Next-generation sequencing technologies are being rapidly adopted as a tool of choice for diagnostic and outbreak investigation in public health laboratories. However, costs of operation and the need for specialized staff remain major hurdles for laboratories with limited resources for implementing these technologies. This project aimed to assess the feasibility of using Oxford Nanopore MinION whole-genome sequencing data of Mycobacterium tuberculosis isolates for species identification, in silico spoligotyping, detection of mutations associated with antimicrobial resistance (AMR) to accurately predict drug susceptibility profiles, and phylogenetic analysis to detect transmission between cases.
KEYWORDS: clinical study, MinION, Mycobacterium tuberculosis, nanopore, public health laboratories, whole-genome sequence
ABSTRACT
Next-generation sequencing technologies are being rapidly adopted as a tool of choice for diagnostic and outbreak investigation in public health laboratories. However, costs of operation and the need for specialized staff remain major hurdles for laboratories with limited resources for implementing these technologies. This project aimed to assess the feasibility of using Oxford Nanopore MinION whole-genome sequencing data of Mycobacterium tuberculosis isolates for species identification, in silico spoligotyping, detection of mutations associated with antimicrobial resistance (AMR) to accurately predict drug susceptibility profiles, and phylogenetic analysis to detect transmission between cases. The results were compared prospectively in real time to those obtained with our current clinically validated Illumina MiSeq sequencing assay for M. tuberculosis and phenotypic drug susceptibility testing results when available. Our assessment of 431 sequenced samples over a 32-week period demonstrates that, when using the proper quality controls and thresholds, the MinION can achieve levels of genotyping analysis and phenotypic resistance predictions comparable to those of the Illumina MiSeq at a very competitive cost per sample. Our results indicate that nanopore sequencing can be a suitable alternative to, or complement, currently used sequencing platforms in a clinical setting and has the potential to be widely adopted in public health laboratories in the near future.
INTRODUCTION
Use of next-generation sequencing (NGS) technologies in public health laboratories and other agencies worldwide has significantly increased in the last few years (1, 2). These high-throughput sequencing platforms are now partly or completely replacing many conventional biochemical or molecular methods, as they generate comprehensive data with an unsurpassed level of accuracy (3; https://www.cdc.gov/amd/whats-new/pulsenet-transition.html). They are now used on a routine basis for disease diagnostics, pathogen identification, antibiotic resistance determination, and outbreak investigations. However, initial capital costs to acquire NGS platforms are still high and often out of reach for laboratories with limited resources. The need for highly trained staff to operate and maintain equipment can also be a hindrance.
In this regard, the MinION, a portable NGS device developed by Oxford Nanopore, is an enticing alternative (4). The device is capable of generating long sequencing reads with yields averaging between 10 to 30 gigabases per 48 hours of runtime, comparable with throughputs of the Illumina MiSeq platform. The sequencing reads generated by the MinION can be analyzed in real time, allowing the user to evaluate the data and report results before the full completion of a sequencing run (5–7). While there are many benefits to short-read sequencing, situations occur where long-read sequencing is necessary. These are most notably for genomes with long repetitive regions, copy number alterations, and complex structural variations along with the need to assess plasmids and improve de novo assemblies (8). Recent advances in nanopore sequencing technologies, including reducing error rates, updated flow cells, and availability of analytic software, has renewed interest in the utilization of the MinION device for clinical applications (9–12), but long-term prospective studies are still lacking. This project aims at providing the first comprehensive assessment of the MinION performance for a real-time NGS application in a public health laboratory.
In 2016, the Wadsworth Center, New York State Department of Health, implemented the first validated and New York State (NYS)-approved MiSeq NGS test for predicting Mycobacterium tuberculosis (MTB) antimicrobial resistance (AMR) for nine drugs/drug classes, identifying the different species of the M. tuberculosis complex and their specific geographical lineages, as well as genotyping by generating in silico spoligotypes that can be compared to historical isolates (3). The overall positive and negative predictive values of this test were 93% and 96%, respectively, with an overall concordance of 96% with drug susceptibility testing (DST). All MTB isolates from diagnosed cases in NYS, including New York City (NYC), undergo whole-genome sequencing using the Illumina MiSeq workflow for in silico genotyping, AMR prediction, and assessing potential transmission links among cases. The final results are communicated to physicians and either the NYS Bureau of Tuberculosis Control or NYC Bureau of Tuberculosis Control to help direct proper treatment regimens and with case management. In October 2018, Wadsworth Center updated its testing algorithm to a reduced phenotyping model, wherein culture-based DST is no longer performed on isolates predicted to be pan-susceptible by whole-genome sequencing (WGS). Phenotypic testing is only performed for specimens where drug resistance is predicted by WGS or when mutations of unknown significance are detected in relevant target genes and drug resistance cannot be ruled out. This testing algorithm was only applied to isolates from New York State patients; isolates originating from NYC patients still have universal phenotypic DST performed at the New York City Public Health Laboratory, regardless of WGS results. This validated workflow was used as a basis of comparison to evaluate the capability and practicality of the MinION as a platform for routine clinical NGS testing in our laboratory. Over a period of 32 weeks, a total of 431 DNA extracts from early positive mycobacterial growth indicator tube (MGIT) cultures were used for library preparation and sequenced in parallel on MiSeq and MinION. A bioinformatic pipeline was built using the same analytic backbone as our MiSeq pipeline (3), with some modifications to accommodate the nanopore reads and higher error rates. More specifically, we wanted to evaluate the overall sample and drug target fail rates, sensitivity, specificity, positive and negative predictive values of the MinION data, and concordance with MiSeq data and DST results when available. We also assessed the turnaround time (TAT) of the MinION workflow and total cost per sample.
MATERIALS AND METHODS
DNA extraction and NGS sequencing.
