Abstract
Purpose:
Physicians are expected to assess prognosis both for patient counseling and for determining suitability for clinical trials. Increasingly, cell-free circulating tumor DNA (cfDNA) sequencing is being performed for clinical decision-making. We sought to determine whether variant allele frequency (VAF) in cfDNA is associated with prognosis.
Experimental Design:
We performed a retrospective analysis of 298 patients with metastatic disease who underwent clinical comprehensive cfDNA analysis and assessed association between VAF and overall survival.
Results:
cfDNA mutations were detected in 240 (80.5%) patients. Median overall survival (OS) was 11.5 months. cfDNA mutation detection and number of nonsynonymous mutations (NSM) significantly differed between tumor types; being lowest in appendiceal cancer and highest in colon cancer. Having more than one NSM detected was associated with significantly worse OS (HR = 2.3, p < 0.0001). VAF was classified by quartiles, Q1 lowest, Q4 highest VAF. Higher VAF levels were associated with a significantly worse overall survival (VAF Q3 HR 2.3, p = 0.0069: VAF Q4 HR = 3.8, p < 0.0001) on univariate analysis. On multivariate analysis, VAF Q4, male sex, albumin level <3.5 g/dL, number of non-visceral metastatic sites >0 and number of prior therapies >4 were independent predictors of worse OS.
Conclusions:
Higher levels of cfDNA VAF and higher number of NSM were associated with worse OS in patients with metastatic disease. Further study is needed to determine optimal VAF thresholds for clinical decision-making and the utility of cfDNA VAF as a prognostic marker in different tumor types.
Keywords: Cell-free DNA, Circulating tumor DNA, Overall survival, Prognosis, Metastatic Cancer
INTRODUCTION
There is an increasing emphasis on optimal cancer management and the importance of prognostication in routine cancer care. The ability to assess prognosis is also very important for research-driven care. Most clinical trials for advanced/metastatic disease list an expected survival of 3 months or longer as an eligibility criterion. However, Arkenau et al described that 18% of patients treated on Phase I trials died within 90 days of starting therapy. Although a variety of clinical factors have been defined as predictors of early mortality and overall survival (1–3), the ability to assess prognosis in the clinic remains challenging. There remains a need for objective biomarkers for prognosis.
Cell-free circulating DNA (cfDNA) is a minimally invasive way to obtain a global picture of mutational status in metastatic cancer patients. Somatic mutations in cfDNA originate from primary tumor, and/or metastases, and is thought to represent a more global picture of the heterogeneity of the tumor (4). Clonal hematopoiesis may also contribute somatic mutations to cfDNA. cfDNA testing is increasingly being explored in precision oncology as a minimally invasive and rapid approach to perform genomic profiling in the setting of advanced/metastatic disease for treatment selection (5), as an early marker of response, and to track mechanisms of acquired resistance (6).
Variant allele frequency (VAF) is the number of mutant molecules over total number of wild-type molecules at a specific location in the genome. As suggested by preliminary data we hypothesize that VAF in part would act as surrogate of tumor burden and therefore maximum VAF (MaxVAF) would negatively correlate with prognosis and overall survival (OS) (7–10). Additionally, we hypothesize that the increasing complexity of the tumor may worsen prognosis, and thus increasing number of genomic alterations would be associated with worse prognosis.
MATERIALS AND METHODS
Patient cohort:
Total of 298 patients with advanced cancers treated at the University of Texas MD Anderson Cancer Center were prospectively enrolled on a molecular testing protocol (NCT01772771) across multiple disease centers. The study was performed after approval by the Institutional review board. This study was conducted in accordance to the U.S Common Rule. Investigators obtained written informed consent from the patients for participation in the study. Patients were eligible for the study if they had active metastatic/local inoperable advanced cancer, and they were considering trial enrollment within the next two lines of therapy. cfDNA analysis was performed for patients without available archival tissue block for genomic testing, or archival tissue >1-year, or for patients who progressed on intervening therapy. We performed a retrospective review of medical records for clinical and pathological variable associated with prognosis.
cfDNA testing protocol:
Plasma was analyzed using targeted comprehensive cfDNA Next-Generation Sequencing (NGS) platform and performed in a Clinical laboratory improvement Amendments (CLIA) certified, College of American Pathologist (CAP)-accredited, and New York State Department of Health-approved laboratory (Guardant Health, Redwood City, CA). All samples were collected and processed with standard operating procedures in accordance with CLIA/CAP guidelines (11, 12). Each patient was assigned two 10 mL Streck tubes containing whole blood. Plasma was stored over a period of 24 to 48 hours before final analysis was performed.
cfDNA was extracted from plasma using a Qiagen circulating nucleic acid kit and DNA in 5–30 ng quantities was exposed to barcoding to be processed and sequenced into a digital library as described earlier (13–15). During the course of this study, the Guardant360 sequencing ranged from 54 to 73 genes to detect genomic alterations including: single nucleotide variations, amplifications, fusions (in ALK, FGFR2, FGFR3, RET, ROS1, and NTRK1 genes), short insertions/deletions, and splice site-disrupting events. One patient underwent 54 gene panel testing, 164 patients underwent 70 gene testing, and 133 patients completed 73 gene testing. Target genes were sequenced by using Illumina HiSeq 2500 or Illumina NextSeq 500 platforms and hq19 as the reference genome. The VAF was defined as number of mutant molecules at a specific nucleotide location over total number of molecules present in the background at a specific given genomic location. VAF levels needed to be detected for indel, fusions and SNVs by Guardant360 panel is 0.1%. Clonal hematopoiesis of indeterminate potential (CHIP) alterations were not filtered given these variants are typically observed at low VAF (e.g. <1%) and unlikely to influence the calculations on maximum VAF in the sample.
Statistical analysis:
Our primary outcome measure was to determine the influence of maximum VAF on OS. Secondary objectives included association of maximum VAF and alteration detection with nonsynonymous mutations (NSM), OS, and site of primary malignancy.
Cox proportional hazards regression methodology was utilized to evaluate hazard ratios for OS. OS was determined by using Kaplan-Meier method from time of blood draw until date of last follow-up or death. All statistical analyses were performed using S-plus for Windows version 8.0 (TIBCO Software, inc). Quartiles divided the patients into 4 equally sized groups of increasing average. Maximum VAF was analyzed on the log scale as a continuous variable. Quartiles were defined as 25th, 50th and 75th percentiles. Groups formed by quartiles were: Q1 = 0–25th, Q2 = 25th*−50th, Q3 = 50th*−75th, Q4 = 75th*-max percentile where * denote the values just above the quartiles. Kruskal-Wallis test was performed to determine if there were statistically significant differences of an independent variable between several groups.
Spearman’s rank correlation coefficient was used to measure the strength of association between two variables.
