5-fold serial dilutions for a maximum concentration of 100 μM. The cells were incubated with both drugs for 72 hours and 5 μL CellTiter-Blue reagent (Promega, Madison, WI) was added to each well. Fluorescence was read at 579 nm Ex/584 nm Em using a microplate reader (Synergy 4, Bio-Tek Instruments, Winooski, VT). Inhibitory concentrations of half maximal inhibitory concentration (IC50) were determined using a sigmoidal equilibrium model fit (XLfit 5.2, ID Business Solutions, Alameda, CA). The IC50 was defined as the concentration of drug required for a 50% reduction in growth/viability. IC50 levels of each cell line are summarized in Table 1.
Table 1. —
Results of the Cell Viability Assays
| Treatment | Time 1, h | Cell Line | Average of IC50, E-6 | SD of IC50, E-6 |
|---|---|---|---|---|
| Carboplatin | 72 | A2780S | 20.7 | 23.5 |
| HeyA8 | 295.9 | 94.0 | ||
| IGR-OV1 | 60.8 | 45.2 | ||
| M41 | 43.3 | 37.0 | ||
| M41CSR | 48.2 | 26.2 | ||
| OVCA420 | 165.8 | 33.0 | ||
| OVCA432 | 63.1 | 11.7 | ||
| OVCA433 | 270.2 | 137.6 | ||
| OVCAR5 | 89.0 | 25.0 | ||
| OVCAR8 | 60.7 | 10.6 | ||
| PEO1 | 22.9 | 16.9 | ||
| TOV-21G | 38.6 | 25.3 | ||
| TYK-nu | 9.7 | 4.8 | ||
| TYKnuCisR | 63.7 | 52.8 | ||
| Cisplatin | 72 | A2780S | 4.7 | 6.7 |
| HeyA8 | 33.1 | 45.0 | ||
| IGR-OV1 | 5.7 | 8.6 | ||
| M41 | 16.8 | 5.8 | ||
| M41CSR | 13.5 | 7.1 | ||
| OVCA420 | 13.7 | NA | ||
| OVCA432 | 1.4 | NA | ||
| OVCA433 | 8.8 | NA | ||
| OVCAR5 | 12.7 | 12.7 | ||
| OVCAR8 | 48.1 | 62.5 | ||
| PEO1 | 2.7 | 1.7 | ||
| TOV-21G | 23.1 | 26.7 | ||
| TYK-nu | 4.8 | 546.9a | ||
| TYKnuCisR | 17.7 | 11.4 |
E-9.
IC50 = half maximal inhibitory concentration, NA = not applicable, SD = standard deviation.
Immunohistochemistry
Immunostaining for ERBB4 was performed using a Ventana Discovery XT Automated System (Ventana Medical Systems, Tucson, AZ). Briefly, slides were deparaffinized on the automated system with a preparatory solution. A heat-induced antigen retrieval method was used in cell conditioning 1. A mouse monoclonal antibody that reacts to ERBB4, NB100–2662 (Novus, Littleton, CO), was used at a 1:25 concentration in Antibody Diluent (Dako, Carpenteria, CA) and incubated for 60 minutes. The Ventana OmniMap Anti-Mouse Secondary Antibody (Ventana Medical Systems) was incubated for 16 minutes and detected using a ChromoMap kit (Ventana Medical Systems). Slides were then counterstained with hematoxylin and eosin.
Immunohistochemistry Scoring by Light Microscopy
Samples were considered positive for ERBB4 expression if nuclear, cytoplasmic, or membranous reactivity of any intensity was observed. Levels of ERBB4 expression were scored utilizing a modified H score, wherein a manual assessment of intensity of staining (0 = no stain, 1+ = weak stain, 2+ = moderate stain, 3+ = strong stain) and percentage of positive cells (0%–100%) were taken into account to yield a composite expression score of 0 to 300. Expression scores were classified into low (0–100), intermediate (101–200), and high (201–300) categories. The samples were then independently reviewed and a final concordant decision was made for the discrepant interpretations.
Digital Microscopy
ERBB4-stained tissue microarray slides were scanned using an Aperio AT Turbo whole-slide scanner (Leica Biosystems, Nussloch, Germany) at ×20 magnification with 0.5 numerical aperture. Images were stored in an Aperio eSlide Manager database system (Leica Biosystems). The tissue microarray laboratory module of the database software was used to extract individual core images from the whole-slide images, and each core was labeled with the appropriate core identification number generated by the Tissue Core at Moffitt Cancer Center.
Each tissue microarray core image was analyzed using version 9 of the Aperio Positive Pixel Count algorithm (Leica Biosystems) at default settings to determine the extent of ERBB4 staining. The algorithm bins pixels into 4 categories (0, 1+, 2+, and 3+) based on the extent of diaminobenzidine staining. This result was used to calculate the H score ([3 × percentage of 3+ pixels] + [2 × percentage of 2+ pixels] + [1 × percentage of 1+ pixels]) for each individual core.
