Abstract
Purpose
Oral squamous cell carcinoma (OSCC) is a challenging disease to treat. Up to 50% of OSCC patients with advanced disease develop recurrences. Elucidation of key molecular mechanisms underlying OSCC development may provide opportunities to target specific genes and, thus, to improve patient survival. In this study, we examined the expression and functional role of interferon transmembrane protein 3 (IFITM3) in OSCC development.
Methods
The expression of IFITM3 in OSCC and normal oral mucosal tissues was assessed by qRT-PCR and immunohistochemistry. The role of IFITM3 in driving OSCC cell proliferation and survival was examined using siRNA-mediated gene knockdown, and the role of IFITM3 in driving cell cycle regulators was examined using Western blotting.
Results
We found that IFITM3 is overexpressed in more than 79% of primary OSCCs. We also found that IFITM3 knockdown led to impaired OSCC cell growth through inhibition of cell proliferation, induction of cell cycle arrest, senescence and apoptosis. In addition, we found that IFITM3 knockdown led to reduced expressions of CCND1 and CDK4 and reduced RB phosphorylation, leading to inhibition of OSCC cell growth. This information may be instrumental for the design of novel targeted therapeutic strategies.
Conclusions
From our data we conclude that IFITM3 is overexpressed in OSCC and may regulate the CCND1-CDK4/6-pRB axis to mediate OSCC cell growth.
Keywords: Oral squamous cell carcinoma, IFITM3, Proliferation, CCND1, CDK4
1. Introduction
The interferon-inducible transmembrane (IFITM) proteins belong to the small interferon-stimulated family with molecular masses ranging from 10 to 20 kDa [1]. The members of this family include IFITM1, IFITM2, IFITM3, IFITM5 and IFITM10 [1, 2]. The IFITM proteins are well known for their role in modulating the interferon-mediated innate immune system to inhibit the invasion and replication of pathogenic viruses [3, 4]. In addition to immune cell regulation [5, 6], IFITM proteins are known to be involved in germ cell homing and maturation during embryonic development [7–10]. More recently, emerging data have revealed an oncogenic role of the IFITM proteins in cancer development. Overexpression of IFITM1 and IFITM3 has, for example, been observed in glioma[11, 12], head and neck cancer [13], lung cancer [14] and breast cancer [15, 16]. Notably, overexpression of IFITM1 and IFITM3 have repeatedly been reported to be associated with the progression of gastrointestinal cancers [17–27], and the expression of these proteins could also be readily detected in premalignant lesions such as ulcerative colitis and inflammatory bowel disease [28–30]. In head and neck cancers, IFITM1, 2 and 3 have been found to be up-regulated in a major proportion of the cases [13, 31–33]. Specifically, IFITM1 has been found to be overexpressed in the invasive front of head and neck tumours and to activate matrix metalloproteinases MMP12 and MMP13, resulting in degradation of the extracellular matrix and subsequent tumour invasion [13]. As yet, however, the role of IFITM2 and IFITM3 in driving head and neck cancer development has remained largely unexplored. Previously, we reported that IFITM3 is overexpressed in oral squamous cell carcinoma (OSCC) compared to normal oral mucosa, regardless of patient risk habits [33]. Here, we sought to investigate the functional impact of IFITM3 on OSCC development and progression in further detail. Interestingly, we found that siRNA-mediated IFITM3 expression knockdown markedly inhibited OSCC cell proliferation through apoptosis and cellular senescence induction. In addition, we found that inhibition of OSCC cell proliferation through IFITM3 knockdown is mediated by reductions in CCND1 and CDK4 expression, which are key cell cycle regulators. Specifically, dysregulation of CCND1 is a frequent event in OSCC and often leads to disruption of the cell cycle [34]. Taken together, our data suggest that IFITM3 may confer a survival advantage to OSCC cells through the regulation of CCND1 and CDK4. This information provides further insight into the molecular perturbations underlying OSCC.
2. Materials and methods
2.1. Human specimens
Forty-seven fresh frozen OSCC surgical tissue samples and 18 normal oral mucosa (NOM) tissue samples obtained from gingiva of healthy individuals who had undergone wisdom tooth removal were used in this study. The tissue samples were collected after informed consent was obtained. In addition, formalin-fixed paraffin-embedded (FFPE) tissue samples, including 43 OSCC, 10 fibroepithelial polyps (FEP) and 13 NOM were used. The diagnoses of all specimens were histology-confirmed by an oral pathologist (RBZ) certified by the Malaysian National Specialists Register. The socio-demographic information of the patients included here were obtained from the Malaysian Oral Cancer Database and Tissue Bank System (MOCDTBS) [35] and is summarized in Table 1. This study was approved by the ethical review board at the Faculty of Dentistry, University of Malaya (Ethical approval code: DFOP0703/0017).
Table 1.
Demographic data of 43 OSCC cases used for IFITM3 IHC analysis
| Variable | OSCC |
||
|---|---|---|---|
| n | % | ||
| Gender | Male | 12 | 27.9 |
| Female | 31 | 72.1 | |
| Age (year) | Range: 38–80 | ||
| Median: 62 | |||
| Primary site | Buccal mucosa & gum | 37 | 86.0 |
| Tongue & floor of mouth | 6 | 14.0 | |
| Ethnicity | Malay | 5 | 11.6 |
| Chinese | 4 | 9.3 | |
| Indian | 33 | 76.7 | |
| Indigenous population | 1 | 2.3 | |
| Risk habit | No habit | 5 | 11.6 |
| Betel quid | 23 | 53.5 | |
| Smoking | 4 | 9.3 | |
| > 1 habit | 11 | 25.6 | |
2.2. qRT-PCR
Fresh frozen tissues were embedded in OCT (Leica, Heidelberger, Germany), sectioned, stained with haematoxylin and histopathologically confirmed to contain at least 70% tumour cells (OSCC specimens) or normal epithelial cells (for NOM). Total RNA was isolated from tissue cryosections collected in microfuge tubes (~500 μm) using a RNeasy Micro kit (Qiagen, Germany) according to the manufacturer’s instructions. The quality and concentration of RNA were determined using a RNA 6000 Nano kit on an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). cDNAwas synthesized from 2 μg total RNA using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems, CA, USA) in a total volume of 100 μl Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using a standard SYBR Green protocol in conjunction with an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems, Germany), as described previously [33]. Briefly, reactions were carried out by performing pre-incubation for 10 min at 95 °C, followed by 40 amplification cycles at 95 °C for 15 s and 60 °C for 1 min. The primer sequences used were IFITM3 (sense: 5’-GACCATTCTGCTCATCGTCATC-3′ and antisense: 5’-AGCCAGACCCTCCCAATGTT-3′), IFITM1 (sense: 5’-TCGCCTACTCCGTGAAGTCT-3′ and antisense: 5’-TGTCACAGAGCCGAATACCA-3′), IFITM2 (sense: 5’-CGATAGATCAGGAGGCATCA-3′ and antisense: 5′- GATAAAGGGCTGATGCAGGA-3′) and GAPDH (sense: 5’-GAAGGTGAAGGTCGGAGTC-3′, antisense: 5′-GAAGATGGTGATGGGATTTC-3′. Relative quantification was performed using the comparative Ct method as described previously [36]. Briefly, the expression of IFITM3 in each sample was normalized to the expression of GAPDH and, subsequently, fold-changes of IFITM3 mRNA in the tumour tissues were compared to those of unmatched NOM tissues. A fold-change value ≥ 2 was considered as overexpression.
2.3. Immunohistochemistry
Immunohistochemistry (IHC) was carried out to examine the expression of IFITM3 and Ki-67 in both OSCC and NOM FFPE samples using a method described previously [37]. Briefly, FFPE tissue sections were incubated at 65 °C for 10 min, rehydrated in decreasing grades of ethanol (100%, 95%, 70%) and finally immersed in water as described previously [36]. Next, the tissue sections were subjected to antigen retrieval by boiling in citrate buffer (pH 6.0) for 10 min and endogenous enzyme blocking was performed by incubating the tissue sections with peroxidase blocking reagent (Dako, Glostrup, Denmark) at room temperature for 10 min. The tissue sections were subsequently incubated with primary antibodies for 90 min in a humidified chamber at room temperature. A polyclonal antibody directed against IFITM3 (ab15592; Abcam, Cambridge, UK) was used at 1:75 dilution. Detection was carried out using a Dako REAL™ ENVISION™ Detection System, Peroxidase/DAB+, Rabbit/Mouse kit (Dako, Glostrup, Denmark). Staining was developed using 3, 3′-diaminobenzidine (DAB) and evaluated microscopically. Tissue sections were counterstained with haematoxylin for 10 s, dehydrated in increasing grades of ethanol (70%, 95% and 100%) and finally immersed in histolene before being mounted with DPX mountant (Fluka, Steinheim, Germany). The immunoreactivity of epithelial cells (cancer and normal) was evaluated independently by a pathologist (RBZ) and scored as described by others: 0 = negative, 1 = weak positive, 2 = moderate positive and 3 = strong positive [22]. The receiver operating characteristic (ROC) curve method was used to identify the best cut-off points in scoring the expression of IFITM3 for specificity and sensitivity, and following this, intensity scores of 1 or 2 were grouped as low expression, and 3 or 4 as high expression.
