LETTER
Whole-genome sequencing (WGS) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to a better understanding of the virus’s origin, transmission, and evolution (1–6). Multiplex amplicon sequencing for viral WGS is a preferred approach to library preparation since it is simple, sensitive, cost-effective, and scalable (7, 8). However, balancing multiplexed primers to achieve high sensitivity and even coverage can be difficult (8), and superlative analytical sensitivity is not assured. Here, we evaluated the Swift Biosciences’ single-tube SARS-CoV-2 multiplex amplicon sequencing panel for the recovery of genomes from low-viral-load samples (threshold cycle [CT] > 26 on a Hologic Panther Fusion system).
This work was approved by the University of Washington Institutional Review Board (proposal no. STUDY00000408). Libraries were constructed from double-stranded cDNA using Swift Biosciences’ Normalase amplicon SARS-CoV-2 panel. The resulting libraries were sequenced on 2× 300-bp MiSeq runs, and a median of 605,654 reads were obtained for each library. Genomes were assembled with a custom pipeline, TAYLOR (https://github.com/greninger-lab/covid_swift_pipeline). Briefly, sequence reads were trimmed using Trimmomatic v0.38, aligned to the Wuhan-Hu-1 genome (NCBI accession no. NC_045512.2) using BBMap v38.70 (https://sourceforge.net/projects/bbmap/), and trimmed of PCR primers using Primerclip (https://github.com/swiftbiosciences/primerclip). Consensus genomes were called bcftools v1.9.
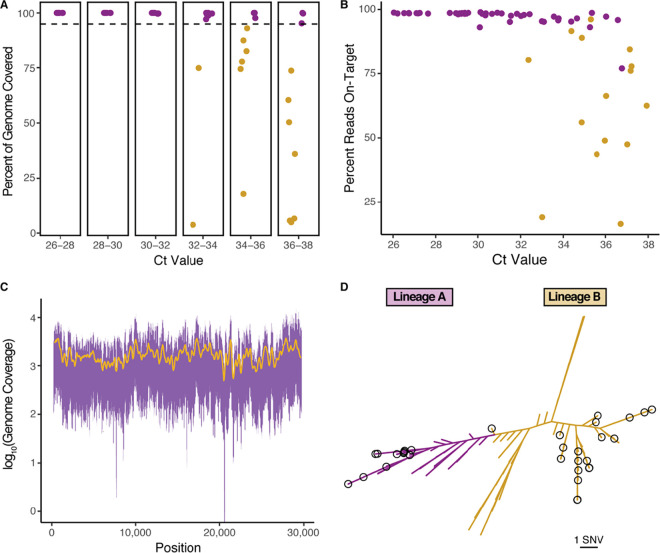
The 61 samples sequenced had CT values ranging from 26.04 to 37.93. We recovered genomes from all samples with a CT of ≤32.16 and from a sample with a CT value of 36.77, equivalent to approximately 4.24 copies input (Fig. 1A) (9–11). For samples with a CT value between 32.01 and 34.00, we recovered genomes from 8/10 (80%) of the samples, and for samples with a CT value between 34.01 and 36.00, we recovered genomes from 4/10 (40%) of the samples. While we recovered genomes from just 3 of the samples with a CT value between 36.01 and 38.00, we were able to recover partial genomes for the other 7 samples (median genome covered, 36.0%; range, 4.9 to 73.7%).
FIG 1.
Evaluation of the Swift Biosciences’ SARS-CoV-2 multiplex amplicon sequencing panel. (A) Complete genomes were recovered from all samples with a CT value of ≤32.16 and a CT value as high as 36.77. Samples for which complete genomes (>95% genome coverage) were recovered are highlighted in purple. Partial genomes are highlighted in gold. (B) SARS-CoV-2 sequences were highly enriched in the sequencing libraries as measured by the percentage of reads mapping to the reference genome for SARS-CoV-2 (NCBI accession no. NC_045512.2). Complete genomes were recovered for samples highlighted in purple, while partial genomes were recovered for those highlighted in gold. (C) The genome coverage between nucleotides 201 and 29741 of the SARS-CoV-2 reference genome is even. The 5th and 95th percentiles of coverage at each position across the 41 samples with a mean depth of >100× are plotted in purple. A 250-nucleotide window moving average is represented in gold. (D) The 46 SARS-CoV-2 samples with complete genomes belong to both major SARS-CoV-2 lineages. A phylogenetic tree with the 46 SARS-CoV-2 genomes recovered in this report and 109 other global strains was constructed with FastTree version 2.1.1. Strains belonging to lineage A are highlighted in purple, while those belonging to lineage B are highlighted in gold. Those genomes sequenced in this report are circled in black. SNV, single nucleotide variation.