Each core image was visually checked to determine if the core was missing or damaged. Data from missing or damaged cores were not considered in the final result.
Statistical Analyses
An unpaired t test was performed to detect the difference of ERBB4 expression among tumor tissue, control tissue, and ovarian cancer cell line samples. Separately, ERBB4 expression in CR, IR, and disease stage was compared using unpaired t tests. Chi-square tests were performed on low (0–100), intermediate (101–200), and high (201–300) ERBB4 expression for tumor and control tissues as well as the CR and IR samples. Multivariate survival analyses using a Cox proportional model and log-rank tests were performed on ERBB4, disease stage, and platinum response to determine whether ERBB4 independently affects rates of overall survival (OS).
Software R v3.2.2 (R Foundation for Statistical Computing, Vienna, Austria) and GraphPad (GraphPad Software, La Jolla, CA) were used for the data analyses. In addition, results obtained from digital microscopy were compared with the H scores given by the pathologist using Pearson correlation tests.
Statistical significance was defined as a P value of less than .05.
Results
In total, 74 ovarian serous carcinoma specimens (33 CRs and 41 IRs), 30 normal tissue specimens, and 16 ovarian cancer cell lines were evaluable. Specimens that showed nonspecific staining, fixation artifacts, or loss of material in deeper levels after processing were considered not evaluable. The mean age of study patients was 61 years.