2.4. In silico gene expression analysis
Using The Cancer Genome Altas (TCGA) database for head and neck squamous cell carcinoma (HNSCC), we identified 43 matched tumour-normal sample pairs. The gene expression profiles of the three IFITM family members (IFITM1, IFITM2 and IFITM3) in the form of Fragments Per Kilobase of transcript per Million mapped reads Upper Quartile (FPKM-UQ) were extracted from the National Cancer Institute Genomic Data Commons (GDC) (https://portal.gdc.cancer.gov/). The relative gene expression levels of these three IFITM members were normalized against TBP (TATA-box binding protein) expression in each tumour-normal sample pair, respectively, and presented as log2 scale.
2.5. Cell culture
Cell lines ORL-48, ORL-115, ORL-150, ORL-153, ORL-156 and ORL-204, derived from human OSCC primary tumours, were maintained in DMEM-F12 medium (Hyclone, Utah, USA) supplemented with 10% Fetal Bovine Serum (FBS; Gibco, Auckland, NZ), 1.2 g/l sodium bicarbonate (Merck, Darmstadt, Germany) and 2 μg/ml hydrocortisone (Sigma, MO, USA) [38]. The cell lines were grown in a humidified CO2 incubator at 37 °C. The cell lines were authenticated using a Geneprint 10 system kit (Promega, Madison, WI), and the short tandem repeat (STR) profiles of the cell lines were compared to those of the tumour and blood samples of matched donors genotyped previously [39].
2.6. IFITM3 knockdown by siRNA
Synthetic oligonucleotides ON-TARGET plus siRNA AUGGAUAGAUCAGGAGGCA (si18, J-014116-18) and TGCTGATCTTCCAGGCCTA (si19, J-014116-19) targeting human IFITM3, and ON-TARGET plus Non-Targeting (NT) siRNA UGGUUUACAUGUCGACUAA were purchased from Thermo Scientific (MA, USA). Two ORL cell lines (ORL-150 and ORL-204) that were found to overexpress IFITM3 were used for the knock-down experiments (Supplementary Fig. 1a). Cells were seeded at a density of 2 × 105/well in a 6-well plate and incubated for 24 h prior to transfection. Cells were transfected with 50 nM siRNA using cationic lipid DharmaFECT (Thermo Scientific, MA, USA) and incubated for an additional 48 h prior to harvesting for analysis. The level of IFITM3 expression after siRNA knockdown was determined by qRT-PCR and Western blotting.
2.7. Protein extraction and Western blotting
48 h post-siRNA transfection, cells were trypsinized and reseeded at a density of 2 × 105/well in a 6-well plate. Next, the cells were harvested at days 2, 4 and 6, and lysed on ice using a lysis buffer (0.5% NaDOC, 0.1% SDS, 25 mM HEPES pH 7.5, 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1% Triton X-100, 0.5 mM DTT, 20 mM β-glycerophosphate, 0 .1mM Na3VO4) supplemented with 1x HALT Protease and Phosphatase Inhibitor Cocktail (Pierce Biotechnology, IL, USA). The resulting cell lysates were collected after centrifugation at 14000 rpm for 5 min at 4 °C and protein concentrations were determined by a Bradford assay using Coomassie Plus reagent (Pierce Biotechnology, IL, USA). Next, the absorbance was measured at 595 nm wavelength in a microplate reader (Thermo Scientific, MO, USA) and 50 μg protein extracts were resolved in 12% SDS-PAGE gels and transferred to Immobilon-P membranes (Millipore, MA, USA). The resulting blots were blocked with 5% skimmed milk or bovine serum albumin (TBS with 0.1% Tween-20) for 1 h and next incubated with the following primary antibodies overnight at 4 °C: anti-IFITM3 (M01), clone 4C8-1B10 (H00010410-M01, Abnova, Taipei, Taiwan, 1:1000), anti-TP53 (sc126; Santa Cruz, CA, US, 1:1000), anti-p16 (sc56330 Santa Cruz, CA, US, 1:200), anti-p21 Waf1/Cip1 (12D1) (Cell Signaling Technology, USA, 1:1000), anti-CCND 1 [SP4] (ab16663; Abcam, 1:500), anti-CCNE1 (HE12) (Cell Signaling Technology, USA, 1:1000), anti-CDK2 (78B2) (Cell Signaling Technology, USA, 1:1000), anti-CDK4 (DCS-35) (sc-23,896; Santa Cruz, CA, US, 1:1000), anti-CDK6 (D4S8S) (Cell Signaling Technology, USA, 1:1000) and anti-α-tubulin (T9026; Sigma-Aldrich, MO, USA, 1:1000). Next, the blots were washed (3x for 5 min each) in TBS with 0.1% Tween 20 (TBST) and probed with appropriate secondary antibodies conjugated with horseradish peroxidase (1:10000, Southern Biotech, AL, USA) for 1 h at RT. After washing (TBS-T), detection was performed using an Immobilon Western Chemiluminescent HRP substrate (Millipore, USA) in a FluorChem HD2 imager (Protein Simple, USA).
2.8. Cell proliferation assay
Cell proliferation assessment was conducted as described previously [36]. Briefly, 48 h post siRNA transfection, 2 × 104 cells/well were seeded into 6-well plates and cultured for up to 7 days and, during this period, cells were trypsinized and counted every 24 h using a CASY cell Counter (Innovartis AG, Reutlingen, Germany). Experiments were conducted in triplicate, and viable cell numbers were recorded. Mean cell counts were compared to the respective non-targeting (NT) siRNA counts across different time intervals. Population doubling times were calculated according to the formula: DT = T ln2/ln(Xe/Xb), whereby T is the time interval, Xb is the cell number at the beginning of the incubation time and Xe is the cell number at the end of incubation time.
2.9. Clonogenic assay
Clonogenic assays were performed as described previously [40]. Briefly, cells were seeded into a 6-well plate based on their seeding efficiency 48 h post-siRNA transfection. ORL-150 and ORL-204 were seeded at 800 cells/well and 200 cells/well, respectively, and grown for 12 days. Culture media were replaced every 3 days. At the end of the experiment, cells were fixed with 4% formaldehyde for 15 min and stained with 0.2% crystal violet for 10 min. Colonies consisting of more than 50 cells were counted and colony forming abilities were compared to those of cells transfected with NT siRNA.
2.10. Soft agar assay
Soft agar assays were performed as described previously with minor modifications [38]. A base agarose layer was prepared by mixing 1% low melting point agarose (w/v) (Sigma-Aldrich, Saint Louis, USA) with DMEM-F12 complete medium, and allowed to solidify in the 6-well plates. Next, a single cell suspension containing 1000 cells was prepared in DMEM-F12 complete medium, mixed with 0.5% low melting point agarose and added to the base agarose layer. The cells were incubated for 14 days in a CO2 incubator, and 1 ml medium was added on day 7. Finally, colonies ≥ 100 μm were counted in 10 random microscopic fields (200x objective).
2.11. Cell cycle analysis
The cell cycle status of the cells transfected with IFITM3 and NT siRNAs were determined using propidium iodide (PI) staining and flow cytometry. Briefly, cells were trypsinized at 48 h post-siRNA transfection, washed with cold PBS and fixed with 70% ethanol. Next, the fixed cells were harvested and resuspended in 0.5 ml PBS containing 50 μg/ml PI and 100 μg/ml RNase and incubated at 4 °C for 30 min. Stained cells were analysed on a flow cytometer (FACSCalibur, Becton Dickinson, Heidelberg, Germany).
2.12. Apoptosis assay
Cells were harvested at 48 h post-transfection with siRNA followed by washing (2x) with cold PBS and centrifugation at 1000 rpm for 5 min. Next, 1 × 105 cells were resuspended in 1x binding buffer (10 mM HEPES pH 7.4, 0.14 M NaCl, 2.5 mM CaCl2) containing 2.5 μl Annexin Vand 50 μg/ml PI, incubated in the dark for 15 min at room temperature and resuspended in an additional 400 μl 1x binding buffer. The populations of apoptotic cells were analysed using a flow cytometer (FACSCalibur, Becton Dickinson, Heidelberg, Germany).
2.13. Cellular senescence assay
The induction of cellular senescence was determined using a Senescence Detection Kit (Biovision, CA, USA) with minor modifications to the manufacturer’s instructions. 48 h post-siRNA transfection, cells were seeded at a density of 1000 cells/well in 6-well plates and incubated overnight. Next, the cells were washed with PBS, fixed with a fixative solution and incubated with a staining solution mix for 48 h at 37 °C. Finally, the cells were examined and counted under a microscope at 200x magnification in 4 random fields. The percentage of senescent cells was calculated as the proportion of blue cells against the total number of cells in each field.
