The objective of this study was to characterize the etiological role of human adenovirus (HAdV) serotypes in pediatric gastroenteritis. Using a case-control design, we compared the frequencies of HAdV serotypes between children with ≥3 episodes of vomiting or diarrhea within 24 h and <7 days of symptoms (i.e., cases) and those with no infectious symptoms (i.e., controls). Stool samples and/or rectal swabs underwent molecular serotyping with cycle threshold (Ct) values provided by multiplex real-time reverse transcription-PCR testing.
KEYWORDS: molecular epidemiology, clinical findings, adenovirus, pediatric, emergency, gastroenteritis
ABSTRACT
The objective of this study was to characterize the etiological role of human adenovirus (HAdV) serotypes in pediatric gastroenteritis. Using a case-control design, we compared the frequencies of HAdV serotypes between children with ≥3 episodes of vomiting or diarrhea within 24 h and <7 days of symptoms (i.e., cases) and those with no infectious symptoms (i.e., controls). Stool samples and/or rectal swabs underwent molecular serotyping with cycle threshold (Ct) values provided by multiplex real-time reverse transcription-PCR testing. Cases without respiratory symptoms were analyzed to calculate the proportion of disease attributed to individual HAdV serotypes (i.e., attributable fraction). Between December 2014 and August 2018, adenoviruses were detected in 18.8% (629/3,347) of cases and 7.2% (97/1,355) of controls, a difference of 11.6% (95% confidence interval [CI], 9.6%, 13.5%). In 96% (95% CI, 92 to 98%) of HAdV F40/41 detections, the symptoms could be attributed to the identified serotype; when serotypes C1, C2, C5, and C6 were detected, they were responsible for symptoms in 52% (95% CI, 12 to 73%). Ct values were lower among cases than among controls (P < 0.001). HAdV F40/41, C2, and C1 accounted for 59.7% (279/467), 17.6% (82/467), and 12.0% (56/467) of all typed cases, respectively. Among cases, Ct values were lower for F40/41 serotypes than for non-F40/41 serotypes (P < 0.001). HAdV F40/41 serotypes account for the majority of HAdV-positive gastroenteritis cases, and when detected, disease is almost always attributed to infection with these pathogens. Non-F40/41 HAdV species have a higher frequency of asymptomatic infection and may not necessarily explain gastroenteritis symptoms. Real-time quantitative PCR may be useful in differentiating asymptomatic shedding from active infection.
INTRODUCTION
Human adenoviruses (HAdVs) are nonenveloped, double-stranded DNA viruses classified into seven species (A to G). HAdVs are assigned to over 100 types, many of which were initially defined based on neutralizing serology typing. The spectrum of HAdV infection depends on the species and type of agent and the age and immune status of the host, with clinical syndromes ranging from mild to severe respiratory, gastrointestinal, or urinary tract disease to fulminant disseminated infections (1).
Serological surveys show that antibodies to HAdV serotypes 1, 2, and 5 are most common in children. However, it has been over 35 years since F species serotypes were identified as causing acute gastroenteritis (AGE) (2–5). HAdV serotypes belonging to subgroups A, B, C, D, and G have been identified in patients with diarrhea; however, their etiological roles are unclear (3). Molecular test interpretation is further complicated by the high incidence of prolonged fecal shedding (6) and asymptomatic infection by HAdV (6, 7). In a Chinese study that included 273 children with diarrhea and 361 healthy controls, HAdVs were detected in 29% of cases and 7% of controls (3). In another study, conducted in India, in which 1,053 stool specimens from children with diarrhea and 1,035 asymptomatic control specimens were analyzed, 5% and 2% were positive for HAdV 40 and 41, respectively (8). Thus, given these widely varying detection rates in symptomatic and asymptomatic children, a better understanding of the epidemiology of HAdV serotypes associated with pediatric AGE is needed.
MATERIALS AND METHODS
Study design, setting, and population.
Children with AGE (i.e., cases) and asymptomatic controls were recruited as part of a prospective cohort study (9). Research Ethics Board approvals were obtained along with informed consent and assent, as appropriate.
Cases were recruited in the emergency departments (EDs) of two pediatric tertiary care institutions in Alberta, Canada. AGE cases managed at home by caregivers, following a recommendation from a province-wide, triage telephone advice resource operated by nurses (10), were asked if their contact information could be shared with the study team. Those who agreed were contacted and recruited.
Cases met the following criteria: <18.0 years of age, ≥3 episodes of vomiting and/or diarrhea within a 24-h period, and <7 days of symptoms (11). Children were excluded if they had been enrolled during the prior 14 days, presented with an acute mental health concern, had a known neutrophil count of <1.0 × 109 neutrophils/liter, or needed emergent medical interventions.
Asymptomatic controls were recruited in the same EDs and in a public health clinic. Eligible controls were <18.0 years old, agreed to provide a stool specimen, and were free of the following symptoms for >7 days: vomiting, diarrhea, fever, rhinorrhea, and cough. Data were collected by research assistants who administered a structured questionnaire at enrollment and again 14 days later.
Objectives and outcome measures.
The study’s primary objectives were to identify and compare the frequencies of HAdV types detected between cases and controls and to assess the likelihood that case AGE symptoms are due to the HAdV identified (i.e., attributable fraction [AF]). The AF quantifies the proportion of patients in a cohort in whom a specific HAdV type was detected, whose AGE symptoms are in fact due to that specific HAdV type. Secondary objectives were to compare stool viral concentrations, codetected pathogens, and illness severity between HAdV 40/41 and non-40/41 cases.
Definitions.
The estimation of the viral concentration in stool was based on the cycle threshold (Ct) values determined using an in-house real-time reverse transcription-quantitative PCR (RT-qPCR) gastroenteritis virus panel (GVP) (12). Illness severity was quantified by employing the total-illness modified Vesikari scale (MVS) score, which ranges from 0 to 20, with higher scores indicating more severe disease (13, 14).
Specimen collection and laboratory methodology. (i) ED cases.
Two flocked swabs (FLOQSwab; Copan Italia, Italy) were inserted sequentially into the rectum and rotated 360° once. One swab was placed into a dry, sterile tube, and the other was inserted into 2 ml of modified Cary-Blair transport medium (FecalSwab; Copan Italia, Italy). Stool specimens were collected in sterile containers (catalog number V302-F; Starplex Scientific Inc., Ontario, Canada). If a stool specimen was not provided before ED discharge, caregivers obtained stool at home and samples were transported to the laboratory by couriers within 24 hours of collection.
