Abstract
Immunoglobulin heavy-chain variable region (IGHV) mutational status and karyotype abnormalities are important prognostic factors in chronic lymphocytic leukemia (CLL). The goal was to assess the impact of IGHV in CLL patients with isolated favorable genetic aberrations (del13q, trisomy 12, or negative fluorescence in situ hybridization [FISH]). We studied 273 CLL patients with both IGHV mutational status and cytogenetic information: 145 with isolated del13q 49 with sole trisomy 12 and 79 with negative FISH. After a median follow-up of 7.8 years, patients with del13q-unmutated IGHV had a shorter time to first treatment (TFT) (2.98 vs. 17.44 years; p<.001) and shorter overall survival (10.45 years vs. not reached; p=.0026). Patients with negative FISH-unmutated IGHV had shorter TFT (p=.02) (3.10 vs. 9.75 years, p=.053). IGHV status did not influence clinical outcomes in trisomy 12 CLL. In conclusion, IGHV mutational status shows prognostic impact in CLL patients with good prognosis genomic features.
Keywords: Chronic lymphocytic leukemia, IGHV, genomic abnormalities, survival
Introduction
With over 14,600 new cases reported in 2015, chronic lymphocytic leukemia (CLL) is the most common leukemia diagnosed in both the United States and western hemisphere [1,2]. Although CLL has classically been characterized as an ‘indolent’ B-cell malignancy, it has a heterogeneous clinical course and survival outcomes vary among different patients [2-4]. Staging systems focused on readily available clinical parameters have revealed a strong correlation between disease burden and overall survival (OS) (Rai and Binet staging) [5-7]. Both staging systems are still widely used in clinical practice and implemented in patient stratification for clinical trials. Nonetheless, clinical staging falls short of differentiating between rapidly evolving cases requiring earlier treatment and asymptomatic cases that remain quiescent for decades without active therapies [3,7]. Ways to analyze the mutational status of the immunoglobulin heavy-chain variable region (IGHV) and the availability of fluorescence in situ hybridization (FISH) to evaluate genomic alterations in CLL cells have improved the ability to prognosticate outcomes in this disease.
The presence of heavy-chain variable region mutations has a well-established prognostic role in CLL, with patients harboring a non-homologous IGHV sequence of >2% from the germline gene (mutated IGHV) having longer treatment-free intervals and better OS than their unmutated counterparts [8-11]. CLL patients with unmutated IGHV also have shorter disease control than CLL patients with mutated IGHV when treated with fludarabine, cyclophosphamide, and rituximab [12]. Lastly, unmutated IGHV CLL status has been correlated with higher prevalence of poor-risk features such as elevated ZAP-70 and CD38 and unfavorable mutations (i.e. del17p) [8,10,11]. More recently, the relevance of IGHV mutational status in CLL risk stratification was further bolstered when it was integrated in the CLL international prognostic score (CLL-IPI) [13]. A multivariate analysis of various established CLL prognostic factors, in 3472 treatment-naive CLL patients, identified five prognostic variables capable of strongly predicting five-year OS, with del17p/TP53 mutations, IGHV mutational status, and β2-microglobulin (β2M) having the highest impact. Importantly, patients with the IGHV 3-21 gene segment were the exception regarding the prognostic influence of IGHV status, with disease behavior similar to that shown with an unmutated clone, including higher prevalence of associated TP53 mutations and inferior survival [14-17].
Döhner et al. [18] used FISH to demonstrate that >80% of CLL patients had at least one genomic alteration at diagnosis. It was established that patients with sole del13q, trisomy 12, and normal FISH had a better OS (133, 114 and 111 months, respectively) than patients with del17p or del11q (OS of 32 and 79 months, respectively) [18]. Clinical information regarding CLL prognosis in patients with sole favorable genetic alterations and IGHV mutational status is scarce. In a retrospective analysis [4], 13 CLL patients with sole del13q mutation and unmutated IGHV had higher rates of progression, were more likely to receive CLL-directed therapies, and had inferior OS compared with patients with mutated IGHV and sole del13q. The prognostic impact of trisomy 12 as the single cytogenetic aberrancy in CLL patients, in relation to their IGHV mutational status, is uncertain. Although Döhner et al. [16] correlated this genetic aberration with a favorable treatment-free interval and prolonged survival, only 42% of patients in this genetic group harbored trisomy 12 as the only genetic aberration. In an Italian retrospective analysis, 70% of patients harboring trisomy 12 mutations showed isolated trisomy 12 (through FISH at diagnosis) as the sole genetic abnormality [19]. Interestingly, mutations in the NOTCH1 gene, which are associated with poor clinical outcomes in CLL [20,21], were significantly more prevalent in patients with isolated trisomy 12 than in those having trisomy 12 and other concomitant cytogenetics lesions (30% vs. 6%; p=.008) [19]. Also, NOTCH1 mutations were associated with unmutated IGHV genes (84%; p=.003) [17].
At diagnosis, approximately 18% of CLL patients do not have identifiable cytogenetic abnormalities [18]. Similar to trisomy 12, this CLL population has been previously classified as having an intermediate clinical prognosis [18]; however, the influence of IGHV mutational status in their prognosis has not been evaluated. Here, our aim was to analyze the prognostic impact of IGHV mutational status in newly diagnosed CLL patients with del13q, trisomy 12, or negative FISH as sole genetic aberrations.