The libraries produced with the Swift SARS-CoV-2 amplicon panel were highly enriched for SARS-CoV-2 reads. Samples with CT values ranging from 26.01 to 32.00 had a median on-target percentage of 98.5% (range, 93.1 to 99.0%) after removal of reads attributed to primer dimer formation (Fig. 1B). Among samples with CT values from 32.01 to 38.00, the median on-target percentage was 92.4% (range, 16.5 to 98.7) (Fig. 1B).
We also assessed the coverage distribution from samples with an average depth of >100×. The coverage across the genome for the 41 samples analyzed was highly even (Pielou’s evenness, 0.988) (Fig. 1C). To assess reproducibility, we performed 8 separate library preparations on a single sample. All 8 preparations yielded identical consensus sequences, demonstrating the high reproducibility of the Swift SARS-CoV-2 amplicon panel.
Lastly, we performed a phylogenetic analysis of the 46 strains with complete genomes and 109 randomly selected global SARS-CoV-2 strains. The 46 strains belonged to both major lineages defined by pangolin (https://github.com/cov-lineages/pangolin) (12) and reflected the genomic diversity currently circulating in the SARS-CoV-2 population (Fig. 1D).
In summary, the Swift SARS-CoV-2 amplicon panel is a simple, highly sensitive approach for recovering SARS-CoV-2 genomes. The panel has allowed for the study of genomic rearrangements and mutations that are uniquely associated with low-viral-load samples (13, 14).
Data availability.
Sequencing data are available under NCBI BioProject no. PRJNA610428 (Table 1). Code for assembling consensus FASTA genomes from FASTQ files is also available online (https://github.com/greninger-lab/covid_swift_pipeline).
TABLE 1.
Assembly and sequencing read accession numbers for strains sequenced in this studya
| Strain | GISAID accession no. | SRA accession no. |
|---|---|---|
| WA-UW-4544 | NA | SRR12473512 |
| WA-UW-4545 | EPI_ISL_460621 | SRR11939565 |
| ID-UW-4550 | EPI_ISL_460622 | SRR11939564 |
| WA-UW-4554 | NA | SRR12473511 |
| WA-UW-4555 | EPI_ISL_460623 | SRR11939553 |
| WA-UW-4562 | EPI_ISL_460624 | SRR11939542 |
| WA-UW-4563 | EPI_ISL_515271 | SRR11939540 |
| WA-UW-4569 | EPI_ISL_515272 | SRR11939539 |
| WA-UW-4570 | EPI_ISL_460625 | SRR11939538 |
| WA-UW-4572 | EPI_ISL_497872 | SRR11939537 |
| WA-UW-4581 | EPI_ISL_515273 | SRR11939536 |
| WA-UW-4585 | EPI_ISL_515274 | SRR11939535 |
| WA-UW-4588 | EPI_ISL_460626 | SRR11939563 |
| WA-UW-4589 | EPI_ISL_515275 | SRR11939562 |
| WA-UW-4591 | EPI_ISL_460627 | SRR11939561 |
| WA-UW-4592 | EPI_ISL_515276 | SRR11939560 |
| ID-UW-4597 | EPI_ISL_460628 | SRR11939559 |
| WA-UW-4599 | EPI_ISL_515277 | SRR11939558 |
| ID-UW-4600 | EPI_ISL_460629 | SRR11939557 |
| ID-UW-4601 | EPI_ISL_515270 | SRR11939556 |
| WA-UW-4603 | EPI_ISL_515278 | SRR11939555 |
| WA-UW-4607 | EPI_ISL_513632 | SRR12515115 |
| WA-UW-4608 | EPI_ISL_515279 | SRR11939554 |
| WA-UW-4610 | EPI_ISL_515286 | SRR12515114 |
| WA-UW-4614 | EPI_ISL_515280 | SRR11939552 |
| WA-UW-4615 | EPI_ISL_515281 | SRR11939551 |
| WA-UW-4616 | EPI_ISL_515282 | SRR11939550 |