(ii) Home care cases.
Stool collection kits containing two rectal swabs, a stool container, and instructions were couriered to the home. Caregivers collected the first stool sample produced and performed the rectal swabs at the same time.
(iii) Controls.
Stool specimens were collected in sterile containers (catalog number V302-F; Starplex Scientific Inc., Ontario, Canada). If a stool specimen was not provided before ED/clinic discharge, caregivers were requested to collect a specimen at home.
Specimen processing.
All testing was performed by staff who were blind to clinical data.
(i) Culture.
Upon receipt at the laboratory, enteric bacterial culture was performed on the stool specimens and rectal swab specimens collected in modified Cary-Blair transport medium as previously described (15).
(ii) Molecular diagnostics.
Stool specimens were stored at −80°C until tested. Immediately before nucleic acid extraction, stool specimens were thawed and aliquoted. Specimen preparation and nucleic acid extraction were subsequently performed as previously described (16). Molecular testing was then performed by employing the Luminex xTAG gastrointestinal pathogen panel (GPP) (Luminex Molecular Diagnostics, Austin, TX, USA) (17) and an in-house RT-qPCR GVP as previously described (16).
The GPP is designed to detect adenovirus 40/41, Campylobacter, Clostridioides difficile, Cryptosporidium, Entamoeba histolytica, Escherichia coli O157, enterotoxigenic Escherichia coli, Giardia, norovirus GI and GII, rotavirus, Salmonella, Shiga toxin-producing Escherichia coli, Shigella, Vibrio cholerae, and Yersinia enterocolitica. The in-house two-step RT-qPCR GVP includes a reverse transcription step using 5 μl of nucleic acid extracts and SuperScript II (Thermo Fisher Scientific, Burlington, ON, Canada), followed by three simultaneous duplex qPCRs (the duplex format for generic HAdV and rotavirus, norovirus GI and GII, and astrovirus and sapovirus) on a single run (18). This approach has high accuracy for a broad range of HAdV serotypes with the exception of serotype 31 when it is present at low concentrations (19).
(iii) Adenovirus amplification and typing.
All HAdV GPP-positive specimens were assumed to be F40/41 (20) according to the manufacturer’s insert. Samples that tested HAdV positive by the GVP assay and negative by the GPP were serotyped, as they were presumed to be non-F40/41. A primer set (21) targeting hypervariable region 7 of the hexon gene that detects all known HAdV serotypes and allows molecular serotype determination by sequence analysis was used for PCR amplification. The AmpliTaq Gold fast PCR master mix (Thermo Fisher Scientific, USA) was combined with a final concentration of 0.5 μM forward and reverse primers and 5 μl of the template. Amplification was performed in a Veriti fast thermal cycler (Applied Biosystems, USA) using a denaturation step at 95°C for 10 min, followed by 37 cycles of denaturation at 96°C for 3 s, annealing at 51°C for 3 s, and amplification at 68°C for 5 s. The final extension step was performed at 72°C for 5 min. After gel electrophoresis and cleaning of the 640-bp PCR product using the E.Z.N.A. gel extraction kit or the Cycle Pure kit (Omega Bio-Tek, Norcross, GA, USA), as appropriate, we performed Sanger sequencing using the BigDye Terminator v3.1 cycle sequencing kit and the BigDye XTerminator purification kit (Life Technologies) on the 3500xl genetic analyzer (ABI).
(iv) Bioinformatic and phylogenetic analyses.
Sequence quality was analyzed using Sequencing Analysis v6.0 (Applied Biosystems, USA), and sequences were compared to those in the nonredundant NCBI database using BLAST (22) for species determination.
Statistical analysis.
Participants were considered positive for a given pathogen if either the swab or stool specimen tested positive using any assay. All positive results were assumed to be true positives, except for Clostridioides difficile when detected in children <2 years of age, for which they were classified as negative.
We performed multivariable logistic regression on case-control data to generate odds ratios (ORs) for HAdV serotypes grouped by species to examine the association of detection with AGE symptoms. The OR for each adenovirus species was adjusted for other adenovirus species detected in cases and controls, adenovirus detections for which typing was not available (i.e., unsuccessful due to an inadequate viral load in the specimen), codetected pathogens (astrovirus, norovirus, rotavirus, sapovirus, C. difficile, and other bacteria and parasites), participant age (using standard and quadratic terms), geographic location (Calgary, Edmonton, or other), and year and season of enrollment. Because many adenovirus serotypes can cause respiratory symptoms, and since control participants were required to have no infectious symptoms, cases with rhinorrhea or cough (i.e., respiratory symptoms) were excluded from this analysis. For each detected adenovirus species, the AFs and 95% confidence intervals (CIs) were calculated by employing the ORs derived from the above-mentioned model.
Two-tailed P values of <0.05 were considered statistically significant. Analyses were conducted using SPSS version 25.0 (IBM Corp., Armonk, NY).
RESULTS
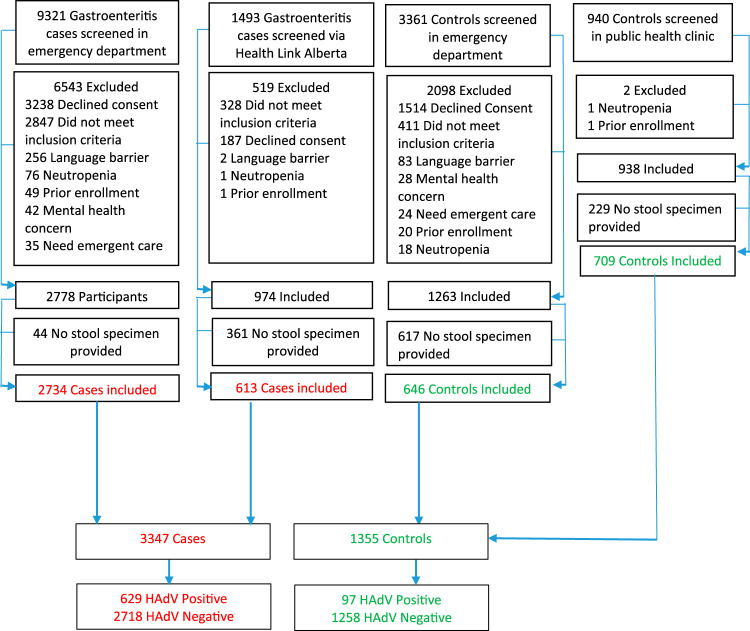
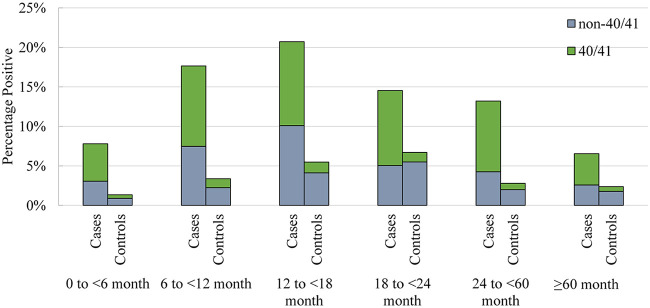
Between December 2014 and August 2018, 1,355 controls and 3,347 cases were enrolled (Fig. 1). Overall, 18.8% (629/3,347) of the cases and 7.2% (97/1,355) of the controls were positive for HAdV (Table 1). The agreement between stool specimens and rectal swabs among AGE cases, assessed using the kappa statistic, was substantial (0.78 [95% CI, 0.75, 0.82]) (23). Positivity peaked among AGE cases between 12 and 18 months and among controls between 18 and 24 months of age (Fig. 2).