ERBB4 expression was not correlated with age when the cut-off was set at 65 years. The pathological stage was documented in 62 cases. In total, 46 of 62 cases had stage III disease at the time of diagnosis and 16 cases had stage IV disease. The mean rates of ERBB4 expression were 199.5 for stage IV disease and 170.9 for stage III disease. Although the difference was apparent (P = .08), it did not reach significance. Most cases were high-grade ovarian serous carcinoma (n = 71); few were low-grade cancers (n = 3).
ERBB4 expression was predominantly observed in the cytoplasm of tumor cells, and it was membranous and cytoplasmic in the ovarian cancer cell lines. Statistical analyses were not performed in the ovarian cancer cell lines due to their small sample size (n = 16). The mean scores of ERBB4 were significantly different for tumor tissue compared with control tissue compared with ovarian cancer cell lines.
The highest level of expression occurred in the ovarian cancer cell lines (P < .001; Fig 1). Expression at H score levels of 100 or higher was observed in 81% of cancer specimens. Expression of ERBB4 was significantly increased in the ovarian serous carcinoma specimens compared with control tissue. High-level ERBB4 expression (201–300) was observed in 37% of ovarian serous carcinoma specimens compared with 10% of control tissue (P = .013). Low-level expression (0–100) was observed in 53% of control tissue compared with 19% of tumor specimens (P = .001). Mean ERBB4 expression levels were 91.43, 173.64, and 227.19 for low-, intermediate-, and high-level expression groups, respectively (Fig 2).
