Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infection among infants and young children, resulting in annual epidemics worldwide. INFORM-RSV is a multiyear clinical study designed to describe the global molecular epidemiology of RSV in children under 5 years of age by monitoring temporal and geographical evolution of current circulating RSV strains, F protein antigenic sites, and their relationships with clinical features of RSV disease. During the pilot season (2017–2018), 410 RSV G-F gene sequences were obtained from 476 RSV-positive nasal samples collected from 8 countries (United Kingdom, Spain, The Netherlands, Finland, Japan, Brazil, South Africa, and Australia).
KEYWORDS: evolution, genetic variation, molecular epidemiology, resistance, respiratory syncytial virus, surveillance
ABSTRACT
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infection among infants and young children, resulting in annual epidemics worldwide. INFORM-RSV is a multiyear clinical study designed to describe the global molecular epidemiology of RSV in children under 5 years of age by monitoring temporal and geographical evolution of current circulating RSV strains, F protein antigenic sites, and their relationships with clinical features of RSV disease. During the pilot season (2017–2018), 410 RSV G-F gene sequences were obtained from 476 RSV-positive nasal samples collected from 8 countries (United Kingdom, Spain, The Netherlands, Finland, Japan, Brazil, South Africa, and Australia). RSV B (all BA9 genotype) predominated over RSV A (all ON1 genotype) globally (69.0% versus 31.0%) and in all countries except South Africa. Geographic clustering patterns highlighted wide transmission and continued evolution with viral spread. Most RSV strains were from infants of <1 year of age (81.2%), males (56.3%), and patients hospitalized for >24 h (70.5%), with no differences in subtype distribution. Compared to 2013 reference sequences, variations at F protein antigenic sites were observed for both RSV A and B strains, with high-frequency polymorphisms at antigenic site Ø (I206M/Q209R) and site V (L172Q/S173L/K191R) in RSV B strains. The INFORM-RSV 2017–2018 pilot season establishes an important molecular baseline of RSV strain distribution and sequence variability with which to track the emergence of new strains and provide an early warning system of neutralization escape variants that may impact transmission or the effectiveness of vaccines and MAbs under development.
INTRODUCTION
Respiratory syncytial virus (RSV) is the leading cause of acute lower respiratory tract infection (LRTI) among infants and young children worldwide (1, 2). Most infections occur seasonally during the winter months in temperate regions, but with greater variability throughout the year in the tropics (3, 4). In 2015, RSV was associated with 33.1 million episodes of LRTI, 3.2 million RSV-related hospital admissions, and 118,000 deaths in children less than 5 years of age, predominantly in developing countries (2). Although prematurity and congenital lung or heart conditions are well-known risk factors for severe RSV LRTI, characterized by bronchiolitis and pneumonia, all children are at risk for RSV LRTI with primary RSV infection during infancy (2, 5).
Prevention of RSV LRTI in all infants is a major public health priority; however, despite many years of attempted vaccine development, there are no licensed vaccines (6). While palivizumab (Synagis) is the only approved passive monoclonal antibody approach for prophylaxis of RSV disease, it is recommended for use only with high-risk infants and children (7). Because there is no approved RSV prophylaxis for the broader population of healthy infants, more than 20 vaccine candidates and monoclonal antibodies (MAbs) are currently in clinical development (8). The most advanced candidate is nirsevimab—a potent, extended half-life MAb recently shown to significantly reduce medically attended RSV LRTI and hospitalization throughout the RSV season in healthy preterm infants in a phase 2b trial (9).
RSV is a nonsegmented, single-stranded, negative-sense RNA Orthopneumovirus belonging to the Pneumoviridae family (10). The attachment (G) and fusion (F) surface glycoproteins mediate viral entry and are both important antigenic targets for virus-neutralizing antibodies. Based on the genetic variability of the second hypervariable 2 region (HVR2) of the G gene, RSV strains are classified into subtype A or B and further characterized into different genotypes (11). In contrast, the F protein exhibits relative genetic and antigenic stability (12), making it a major target for vaccine and MAb development. The extracellular region of the mature F protein is a trimer of F1 and F2 subunits produced by cleavage of an inactive precursor F0 and exists in prefusion and postfusion conformations. Six antigenic sites (Ø and I to V) have been identified in prefusion and/or postfusion F proteins (13) with target epitopes for prophylactic neutralizing MAbs, including: palivizumab (site II), nirsevimab (site Ø), suptavumab (site V), and MK-1654 (site IV) (14).
As RSV immunization candidates reach the final stages of clinical development, the need for global monitoring of RSV molecular epidemiology becomes increasingly important to ensure their effectiveness during licensure and use. While prophylactic approaches invariably rely on conservation of neutralizing epitopes, RSV replication is inherently error-prone, resulting in natural polymorphisms (15). Selective immune pressure may further result in the emergence and spread of neutralization escape variants, allowing for immune and/or prophylaxis resistance. Finally, evolutionary dynamics of RSV genotypes may correlate with transmission between seasons (16) and disease severity among patient types (17).
The International Network For Optimal Resistance Monitoring of RSV (INFORM-RSV) study was established to describe global molecular epidemiology of RSV by monitoring temporal and geographical distribution of current circulating strains, with a focus on antigenic site changes that may confer selective advantages in transmission or resistance. Here, we describe geographic, demographic, and clinical distribution of RSV strains and sequence diversity of G genes and F proteins collected from mostly hospitalized infants in 8 countries across 4 continents during the pilot 2017–2018 RSV season.
MATERIALS AND METHODS
Study design.
INFORM-RSV is a prospective, multicenter, global molecular epidemiology study to investigate temporal and geographic diversity of RSV isolates collected from children less than 5 years of age who are admitted to the hospital or visiting the outpatient clinic and are not using preventive or treatment medication for RSV. Over the course of a 5-year period (2017–2022), 10 to 20 RSV-positive nasal samples will be collected per month per site each RSV season. Informed consent is obtained from parent(s)/legal representative(s) in accordance with the International Conference on Harmonization Guideline on Good Clinical Practice E6 (ICH-GCP) and applicable national and international regulatory requirements (18).
Sample collection.
The INFORM-RSV study was initiated in 2017–2018 in 8 countries (United Kingdom [GBR], Spain [ESP], The Netherlands [NLD], Finland [FIN], Japan [JPN], Brazil [BRA], South Africa [ZAF], and Australia [AUS]) with an aim to expand to other countries where disease burden studies are ongoing (Fig. 1). RSV-positive nasal samples were collected in Universal Transport Medium from hospital-based laboratories as part of routine clinical care or specifically for research purposes and shipped to the University Medical Centre Utrecht for sequencing. Individual patient data collected included: location, sample date, age, gender, referring department, and length of hospital stay (18).