Materials and methods
Using the total cancer care (TCC) and Moffitt cancer center (MCC) malignant hematology databases, we retrospectively identified all patients with established CLL diagnosis from January 2000 to December 2013. Data included were CD38 and ZAP-70 expression, β2M level (the upper limit of normal of β2M at MCC was 2.2 mg/dL), conventional cytogenetics, interphase FISH and IGHV mutational status, and patient demographics. IGHV status was assessed at the patient’s first evaluation at MCC. We included only patients with favorable genetic information (del13q, trisomy 12, and negative FISH) and IGHV mutational status. This study was approved by the Institutional Review Board of the University of South Florida. For this Health Insurance Portability and Accountability Act-compliant study, informed consent was not required, although patient consent was obtained prospectively prior to database inclusion.
FISH and IGHV analyses
Fluorescence in situ hybridization was done by interphase analysis utilizing probes for five centromeres: 11cen/11q22.3 (ATM), 12cen/12q15 (MDM2), 13q14 (D13S319)/13q34 (LAMP1), 17cen/17p13.1 (p53), and 11q13 (CCND1-XT)/14q32 (IGH-XT). IGHV mutational status was performed using cycle sequencing analysis. Mutated IGHV and unmutated IGHV were defined as non-homologous immunoglobulin sequence of ≥2% and <2% from the baseline sequence, respectively [9,22-24].
Statistical analyses
Patient demographic data were analyzed using descriptive statistics. Chi-square test was used to compare categorical variables between patient groups. Time-to-first treatment (TFT) was calculated as time from diagnosis to date of first prescribed antineoplastic therapy, and OS was calculated from diagnosis to the time of death or last recorded follow-up. Event time distributions were estimated by the Kaplan-Meier estimator and compared using log-rank test. Cox proportional hazard models were used to estimate hazard ratios and p values. We used SAS 9.4 software (Cary, NC) for statistical analyses.
Results
Demographics and clinical findings
Between 2000 and 2013, 1267 CLL patients were seen at our institution. Cytogenetic data were available for 601 (47%). Moreover, 338 individuals (26.5%) had IGHV mutational analyses performed and readily available. Of these, 273 CLL patients had both cytogenetic and IGHV mutational status information available: 145 patients (53.1%) had isolated del13q, 49 patients (17.9%) had isolated trisomy 12, and 79 patients (28.9%) had a negative FISH (Figure S1 in the Supplementary material). Patient demographics and clinical profile based on their IGHV status are summarized in Tables 1-3. Of 273 patients included in our study, 173 were men (63.3%) and 100 were women (36.6%) with overall median age at diagnosis of 59 years (34–83; 52% were younger than 60 years), which was similar to the median age for the whole cohort (60 years; 23–89 years). Long-term follow-up (median 7.8 years; 0.04–28.5) showed that more than half of survivors in our favorable genomic status patient group did not require CLL-directed treatment, underscoring the overall benign risk profile of this population.
Table 1.
Patient characteristics with isolated del13q (overall and per IGVH status).
| Del13q |
||||
|---|---|---|---|---|
| Overall (n=145) | IGHV<2% (n=49) | IGHV≥2% (n=96) | p Value* | |
| Age at diagnosis, median (range), years | 59 (36–83) | 58 (37–76) | 60 (36–83) | |
| Age | ||||
| Age >60 years | 71 (49.0%) | 23 (47.0%) | 48 (50.0%) | .86 |
| Age <60 years | 74 (51.0%) | 26 (53.1%) | 48 (50.0%) | |
| Sex | ||||
| Male | 91 (62.8%) | 30 (61.2%) | 61 (63.5%) | .85 |
| Female | 54 (37.2%) | 19 (38.7%) | 35 (36.4%) | |
| Modified Rai staging | ||||
| Rai 0–II | 109 (75.1%) | 30 (61.2%) | 79 (82.2%) | .063 |
| Rai III–IV | 27 (18.6%) | 13 (26.5%) | 14 (14.5%) | |
| Unknown | 9 (6.2%) | 8 (16.3%) | 12 (12.5%) | |
| ZAP-70 | ||||
| >20% | 34 (23.4%) | 21 (42.8%) | 13 (13.5%) | <.001 |
| <20% | 70 (48.2%) | 17 (34.6%) | 53 (55.2%) | |
| Unknown | 41 (28.2%) | 11 (22.4%) | 30 (31.2%) | |
| CD38 | ||||
| >30% | 22 (15.1%) | 12 (24.4%) | 10 (10.4%) | .043 |
| <30% | 92 (63.4%) | 27 (55.1%) | 65 (67.7%) | |
| Unknown | 31 (21.3%) | 10 (20.4%) | 21 (21.8%) | |
| β2M | ||||
| Elevated | 36 (24.8%) | 14 (28.5%) | 22 (22.9%) | .362 |
| Not elevated | 52 (35.8%) | 15 (30.6%) | 37 (38.5%) | |
| Unknown | 57 (39.3%) | 20 (40.8%) | 37 (38.5%) | |
| CLL treatment | ||||
| No | 80 (55.2%) | 15 (30.6%) | 65 (67.7%) | <.001 |
| Yes | 65 (44.8%) | 34 (69.3%) | 31 (32.2%) | |
| Unknown | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
β2M: β2-microglobulin; CLL: chronic lymphocytic leukemia; IGVH: immunoglobulin variable heavy chain.