| WA-UW-4618 | EPI_ISL_515283 | SRR11939549 |
| WA-UW-4619 | EPI_ISL_460630 | SRR11939548 |
| WA-UW-4620 | EPI_ISL_515284 | SRR11939547 |
| WA-UW-4623 | EPI_ISL_515285 | SRR11939546 |
| WA-UW-4627 | EPI_ISL_513633 | SRR12515113 |
| WA-UW-4632 | EPI_ISL_460631 | SRR11939545 |
| WA-UW-4633 | EPI_ISL_460632 | SRR11939544 |
| WA-UW-4636 | NA | SRR12473505 |
| WA-UW-4641 | NA | SRR12473504 |
| WA-UW-4643 | EPI_ISL_460633 | SRR11939543 |
| WA-UW-4646 | EPI_ISL_460634 | SRR11939541 |
| WA-UW-4648 | NA | SRR12473503 |
| WA-UW-4657 | EPI_ISL_461399 | SRR11940010 |
| WA-UW-4660 | EPI_ISL_513634 | SRR11940009 |
| WA-UW-4663 | EPI_ISL_461400 | SRR11939943 |
| WA-UW-4664 | EPI_ISL_513635 | SRR11939932 |
| WA-UW-4665 | NA | SRR12473502 |
| WA-UW-4684 | EPI_ISL_461401 | SRR11939921 |
| WA-UW-4687 | NA | SRR12473501 |
| WA-UW-4698 | EPI_ISL_461402 | SRR11939974 |
| WA-UW-4731 | EPI_ISL_513636 | SRR12515112 |
| WA-UW-4735 | NA | SRR12473500 |
| WA-UW-4738 | NA | SRR12473499 |
| WA-UW-4750 | NA | SRR12473498 |
| WA-UW-4758 | EPI_ISL_513637 | SRR11939963 |
| WA-UW-4761 | NA | SRR12473510 |
| WA-UW-4765 | EPI_ISL_461403 | SRR11939952 |
| WA-UW-4881 | NA | SRR12473509 |
| WA-UW-4884 | NA | SRR12473508 |
| WA-UW-4888 | NA | SRR12473507 |
| WA-UW-4898 | EPI_ISL_513638 | SRR12515111 |
| WA-UW-4903 | NA | SRR12473506 |
| WA-UW-4917 | EPI_ISL_461404 | SRR11939996 |
| WA-UW-4931 | EPI_ISL_461405 | SRR11939985 |
Samples without a GISAID accession number were not considered complete genomes. NA, not applicable.
Supplementary Material
ACKNOWLEDGMENT
Swift Biosciences provided reagent for optimization of this protocol but had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat Med 26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng X, Gu W, Federman S, Du Plessis L, Pybus OG, Faria NR, Wang C, Yu G, Bushnell B, Pan C-Y, Guevara H, Sotomayor-Gonzalez A, Zorn K, Gopez A, Servellita V, Hsu E, Miller S, Bedford T, Greninger AL, Roychoudhury P, Starita LM, Famulare M, Chu HY, Shendure J, Jerome KR, Anderson C, Gangavarapu K, Zeller M, Spencer E, Andersen KG, MacCannell D, Paden CR, Li Y, Zhang J, Tong S, Armstrong G, Morrow S, Willis M, Matyas BT, Mase S, Kasirye O, Park M, Masinde G, Chan C, Yu AT, Chai SJ, Villarino E, Bonin B, Wadford DA, Chiu CY. 2020. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science 369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauver JR, Petrone ME, Hodcroft EB, Shioda K, Ehrlich HY, Watts AG, Vogels CBF, Brito AF, Alpert T, Muyombwe A, Razeq J, Downing R, Cheemarla NR, Wyllie AL, Kalinich CC, Ott IM, Quick J, Loman NJ, Neugebauer KM, Greninger AL, Jerome KR, Roychoudhury P, Xie H, Shrestha L, Huang M-L, Pitzer VE, Iwasaki A, Omer SB, Khan K, Bogoch II, Martinello RA, Foxman EF, Landry ML, Neher RA, Ko AI, Grubaugh ND. 2020. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell 181:990–996.E5. doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z, Cui J, Lu J. 2020. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev 7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, Keeley AJ, Lindsey BB, Parsons PJ, Raza M, Rowland-Jones S, Smith N, Tucker RM, Wang D, Wyles MD. