The myelodysplastic syndromes (MDS) are haematopoietic stem cell malignancies with recurring myeloid specific mutations that may instruct poor prognosis (Steensma et al, 2015). EZH2 (Enhancer of Zester homolog 2) encodes the catalytic subunit of the PRC2 complex methylating H3K27 influencing stem cell renewal by epigenetic transcriptional repression. EZH2 is mutated in ~10% of MDS or myelodysplastic/myeloproliferative neoplasms (MPN) with unknown function, demonstrating a need for characterization in disease modulation or prognostication (Xu & Li, 2012; Karoopongse et al, 2014; Sashida et al, 2014). We explored EZH2 protein expression by immunohistochemistry (IHC) in patient bone marrow (BM) samples with regard to mutation status, clinical characteristics and outcome.
Patients with MDS, MDS/MPN or acute myeloid leukaemia with myelodysplasia-related changes (AML-MRC) with available myeloid-targeted, next generation sequencing (NGS) and corresponding BM biopsy were retrieved. NGS included a gene panel of up to 54 genes with 5% variant allele frequency (VAF) threshold and 500X minimum coverage depth. EZH2 antibody (catalogue number 790-4651, Ventana Medical Systems, Oro Valley, AZ, USA) was used for IHC with tonsil, reactive germinal centre cells as positive controls and tonsillar interfollicular T-cells with undetectable EZH2 or no antibody staining as negative controls. Nuclear EZH2 expression was assessed semi-quantitatively using an IHC score calculated by haematopoietic cell signal strength (0–3+) times percent positivity. Means were compared using chi-square, Student t-test, or ANOVA. Correlations were analysed by Pearson’s coefficient. Overall survival (OS) was defined as the time between date of diagnosis (or NGS test date) and death/last follow -up by Kaplan–Meier method with log rank. P-values <0·05 were considered statistically significant.
Sixty-one biopsies [33 MDS, 18 MDS/MPN, 10 AML-MRC], were compared to 29 wild type (WT), 19 MDS, 6 MDS/MPN and 4 AML-MRC. Five haematologically normal BM without EZH2 or other myeloid-specific mutations were included as normal controls. While the mean EZH2 IHC score was reduced in MDS cases [n = 90, 0·813 ± 0·073 (mean ± standard error)] compared to normal controls (n = 5, 1·025 ± 0·140), statistical significance was not reached, probably due to low control numbers (P = 0·223). Importantly, expression was significantly decreased in mutant compared to WT samples (0·617 ± 0·081 versus 1·23 ± 0·12, respectively, P < 0·001). Furthermore, only mutated cases significantly differed from normal controls (P = 0·040) (Fig 1A). Notably, EZH2 expression did not correlate with blasts (P = 0·16), haemoglobin (P = 0·15), neutrophils (P = 0·99), platelets (P = 0·358), International Prognostic Scoring System (IPSS) (P = 0·709) or revised IPSS (P = 0·871). Expectedly, chromosomal 7(chr7) abnormality [−7 or del(7q)] was over represented in EZH2 mutant cases (3·4% of WT, 20·7% of mutated, P = 0·041) and EZH2 expression was significantly lower in −7/del(7q) cases compared to those without (0·336 ± 0·104 vs. 0·894 ± 0·080, respectively, P < 0·001). IHC score and EZH2 VAF were negatively correlated but not statistically significant (r = −0·264, P = 0·138 mutated cases; r = −0.264, P = 0·100 mutated cases excluding chr7 abnormalities). Prior studies demonstrate mutant EZH2 negatively impacts OS, which we confirmed (Fig 1C) [WT median OS not reached versus mutant OS 29·0 ± 2·2, 95% confidence interval (CI): 25·0–33·0 months, P = 0·003] (Bejar et al, 2011). We also confirmed that chr7 abnormality negatively impacts OS (no abnormality OS, 61·0 ± 21·8, 95% CI: 18·3–103·7 months versus OS 14·0 ± 2·9, 95% CI: 8·32–19·7 months with chr7 abnormality, P < 0·001) (Fig 1D). Differences in OS (using NGS date) were not detected using an IHC cutoff score of 0·5 in all cases, mutant, or mutant excluding −7/del(7q) (P = 0·131, 0·761, 0·811, respectively) (Fig 1E for the latter). A 1·0 cutoff was similar. There was no significant difference in OS or EZH2 expression in cases with EZH2 + TET2, EZH2 + RUNX1, EZH2 + ASXL1 mutations, or in cases with EZH2 plus ≥2 mutations. Previous reports demonstrated that splicing mutations decrease EZH2 expression (Kim et al, 2015), however, we did not observe this (P = 0·896) which may be attributed to low case numbers. Importantly, 6 sequential BM from mutant EZH2 patients with progression defined by increased blasts and worsening cytopenias were investigated. Four progressed from MDS to AML-MRC with decreased IHC expression upon transformation (mean pre- and postexpression was 0·875 ± 0·09 and 0·281 ± 0·06, respectively). One case progressed from MDS with excess blasts, type I (MDS-EB-I) to MDS-EB-II and had no change in expression. Another case progressed from MDS with multilineage dysplasia to MDS-EB-II with a slight increase in EZH2 expression (0·15–0·45).