2.14. Correlation co-expression analysis of genes interacting with IFITM3
We used the STRING v10 database (https://string-db.org) [41] to identify possible interactions between IFITM3 and CCND1. By setting the interaction score of the highest confidence (0.9), we identified 50 genes interacting with IFITM3 and CCND1. Next, we examined the expression of these 50 genes in a subset of OSCC cell lines enriched in cell cycle signalling [39]. Pearson correlation analysis was used to determine the co-expression of genes. Genes co-expressed with IFITM3 with a positive correlation score of > 0.5 were used for the construction of a gene co-expression network using Cytoscape version 3.2.1 (http://cytoscape.org) [42].
2.15. Statistical analysis
SPSS 16 (SPSS Inc., Chicago, IL, USA) software was used to assess the qRT-PCR and IHC results. IFITM3 mRNA levels in OSCC and NOM tissues were compared using the Mann-Whitney U test. The receiver operating characteristic (ROC) curve method was used to determine the cut-off point for IHC scoring of IFITM3 expression to differentiate tumours with a low and a high expression. Subsequently, IFITM3 protein expression in OSCC tumours and NOM tissues was compared using a Chi-square test. The Fisher-exact test was used to assess associations between IFITM3 protein expression in OSCCs with various clinicopathological parameters. GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA) was used to examine the results from in silico and in vitro functional assays. A two-tailed paired t-test was used to assess in silico differential gene expression of three IFITM members in matched tumour-normal tissue sample pairs. Phenotypic differences between IFITM3 knockdown and NT siRNA cells were analysed using a two-tailed independent t-test. A p value of < 0.05 was considered statistically significant.
3. Results
3.1. IFITM3 is overexpressed in OSCC
Previously, we reported that IFITM3 mRNA levels are up-regulated by more than 2-fold in OSCC [33]. Here, we validated these observations in an independent set of tissue samples and assessed whether high IFITM3 expression levels have an impact on tumour progression. To this end, IFITM3 mRNA levels were examined in 46 OSCC and 18 NOM tissues by qRT-PCR. We found that the IFITM3 mRNA levels were significantly up-regulated in OSCC tissues, with a mean 2.9 ± 0.46-fold increase, compared to NOM tissues (p = 0.003; Fig. 1a), where 21/46 (46%) of OSCC tissues exhibited a ≥ 2-fold increase, indicating overexpression of IFITM3 in this cancer type. Consistently, IHC analysis of 43 OSCC, 10 FEP and 13 NOM tissues revealed that the IFITM3 protein levels were elevated in the OSCC tissues (p < 0.001; Fig. 1b), where 34/43 (79.1%) of these tissues exhibited intensity scores of 2 or 3 (Fig. 1c i–ii). We found the IFITM3 expression to be low in the epithelial compartment of non-cancerous tissues including FEP and those from NOM, whereby 19/23 (82.6%) of the FEP and NOM tissues exhibited low IFITM3 levels (intensity score 0–1; Fig.1c iii–iv). While IFITM3 was found to be differentially expressed between OSCC and NOM, no association was observed between IFITM3 expression and any of the clinico-pathological OSCC parameters evaluated (Supplementary Table 1). Further analysis of a dataset in TCGA revealed that IFITM1 and IFITM3 are significantly overexpressed in HNSCC tumours compared to matched normal tissues (Supplementary Fig. 1).
Fig. 1. IFITM3 is overexpressed in OSCC but not in normal oral mucosa (NOM).
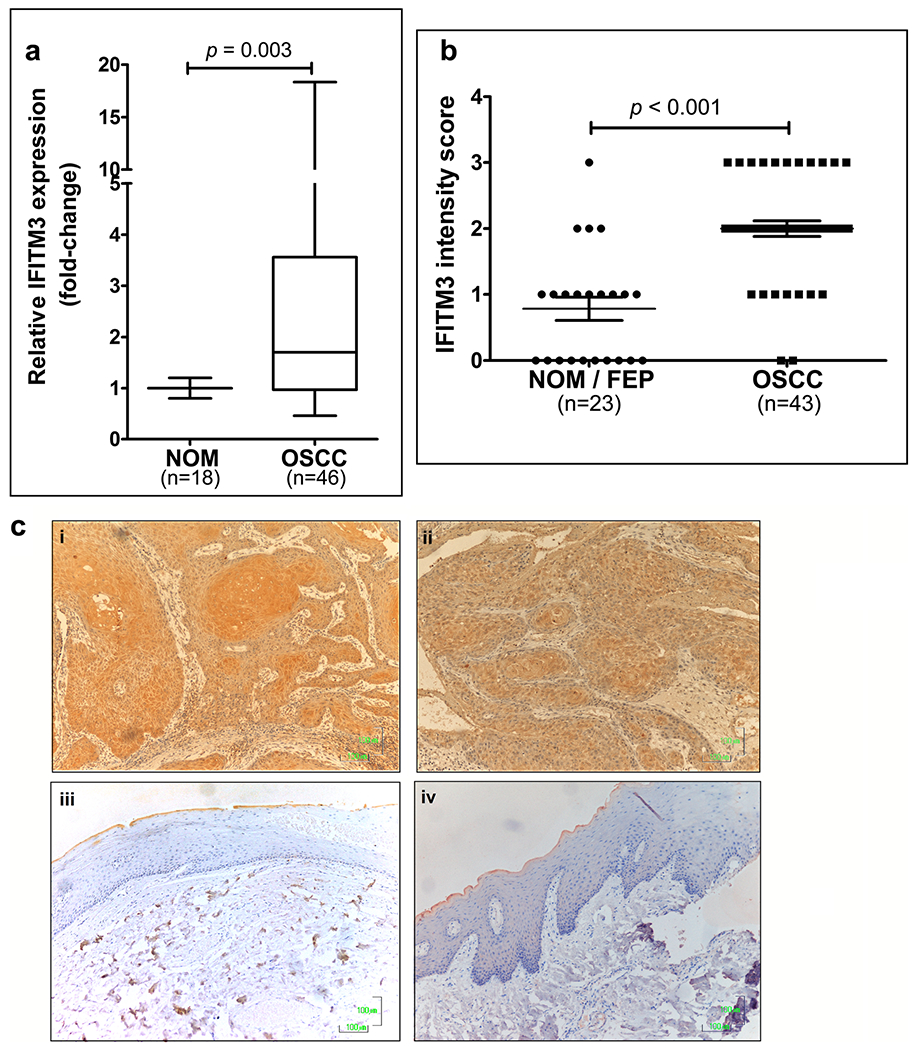
a Quantitative RT-PCR (qRT-PCR) analysis showing that IFITM3 mRNA levels are significantly up-regulated in OSCC tissues. As indicated by the box and whiskers plot, OSCC tissues exhibited a mean 2.9 ± 0.46-fold increase in IFITM3 expression, with a maximum level of 18-fold overexpression compared to NOM tissues. b IHC staining analysis showing that IFITM3 is significantly overexpressed in primary tumours compared to NOM tissues. c Representative IHC images showing that IFITM3 is expressed at high levels in primary OSCC tissues (i & ii), while the NOM tissues do not exhibit IFITM3 expression (iii & iv). Original magnification: 200x
3.2. IFITM3 knockdown in OSCC cell lines
To determine the functional role of IFITM3 in OSCC, we first assessed the endogenous level of IFITM3 in 6 OSCC cell lines (Supplementary Fig. 2a). We found that ORL-150 and ORL-204 exhibited high IFITM3 expression levels. These cell lines were, therefore, selected for IFITM3 knockdown using 2 different siRNAs (si18 and si19). Subsequent Western blot analysis showed that the IFITM3 protein levels were reduced in the OSCC cells transfected with si18 or si19 compared to the respective NT siRNA transfected control cells (Fig. 2a). We selected si18 for further experiments since the knockdown was more robust, i.e., the IFITM3 protein level was markedly reduced at day 2 post-transfection and the knockdown persisted through day 4 in both ORL-150 and ORL-204 cells. Since IFITM1, IFITM2 and IFITM3 exhibit high sequence homologies, we examined the effect of si18 on the mRNA levels of the different IFITM members in ORL-150 and ORL-204 cells. Our data showed that IFITM3 mRNA levels were reduced by > 70% in both cell lines until day 6 post-transfection compared to the other 2 family members, indicating that si18 specifically targets IFITM3 in OSCC cells (Supplementary Fig. 2b). Hence, ORL-150 and ORL-204 cells transfected with si18 were used in subsequent functional experiments.
Fig. 2. siRNA knockdown of IFITM3 inhibits OSCC cell growth.