FIG 1.
Study participants.
TABLE 1.
Summary information on study participants based on acute gastroenteritis case and control statusa
| Parameter | Value |
|||||
|---|---|---|---|---|---|---|
| HAdV-negative cases (n = 2,718) | HAdV-positive cases (n = 629) | P value comparing cases | HAdV-negative controls (n = 1,258) | HAdV-positive controls (N = 97) | P value comparing controls | |
| Median age (mo) (IQR) | 19.7 (10.1, 45.2) | 16.5 (11.1, 30.6) | 0.002 | 12.1 (4.1, 31.1) | 18.2 (12.1, 38.3) | <0.001 |
| No. of males (%) | 1,437 (52.9) | 369 (58.7) | 0.009 | 665 (52.9) | 57 (58.8) | 0.29 |
| No. of cases in enrollment season (%) | 0.006 | 0.20 | ||||
| January–March | 686 (25.2) | 142 (22.6) | 0.17 | 222 (17.6) | 25 (25.8) | 0.06 |
| April–June | 879 (32.3) | 172 (27.3) | 0.02 | 298 (23.7) | 22 (22.7) | 0.90 |
| July–September | 536 (19.7) | 144 (22.9) | 0.08 | 380 (30.2) | 23 (23.7) | 0.21 |
| October–December | 617 (22.7) | 171 (27.2) | 0.02 | 358 (28.5) | 27 (27.8) | 0.91 |
| No. of HAdV-positive cases (%) | ||||||
| Overall | NA | 629 (100) | NA | 97 (100) | NA | |
| GPP positive | NA | 271 (43.1) | NA | 10 (10.3) | NA | |
| GVP positive | NA | 629 (100) | NA | 97 (100) | NA | |
| GPP positive only | NA | 0 (0) | NA | 0 (0) | NA | |
| GVP positive only | NA | 358 (56.9) | NA | 87 (89.7) | NA | |
| Both GPP and GVP positive | NA | 271 (43.1) | NA | 10 (10.3) | NA | |
| Median Ct value (IQR) | NA | 22.8 (14.5, 32.7) | NA | 32.3 (29.5, 34.2) | NA | |
HAdV, human adenovirus; GPP, gastrointestinal pathogen panel; GVP, gastroenteritis virus panel; NA, not applicable; Ct, cycle threshold.
FIG 2.
Human adenovirus distributions across age groupings in children with acute gastroenteritis (cases and healthy controls). Numbers of participants in each age group are as follows: 0 to <6 months, 359 cases and 448 controls; 6 to <12 months, 669 cases and 178 controls; 12 to <18 months, 584 cases and 146 controls; 18 to <24 months, 337 cases and 164 controls; 24 to <60 months, 894 cases and 251 controls; ≥60 months, 504 cases and 168 controls. The denominator reflects the entire cohort of controls and cases tested for human adenovirus.
Primary outcome: adenovirus typing.
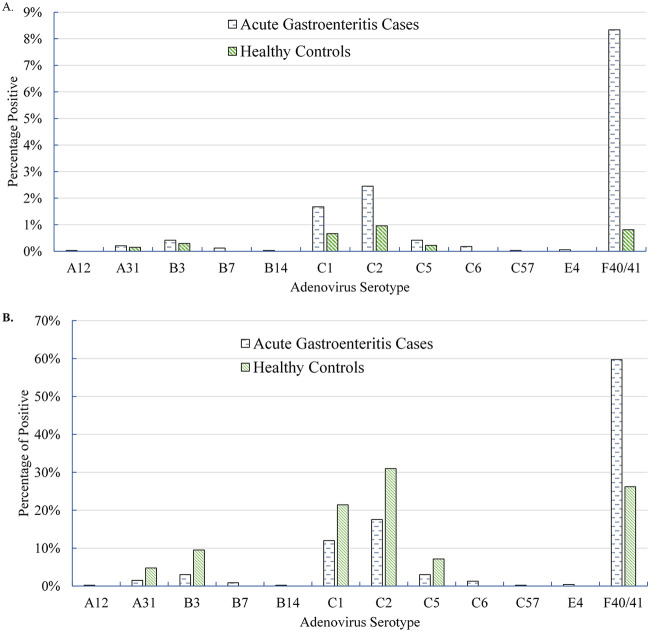
Typing results were available for 74.2% (467/629) of positive cases, which exceeded the rate among controls (74.2% versus 43.3% [P < 0.001]). HAdV F40/41 accounted for the majority (59.7%; 279/467) of all typed adenovirus AGE cases (Table 2). The most common typeable HAdV serotype among controls was C2 (13/42; 31.0%). The only other common serotypes among cases were C1 and C2, both of which were more commonly detected in the controls (Fig. 3).
TABLE 2.