Fig 1.
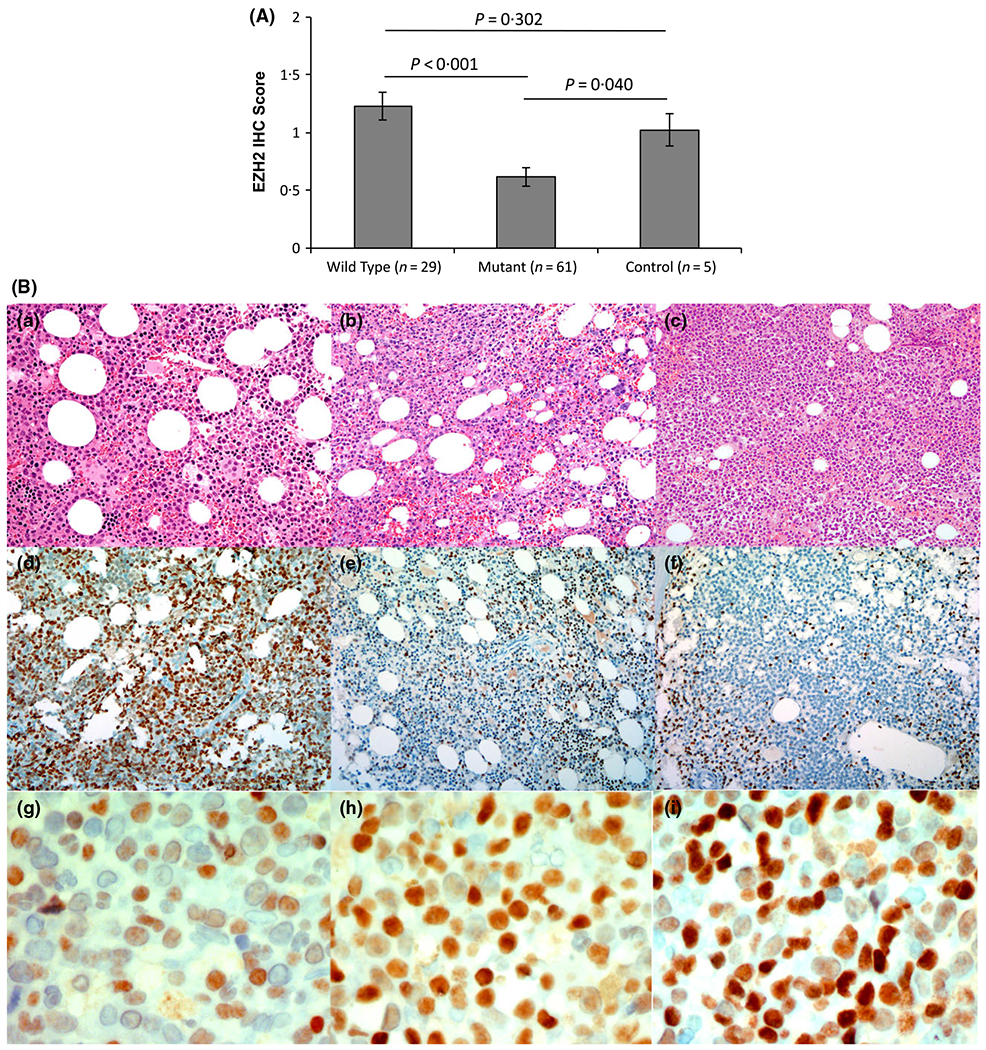
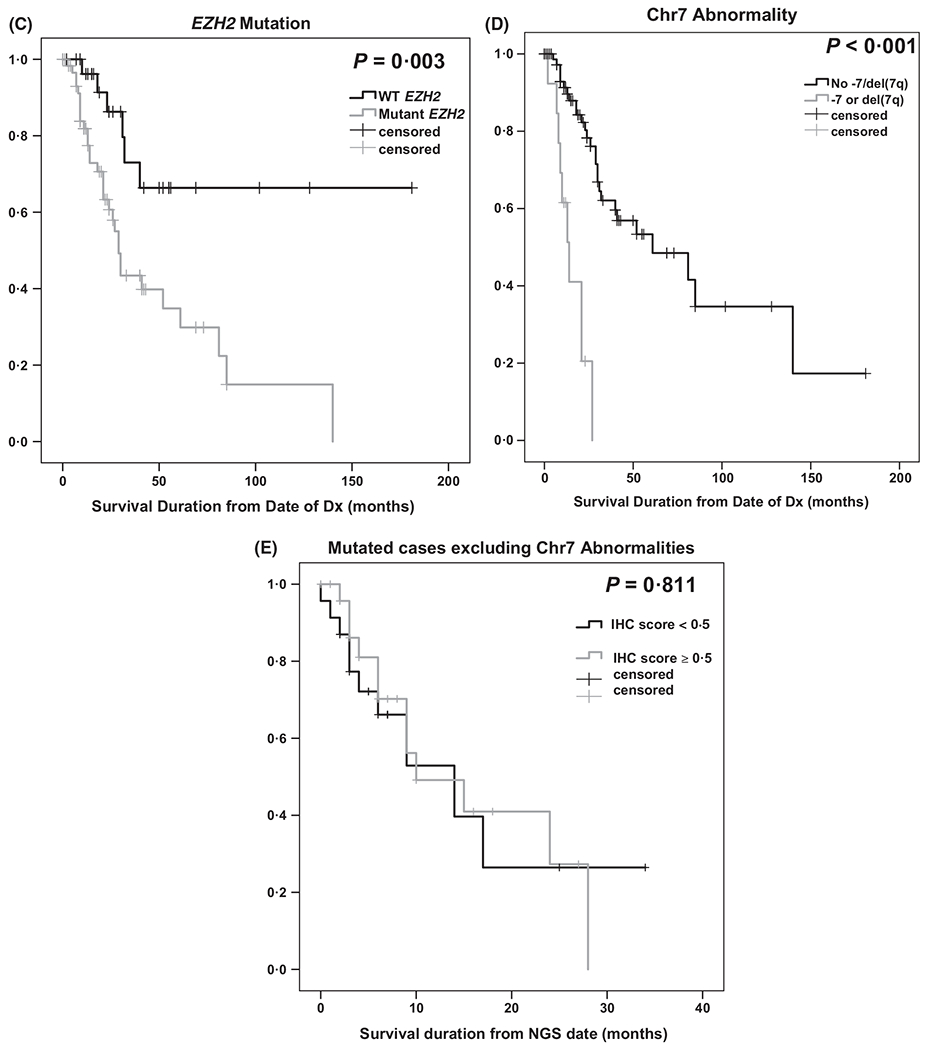
(A) Differences in EZH2 protein expression by immunohistochemistry (IHC) in mutated cases compared to either wild type cases or haematologically normal controls. (B) Immunohistochemical staining. (a and d) Bone marrow biopsy from a patient with low grade myelodysplastic syndrome (MDS with multilineage dysplasia, MDS-MLD) and high EZH2 expression; (b and e) bone marrow core biopsy from a patient with high grade MDS (MDS with excess blasts, MDS-EB) who harboured an EZH2 mutation, del(7q), and lower EZH2 expression; (c and f) bone marrow core biopsy from the same patient in b at the time of transformation to acute myeloid leukaemia, showing significantly reduced EZH2 expression on leukaemic blasts. Signal intensity was scored as 1 + (g), 2 + (h), or 3 + (i) (a-c, haematoxylin-eosin, x200; d-f, immunoperoxidase, x200; g-i, immunoperoxidase x600). (C) Kaplan–Meier plot of overall survival, defined as the duration between the date of diagnosis (Dx) and death or last follow-up, based on EZH2 mutation status is significantly decreased in EZH2 mutant cases compared to wild-type (WT) EZH2 cases. (D) Overall survival, defined as in (C), by chromosome 7 abnormality (Chr7 abn), is significantly decreased in cases with −7 or del(7q). (E) No difference in overall survival in mutated cases excluding those with −7 or del(7q) using an IHC cut-off score of 0·5.