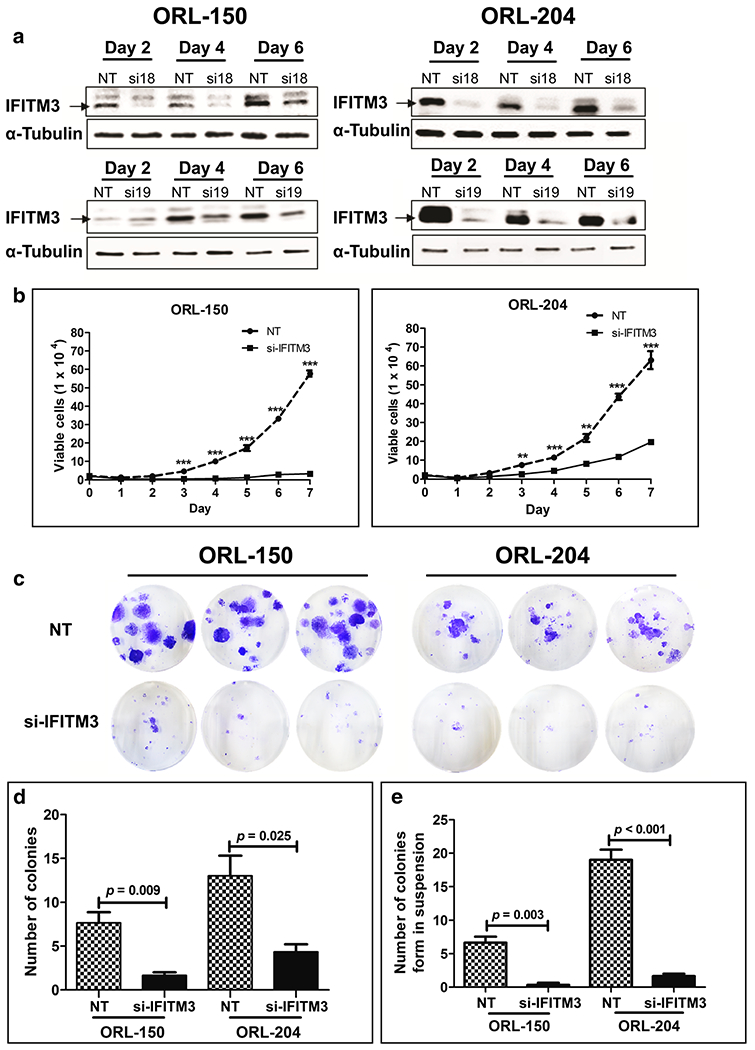
a Western blot showing reduction in IFITM3 expression after siRNA transfection in ORL-150 and ORL-204 cells. b siRNA knockdown of IFITM3 in OSCC cells significantly inhibits cell proliferation in comparison to cells transfected with non-targeting (NT) siRNA (p value: *** ≤ 0.001, ** ≤ 0.001, * ≤0.05). c Colony formation ability of cells transfected with siRNA targeting IFITM3 or NT siRNA. Cells were stained with crystal violet after 12 days of incubation, after which colonies of more than 50 cells were counted. d Significantly reduced colony formation of IFITM3 knockdown cells compared to cells transfected with NT siRNA. e siRNA-mediated knockdown of IFITM3 inhibits OSCC colony formation in semi-solid media
3.3. IFITM3 knockdown suppresses OSCC cell proliferation
Following siRNA transfection, we found that IFITM3 knockdown impacted ORL-150 and ORL-204 cell growth, i.e., confluency was achieved at a markedly slower rate than that of NT controls even though all cells were seeded at equal densities (Supplementary Fig. 3a). Based on this observation, we next examined whether IFITM3 influences cell proliferation by determining cell numbers every 24 h for 7 days. Indeed, we found that IFITM3 knockdown significantly inhibited OSCC cell proliferation (Fig. 2b). This inhibition was most prominent in ORL-150 cells after IFITM3 knockdown. Both ORL-150 and ORL-204 cells have a population doubling time of ~5.6 h. However, after IFITM3 knockdown, ORL-150 cells showed a prolonged population doubling time of 39.1 h, whereas the population doubling time of ORL-204 following IFITM3 knockdown increased to 8.1 h. These findings indicate that reduction of IFITM3 expression may lead to inhibition of OSCC cell proliferation.
Next, we examined the long-term effects of IFITM3 knockdown on the proliferation of OSCC cells using a clonogenic assay. Interestingly, we found that the ability of single cells to form colonies was significantly suppressed after IFITM3 knockdown compared to NT control cells (Fig. 2c, d). These results indicate that IFITM3 knockdown leads to a loss of proliferative ability of OSCC cells.
In addition, soft agar assays were performed to assess the effect of IFITM3 knockdown on anchorage-independent growth. By doing so, we found that IFITM3 knockdown in ORL-150 and ORL-204 cells resulted in significant reductions in the numbers of colonies formed compared to those by NT control cells (Fig. 2e). The majority of IFITM3 knockdown cells remained as single cells after a 14 day incubation period (Supplementary Fig. 3b). This finding indicates that OSCC cells lose the ability of anchorage-independent growth when IFITM3 is inhibited, and suggest that inhibition of IFITM3 may suppress OSCC cell transformation.
3.4. IFITM3 knockdown affects OSCC cell survival
In conjunction with the inhibition of cell proliferation, we also noted the presence of floating cells following IFITM3 knockdown. This observation led us to examine the impact of IFITM3 knockdown on cell survival by flow cytometry. We observed an increase in Annexin V positive and PI negative cell populations (lower right quadrant), indicating that these cells were undergoing early apoptosis (Fig. 3a). In comparisons to NT siRNA, IFITM3 knockdown significantly induced apoptosis in ORL-150 (17.3%) and ORL-204 (19.6%) cells (p < 0.001; Fig. 3b). In addition, when examining cells that did not undergo cell death following IFITM3 knockdown, we noted that the cell morphology appeared to be flattened with an enlarged cytoplasm as well as an accumulation of vacuoles. This observation prompted us to assess these cells for senescence-associated β-galactosidase activity. We found that these flattened cells readily stained blue, indicating expression of senescence-associated β-galactosidase (Fig. 3c). This significant increase in senescence-associated β-galactosidase staining indicates that IFITM3 knockdown induces senescence in ORL-150 (48%) and ORL-204 (14.2%) cells compared to the respective NT control cells (Fig. 3d). We also noticed that the induction of senescence by IFITM3 knockdown was most prominent in ORL-150 cells.
Fig. 3. OSCC growth inhibition by IFITM3 knockdown is mediated by induction of apoptosis, cell senescence and cell cycle arrest.
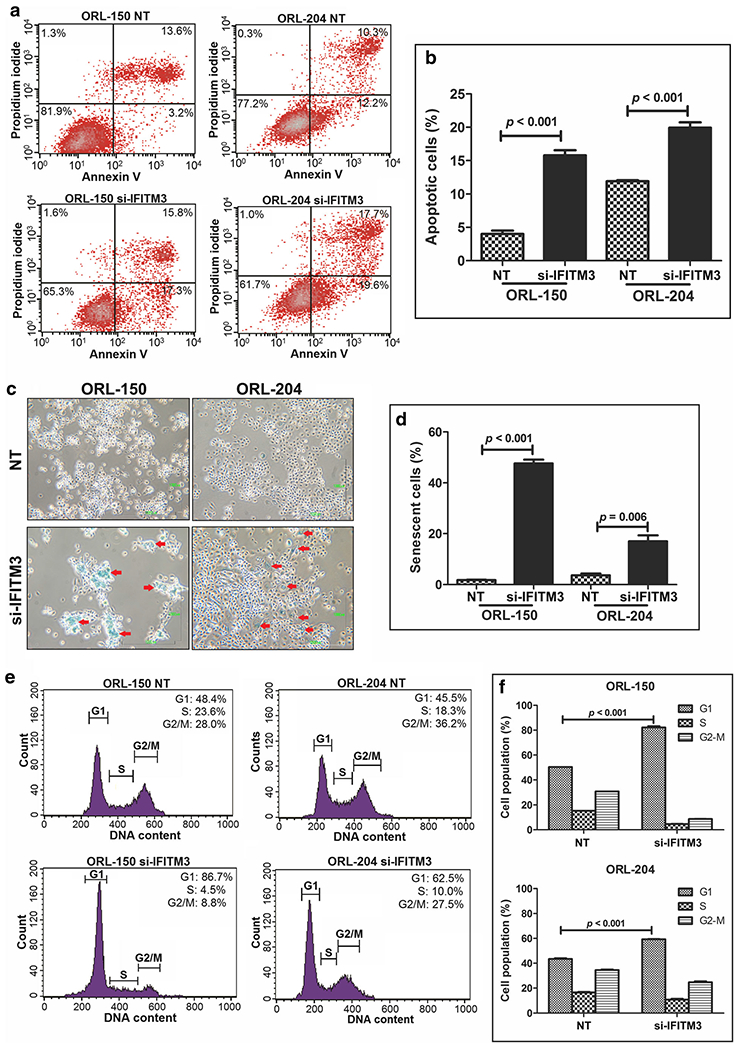
a Apoptosis was assessed by staining OSCC cells with Annexin V and PI and analysis by flow cytometry. Apoptotic cells are positive for Annexin V as indicated by the cells in the lower right quadrant of the scatter plot. b Increased populations of apoptotic cells after IFITM3 knockdown. c Senescent cells stained in blue (arrows) indicating activation of senescent-associated β-galactosidase activity (magnification: 200x). d IFITM3 knockdown significantly induces cellular senescence. e Cell cycle analysis using propidium iodide staining. Histograms indicate the distribution of cells in different stages of the cell cycle. f IFITM3 knockdown significantly arrests OSCC cells at the G1-S phase of the cell cycle
As cellular senescence is defined by an irreversible arrest in the G1 phase of the cell cycle, we next sought to analyse the cell cycle distribution in ORL-150 and ORL-204 cells following IFITM3 knockdown by flow cytometry. By doing so, we indeed observed a significant increase of cells in the G1 phase following IFITM3 knockdown in ORL-150 (86.7%) and ORL-204 (62.5%) cells compared to NT control cells (Fig. 3e, f), thus confirming that IFITM3 knockdown induces senescence and accumulation of cells in the G1 cell cycle phase. These observations are in line with growth inhibition as determined above by the clonogenic assay. Taken together, our data show that IFITM3 knockdown induces apoptosis and causes G1 cell cycle arrest, indicating that IFITM3 may play an important role in mediating cell survival and growth.