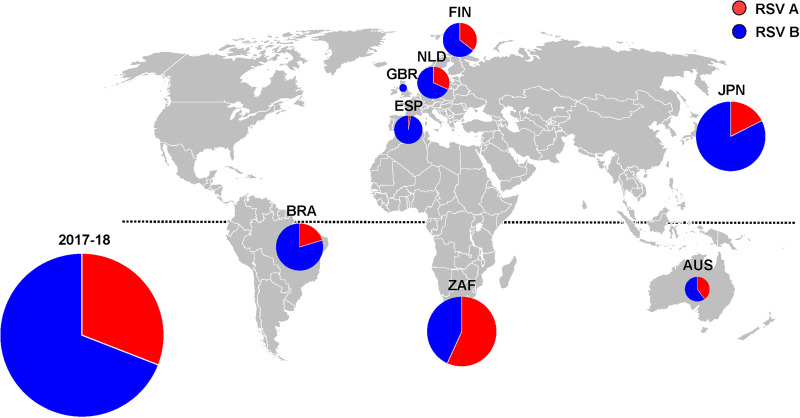
FIG 1.
Geographic distribution of RSV A (n = 128) and RSV B (n = 283) subtypes, 2017–2018 (n = 8 countries). Overall size of the pies is proportional to the number of RSV isolates and the segments of the pies are proportional to the frequency of subtype A (red) and subtype B (blue) (Table 1). Northern hemisphere: GBR, United Kingdom (n = 2); ESP, Spain (n = 36); NLD, The Netherlands (n = 43); FIN, Finland (n = 45); JPN, Japan (n = 91). Southern hemisphere: BRA, Brazil (n = 64); ZAF, South Africa (n = 95); AUS, Australia (n = 34). (The figure was created with Microsoft PowerPoint.)
RNA extraction, subtyping, RSV genome amplification, and next-generation sequencing.
Nucleic acids were extracted from RSV-positive nasal specimens using the MagNA Pure LC kit (Roche Diagnostics, Mannheim, Germany) as previously described (18). RSV subtyping and quantification were performed by multiplexed TaqMan RT-PCR analysis of the RSV N gene using RSV A and RSV B specific primer/probe mixes. Subsequently, subtype-specific RT-PCR was performed using the SuperScript IV one-step RT-PCR system (Invitrogen, Carlsbad, CA USA) to amplify 4 overlapping fragments covering the full RSV genome. The resultant 3.5 to 5.0 kb amplicons were pooled, purified from 1% agarose gels, used to construct libraries by means of the Nextera XT DNA Library Prep kit, and sequenced on a NextSeq 500 system (Illumina, San Diego, CA USA) (18).
Sequence assembly and genotyping analysis.
Assembly of next-generation sequencing (NGS) reads into RSV G-F contigs was performed using AstraZeneca’s open-source NGS-Microbial Sequencing Toolbox, as previously described (18, 19). Alignment of RSV G HVR2 and full-length nucleotide sequences was performed in MUSCLE and evolutionary analyses of full-length RSV G sequences were conducted in MEGA7. Assignment of RSV genotypes was performed by phylogenetic clustering of RSV G HVR2 nucleotide sequences using a previously described 2014 reference database (11).
Amino acid sequence variation analysis of RSV F proteins.
The RSV A and RSV B F sequences in FASTA format were translated into amino acid sequences and aligned against reference F sequences derived from year 2013 Netherlands RSV A/13-005275 (GenBank accession no: KX858757) and RSV B/13-001273 (GenBank accession no: KX858756) reference strains, respectively. Amino acid variation per position was determined and reported from pairwise alignments as previously described (18).
Structural visualization of RSV F protein antigenic sites.
The 3D structures of prefusion and postfusion RSV F protein trimers were visualized with PyMOL molecular Graphics System, v2.2.2 (Schrödinger, LLC) using PDB 5UDE (12) and PDB 3RRR (20), respectively. Antigenic sites were defined using the six antibody epitopes (Ø and I to V) previously described (13).
Statistical analyses.
A two-sided Fisher’s exact test was used to assess statistical significance of global subtype distribution among demographic categories and to compare proportions of amino acid changes between antigenic sites.
RESULTS
Geographic and demographic distribution of RSV A and B subtypes and genotypes.
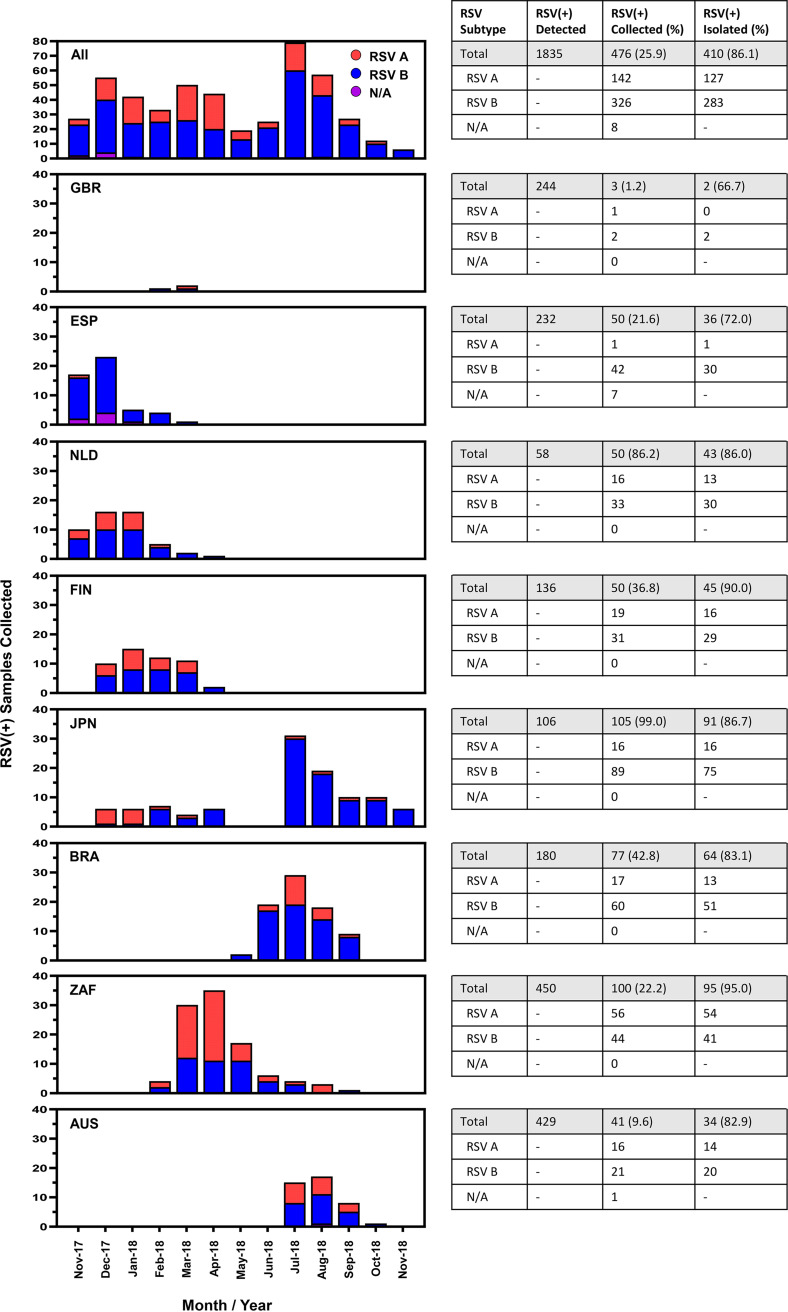
Between November 2017 and November 2018, 1,835 nasal samples tested RSV-positive among participating sites in 8 countries. Among the RSV-positive detections, 476 (25.9%) nasal samples were collected for inclusion in the INFORM-RSV study. The frequency and monthly pattern of RSV-positive samples collected from each country are shown in Fig. 2. Delayed study initiation resulted in fewer than the targeted 50 RSV-positive samples collected in 5 of the 8 countries. With some exceptions, the peak period for RSV-positive sample collection occurred from December to January and July to August in northern and southern hemisphere countries, respectively. Sequencing and assembly of full-length RSV G-F sequences was successful for 410 of the 476 (86.1%) RSV-positive samples, with even distribution between northern (52.9%; 217 of 410) and southern (47.1%; 193 of 410) hemispheres. The remaining 66 of 476 (13.9%) RSV-positive nasal samples failed sequencing due to unsuccessful RT-PCR amplification, insufficient sequencing depth, or low read quality. Among the 410 RSV strains with G-F sequence data, 127 (31.0%) were subtype A and 283 (69.0%) were subtype B. Overall, the proportion of RSV subtypes differed by country (P < 0.001), as RSV B was more prevalent than RSV A in 7 of 8 countries studied, with the exception being South Africa (Fig. 1 and Table 1). Finally, genotype determination revealed that all RSV A strains were of the Ontario 1 (ON1) genotype and all RSV B strains were of the Buenos Aires 9 (BA9) genotype.