RESULTS
Patient Characteristics and cfDNA Detection:
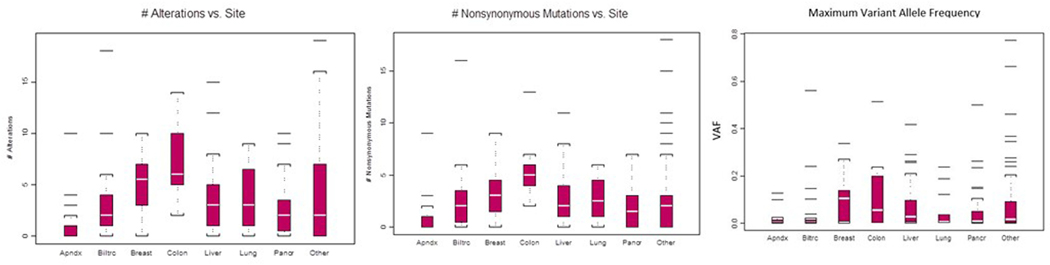
Patient characteristics and tumor types can be found in Table 1. Two hundred and forty (80.5%) of 298 patients had genomic alterations (copy number or mutation changes) detected during cfDNA analysis. We found that the median number of alterations varied by disease (Kruskal-Wallis test p=0.0001, Figure 1A). The highest median number of alterations were in patients with colorectal cancer, while lowest levels were found in appendiceal carcinoma. The total number of NSM detected was 752, and patients with colorectal cancer had the highest number. The maximum VAF statistically varied by tumor type (Kruskal-Wallis test p=0.0013, Figure 1C). The median maximum VAF was highest in breast cancer patients and lowest amongst patients with appendiceal carcinoma.
Table 1.
Patient characteristics n=298
| Characteristic | N (%) |
|---|---|
| Age, y, median | 59 |
| Range | 8–84 |
| Sex | |
| Female | 123 (41%) |
| Male | 175 (59%) |
| Primary site of malignancy | |
| Liver | 61 (20%) |
| Biliary tract | 36 (12%) |
| Appendix | 24 (8%) |
| Breast | 22 (7%) |
| Colorectal | 13 (4%) |
| Pancreas | 52 (17%) |
| Lung | 16 (6%) |
| Other | 74 (26%) |
| Patients with alterations detected | 240 (80%) |
| Platelets >198 | 143 (47 %) |
| WBC >5.9 | 139 (46%) |
| HGB >11.9 | 141 (47 %) |
| Albumin <3.5 | 50 (16%) |
| ECOG >1 | 20 (7%) |
| Number of prior therapies >4 | 59 (20%) |
| History of Thromboembolism | 31 (10%) |
| History of radiation | 123 (41%) |
| Liver metastasis | 76 (25%) |
| Number of metastasis >2 | 51 (17%) |
| Number of non-visceral metastasis >0 | 123 (41%) |
| Number of visceral metastasis >0 | 181 (60%) |
Figure 1. cfDNA Alterations by Site of Malignancy.
A. Number of alterations vs primary malignancy site. B. Number of nonsynonymous vs primary malignancy site. C. Maximum variant allele frequency (VAF) vs site of primary malignancy
Association of cfDNA characteristics and Overall Survival
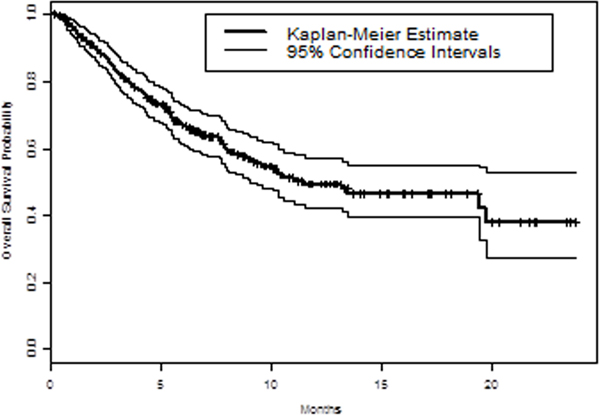
Median OS was 11.5 months, and median follow up time was 8.4 months. Kaplan Meier estimate curve with 95% confidence intervals were plotted in Figure 2A. Out of the 298 patients, 116 (38%) deaths were observed. At 3 months 83% (79–88%), 6 months 67% (61–73%), and at 12 months 49% (42–57%) of patients were still alive.
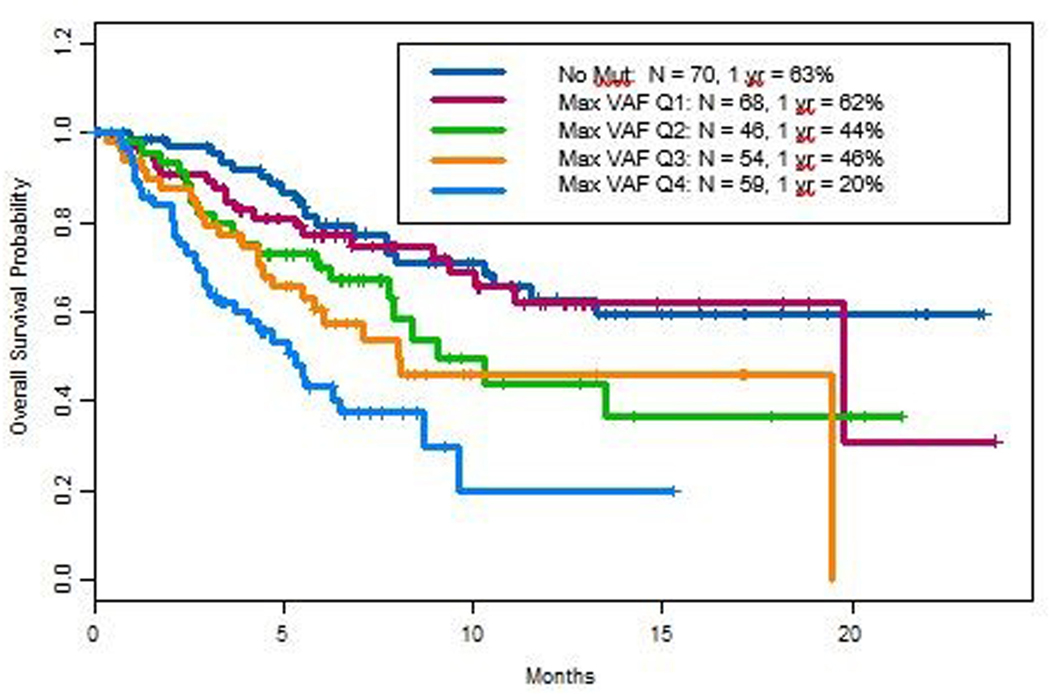
Figure 2. Variant allele frequency (VAF) and Overall Survival (OS).
A. Kaplan Meier estimate curve with 95% confidence intervals were plotted for OS of the study cohort. B. Kaplan Meier analysis of OS by maximum VAF. Maximum VAF was analyzed by quartiles, with Q4 and Q1 representing the highest and slowest quartile respectively.
We used Kaplan-Meier analysis to determine the association of OS with maximum VAF (MaxVAF) by quartiles, with Q4 and Q3 representing the highest and second highest quartile respectively (Figure 2B). maxVAF ranges from 0.1% to 77.3%. MaxVAF was 0.3% for Q1, 1.3% for Q2, 8.6% for Q3. One year OS of patients with no mutations detected and those with lowest quartile of maximum VAF were 63% and 62% respectively. In contrast, patients with maxVAF Q4 had a 1-year OS of 20%. Supplemental Table 1 demonstrates an increasing number of alterations with increasing levels of maxVAF.