Fig 1. —
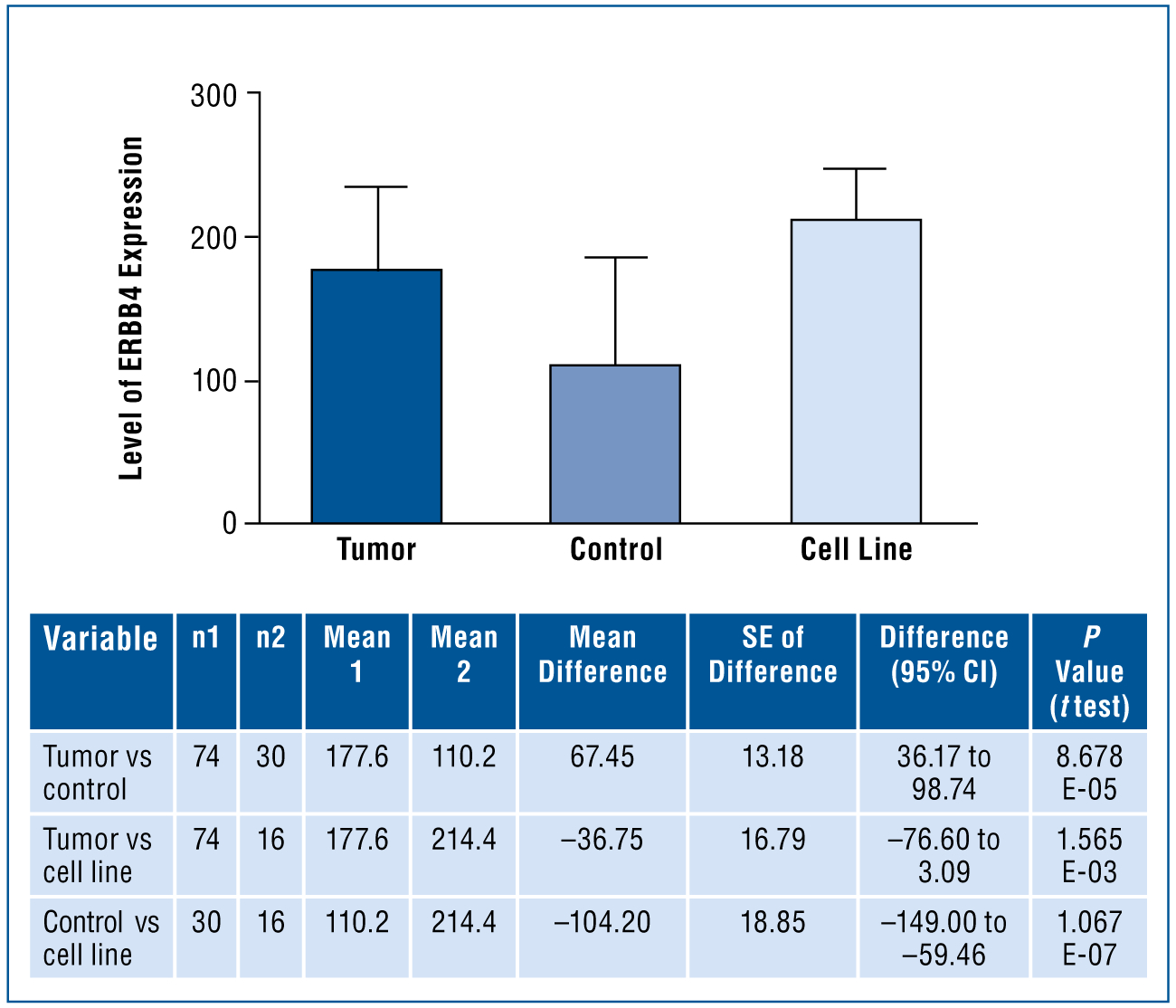
Mean level of ERBB4 expression in normal controls, tumor tissue, and ovarian cancer cell lines. Analysis of variance was statistically significant (P < .001). The mean scores of ERBB4 are significantly different for tumors vs controls vs cell lines, with cell lines greater than tumors and greater than controls.
CI = confidence interval, ERBB4 = erb-b2 receptor tyrosine kinase 4, SE = standard error.
Fig 2A–H. —
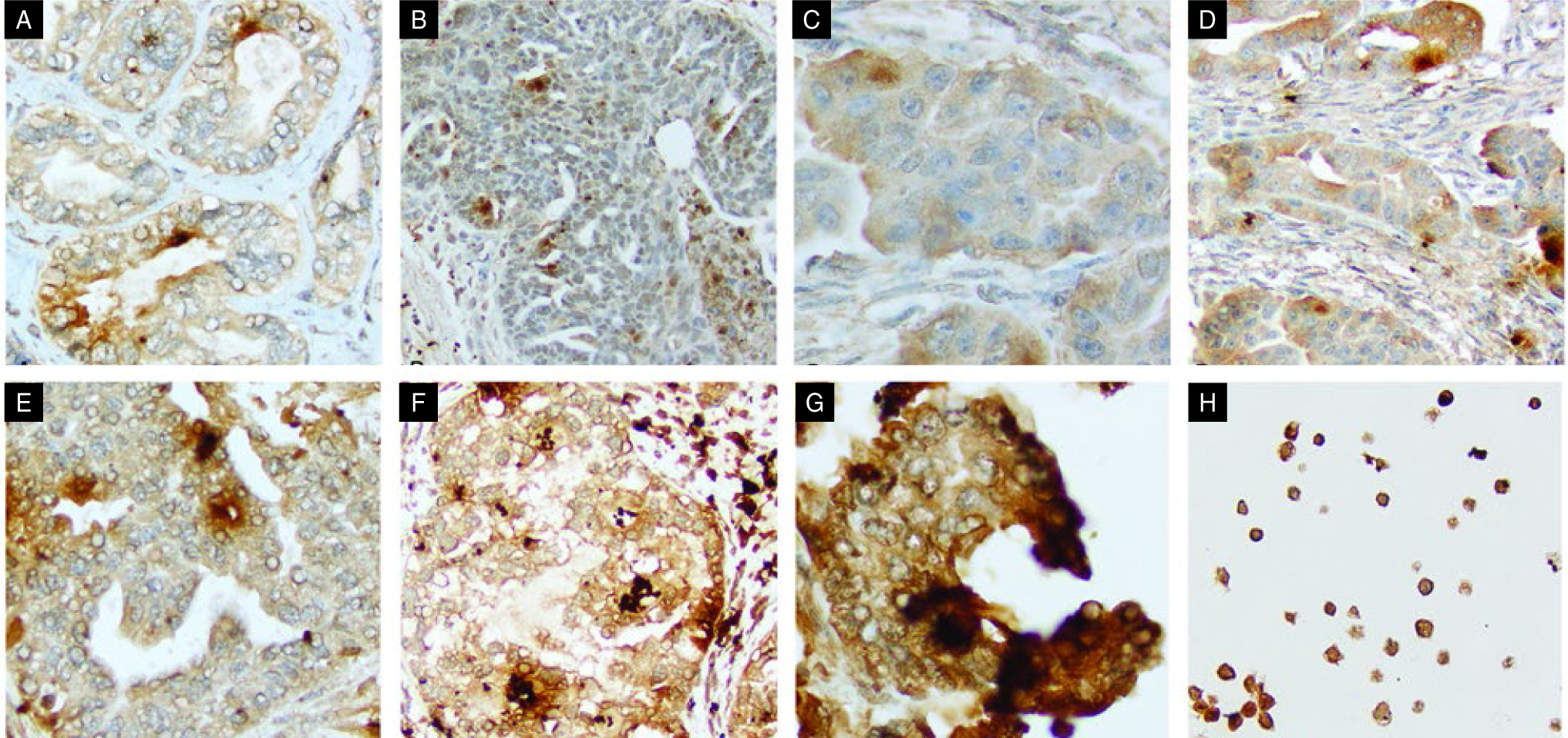
ERBB4 expression by immunohistochemistry. Shown are (A) rare membranous, (B) nuclear staining, (C–G) increasing H scores from less than 50 to 300 for particular fields, and (H) ERBB4 staining in cells lines.
ERBB4 = erb-b2 receptor tyrosine kinase 4.
Expression of ERBB4 was inversely correlated with sensitivity to chemotherapy (P = .033; Fig 3). Within the ovarian serous carcinoma cohort, high levels of ERBB4 expression were observed in 21% of CR specimens compared with 49% of IR specimens. Low levels of ERBB4 expression were observed in 27% of CR specimens and 12% of IR specimens (P = .178; Table 2).
Fig 3. —
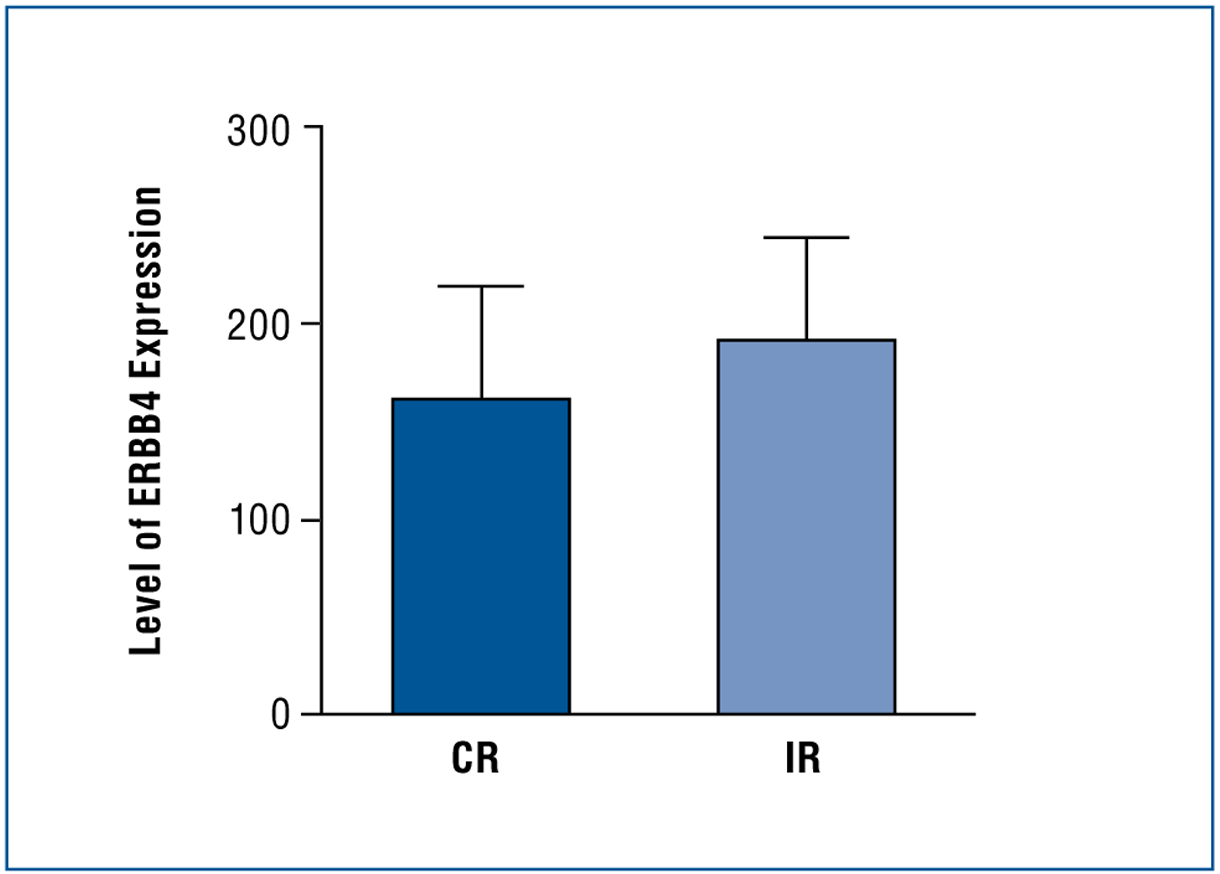
Using an unpaired t test with Welch correction, ERBB4 expression was higher patients with an IR than CR to platinum-based therapy (P = .033).