3.5. IFITM3 knockdown induces growth arrest by reducing CCND1-CDK4-pRB signalling
Since the above data indicated that IFITM3 knockdown led to decreased proliferative and survival capabilities of OSCC cells, we next set out to investigate the molecular mechanisms underlying these effects. To this end, we first assessed two major pathways known to regulate cell growth, i.e., the TP53/p21 and the p16 pathways, of which key regulators were evaluated by Western blotting. Activation of TP53 is known to result in p21 upregulation and subsequent cell cycle arrest. Of note, both ORL-150 and ORL-204 cells inherently harbour inactivating TP53 mutations [39]. Concordantly, we found that TP53 expression was not detectable in ORL-150 cells due to the presence of a deleterious A138fs*31 mutation, which leads to a frameshift and subsequent truncation of the protein (Fig. 4a). ORL-204 cells carry a R273C missense mutation in TP53, which is a hotspot mutation in its DNA binding domain. Substitution of R273 by cysteine causes a dramatic reduction in the DNA binding affinity, even though the protein retains its wild-type stability and, thus, can be detected by Western blotting [43]. Importantly, the TP53 gene is dysfunctional in both cell lines regardless of whether the protein could be detected by Western blotting. We continued to examine the expression of p21, the primary downstream target of TP53. While constitutive expression of TP53 and p21 are typically required for cell cycle arrest, we did not find that the expression of p21 was affected by IFITM3 knockdown. Hence, it appears that the observed growth inhibition in response to IFITM3 knockdown is not dependent on the TP53/p21 pathway.
Fig. 4. IFITM3 knockdown modulates levels of cell cycle proteins.
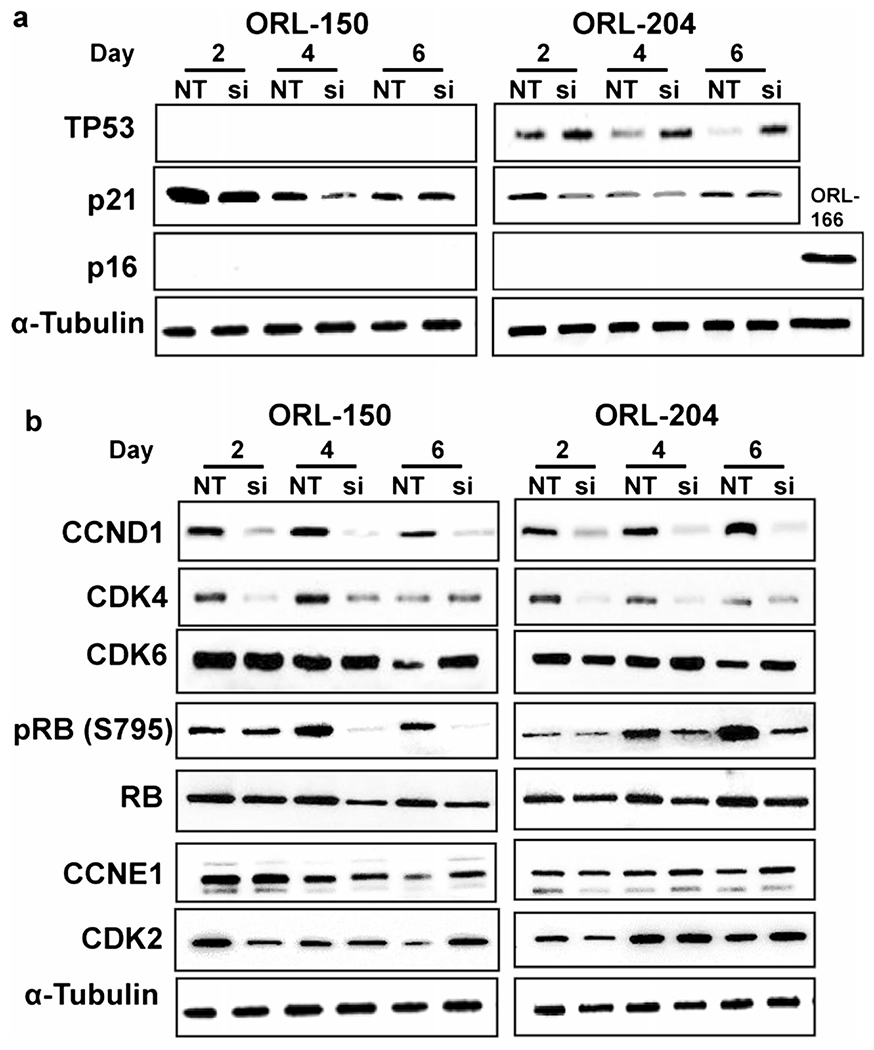
Western blots showing the expression of cell cycle markers in OSCC cells post-transfection with siRNA for IFITM3. a TP53 is not detected in ORL-150 cells due to presence of an inactivating mutation, while ORL-204 cells show up-regulation of TP53 upon IFITM3 knockdown. ORL-166 cells were used as positive control for p16 expression. b Reduced CCND1, CDK4 and pRB expression in IFITM3 knockdown cells
Next to TP53/p21, p16 is known to act as a key molecule regulating the cell cycle and is widely used as a senescence biomarker. However, both ORL-150 and ORL-204 cells fail to express this protein due to the presence of an inherent inactivating pArg80X mutation that results in a premature stop codon and protein truncation [39]. Since both the TP53/p21 and p16 pathways converge to the downstream cyclin/CDK/RB pathway, we next examined the effects of IFITM3 on the key proteins of this pathway. Interestingly, we found that the expression of CCND1 was markedly reduced following IFITM3 knockdown in both cell lines (Fig. 4b). Similarly, we found that the CDK4 expression levels were reduced, but recovered at day 6 post-transfection when IFITM3 levels increased again (Fig. 2a). The reductions in both CCND1 and CDK4 following IFITM3 knockdown eventually resulted in a reduction in RB phosphorylation (pRB) and to a lesser extent in total RB expression. The expression ofCDK2 was found to reduce slightly on day 2 after IFITM3 knockdown, but rapidly recovered on day 4. On the other hand, we found that IFITM3 knockdown did not have a pronounced effect on the CDK6 and CCNE1 expression levels. Taken together, our data indicate that IFITM3 knockdown may inhibit OSCC cell growth through the CCND1-CDK4-pRB axis, despite the presence of deleterious TP53 and p16 mutations in these cells.
Since the molecular mechanisms underlying the effect of IFITM3 knockdown on reduced cell cycle marker expression have so far not been reported, we resorted to the STRING database (a biological database and web resource of known and predicted protein–protein interactions) to assure that our Western blot observations are valid. Notably, we identified 50 genes interacting with IFITM3 and CCND1. Subsequent functional analysis of the IFITM3 interaction network revealed that these genes are enriched in cell cycle and cancer pathways (Fig. 5a). As both ORL-150 and ORL-204 cells have gene signatures that are cell cycle enriched [39], we next conducted Pearson correlation to assess co-expression of the 50 genes identified in the STRING database in a subset of 5 OSCC cell lines enriched in cell cycle pathways (including ORL-150 and ORL-204). We found that 70% (14/20) of the genes co-expressed with IFITM3 are known to be involved in the regulation of cell growth, including CCND1 and CDK4 (Fig. 5b). These data substantiate our Western blot observations indicating that IFITM3 down-regulation markedly reduces CDK4 and CCND1 expressions.
Fig. 5. Bioinformatics analysis of IFITM3 associations with cell cycle pathways.