Table 2 demonstrates both univariate and multivariate hazard ratios after adjustment for specific variables. On univariate analysis, MaxVAF Q3 (HR 2.3, p=0.0069) and MaxVAF Q4 (HR 3.8, p<0.0001) were both significantly associated with worse OS. On multivariate analysis, top quartile (MaxVAF Q4), being a male, albumin less than 3.5 gm/dL, more than 4 prior therapies and presence of non-visceral metastasis were independently correlated with worse OS (Table 3).
Table 2.
Univariate analysis of Maximum VAF
| UNIVARIATE | ||
|---|---|---|
| Group | HR (95% CI) | p-value |
| No mutation | 1.0 | -- |
| Max VAF Q1 | 1.2 (0.6, 2.2) | 0.58 |
| Max VAF Q2 | 1.8 (1.0, 3.4) | 0.057 |
| Max VAF Q3 | 2.3 (1.3, 4.2) | 0.0069 |
| Max VAF Q4 | 3.8 (2.2, 6.8) | <0.0001 |
Table 3.
Prognostic Variables by Multivariate analysis
| Multivariate (n=209)* | ||
|---|---|---|
| Group | HR (95% CI) | p-value |
| Max VAF Q1 | 1.3 (0.6, 2.7) | 0.48 |
| Max VAF Q2 | 1.2 (0.6, 2.6) | 0.56 |
| Max VAF Q3 | 1.5 (0.7, 3.1) | 0.25 |
| Max VAF Q4 | 2.6 (1.2, 5.2) | 0.011 |
| Age >60 | 1.2 (0.8, 2.0) | 0.39 |
| Male Sex | 1.7 (1.1, 2.7) | 0.025 |
| Platelets >198 | 0.8 (0.5, 1.3) | 0.34 |
| ECOG >1 | 1.5 (0.8, 2.9) | 0.20 |
| WBC >5.9 | 1.2 (0.8, 1.8) | 0.44 |
| Hgb >11.9 | 0.9 (0.6, 1.5) | 0.73 |
| LDH >618 | 1.6 (0.9, 2.7) | 0.085 |