CR = complete response, ERBB4 = erb-b2 receptor tyrosine kinase 4, IR = incomplete response.
Table 2. —
Multivariate Survival Analysis Using a Cox Proportional Hazard Model
| Variable | Coefficient | HR | SE (Coefficient) | Lower 95% | Upper 95% | Z score | P value |
|---|---|---|---|---|---|---|---|
| IR | 2.4029 | 11.0550 | 0.4299 | 4.7604 | 25.673 | 5.590 | 2.27 E-08 |
| Stage IV | 0.3268 | 1.3865 | 0.3092 | 0.7563 | 2.542 | 1.057 | .2906 |
| ERBB4 | |||||||
| 101–200 | 0.8516 | 2.3434 | 0.4127 | 1.0436 | 5.262 | 2.063 | .0390 |
| 201–300 | 1.0985 | 2.9995 | 0.4230 | 1.3091 | 6.873 | 2.597 | .0094 |
When treating ERBB4 as a categorical variable (0–100, 101–200, or 201–300), its presence significantly affected OS.
ERBB4 = erb-b2 receptor tyrosine kinase 4, HR = hazard ratio, IR = incomplete response, OS = overall survival, SE = standard error.
In multivariate analysis using a Cox proportional hazard model when therapeutic response, disease stage, and ERBB4 expression were used as parameters, a significant difference in OS was observed between high and low levels of ERBB4 expression (P = .009) and intermediate and low levels of ERBB4 expression (P = .039). The difference was also observed when ERBB4 was used as a continuous variable (P = .071), although it did not reach statistical significance.
Among ovarian serous carcinoma specimens, OS was inversely correlated with ERBB4 expression levels. Median rates of OS were observed to be 18 months (95% confidence interval [CI]: 12–23) for high-level expressing tumors, 22 months (95% CI: 16–14) for intermediate-level expressing tumors, and 58 months (95% CI: 30–100) for low-level expressing tumors. Log-rank tests were performed in 3 different settings, including ERBB4 expression vs OS, CR vs OS, and ERBB4 expression and CR vs OS. When ERBB4 expression was used as a categorical variable, OS was significantly different for the high-level group (H score: 200–300) compared with the low-level group (H score: 0–100) in univariate analysis (P = .020; Fig 4). However, a significant OS difference was observed in log-rank analyses among the CR group without ERBB4 expression. In addition, no survival difference was observed among low- and high-level ERBB4-expressing tumors within the CR and IR groups.