FIG 2.
Monthly collection of RSV-positive(+) samples by country and overall number of RSV(+) detected, collected, and isolated/sequenced for RSV G-F gene analysis.
TABLE 1.
Frequency of RSV A (n = 127) and RSV B (n = 283) subtypes by demographic and clinical characteristics and country
| Countrya | No. total (%) | Gender |
Age |
Length of Stay |
Referring dept.b
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. male (%) | No. female (%) | No. <1 yr (%) | No. 1 to <2 yrs (%) | No. 2 to <5 yrs (%) | No. ≥24 h (%) | No. <24 h (%) | No. ER/ED (%) | No. PICU (%) | No. PW (%) | No. other (%) | ||
| All | 410 | 231 | 179 | 333 | 59 | 18 | 289 | 121 | 25 | 39 | 74 | 272 |
| RSV A | 127 (31.0) | 82 (35.5) | 45 (25.1) | 104 (31.2) | 21 (35.6) | 2 (11.1) | 88 (30.4) | 39 (32.2) | 3 (12.0) | 8 (20.5) | 19 (25.7) | 97 (35.7) |
| RSV B | 283 (69.0) | 149 (64.5) | 134 (74.9) | 229 (68.8) | 38 (64.4) | 16 (88.9) | 201 (69.6) | 82 (67.8) | 22 (88.0) | 31 (79.5) | 55 (74.3) | 175 (64.3) |
| GBR | 2 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 |
| RSV A | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RSV B | 2 (100.0) | 0 | 2 (100.0) | 2 (100.0) | 0 | 0 | 2 (100.0) | 0 | 0 | 1 (50.0) | 1 (50.0) | 0 |
| ESP | 36 | 20 | 16 | 31 | 3 | 2 | 31 | 5 | 5 | 0 | 0 | 31 |
| RSV A | 1 (2.8) | 1 (5.0) | 0 | 1 (3.2) | 0 | 0 | 1 (3.2) | 0 | 0 | 0 | 0 | 1 (3.2) |
| RSV B | 35 (7.2) | 19 (95.0) | 16 (100.0) | 30 (96.8) | 3 (100.0) | 2 (100.0) | 30 (96.8) | 5 (100.0) | 5 (100.0) | 0 | 0 | 30 (96.8) |
| NLD | 43 | 23 | 20 | 40 | 1 | 2 | 39 | 4 | 1 | 22 | 19 | 1 |
| RSV A | 13 (30.2) | 7 (30.4) | 6 (30.0) | 12 (30.0) | 1 (100.0) | 0 | 11 (28.2) | 2 (50.0) | 0 | 4 (18.2) | 8 (42.1) | 1 (100.0) |
| RSV B | 30 (69.8) | 16 (69.6) | 14 (70.0) | 28 (70.0) | 0 | 2 (100.0) | 28 (71.8) | 2 (50.0) | 1 (100.0) | 18 (81.8) | 11 (57.9) | 0 |
| FIN | 45 | 27 | 18 | 45 | 0 | 0 | 4 | 41 | 0 | 0 | 0 | 45 |
| RSV A | 16 (35.6) | 11 (40.7) | 5 (27.8) | 16 (35.6) | 0 | 0 | 1 (25.0) | 15 (36.6) | 0 | 0 | 0 | 16 (35.6) |
| RSV B | 29 (64.4) | 16 (59.3) | 13 (72.2) | 29 (64.4) | 0 | 0 | 3 (75.0) | 26 (63.4) | 0 | 0 | 0 | 29 (64.4) |
| JPN | 91 | 43 | 48 | 46 | 33 | 12 | 43 | 48 | 5 | 0 | 54 | 32 |
| RSV A | 16 (17.6) | 8 (18.6) | 8 (16.7) | 6 (13.0) | 9 (27.3) | 1 (8.3) | 2 (4.7) | 14 (29.2) | 0 | 0 | 11 (20.4) | 5 (15.6) |
| RSV B | 75 (82.4) | 35 (81.4) | 40 (83.3) | 40 (87.0) | 24 (72.7) | 11 (91.7) | 41 (95.3) | 34 (70.8) | 5 (100.0) | 0 | 43 (79.6) | 27 (84.4) |
| BRA | 64 | 42 | 22 | 59 | 5 | 0 | 44 | 20 | 14 | 13 | 0 | 37 |
| RSV A | 13 (20.3) | 9 (21.4) | 4 (18.2) | 12 (20.3) | 1 (20.0) | 0 | 7 (15.9) | 6 (30.0) | 3 (21.4) | 2 (15.4) | 0 | 8 (21.6) |
| RSV B | 51 (79.7) | 33 (78.6) | 18 (81.8) | 47 (79.7) | 4 (80.0) | 0 | 37 (84.1) | 14 (70.0) | 11 (78.6) | 11 (84.6) | 0 | 29 (78.4) |
| ZAF | 95 | 55 | 40 | 83 | 11 | 1 | 92 | 3 | 0 | 0 | 0 | 95 |
| RSV A | 54 (56.8) | 35 (63.6) | 19 (47.5) | 47 (56.6) | 6 (54.5) | 1 (100.0) | 52 (56.5) | 2 (66.6) | 0 | 0 | 0 | 54 (56.8) |
| RSV B | 41 (43.2) | 20 (36.4) | 21 (52.5) | 36 (43.4) | 5 (45.5) | 0 | 40 (43.5) | 1 (33.3) | 0 | 0 | 0 | 41 (43.2) |
| AUS | 34 | 21 | 13 | 27 | 6 | 1 | 34 | 0 | 0 | 3 | 0 | 31 |
| RSV A | 14 (41.2) | 11 (52.4) | 3 (23.1) | 10 (37.0) | 4 (66.7) | 0 | 14 (41.2) | 0 | 0 | 2 (66.7) | 0 | 12 (38.7) |
| RSV B | 20 (58.8) | 10 (47.6) | 10 (76.9) | 17 (63.0) | 2 (33.3) | 1 (100.0) | 20 (58.8) | 0 | 0 | 1 (33.3) | 0 | 19 (61.3) |
GBR, United Kingdom; ESP, Spain; NLD, The Netherlands; FIN, Finland; JPN, Japan; BRA, Brazil; ZAF, South Africa; AUS, Australia.