DNA extractions from cultures that were within 1 to 7 days of MGIT positivity and library preparations for sequencing on the Illumina MiSeq platform were performed as described by Shea et al. (3). An aliquot from the same DNA extract used for MiSeq sequencing was employed to prepare MinION libraries. Libraries were prepared using the ligation sequencing kit (Oxford Nanopore; catalog no. SQK-LSK109) according to the manufacturer’s protocol with the following modifications. DNA was end repaired and dA tailed for 15 min at 20°C followed by 15 min at 65°C. Barcode adapter ligation was carried out for 15 min at room temperature. For the barcoding PCR (Oxford Nanopore; catalog no. EXP-PBC096), a 50-μl reaction was set up using Q5 high-fidelity polymerase (New England Biolabs; catalog no. M0491) that included the High GC enhancer, a maximum of 20 ng DNA as the template, and 2 μl barcode. Following an initial denaturation step of 2 min at 98°C, the PCR went through 17 cycles of 10 s at 98°C, 25 s at 67°C, and then 3 min at 72°C. The final extension step was 2 min at 72°C. Samples were DNA repaired and end prepped for 15 min at 20°C followed by 15 min at 65°C. The sequencing adapter was ligated for 15 min at room temperature. For all cleanup steps, Agencourt AMPure XP beads (Beckman Coulter; catalog no. A63881) were added based on 0.45× sample volume and incubated for 10 min at room temperature. Samples were placed on a magnetic stand for 3 min, and then supernatant was removed. Samples were washed 2 times, each time by adding 200 μl 70% EtOH, incubating for 30 s, and then removing the 70% EtOH by pipetting. Beads were dried for 1 min to 1 min 30 s and then eluted in water or elution buffer (after sequencing adapter ligation only). Approximately 130 fmol of the final libraries were loaded on the MinION flow cell version R9.4.1 (Oxford Nanopore) according to the manufacturer’s instructions, and sequencing was started with the default parameters using the MinKNOW interface. Base calling was performed either on a Nanopore MinIT (Oxford Nanopore) or a Unix laptop equipped with a graphic processing unit (GPU; Nvidia Quadro P2000), using Guppy basecaller with the flip-flop fast algorithm. For this study, we preordered 48 flow cells, scheduled to be delivered at a rate of 8 flow cells every 2 months at a cost of $500 per flow cell, while associated kits (ligation, wash, etc.) were ordered on an as-needed basis, keeping one or two extra kits on hand at any given time. MiSeq libraries were prepared using the Illumina Nextera XT 250-bp paired-end library protocol using the manufacturer’s recommendation with the exception of carrying out 15 PCR cycles at the indexing step. The Illumina MiSeq runs were limited to a maximum of 16 MTB samples per flow cells.
Culture-based drug susceptibility testing.
Phenotypic DST was determined using the liquid MGIT 960 system (Bactec MGIT SIRE and PZA package inserts; Becton, Dickinson) and solid 7H10 agar proportion method following the Clinical and Laboratory Standards Institute's recommendations (13). First-line DST includes rifampin (RIF) (1.0 μg/ml), isoniazid (INH) (0.1, 0.4 μg/ml), ethambutol (EMB) (5.0 μg/ml), streptomycin (SM) (1.0 μg/ml), and pyrazinamide (PZA) (100 μg/ml), while second-line DST includes RIF (1.0 μg/ml), INH (0.2 and 1.0 μg/ml), EMB (5.0 and 10.0 μg/ml), SM (2.0 and 10.0 μg/ml), ethionamide (ETH) (5.0 μg/ml), kanamycin (KAN) (5.0 μg/ml), and ofloxacin (OFL) (1.0, 2.0, and 4.0 μg/ml). Ofloxacin is used in our laboratory as a representative of the fluoroquinolone (FLQ) drug class.
Bioinformatics pipeline.
(i) MiSeq workflow. For the MiSeq NGS data analyses and susceptibility predictions, we used the pipeline previously described by Shea et al. (3). More specifically, this pipeline uses Kraken (14) and single-nucleotide polymorphism (SNP) markers to conduct species identification and contaminant detections. In silico spoligotyping is performed by looking for the presence/absence of specific k-mers in the read files associated with the 43 CRISPR spacers (Table S1 in the supplemental material). The presence of larger genomic deletions is screened using lumpy-SV (15), and SNP calling is performed using the Genome Analysis Toolkit (GATK) (16) with a minimum depth of 10× and diploid mode to allow for the detection of heteroresistance.
(ii) MinION quality control. Raw reads were demultiplexed with QCAT v1.1.0 (https://github.com/nanoporetech/qcat), using the “–detect-middle” option to detect adaptors in the middle of reads and the “–trim” option to trim adaptors and barcodes. Filtlong v0.2.0 (https://github.com/rrwick/Filtlong) was used to downsample read files when necessary to a maximum of 200× depth of coverage. Kraken2 v2.0.7-beta (17), along with the minikraken2 database, was used to determine the taxonomic content of the reads and assess the presence of contaminating species or host DNA in the sample.
(iii) Spoligotyping. In silico spoligotyping, i.e., determining which of 43 CRISPR spacers are present in the samples, was done by first mapping reads over a synthetic sequence containing all 43 spacers interspersed with the conserved repeats (Table S1). Read mapping was performed with minimap2 v2.16-r922 (https://github.com/lh3/minimap2), and only reads that mapped over the CRISPR sequence were extracted with SAMtools v1.9 (samtools view, F4 option) (18) and used for spacer screening. Agrep (approximate grep) (https://github.com/Wikinaut/agrep) was used to determine the presence/absence of each spacer in the mapped read set by allowing up to two mismatches in the spacer sequence, except for spacer number three, where only one mismatch was allowed to occur to limit the possibility of false-positive hits sometimes associated with this specific spacer. A spacer was deemed present in the data set if a match was found in at least five reads. Discrepant samples were compared to the spoligotype result found in the Tuberculosis Genotyping Information Management System (TB GIMS), Centers for Disease Control and Prevention.
SNP calling.
The reads were mapped to the M. tuberculosis H37Rv genome sequence (NC_000962.3) with BWA mem v0.7.17-r1188 (https://github.com/lh3/bwa) using the default parameters, and read duplicates were removed using samtools rmdup. Ninety-one genomic regions totaling 2.55% of the reference genome (112,398 nucleotides [nt]), which contains insertion sequence (IS) elements, genomic duplications, and anomalous read mapping, were masked after mapping to minimize any artefactual SNP positions (Table S2). A sequence pileup was then created from the aligned reads with samtools mpileup for all positions (-aa), using a minimum base quality of 7 (-Q 7), mapping quality of 20 (-q 20), and per-base alignment quality disabled (-B). The pileup file was parsed to assess the depth of coverage and allele frequencies of all bases/insertions/deletions at every position and generate a high-quality consensus sequence. Variant and invariant single-nucleotide positions were determined using a minimum of 10× depth of coverage and 70% allele agreement. Positions failing these thresholds we classified as undetermined and set as unknown state positions (Ns) in the consensus sequence and for variant annotations. Insertions and deletions were called using a minimum allele agreement of 60% but with a minimum depth of 20× and 30× for insertions and deletions, respectively. In addition, any insertion or deletion detected in homopolymeric regions of three nucleotides or more were ignored. These thresholds listed above were empirically determined by resequencing samples already present in our database with MinION and comparing the final consensus sequences with the one obtained with MiSeq. Our aim was to find a set of thresholds that would minimize the number of differences between the consensus sequences and failed positions while maximizing the number of high-confidence positions assessable for susceptibility predictions. For example, using a lower allele frequency threshold to build out consensus sequences introduced too many unreliable called positions, increasing the overall number of differences between the MinION and MiSeq consensus sequences. At the inverse, using a higher allele frequency threshold creates a higher fidelity of consensus sequences but at the expense of decreasing the overall percentage of genome positions that can be assessed. SNP distance matrices were built using snp-dist v0.6.3 (github.com/tseemann/snp-dists). Larger genomic deletions were confirmed by manual inspection of the alignments reads when partial or complete target failure was reported. Final SNP annotations and AMR predictions were done using in-house-developed Perl scripts as described in reference 3. Confirmation of the species identification and specific MTB lineage assignment was conducted by the presence of variant codons or nucleotides at specific locations or loci on the high-quality consensus sequence (Table S3). This list of lineage-specific variants was compiled based on an approach described by Feuerriegel et al. (19), using an internal collection of ∼2,000 whole-genome-sequenced strains and identifying unique variations of combinations of SNPs specific to each lineage or species of interest.