A p<.05 was considered a statistical difference.
Table 3.
Patient characteristics with normal karyotype; overall and per IGVH status.
| Normal karyotype |
||||
|---|---|---|---|---|
| Overall (n=79) | IGHV<2% (n=38) | IGHV≥2% (n=41) | p Value* | |
| Age at diagnosis, median (range), years | 57 34–74 |
58 34–74 |
55 34–71 |
|
| Age | ||||
| Age >60 years | 31 (39.2%) | 17 (44.7%) | 14 (34.1% | .365 |
| Age <60 years | 48 (60.7%) | 21 (55.2%) | 27 (65.8%) | |
| Gender | ||||
| Male | 51 (64.5%) | 25 (65.7%) | 26 (63.4%) | 1 |
| Female | 28 (35.4%) | 13 (34.2%) | 15 (36.5%) | |
| Modified Rai staging | ||||
| Rai 0–II | 63 (78.4%) | 29 (76.3%) | 34 (82.9%) | .72 |
| Rai III–IV | 9 (10.1%) | 5 (13.1%) | 4 (9.7%) | |
| Unknown | 7 (11.4%) | 4 (10.5%) | 3 (7.3%) | |
| ZAP-70 | ||||
| >20% | 23 (27.8%) | 12 (31.5%) | 11 (26.8%) | .44 |
| <20% | 41 (41.7%) | 17 (44.7%) | 24 (58.5%) | |
| Unknown | 16 (30.3%) | 9 (23.6%) | 6 (14.6%) | |
| CD38 | ||||
| >30% | 20 (25.3%) | 18 (47.3%) | 12 (29.2%) | .08 |
| <30% | 37 (46.8%) | 14 (36.8%) | 23 (56.0%) | |
| Unknown | 12 (15.1%) | 6 (15.7%) | 6 (14.6%) | |
| β2M | ||||
| Elevated | 16 (20.2%) | 6 (15.7%) | 10 (24.3%) | .54 |
| Not elevated | 31 (39.2%) | 15 (39.4%) | 16 (39.0%) | |
| Unknown | 32 (40.5%) | 17 (44.7%) | 15 (36.5%) | |
| CLL treatment | ||||
| No | 40 (50.6%) | 14 (36.8%) | 26 (63.4%) | .02 |
| Yes | 39 (49.3%) | 24 (63.1%) | 15 (36.5%) | |
| Unknown | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
β2M: β2-microglobulin; CLL: chronic lymphocytic leukemia; IGVH: immunoglobulin variable heavy chain.
p<.05.
In the isolated del13q (favorable cytogenetic risk) group, 49 patients (33.7%) harbored unmutated IGHV and 96 (66.2%) had mutated status. A larger proportion of patients with unmutated IGHV had ZAP-70 (42.8% vs. 13.5% with mutated; p<.001) and CD38 overexpression (24.4% vs. 10.4% with mutated; p=.04) and showed greater likelihood to require CLL-directed therapy (69.3% vs. 32.2% with mutated; p≤.001). However, patients with mutated IGHV had a trend (p=.06) toward having a lower-risk disease by Rai staging (i.e. Rai stage 0–II; Table 1). In the isolated trisomy 12 cohort, 32 patients (65%) had unmutated and 17 patients (34.6%) had mutated IGHV, with no significant differences. In this cohort, more patients with unmutated IGHV genes had ZAP-70 overexpression at initial diagnosis, but both populations were otherwise similar (Table 2).
Table 2.
Patient characteristics with isolated trisomy 12 (overall and per IGVH status).
| Trisomy 12 |
||||
|---|---|---|---|---|
| Overall (n=49) | IGHV<2% (n=32) | IGHV≥2% (n=17) | p Value* | |
| Age at diagnosis, median (range), years | 57 34–74 |
58 34–74 |
55 34–71 |
|
| Age | ||||
| Age >60 years | 31 (39.2%) | 17 (44.7%) | 14 (34.1% | .365 |
| Age <60 years | 48 (60.7%) | 21 (55.2%) | 27 (65.8%) | |
| Gender | ||||
| Male | 51 (64.5%) | 25 (65.7%) | 26 (63.4%) | 1 |
| Female | 28 (35.4%) | 13 (34.2%) | 15 (36.5%) | |
| Modified Rai staging | ||||
| Rai 0–II | 63 (78.4%) | 29 (76.3%) | 34 (82.9%) | .72 |
| Rai III–IV | 9 (10.1%) | 5 (13.1%) | 4 (9.7%) | |
| Unknown | 7 (11.4%) | 4 (10.5%) | 3 (7.3%) | |
| ZAP-70 | ||||
| >20% | 23 (27.8%) | 12 (31.5%) | 11 (26.8%) | .44 |
| <20% | 41 (41.7%) | 17 (44.7%) | 24 (58.5%) | |
| Unknown | 16 (30.3%) | 9 (23.6%) | 6 (14.6%) | |
| CD38 | ||||
| >30% | 20 (25.3%) | 18 (47.3%) | 12 (29.2%) | .08 |
| <30% | 37 (46.8%) | 14 (36.8%) | 23 (56.0%) | |
| Unknown | 12 (15.1%) | 6 (15.7%) | 6 (14.6%) | |
| β2M | ||||
| Elevated | 16 (20.2%) | 6 (15.7%) | 10 (24.3%) | .54 |
| Not elevated | 31 (39.2%) | 15 (39.4%) | 16 (39.0%) | |
| Unknown | 32 (40.5%) | 17 (44.7%) | 15 (36.5%) | |
| CLL treatment | ||||
| No | 40 (50.6%) | 14 (36.8%) | 26 (63.4%) | .02 |
| Yes | 39 (49.3%) | 24 (63.1%) | 15 (36.5%) | |
| Unknown | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
β2M: β2-microglobulin; CLL: chronic lymphocytic leukemia; IGVH: immunoglobulin variable heavy chain.
p<.05.