ER/ED, emergency room/department; PICU, pediatric intensive care unit; PW, pediatric ward; Other, other/undefined location.
Distribution of RSV strains by gender, age, and length of hospital stay was also determined. The median age of RSV-positive individuals was 5 months (interquartile range [IQR], 2 to 9 months) and 81.2% (333 of 410) were aged less than 1 year; 56.3% (231 of 410) were males; and 70.5% (289 of 410) were hospitalized for ≥24 h. RSV isolates from outpatients, characterized by a length of hospital stay of <24 h, were mostly derived from 3 countries (Finland, Japan, and Brazil) and accounted for 29.5% (121 of 410) of the total. Stratification by referring department revealed that most RSV isolates came from other/undefined locations (66.3%; 272 of 410), followed by the pediatric ward (PW) (18.0%; 74 of 410), emergency room/department (ER/ED) (6.1%; 25 of 410), and pediatric intensive care unit (PICU) (9.5%; 39 of 410) (Table 1). Overall, RSV B was more frequent than RSV A in all categories and there were no significant differences in the global proportion of subtypes by age group (P = 0.141) or length of hospital stay (P = 0.722). While a significantly higher proportion of RSV B cases were observed globally in females compared to males (P = 0.0311), no gender differences were observed within individual countries.
Global analysis of RSV genetic variability.
To understand genetic variability of the 2017−2018 RSV strains, we performed a phylogeographic analysis of G gene sequences by country. Within both RSV A (all ON1 genotype) and RSV B (all BA9 genotype) phylogenies, some sequences clustered within a country, suggesting microevolution, while other clusters contained sequences from multiple countries (Fig. 3). These data show that RSV A ON1 and RSV B BA9 strains from 2017–2018 were genetically diverse by geographic locale, consistent with wide transmission and continued evolution.
FIG 3.
RSV A ON1 (n = 127) and RSV B BA9 (n = 283) G-based clades by country.
Evidence for evolution of the RSV F protein.
To assess recent evolution of the fusion protein, 2017–2018 RSV A F and B F protein sequences were compared to year 2013 RSV A/13-005275 and RSV B/13-001273 reference strains, respectively. Overall, diversity of RSV F sequences was low, with mostly conserved amino acid changes detected at 45 of 574 positions (7.8%) in RSV A F and at 62 of 574 positions (10.8%) in RSV B F (Fig. 4). Only 2 amino acid changes in RSV A F were highly polymorphic: A23T (17.3%) in the signal peptide and T122A (11.8%) in the fusion peptide. In contrast, 7 amino acid changes in RSV B F were detected in a majority of sequences as follows: F15L (99.6%) in the signal peptide, A103V (100%) in F2, and L172Q (100%), S173L (99.6%), K191R (74.2%), I206M (77.4%), and Q209R (76.3%) in F1.
FIG 4.
Individual frequency and structural location of amino acid polymorphisms in RSV A F (n = 127) and RSV B F (n = 283) protein sequences. (Top) Major structural features of full-length RSV F protein (amino acids [AA] 1–574), including the extracellular region (F2: AA 24–109 and F1: 137-524); SP, signal peptide; p27 peptide; FP, fusion peptide; HR, heptad repeats; and TM/CT, transmembrane/carboxy terminus. (Middle) Linear plot of individual amino acid variation frequency in full-length RSV A F (red) and RSV B F (blue) protein sequences compared to year 2013 RSV A/13-005275 and RSV B/13-001273 reference strains, respectively. Amino acid polymorphisms detected at ≥10% frequency (Table 2) are denoted. (Bottom) Proximal locations of amino acid polymorphisms in antigenic sites of mature prefusion and postfusion RSV F protein trimers. Previously defined antigenic sites (Ø and I to V) (13) are delineated in color. Amino acid positions at which polymorphisms were detected at ≥1% frequency (Table 2) are highlighted in black with adjoining arrows. A and B superscripts denote subtype A and B, respectively.
Amino acid variation was further examined in each antigenic site (Ø and I to V) by geography (Table 2) and depicted on prefusion and postfusion F protein trimer structures (Fig. 4). No statistical differences in the global proportion of amino acid changes were observed between antigenic sites (data not shown) and some changes occurred in both RSV A F and B F at the same positions (Y33, I206, S255, and S276). Overall, 11 amino acid changes were detected in 4 of 6 antigenic sites for RSV A F, with frequencies ranging from 0.8 to 9.4%, and 32 amino acid changes were detected in 6 of 6 antigenic sites for RSV B F, with frequencies ranging from 0.4 to 100.0%. Only 5 of the 32 antigenic site changes in RSV B F were highly polymorphic and detected in all countries: I206M (77.0%) and Q209R (76.3%) in site Ø and L172Q (100.0%), S173L (99.6%), and K191R (74.2%) in site V. With few exceptions, antigenic site changes of intermediate polymorphic frequency (≥1% and <10%) were detected in multiple countries. These results indicate that F protein sequences and antigenic sites from 2017–2018 were generally well-conserved compared to year 2013 reference strains, although RSV B strains exhibited greater variability.
TABLE 2.
Global frequency of amino acid polymorphisms in antigenic sites of RSV A F (n = 127) and RSV B F (n = 283) protein sequences and countries of detection
| Site | Amino acid positionsa | RSV A (n = 127) |
RSV B (n = 283) |
||||
|---|---|---|---|---|---|---|---|
| Changeb | No. (%) | Countryc | Changeb | No. (%) | Countryc | ||
| Ø | 62–96; 195–227 | T72A | 2 (1.6) | ESP, NLD | K68R | 1 (0.4) | AUS |
| N88T | 4 (3.1) | FIN | K68Q | 1 (0.4) | JPN | ||
| I206T | 1 (0.8) | FIN | N201S | 1 (0.4) | NLD | ||
| I206M | 219 (77.4) | All | |||||
| Q209K | 1 (0.4) | NLD | |||||
| Q209L | 2 (0.7) | BRA | |||||
| Q209R | 216 (76.3) | All | |||||
| I | 27–45; 312–318; 378–389 | Y33H | 1 (0.8) | ZAF | Y33F | 1 (0.4) | ZAF |
| I384T | 12 (9.4) | ZAF | P312H | 1 (0.4) | NLD | ||
| S380N | 2 (0.7) | BRA | |||||
| L381I | 1 (0.4) | FIN | |||||
| S389F | 1 (0.4) | ZAF | |||||
| S389P | 3 (1.1) | BRA | |||||
| II | 254–277 | S255N | 1 (0.8) | ZAF | S255G | 1 (0.4) | ESP |
| S276N | 1 (0.8) | ZAF | M264I | 1 (0.4) | FIN | ||
| S276R | 1 (0.8) | FIN | K272N | 1 (0.4) | BRA | ||
| L273I | 1 (0.4) | ZAF | |||||
| S276N | 25 (8.8) | BRA, ESP, FIN, GBR, NLD | |||||
| III | 46–54; 301–311; 345–352; 367–378 | L303I | 2 (0.7) | NLD | |||
| I305T | 1 (0.4) | ESP | |||||
| V349A | 1 (0.4) | JPN | |||||
| N371S | 3 (1.1) | JPN | |||||
| IV | 422–471 | S425T | 1 (0.8) | ZAF | K433R | 1 (0.4) | AUS |
| S466N | 3 (2.4) | ZAF, NLD, JPN | L462Q | 1 (0.4) | NLD | ||
| L467I | 6 (4.7) | ZAF, BRA | E463D | 24 (8.5) | BRA, ESP, FIN, NLD | ||
| V | 55–61; 146–194; 287–300 | L172Q | 283 (100.0) | All | |||
| S173L | 282 (99.6) | All | |||||
| K176R | 1 (0.4) | AUS | |||||
| V179I | 1 (0.4) | JPN | |||||
| S190N | 10 (3.5) | BRA, ESP, FIN, JPN | |||||
| K191R | 210 (74.2) | All | |||||
| V300I | 1 (0.4) | FIN | |||||
Amino acid positions that define antigenic sites Ø and I–V (13).