Susceptibility predictions.
Antimicrobial resistance (AMR) determination to rifampin, isoniazid, pyrazinamide, ethambutol, streptomycin, kanamycin, amikacin, fluoroquinolones, and ethionamide was performed by screening 16 loci of interest for the presence of variant positions and/or indels and determining the presence of 70 well-characterized (high-confidence) mutations associated with resistance. In addition, any variants in pncA and pncA promoters not previously confirmed as nonresistant-causing phenotypes (Table S4) are de facto predicted to cause a phenotype of resistance to PZA (R). This list of high-confidence mutations was compiled using a combination of literature review, public databases, and internal susceptibility data. An unknown prediction (U) means a novel mutation, not part of our catalog of high-confidence mutations or neutral mutations (Table S5), that was identified. Neutral mutations are well-characterized mutations that have been found to have no impact on resistance and are often phylogenetic in nature. A WGS-susceptible prediction (S) means no high-confidence mutation and no mutations of unknown significance are present in the screened loci for that specific drug. A specific position known to be associated with resistance that failed to pass the required quality control (QC) thresholds will result in a not determined (ND) prediction if no other resistant mutation(s) associated with that drug is present.
Data availability.
The MinION sequence reads can be accessed at the NCBI Sequence Read Archives under BioProject accession no. PRJNA650381.
RESULTS
Sequencing statistics.
We sequenced a total of 431 samples over a period of 32 weeks using the Oxford Nanopore MinION platform (Table 1). The number of samples sequenced in multiplex ranged from 3 to 28 samples, with an average of 14 samples per week. About 36% (8 out of 22) of the flow cells were used twice or three times when enough pores remained active after cleanup without noticeable adverse effects on the quality of the data. Sequencing time on flow cells ranged from 5 to 72 hours, depending on the number of samples multiplexed and the total yield achieved during the sequencing run. The total yields of each flow cell ranged from ∼11 gigabases (Gb) per 24 hours to >16 Gb per 48 hours of sequencing, aiming for read depth of at least 100× per sample during the first 15 weeks and increasing to at least 150× in the subsequent weeks in order to reduce the number of allele dropouts in the rrs loci. One or two rounds of ATP refueling of the flow cell were performed for 11 of the runs to provide extra data yields when the pores’ translocation speed fell below the recommended threshold. Our average read size was ∼1,550 bp, which is well below the capabilities of nanopore sequencing, where reads can exceed 1 megabase (Mb) in length. The reason for producing short reads was the inherent difficulty to extract MTB DNA and the need to use harsh methods, leading to increased DNA fragmentation. The shorter reads prevented us from being able to fully assemble the MTB genomes into single contigs; thus, we decided to rely on read mapping with a reference genome for SNP calling. Of all samples received for MinION sequencing, a total of nine samples (2.1%) failed (less than 40× of sequencing depth) to generate usable data due to low DNA concentration, low sequencing depth, or technical error versus 15 for the MiSeq (3.5%), including four samples failing on both platforms. Of the remaining 411 samples, one sample was also eliminated because it was a control BCG sample, not part of this study, and three were discarded due to the presence of foreign contaminating DNA, leaving a total of 407 samples that passed both MiSeq and MinION minimum sequencing depth requirements. Sample IDR1900042441 was sequenced twice but only counted once, as shown in Table 2.
TABLE 1.
MinION sequencing metrics for clinical Mycobacterium tuberculosis NGS performed weekly
| Run date (mo/day/yr) | No. of samples | Failed samples (<40×) | Run time (hrs) | Starting no. of pores | Basecall yield (Gb) | Reused cell | Times refueled |
|---|---|---|---|---|---|---|---|
| 6/20/19 | 10 | 2 | 48 | 1,162 | 16.07 | 1 | |
| 6/27/19 | 22 | 0 | 24 | 1,627 | 13.61 | ||
| 7/4/19 | 10 | 0 | 24 | 1,187 | 13.37 | ||
| 7/10/19 | 4 | 0 | 5 | 840 | 2.51 | Yes | |
| 7/18/19 | 7 | 0 | 8 | 1,498 | 4.29 | ||
| 7/26/19 | 28 | 0 | 66 | 1,689 | 25.27 | 1 | |
| 8/1/19 | 15 | 0 | 23 | 918 | 9.05 | Yes | |
| 8/8/19 | 13 | 3 | 20.5 | 1,528 | 11.46 | ||
| 8/15/19 | 11 | 0 | 21.5 | 1,645 | 11.3 | ||
| 8/22/19 | 13 | 1 | 24 | 1,300 | 9.7 | Yes | |
| 8/29/19 | 12 | 0 | 24 | 1,367 | 9.83 | Yes | |
| 9/5/18 | 25 | 0 | 48 | 1,606 | 19.8 | 1 | |
| 9/12/18 | 14 | 1 | 26 | 1,347 | 10.26 | 1 | |
| 9/19/19 | 9 | 0 | 23.5 | 967 | 11.11 | Yes | |
| 9/26/19 | 16 | 0 | 23 | 1,183 | 10.41 | ||
| 10/3/19 | 9 | 0 | 22.5 | 1,378 | 12.57 | ||
| 10/11/19 | 17 | 2 | 48 | 1,072 | 17.53 | Yes | |
| 10/17/19 | 8 | 0 | 23.3 | 1,270 | 12.31 | ||
| 10/24/19 | 24 | 0 | 48 | 1,712 | 22.83 | ||
| 10/31/19 | 16 | 0 | 28 | 1,469 | 15.49 | ||
| 11/7/19 | 8 | 0 | 28 | 987 | 10.06 | Yes | 1 |
| 11/14/19 | 5 | 0 | 23 | 774 | 7.76 | Yes | |
| 11/21/19 | 17 | 0 | 24 | 1,229 | 21.61 | 1 | |
| 12/2/19 | 12 | 0 | 26 | 1,402 | 14.17 | ||
| 12/5/19 | 6 | 0 | 21 | 1,246 | 11.07 | ||
| 12/12/19 | 19 | 0 | 48 | 1,589 | 17.27 | 2 | |
| 12/19/19 | 13 | 0 | 48 | 1,494 | 19.83 | 1 | |
| 1/6/20 | 3 | 0 | 24 | 733 | 8.57 | Yes | |
| 1/6/20 | 23 | 0 | 24 | 1,562 | 23.67 | 1 | |
| 1/9/20 | 4 | 0 | 22 | 893 | 8.16 | Yes | |
| 1/16/20 | 3 | 0 | 24 | 604 | 619 | Yes | |
| 1/23/20 | 18 | 0 | 48 | 1,599 | 20.81 | 2 | |
| 1/23/20 | 17 | 0 | 72 | 1,495 | 24.15 | 2 | |
| Total | 431 | 9 |
TABLE 2.