Fig 4. —
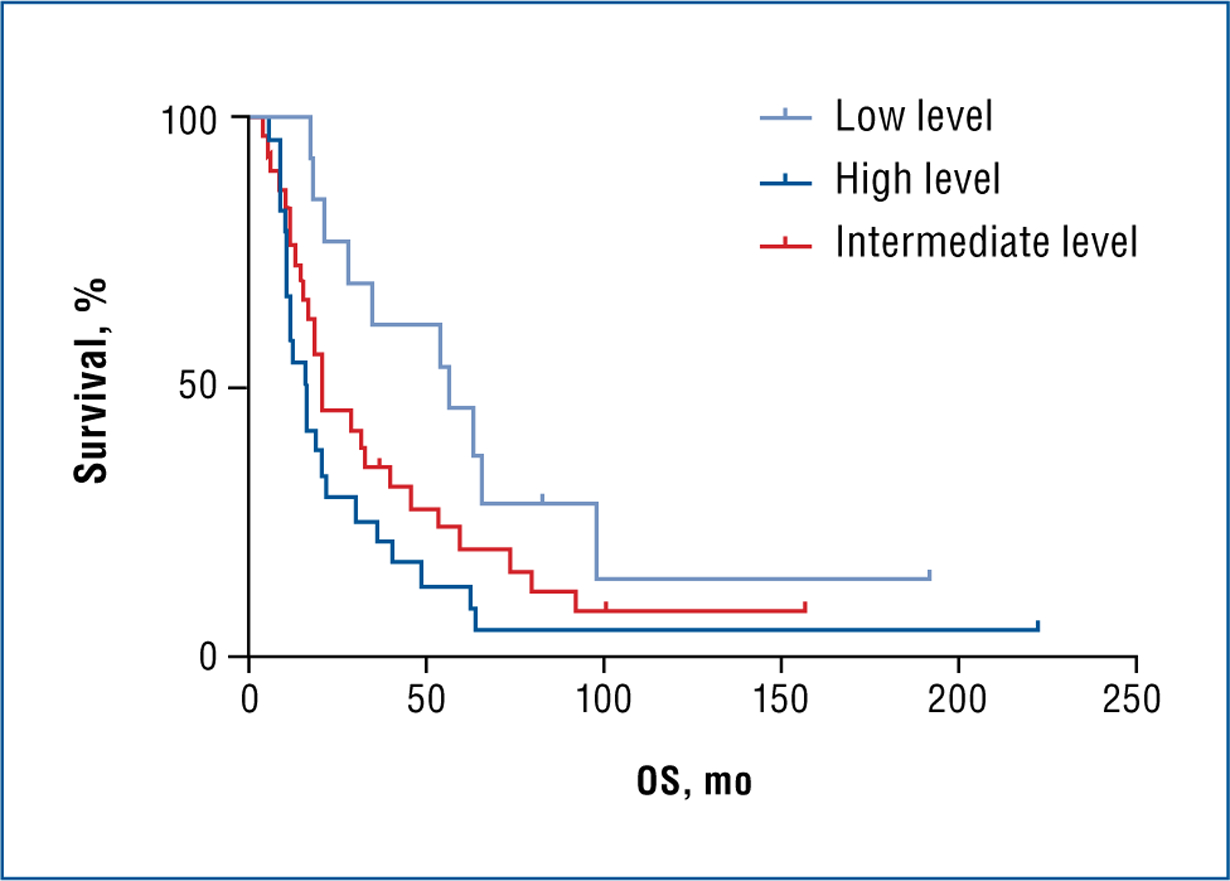
Log-rank test results illustrating ERBB4 expression and OS. Probability values were as follows: all 3 levels, P = .020; low vs intermediate, P = .080; low vs high, P = .004; and intermediate vs high, P = .201.
ERBB4 = erb-b2 receptor tyrosine kinase 4, OS = overall survival.
H scores obtained by light microscopy were correlated with that of analytic microscopy. By digital microscopy, the mean score of ERBB4 was significantly higher in the IR group (190.6) compared with the CR group (161.5; P = .033). The IR group had significantly worse rates of OS compared with the CR group when ERBB4 expression was used as either a categorical or continuous variable in the multivariate survival analysis using a Cox proportional hazard model.
Discussion
The role of ERBB4 in cancer biology is controversial.18–21 An explanation for conflicting observations may be that different isoforms and the subcellular localizations of ERBB4 differ in function. Analyzing functionally dissimilar isoforms can complicate the cancer biology of ERBB4.14,22 ERBB4 expression was observed to be an adverse prognostic factor in some studies but a favorable factor in others, even when studying the same tumor type.18,23,24 In addition, some researchers have not distinguished between the membranous, cytoplasmic, and nuclear expression of ERBB4 in evaluating clinical outcomes.25 In the present study, ERBB4 expression was predominantly localized to the cytoplasm in all ovarian serous carcinoma samples. Observed nuclear positivity within the tumor tissue was rare. Therefore, we did not evaluate ERBB4 expression in separate compartments of the tumor cells. All H scores represent cytoplasmic staining in clinical samples.
In vitro data describing the role of ERBB4 in the regulation of ovarian cancer cell growth is also contradictory, because both growth-promoting and growth-suppressing activities have been reported.26,27 The role of ERBB4 was analyzed in the development of chemoresistance in ovarian and other cancer cell lines, and reportedly high levels of ERBB4 expression were associated with cisplatin resistance within ovarian cancer cell lines.25 In addition, the insertion and overexpression of 4ICD has been shown to suppress the cell cycle and proliferation in neuroblastoma cell lines.28 ERBB4 functioned as a cell-cycle suppressor, maintaining resistance to cellular stress, including chemotherapeutic agents. All ovarian cancer cell lines in our cohort showed the highest level of ERBB4 expression, which is comparable with the literature.25 However, different drug inhibitory levels (IC50 levels) and the absence of an in vivo tumor microenvironment are limitations faced when comparing results from ovarian cancer cell lines and clinical samples.