Amino acid changes compared to year 2013 reference sequences; high-frequency polymorphisms (≥10%) are indicated in boldface type.
GBR, United Kingdom; ESP, Spain; NLD, The Netherlands; FIN, Finland; JPN, Japan; BRA, Brazil; ZAF, South Africa; AUS, Australia.
DISCUSSION
RSV A and B cocirculate during seasonal epidemic periods with alternating patterns of predominance over time (21). However, little is known about temporal evolution of RSV strains, global spread of unique genotypes, or how these factors relate to disease severity. Also important to the development of vaccines and MAbs is the need to identify and track patterns of F protein antigenic site changes, which may confer selective advantages in transmission or resistance. The INFORM-RSV study aims to describe global molecular evolution and epidemiology of RSV by prospectively monitoring temporal and geographical distribution of currently circulating strains. At the time of writing, the INFORM-RSV study has been ongoing for 3 years and is currently being conducted in 17 countries across 5 continents. The results herein provide baseline information on RSV strain distribution associated with different clinical parameters of disease severity and genetic variation of RSV G and F from 8 countries (GBR, ESP, NLD, FIN, JPN, BRA, ZAF, and AUS) across 4 continents during the 2017–2018 pilot season.
Genomic variation and evolutionary dynamics of RSV may affect its geographic, demographic, and clinical transmission behavior with important implications. During the INFORM-RSV 2017–2018 season, RSV B predominated over RSV A in all countries except South Africa, which may be attributed to virulence and local spread of RSV A strains specific to South Africa. Recent reports from North America (USA, 2015–2017 [22, 23]; Mexico, 2003–2015 [24]), Africa (ZAF, 2015–2017 [25]); [Kenya, 2000–2012 (26)], Asia (China, 2007–2015 [21]), and Australia (AUS, 2010–2016 [27]) describe alternating periodicity of RSV subtype prevalence, dominated by RSV A ON1 and RSV B BA9 genotypes. Consistent with these reports, RSV A ON1 and RSV B BA9 were the predominant genotypes of circulating RSV strains during the 2017–2018 RSV season. Geographic clustering patterns further suggest RSV transmission is characterized by continued genotype diversification during local spread and global dissemination.
Because the impact of viral factors on clinical parameters of disease severity has remained inconclusive (28), it was important to understand the distribution of RSV strains among demographic and clinical characteristics. Ultimately, most RSV strains were collected from hospitalized male infants aged less than 1 year, consistent with estimates of incidence and hospitalization rates (29), known risk factors, and the anatomic nature of shorter and narrower airways in infant males who are more likely to develop bronchial obstruction due to RSV infection (5). Unfortunately, the outpatient burden of RSV on health care resources has not been well defined (1, 2, 30) and few INFORM-RSV countries collected RSV-positive samples from outpatients who were medically managed without hospital admission. While hospital-based laboratory data on RSV infections may markedly underestimate the global burden of RSV disease, nevertheless, we observed no significant or meaningful differences in subtype/genotype distribution on clinical features of disease severity as assessed by gender, age group, or length of hospital stay.
The RSV F protein has historically been relatively well conserved, yet continues to evolve (12, 31). To that end, data from the INFORM-RSV 2017–2018 pilot season establishes an important molecular baseline of RSV F protein sequence and antigenic site variation from which to track frequency, geography, and evolutionary trajectory of potential neutralization escape variants as an early warning for vaccines and MAbs in development. Although the observed variability of the 2017–2018 RSV F sequences was low, with no differences in the proportion of amino acid changes between antigenic sites, the frequency and geographical distribution of some variants suggest a recent positive selection of favorable amino acid changes. Indeed, RSV B strains containing Q209R (site Ø) and L172Q/S173L (site V), first reported in China (2014–2016) (32), have recently emerged as dominant variants, with the addition of the I206M (site Ø) and K191R (site V) changes detected in the United States (2015–2019) (22, 33). These additional changes are possibly due to natural selective pressure from maternal or host neutralizing antibodies. Since site Ø and V elicit the greatest frequency of high-potency antibodies (34) in a structural area requiring a great deal of flexibility (13), these sites may tolerate greater amino acid variation than others. Additional, less frequent amino acid changes detected during the INFORM-RSV 2017–2018 study were frequent enough to be resampled in multiple countries but have yet to spread globally.
While the impact that widespread use of anti-RSV F MAbs will have on the emergence and transmission of resistant variants is unknown, these variants may also arise naturally in the absence of drug selection pressure. To date, palivizumab resistance-associated polymorphisms have been rarely observed in circulating RSV strains (35). Consistent with these reports, the restricted use of palivizumab (Synagis) (7), and the growth disadvantage of resistant variants in the absence of palivizumab selective pressure (36), we observed no known palivizumab target site II polymorphisms among 2017–2018 RSV strains. Also consistent with the rapid emergence and outgrowth of a RSV B strains containing L172Q/S173L in the United States (2015–2019) (22, 33), these nonconservative polymorphisms in suptavumab target site V were detected in 100% of global 2017–2018 RSV strains and coincide with clinical resistance and the recent failure of suptavumab to reduce overall RSV hospitalizations or outpatient LRTI in preterm infants in a phase 3 trial (6, 37). Finally, conservative I206M/Q209R polymorphisms in nirsevimab target site Ø were detected in 77% of RSV B strains but have been shown to retain susceptibility to neutralization by nirsevimab (38). Accordingly, despite the recent emergence of these polymorphisms, nirsevimab significantly reduced medically attended RSV LRTI in healthy preterm infants in a recent Phase 2b trial (9).