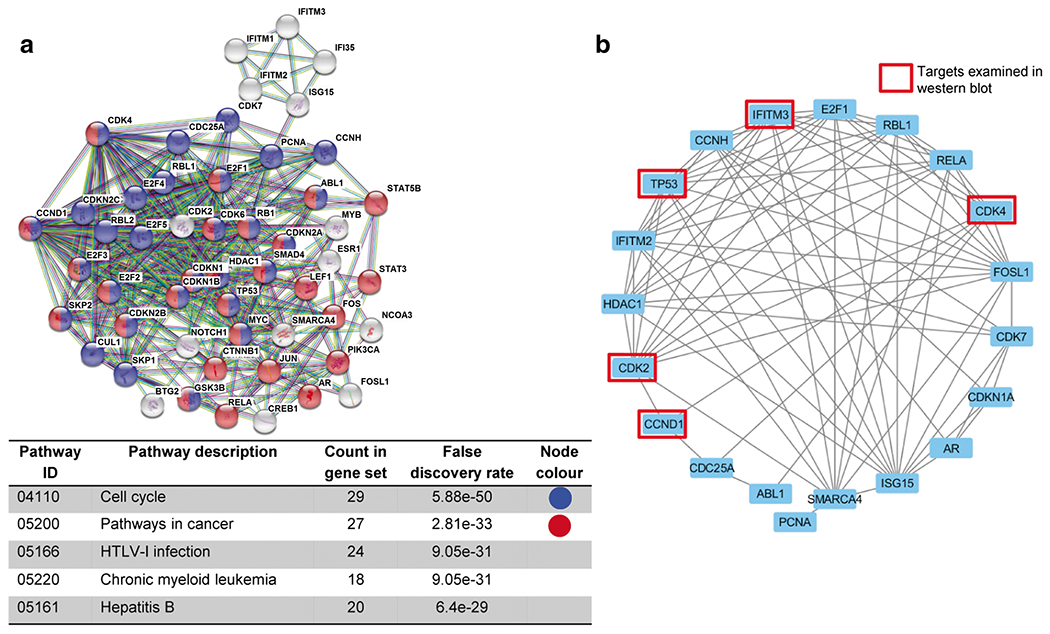
a Data mining from the STRING database indicate that IFITM3 is associated with genes involved in KEGG cell cycle and cancer pathways. b Gene co-expression network depicting genes positively co-expressed with IFITM3 in ORL cell lines enriched in cell cycle signalling pathways
4. Discussion
OSCC represents a subset of head and neck squamous cell carcinoma (HNSCC) that is challenging to treat. Patients usually experience a poor quality of life. Moreover, up to 50% of patients with advanced disease experience recurrence, despite advancements that have been made in treatment modalities [44]. Understanding the molecular mechanisms underlying this disease may lead to the identification of novel biomarkers associated with OSCC development, and result in the development of new treatment modalities. In our effort to identify molecular targets involved in OSCC development, we found that IFITM3 was significantly overexpressed in tumour tissues compared to NOM tissues regardless risk habits of the patients [33]. Others have examined differentially expressed genes between OSCC and matched normal oral tissues in patients chewing tobacco and also found that IFITM3 was overexpressed in the OSCC tissues [31]. Here, we further validated IFITM3 overexpression in a high percentage of patients in an independent OSCC cohort at both the mRNA and protein levels. Concordantly, we found after analysis of specimens deposited in TCGA that IFITM3 is overexpressed in HNSCC tumour tissues compared to matched normal tissues.
Whilst information on the role of IFITM family members in tumourigenesis is emerging, information on the role of these proteins in HNSCC has so far remained limited. IFITM1 has been reported to be overexpressed in invasive HNSCC tumours, and to promote cancer cell invasion through up-regulation of MMPs [13]. A transcriptional analysis conducted on nasopharyngeal cancers revealed that IFITM2 is up-regulated in tumour tissues compared to normal nasopharyngeal tissues, but further information on the role of IFITM2 in these tumours is currently lacking [32]. IFITM3 overexpression has been consistently reported in OSCC, suggesting an oncogenic role for IFITM3 in OSCC development [31, 33]. Since the functional role and mode of action of IFITM3 in OSCC development have so far not been determined, we sought to investigate this here. We found that IFITM3 knockdown markedly hampered the proliferative capability of OSCC cells, as indicated by induction of apoptosis, cell cycle arrest and senescence. The growth inhibitory effect of IFITM3 knockdown has also been reported in other malignancies including breast, gastric, glioma and lung cancers [12, 14, 16, 27]. Furthermore, IFITM family members may have inherent properties of driving cell proliferation and maintaining pluripotency as they have been shown to be expressed in murine pluripotent embryonic stem cells and in germ cell precursors [8, 44]. Andreu and colleagues found that IFITM members are expressed in stem cell compartments and that their induction results in a rapid transformation of adenomas in a conditional adenomatous polyposis coli (Apc) inactivation mouse model [19]. Although it is challenging to distinguish functional differences between IFITM members due to their high sequence homology, we found that the siRNA used in this study could specifically and sustainably knockdown IFITM3 in OSCC cells up to day 6. The growth of these cells was consistently inhibited as revealed by cell proliferation and colony formation assays, despite the expressions of IFITM1 and IFITM2. Taken together, these data indicate that IFITM3 may play an important role in OSCC growth, rather than IFITM1 and IFITM2.
Further analysis of the CCND1-CDK4/6-pRB axis revealed that expression of its rate-limiting proteins CCND1 and CDK4 was reduced after IFITM3 knockdown. We noted that CDK6 remained expressed when the CCND1 and CDK4 levels were reduced after IFITM3 knockdown. Both CDK4 and CDK6 are closely associated with CCND1 in regulating cellular growth, and it is possible that CDK6 remained expressed in order to compensate the function of CDK4 in an attempt to rescue cell cycle arrest. Alternatively, CDK6 may have a unique function that is distinct from CDK4 in regulating the cell cycle. Expression of CDK6 may, for example, restrain the proliferation of certain cell types, although the molecular mechanisms involved are as yet not fully understood [45–47]. Importantly, we found that CDK4 and CCND1 reduction after IFITM3 knockdown led to dephosphorylation of RB, indicating that IFITM3 knockdown indeed results in cell cycle arrest.
Other members of the IFITM family, in particular IFITM1, have also been shown to regulate the cell cycle. It has been reported that IFITM1 silencing leads to CCND1 expression downregulation and inhibition of cancer cell proliferation [11]. Our findings on OSCC cells are similar to those reported for glioma cells harbouring TP53 inactivating mutations, whereby IFITM1 knockdown led to decreases in CCND1 and CDK2 expression and, subsequently, G1 cell cycle arrest [11]. Notably, loss of G1 cell cycle regulation has been reported to be a fundamental event in HNSCC development [34, 48] and genomic HNSCC studies have revealed that TP53 mutations (66–87%), p16 alterations (22–74%) and CCND1 amplifications (23–31%) leading to uncontrolled cell cycle progression are common events, particularly in human papillomavirus (HPV)-negative tumours [34, 48–50]. Therefore, we conclude that our observation that IFITM3 may modulate the expression of CCND1 and CDK4 in OSCC cells supports a role of IFITM3 in regulating the cell cycle in these tumours.
In summary, we show that IFITM3 knockdown affects the expression of key cell cycle regulators in OSCC cells. Several clinical trials targeting the cell cycle are underway for HNSCC [51]. Additional studies aimed at the intricacies of IFITM3 overexpression and how this expression can be inhibited may be instrumental for the design of novel targeted strategies to control OSCC.
Supplementary Material
Acknowledgments
We would like to thank all consented patients who participated in this study. This study was funded by Ministry of Science, Technology and Innovation (MOSTI 06-00-00-0000), University of Malaya (UM.C/625/1/HIR/MOHE/DENT-03) and sponsors of Cancer Research Malaysia. Cancer Research Malaysia is a non-profit research organization. We are committed to an understanding of cancer prevention, diagnosis and treatment through a fundamental research program.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s13402-019-00437-z) contains supplementary material, which is available to authorized users.
Conflict of interest All authors declared that they have no conflict of interest.