Correlation of antimicrobial resistance predictions for nine currently reported drugsa between the MiSeq and MinION workflows
| MinION result | MiSeq result (no. of isolates) |
|||
|---|---|---|---|---|
| Susceptible | Resistant | Unknown | NDb | |
| Susceptible | 3,377 | 4 | 6 | 13 |
| Resistant | 0 | 146 | 0 | 0 |
| Unknown | 0 | 0 | 63 | 0 |
| ND | 38 | 7 | 0 | 0 |
Includes rifampin, isoniazid, pyrazinamide, ethambutol, streptomycin, kanamycin, amikacin, fluoroquinolones, and ethionamide.
ND, not determined.
Classifications of the MiSeq and MinION reads using Kraken and Kraken2, respectively, were 98.5% concordant for most samples, except for three cases where Kraken2 classified M. tuberculosis samples as Mycobacterium bovis bacillus Calmette-Guérin (BCG), two cases of samples identified as M. tuberculosis instead of M. bovis, and one instance of a Mycobacterium africanum sample mistakenly classified as M. tuberculosis. We suspect the MinION platform had a slightly higher fail rate in differentiating between very close species/subspecies due to the higher intrinsic error rates of the raw reads. MTB lineage assignments based on specific lineage nucleotide variants (Table S3 in the supplemental material) were 99.5% concordant between the MiSeq and the MinION. In only two cases, the MinION failed to assign an SNP-based lineage, while they were identified as members of lineage 2 (Beijing) by the MiSeq workflow. Of the 418 samples with spoligotype information, 391 (94%) of the MiSeq and 410 (98%) of MinION in silico-derived spoligotypes were in agreement with each other, and any discrepant results were verified against the Tuberculosis Genotyping Information Management System (TB GIMS), Centers for Disease Control and Prevention. Most of the discrepancies were either associated with the presence of contaminating sequences in the data set or single/multiple spacer dropouts. Two samples with discrepant results did not have TB GIMS information; therefore, we could not determine which platform was correct.
Out of the total 3,654 AMR NGS predictions for nine drug resistance predictions, we found a 98.1% agreement between the MiSeq and MinION (Table 2; Table S6). Of these, 92.4% predicted a susceptible phenotype, 4.00% a resistant phenotype, and the remaining 1.72% did not predict a phenotype due to the presence of mutations of unknown significance. For the 1.86% of the predictions that did not agree, 13 were allele dropouts in the MiSeq that were predicted to be susceptible by the MinION, and 38 were predicted to be susceptible by the MiSeq but failed drug predictions by the MinION. All of the 38 instances with MinION were related to failed position(s) in the 16s rRNA loci (rrs), while the 13 allele dropouts for the MiSeq were scattered between the katG, inhA, rrs, and rpsL loci. We found 6 instances of novel mutations (mutations of unknown significance) detected by the MiSeq workflow that were missed using the MinION. In all cases, the mutations present were at heterozygous positions, and the minor populations in the samples were carrying the variant allele or missed deletion events. We also found seven events of predicted resistance that were detected by the MiSeq but failed our QC threshold in the MinION workflow (one at rpsL codon 43, three at rpsL codon 88, two at katG codon 315, and one rpoB codon 511) and flagged as not determined (ND) for resistance prediction to the assessed drug. All but three of these mutations were from mixed genotypes. Finally, there were four instances of samples predicted to be resistant with the MiSeq but predicted to be susceptible using the MinION. Two of these cases were due to the presence of heterozygous mutations in the gyrA locus, and two were caused by the presence of single-nucleotide frameshift mutation in ethA. When comparing the MinION and MiSeq NGS predictions with drug susceptibility testing (DST) results when available, we obtained overall sensitivity (76% versus 80%), specificity (99% versus 98%), positive predictive values (PPV; 86% versus 87%), and negative predictive values (NPV; 97% versus 97%) (Table 3). The total concordance of the predictions with DST for both platforms was found to be 96%. No major differences were noted when looking at each individual drug except for fluoroquinolones, where the sensitivity of prediction was 40% using the MinION versus 80% for the MiSeq. This difference in sensitivity was caused by a low sample size of fluoroquinolone-resistant samples in our data set and the presence of heteroresistance missed by the MinION workflow in some samples. The sensitivity of detection of amikacin and ethionamide resistance was at or below 50% using both platforms. This was caused by a very small sample size of drug-resistant samples in the case of amikacin and still a lack of knowledge of many resistance-causing mutations for ethionamide.
TABLE 3.
Correlation between NGS drug susceptibility prediction and drug susceptibility test results when availablea
| Test result | No. of susceptible isolates for drug: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RIF | ISO | PZA | ETH | STR | AMI | KAN | FLQ | ETH | Total | |
| MiSeq | ||||||||||
| TP | 14 | 42 | 16 | 4 | 26 | 1 | 3 | 4 | 13 | 123 |
| TN | 224 | 191 | 219 | 216 | 37 | 65 | 64 | 65 | 72 | 1,153 |
| FP | 4 | 0 | 8 | 0 | 3 | 0 | 1 | 0 | 2 | 18 |
| FN | 0 | 4 | 5 | 1 | 6 | 1 | 0 | 1 | 13 | 31 |
| Total | 242 | 237 | 248 | 221 | 72 | 67 | 68 | 70 | 100 | 1,325 |
| Sensitivity (%) | 100 | 91 | 76 | 80 | 81 | 50 | 100 | 80 | 50 | 80 |
| Specificity (%) | 98 | 100 | 96 | 100 | 93 | 100 | 98 | 100 | 97 | 98 |
| PPV (%) | 78 | 100 | 67 | 100 | 90 | 100 | 75 | 100 | 87 | 87 |
| NPV (%) | 100 | 98 | 98 | 100 | 86 | 98 | 100 | 98 | 85 | 97 |
| Concordance (%) | 98 | 98 | 95 | 100 | 88 | 99 | 99 | 99 | 85 | 96 |
| MinION | ||||||||||
| TP | 13 | 40 | 16 | 4 | 21 | 1 | 3 | 2 | 11 | 111 |
| TN | 228 | 200 | 224 | 221 | 39 | 64 | 62 | 66 | 75 | 1,179 |
| FP | 4 | 0 | 8 | 0 | 3 | 0 | 1 | 0 | 2 | 18 |
| FN | 0 | 4 | 5 | 1 | 6 | 1 | 0 | 3 | 15 | 35 |
| Total | 245 | 244 | 253 | 226 | 69 | 66 | 66 | 71 | 103 | 1,343 |
| Sensitivity (%) | 100 | 91 | 76 | 80 | 78 | 50 | 100 | 40 | 42 | 76 |
| Specificity (%) | 98 | 100 | 97 | 100 | 93 | 100 | 98 | 100 | 97 | 98 |
| PPV (%) | 76 | 100 | 67 | 100 | 88 | 100 | 75 | 100 | 85 | 86 |
| NPV (%) | 100 | 98 | 98 | 100 | 87 | 98 | 100 | 96 | 83 | 97 |
| Concordance (%) | 98 | 98 | 95 | 100 | 87 | 98 | 98 | 96 | 83 | 96 |
Includes a detailed list of true-positive (TP), true-negative (TN), false-positive (FP), and false-negative (FV) predictions, as well as sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and concordance for both the MiSeq and MinION platforms.