MDS classification and prognostication has advanced from historic morphological characterization to more molecularly focused systems, including identifying myeloid-specific acquired somatic mutations that may adversely impact outcome. Often, patients who harbour such mutations transit from clinically-defined risk groups to the next higher risk category despite displaying similar cytopenic and morphological characteristics (Bejar et al, 2011). The functional consequences of these mutations may be equally important, particularly for elusive genes like EZH2. We show decreased EZH2 expression is associated with EZH2 gene mutation, although not VAF, and chr7 abnormalities. Our prior study demonstrated P53 IHC expression directly correlated with TP53 VAF where the majority of mutations result in stabilization and up-regulation of the protein (McGraw et al, 2016). Alternatively, the functional consequences of EZH2 gene mutations are largely unknown in MDS and, as the clinical and pathological significance of certain EZH2 mutations identified in our cohort have not been reported (Table I), a further functional examination of these variants is required to correlate IHC with VAF. We examined expression by nonsense vs missense or known clinical significance vs unknown mutations, however, the number of cases was too limited to draw any conclusions.
Table I.
EZH2 mutation description
| Case | Subtype | Monosomy 7 or del(7q) | IHC Score | EZH2 mutation | Missense/Nonsense | Previously reported | Clinical significance |
|---|---|---|---|---|---|---|---|
| 1 | AML-MRC | Yes | 0 | c.1555 + 1G>T | Missense | Yes | Yes, Significant |
| 2 | Other | No | 0·75 | c.2187_2188insT; p.D730 fs | Missense | Yes | Yes, Significant |
| 3 | Other | No | 0·15 | c.862C>T; c.1852_6C>T and c.2007C>A; p. S669R | Yes | Yes, Significant | |
| 4 | MDS/MPN | No | 2·125 | c.2196_2A>T | Yes | Yes, Significant | |
| 5 | AML-MRC | No | 0 | c.1505G>A; R502Q and c.1694G>C; C565S | Missense | Yes | Yes, Significant |
| 6 | Other | No | 0·6 | c.71T>C; p.M24T | Missense | Yes | Yes, Significant |
| 7 | AML-MRC | No | 1·5 | p.A656V | Missense | Yes | Yes, Significant |
| 8 | AML-MRC | Yes | 1·25 | c.2080C>T; p.H694Y | Missense | Yes | Yes, Significant |
| 9 | MDS/MPN | Yes | 0·1 | c.1673delG | No | Yes, Significant | |
| 10 | Other | No | 0·875 | c.490G>A; p. G164R,c.492delG and c.2187delT | No | Yes, Significant | |
| 11 | MDS-EB-I | No | 2·125 | p.R690H | Missense | Yes | Yes, Significant |
| 12 | MDS/MPN | No | 1·5 | p.R690H | Missense | Yes | Yes, Significant |
| 13 | Other | No | 0·525 | p.A685T (c.2053G>A) and p.K571E (c.1711A>G) | Missense | Yes | Yes, Significant |
| 14 | MDS/MPN | No | 1·05 | p.R690H | Missense | Yes | Yes, Significant |
| 15 | Other | Yes | 0·8 | c.2139A>G; pl713M | Missense | Yes | Probably Significant |
| 16 | MDS/MPN | No | 0·15 | p.D185H | Missense | Yes | Probably Significant |
| 17 | Other | No | 2 | D432EFS, Y741H | Missense | No | Probably Significant |
| 18 | AML-MRC | No | 1·625 | p.G663R (c.1987G>A); g.148507476C>T | Yes | Probably Significant | |
| 19 | AML-MRC | Yes | 0·27 | c.923C>G; p.P308R | Missense | No | Probably Significant |
| 20 | Other | No | 0·3 | p.D185H | 0 | Yes | Probably Significant |
| 21 | Other | No | 0·1125 | 2111-2A>G | Nonsense | Yes | Probably Significant |
| 22 | AML-MRC | No | 0·75 | p.D185H | Missense | Yes | Probably Significant |