References
- 1.Martensen PM, Justesen J, Small ISGs coming forward. J Interf Cytokine Res 24, 1–19 (2004) [DOI] [PubMed] [Google Scholar]
- 2.Siegrist F, Ebeling M, Certa U, The small interferon-induced transmembrane genes and proteins. J Interf Cytokine Res 31, 183–197 (2011) [DOI] [PubMed] [Google Scholar]
- 3.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, Digard P, Anttila V, Baillie JK, Walsh TS, Hume DA, Palotie A, Xue Y, Colonna V, Tyler-Smith C, Dunning J, Gordon SB, Smyth RL, Openshaw PJ, Dougan G, Brass AL, Kellam P, IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ, The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans SS, Collea RP, Leasure JA, Lee DB, IFN-alpha induces homotypic adhesion and Leu-13 expression in human B lymphoid cells. J Immunol 150, 736–747 (1993) [PubMed] [Google Scholar]
- 6.Smith RA, Young J, Weis JJ, Weis JH, Expressionofthe mouse fragilis gene products in immune cells and association with receptor signaling complexes. Genes Immun 7, 113–121 (2006) [DOI] [PubMed] [Google Scholar]
- 7.Lange UC, Saitou M, Western PS, Barton SC, Surani MA, The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol 3, 1 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saitou M, Payer B, Lange UC, Erhardt S, Barton SC, Surani MA, Specification of germ cell fate in mice. Philos Trans R Soc Lond Ser B Biol Sci 358, 1363–1370 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka SS, Nagamatsu G, Tokitake Y, Kasa M, Tam PP, Matsui Y, Regulation of expression of mouse interferon-induced transmembrane protein like gene-3, Ifitm3 (mil-1, fragilis), in germ cells. Dev Dyn 230, 651–659 (2004) [DOI] [PubMed] [Google Scholar]
- 10.Tanaka SS, Yamaguchi YL, Tsoi B, Lickert H, Tam PP, IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev Cell 9, 745–756 (2005) [DOI] [PubMed] [Google Scholar]
- 11.Yu F, Ng SS, Chow BK, Sze J, Lu G, Poon WS, Kung HF, Lin MC, Knockdown of interferon-induced transmembrane protein 1 (IFITM1) inhibits proliferation, migration, and invasion of glioma cells. J Neuro-Oncol 103, 187–195 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B, Wang H, Zong G, Li P, The role of IFITM3 in the growth and migration of human glioma cells. BMC Neurol 13, 210 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatano H, Kudo Y, Ogawa I, Tsunematsu T, Kikuchi A, Abiko Y, Takata T, IFN-induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression. Clin Cancer Res 14, 6097–6105 (2008) [DOI] [PubMed] [Google Scholar]
- 14.Zhang D, Wang H, He H, Niu H, Li Y, Interferon induced transmembrane protein 3 regulates the growth and invasion of human lung adenocarcinoma. Thorac Cancer 8, 337–343 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lui AJ, Geanes ES, Ogony J, Behbod F, Marquess J, Valdez K, Jewell W, Tawfik O, Lewis-Wambi J, IFITM1 suppression blocks proliferation and invasion of aromatase inhibitor-resistant breast cancer in vivo by JAK/STAT-mediated induction of p21. Cancer Lett 399, 29–43 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang M, Gao H, Chen P, Jia J, Wu S, Knockdown of interferon-induced transmembrane protein 3 expression suppresses breast cancer cell growth and colony formation and affects the cell cycle. Oncol Rep 30, 171–178 (2013) [DOI] [PubMed] [Google Scholar]
- 17.Sari IN, Yang YG, Phi LT, Kim H, Baek MJ, Jeong D, Kwon HY, Interferon-induced transmembrane protein 1 (IFITM1) is required for the progression of colorectal cancer. Oncotarget 7, 86039–86050 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu F, Xie D, Ng SS, Lum CT, Cai MY, Cheung WK, Kung HF, Lin G, Wang X, Lin MC, IFITM1 promotes the metastasis of human colorectal cancer via CAV-1. Cancer Lett 368, 135–143 (2015) [DOI] [PubMed] [Google Scholar]
- 19.Andreu P, Colnot S, Godard C, Laurent-Puig P, Lamarque D, Kahn A, Perret C, Romagnolo B, Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res 66, 1949–1955 (2006) [DOI] [PubMed] [Google Scholar]
- 20.Fan J, Peng Z, Zhou C, Qiu G, Tang H, Sun Y, Wang X, Li Q, Le X, Xie K, Gene-expression profiling in Chinese patients with colon cancer by coupling experimental and bioinformatic genomewide gene-expression analyses: Identification and validation of IFITM3 as a biomarker of early colon carcinogenesis. Cancer 113, 266–275 (2008) [DOI] [PubMed] [Google Scholar]
- 21.Tirosh B, Daniel-Carmi V, Carmon L, Paz A, Lugassy G, Vadai E, Machlenkin A, Bar-Haim E, Do MS, Ahn IS, Fridkin M, Tzehoval E, Eisenbach L, ‘1-8 interferon inducible gene family’: Putative colon carcinoma-associated antigens. Br J Cancer 97, 1655–1663 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Peng Z, Tang H, Wei P, Kong X, Yan D, Huang F, Li Q, Le X, Xie K, KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin Cancer Res 17, 3558–3568 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia Y, Zhang M, Jiang W, Zhang Z, Huang S, Wang Z, Overexpression of IFITM3 predicts the high risk of lymphatic metastatic recurrence in pN0 esophageal squamous cell carcinoma after Ivor-Lewis esophagectomy. PeerJ 3, e1355 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borg D, Hedner C, Gaber A, Nodin B, Fristedt R, Jirstrom K, Eberhard J, Johnsson A, Expression of IFITM1 as a prognostic biomarker in resected gastric and esophageal adenocarcinoma. Biomark Res 4, 10 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Lee JH, Kim KY, Song HK, Kim JK, Yoon SR, Cho D, Song KS, Lee YH, Choi I, The interferon-inducible 9-27 gene modulates the susceptibility to natural killer cells and the invasiveness of gastric cancer cells. Cancer Lett 221, 191–200 (2005) [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Goh SH, Song N, Hwang JA, Nam S, Choi IJ, Shin A, Kim IH, Ju MH, Jeong JS, Lee YS, Overexpression of IFITM1 has clinicopathologic effects on gastric cancer and is regulated by an epigenetic mechanism. Am J Pathol 181, 43–52 (2012) [DOI] [PubMed] [Google Scholar]
- 27.Hu J, Wang S, Zhao Y, Guo Q, Zhang D, Chen J, Li J, Fei Q, Sun Y, Mechanism and biological significance of the overexpression of IFITM3 in gastric cancer. Oncol Rep 32, 2648–2656 (2014) [DOI] [PubMed] [Google Scholar]
- 28.Hisamatsu T, Watanabe M, Ogata H, Ezaki T, Hozawa S, Ishii H, Kanai T, Hibi T, Interferon-inducible gene family 1-8U expression in colitis-associated colon cancer and severely inflamed mucosa in ulcerative colitis. Cancer Res 59, 5927–5931 (1999) [PubMed] [Google Scholar]
- 29.Wu F, Dassopoulos T, Cope L, Maitra A, Brant SR, Harris ML, Bayless TM, Parmigiani G, Chakravarti S, Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: Insights into distinctive pathogenesis. Inflamm Bowel Dis 13, 807–821 (2007) [DOI] [PubMed] [Google Scholar]
- 30.Seo GS, Lee JK, Yu JI, Yun KJ, Chae SC, Choi SC, Identification of the polymorphisms in IFITM3 gene and their association in a Korean population with ulcerative colitis. Exp Mol Med 42, 99–104 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arora S, Matta A, Shukla NK, Deo SV, Ralhan R, Identification of differentially expressed genes in oral squamous cell carcinoma. Mol Carcinog 42, 97–108 (2005) [DOI] [PubMed] [Google Scholar]
- 32.Fang W, Li X, Jiang Q, Liu Z, Yang H, Wang S, Xie S, Liu Q, Liu T, Huang J, Xie W, Li Z, Zhao Y, Wang E, Marincola FM, Yao K, Transcriptional patterns, biomarkers and pathways characterizing nasopharyngeal carcinoma of southern China. J Transl Med 6, 32 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheong SC, Chandramouli GV, Saleh A, Zain RB, Lau SH, Sivakumaren S, Pathmanathan R, Prime SS, Teo SH, Patel V, Gutkind JS, Gene expression in human oral squamous cell carcinoma is influenced by risk factor exposure. Oral Oncol 45, 712–719 (2009) [DOI] [PubMed] [Google Scholar]