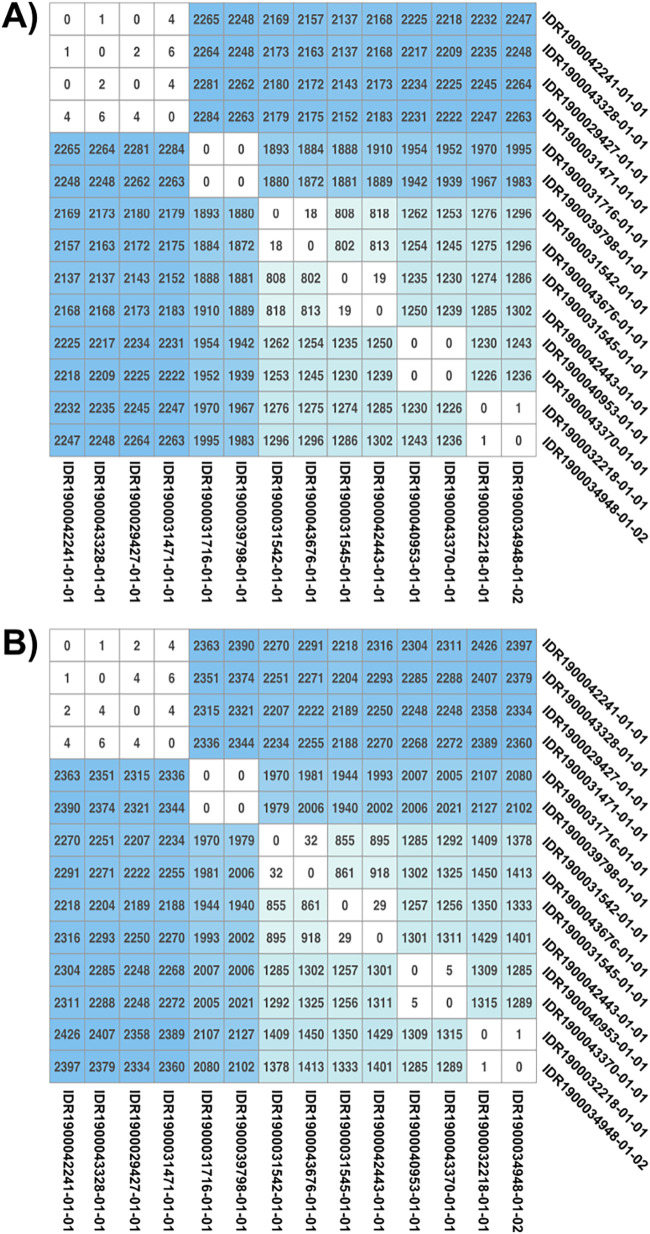
Comparison of the number of differences between MinION- and MiSeq-generated consensus sequences reveals very few site disagreements, with an average number of 1.36 differences over more than 95% of the genomes, or 1 disagreement per 2.87 Mb. The range of sites in disagreement varied from 0 to 8, with two outlier samples showing a total of 23 and 19 differences with their corresponding MiSeq consensus sequences. However, both outlier samples showed signs of low-level contamination in the MiSeq runs that may explain the higher level of SNP differences between both workflows. A semirandom subset of samples was selected to create a pairwise SNP matrix to determine if SNP distance analysis can be performed using the MinION platform for contact tracing purposes. Results showed broad agreement between the two workflows and demonstrated their ability to identify potential outbreak clusters and potential transmission links between samples as represented by clusters with limited numbers of SNP differences (Fig. 1).
FIG 1.
MiSeq and MinION single-nucleotide polymorphism (SNP) matrices. A subset of samples from the MiSeq (A) and MinION (B) workflows were used to determine the number of SNP differences across the consensus sequences. Sites with unknown nucleotides (N) in either compared sequences were disregarded. Clusters of samples with potential transmission links as determined by low SNP counts are colored white, while unrelated samples are colored a shade of blue, proportional with the total number of differences found between the two samples.
DISCUSSION
Logistics.
Some important qualities required for an NGS diagnostic test to be appropriate for routine clinical testing are the reliability of the platform, consistencies between flow cells and sequencing kits, and dependability of the manufacturer supply chain. Breakdown in any of these features can lead to negative consequences, such as lack of reliability in the results and delay in reporting. One aspect of this study was to assess the reliability of nanopore sequencing and its supply chain by performing prospective real-time sequencing of clinical samples and to determine if it could be used as a dependable sequencing platform for routine testing in a public health laboratory. During the course of the study, we always had two MinION sequencers available in the event of instrument failure or for the anticipated cases where too many samples were received during a week and needed to be sequenced on two flow cells concurrently. In addition, for redundancy, we had, in our possession, both an Oxford Nanopore MinIT unit for sequencing and base calling, as well as a high-end laptop equipped with a GPU that could also be used for sequencing and base calling, if needed. Over the course of 32 weeks of consecutive sequencing, we reliably received all our scheduled shipments of flow cells and sequencing kits without any delays. MinION flow cell costs can also vary significantly depending on whether purchasing individual flow cells ($900) or bulk ordering multiple flow cells (48 cells for $500 each), with the option of staggering shipments over several months at a significant discount.
Sequencing run QC.