Human adenovirus serotypes detected in AGE cases without respiratory symptoms and control study participantsa
| Species | Type | AGE cases—all |
AGE cases—no respiratory symptoms |
Asymptomatic controls |
OR (95% CI) for AGE (no respiratory symptoms) vs controlsd | Attributable fraction exposed (%) (95% CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Proportion positive (%)b (n = 467) | Cohort positive (%)c (n = 3,347) | No. | Proportion positive (%)b (n = 171) | Cohort positive (%)c (n = 1,363) | No. | Proportion positive (%)b (n = 42) | Cohort positive (%)c (n = 1,355) | ||||
| A | All | 8 | 1.7 | 0.2 | 4 | 2.3 | 0.3 | 2 | 4.8 | 0.1 | 2.1 (08, 5.4) | 51.2 (−23.0, 81.6) |
| 12 | 1 | 0.2 | 0.03 | 0 | 0 | |||||||
| 31 | 7 | 1.5 | 0.2 | 4 | 2.3 | 0.3 | 2 | 4.8 | 0.1 | |||
| B | All | 19 | 4.1 | 0.6 | 8 | 4.7 | 0.6 | 4 | 9.5 | 0.3 | 3.5 (0.9, 13.5) | 71.3 (−11.1, 92.6) |
| 3 | 14 | 3.0 | 0.4 | 6 | 3.5 | 0.4 | 4 | 9.5 | 0.3 | |||
| 7 | 4 | 0.9 | 0.1 | 1 | 0.6 | 0.1 | 0 | |||||
| 14 | 1 | 0.2 | 0.03 | 1 | 0.6 | 0.1 | 0 | |||||
| C | All | 159 | 34.0 | 4.8 | 45 | 26.3 | 3.3 | 24 | 57.1 | 1.8 | 2.1 (1.1, 3.8) | 51.7 (11.5, 73.4) |
| 1 | 56 | 12.0 | 1.7 | 15 | 8.8 | 1.1 | 9 | 21.4 | 0.7 | |||
| 2 | 82 | 17.6 | 2.4 | 24 | 14.0 | 1.8 | 13 | 31.0 | 1.0 | |||
| 5 | 14 | 3.0 | 0.4 | 4 | 2.3 | 0.3 | 3 | 7.1 | 0.2 | |||
| 6 | 6 | 1.3 | 0.2 | 2 | 1.2 | 0.2 | 0 | |||||
| 57 | 1 | 0.2 | 0.03 | 0 | 0 | |||||||
| E | 4 | 2 | 0.4 | 0.06 | 0 | 0 | ||||||
| F | 40/41 | 279 | 59.7 | 8.3 | 114 | 66.7 | 8.4 | 11 | 26.2 | 0.8 | 24.9 (13.0, 47.7) | 96.0 (92.3, 97.7) |
AGE, acute gastroenteritis.
The denominator reflects the number of participants positive for any adenovirus and the serotype available, within the respective cohort.
The denominator reflects the entire cohort tested for human adenovirus, within the respective cohort.
OR, odds ratio calculated by performance of multivariable logistic regression analysis employing HAdV serotypes grouped by species (exposure) to determine the association of detection with AGE symptoms (outcome). The OR for each adenovirus species was adjusted for other adenovirus species detected in both cases and controls, adenovirus detections for which typing was not available, codetected pathogens (astrovirus, norovirus, rotavirus, sapovirus, C. difficile, and other bacteria and parasites), participant age (using standard and quadratic terms), geographic location (Calgary, Edmonton, or other), and year (2014 to 2018) and season (winter, spring, summer, or fall) of enrollment. Because many adenovirus serotypes can cause respiratory symptoms, and since control participants were required to have no infectious symptoms, the multivariable logistic regression analysis included only cases without rhinorrhea or cough (i.e., respiratory symptoms). For each detected adenovirus species, the AF and 95% confidence intervals (CIs) were calculated by employing the ORs and 95% CIs derived in the multivariable logistic regression model.
FIG 3.
Adenovirus serotype distribution. (A) Distribution of adenovirus serotypes among all included acute gastroenteritis cases (n = 3,347) and controls (n = 1,355). (B) Distribution of adenovirus serotypes among typeable acute gastroenteritis cases (n = 467) and controls (n = 42) who tested positive for adenovirus.
Respiratory symptoms were present in 50.0% (1,365/2,728) of the AGE cases for whom they were recorded. Among those without respiratory symptoms, 24.6% (335/1,363) were positive for HAdV, and serotyping was available for 51.0% (171/335) (Table 2). Multivariable logistic regression demonstrated that HAdV species F (40/41) and C (1/2/5/6) detection was associated with AGE (ORs, 24.9 [95% CI, 13.0, 47.7] and 2.1 [95% CI, 1.1, 3.8], respectively). The AF for F40/41 was 96% (95% CI, 92.3 to 97.7%), whereas that for C1/2/5/6 was 51.7% (95% CI, 11.5 to 73.4%). An association between AGE and detection of species A and B could not be confirmed.
Secondary outcomes.
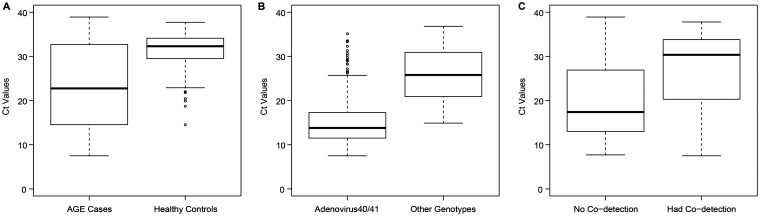
Stool Ct values were lower among HAdV-positive cases (median, 22.8 [interquartile range {IQR}, 14.5, 32.7]) than among HAdV-positive controls (median, 32.3 [IQR, 29.5, 34.2] [P < 0.001]) (Fig. 4A). The median Ct values of the positive case and control specimens that could not be typed were higher than those of specimens that were typeable (34.4 [IQR, 32.9, 35.7] versus 18.3 [IQR, 13.2, 26.0] [P < 0.001] among cases and 34.0 [IQR, 32.4, 35.1] versus 29.5 [IQR, 25.4, 31.3] [P < 0.001] among controls). Among all cases (n = 629), the median Ct values were lower among HAdV F40/41-positive specimens than among non-F40/41-positive specimens (Fig. 4B).
FIG 4.