Regarding the negative FISH CLL cohort, 38 patients (48%) had unmutated and 41 patients (51.8%) had mutated IGHV status. Those with an unmutated IGHV status had a higher frequency of CLL-directed therapies (63.1%) than those with mutated IGHV status (36.5%; p=.02) and a proportionally higher trend (47.3%) of CD38 overexpression at baseline than those with unmutated status (29.2%), albeit not statistically significant (p=.08; Table 3). Median TFT and OS Kaplan-Meir curves for each FISH genetic category are noted in Figure S2.
IGHV status as a prognostic factor in CLL patients with isolated del13q
The median number of therapies among unmutated IGHV patients was 1 (0–4), with allogeneic hematopoietic cell transplantation (allo-HCT) used in four patients. Patients with mutated IGHV received 0.5 median treatments (0–4), with three patients requiring allo-HCT. The median TFT of patients harboring an isolated de13q was 6.4 years (95% CI, 4.51–13.06), with TFT being significantly shorter for patients with unmutated IGHV (2.98 years; 95% CI, 1.34–4.13) than with mutated IGHV genes (17.44 years; 95% CI, 6.86 to not reached; p<.001), with a hazard ratio (HR) of 3.41 (95% CI, 2.07–5.26, p<.001) (Figure 1 and Table S1 in the Supplementary material). In a preplanned multivariable analysis of different baseline prognostic risk factors, unmutated IGHV status prevailed as an independent risk factor for shorter TFT in CLL patients (HR 10.7; 95% CI, 2.97–38.6; p<.001). Other risk factors that significantly correlated with earlier treatment were CD38 (p=.003) and ZAP-70 positive (p=.024) (Tables S2 and S3 in the Supplementary material).
Figure 1.
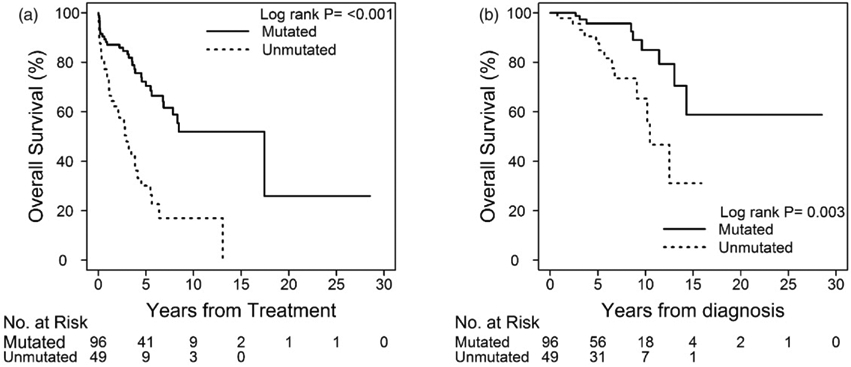
Time to first treatment and overall survival of patients with isolated del13q based on IGVH mutational status.
In patients harboring sole del13q, the median OS for the whole group was 14.32 years (95% CI, 12.50 to not reached); a higher OS was shown in patients with mutated IGHV, with OS not reached at the time of this analysis (95% CI, 13.04 years to not reached), versus that shown in patients with unmutated IGHV (10.45 years; 95% CI, 9.10 to not reached; p=.0026) (Figure 1). A univariate analysis showed that unmutated IGHV status was strongly associated with worse survival in patients with isolated del13q (HR for unmutated IGHV status was 3.48; 95% CI, 1.469–8.240; p=.0046) (Table S1 in the Supplementary material). Nonetheless, when several baseline CLL prognostic risk factors were analyzed in a multivariate model, none of the variables correlated with OS. Of the patients who died in this group, 13 had unmutated IGHV and 9 had mutated IGHV, although none harbored IGHV 3–21 mutations [14,15]. All patients who died received at least one line of CLL-directed therapy, and none of them had Richter transformation. However, most patients had documented disease progression at time of death.
IGHV mutation as a prognostic factor for CLL with isolated trisomy 12 and negative FISH
We grouped CLL patients with either isolated trisomy 12 or negative FISH under the intermediate cytogenetic risk category based on the established prognostic role of these genetic abnormalities in this disease [7,18]. Among patients with sole trisomy 12, the median number of CLL-directed treatments was similar in both IGHV groups (median of 1; 0–3), and none with sole trisomy 12 underwent salvage allo-HCT or experienced Richter transformation. The median TFT for patients with sole trisomy 12 was 3.74 years (95% CI, 2.17–7.95). Of note, in this isolated abnormality group, there were no significant differences in TFT between those with unmutated (6.06 years; 95% CI, 1.88–7.70) versus those with mutated IGHV (3.74 years; 95% CI, 0.28 to not reached; p=.551), with a HR of 1.29 (95% CI, 0.56–2.95) for unmutated IGHV (Figure 2). In addition, median OS for patients with sole trisomy 12 was 12.78 years (95% CI, 11.50 years to not reached) without statistical differences between unmutated (median OS not reached) and mutated IGHV CLL (median OS of 12.78 years; p=.28). Univariate analysis did not show any significant differences in TFT or OS based on the IGHV mutational status (Table S1 in the Supplementary material). Nine patients in this cytogenetic category succumbed to their disease (five with unmutated and four with mutated IGHV, although none with IGHV 3–21 mutations) [14,15].