Each MinION flow cell contains a theoretical maximum of 2,048 available and active sequencing pores, with a guarantee of at least 800 active pores per newly purchased flow cells. The number of active pores and their cumulative individual sequencing capabilities dictate the final throughput of a sequencing run. We noticed some variation in the available number of sequencing pores of the unused flow cells, with the number of pores ranging from 1,162 to 1,712 and an average of 1,441 sequencing pores available per flow cell. To ensure sufficient throughput for our weekly samples, we performed quality controls of our flow cells at the time of receiving and reserved cells with higher pore counts for weeks with greater numbers of samples. We also had the possibility of using two flow cells concurrently for sequencing when the number of samples for a given week exceeded the capacity of a single flow cell. Although the sequencing throughput is not quite linear and varies between flow cells, the theoretical throughput of a flow cell can be calculated using the average yields per pore (AYP) obtained from previous runs (AYP = final base call yields/starting number of pores/run time). In our cases, for over 32 runs, we obtain an AYP of 0.368 Mb of data per pores per hour. For a typical unused flow cell with 1,500 starting pores, our expected throughput would be ∼13 Gb and ∼26.5 Gb for sequencing runs of 24 hours and 48 hours, respectively. We also noticed some inconsistencies in the capacity of some flow cells to retain their optimal DNA translocation speed, i.e., the speed at which the DNA strand travels through the pores throughout a run. A slowdown of translocation speed is often associated with a depletion of ATP used by the motor protein on the sensor array. Because the cost associated with ATP refueling is not significant ($9), one could systematically refuel all flow cells at every 24-hour mark. The barcode sequencing kit was also proven to be reliable in terms of amplification performances and consistencies of multiplexing. We did not find any biases in any of the 96 possible barcodes utilized in this study, resulting in approximately equal representation of reads per sample per pool. The number of reads with undetermined barcode sequences for each run ranged from 10% to 15% of the total pool of reads. We did not have any complete sequencing run failures and only had nine samples (2.1%) either failing the library preparation step or that did not generate enough sequencing depth for reliable analyses. This failure rate was slightly below the MiSeq sample failure rate of 3.1%. The nine failed samples were not resequenced on the MinION. Sequencing failure or discrepancies in the results were not associated with flow cell reutilization, refueling, or number of available pores. Overall, these results demonstrate the reliability of the MinION for routine sequencing of clinical isolates. We recommend using at least 30 ng of TB genomic DNA for MinION sequencing in order to minimize sample dropouts. We sequenced ATCC 35734 Mycobacterium bovis strain BCG Pasteur 1173P2 as a control on both platforms to compare with the genome sequence deposited at the NCBI. A total of seven SNP differences were detected between the reference genome and the MinION consensus sequence generated with our pipeline, and the same seven SNPs were also present on the MiSeq data set. We believe that this discrepancy is either due to sequencing errors in the sequence deposited at the NCBI in 2007 or that we sequenced a slightly different genomovar of BCG Pasteur 1173P2. Nevertheless, the perfect correlation with the MiSeq sequence demonstrates that nanopore sequencing can generate highly accurate data with an appropriate analytic workflow. Furthermore, we also added in our data set a sample from the same patient (IDR1900042441-01-01 and IDR1900042441-01-02) that was sequenced twice on two different runs with two different library preparations. This can be used to verify sequence variation and susceptibility prediction differences from run to run. No differences in the mapped sequences and AMR prediction were found between the two samples, showing reproducible sequencing on the MinION platform.
Taxonomic identification.
Using a combination of k-mer matching (Kraken) and presence of specific genomic markers, we were able to correctly identify to the species level in 98.5% of the samples and the specific lineage of M. tuberculosis in 99.5% of the samples. The few species misidentifications were due to difficulty in differentiating between M. tuberculosis and M. africanum (1 case) or M. bovis/M. bovis BCG (5 cases). All in all, these results show that nanopore sequencing can be used with great accuracy for taxonomic identification as long as the reference database contains the organism(s) of interest to find matches. We believe that a few simple changes in our identification algorithm, such as including new strain-specific markers, would increase our ability to distinguish between M. tuberculosis/M. africanum and M. bovis/M. bovis BCG. Genotyping performed by detecting the presence of specific genomic markers, in this case, CRISPR spacers, was also very efficient, with 98% of the correct spoligotypes identified using the MinION workflow, surpassing the 94% accuracy achieved by MiSeq. We found that because of the longer reads generated, the MinION platform was less affected by the presence of contaminating sequences in a data set, as the contaminating reads were simply discarded by the mapping software and not carried forward in the workflow. Differences in the algorithm and thresholds cutoff may have also contributed to the difference in accuracy between the workflows.
Drug susceptibility predictions.
Agreement between MiSeq and MinION phenotypic resistance predictions for nine drugs was 98.1% (Table 2). Most of the discrepancies did not lead to wrongly predicting susceptibility or resistance to a specific drug but occurred when one workflow predicted a susceptible phenotype while the other could not reach a prediction. In addition, many of the discrepant results were caused by the presence of a second subpopulation of MTB in the sample, resulting in some genomic sites being heterozygous. This heterozygosity can be correctly called by the MiSeq workflow if present in at least 10% to 20% of reads but is difficult to assess with high accuracy using the MinION due to higher error rates and, thus, is ignored by the MinION pipeline. It was also common to obtain a significantly lower coverage for the 16S rRNA locus (rrs) on the MinION platform compared to the other loci, leading to our inability to make a resistance prediction for streptomycin, amikacin, or kanamycin in 12.9% of the samples. To minimize the number of failures with the rrs locus, we increased the required throughput of a sequencing run from ∼750 Mb to ∼1 Gb of base call nucleotides per sequenced samples, resulting in a decrease to 1% of unsuccessful resistance prediction for the rrs locus in subsequent weeks. Other discrepancies between the two workflows were mostly caused by the difficulty for the MinION to accurately identify indels. MiSeq will keep outperforming nanopore sequencing for those particular cases until read accuracies and allele frequency consistencies of the MinION improve significantly. Lastly, three occurrences of the same high-confidence mutation in codon 88 of rpsL conferring resistance to streptomycin were missed by the MinION, resulting in a nondetermined prediction. In these cases, the result would prompt phenotypic testing to determine the final DST result. A manual inspection revealed that these mutations were indeed present in the MinION data but at an allele frequency slightly lower (∼65%) than the required threshold of 70% for detection. The reason for the difficulty in detecting this specific mutation with the MinION is unknown, but we suspect that the combination of genomic context at this location and shortcomings in the training of the base caller might be the cause. Nevertheless, the detection of high-confidence mutations known to cause drug resistance, without taking into account phenotypic results, was highly accurate, with a final concordance with DST results of 96.0% for MinION versus 96.2% for the MiSeq platform (Table 4).
TABLE 4.
Performance comparisons between MiSeq and MinION sequencing platforms
| Characteristic | MiSeq | MinION |
|---|---|---|
| Sample failures (no. [%]) | 15 (3.1) | 9 (2.1) |
| Spoligotyping assignation (%) | 95.3 | 98.6 |
| Total NGS/DST concordance (%) | 96.2 | 96.0 |
| Turnaround time (days) | 3 | 2–4 |
| Avg cost per sample ($) | 130 | 63 |
When comparing the WGS predictions for MiSeq and MinION with the DST results, we found overall positive predictive values of 87% and 86% for the MiSeq and MinION, respectively, and a negative predictive value of 97% for both platforms (Table 3). Most of the discrepancies with DST results were either caused by the presence of unknown resistance mechanisms for ethionamide and streptomycin or by overcalling resistance-causing mutations in the pncA locus (pyrazinamide resistance). This is an indication that we still have to refine our knowledge about alternate mechanisms of resistance to some of the commonly used TB drugs. Most of the discrepancies between phenotypic and WGS results had an explanation. For example, we found that all four false-positive samples (Table 3) for rifampin resistance shared an rpoB L511P mutation that is known to confer low-level resistance to rifampin (20, 21). Strains harboring this mutation are consistently found to be susceptible to rifampin when tested with standard DST methods such as MGIT or agar proportion but exhibit an elevated MIC over fully susceptible strains. This is an example where WGS can outperform traditional phenotypic testing. Another example is the five false-negative isoniazid (INH) samples. Two were true discrepancies with no known resistance-causing mutations, one had discordant MGIT DST results and could not conclusively be determined to be truly resistant, and one contained a katG silent mutation at codon 1 (GTG to GTA) that clearly has the potential to disrupt the transcription of the gene and cause resistance. This mutation was detected by both platforms but was reported as sensitive by the bioinformatics pipeline. A simple fix to the algorithm would prevent missing these types of mutations in the future. It is important to note that the issue of the MinION platform with accurately detecting heteroresistance or detecting the presence of some indel positions can result in erroneous reporting. Releases of upgraded flow cells, updated base callers, and better bioinformatics algorithms might help alleviate these deficiencies in the future.