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.E19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedford T, Greninger AL, Roychoudhury P, Starita LM, Famulare M, Huang M-L, Nalla A, Pepper G, Reinhardt A, Xie H, Shrestha L, Nguyen TN, Adler A, Brandstetter E, Cho S, Giroux D, Han PD, Fay K, Frazar CD, Ilcisin M, Lacombe K, Lee J, Kiavand A, Richardson M, Sibley TR, Truong M, Wolf CR, Nickerson DA, Rieder MJ, Englund JA, Hadfield J, Hodcroft EB, Huddleston J, Moncla LH, Müller NF, Neher RA, Deng X, Gu W, Federman S, Chiu C, Duchin JS, Gautom R, Melly G, Hiatt B, Dykema P, Lindquist S, Queen K, Tao Y, Uehara A, Tong S, MacCannell D, Armstrong GL, Seattle Flu Study Investigators, et al. 2020. Cryptic transmission of SARS-CoV-2 in Washington state. Science. doi: 10.1126/science.abc0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyson JR, James P, Stoddart D, Sparks N, Wickenhagen A, Hall G, Choi JH, Lapointe H, Kamelian K, Smith AD, Prystajecky N, Goodfellow I, Wilson SJ, Harrigan R, Snutch TP, Loman NJ, Quick J. 2020. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv 10.1101/2020.09.04.283077. [DOI]
- 8.Itokawa K, Sekizuka T, Hashino M, Tanaka R, Kuroda M. 2020. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS One 15:e0239403. doi: 10.1371/journal.pone.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman JA, Pepper G, Naccache SN, Huang M-L, Jerome KR, Greninger AL. 2020. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol 58:e00821-20. doi: 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nalla AK, Casto AM, Huang M-LW, Perchetti GA, Sampoleo R, Shrestha L, Wei Y, Zhu H, Jerome KR, Greninger AL. 2020. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J Clin Microbiol 58:e00557-20. doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perchetti GA, Nalla AK, Huang M-L, Zhu H, Wei Y, Stensland L, Loprieno MA, Jerome KR, Greninger AL. 2020. Validation of SARS-CoV-2 detection across multiple specimen types. J Clin Virol 128:104438. doi: 10.1016/j.jcv.2020.104438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 15 July 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Addetia A, Xie H, Roychoudhury P, Shrestha L, Loprieno M, Huang M-L, Jerome KR, Greninger AL. 2020. Identification of multiple large deletions in ORF7a resulting in in-frame gene fusions in clinical SARS-CoV-2 isolates. J Clin Virol 129:104523. doi: 10.1016/j.jcv.2020.104523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Addetia A, Lieberman NAP, Phung Q, Xie H, Roychoudhury P, Shrestha L, Loprieno M, Huang M-L, Jerome KR, Greninger AL. 2020. SARS-CoV-2 ORF6 disrupts nucleocytoplasmic transport through interactions with Rae1 and Nup98. bioRxiv https://www.biorxiv.org/content/10.1101/2020.08.03.234559v1.full. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available under NCBI BioProject no. PRJNA610428 (Table 1). Code for assembling consensus FASTA genomes from FASTQ files is also available online (https://github.com/greninger-lab/covid_swift_pipeline).
TABLE 1.