Box plot of stool cycle threshold (Ct) values of adenovirus. (A) Adenovirus-positive acute gastroenteritis cases (n = 516) (median, 22.8 [IQR, 14.6, 32.7]) versus adenovirus-positive controls (n = 97) (median, 32.3 [IQR, 29.5, 34.1]). Significance was tested by a Mann-Whitney U test (two-sided unadjusted P value of <0.001). (B) Adenovirus F40/41-positive acute gastroenteritis cases (n = 229) (median, 13.8 [IQR, 11.5, 17.3]) versus non-F40/41-positive cases (n = 153) (median, 25.8 [IQR, 20.9, 30.9]). Significance was tested by a Mann-Whitney U test (two-sided unadjusted P value of <0.001). (C) Adenovirus-positive acute gastroenteritis cases with monodetections (n = 250) (median, 17.4 [IQR, 13.0, 26.9]) versus adenovirus-positive cases with codetections (n = 266) (median, 30.4 [IQR, 20.3, 33.7]). Significance was tested by a Mann-Whitney U test (two-sided unadjusted P value of <0.001).
Codetection of adenovirus and other pathogens occurred in 16.8% (47/279) of cases in whom F40/41 was detected and in 50.5% (95/188) of those with non-F40/41 HAdV (difference, −33.7% [95% CI of the difference, −42.1%, −24.8%]) (Table 3). The overall codetection rate was higher among cases (250/629; 39.8%) than among controls (21/97; 21.7%) (difference, 18.1% [95% CI of the difference, 7.6%, 26.5%]).
TABLE 3.
Codetection among human adenovirus 40/41 and non-40/41 acute gastroenteritis casesa
| Codetected pathogen | No. of cases (%) |
Difference (%) (95% CI)d | P valued | ||
|---|---|---|---|---|---|
| Overallc (n = 629) | F40/41 (n = 279) | Non-F40/41 (n = 188) | |||
| Any codetection | 250 (39.7) | 47 (16.8) | 95 (50.5) | −33.7 (−42.1, −24.8) | <0.001 |
| Any virus codetection | 225 (35.8) | 36 (12.9) | 89 (47.3) | −34.4 (−42.6, −25.8) | <0.001 |
| Norovirus | 119 (18.9) | 14 (5.0) | 51 (27.1) | −22.1 (−29.5, −15.1) | <0.001 |
| Sapovirus | 62 (9.9) | 15 (5.4) | 27 (14.4) | −10.4 (−16.7, −4.9) | 0.001 |
| Rotavirus | 51 (8.1) | 9 (3.2) | 16 (8.5) | −5.3 (−10.7, −0.7) | 0.02 |
| Astrovirus | 15 (2.4) | 2 (0.7) | 6 (3.2) | −2.5 (−6.5, 0.4) | 0.07 |
| Any bacterium codetection | 31 (4.9) | 10 (3.6) | 9 (4.8) | −1.2 (−5.9, 2.8) | 0.63 |
| Clostridioides difficileb | 10 (1.6) | 5 (1.8) | 2 (1.1) | 0.7 (−3.5, 2.6) | 0.71 |
| Campylobacter spp. | 2 (0.3) | 1 (0.4) | 0 (0) | 0.4 (−2.2, 2.3) | >0.99 |
| Enterotoxigenic Escherichia coli | 2 (0.3) | 1 (0.4) | 1 (0.5) | −0.1 (−3.0, 1.8) | >0.99 |
| Salmonella spp. | 3 (0.5) | 1 (0.4) | 2 (1.1) | −0.7 (−3.9, 1.4) | 0.57 |
| Shiga toxin-producing E. coli | 3 (0.5) | 1 (0.4) | 0 (0) | 0.4 (−2.2, 2.3) | >0.99 |
| Shigella spp. | 1 (0.2) | 1 (0.4) | 0 (0) | 0.4 (−2.2, 2.3) | >0.99 |
| Aeromonas spp. | 6 (1.0) | 0 (0) | 5 (2.7) | −2.7 (−6.4, −0.3) | 0.01 |
| Yersinia spp. | 1 (0.2) | 0 (0) | 0 (0) | NA | NA |
| Any parasite codetection | 4 (0.6) | 2 (0.7) | 0 (0) | 0.7 (−1.9, 2.9) | 0.52 |
| Cryptosporidium | 1 (0.2) | 0 (0) | 0 (0) | NA | NA |
| Entamoeba | 1 (0.2) | 1 (0.4) | 0 (0) | 0.4 (−2.2, 2.3) | >0.99 |
| Giardia | 2 (0.3) | 1 (0.4) | 0 (0) | 0.4 (−2.2, 2.3) | >0.99 |
Participants may have had more than 1 codetected pathogen.
Children <2.0 years of age in whom C. difficile was detected were considered negative and are not reported in this table.
The denominator included all cases that tested positive for adenovirus.
Comparison of serogroup F40/41 versus non-F40/41 HAdVs.
Symptom severity.
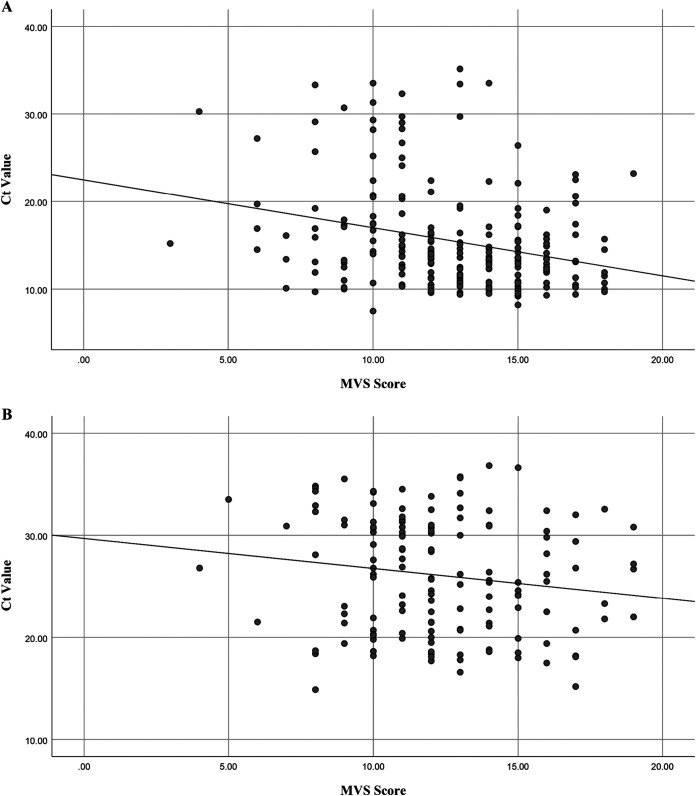
Among HAdV cases (n = 629), the total illness severity scores did not differ between those with HAdV F40/41 and those with non-F40/41 (P = 0.17) (Table 4). For both HAdV F40/41 and non-F40/41 cases, total MVS scores correlated inversely with Ct values (lower values indicate higher viral loads), indicating that children with higher viral loads had more severe disease (P < 0.0001 and P = 0.06, respectively) (Fig. 5).