In prior studies, overexpression of ERBB4 was documented in ovarian cancer without any clear clinical associations.25,29 In 1 study, approximately 90% of ovarian cancers — predominantly of the serous sub-type — expressed ERBB4.30 In our cohort, all clinical ovarian serous carcinoma samples except 1 expressed ERBB4 in varying H scores. We also observed a significant difference in ERBB4 expression between control tissue and ovarian serous carcinoma cases. In all clinical samples, ERBB4 expression was significantly higher in the IR group compared with the CR group. Expression of ERBB4 was inversely correlated to response to therapy. Our results can be disputed due to semiquantitative and subjective classifications of scoring. However, H scores obtained by light microscopy were supported by our analytical microscopy findings. ERBB4 expression was also higher in tumor samples compared with normal control tissue and in IR vs CR samples by analytic microscopy.
High ERBB4 expression was also correlated with a low mitosis/karyorrhexis index, which is an indicator of mitosis activity, and also in clinical high-risk groups, metastasis, and poor rates of survival.31 These results may lead some to conclude that tumors growing more slowly are more difficult to treat and ERBB4 overexpression may reduce proliferation, thus rendering a refractory phenotype. It may also be the explanation for the high level of ERBB4 expression observed in our tumor samples obtained from patients with IR to therapy. Among ovarian serous carcinoma specimens, OS was inversely correlated with ERBB4 expression levels when ERBB4 was used as a categorical variable. Analytic microscopy results were also concordant with results seen on light microscopy. The IR group had significantly worse rates of OS compared with the CR group when ERBB4 expression was used as either a categorical or continuous variable in the multivariate survival analysis.
To our knowledge, this is the first report in the literature showing the relationship between platinum resistance and ERBB4 expression in patients with ovarian serous carcinoma and adverse prognoses.
It is possible that the difference observed in the survival rates between the CR and IR groups is independent of ERBB4 expression. The significance levels did not persist within the CR and IR groups between low and high levels of ERBB4-expressing tumors. Lack of early-stage disease (stages I/II) and a small number of low-grade tumors are other limitations of our cohort. However, both early-stage disease and low-grade ovarian serous carcinoma make up a small percentage of clinical cases. Low-grade ovarian serous carcinoma is also known to be more resistant to chemotherapy than high-grade disease.32 The retrospective nature of our data collection for disease outcomes without discriminating between short-term and long-term response to therapy is another shortcoming of our data set.
Conclusions
Considering the limited therapeutic options for patients with ovarian carcinoma whose disease is resistant to platinum-based chemotherapy, a need exists to explore second-line treatment modalities. The addition of pan-tyrosine kinase blockers, such as afatinib, to cisplatin has significantly enhanced platinum-based therapies in head and neck cancers.33 Thus, it may be an option in platinum-resistant ovarian carcinoma. Our preliminary results require further study to validate the prognostic and potential treatment in a larger cohort of ovarian serous carcinoma samples.
Acknowledgments
This work is supported in part by the Tissue Core Facility at the H. Lee Moffitt Cancer Center & Research Institute (P30-CA076292).
Footnotes
No significant relationships exist between the authors and the companies/organizations whose products or services may be referenced in this article.
References
- 1.American Cancer Society. Cancer Facts & Figures. Atlanta, GA: American Cancer Society; 2016. http://www.cancer.org/acs/groups/content/@research/documents/document/acspc-047079.pdf. Accessed January 18, 2017. [Google Scholar]
- 2.Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet. 2014;384(9951):1376–1388. [DOI] [PubMed] [Google Scholar]
- 3.Bast RC Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9(6):415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaughan S, Coward JI, Bast RC Jr, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11(10):719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351(24): 2519–2529. [DOI] [PubMed] [Google Scholar]
- 6.Staropoli N, Ciliberto D, Chiellino S, et al. Is ovarian cancer a targetable disease? A systematic review and meta-analysis and genomic data investigation. Oncotarget. 2016;7(50):82741–82756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Fu S. An overview of tyrosine kinase inhibitors for the treatment of epithelial ovarian cancer. Expert Opin Investig Drugs. 2016;25(1):15–30. [DOI] [PubMed] [Google Scholar]
- 8.Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12(8):553–563. [DOI] [PubMed] [Google Scholar]