| Albumin <3.5 | 2.4 (1.4, 4.1) | 0.00011 |
| History of Thromboembolism | 1.0 (0.5, 2.0) | 0.94 |
| History of radiation | 0.7 (0.5, 1.2) | 0.26 |
| Liver metastasis | 1.1 (0.6, 2.0) | 0.71 |
| Number of prior therapies >4 | 0.5 (0.3, 1.0) | 0.044 |
| Number of alterations 3–5 vs <3 | 0.8 (0.4, 1.6) | 0.57 |
| >5 vs <3 | 1.6 (0.9, 2.8) | 0.12 |
| Number metastasis >2 | 1.7 (0.9, 3.2) | 0.11 |
| Number of Visceral metastasis >0 | 1.6 (0.9, 2.7) | 0.085 |
| Number of non-Visceral metastasis >0 | 2.0 (1.2, 3.2) | 0.0046 |
Only 209 patients were analyzed due to missing variables
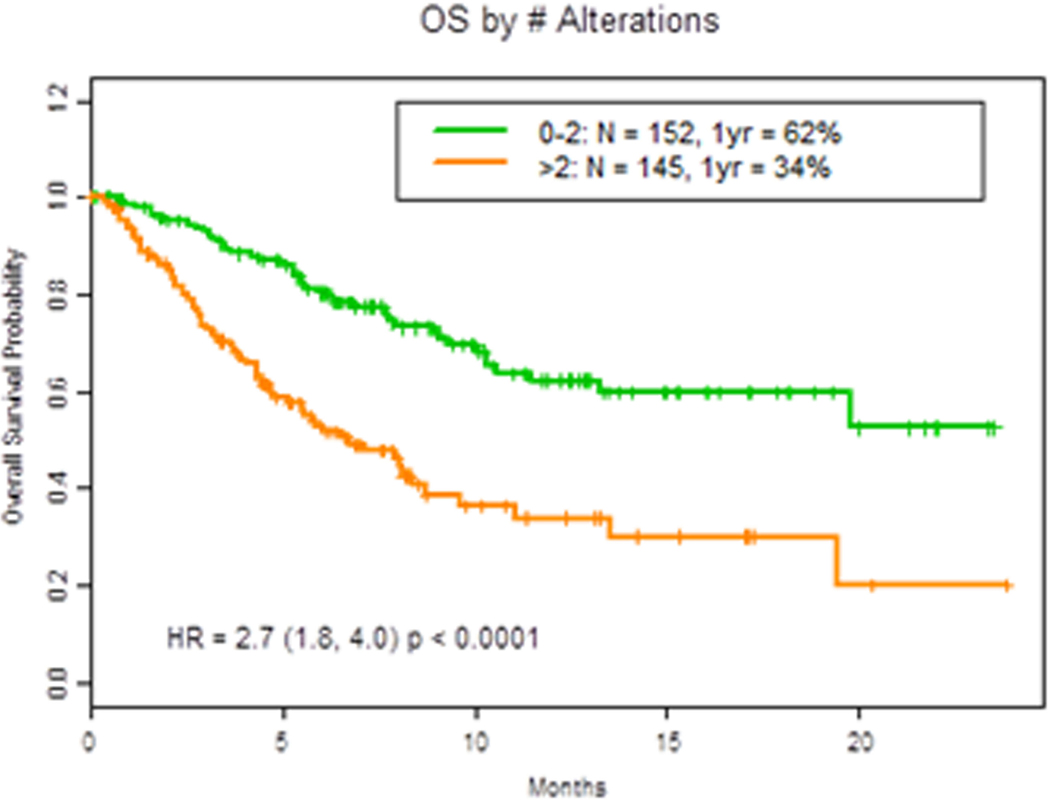
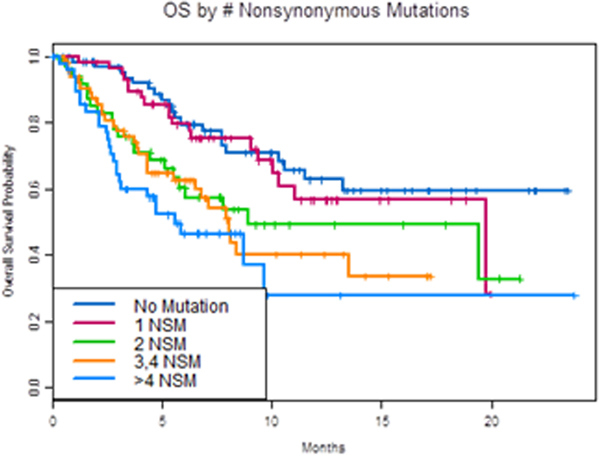
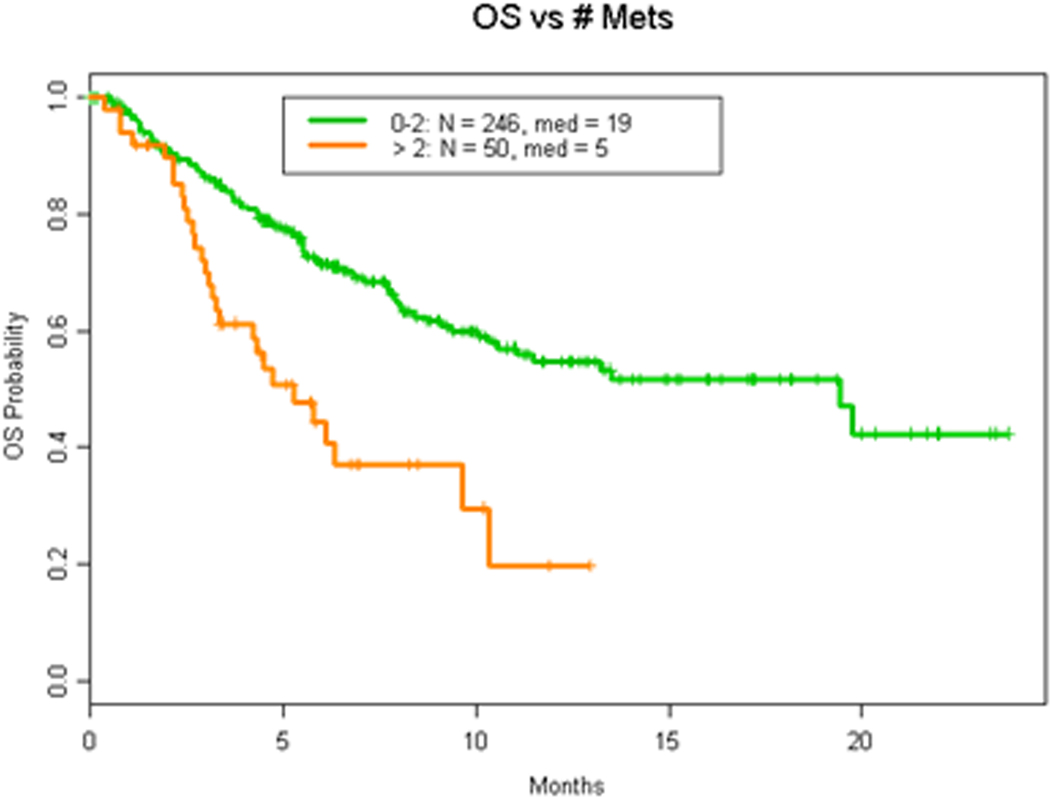
We next analyzed whether the number of mutations detected affected prognosis. Indeed, we found that patients with more than two genomic alterations (mutations or copy number changes) were associated with worse OS (HR = 2.7 (1.8, 4.0) p <0.0001; Figure 3A). Having more than one NSM (HR 2.3 (1.6, 3.4, p<0.0001) was found to negatively impact OS, as demonstrated in Figure 3B.
Figure 3. OS by number of alterations.
A. OS in patients with 2 or less vs more than 2 cfDNA alterations. B. OS by number of nonsynonymous mutations (NSM).
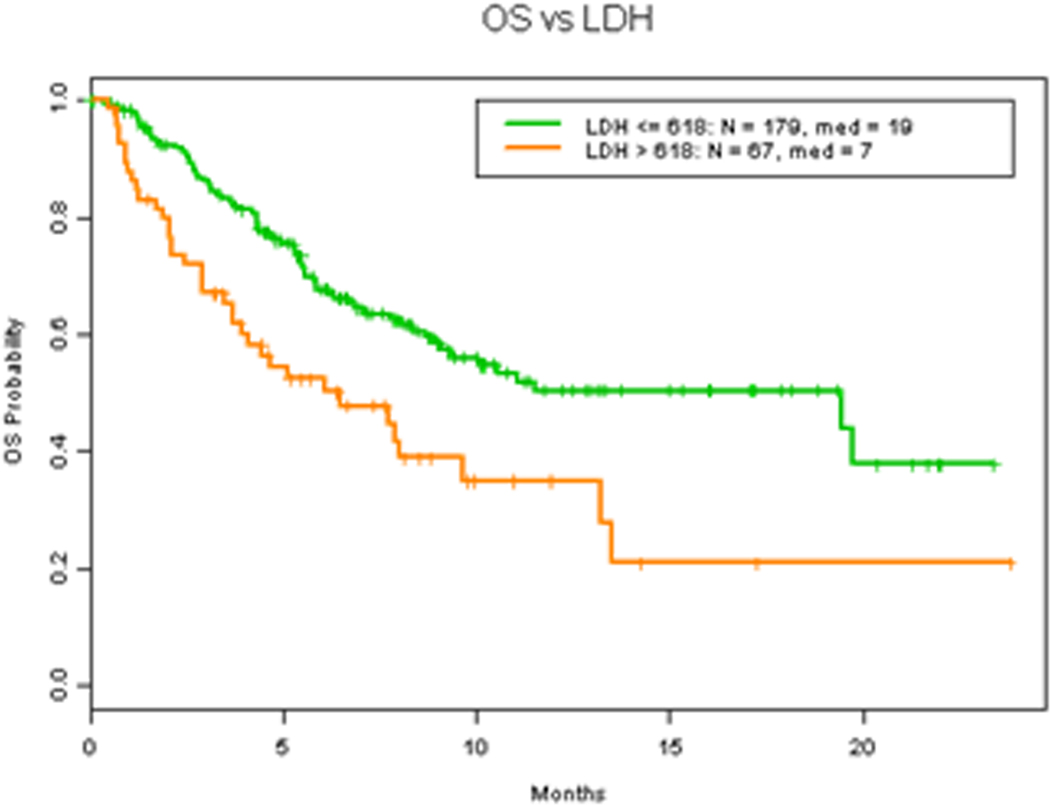
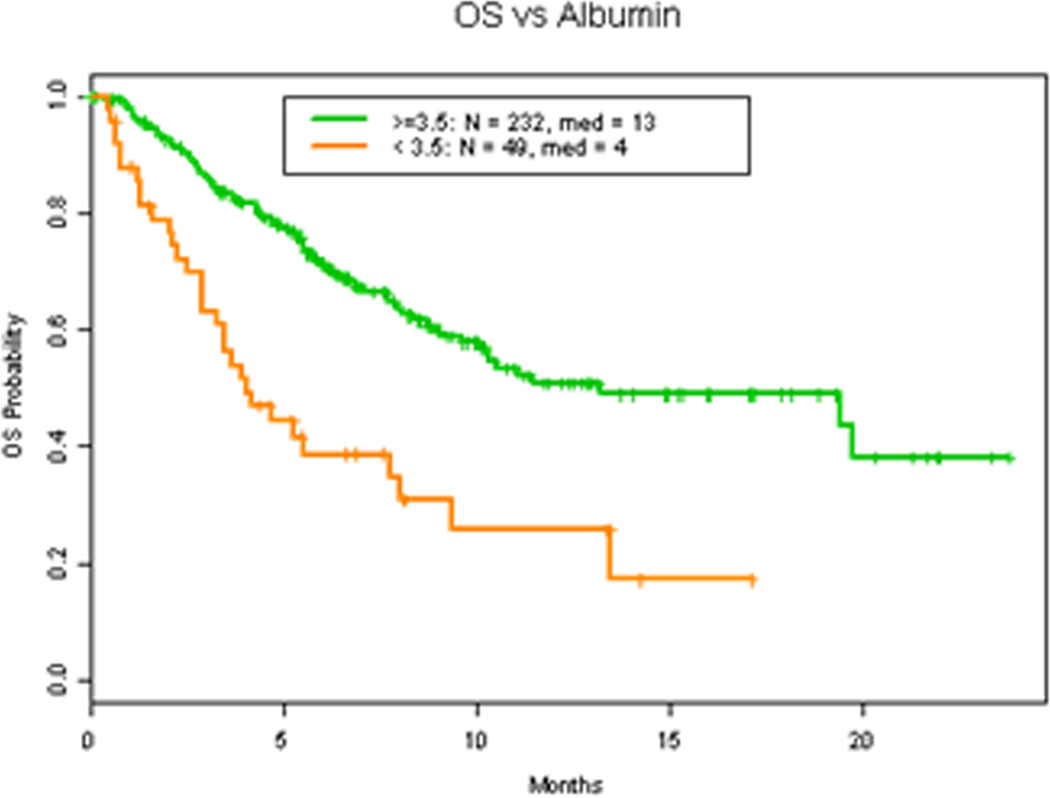