Figure 2.
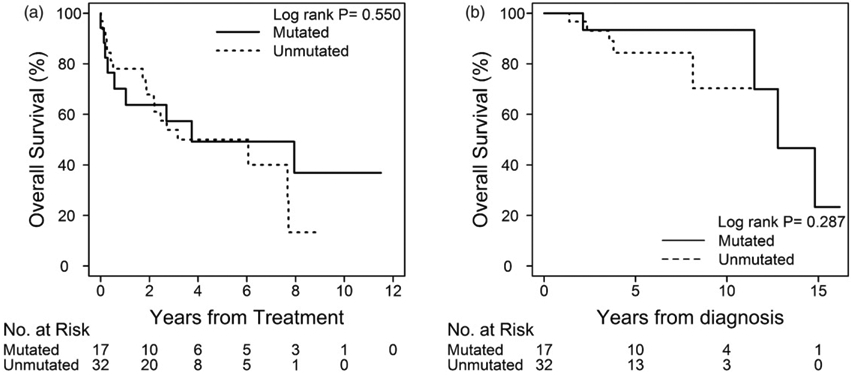
Time to first treatment and overall survival of patients with isolated trisomy 12q based on IGVH mutational status.
The median number of CLL treatments received by patients without identifiable cytogenetic abnormalities by FISH (isolated normal karyotype) was 1 (1–4 treatments). Within this group, four patients underwent allo-HCT (two with unmutated and two with mutated IGHV). Three of these patients were alive at the time of our data analyses. Of note, the patient who died after allo-HCT harbored unmutated IGHV and experienced clonal evolution by acquiring del17p mutation after the first administered chemoimmunotherapy. The median TFT of patients with negative FISH was 4.20 years (95% CI, 3.01–10.16 years), with shorter intervals in patients with unmutated (3.10 years, 95% CI, 2.37–4.42) versus mutated IGHV (9.75 years, 95% CI, 3.27–21.26; p=.053). Univariate analysis showed HR of 1.909 (95% CI, 0.979–3.720) for unmutated IGHV (Figure 3 and Table S1 in the Supplementary material). Multivariate analysis showed that CD38 overexpression and low serum albumin levels at diagnosis (defined as ≤3.7 mg/dL) were independent risk factors for shorter TFT in this group; however, unmutated IGHV was not identified as an individual prognostic variable in this model (Tables S2 and S3 in the Supplementary material). The median OS for these patients had not been reached at the time of data analyses, although the 10-year OS of those with mutated IGHV (88%) versus unmutated IGHV (78%) in this genetic subgroup did not differ significantly (HR of 3.13, 95% CI, 0.631–15.5; p=.162). Of the nine patients who died in this group, two had mutated IGHV, although without IGHV 3–21 [14,15]. Two patients never received CLL treatment before they died (one from stage IV non-small cell lung carcinoma and one from unknown reasons). Also, none of the patients in this group had documented Richter transformation.
Figure 3.
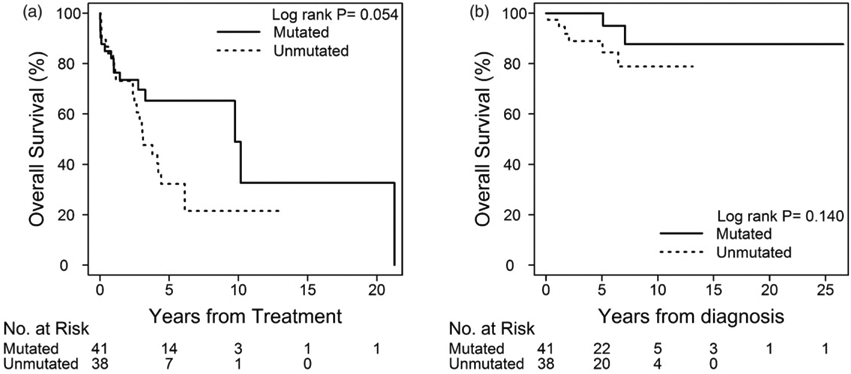
Time to first treatment and overall survival of patients with normal karyotype based on IGVH mutational status.
Discussion
During the past decades, the prognostication of CLL has evolved, especially with the integration of traditional prognostic markers with modern technologies (i.e. genomics) [3,5,6,9,18,24,25]. CLL cytogenetics, classically assessed by FISH and karyotyping, and IGHV mutational status determined by PCR are probably the most powerful and validated clinical prognostic biomarkers used in our daily practice [2,3,18,23]. CLL patients with unmutated IGHV usually have concomitant cytogenetic aberrations that have been strongly associated with dismal CLL-related outcomes, such as del11q and del17p [18,23,26]. On the other hand, del13q has classically been correlated with mutated IGHV [4,23,26].