There are some limitations to the INFORM-RSV study. Key challenges to temporal analyses between geographies include adequate country representation and timing of RSV epidemics by season and location. Although low rates of RSV A and B coinfection (<2%) have been reported (22, 39), the use of subtype-specific primers/probes in the INFORM-RSV study did not permit detection of RSV A and B coinfection. Data on patients’ viral load are unavailable and therefore additional phylodynamic evolutionary and viral spread analyses are not possible. Since our data are heavily weighted toward infants with severe RSV disease that required hospitalization, we do not know about trends and molecular analyses of RSV from children who were medically managed as outpatients or were asymptomatic and did not seek medical attention. Our use of a 2014 RSV G HVR2 reference database (11) to genotype contemporary isolates has limitations as RSV continues to evolve. Accordingly, an extensible, centralized, curated, open database of reference sequences is needed to standardize genotyping and allow comparability across studies. Finally, future phenotypic susceptibility data would help to understand the functional impact of F protein antigenic site changes against anti-RSV F MAbs.
The strength of the INFORM-RSV study is reflected in its prospective design to characterize temporal and geographic trends in RSV diversity and to progress for several years with widespread global participation. Historically, RSV molecular epidemiology studies have been retrospective, focused exclusively on G gene diversity, and/or have been limited by geographical and low sampling effort constraints (15, 26, 40, 41). While global RSV surveillance is conducted by the European Influenza Surveillance Network (4) and the World Health Organization (42), none provide subtype differentiation or sequence analyses when reporting patterns of circulation. Findings from the INFORM-RSV study may have important implications in understanding the impact of RSV evolution on transmission, pathogenesis, and prophylaxis effectiveness. Tracking the frequency, recurrence, and distribution of amino acid changes that may confer selective advantages is a key focus of INFORM-RSV. Recent strains and dominant genotypes have genetic differences from the prototype virus strain used in most vaccine research (43). Since antigenic site changes could alter viral antigenicity for vaccines and affect their susceptibility to MAbs, novel agents for prophylaxis cannot afford to miss their contemporary targets when they are eventually deployed.
In conclusion, ongoing surveillance of global molecular epidemiology of RSV is important for detecting the emergence and spread of new strains, predicting their clinical impact, and providing an early warning system of antigenic changes that may affect the effectiveness of vaccines and MAbs. To that end, the INFORM-RSV 2017–2018 pilot season establishes an important molecular baseline of RSV strain distribution and sequence variability among hospitalized infants from which to investigate temporal and geographic relationships in the years ahead.
ACKNOWLEDGMENTS
We gratefully thank study participants and their families; and Mike McCarthy and Tonya Villafana for critical review of the manuscript.
The INFORM-RSV Study Group in 2017–2018 were: Christina Naaktgeboren, Anouk Evers, Marco C. Viveen, Joanne G. Wildenbeest, Frank E. J. Coenjaerts, Marije P. Hennus (University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands); Anne Greenough (ReSViNET Foundation, Zeist, The Netherlands, and King’s College London, London, United Kingdom); Terho Heikkinen, (ReSViNET Foundation, Zeist, The Netherlands, and University of Turku and Turku University Hospital, Turku, Finland); Renato Tetelbom Stein (Pontificia Universidade Catolica de Rio Grande do Sul, Porto Alegre, Brazil); Peter Richmond (The University of Western Australia, Perth, Australia); Federico Martinón-Torres (Hospital Clínico Universitario de Santiago, Galicia, Spain); Marta Nunes (ReSViNET Foundation, Zeist, The Netherlands, and University of the Witwatersrand, Johannesburg, South Africa); Mitsuaki Hosoya (Fukushima Medical University School of Medicine, Fukushima, Japan); Christian Keller (University Hospital Giessen and Marburg, Marburg, Germany); Robert Cohen (Université Paris XII, Créteil, France); Jesse Papenburg (McGill University Health Centre, Montreal, Canada); Jeffrey Pernica (McMaster University, Hamilton, Canada).
We declare that the planning, conduct, and reporting from this study was in line with the Declaration of Helsinki, as revised in 2013. The Medical Research Ethics Committee of the UMC Utrecht confirmed in their letter of 31 May 2017 (reference number WAG/mb/17/016170) that the Medical Research Involving Human Subjects Act (WMO) does not apply to the present study and therefore an official approval of this study by the MREC UMC Utrecht was not required under the WMO. Informed consent was obtained from parent(s) or legal representative(s) prior to sample collection in accordance with the International Conference on Harmonization Guideline on Good Clinical Practice E6 (ICH-GCP) and applicable national and international regulatory requirements.
The INFORM-RSV study received funding from AstraZeneca, Sanofi Pasteur, and Julius Clinical.
D. E. Tabor, F. Fernandes, D. Wilkins, A. Tovchigrechko, A. Ruzin, H. Jin, M. T. Esser, and M. E. Abram are employees of AstraZeneca and own stock. L. J. Bont, affiliated with the University Medical Centre Utrecht (UMCU) and founding chairman of the ReSViNET Foundation, has not received personal fees or other personal benefits. UMCU has received funding from Abbvie, AstraZeneca, Janssen, The Bill and Melinda Gates Foundation, Nutricia (Danone), and MeMed Diagnostics. UMCU has received major cash or in kind funding as part of the public private partnership IMI-funded RESCEU project from GSK, Novavax, Janssen, AstraZeneca, Pfizer and Sanofi. UMCU has received major funding from Julius Clinical for participating in the INFORM-RSV study sponsored by AstraZeneca. UMCU has received minor funding for participation in trials by Regeneron and Janssen from 2015 to 2017. UMCU received minor funding for consultation and invited lectures by AbbVie, AstraZeneca, Ablynx, Bavaria Nordic, MabXience, Novavax, Pfizer, and Janssen.
Contributor Information
Yi-Wei Tang, Cepheid.