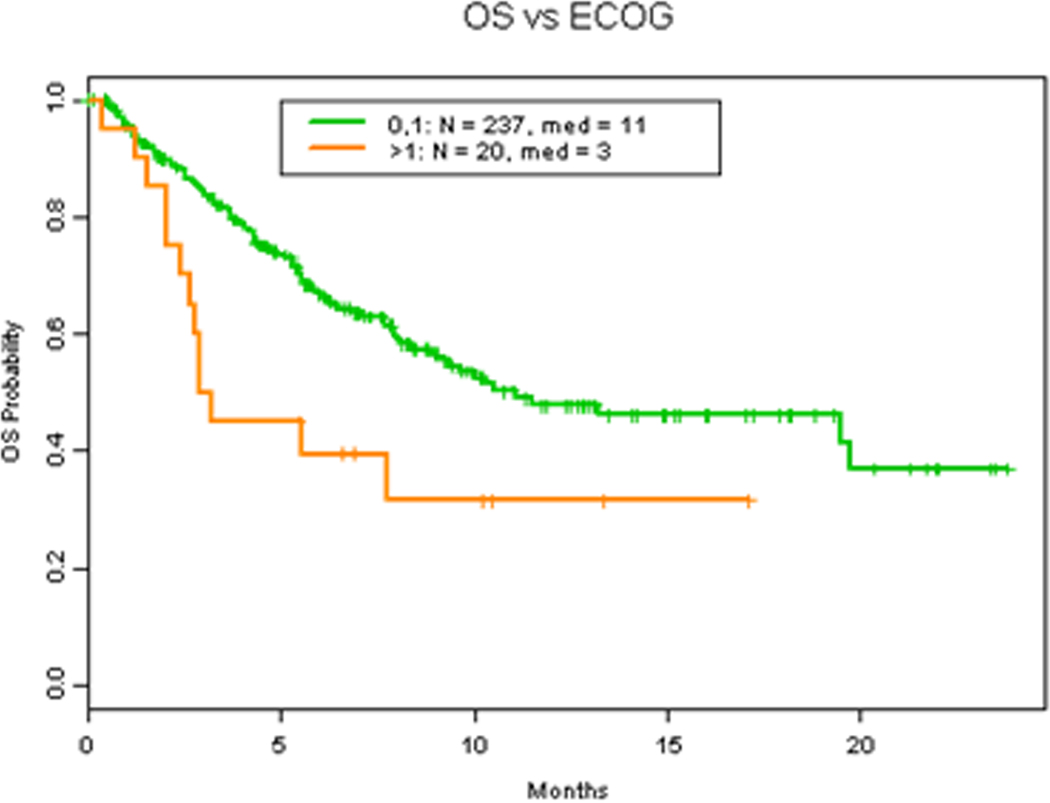
Albumin, LDH level and number of metastasis have all been demonstrated to be main prognostic parameters in patients with advanced malignancy associated with worse overall survival (3, 16–19). Consistent with these findings, our results demonstrates that LDH level higher than 618 IU/L (Figure 4A) (HR 2.0 (1.3, 2.9) p=0.0015), albumin level less than 3.5 gm/dL (Figure 4B) (HR 2.6 (1.8, 4.0) p=0.0001) and more than 2 metastasis (Figure 5A) (HR 2.5 (1.6, 3.8) p=0.0002) were all associated with worse OS. Additionally, Eastern Cooperative Oncology Group (ECOG) status, which also has previously been described a prognostic factor of survival which led to the expansion of the RMH score and development of the MD Anderson Cancer Center (MDACC) Prognostic scoring system (20), was found to be associated with worse OS survival when ECOG >1 (Figure 4C) (HR 2.1 (1.2, 3.7) p=0.024). However, hemoglobin level was not related to survival (Figure S1A) (HR 0.8 (0.5, 1.1) p=0.17).
Figure 4. Clinical factors and OS.
A. OS in patients with 618 IU/L or less vs more than 618 IU/L LDH level. B. OS in patients with 3.5 gm/dL or more vs less than 3.5 gm/dL albumin level. C. OS in patients with ECOG 1 or less vs more than 1.
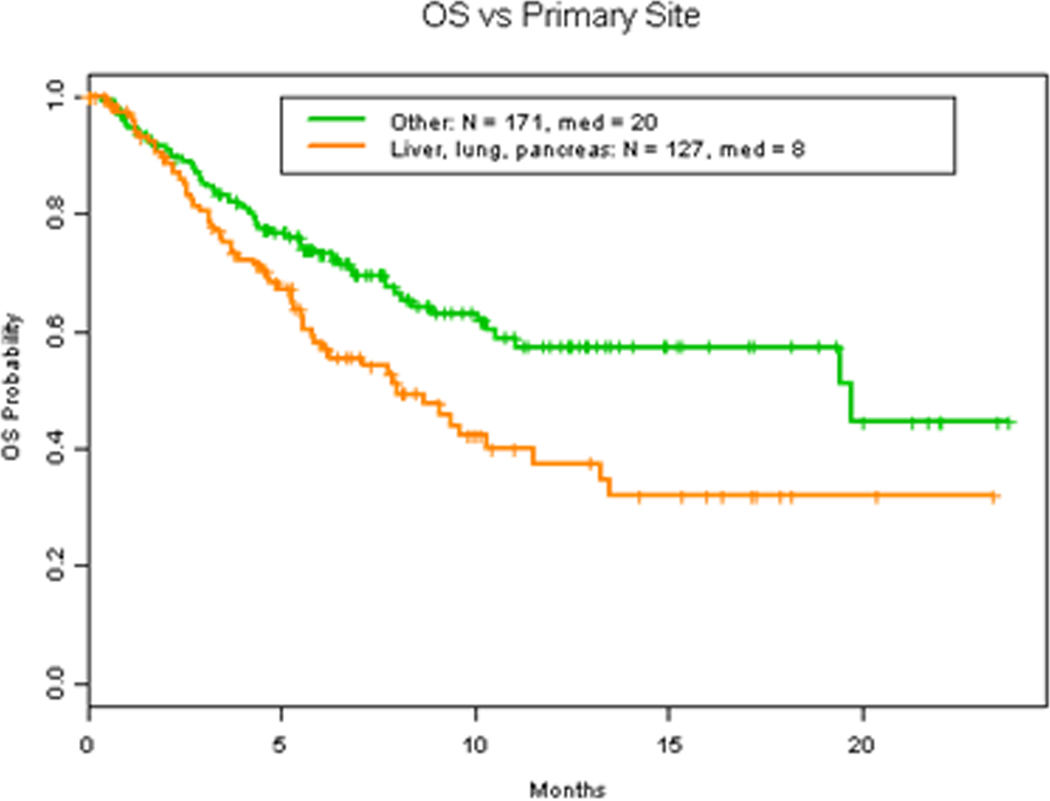
Figure 5. Malignancy and OS.