- 9.Reyes HD, Thiel KW, Carlson MJ, et al. Comprehensive profiling of EGFR/HER receptors for personalized treatment of gynecologic cancers. Mol Diagn Ther. 2014;18(2):137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5(5):341–354. [DOI] [PubMed] [Google Scholar]
- 11.Muraoka-Cook RS, Sandahl MA, Strunk KE, et al. ErbB4 splice variants Cyt1 and Cyt2 differ by 16 amino acids and exert opposing effects on the mammary epithelium in vivo. Mol Cell Biol. 2009;29(18):4935–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veikkolainen V, Vaparanta K, Halkilahti K, et al. Function of ERBB4 is determined by alternative splicing. Cell Cycle. 2011;10(16):2647–2657. [DOI] [PubMed] [Google Scholar]
- 13.Rokicki J, Das PM, Giltnane JM, et al. The ERalpha coactivator, HER4/4ICD, regulates progesterone receptor expression in normal and malignant breast epithelium. Mol Cancer. 2010;9:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundvall M, Veikkolainen V, Kurppa K, et al. Cell death or survival promoted by alternative isoforms of ErbB4. Mol Biol Cell. 2010;21(23):4275–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang HG, Jenabi JM, Zhang J, et al. E-cadherin cell-cell adhesion in Ewing tumor cells mediates suppression of anoikis through activation of the ErbB4 tyrosine kinase. Cancer Res. 2007;67(7):3094–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Wang W, Wang C, et al. Characterization of EGFR family gene aberrations in cholangiocarcinoma. Oncol Rep. 2014;32(2):700–708. [DOI] [PubMed] [Google Scholar]
- 17.Zhao WY, Zhuang C, Xu J, et al. HER4 is a novel prognostic biomarker in gastrointestinal stromal tumor specifically originated from stomach. Am J Cancer Res. 2014;4(6):838–849. [PMC free article] [PubMed] [Google Scholar]
- 18.Hollmen M, Maatta JA, Bald L, et al. Suppression of breast cancer cell growth by a monoclonal antibody targeting cleavable ErbB4 isoforms. Oncogene. 2009;28(10):1309–1319. [DOI] [PubMed] [Google Scholar]
- 19.Sartor CI, Zhou H, Kozlowska E, et al. Her4 mediates ligand-dependent antiproliferative and differentiation responses in human breast cancer cells. Mol Cell Biol. 2001;21(13):4265–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundvall M, Iljin K, Kilpinen S, et al. Role of ErbB4 in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13(2):259–268. [DOI] [PubMed] [Google Scholar]
- 21.Thor AD, Edgerton SM, Jones FE. Subcellular localization of the HER4 intracellular domain, 4ICD, identifies distinct prognostic outcomes for breast cancer patients. Am J Pathol. 2009;175(5):1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mill CP, Gettinger KL, Riese DJ II. Ligand stimulation of ErbB4 and a constitutively-active ErbB4 mutant result in different biological responses in human pancreatic tumor cell lines. Exp Cell Res. 2011;317(4):392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naresh A, Long W, Vidal GA, et al. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006;66(12):6412–6420. [DOI] [PubMed] [Google Scholar]
- 24.Vidal GA, Clark DE, Marrero L, et al. A constitutively active ERBB4/HER4 allele with enhanced transcriptional coactivation and cell-killing activities. Oncogene. 2007;26(3):462–466. [DOI] [PubMed] [Google Scholar]
- 25.Gilmour LM, Macleod KG, McCaig A, et al. Expression of erbB-4/HER-4 growth factor receptor isoforms in ovarian cancer. Cancer Res. 2001;61(5):2169–2176. [PubMed] [Google Scholar]
- 26.Campiglio M, Ali S, Knyazev PG, et al. Characteristics of EGFR family-mediated HRG signals in human ovarian cancer. J Cell Biochem. 1999;73(4):522–532. [PubMed] [Google Scholar]
- 27.Gilmour LM, Macleod KG, McCaig A, et al. Neuregulin expression, function, and signaling in human ovarian cancer cells. Clin Cancer Res. 2002;8(12):3933–3942. [PubMed] [Google Scholar]
- 28.Hua Y, Gorshkov K, Yang Y, et al. Slow down to stay alive: HER4 protects against cellular stress and confers chemoresistance in neuroblastoma. Cancer. 2012;118(20):5140–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pejovic T, Pande NT, Mori M, et al. Expression profiling of the ovarian surface kinome reveals candidate genes for early neoplastic changes. Transl Oncol. 2009;2(4):341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paatero I, Lassus H, Junttila TT, et al. CYT-1 isoform of ErbB4 is an independent prognostic factor in serous ovarian cancer and selectively promotes ovarian cancer cell growth in vitro. Gynecol Oncol. 2013;129(1):179–187. [DOI] [PubMed] [Google Scholar]
- 31.Izycka-Swieszewska E, Wozniak A, Drozynska E, et al. Expression and significance of HER family receptors in neuroblastic tumors. Clin Exp Metastasis. 2011;28(3):271–282. [DOI] [PubMed] [Google Scholar]
- 32.Gershenson DM. Low-grade serous carcinoma of the ovary or peritoneum. Ann Oncol. 2016;(27 suppl 1):i45–i49. [DOI] [PubMed] [Google Scholar]
- 33.Brands RC, Muller-Richter UD, De Donno F, et al. Co-treatment of wildtype EGFR head and neck cancer cell lines with afatinib and cisplatin. Mol Med Rep. 2016;13(3):2338–2344. [DOI] [PubMed] [Google Scholar]