Assembly and sequencing read accession numbers for strains sequenced in this studya
| Strain | GISAID accession no. | SRA accession no. |
|---|---|---|
| WA-UW-4544 | NA | SRR12473512 |
| WA-UW-4545 | EPI_ISL_460621 | SRR11939565 |
| ID-UW-4550 | EPI_ISL_460622 | SRR11939564 |
| WA-UW-4554 | NA | SRR12473511 |
| WA-UW-4555 | EPI_ISL_460623 | SRR11939553 |
| WA-UW-4562 | EPI_ISL_460624 | SRR11939542 |
| WA-UW-4563 | EPI_ISL_515271 | SRR11939540 |
| WA-UW-4569 | EPI_ISL_515272 | SRR11939539 |
| WA-UW-4570 | EPI_ISL_460625 | SRR11939538 |
| WA-UW-4572 | EPI_ISL_497872 | SRR11939537 |
| WA-UW-4581 | EPI_ISL_515273 | SRR11939536 |
| WA-UW-4585 | EPI_ISL_515274 | SRR11939535 |
| WA-UW-4588 | EPI_ISL_460626 | SRR11939563 |
| WA-UW-4589 | EPI_ISL_515275 | SRR11939562 |
| WA-UW-4591 | EPI_ISL_460627 | SRR11939561 |
| WA-UW-4592 | EPI_ISL_515276 | SRR11939560 |
| ID-UW-4597 | EPI_ISL_460628 | SRR11939559 |
| WA-UW-4599 | EPI_ISL_515277 | SRR11939558 |
| ID-UW-4600 | EPI_ISL_460629 | SRR11939557 |
| ID-UW-4601 | EPI_ISL_515270 | SRR11939556 |
| WA-UW-4603 | EPI_ISL_515278 | SRR11939555 |
| WA-UW-4607 | EPI_ISL_513632 | SRR12515115 |
| WA-UW-4608 | EPI_ISL_515279 | SRR11939554 |
| WA-UW-4610 | EPI_ISL_515286 | SRR12515114 |
| WA-UW-4614 | EPI_ISL_515280 | SRR11939552 |
| WA-UW-4615 | EPI_ISL_515281 | SRR11939551 |
| WA-UW-4616 | EPI_ISL_515282 | SRR11939550 |
| WA-UW-4618 | EPI_ISL_515283 | SRR11939549 |
| WA-UW-4619 | EPI_ISL_460630 | SRR11939548 |
| WA-UW-4620 | EPI_ISL_515284 | SRR11939547 |
| WA-UW-4623 | EPI_ISL_515285 | SRR11939546 |
| WA-UW-4627 | EPI_ISL_513633 | SRR12515113 |
| WA-UW-4632 | EPI_ISL_460631 | SRR11939545 |
| WA-UW-4633 | EPI_ISL_460632 | SRR11939544 |
| WA-UW-4636 | NA | SRR12473505 |
| WA-UW-4641 | NA | SRR12473504 |
| WA-UW-4643 | EPI_ISL_460633 | SRR11939543 |
| WA-UW-4646 | EPI_ISL_460634 | SRR11939541 |
| WA-UW-4648 | NA | SRR12473503 |
| WA-UW-4657 | EPI_ISL_461399 | SRR11940010 |
| WA-UW-4660 | EPI_ISL_513634 | SRR11940009 |
| WA-UW-4663 | EPI_ISL_461400 | SRR11939943 |
| WA-UW-4664 | EPI_ISL_513635 | SRR11939932 |
| WA-UW-4665 | NA | SRR12473502 |
| WA-UW-4684 | EPI_ISL_461401 | SRR11939921 |
| WA-UW-4687 | NA | SRR12473501 |
| WA-UW-4698 | EPI_ISL_461402 | SRR11939974 |
| WA-UW-4731 | EPI_ISL_513636 | SRR12515112 |
| WA-UW-4735 | NA | SRR12473500 |
| WA-UW-4738 | NA | SRR12473499 |
| WA-UW-4750 | NA | SRR12473498 |
| WA-UW-4758 | EPI_ISL_513637 | SRR11939963 |
| WA-UW-4761 | NA | SRR12473510 |
| WA-UW-4765 | EPI_ISL_461403 | SRR11939952 |
| WA-UW-4881 | NA | SRR12473509 |
| WA-UW-4884 | NA | SRR12473508 |
| WA-UW-4888 | NA | SRR12473507 |
| WA-UW-4898 | EPI_ISL_513638 | SRR12515111 |
| WA-UW-4903 | NA | SRR12473506 |
| WA-UW-4917 | EPI_ISL_461404 | SRR11939996 |
| WA-UW-4931 | EPI_ISL_461405 | SRR11939985 |
Samples without a GISAID accession number were not considered complete genomes. NA, not applicable.