| 23 | MDS-EB-II | No | 1 | p.D185H | Missense | Yes | Probably Significant |
| 24 | Other | No | 1 | p.D185H | Missense | Yes | Probably Significant |
| 25 | MDS-EB-I | No | 0·9 | p.D185H | Missense | Yes | Probably Significant |
| 26 | Other | No | 0·15 | p.D185H (c.553G>C) | Missense | Yes | Probably Significant |
| 27 | Other | No | 0·06 | p.Q55* (c.163C>T), | Nonsense | No | Probably Significant |
| 28 | MDS/MPN | No | 0·675 | p.N699H c.2095A>C and p.R684C c.2050C>T | Missense | Yes | Probably Significant |
| 29 | MDS/MPN | No | 1·4 | p.Q273* | Missense | Yes | Probably Significant |
| 30 | CMML | No | 0·15 | p.R313Q | Missense | Yes | Probably Significant |
| 31 | MDS/MPN | No | 0·15 | p.R690H and chr7:g148598715A>T | Missense | Yes | Probably Significant |
| 32 | Other | No | 0·3 | c.1852_6C>T | Yes | Probably not significant | |
| 33 | MDS/MPN | No | 0·45 | c.2110G>T; V704F | Missense | No | Unknown |
| 34 | Other | No | 1·5 | D730FS | Yes | Unknown | |
| 35 | AML-MRC | No | 0·075 | c.1160_1163del (p.D387Vfs*39) | Missense | No | Unknown |
| 36 | Other | No | 0·625 | p.D185H (c.553G>C) | Missense | Yes | Unknown |
| 37 | MDS/MPN | No | 0 | p.Y153* c.459T>A | No | Unknown | |
| 38 | MDS-EB-I | Yes | 0·05 | c.1547·3C>G | Unknown | ||
| 39 | MDS-EB-II | Yes | 0·7 | R882H | Unknown | ||
| 40 | Other | No | 0·8 | c.2097C>G, p.N699K | Nonsense | No | Unknown |
| 41 | Other | Yes | 0·15 | p.R33Afs*12 (c.96_97insG), | Missense | No | Unknown |
| 42 | MDS-EB-II | No | 0·1 | c.1643G>T; C548F | Missense | Yes | Unknown |
| 43 | MDS/MPN | No | 0·0125 | IVS1410 + 2T>A | No | Unknown | |
| 44 | Other | No | 0·225 | c.485·1G>C; and c.1957C>A; Q653K | No | Unknown | |
| 45 | MDS/MPN | No | 1·625 | c.2050C>T; R684C | Missense | Yes | Unknown |
| 46 | MDS-EB-II | Yes | 0·00525 | p.D193H | No | Unknown | |
| 47 | Other | No | 0 | chr7:g.148511050C>T | Yes | Unknown | |
| 48 | Other | No | 0·975 | p.Q648E and p.R502Q | Missense | Yes | Unknown |
| 49 | MDS/MPN | No | 0·01 | p.Y677H and p.F627L | Missense | No | Unknown |
| 50 | Other | Yes | 0·3 | p.D176H | Missense | Yes | Unknown |
| 51 | Other | No | 0·03 | c.293_294insT | Missense | Yes | Unknown |
| 52 | AML-MRC | Yes | 0·1875 | p.R207 | Nonsense | Yes | Unknown |
| 53 | AML-MRC | No | 1·35 | p.V674M and p.C443Y | Missense | Yes | Unknown |
| 54 | MDS/MPN | Yes | 0·1 | p.T283R | Missense | No | Unknown |
| 55 | MDS/MPN | No | 0 | c.1663del(p.C555Vfs* 123) | No | Unknown | |
| 56 | MDS-EB-I | No | 0·0875 | p.I646T (c.1937T>C), | Missense | Yes | Unknown |
| 57 | Other | No | 0·6 | p.T283M and p.V662Cfs*13 | Missense | Yes | Unknown |
| 58 | MDS/MPN | No | 0·3 | p.Y663C | No | Unknown | |
| 59 | MDS/MPN | No | 0·8 | p.S669G | No | Unknown | |
| 60 | MDS-EB-I | No | 2·25 | p.A702P | No | Unknown | |
| 61 | MDS-EB-I | No | 0·015 | p.R360Tfs*5 | No | Unknown |
AML-MRC, acute myeloid leukemia with myelodysplasia-related changes
CMML, chronic myelomonocytic leukaemia; IHC, immunohistochemistry; MDS-EB-I/II, myelodysplastic syndrome with excess blasts type I/II; MDS, myelodysplastic syndromes; MPN, myeloproliferative neoplasm; Other: MDS unclassifiable (MDS-U), MDS with multilineage dysplasia (MDS-MLD), MDS with ring sideroblasts and multilineage dysplasia (MDS-RS-MLD), MDS with single lineage dysplasia (MDS-SLD), MDS with ring sideroblasts and single lineage dysplasia (MDS-RS-SLD), therapy-related MDS (tMDS).