TABLE 4.
Human adenovirus serotypes and acute gastroenteritis illness severitya
| Parameter | HAdV F40/41 (n = 279) |
HAdV non-F40/41 (n = 188) |
% difference (95% CI) | P value | ||
|---|---|---|---|---|---|---|
| No. of cases in analysis | Value | No. of cases in analysis | Value | |||
| Median age (mo) (IQR) | 279 | 17.1 (10.8, 30.7) | 188 | 15.3 (10.5, 25.4) | 1.5 (0.4, 3.5) | 0.11 |
| No. of males (%) | 279 | 170 (60.9) | 188 | 109 (58.0) | 3.0 (−6.3, 12.3) | 0.56 |
| No. of children with vomiting (%) | 273 | 249 (91.2) | 182 | 152 (83.5) | 7.7 (1.2, 14.7) | 0.02 |
| Median no. of vomiting episodes (IQR)b | 257 | 4 (3, 7) | 174 | 4 (2, 7) | 0 (−1, 1) | 0.97 |
| Median vomiting duration (h) (IQR) | 257 | 105.1 (54.3, 158.9) | 174 | 60.4 (11.8, 111.5) | 38.3 (24.6, 52.3) | <0.001 |
| No. of cases with diarrhea (%) | 274 | 262 (95.6) | 184 | 155 (84.2) | 11.4 (5.5, 18.0) | <0.001 |
| Median no. of diarrhea episodes (IQR)b | 257 | 6 (4, 9) | 173 | 4 (2, 6) | 2 (1, 3) | <0.001 |
| Median diarrhea duration (h) (IQR) | 257 | 151.1 (94.4, 198.0) | 174 | 115.4 (48.0, 192.0) | 31.7 (12.0, 50.4) | 0.001 |
| No. of cases with isolated vomiting (%) | 273 | 12 (4.4) | 182 | 29 (15.9) | −11.5 (−18.2, −5.7) | <0.001 |
| No. of cases with isolated diarrhea (%) | 274 | 24 (8.8) | 184 | 30 (16.3) | −7.6 (−14.5, −1.1) | 0.02 |
| No. of cases with both vomiting and diarrhea (%) | 269 | 233 (86.6) | 178 | 119 (66.9) | 19.8 (11.5, 28.1) | <0.001 |
| No. of cases with fever (%)c | 267 | 88 (33.0) | 180 | 131 (72.8) | −39.8 (−48.2, −30.4) | <0.001 |
| No. of cases with respiratory symptoms (%) | 235 | 121 (51.5) | 155 | 98 (63.2) | −11.7 (−21.7, −1.3) | 0.03 |
| No. of cases with ED visit after enrollment (%) | 257 | 31 (12.1) | 174 | 21 (12.1) | 0.01 (−6.4, 7.0) | >0.99 |
| No. of cases receiving intravenous fluid (%) | 260 | 44 (16.9) | 177 | 31 (17.5) | −0.6 (−8.4, 6.8) | 0.90 |
| No. of hospitalizations (%) | 258 | 14 (5.4) | 175 | 12 (6.9) | −1.4 (−7.0, 3.4) | 0.68 |
| Median MVS score (IQR) | 257 | 13.0 (11.0, 15.0) | 173 | 12.0 (10.0, 15.0) | 0 (0, 1.0) | 0.17 |
ED, emergency department; MVS, modified Vesikari scale; HAdV, human adenovirus.
Maximal number of episodes in a given 24 h.
Fever was defined as a rectal temperature of ≥38.0°C. Temperatures were adjusted for the location of measurement: 1.1°C was added to axillary temperatures, and 0.6°C was added to oral temperatures.
FIG 5.
Scatterplots showing the relationship of adenovirus stool cycle threshold (Ct) values and modified Vesikari scale scores with a fitted linear regression line. (A) Adenovirus serotype F40/41 (n = 218) (Pearson correlation coefficient, −0.28; P < 0.0001). (B) Adenovirus non-F40/41 serotypes (n = 144) (Pearson correlation coefficient, −0.16; P = 0.06).
DISCUSSION
This study confirms the importance of HAdV as an enteropathogen and the role of HAdV F40/41 serotypes, which accounted for nearly 60% of all HAdV-positive cases. Among AGE cases who had HAdV species F40/41 detected, disease could almost always (AF = 96%) be attributed to infection with these pathogens. When HAdV species C serotypes were detected, only half (AF = 52%) of illnesses could be attributed to them. We could not confirm any association between AGE symptoms and the detection of HAdV species A or B. When combined with Ct data showing overall lower HAdV concentrations in controls than in cases, our findings demonstrate the need for caution when attributing gastroenteritis symptoms to HAdV species C serotypes.
We detected HAdV F40/41 in 8.3% of all gastroenteritis cases. This finding confirms previous reports (16, 24); however, these studies lacked a control cohort to differentiate infection from detection. In a multisite U.S. study that employed a control cohort, HAdV was identified in 12% of children with AGE and just 2% of controls (25). The low detection rate among controls relative to our study may reflect their detection of only HAdV 41, which accounted for a minority of detections among our controls. Although a U.S.-based study focused on children <2 years of age that tested only for HAdV F40/41 reported no statistically significant difference between AGE cases (23%) and controls (16%) (26), these results were skewed by a single site that detected adenovirus in >40% of cases and controls.
Few studies have specified HAdV subgroups and serotypes among children with AGE. A study of children in Thailand most commonly identified subgroups C (41%), F (29%), and B (21%) (2). HAdV F41 was the predominant serotype (22%), followed by HAdV C2 (18%), B3 (15%), C1 (13%), and F40 (6%). In our study, HAdV subgroup C was the leading non-F40/41 HAdV subgroup detected, but the role of serotypes in this subgroup warrants further evaluation considering the low AF. While we also could not confirm any association between AGE and detection of HAdV species A or B, our analysis was impacted by the small number of such detections.