A. OS in patients with 2 or less vs more than 2 number of metastasis. B OS in patients with lung, liver and pancreas as site of primary malignancy vs other sites of primary malignancy.
LDH was found to be correlated with survival as an independent clinical factor (Table 3). However, when evaluating correlation of LDH to median VAF, they were not correlated. (Spearman rank correlation coefficient = 0.10, 95% CI = (−0.03, 0.22) p=0.23) (Figure S1B).
DISCUSSION
cfDNA offers a non-invasive approach to assess patients’ overall mutational picture. It is increasingly being used in precision oncology, with a focus on therapy selection, response assessment and identifying mechanism of acquired resistance. We here demonstrate that higher levels of cfDNA VAF, higher number of NSM and genomic alterations were associated with worse OS in patients with metastatic disease.
There is growing interest in advanced care planning in order to ensure patients with advanced disease and their caregivers are appropriately prepared for transition of care from curative/therapeutic intent to palliative intent. Prognostication has been also a challenge in patient selection for clinical trials. Enrollment of patients with short expected life expectation has been especially problematic in Phase I trials, where early progression within the dose-limiting toxicity window, would result in inability to assess the safety and tolerability of the investigational agent. In our study, we sought to enroll patients that may benefit from genomic profiling with expectation that they would be considered for genomically-informed trials. However, 17% of the patients passed away within 3 months of cfDNA testing. Our finding is similar to that reported by Arkenau et al; although most Phase I trials require a life expectancy of 3 months or longer, 18% of patients treated on Phase I trials died within 90 days of starting therapy (1). These findings highlight the difficulty of prognostication, especially in patients with advanced, heavily pretreated disease. Although a variety of clinical factors have been defined as predictors of early mortality and OS (1–3), further refinements are required. Objective measures such as maximum VAF may therefore hold promise.
There have been emerging data demonstrating the prognostic role of liquid biopsies across multiple different malignancies. cfDNA was found to be predictive of recurrence free survival (RFS) in a prospective cohort of 230 patients with stage II colon cancer after curative surgery. Patients with positive cfDNA status postoperatively had a markedly reduced RFS compared to the negative status group (21). In stage III/IV resected melanoma patients, cfDNA was predictive of disease-free interval, distant metastasis interval and OS rate in patients with detectable cfDNA (HR 2.63; 95% CI 1.40–4.96); P = 0.003) (22).
There have been several recent reports that cfDNA is prognostic in the advanced/metastatic setting (9, 10, 23). Detection of BRAF-mutant specific VAF levels has been reported as poor prognostic marker in patients with metastatic melanoma, and also the lack thereof has also been reported as a positive prognostic marker of overall survival (8, 24). VAF has been associated with OS in patients with metastatic colorectal cancer, with a VAF level <10% associated with statistically improved OS (25). Similarly, Mohrmann et al reported that high amount of exosomal nucleic acid is associated with lower shorter median survival (26). Our results are consistent with these studies, and demonstrates that being in the top quartile of maximum VAF is associated with worse OS.
Interestingly, in the PALOMA-3 study of fulvestrant with or without CDK4/6 inhibitor palbociclib, baseline levels of PIK3CA mutations in the cfDNA did not correlate with progression-free survival (PFS)(27) However, compared with treatment with fulvestrant alone, treatment with the fulvestrant+ palboclib combination was associated with a more marked fall in PIK3CA mutations detected in cfDNA at day 15 compared to baseline. Further, the relative change in the circulating PIK3CA mutation ratio after 15 days strongly predicted PFS on fulvestrant+ palboclib. This is consistent with several recent reports that cfDNA may also have a role in assessing response to therapy (28–30). For example, Janku et al reported that in patients with advanced disease undergoing systemic therapy, radiological response was positively associated with changes in cfDNA VAF (P = 0.02), and compared with unchanged/increased mutant cfDNA MAF, decreased cfDNA VAF was associated with longer time to treatment failure (P = 0.03) (10). A drop in mutations being targeted has been reported in clinical trials with neratinib targeting HER2 mutations, and AZD5363 targeting AKT1 mutations (31, 32). Taken together these data suggest cfDNA dynamics may be reflect early response in genotype-matched as well as unmatched therapies. In our study, patients received a variety of different therapies and unfortunately we did not have the opportunity to assess the predictive value of longitudinal sampling. This is an important area of future study.
Previous studies have demonstrated worse overall survival of clinical markers including lower albumin level, elevated LDH level and increasing number of metastasis (3, 16–19). Our results further echoes these findings and they remain to be important markers when evaluating patients, A meta-analysis by Gao et al suggested an association between OS and LDH serum concentration in patients with advanced melanoma. They observed worse overall survival in patients with elevated LDH levels (33). This was further observed in small cell lung cancer (34). Increasing number of metastasis remains an important clinical parameter in order to predict prognosis. Patients with metastatic renal cell carcinoma was found to have better survival when only bone or lung was involved (median survival 27 months) vs several organ involvement (median survival 11 months). This was found to be an independent predictor of survival as well on multivariate analysis (HR 2.05 p<0.001)(35). Gibson et al discovered a similar association with survival in patients with stage 4B NSCLC on multivariate analysis (HR 1.5 (1.3, 1.7) p<0.001)(36). This finding was consistent with our analysis, where more than 2 metastases (Figure 5A) (2.5 (1.6, 3.8) p=0.0002) was found to be a worse predictor of survival.
There are several limitations to this study. Our patients were treated with a variety of therapies and had multiple tumor types (Table 1 and Supplemental Table 2). We found that detection of any alteration, VAF, as well as NSM differed across tumor histologies. Thus tumors may have had uneven shedding of cfDNA, and different tumors likely varied in their genomic alterations and thus the ability to detect alterations with our pan-cancer cfDNA panel may have differed. Patients were undergoing clinical genomic testing over a two-year period and the panel evolved from a 54 gene panel to a 70 gene and then a 73 gene panel; however, all but one patient had a 70 or 73 gene panel test. The validation of the Guardant360 has previously been published by Lanman et al, Odegaard et al, and Zill et al (11, 12, 14). The panel versions differ in number of genes reported (Supplemental Table 4). Because of the small size of the study population, confounding from other variables could not be determined. Amplification of altered oncogenes has been demonstrated to raise VAF(12, 37), and a method to adjust VAF for copy number amplification has been described (12), however copy number was not assessed for all the genes in the assay used herein, and thus we could not adjust VAF based on copy number. Clonal hematopoiesis of indeterminant potential (CHIP) was initially proposed by Steensma et al, who described it as a population of clonal cells from hematopoietic source with a known driver mutation that has a VAF ≥ 2%, without a WHO-defined disorder and cytopenias. Patients with CHIP alterations were found to have elevated probability of disease development of hematological malignancy compared to patients without any mutations (38). Due to the possible contribution of white blood cells to cfDNA, we cannot exclude CHIP as a confounder of cfDNA results.