Cluster identification for epidemiological applications.
In addition to clinical diagnostic applications, we also wanted to determine our capabilities of inferring possible transmission links between patients by performing a pairwise comparison of the consensus sequences to generate SNP matrices (Fig. 1). Because mutations independently accumulate over time in genomes from daughter cells derived from the same common ancestor, the closer a group of samples is related in time epidemiologically, the fewer number of SNP differences they will have. A semirandom subset of potentially related and unrelated samples was selected. Six possible transmission clusters were visible in both matrices generated using data from MiSeq and MinION, indicating the potential of utilizing the MinION for disease outbreak investigations. The only difference observed between the workflows was the slightly higher number of SNPs identified using the MinION. This could be the result of a combination of factors such as the algorithm used to call SNPs, overall genome coverage sequencing depth, error rates, and differences in genome masking. Regardless, SNP thresholds to define clusters can be adjusted and reevaluated based on epidemiologically relevant information. Another potential source of errors in performing SNP-based outbreak tracking is the presence of unknown state positions (Ns) in the genomic sequences. The greater the amount of Ns, the lower the number of comparable positions between two sequences, increasing the risk of obtaining lower SNP distances than reality. Using the MinION workflow, we were able to assess, on average, 96.3% of the reference genome (including the masked regions), which is sufficient to achieve accurate SNP counts and is equivalent to those obtained with the MiSeq platform.
Sequencing costs.
The cost per sample was calculated by taking into account flow cell costs, sequencing kits, wash kits, and ATP refueling costs when applicable, and also includes all other costs pre- and postpooling of sample preparation and purification prior to sequencing (Table S7 in the supplemental material). The final costs per sample minus labor, assuming full runs of 18 samples per sequencing run, totaled ∼$63 per sample without flow cell reuse. The costs per sample when reusing a flow cell varied depending on the number of samples sequenced on the same flow cell but averaged ∼$57 per sample. Compared to the MiSeq workflow with average costs of $130 per sample (for a full run of 16 samples), adopting the MinION platform in a public health laboratory offers a substantial financial incentive with minimum impact on the overall assay performances and accuracies.
Hands-on time and ease of use.
One advantage of nanopore sequencing using the MinION platform is its inherent simplicity. From the time you take possession of this sequencer, you can be ready to generate sequences in less than a day. There is no maintenance or calibration associated with sequencing on this platform. As long as the DNA preparation is of good quality, i.e., does not contain any proteins or solvent, not much can go wrong. The only step where the user has to be careful is when loading the library on the flow cells to avoid introducing air bubbles over the sensor array, which will automatically strip away any pores it comes in contact with and cripple the throughput of the flow cell. For this step, it is beneficial for any new users to practice beforehand on older flow cells or attend one of the hands-on trainings offered by Oxford Nanopore. The time needed for the library preparation depends on the protocol chosen and starting material types (DNA, RNA, or amplicons). The total library preparation time can vary from ∼60 min for transposase-based adaptor ligation to up to a few hours or a day for direct ligation protocols with PCR. The final expected yields will also vary depending on the library protocol, with direct ligation having the highest yields of to up to 30 megabases or more. Overall, most protocols are straightforward and do not require sophisticated equipment or extensive training. We estimate that two or three supervised library preparation and flow cell handling training sessions would be sufficient for a new staff member to be proficient using the MinION platform. For this study, we used a direct ligation protocol of barcodes and sequencing adapters. The total time required from receiving DNA to sequencing was ∼10 hours (split over 1.5 days), with most of the hands-on time (∼5.5 hours) spent on the numerous (5 times) magnetic bead cleanups. The rest of the time is spent on incubation times, PCR setup, library quantifications, and flow cell loading. The time for sequencing library preparation from receiving sample DNA was 1.5 days, with sequencing time varying from 5 to 72 hours, depending on the number of samples sequenced and flow cell total throughput needed for analyses. In comparison, the MiSeq Nextera XT protocol requires a total of about 6 hours from sample received to loading, with a total hands-on time of ∼2.5 hours. The sequencing time itself on the MiSeq platform is fixed to 48 hours, while it is variable for nanopore sequencing and can be stopped at any time when sufficient throughput has been achieved for analyses. In all, the turnaround time for WGS MTB sequencing on the MiSeq is about 3 days, while it ranges from 2 to 4 days on the MinION.
Lastly, due to the increasing popularity of nanopore sequencing among laboratories worldwide, the availability of dedicated bioinformatics software for nanopore reads is rapidly expanding. Oxford Nanopore is offering, free of charge, an online suite of analytic programs (EPI2ME) that can help perform some real-time analyses of data; however, this is still in active development, and the current list of available workflows is limited. Many publicly available software applications specialized for nanopore sequencing reads have also been released in the past few years, but most laboratories would still need bioinformatics on staff to develop pipelines using this technology. Other popular applications, previously developed for short-read sequencing, are also starting to be upgraded to accept long-read technology such as Oxford Nanopore. One particular application worth mentioning and related to TB sequencing is TBprofiler (22), a popular online tool that has been recently updated to accept nanopore data for AMR predictions and genotyping of Mycobacterium tuberculosis directly from sequencing reads.
Possible impact on clinical regimens.
The direct impact of MTB MinION sequencing on patients’ treatment is yet to be determined. However, based on our 4 years of experience in reporting back MiSeq WGS results to health providers, this type of approach has proven to be highly beneficial and widely accepted among physicians and our TB control bureau. Whole-genome sequencing is an improvement over other molecular methods such as pyrosequencing and the commercially available Xpert MTB/RIF test from Cepheid for rifampin resistance specifically. Most of these assays are only targeting the rifampin resistance-determining region (RRDR). Since some mutations can be found outside the RRDR or are silent mutations, such mutations would be missed by targeted sequencing and RRDR mutations or mistakenly reported as positive (silent mutations) with Xpert MTB/RIF. One particular case that illustrates this was when an rpoB I572F mutation was found by WGS and no other mutations were detected. This specimen would have likely been susceptible by DST due to this mutation being associated with low-level RIF resistance. Given this information, treatment was changed from rifampin to other drugs by the clinician. Sequencing results have also been used repeatedly and successfully in false-positive investigations and provide evidence for epidemiological links (23, 24).