In Fig. 4, we used Ct values to clarify the contributions of specific pathogens. RT-qPCR Ct values represent the number of PCR cycles required for the amplification curve to intersect the threshold line, as a relative measure of the target concentration in the PCR. The higher the starting concentration of the target, the lower the Ct value. Thus, Ct values are inversely proportional to viral loads and provide a semiquantitative measure of the viral concentration in a sample. However, the assay itself and other factors within the sample matrix can affect RT-qPCR efficiency, thereby influencing the Ct values. Although a case-control study conducted in China reported no significant differences in HAdV concentrations between cases and controls, suggesting that this value alone cannot differentiate between infection and asymptomatic shedding, the study was limited by a very small sample size (3). In a larger U.S.-based study, Ct values differed between groups (P = 0.0001), particularly among those with HAdV as the only detected pathogen (26). As depicted in Fig. 4, we found that Ct values, when HAdVs were detected, were lower among cases than among controls (Fig. 4A), lower for HAdV F40/41 than for non-40/41 HAdV (Fig. 4B), and lower for monodetections than for codetections (Fig. 4C).
Our study cannot rule out the possibility that detection represents long-term intermittent or asymptomatic shedding of HAdV species (6, 7), and thus, we cannot confirm with certainty the etiological role in childhood gastroenteritis of the HAdV species detected. However, our findings indicate that when HAdV F40/41 species are detected in children with AGE, the symptoms can be attributed to these pathogens with reasonable confidence, which would be improved if the signal generated has a low Ct value and if other plausibly causative agents are absent. On the other hand, our results support the notion that when non-F40/41 HAdVs are detected in children with AGE, they should not routinely be assumed to be causative. When clinically indicated, the use of quantitative PCR may prove to be a useful adjunct to clarify the role played by HAdV in children with AGE by differentiating asymptomatic shedding from active infection.
ACKNOWLEDGMENTS
We thank Bryanne Crago, Christina Ferrato from the Provincial Laboratory for Public Health (ProvLab), Judy Qiu from the Department of Laboratory Medicine and Pathology, University of Alberta, and the staff from DynaLIFE Dx Diagnostic Laboratory Services, Calgary Laboratory Services, community laboratories, as well as ProvLab Edmonton and Calgary, Alberta, for their assistance with specimen receiving, handling, and processing; the ED research nurses and the Pediatric Emergency Medicine Research Associate Program (PEMRAP) at the Alberta Children’s Hospital for recruiting study participants; the ED bedside nurses for assisting with obtaining rectal swabs; Nadia Dow and Manasi Rajagopal as well as the research assistants, research nurses, and the Little Bit of Help (LBoH) research volunteer program for their assistance with participant recruitment at the Stollery Children’s Hospital; and the nurses at Health Link Alberta who respond to calls from across the province for their assistance with participant recruitment. We thank Laurel Ryan for her role as patient advisor. We thank Anita Wong for her organizational and technical assistance.
This work was supported by the Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE), which is funded by a grant from the Alberta Innovates-Health Solutions Team Collaborative Innovation Opportunity. APPETITE is also supported by the Alberta Children’s Hospital Research Institute (Calgary, Alberta) and the Women and Children’s Partnership Award Health Research Institute (Edmonton, Alberta). Stephen B. Freedman is supported by the Alberta Children’s Hospital Foundation Professorship in Child Health and Wellness. The Pediatric Emergency Medicine Research Associate Program is supported by a grant from the Alberta Children’s Hospital Foundation. In-kind support to enable the conduct of this study is provided by Calgary Laboratory Services, ProvLab Alberta, Luminex Corporation, and Copan Italia.
We have no potential conflicts of interest to declare.
Kanti Pabbaraju and Stephen B. Freedman conceptualized and designed the study, carried out the initial analyses, drafted the initial manuscript, and reviewed and revised the manuscript. Jianling Xie and Anna Funk designed the statistical analysis plan and critically reviewed the manuscript for important intellectual content. Kanti Pabbaraju, Raymond Tellier, Xiao-Li Pang, Bonita E. Lee, Linda Chui, and Ran Zhuo designed the study, conducted the microbiological analyses, and critically reviewed the manuscript for important intellectual content. Otto G. Vanderkooi, Samina Ali, and Phillip I. Tarr conceptualized and designed the study, designed the data collection instruments, and reviewed and revised the manuscript. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
REFERENCES
- 1.Ison MG, Hayden RT. 2016. Adenovirus. Microbiol Spectr 4:DMIH2-0020-2015. doi: 10.1128/microbiolspec.DMIH2-0020-2015. [DOI] [PubMed] [Google Scholar]
- 2.Kumthip K, Khamrin P, Ushijima H, Maneekarn N. 2019. Enteric and non-enteric adenoviruses associated with acute gastroenteritis in pediatric patients in Thailand, 2011 to 2017. PLoS One 14:e0220263. doi: 10.1371/journal.pone.0220263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu FZ, Shen XX, Li GX, Zhao L, Chen C, Duan SX, Guo JY, Zhao MC, Yan TF, Qi JJ, Wang L, Feng ZS, Ma XJ. 2018. Adenovirus associated with acute diarrhea: a case-control study. BMC Infect Dis 18:450. doi: 10.1186/s12879-018-3340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu L, Zhong H, Su L, Cao L, Xu M, Dong N, Xu J. 2017. Detection and molecular characterization of human adenovirus infections among hospitalized children with acute diarrhea in Shanghai, China, 2006-2011. Can J Infect Dis Med Microbiol 2017:9304830. doi: 10.1155/2017/9304830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiba S, Nakata S, Nakamura I, Taniguchi K, Urasawa S, Fujinaga K, Nakao T. 1983. Outbreak of infantile gastroenteritis due to type 40 adenovirus. Lancet ii:954–957. doi: 10.1016/s0140-6736(83)90463-4. [DOI] [PubMed] [Google Scholar]