As previously noted, reports are scarce on the prognostic relation between isolated good (del13q) and intermediate-risk cytogenetic aberrations (trisomy 12 or negative FISH) and the mutational changes of the IGHV homology sequence in CLL. To our knowledge, only one retrospective study has explored the prognostic relevance of IGHV mutational changes in CLL with sole del13q. Gladstone et al. retrospectively identified 47 CLL patients with concomitant isolated del13q and available IGHV mutational status (34 with mutated and 13 with unmutated IGHV) [4]. Our report, which comprised three times as many patients with the above characteristics, demonstrated prevalence of unmutated IGHV of 33% (49/145), which is slightly higher than previously reported [4,26]. Importantly, although this cohort of patients had a fairly indolent disease course, those with unmutated IGHV were more likely to receive treatment for CLL and treatment was initiated significantly earlier, as previously reported [4]. The most clinically relevant association observed in this population was the dismal prognosis that unmutated IGHV confers on patients with isolated del13q. Indeed, 8 of 10 CLL patients with this mutational aberration harboring a mutated IGHV gene were alive 10 years after diagnosis, whereas more than half of those with unmutated IGHV had died. Although our study had a longer follow-up, our findings are in accordance with those previously observed [4]. Nonetheless, we could only identify IGHV as an independent prognostic factor for TFT but not for OS. This is in line with findings reported by other groups that found that IGHV mutational status in CLL patients also failed to independently prognosticate OS [4]. Although elevated CD38 overexpression and advanced Rai stage at diagnosis were strongly associated with a shorter TFT, no other baseline variable independently predicted mortality in patients harboring del13q. As such, patients with del13q included in our study were younger at diagnosis than usual; albeit not significantly, with the proportion of unmutated IGHV patients with isolated del13q who were <60 years old being slightly higher. This observation, which was also noted in a previous retrospective analysis, may be explained by referral bias [4,26]. At least one large report described a significantly higher incidence of unmutated IGHV in CLL patients 55 years or younger who also had a worse OS compared with a normal population group matched by sex and age [1,4,27,28]. These observations are pertinent because, despite those with isolated del13q CLL carrying a better clinical outlook as a whole, younger patients may be prone to having more genotoxic treatments in the pursuit of long-lasting remission [29,30]. In turn, these patients could develop a molecularly unstable disease, thus perhaps generating high-risk CLL clones linked to dismal outcomes [31-33].
Although controversial, CLL patients with sole trisomy 12 or without identifiable cytogenetic changes by FISH are classified under an intermediate-risk group based on the seminal work of Döhner et al., which was reiterated in a more contemporary analysis by Rossi et al. [18,34]. The prognostic influence of the IGHV mutational status in these specific cytogenetic risk subgroups has not been previously reported. While the sole trisomy 12 group was acknowledged as the smallest one in sample size, the prevalence of this isolated mutation (18%) was similar to that reported in previous larger studies of newly diagnosed CLL patients [35]. Although patients with unmutated IGHV phenotype expressing higher ZAP-70 levels at diagnosis were slightly more prevalent in this cohort, there were no significant differences among the groups. Nonetheless, patients with unmutated or mutated IGHV genes harboring trisomy 12 as the only CLL-related genetic aberration showed a fairly indolent disease, with OS in excess of 10 years.
It is possible that outcomes of this CLL population might be dictated by more complex biologic factors other than IGHV mutational status. The prognosis of trisomy 12 CLL may also be influenced by its allele the burden. It seems that worse outcomes are seen if more than 60% CLL cells harbor trisomy 12, especially in the presence of 11q deletion [36]. For instance, NOTCH1 mutations occur in approximately 30% of CLL patients harboring an isolated trisomy 12 [19]. Mutations in the PEST domain of the key ligand-activated transcription factor of the NOTCH signaling pathway, NOTCH1, were among the first novel molecular aberrations discovered by next-generation sequencing (NGS) in CLL [25,37], and it correlates with early disease progression, rituximab-based therapy refractoriness, Richter transformation, and shorter survival [21,37-40].
Patients clustered on the other arm of the intermediate-risk subgroup had no identifiable CLL-related cytogenetic aberrations. Interestingly, they had the longest survival of the three groups. Moreover the IGHV mutational profile had no prognostic influence on survival of these patients. Conversely, patients with negative FISH CLL harboring unmutated IGHV genes tended to have earlier disease progression and were more likely to require specific CLL treatment faster. Using FISH, which is currently recommended by several CLL clinical guidelines to assess baseline cytogenetics [2,3,41,42], approximately 20% of newly diagnosed patients lack mutational aberrations [18]. Apparently, the prognosis of these patients is probably determined by other molecular signatures. Thus, Jeromin et al. [43] analyzed a large cohort of untreated CLL patients for several genetic markers (including SF3B1, NOTCH1, FBXW7, MYD88) using direct Sanger sequencing and NGS. In patients with negative FISH only, mutations in the spliceosome protein SF3B1 were frequent (15% of patients), whereas NOTCH1 mutations rarely occurred. In multivariate analyses both SF3B1 mutations and IGHV mutational status had independent prognostic impact in TFT and OS. Further investigations into the prognostic influence of IGHV mutational status in CLL patients carrying novel described mutations (i.e. BIRC3) may help in understanding the prognosis of this patient subgroup.