the INFORM-RSV Study Group:
Christina Naaktgeboren, Anouk Evers, Marco C. Viveen, Joanne G. Wildenbeest, Frank E.J. Coenjaerts, Marije P. Hennus, Anne Greenough, Terho Heikkinen, Renato Tetelbom Stein, Peter Richmond, Federico Martinón-Torres, Marta Nunes, Mitsuaki Hosoya, Christian Keller, Robert Cohen, Jesse Papenburg, and Jeffrey Pernica
REFERENCES
- 1.Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, Grijalva CG, Zhu Y, Szilagyi P. 2009. The burden of respiratory syncytial virus infection in young children. N Engl J Med 360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi T, McAllister DA, O'Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A, Antonio M, Awasthi S, Awori JO, Azziz-Baumgartner E, Baggett HC, Baillie VL, Balmaseda A, Barahona A, Basnet S, Bassat Q, Basualdo W, Bigogo G, Bont LJ, Breiman RF, Brooks WA, Broor S, Bruce N, Bruden D, Buchy P, Campbell S, Carosone-Link P, Chadha M, Chipeta J, Chou M, Clara W, Cohen C, de Cuellar E, Dang D-A, Dash-Yandag B, Deloria-Knoll M, Dherani M, Eap T, Ebruke BE, Echavarria M, de Freitas Lázaro Emediato CC, Fasce RA, Feikin DR, Feng L, et al. 2017. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obando-Pacheco P, Justicia-Grande AJ, Rivero-Calle I, Rodriguez-Tenreiro C, Sly P, Ramilo O, Mejias A, Baraldi E, Papadopoulos NG, Nair H, Nunes MC, Kragten-Tabatabaie L, Heikkinen T, Greenough A, Stein RT, Manzoni P, Bont LJ, Martinon-Torres F. 2018. Respiratory syncytial virus seasonality: a global overview. J Infect Dis 217:1356–1364. doi: 10.1093/infdis/jiy056. [DOI] [PubMed] [Google Scholar]
- 4.Broberg EK, Waris M, Johansen K, Snacken R, Penttinen P, Network EIS. 2018. Seasonality and geographical spread of respiratory syncytial virus epidemics in 15 European countries, 2010 to 2016. Euro Surveill 23:17-00284. doi: 10.2807/1560-7917.ES.2018.23.5.17-00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simoes EAF. 2003. Environmental and demographic risk factors for respiratory syncytial virus lower respiratory tract disease. The J Pediatrics 143:118–126. doi: 10.1067/S0022-3476(03)00511-0. [DOI] [PubMed] [Google Scholar]
- 6.Mazur NI, Higgins D, Nunes MC, Melero JA, Langedijk AC, Horsley N, Buchholz UJ, Openshaw PJ, McLellan JS, Englund JA, Mejias A, Karron RA, Simões EAF, Knezevic I, Ramilo O, Piedra PA, Chu HY, Falsey AR, Nair H, Kragten-Tabatabaie L, Greenough A, Baraldi E, Papadopoulos NG, Vekemans J, Polack FP, Powell M, Satav A, Walsh EE, Stein RT, Graham BS, Bont LJ, Respiratory Syncytial Virus Network (ReSViNET) Foundation. 2018. The respiratory syncytial virus vaccine landscape: lessons from the graveyard and promising candidates. Lancet Infect Dis 18:e295–e311. doi: 10.1016/S1473-3099(18)30292-5. [DOI] [PubMed] [Google Scholar]
- 7.American Academy of Pediatrics Committee on Infectious Diseases, American Academy of Pediatrics Bronchiolitis Guidelines Committee. 2014. Updated guidance for palivizumab prophylaxis among infants and young children at increased risk of hospitalization for respiratory syncytial virus infection. Pediatrics 134:e620-38. doi: 10.1542/peds.2014-1666. [DOI] [PubMed] [Google Scholar]
- 8.PATH. 2020. RSV vaccine and mAb snapshot. [Accessed March 12, 2020]. https://vaccineresources.org/details.php?i=1562.
- 9.Griffin MP, Yuan Y, Takas T, Domachowske JB, Madhi SA, Manzoni P, Simões EAF, Esser MT, Khan AA, Dubovsky F, Villafana T, DeVincenzo JP. 2020. Single dose nirsevimab for prevention of RSV disease in preterm infants. N Engl J Med 383:415–425. doi: 10.1056/NEJMoa1913556. [DOI] [PubMed] [Google Scholar]
- 10.Rima B, Collins P, Easton A, Fouchier R, Kurath G, Lamb RA, Lee B, Maisner A, Rota P, Wang L, ICTV Report Consortium. 2017. ICTV virus taxonomy profile: Pneumoviridae. J Gen Virol 98:2912–2913. doi: 10.1099/jgv.0.000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabatabai J, Prifert C, Pfeil J, Grulich-Henn J, Schnitzler P. 2014. Novel respiratory syncytial virus (RSV) genotype ON1 predominates in Germany during winter season 2012-13. PLoS One 9:e109191. doi: 10.1371/journal.pone.0109191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Q, McLellan JS, Kallewaard NL, Ulbrandt ND, Palaszynski S, Zhang J, Moldt B, Khan A, Svabek C, McAuliffe JM, Wrapp D, Patel NK, Cook KE, Richter BWM, Ryan PC, Yuan AQ, Suzich JA. 2017. A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci Transl Med 9:eaaj1928. doi: 10.1126/scitranslmed.aaj1928. [DOI] [PubMed] [Google Scholar]
- 13.Gilman MS, Castellanos CA, Chen M, Ngwuta JO, Goodwin E, Moin SM, Mas V, Melero JA, Wright PF, Graham BS, McLellan JS, Walker LM. 2016. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci Immunol 1:eaaj1879. doi: 10.1126/sciimmunol.aaj1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broadbent L, Groves H, Shields MD, Power UF. 2015. Respiratory syncytial virus, an ongoing medical dilemma: an expert commentary on respiratory syncytial virus prophylactic and therapeutic pharmaceuticals currently in clinical trials. Influenza Other Respir Viruses 9:169–178. doi: 10.1111/irv.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bose ME, He J, Shrivastava S, Nelson MI, Bera J, Halpin RA, Town CD, Lorenzi HA, Noyola DE, Falcone V, Gerna G, De Beenhouwer H, Videla C, Kok T, Venter M, Williams JV, Henrickson KJ. 2015. Sequencing and analysis of globally obtained human respiratory syncytial virus A and B genomes. PLoS One 10:e0120098. doi: 10.1371/journal.pone.0120098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elawar F, Griffiths CD, Zhu D, Bilawchuk LM, Jensen LD, Forss L, Tang J, Hazes B, Drews SJ, Marchant DJ. 2017. A virological and phylogenetic analysis of the emergence of new clades of respiratory syncytial virus. Sci Rep 7:12232. doi: 10.1038/s41598-017-12001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Fernandez R, Tapia LI, Yang CF, Torres JP, Chavez-Bueno S, Garcia C, Jaramillo LM, Moore-Clingenpeel M, Jafri HS, Peeples ME, Piedra PA, Ramilo O, Mejias A. 2017. Respiratory syncytial virus genotypes, host immune profiles, and disease severity in young children hospitalized with bronchiolitis. J Infect Dis 217:24–34. doi: 10.1093/infdis/jix543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langedijk AC, Lebbink RJ, Naaktgeboren C, Evers A, Viveen MC, Greenough A, Heikkinen T, Stein RT, Richmond P, Martinón-Torres F, Nunes M, Hosoya M, Keller C, Bauck M, Cohen R, Papenburg J, Pernica J, Hennus MP, Jin H, Tabor DE, Tovchigrechko A, Ruzin A, Abram ME, Wilkins D, Wildenbeest JG, Kragten-Tabatabaie L, Coenjaerts FEJ, Esser MT, Bont LJ. 2020. Global molecular heterogeneity of RSV—the “INFORM RSV” study. BMJ Infect Dis 20:450. doi: 10.1186/s12879-020-05175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tovchigrechko A, Aspinal E, Slidel T, Liu H, Lu B, Ruzin A, Lebbink RJ, Jin H, Abram ME, Esser MT, Tabor DE. 2020. NGS-MSTB: Next Generation Sequencing Microbial Surveillance Toolbox for scalable de-novo assembly of viral genomes and bacterial genes Zenodo doi: 10.5281/zenodo.3619527. [DOI] [Google Scholar]
