Abstract
This paper summarizes the methodology for the registry of suspected COVID‐19 in emergency care (RECOVER), a large clinical registry of patients from 155 United States (US) emergency departments (EDs) in 27 states tested for SARS‐CoV‐2 from March–September 2020. The initial goals are to derive and test: (1) a pretest probability instrument for prediction of SARS‐CoV‐2 test results, and from this instrument, a set of simple criteria to exclude COVID‐19 (the COVID‐19 Rule‐Out Criteria—the CORC rule), and (2) a prognostic instrument for those with COVID‐19. Patient eligibility included any ED patient tested for SARS‐CoV‐2 with a nasal or oropharyngeal swab. Abstracted clinical data included 204 variables representing the earliest manifestation of infection, including week of testing, demographics, symptoms, exposure risk, past medical history, test results, admission status, and outcomes 30 days later. In addition to the primary goals, the registry will provide a vital platform for characterizing the course, epidemiology, clinical features, and prognosis of patients tested for COVID‐19 in the ED setting.
Keywords: COVID‐19, decision making, diagnosis, probability, prognosis, pulmonary embolism, registries, risk, SARS‐CoV‐2, venous thromboembolism, venous thrombosis
1. BACKGROUND
The COVID‐19 pandemic caused by the novel coronavirus SARS‐CoV‐2 has impacted citizens internationally. As of October 12, 2020, there have been over 7 million infections and 215,000 deaths across the United States (US), with the number of cases growing. 1 Much remains unknown regarding the actual number of cases and evolving epidemiology of COVID‐19 in the United States. 2 , 3 , 4 , 5 Factors limiting current knowledge include the current lack of systematically collected data from patients with atypical symptoms and problems associated with diagnostic test availability and accuracy. 2 , 4 , 6 , 7 In addition, the clinical picture of SARS‐CoV‐2 infection has a wide range of presenting symptoms, followed by a wide spectrum of associated complications and outcomes. 8 , 9 , 10 , 11 , 12
The emergency department (ED) is a key location for disease surveillance and collection of data to answer critically important questions about COVID‐19 prediction, prognosis, and outcome. Furthermore, the ED is the catchment location for the population who may not have access to routine medical care and may use the ED exclusively for on‐demand care and may be most threatened by this outbreak. Toward that goal, this report describes the methodology for the creation and execution of the registry of suspected COVID‐19 in emergency care (RECOVER), a national registry of ED patients tested for severe acute respiratory syndrome, coronavirus‐2 (SARS‐CoV‐2). The primary goals of this work involve the prediction of positive testing for SARS‐CoV‐2 and prognosis of patients with infection. RECOVER will also provide the platform for answering myriad other COVID‐19 questions.
2. METHODS
2.1. History and rationale for the registry
The RECOVER registry is the result of a grass‐roots effort of a consortium of 45 emergency medicine clinician investigators from 27 US states representing both community and academic centers. The registry had 2 preliminary aims. First, creation of a quantitative pretest probability scoring system to predict a positive SARS‐CoV‐2 test, and relatedly the derivation and internal validation of a prediction rule to identify those at very low probability of disease (COVID‐19 Rule Out Criteria‐CORC rule). The second aim is the derivation and internal validation of a set of criteria to predict the development of severe COVID‐19. Rationale for the RECOVER registry hinges on the fact that the ED represents a pivotal site for syndromic surveillance because of the high volume of undifferentiated patients and the spectrum of illness that can be captured early in the disease course. In 2016, US EDs had >145 million encounters each year, and we anticipated that millions of ED patients would be evaluated for suspected COVID‐19 in 2020. 13 Additionally, because the ED interfaces with both outpatient and inpatient medical care, the critical question of SARS‐CoV‐2 infection status affects decisions to admit or discharge the patient, return to work, need for home isolation, and the location of hospital admission.
Recognizing in March of 2020 that a vaccine was at least 12 months away, and SARS‐CoV‐2 would likely remain endemic in the years to come, the first aim focused on developing simple clinical criteria to exclude COVID‐19 at the bedside, without the need for blood or radiographic testing. In many EDs, the turnaround time for laboratory testing for SARS‐CoV‐2 nucleic acid takes longer than 24 hours and rapid point‐of‐care assays have been hampered by low sensitivity, with one systematic review finding the sensitivity in pooled data at only 56% for antigen tests. 7 Reports of low test sensitivity for swab reverse transcriptase polymerase chain reaction (rtPCR) tests, and tests that use other nucleic amplification techniques, have raised concern. 4 , 6 , 14 Without specific reliable exclusionary criteria, emergency care clinicians cannot make expeditious decisions for each of the tens of millions of ED patients with symptoms consistent with COVID‐19, nor can the emergency care system operate efficiently with the potential need to order a SARS‐CoV‐2 test for each of these patients.
2.2. Design and setting
The registry is being completed by a network of 45 site investigators from 27 states listed in Figure 1. Most of the sites are part of hospital systems, and as a result, data are derived from patients seen at 155 hospitals. The majority of hospitals are located in metropolitan areas, and ≈60% are community hospitals without residency programs.
FIGURE 1.