In conclusion, this study demonstrates that the adoption of MinION Nanopore sequencing for routine clinical diagnostic testing in a public health laboratory setting can be a suitable alternative to the Illumina MiSeq platform. The instruments were reliable, and kit and flow cell performance were consistent across lots, with sequencing cost per sample at a fraction of the MiSeq cost for whole-genome sequencing. The quality of the sequencing data was sufficient to allow for accurate species identification, in silico genotyping, drug resistance predictions, and phylogenetic analysis to assess transmission, with an equal or faster TAT. Some of the shortcomings of the MinION included the difficulty to accurately ascertain small insertions and deletions, as well as heterozygous variants of minor population in a sample and the systematic failure to call the presence of variant bases at certain specific positions in the genome. Active development of the hardware and software of nanopore technology is addressing these shortcomings and promises better single-read accuracy in the near future. Although the current lack of clinically validated nanopore platforms and applications utilizing nanopore sequencing may render its adoption for routine diagnostics challenging in the short term, we believe that this type of long-read technology will be soon widely adopted for clinical diagnostics and may eventually replace, or at least complement, current short-read platforms.
Supplementary Material
ACKNOWLEDGMENTS
This research project was supported by a grant from the National Institute of Allergy and Infectious Diseases (5R21AI13985602).
We also want to acknowledge the Wadsworth Center Advanced Genomic Technologies core for their support throughout this research project.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E. 2018. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect 24:335–341. doi: 10.1016/j.cmi.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Goethem N, Descamps T, Devleesschauwer B, Roosens NHC, Boon NAM, Van Oyen H, Robert A. 2019. Status and potential of bacterial genomics for public health practice: a scoping review. Implement Sci 14:79. doi: 10.1186/s13012-019-0930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shea J, Halse TA, Lapierre P, Shudt M, Kohlerschmidt D, Van Roey P, Limberger R, Taylor J, Escuyer V, Musser KA. 2017. Comprehensive whole-genome sequencing and reporting of drug resistance profiles on clinical cases of Mycobacterium tuberculosis in New York State. J Clin Microbiol 55:1871–1882. doi: 10.1128/JCM.00298-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain M, Olsen HE, Paten B, Akeson M. 2016. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyeon J-Y, Li S, Mann DA, Zhang S, Li Z, Chen Y, Deng X. 2017. Quasimetagenomics-based and real-time-sequencing-aided detection and subtyping of Salmonella enterica from food samples. Appl Environ Microbiol 84:e02340-17. doi: 10.1128/AEM.02340-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards HS, Krishnakumar R, Sinha A, Bird SW, Patel KD, Bartsch MS. 2019. Real-time selective sequencing with RUBRIC: read until with basecall and reference-informed criteria. Sci Rep 9:11475. doi: 10.1038/s41598-019-47857-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanderson ND, Street TL, Foster D, Swann J, Atkins BL, Brent AJ, McNally MA, Oakley S, Taylor A, Peto TEA, Crook DW, Eyre DW. 2018. Real-time analysis of nanopore-based metagenomic sequencing from infected orthopaedic devices. BMC Genomics 19:714. doi: 10.1186/s12864-018-5094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodwin S, McPherson JD, McCombie WR. 2016. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greig DR, Jenkins C, Gharbia S, Dallman TJ. 2019. Comparison of single-nucleotide variants identified by Illumina and Oxford Nanopore technologies in the context of a potential outbreak of Shiga toxin-producing Escherichia coli. Gigascience 8:giz104. doi: 10.1093/gigascience/giz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Votintseva AA, Bradley P, Pankhurst L, Del Ojo Elias C, Loose M, Nilgiriwala K, Chatterjee A, Smith EG, Sanderson N, Walker TM, Morgan MR, Wyllie DH, Walker AS, Peto TEA, Crook DW, Iqbal Z. 2017. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol 55:1285–1298. doi: 10.1128/JCM.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter J-M, Hussain S. 2018. Robust long-read native DNA sequencing using the ONT CsgG Nanopore system. Wellcome Open Res 2:23. doi: 10.12688/wellcomeopenres.11246.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cervantes J, Yokobori N, Hong B-Y. 2020. Genetic identification and drug-resistance characterization of Mycobacterium tuberculosis using a portable sequencing device. A pilot study. Antibiotics 9:548. doi: 10.3390/antibiotics9090548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woods GL, Brown-Elliott BA, Conville PS, Desmond EP, Hall GS, Lin G, Pfyffer GE, Ridderhof JC, Siddiqi SH, Wallace RJ, Warren NG, Witebsky FG. 2011. Susceptibility testing of Mycobacteria, Nocardiae, and other aerobic Actinomycetes, 2nd ed. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 14.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleusener V, Köser CU, Beckert P, Niemann S, Feuerriegel S. 2017. Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools. Sci Rep 7:46327. doi: 10.1038/srep46327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ocheretina O, Shen L, Escuyer VE, Mabou M-M, Royal-Mardi G, Collins SE, Pape JW, Fitzgerald DW. 2015. Whole genome sequencing investigation of a tuberculosis outbreak in Port-au-Prince, Haiti caused by a strain with a “low-level” rpoB mutation L511P – insights into a mechanism of resistance escalation. PLoS One 10:e0129207. doi: 10.1371/journal.pone.0129207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamieson FB, Guthrie JL, Neemuchwala A, Lastovetska O, Melano RG, Mehaffy C. 2014. Profiling of rpoB mutations and MICs for rifampin and rifabutin in Mycobacterium tuberculosis. J Clin Microbiol 52:2157–2162. doi: 10.1128/JCM.00691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phelan JE, O'Sullivan DM, Machado D, Ramos J, Oppong YEA, Campino S, O'Grady J, McNerney R, Hibberd ML, Viveiros M, Huggett JF, Clark TG. 2019. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med 11:41. doi: 10.1186/s13073-019-0650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahuja S, Knorr J, Ramachandran J, Silin M, Meissner JS, Trieu L. 2018. New York City Bureau of Tuberculosis Control annual summary. New York City Department of Health and Mental Hygiene, Queens, NY, https://www1.nyc.gov/assets/doh/downloads/pdf/tb/tb2018.pdf. [Google Scholar]
- 24.Min J, Kim K, Choi H, Kang ES, Shin YM, An JY, Choe KH, Lee KM. 2019. Investigation of false-positive Mycobacterium tuberculosis culture tests using whole genome sequencing. Ann Thorac Med 14:90–93. doi: 10.4103/atm.ATM_184_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MinION sequence reads can be accessed at the NCBI Sequence Read Archives under BioProject accession no. PRJNA650381.