- 34.Iglesias-Bartolome R, Martin D, Gutkind JS, Exploiting the head and neck cancer oncogenome: Widespread PI3K-mTOR pathway alterations and novel molecular targets. Cancer Discov 3, 722–725 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zain RB, Ghani WM, Razak IA, Latifah RJ, Samsuddin AR, Cheong SC, Abdullah N, Ismail AR, Hussaini HB, Talib NA, Jallaludin A, Building partnership in oral cancer research in a developing country: Processes and barriers. Asian Pac J Cancer Prev 10,513–518 (2009) [PubMed] [Google Scholar]
- 36.Gan CP, Patel V, Mikelis CM, Zain RB, Molinolo AA, Abraham MT, Teo SH, Abdul Rahman ZA, Gutkind JS, Cheong SC, Heterotrimeric G-protein alpha-12 (Galpha12) subunit promotes oral cancer metastasis. Oncotarget 5, 9626–9640 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanaruddin SN, Saleh A, Yang YH, Hamid S, Mustafa WM, Khairul Bariah AA, Zain RB, Lau SH, Cheong SC, Four-protein signature accurately predicts lymph node metastasis and survival in oral squamous cell carcinoma. Hum Pathol 44, 417–426 (2013) [DOI] [PubMed] [Google Scholar]
- 38.Hamid S, Lim KP, Zain RB, Ismail SM, Lau SH, Mustafa WM, Abraham MT, Nam NA, Teo SH, Cheong SC, Establishment and characterization of Asian oral cancer cell lines as in vitro models to study a disease prevalent in Asia. Int J Mol Med 19, 453–460 (2007) [PubMed] [Google Scholar]
- 39.Fadlullah MZ, Chiang IK, Dionne KR, Yee PS, Gan CP, Sam KK, Tiong KH, Ng AK, Martin D, Lim KP, Kallarakkal TG, Mustafa WM, Lau SH, Abraham MT, Zain RB, Rahman ZA, Molinolo A, Patel V, Gutkind JS, Tan AC, Cheong SC, Genetically-defined novel oral squamous cell carcinoma cell lines for the development of molecular therapies. Oncotarget 7, 27802–27818 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gan CP, Hamid S, Hor SY, Zain RB, Ismail SM, Wan Mustafa WM, Teo SH, Saunders N, Cheong SC, Valproic acid: Growth inhibition of head and neck cancer by induction of terminal differentiation and senescence. Head Neck 34, 344–353 (2012) [DOI] [PubMed] [Google Scholar]
- 41.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C, The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45, D362–D368 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eldar A, Rozenberg H, Diskin-Posner Y, Rohs R, Shakked Z, Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions. Nucleic Acids Res 41, 8748–8759 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lickert H, Cox B, Wehrle C, Taketo MM, Kemler R, Rossant J, Dissecting Wnt/beta-catenin signaling during gastrulation using RNA interference in mouse embryos. Development 132,2599–2609 (2005) [DOI] [PubMed] [Google Scholar]
- 45.Nagasawa M, Gelfand EW, Lucas JJ, Accumulation of high levels of the p53 and p130 growth-suppressing proteins in cell lines stably over-expressing cyclin-dependent kinase 6 (cdk6). Oncogene 20, 2889–2899 (2001) [DOI] [PubMed] [Google Scholar]
- 46.Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RG, Schafer M, Fajmann S, Schlederer M, Schiefer AI, Reichart U, Mayerhofer M, Hoeller C, Zochbauer-Muller S, Kerjaschki D, Bock C, Kenner L, Hoefler G, Freissmuth M, Green AR, Moriggl R, Busslinger M, Malumbres M, Sexl V, A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell 30, 359–360 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lucas JJ, Domenico J, Gelfand EW, Cyclin-dependent kinase 6 inhibits proliferation of human mammary epithelial cells. Mol Cancer Res 2, 105–114 (2004) [PubMed] [Google Scholar]
- 48.Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, Zhao M, Ortega Alves MV, Chang K, Drummond J, Cortez E, Xie TX, Zhang D, Chung W, Issa JP, Zweidler-McKay PA, Wu X, El-Naggar AK, Weinstein JN, Wang J, Muzny DM, Gibbs RA, Wheeler DA, Myers JN, Frederick MJ, Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov 3, 770–781 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beck TN, Golemis EA, Genomic insights into head and neck cancer. Cancers of the head & neck 1, (2016). 10.1186/s41199-016-0003-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Network TCGA, Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB, Getz G, Ally A, Balasundaram M, Birol I, Bowlby R, Brooks D, Butterfield YSN, Carlsen R, Cheng D, Chu A, Dhalla N, Guin R, Holt RA, Jones SJM, Lee D, Li HI, Marra MA, Mayo M, Moore RA, Mungall AJ, Robertson AG, Schein JE, Sipahimalani P, Tam A, Thiessen N, Wong T, Protopopov A, Santoso N, Lee S, Parfenov M, Zhang J, Mahadeshwar HS, Tang J, Ren X, Seth S, Haseley P, Zeng D, Yang L, Xu AW, Song X, Pantazi A, Bristow CA, Hadjipanayis A, Seidman J, Chin L, Park PJ, Kucherlapati R, Akbani R, Casasent T, Liu W, Lu Y, Mills G, Motter T, Weinstein J, Diao L, Wang J, Fan YH, Liu J, Wang K, Auman JT, Balu S, Bodenheimer T, Buda E, Hayes DN, Hoadley KA, Hoyle AP, Jefferys SR, Jones CD, Kimes PK, Liu Y, Marron JS, Meng S, Mieczkowski PA, Mose LE, Parker JS, Perou CM, Prins JF, Roach J, Shi Y, Simons JV, Singh D, Soloway MG, Tan D, Veluvolu U, Walter V, Waring S, Wilkerson MD, Wu J, Zhao N, Cherniack AD, Hammerman PS, Tward AD, Pedamallu CS, Saksena G, Jung J, Ojesina AI, Carter SL, Zack TI, Schumacher SE, Beroukhim R, Freeman SS, Meyerson M, Cho J, Chin L, Getz G, Noble MS, DiCara D, Zhang H, Heiman DI, Gehlenborg N, Voet D, Lin P, Frazer S, Stojanov P, Liu Y, Zou L, Kim J, Sougnez C, Gabriel SB, Lawrence MS, Muzny D, Doddapaneni H, Kovar C, Reid J, Morton D, Han Y, Hale W, Chao H, Chang K, Drummond JA, Gibbs RA, Kakkar N, Wheeler D, Xi L, Ciriello G, Ladanyi M, Lee W, Ramirez R, Sander C, Shen R, Sinha R, Weinhold N, Taylor BS, Aksoy BA, Dresdner G, Gao J, Gross B, Jacobsen A, Reva B, Schultz N, Sumer SO, Sun Y, Chan TA, Morris LG, Stuart J, Benz S, Ng S, Benz C, Yau C, Baylin SB, Cope L, Danilova L, Herman JG, Bootwalla M, Maglinte DT, Laird PW, Triche T Jr., Weisenberger DJ, Van Den Berg DJ, Agrawal N, Bishop J, Boutros PC, Bruce JP, Byers LA, Califano J, Carey TE, Chen Z, Cheng H, Chiosea SI, Cohen E, Diergaarde B, Egloff AM, El-Naggar AK, Ferris RL, Frederick MJ, Grandis JR, Guo Y, Haddad RI, Hammerman PS, Harris T, Hayes DN, Hui ABY, Lee JJ, Lippman SM, Liu F-F, McHugh JB, Myers J, Ng PKS, Perez-Ordonez B, Pickering CR, Prystowsky M, Romkes M, Saleh AD, Sartor MA, Seethala R Seiwert TY, Si H, Tward AD, Van Waes C, Waggott DM, Wiznerowicz M, Yarbrough WG, Zhang J, Zuo Z, Burnett K, Crain D, Gardner J, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton C, Shelton T, Sherman M, Yena P Black AD, Bowen J, Frick J, Gastier-Foster JM, Harper HA, Leraas K, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Baboud J, Jensen MA, Kahn AB, Pihl TD, Pot DA, Srinivasan D, Walton JS, Wan, Burton RA, Davidsen T, Demchok JA, Eley G, Ferguson ML, Shaw KRM, Ozenberger BA, Sheth M, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Sailer C, Tarvin K, Chen C, Bollag R, Weinberger P, Golusinski W, Golusinski P, Ibbs M, Korski K, Mackiewicz A, Suchorska W, Szybiak B, Wiznerowicz M, Burnett K, Curley E, Gardner J, Mallery D, Penny R, Shelton T, Yena P, Beard C, Mitchell C, Sandusky G, Agrawal N, Ahn J, Bishop J, Califano J, Khan Z, Bruce JP, Hui ABY, Irish J, Liu F-F, Perez-Ordonez B, Waldron J, Boutros PC, Waggott DM, Myers J, William WN Jr., Lippman SM, Egea S, Gomez-Fernandez C, Herbert L, Bradford CR, Carey TE, Chepeha DB, Haddad AS, Jones TR, Komarck CM, Malakh M, McHugh JB, Moyer JS, Nguyen A, Peterson LA, Prince ME, Rozek LS, Sartor MA, Taylor EG, Walline HM, Wolf GT, Boice L, Chera BS, Funkhouser WK, Gulley ML, Hackman TG, Hayes DN, Hayward MC, Huang M, Rathmell WK, Salazar AH, Shockley WW, Shores CG, Thorne L, Weissler MC, Wrenn S, Zanation AM, Chiosea SI, Diergaarde B, Egloff AM, Ferris RL, Romkes M, Seethala R, Brown BT, Guo Y, Pham M, Yarbrough WG, Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michel L, Ley J, Wildes TM, Schaffer A, Robinson A, Chun SE, Lee W, Lewis J Jr., Trinkaus K, Adkins D, Phase I trial of palbociclib, a selective cyclin dependent kinase 4/6 inhibitor, in combination with cetuximab in patients with recurrent/metastatic head and neck squamous cell carcinoma. Oral Oncol 58, 41–48 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