CONCLUSION
Emerging data in the literature increasingly demonstrates the clinical utility of liquid biopsies. In our study we demonstrated the association between VAF, and total number of alterations with overall survival and propose that ctDNA may be useful to determine whether patient prognosis is adequate for clinical trial enrollment. Prospective studies are necessary to optimize and validate thresholds for this clinical application.
Supplementary Material
Statement of Translational Relevance.
This report describes the association between variant allele frequency (VAF) of cell-free circulating tumor DNA and prognosis in patients with advanced malignancy of multiple different cancer histologies. The research presented show that higher VAF levels and multiple other clinical factors including albumin level, higher number of prior therapies, non-visceral metastatic sites and male sex were found to be independent predictors of prognosis and survival. These findings indicate the potential to use VAF as a part of the clinical decision-making process and when discussing prognosis with patients.
ACKNOWLEDGMENTS
This work was supported by the T32 (CA009599 to SSP and FMB), Guardant Health (to NSS, FMB), Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy (NSS,KS, MK, FMB), Cancer Prevention Research Institutive of Texas (CPRIT) Precision Oncology Decision Support Core RP150535 (NSS,KS, MK, FMB), and the MD Anderson Cancer Support Grant (P30 CA016672).
Footnotes
Conflict of Interest:
S. Pairawan, N. Sanchez, S. Damodaran, D. Fogelman, K. Hess, A. Kaseb, and K. Shaw declare no potential conflicts of interest.
S. Fu reports clinical research support from Aprea Therapeutics, Endocyte, AstraZeneca, Novartis, Anaeropharma Science, OncoMed Pharmaceuticals, NeuPharma, Inc., Medivir AB, Lilly, BeiGene, MacroGenics, Huya Bioscience International, BioAtla LLC, Parexel International, LLC, and Takeda.
D. Hong reports research support from AbbVie, Adaptimmune, Amgen, AstraZeneca, Bayer, BMS, Daiichi Sankyo, Eisai, Fate Therapeutics, Genentech, Genmab, Ignyta, Infinity, Kite, Kyowa, Lilly, Loxo, Merck, Medimmune, MiRNA, Mirati, Molecular Templates, Mologen, Novartis, Seattle Genetics, Takeda, and Pfizer. He has served on the advisory board for Adaptimmune, Bayer, Genentech, Infinity, Seattle Genetics, Takeda, Alpha Insights, Axiom, Baxter, GLG, Group H, Guidepoint Global, Janssen, Merrimack, Medscape, Numab, Trieza Therapeutics, Molecular Match, Presagia Inc., and Pfizer. He also served on the Speakers Bureau for Bayer and received compensation for travel from Loxo and miRNA. He is founder of OncoResponse.
F. Janku has research support from Novartis, Genentech, BioMed Valley Discoveries, Plexxikon, Deciphera, Piqur, Symphogen, Bayer, FujiFilm Corporaton, and Usher-Smith Laboratories. He is or has been on the Scientific Advisory Boards of Deciphera, IFM Therapeutics, Synlogic, and Guardant Health; is a paid consultant for Trovagene and Immunomet; and has ownership interests in Trovagene.
M. Javle reports personal fees from Rafael Pharma, Aslan, Incyte Medical Affairs, Mundipharma, Pieris Pharmaceuticals, Merck, Halozyme, Novartis, Seattle Genetics, BeiGene, QED Therapeutics, Bayer, and Taiho.
R. Lanman is an employee and stockholder of Guardant Health. He is also a board member and stockholder of Biolase, Inc. and advisor and stockholder of Forward Medical, Inc.
F. Meric-Bernstam reports receiving commercial research grants from Novartis, AstraZeneca, Calithera, Aileron, Bayer, Jounce, CytoMx, eFFECTOR, Zymeworks, PUMA Biotechnology, Curis, Millennium, Daiichi Sankyo, Abbvie, Guardant Health, Takeda, Seattle Genetics, and GlaxoSmithKline as well as grants and travel related fees from Taiho, Genentech, Debiopharm Group, and Pfizer. She also served as a consultant to Pieris, Dialectica, Sumitomo Dainippon, Samsung Bioepis, Aduro, OrigiMed, Xencor, Jackson Laboratory, Zymeworks, Kolon Life Science, and Parexel International, and advisor to Inflection Biosciences, GRAIL, Darwin Health, Spectrum, Mersana, and Seattle Genetics.
V. Raymond is an employee and stockholder of Guardant Health
V. Subbiah receives research funding for clinical trials from Novartis, Bayer, GlaxoSmithKline, Nanocarrier, Vegenics, Celgene, Northwest Biotherapeutics, Berghealth, Incyte, Fujifilm, Pharmamar, D3, Pfizer, Multivir, Amgen, Abbvie, Alfa-sigma, Agensys, Boston Biomedical, Idera Pharma, Inhibrx, Exelixis, Blueprint Medicines, Loxo Oncology, Takeda and Roche/ Genentech, National Comprehensive Cancer Network, NCI-CTEP and UT MD Anderson Cancer Center. He has also received compensation for travel from Novartis, Pharmamar, Astra Zeneca, and Medimmune.