To apply EZH2 expression as a prognostic co-variate, it is critical to identify an IHC score cut-off point where EZH2 delineates clinical outcome. Analysis of different cut-off points did not discern OS differences; however, a larger cohort, or focusing on CD34 + stem cells might assist in uncovering the prognostic value of EZH2 expression. We demonstrated that EZH2 expression decreases after AML-MRC progression, warranting investigation in a larger cohort of transformed or therapy-related and de novo AML. As IHC is an inexpensive and rapidly discernable alternative to sequencing, we propose that EZH2 IHC should be analysed in a larger cohort to provide further insight into the differences between specific mutations and prognostication, and may prove EZH2 IHC a useful marker for discriminating disease outcome or transformation risk. As MDS prognostication moves from morphological and cytopenic characterization to more molecularly focused determinations, utility of tools such as IHC may offer definitive surrogates.
Acknowledgements
The authors would like to thank the Alpha Omega Alpha Honor Medical Society for support award given to JN. We would also like to acknowledge the histology laboratory at Moffitt Cancer Center, Helen Molina, and Jodi Balasi who assisted with immunohistochemistry.
References
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D & Ebert BL (2011) Clinical effect of point mutations in myelodysplastic syndromes. New England Journal of Medicine, 364, 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoopongse E, Yeung C, Byon J, Ramakrishnan A, Holman ZJ, Jiang PY, Yu Q, Deeg HJ & Marcondes AM (2014) The KDM2B-let-7b -EZH2 axis in myelodysplastic syndromes as a target for combined epigenetic therapy. PLoS ONE, 9, e107817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A, Li Y, Chung YR, Micol JB, Murphy ME, Cho H, Kim MK, Zebari AS, Aumann S, Park CY, Buonamici S, Smith PG, Deeg HJ, Lobry C, Aifantis I, Modis Y, Allain FH, Halene S, Bradley RK & Abdel-Wahab O (2015) SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell, 27, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw KL, Nguyen J, Komrokji RS, Sallman D, Al Ali NH, Padron E, Lancet JE, Moscinski LC, List AF & Zhang L (2016) Immunohistochemical pattern of p53 is a measure of TP53 mutation burden and adverse clinical outcome in myelodysplastic syndromes and secondary acute myeloid leukemia. Haematologica, 101, e320–e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashida G, Harada H, Matsui H, Oshima M, Yui M, Harada Y, Tanaka S, Mochizuki-Kashio M, Wang C, Saraya A, Muto T, Hayashi Y, Suzuki K, Nakajima H, Inaba T, Koseki H, Huang G, Kitamura T & Iwama A (2014) Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its pre-disposition to leukaemic transformation. Nature Communications, 5, 4177. [DOI] [PubMed] [Google Scholar]
- Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP & Ebert BL (2015) Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood, 126, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F & Li X (2012) The role of histone methyltransferase EZH2 in myelodysplastic syndromes. Expert Rev Hematol, 5, 177–185. [DOI] [PubMed] [Google Scholar]