- 20.McLellan JS, Yang Y, Graham BS, Kwong PD. 2011. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol 85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Liu C, Xiao Y, Xiang Z, Zhou H, Chen L, Shen K, Xie Z, Ren L, Wang J. 2019. Respiratory syncytial virus seasonality, Beijing, China, 2007-2015. Emerg Infect Dis 25:1127–1135. doi: 10.3201/eid2506.180532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu B, Liu H, Tabor DE, Tovchigrechko A, Qi Y, Ruzin A, Esser MT, Jin H. 2019. Emergence of new antigenic epitopes in the glycoproteins of human respiratory syncytial virus collected from a US surveillance study, 2015-17. Sci Rep 9:3898. doi: 10.1038/s41598-019-40387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruzin A, Pastula ST, Levin-Sparenberg E, Jiang X, Fryzek J, Tovchigrechko A, Lu B, Qi Y, Liu H, Jin H, Yu L, Hackett J, Villafana T, Esser MT. 2018. Characterization of circulating RSV strains among subjects in the OUTSMART-RSV surveillance program during the 2016-17 winter viral season in the United States. PLoS One 13:e0200319. doi: 10.1371/journal.pone.0200319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comas-Garcia A, Noyola DE, Cadena-Mota S, Rico-Hernandez M, Bernal-Silva S. 2018. Respiratory syncytial virus-A ON1 genotype emergence in central Mexico in 2009 and evidence of multiple duplication events. J Infect Dis 217:1089–1098. doi: 10.1093/infdis/jiy025. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Lu B, Tabor DE, Tovchigrechko A, Wilkins D, Jin H, Madhi SA, Soofie N, Esser MT, Nunes MC. 2020. Characterization of human respiratory syncytial virus (RSV) isolated from HIV-exposed-uninfected and HIV-unexposed infants in South Africa during 2015–2017. Influenza Other Respir Viruses 14:403–411. doi: 10.1111/irv.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otieno JR, Agoti CN, Gitahi CW, Bett A, Ngama M, Medley GF, Cane PA, Nokes DJ. 2016. Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya. J Virol 90:4990–5002. doi: 10.1128/JVI.03105-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Giallonardo F, Kok J, Fernandez M, Carter I, Geoghegan JL, Dwyer DE, Holmes EC, Eden JS. 2018. Evolution of human respiratory syncytial virus (RSV) over multiple seasons in New South Wales, Australia. Viruses 10:476. doi: 10.3390/v10090476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandini S, Biagi C, Lanari M. 2017. Respiratory syncytial virus: the influence of serotype and genotype variability on clinical course of infection. Int J Mol Sci 18:1717. doi: 10.3390/ijms18081717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita CB, Simões EAF, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bont L, Checchia PA, Fauroux B, Figueras-Aloy J, Manzoni P, Paes B, Simoes EA, Carbonell-Estrany X. 2016. Defining the epidemiology and burden of severe respiratory syncytial virus infection among infants and children in western countries. Infect Dis Ther 5:271–298. doi: 10.1007/s40121-016-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hause AM, Henke DM, Avadhanula V, Shaw CA, Tapia LI, Piedra PA. 2017. Sequence variability of the respiratory syncytial virus (RSV) fusion gene among contemporary and historical genotypes of RSV/A and RSV/B. PLoS One 12:e0175792. doi: 10.1371/journal.pone.0175792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Xu B, Guo J, Li C, An S, Zhou Y, Chen A, Deng L, Fu Z, Zhu Y, Liu C, Xu L, Wang W, Shen K, Xie Z. 2018. Genetic variations in the fusion protein of respiratory syncytial virus isolated from children hospitalized with community-acquired pneumonia in China. Sci Rep 8:4491. doi: 10.1038/s41598-018-22826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes F, Tabor DE, Lu B, Layman H, Nair V, Chaudhuri V, Qi Y, Tovchigrechko A, Wilkins D, Ruzin A, Villafana T, Esser MT, Abram ME. 2019. OUTSMART-RSV molecular surveillance in the United States over the 2016–2019 RSV seasons. Poster Presentation at RSVVW'19, Accra, Ghana, 12–14 November 2019.
- 34.Ruckwardt TJ, Morabito KM, Graham BS. 2019. Immunological lessons from respiratory syncytial virus vaccine development. Immunity 51:429–442. doi: 10.1016/j.immuni.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira DB, Iwane MK, Prill MM, Weinberg GA, Williams JV, Griffin MR, Szilagyi PG, Edwards KM, Staat MA, Hall CB, Durigon EL, Erdman DD. 2015. Molecular characterization of respiratory syncytial viruses infecting children reported to have received palivizumab immunoprophylaxis. J Clin Virol 65:26–31. doi: 10.1016/j.jcv.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Q, Patel NK, McAuliffe JM, Zhu W, Wachter L, McCarthy MP, Suzich JA. 2012. Natural polymorphisms and resistance-associated mutations in the fusion protein of respiratory syncytial virus (RSV): effects on RSV susceptibility to palivizumab. J Infect Dis 205:635–638. doi: 10.1093/infdis/jir790. [DOI] [PubMed] [Google Scholar]
- 37.Simoes EAF, Forleo-Neto E, Geba GP, Kamal M, Yang F, Cicirello H, Houghton MR, Rideman R, Zhao Q, Benvin SL, Hawes A, Fuller ED, Wloga E, Pizarro JMN, Munoz FM, Rush SA, McLellan JS, Lipsich L, Stahl N, Yancopoulos GD, Weinreich DM, Kyratsous CA, Sivapalasingam S. 2020. Suptavumab for the prevention of medically attended respiratory syncytial virus infection in preterm infants. Clin Infect Dis doi: 10.1093/cid/ciaa951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Q, Lu B, McTamney P, Palaszynski S, Diallo S, Ren K, Ulbrandt ND, Kallewaard N, Wang W, Fernandes F, Wong S, Svabek C, Moldt B, Esser MT, Jing H, Suzich JA. 2018. Prevalence and significance of substitutions in the fusion protein of respiratory syncytial virus resulting in neutralization escape from antibody MEDI8897. J Infect Dis 218:572–580. doi: 10.1093/infdis/jiy189. [DOI] [PubMed] [Google Scholar]
- 39.Fall A, Dia N, Cisse el HA, Kiori DE, Sarr FD, Sy S, Goudiaby D, Richard V, Niang MN. 2016. Epidemiology and molecular characterization of human respiratory syncytial virus in Senegal after four consecutive years of surveillance, 2012–2015. PLoS One 11:e0157163. doi: 10.1371/journal.pone.0157163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mas V, Nair H, Campbell H, Melero JA, Williams TC. 2018. Antigenic and sequence variability of the human respiratory syncytial virus F glycoprotein compared to related viruses in a comprehensive dataset. Vaccine 36:6660–6673. doi: 10.1016/j.vaccine.2018.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan R, Fan C, Zhang J, Wen B, Lei Y, Liu C, Chen L, Liu W, Wang C, Qu X. 2017. Respiratory syncytial virus subtype ON1/NA1/BA9 predominates in hospitalized children with lower respiratory tract infections. J Med Virol 89:213–221. doi: 10.1002/jmv.24619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirve S, Crawford N, Palekar R, Zhang W, WHO RSV Surveillance Group. 2019. Clinical characteristics, predictors, and performance of case definition-Interim results from the WHO global respiratory syncytial virus surveillance pilot. Influenza Other Respir Viruses doi: 10.1111/irv.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandya MC, Callahan SM, Savchenko KG, Stobart CC. 2019. A contemporary view of respiratory syncytial virus (RSV) biology and strain-specific differences. Pathogens 8:67. doi: 10.3390/pathogens8020067. [DOI] [PMC free article] [PubMed] [Google Scholar]