REFERENCES:
- 1.Arkenau HT, Olmos D, Ang JE, Barriuso J, Karavasilis V, Ashley S, et al. 90-Days mortality rate in patients treated within the context of a phase-I trial: how should we identify patients who should not go on trial? Eur J Cancer. 2008;44(11):1536–40. [DOI] [PubMed] [Google Scholar]
- 2.Chau NG, Florescu A, Chan KK, Wang L, Chen EX, Bedard P, et al. Early mortality and overall survival in oncology phase I trial participants: can we improve patient selection? BMC Cancer. 2011;11:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrido-Laguna I, Janku F, Vaklavas C, Falchook GS, Fu S, Hong DS, et al. Validation of the Royal Marsden Hospital prognostic score in patients treated in the Phase I Clinical Trials Program at the MD Anderson Cancer Center. Cancer. 2012;118(5):1422–8. [DOI] [PubMed] [Google Scholar]
- 4.Polivka J, Jr., Pesta M, Janku F. Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: are we there yet? Expert Rev Mol Diagn. 2015;15(12):1631–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hovelson DH, Liu CJ, Wang Y, Kang Q, Henderson J, Gursky A, et al. Rapid, ultra low coverage copy number profiling of cell-free DNA as a precision oncology screening strategy. Oncotarget. 2017;8(52):89848–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goyal L, Saha SK, Liu LY, Siravegna G, Leshchiner I, Ahronian LG, et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov. 2017;7(3):252–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janku F, Claes B, Huang HJ, Falchook GS, Devogelaere B, Kockx M, et al. BRAF mutation testing with a rapid, fully integrated molecular diagnostics system. Oncotarget. 2015;6(29):26886–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janku F, Huang HJ, Claes B, Falchook GS, Fu S, Hong D, et al. BRAF Mutation Testing in Cell-Free DNA from the Plasma of Patients with Advanced Cancers Using a Rapid, Automated Molecular Diagnostics System. Mol Cancer Ther. 2016;15(6):1397–404. [DOI] [PubMed] [Google Scholar]
- 9.Janku F, Huang HJ, Fujii T, Shelton DN, Madwani K, Fu S, et al. Multiplex KRASG12/G13 mutation testing of unamplified cell-free DNA from the plasma of patients with advanced cancers using droplet digital polymerase chain reaction. Ann Oncol. 2017;28(3):642–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janku F, Zhang S, Waters J, Liu L, Huang HJ, Subbiah V, et al. Development and Validation of an Ultradeep Next-Generation Sequencing Assay for Testing of Plasma Cell-Free DNA from Patients with Advanced Cancer. Clin Cancer Res. 2017;23(18):5648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res. 2018;24(15):3539–49. [DOI] [PubMed] [Google Scholar]
- 12.Zill OA, Banks KC, Fairclough SR, Mortimer SA, Vowles JV, Mokhtari R, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res. 2018;24(15):3528–38. [DOI] [PubMed] [Google Scholar]
- 13.Kim ST, Lee WS, Lanman RB, Mortimer S, Zill OA, Kim KM, et al. Prospective blinded study of somatic mutation detection in cell-free DNA utilizing a targeted 54-gene next generation sequencing panel in metastatic solid tumor patients. Oncotarget. 2015;6(37):40360–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One. 2015;10(10):e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi G, Mu Z, Rademaker AW, Austin LK, Strickland KS, Costa RLB, et al. Cell-Free DNA and Circulating Tumor Cells: Comprehensive Liquid Biopsy Analysis in Advanced Breast Cancer. Clin Cancer Res. 2018;24(3):560–8. [DOI] [PubMed] [Google Scholar]
- 16.Barbot AC, Mussault P, Ingrand P, Tourani JM. Assessing 2-month clinical prognosis in hospitalized patients with advanced solid tumors. J Clin Oncol. 2008;26(15):2538–43. [DOI] [PubMed] [Google Scholar]
- 17.Livingston JA, Hess KR, Naing A, Hong DS, Patel S, Benjamin RS, et al. Validation of prognostic scoring and assessment of clinical benefit for patients with bone sarcomas enrolled in phase I clinical trials. Oncotarget. 2016;7(39):64421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bigot F, Castanon E, Baldini C, Hollebecque A, Carmona A, Postel-Vinay S, et al. Prospective validation of a prognostic score for patients in immunotherapy phase I trials: The Gustave Roussy Immune Score (GRIm-Score). Eur J Cancer. 2017;84:212–8. [DOI] [PubMed] [Google Scholar]
- 19.Arkenau HT, Olmos D, Ang JE, de Bono J, Judson I, Kaye S. Clinical outcome and prognostic factors for patients treated within the context of a phase I study: the Royal Marsden Hospital experience. Br J Cancer. 2008;98(6):1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheler J, Tsimberidou AM, Hong D, Naing A, Falchook G, Piha-Paul S, et al. Survival of 1,181 patients in a phase I clinic: the MD Anderson Clinical Center for targeted therapy experience. Clin Cancer Res. 2012;18(10):2922–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valpione S, Gremel G, Mundra P, Middlehurst P, Galvani E, Girotti MR, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer. 2018;88:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Use of Liquid Biopsies in Clinical Oncology: Pilot Experience in 168 Patients. Clin Cancer Res. 2016;22(22):5497–505. [DOI] [PubMed] [Google Scholar]
- 24.Santiago-Walker A, Gagnon R, Mazumdar J, Casey M, Long GV, Schadendorf D, et al. Correlation of BRAF Mutation Status in Circulating-Free DNA and Tumor and Association with Clinical Outcome across Four BRAFi and MEKi Clinical Trials. Clin Cancer Res. 2016;22(3):567–74. [DOI] [PubMed] [Google Scholar]
- 25.Sanz-Garcia E. Impact in prognosis of circulating tumor DNA mutant allele fraction in RAS mutant metastatic colorectal cancer (mCRC). Annals of Oncology. 2017;28(supple_3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohrmann L, Huang HJ, Hong DS, Tsimberidou AM, Fu S, Piha-Paul SA, et al. Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin Cancer Res. 2018;24(1):181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, Liu Y, Bartlett CH, Koehler M, Cristofanilli M, Garcia-Murillas I, Bliss JM, Turner NC Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018. March 1;9(1):896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7(12):1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khagi Y, Goodman AM, Daniels GA, Patel SP, Sacco AG, Randall JM, et al. Hypermutated Circulating Tumor DNA: Correlation with Response to Checkpoint Inhibitor-Based Immunotherapy. Clin Cancer Res. 2017;23(19):5729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Long GV, Menzies AM, Lo S, Guminski A, Whitbourne K, et al. Association Between Circulating Tumor DNA and Pseudoprogression in Patients With Metastatic Melanoma Treated With Anti-Programmed Cell Death 1 Antibodies. JAMA Oncol. 2018;4(5):717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyman DM, Smyth LM, Donoghue MTA, Westin SN, Bedard PL, Dean EJ, et al. AKT Inhibition in Solid Tumors With AKT1 Mutations. J Clin Oncol. 2017;35(20):2251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma CX, Bose R, Gao F, Freedman RA, Telli ML, Kimmick G, et al. Neratinib Efficacy and Circulating Tumor DNA Detection of HER2 Mutations in HER2 Nonamplified Metastatic Breast Cancer. Clin Cancer Res. 2017;23(19):5687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao D, Ma X. Serum lactate dehydrogenase is a predictor of poor survival in malignant melanoma. Panminerva Med. 2017;59(4):332–7. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Guo M, Fan J, Lv Z, Huang Q, Han J, et al. Prognostic significance of serum LDH in small cell lung cancer: A systematic review with meta-analysis. Cancer Biomark. 2016;16(3):415–23. [DOI] [PubMed] [Google Scholar]
- 35.Han KR, Pantuck AJ, Bui MH, Shvarts O, Freitas DG, Zisman A, et al. Number of metastatic sites rather than location dictates overall survival of patients with node-negative metastatic renal cell carcinoma. Urology. 2003;61(2):314–9. [DOI] [PubMed] [Google Scholar]
- 36.Gibson AJW, Li H, D’Silva A, Tudor RA, Elegbede AA, Otsuka SM, et al. Impact of number versus location of metastases on survival in stage IV M1b non-small cell lung cancer. Med Oncol. 2018;35(9):117. [DOI] [PubMed] [Google Scholar]
- 37.Stover DG, Parsons HA, Ha G, Freeman SS, Barry WT, Guo H, et al. Association of Cell-Free DNA Tumor Fraction and Somatic Copy Number Alterations With Survival in Metastatic Triple-Negative Breast Cancer. J Clin Oncol. 2018;36(6):543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.