- 6.Fox JP, Brandt CD, Wassermann FE, Hall CE, Spigland I, Kogon A, Elveback LR. 1969. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VI. Observations of adenovirus infections: virus excretion patterns, antibody response, efficiency of surveillance, patterns of infections, and relation to illness. Am J Epidemiol 89:25–50. doi: 10.1093/oxfordjournals.aje.a120913. [DOI] [PubMed] [Google Scholar]
- 7.Fox JP, Hall CE, Cooney MK. 1977. The Seattle virus watch. VII. Observations of adenovirus infections. Am J Epidemiol 105:362–386. doi: 10.1093/oxfordjournals.aje.a112394. [DOI] [PubMed] [Google Scholar]
- 8.Dey RS, Ghosh S, Chawla-Sarkar M, Panchalingam S, Nataro JP, Sur D, Manna B, Ramamurthy T. 2011. Circulation of a novel pattern of infections by enteric adenovirus serotype 41 among children below 5 years of age in Kolkata, India. J Clin Microbiol 49:500–505. doi: 10.1128/JCM.01834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman SB, Lee BE, Louie M, Pang X-L, Ali S, Chuck A, Chui L, Currie GR, Dickinson J, Drews SJ, Eltorki M, Graham T, Jiang X, Johnson DW, Kellner J, Lavoie M, MacDonald J, MacDonald S, Svenson LW, Talbot J, Tarr P, Tellier R, Vanderkooi OG. 2015. Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE): epidemiology, emerging organisms, and economics. BMC Pediatr 15:89. doi: 10.1186/s12887-015-0407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letourneau S. 2009. Health Link Alberta: a model for successful health service integration. Healthc Q 13:56–60. doi: 10.12927/hcq.2009.21099. [DOI] [PubMed] [Google Scholar]
- 11.Guarino A, Albano F, Ashkenazi S, Gendrel D, Hoekstra JH, Shamir R, Szajewska H, European Society for Paediatric Gastroenterology, Hepatology, and Nutrition, European Society for Paediatric Infectious Diseases. 2008. European Society for Paediatric Gastroenterology, Hepatology, and Nutrition/European Society for Paediatric Infectious Diseases evidence-based guidelines for the management of acute gastroenteritis in children in Europe. J Pediatr Gastroenterol Nutr 46(Suppl 2):S81–S122. doi: 10.1097/MPG.0b013e31816f7b16. [DOI] [PubMed] [Google Scholar]
- 12.Trang NV, Choisy M, Nakagomi T, Chinh NT, Doan YH, Yamashiro T, Bryant JE, Nakagomi O, Anh DD. 2015. Determination of cut-off cycle threshold values in routine RT-PCR assays to assist differential diagnosis of norovirus in children hospitalized for acute gastroenteritis. Epidemiol Infect 143:3292–3299. doi: 10.1017/S095026881500059X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedman SB, Eltorky M, Gorelick M, Pediatric Emergency Research Canada Gastroenteritis Study Group. 2010. Evaluation of a gastroenteritis severity score for use in outpatient settings. Pediatrics 125:e1278–e1285. doi: 10.1542/peds.2009-3270. [DOI] [PubMed] [Google Scholar]
- 14.Schnadower D, Tarr PI, Gorelick MH, O’Connell K, Roskind CG, Powell EC, Rao J, Bhatt S, Freedman SB. 2013. Validation of the modified Vesikari score in children with gastroenteritis in 5 US emergency departments. J Pediatr Gastroenterol Nutr 57:514–519. doi: 10.1097/MPG.0b013e31829ae5a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellner T, Parsons B, Chui L, Berenger BM, Xie J, Burnham CA, Tarr PI, Lee BE, Nettel-Aguirre A, Szelewicki J, Vanderkooi OG, Pang XL, Zelyas N, Freedman SB. 2019. Comparative evaluation of enteric bacterial culture and a molecular multiplex syndromic panel in children with acute gastroenteritis. J Clin Microbiol 57:e00205-19. doi: 10.1128/JCM.00205-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie J, Nettel-Aguirre A, Lee BE, Chui L, Pang XL, Zhuo R, Parsons B, Vanderkooi OG, Tarr PI, Ali S, Dickinson JA, Hagen E, Svenson LW, MacDonald SE, Drews SJ, Tellier R, Graham T, Lavoie M, MacDonald J, Freedman SB, Pediatric Emergency Research Canada (PERC), Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE Team). 2019. Relationship between enteric pathogens and acute gastroenteritis disease severity: a prospective cohort study. Clin Microbiol Infect 25:454–461. doi: 10.1016/j.cmi.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Tilmanne A, Martiny D, Quach C, Wautier M, Vandenberg O, Lepage P, Hallin M. 2019. Enteropathogens in paediatric gastroenteritis: comparison of routine diagnostic and molecular methods. Clin Microbiol Infect 25:1519–1524. doi: 10.1016/j.cmi.2019.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Pang XL, Preiksaitis JK, Lee BE. 2014. Enhanced enteric virus detection in sporadic gastroenteritis using a multi-target real-time PCR panel: a one-year study. J Med Virol 86:1594–1601. doi: 10.1002/jmv.23851. [DOI] [PubMed] [Google Scholar]
- 19.Wong S, Pabbaraju K, Pang XL, Lee BE, Fox JD. 2008. Detection of a broad range of human adenoviruses in respiratory tract samples using a sensitive multiplex real-time PCR assay. J Med Virol 80:856–865. doi: 10.1002/jmv.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claas EC, Burnham CA, Mazzulli T, Templeton K, Topin F. 2013. Performance of the xTAG gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J Microbiol Biotechnol 23:1041–1045. doi: 10.4014/jmb.1212.12042. [DOI] [PubMed] [Google Scholar]
- 21.Sarantis H, Johnson G, Brown M, Petric M, Tellier R. 2004. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J Clin Microbiol 42:3963–3969. doi: 10.1128/JCM.42.9.3963-3969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Landis JR, Koch GG. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 24.Brown JR, Shah D, Breuer J. 2016. Viral gastrointestinal infections and norovirus genotypes in a paediatric UK hospital, 2014-2015. J Clin Virol 84:1–6. doi: 10.1016/j.jcv.2016.08.298. [DOI] [PubMed] [Google Scholar]
- 25.Chhabra P, Payne DC, Szilagyi PG, Edwards KM, Staat MA, Shirley SH, Wikswo M, Nix WA, Lu X, Parashar UD, Vinje J. 2013. Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008-2009. J Infect Dis 208:790–800. doi: 10.1093/infdis/jit254. [DOI] [PubMed] [Google Scholar]
- 26.Hassan F, Kanwar N, Harrison CJ, Halasa NB, Chappell JD, Englund JA, Klein EJ, Weinberg GA, Szilagyi PG, Moffatt ME, Oberste MS, Nix WA, Rogers S, Bowen MD, Vinje J, Wikswo ME, Parashar UD, Payne DC, Selvarangan R. 2019. Viral etiology of acute gastroenteritis in <2-year-old US children in the post-rotavirus vaccine era. J Pediatr Infect Dis Soc 8:414–421. doi: 10.1093/jpids/piy077. [DOI] [PubMed] [Google Scholar]