Our report has several limitations. First, this was a single center retrospective study, and our results may be affected by an inherent referral bias. However, most patients had an overall low-risk disease profile and were not heavily pretreated before referral. Also, their community oncologists consistently followed them throughout the study period, including having occasional visits to our center. Therefore, our results may more accurately portray ‘real-world’ disease progression, reaffirming the biologic relevant relation between IGHV and these cytogenetic groups, especially for del13q. Also, the relatively small sample size of certain cytogenetic risk groups (i.e. isolated trisomy 12), and thus low incidence of the measured outcomes, may have underestimated some of the studied endpoints. Although missing information is an inherent deficiency of large retrospective cohorts where data from multiple sources is reviewed, it was also quite informative because it gave us a scope of the current prognostic stratification practices in CLL performed both in the community at large and at our own institution. This is much more relevant at the present time with the advent of the CLL-IPI score, which accentuates the prognostic relevance of both IGHV mutational status and β2M level [13].
In conclusion, our study further expands the evidence suggesting that CLL with isolated good cytogenetic aberrations (del13q) and unmutated IGHV could represent a different disease subgroup characterized by worse biologic characteristics and more dreadful clinical outcomes. Although we could only detect a possible association between IGHV mutations and the need for earlier treatment in patients with negative FISH, the biologic effects of the IGHV status did not alter survival outcomes in patients within the intermediate cytogenetic group. Backed by a substantial amount of evidence confirming the prognostic importance of IGHV [13,29,30,44] and the usefulness of β2M [13,45], we propose that both tests should be included in the initial diagnostic work-up of all CLL patients, along with the already recommended FISH-based cytogenetic analysis [13]. The routine use of this prognostic work-up in newly diagnosed CLL patients should be widely expanded to the community, and its cost-effectiveness should be studied in real-world settings. Also, further research on IGHV mutational status distribution and on its prognostic impact in CLL patients with well-established poor prognostic recurrent mutations (including, NOTCH1, TP53, SF3B1, ATM, BIRC3), as detected through NGS, is needed [25]. Expanding this molecular knowledge might be especially relevant in patients with either isolated trisomy 12 or normal cytogenetics. These studies could potentially be carried though multi-institutional collaborations, utilizing prospective comprehensive prognostic information from phase 3 clinical studies and/or robust single center databases.
Supplementary Material
Acknowledgements
We thank Rasa Hamilton (Moffitt Cancer Center) for editorial assistance.
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article online at https://doi.org/10.1080/10428194.2017.1323271.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- [2].Nabhan C, Rosen ST. Chronic lymphocytic leukemia: a clinical review. JAMA. 2014;312:2265–2276. [DOI] [PubMed] [Google Scholar]
- [3].Nabhan C, Raca G, Wang YL. Predicting prognosis in chronic lymphocytic leukemia in the contemporary era. JAMA Oncol. 2015;1:965–974. [DOI] [PubMed] [Google Scholar]
- [4].Gladstone DE, Swinnen L, Kasamon Y, et al. Importance of immunoglobulin heavy chain variable region mutational status in del(13q) chronic lymphocytic leukemia. Leuk Lymphoma. 2011;52:1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rai KR, Sawitsky A, Cronkite EP, et al. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46:219–234.1139039 [Google Scholar]
- [6].Binet JL, Lepoprier M, Dighiero G, et al. A clinical staging system for chronic lymphocytic leukemia: prognostic significance. Cancer. 1977;40:855–864. [DOI] [PubMed] [Google Scholar]
- [7].Shanafelt TD, Geyer SM, Kay NE. Prognosis at diagnosis: integrating molecular biologic insights into clinical practice for patients with CLL. Blood. 2004;103: 1202–1210. [DOI] [PubMed] [Google Scholar]
- [8].Morilla A, Gonzalez de Castro D, Del Giudice I, et al. Combinations of ZAP-70, CD38 and IGHV mutational status as predictors of time to first treatment in CLL. Leuk Lymphoma. 2008;49:2108–2115. [DOI] [PubMed] [Google Scholar]
- [9].Hamblin TJ, Davis Z, Gardiner A, et al. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- [10].Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. [DOI] [PubMed] [Google Scholar]
- [11].Rassenti LZ, Jain S, Keating MJ, et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112:1923–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lin KI, Tam CS, Keating MJ, et al. Relevance of the immunoglobulin VH somatic mutation status in patients with chronic lymphocytic leukemia treated with fludarabine, cyclophosphamide, and rituximab (FCR) or related chemoimmunotherapy regimens. Blood. 2009;113:3168–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].An international prognostic index for patients with chronic lymphocytic leukaemia (CLL–IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016;17:779–790. [DOI] [PubMed] [Google Scholar]
- [14].Tobin G, Thunberg U, Johnson A, et al. Somatically mutated Ig V(H)3-21 genes characterize a new subset of chronic lymphocytic leukemia. Blood. 2002;99:2262–2264. [DOI] [PubMed] [Google Scholar]
- [15].Lin K, Manocha S, Harris RJ, et al. High frequency of p53 dysfunction and low level of VH mutation in chronic lymphocytic leukemia patients using the VH3-21 gene segment. Blood. 2003;102:1145–1146. [DOI] [PubMed] [Google Scholar]
- [16].Gonzalez-Gascon YMI, Hernandez JA, Martin A, et al. Mutation status and immunoglobulin gene rearrangements in patients from northwest and central region of Spain with chronic lymphocytic leukemia. Biomed Res Int. 2014;2014:257517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hernandez JA, Rodriguez AE, Gonzalez M, et al. A high number of losses in 13q14 chromosome band is associated with a worse outcome and biological differences in patients with B-cell chronic lymphoid leukemia. Haematologica. 2009;94:364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. [DOI] [PubMed] [Google Scholar]
- [19].Del Giudice I, Rossi D, Chiaretti S, et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2012;97:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sportoletti P, Baldoni S, Cavalli L, et al. NOTCH1 PEST domain mutation is an adverse prognostic factor in B-CLL. Br J Haematol. 2010;151:404–406. [DOI] [PubMed] [Google Scholar]
- [21].Weissmann S, Roller A, Jeromin S, et al. Prognostic impact and landscape of NOTCH1 mutations in chronic lymphocytic leukemia (CLL): a study on 852 patients. Leukemia. 2013;27:2393–2396. [DOI] [PubMed] [Google Scholar]
- [22].Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- [23].Kharfan-Dabaja MA, Chavez JC, Khorfan KA, et al. Clinical and therapeutic implications of the mutational status of IgVH in patients with chronic lymphocytic leukemia. Cancer. 2008;113:897–906. [DOI] [PubMed] [Google Scholar]
- [24].Trojani A, Montillo M, Nichelatti M, et al. ZAP-70, IgVh, and cytogenetics for assessing prognosis in chronic lymphocytic leukemia. Cancer Biomark. 2010;6:1–9. [DOI] [PubMed] [Google Scholar]
- [25].Guieze R, Wu CJ. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood. 2015;126:445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Krober A, Seiler T, Benner A, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- [27].Van Dyke DL, Shanafelt TD, Call TG, et al. A comprehensive evaluation of the prognostic significance of 13q deletions in patients with B-chronic lymphocytic leukaemia. Br J Haematol. 2010;148:544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Parikh SA, Rabe KG, Kay NE, et al. Chronic lymphocytic leukemia in young (</= 55 years) patients: a comprehensive analysis of prognostic factors and outcomes. Haematologica. 2014;99:140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Thompson PA, Tam CS, O'Brien SM, et al. Fludarabine, cyclophosphamide, and rituximab treatment achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood. 2016;127:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fischer K, Bahlo J, Fink AM, et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood. 2016;127:208–215. [DOI] [PubMed] [Google Scholar]
- [31].Stilgenbauer S, Sander S, Bullinger L, et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92:1242–1245. [DOI] [PubMed] [Google Scholar]
- [32].Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rodriguez D, Bretones G, Arango JR, et al. Molecular pathogenesis of CLL and its evolution. Int J Hematol. 2015;101:219–228. [DOI] [PubMed] [Google Scholar]
- [34].Rossi D, Spina V, Bomben R, et al. Association between molecular lesions and specific B-cell receptor subsets in chronic lymphocytic leukemia. Blood. 2013;121:4902–4905. [DOI] [PubMed] [Google Scholar]
- [35].Dohner H, Stilgenbauer S, Dohner K, et al. Chromosome aberrations in B-cell chronic lymphocytic leukemia: reassessment based on molecular cytogenetic analysis. J Mol Med (Berl). 1999;77:266–281. [DOI] [PubMed] [Google Scholar]
- [36].Gonzalez-Gascon YMI, Hernandez-Sanchez M, Rodriguez-Vicente AE, et al. A high proportion of cells carrying trisomy 12 is associated with a worse outcome in patients with chronic lymphocytic leukemia. Hematol Oncol. 2016;34:84–92. [DOI] [PubMed] [Google Scholar]
- [37].Fabbri G, Rasi S, Rossi D, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208:1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gianfelici V Activation of the NOTCH1 pathway in chronic lymphocytic leukemia. Haematologica. 2012;97:328–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bo MD, Del Principe MI, Pozzo F, et al. NOTCH1 mutations identify a chronic lymphocytic leukemia patient subset with worse prognosis in the setting of a rituximab-based induction and consolidation treatment. Ann Hematol. 2014;93:1765–1774. [DOI] [PubMed] [Google Scholar]
- [40].Stilgenbauer S, Schnaiter A, Paschka P, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123:3247–3254. [DOI] [PubMed] [Google Scholar]
- [41].Eichhorst B, Robak T, Montserrat E, et al. Chronic lymphocytic leukaemia: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26 Suppl 5:v78–v84. [DOI] [PubMed] [Google Scholar]
- [42].Zelenetz AD, Gordon LI, Wierda WG, et al. Chronic lymphocytic leukemia/small lymphocytic lymphoma, version 1.2015. J Natl Compr Canc Netw. 2015;13:326–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jeromin S, Weissmann S, Haferlach C, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28:108–117. [DOI] [PubMed] [Google Scholar]
- [44].Woyach JA. FCR holds up to the test of time: CLL8 follow-up. Blood. 2016;127:172–173. [DOI] [PubMed] [Google Scholar]
- [45].Thompson PA, O'Brien SM, Xiao L, et al. beta2-microglobulin normalization within 6 months of ibrutinib-based treatment is associated with superior progression-free survival in patients with chronic lymphocytic leukemia. Cancer. 2016;122:565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