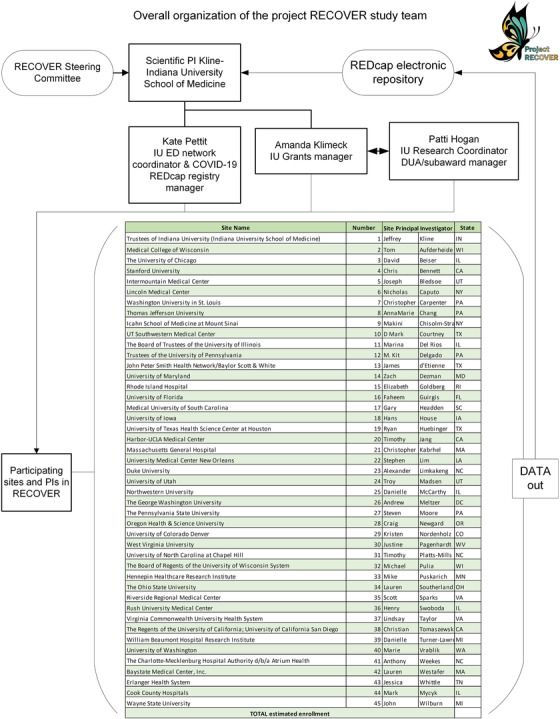
Diagram of administrative and scientific oversight of the RECOVER registry and network
2.3. Registry development
The RECOVER registry was built using RedCAP and includes 204 explicitly defined questions resulting in 360 discrete possible answers among seven domains: (1) Visit Information (18 questions), (2) Demographics, Symptoms and Risk Factors (11 questions), (3) Vital Signs (16 questions), (4) Past Medical History (39 questions), (5) Current Medications (18 questions), (6) Test Results (47 questions), and (7) Outcomes (48 questions). Trained abstractors obtained data from the local electronic medical record. Using a combination of computer scripts and the real‐time work of trained research associates, the RECOVER registry transforms information stored in the electronic medical record into a REDCap (http://www.projectredcap.org) database suitable for further transformation into research manuscripts. Using the first ED visit that occurred in the 14 days before testing (the maximum probable incubation period), allows the data set to collect atypical and early symptoms.
To accelerate the time frame to completion, the data collection form was designed and tested to allow completion by research personnel with basic medical knowledge, with minimal supervision. We used branching logic where possible to improve efficiency (eg, only show a list of possible statins if previous answer to statin use is “yes”). The form includes features to avoid protected health information while still record the timing of enrollment, tests, and outcomes. For example, the form collects the week, month, and year of the index ED visit, as well as the first 4 digits of the patients’ ZIP code. These are not protected health information but allow a close estimate to when the patient was enrolled and their region of residence, which is important for epidemiological surveillance.
This form was pilot‐tested at 19 sites before network‐wide implementation. This pilot testing resulted in multiple changes to clarify fields, alter input ranges, and methods of questioning, and response options. As one example, this process resulted in adding more detail on timing of all tests relative to the index visit and results of antibody testing for SARS‐CoV‐2. Additionally, in April 2020 at Indiana University, using a convenience sample of 50 charts, 2 abstractors, each of whom had experience using REDcap and with 1 hour of training from the site principal investigator, double coded 2 REDcap forms for each of the 50 patients and compared results for the questions in Table 1, which indicated 98% homology in answers (2% variation occurring in the community risk question). The entire contents of the RECOVER registry and data dictionary can be seen at this link: https://indiana-my.sharepoint.com/:b:/g/personal/jefkline_iu_edu/EdOh2z7c07pMtn2Eh5sJhMQBjl4Z3rlvwz5edpT3qG8sXw?e=6fNTkm.
TABLE 1.
Symptoms and risk factors for severe acute respiratory novel coronavirus 2 infection recorded in the registry*
| Symptoms | Risk factors |
|---|---|
| Ear pain | None |
| Rhinorrhea | Travel to the United States from a country with known endemic disease |
| Sore throat | Sick contacts without confirmed COVID‐19 |
| Cough without sputum (dry) | Unemployed or retired, social contact with friends, family and/or general public |
| Cough with sputum production (wet) | Employed, non‐health care worker, contact with family, and/or friends, and/or general public |
| Bloody sputum/hemoptysis | Employed, health care worker with patient contact |
| Chest pain | Caretaker, partner, or family member in close contact with a person with known or suspected COVID‐19 |
| Wheezing | Person experiencing homelessness |
| Shortness of breath (dyspnea) | Institutional exposure: hospital |
| Respiratory distress/failure | Institutional exposure: nursing home |
| History of fever | Institutional exposure: assisted living facility |
| Fatigue/malaise | Institutional exposure: prison/jail or other correctional facility |
| Muscle aches (myalgia) | Group home |
| Joint pain (arthralgia) | Other risk exposure |
| Headache | |
| Altered mental status/confusion | |
| Seizures | |
| Abdominal pain | |
| Vomiting/nausea | |
| Diarrhea | |
| Conjunctivitis | |
| Skin rash | |
| Skin ulcers | |
| Lymphadenopathy | |
| Bleeding (hemorrhage) | |
| Olfactory/taste disturbance | |
| Syncope | |
| Cardiac arrest |
*Examples of answer options to 2 of 204 questions in the registry.
Each site investigator was given an extensive written manual of operations describing patient eligibility and the goals of the registry with specific instructions on each form and field. The registry uses REDcap features to force data entry for critical fields, perform error checking, forcing functions ensure correct alpha‐numeric content, and ranges for numeric data. By rule, if the answer to a question was not documented, the field is coded as negative—the same as if the chart had deliberate statement about the absence of the datum.
The RECOVER registry, and its interaction with 45 contributing sites, are overseen by 2 cores: the administrative core (3 individuals, including a program manager, service and data use agreement coordinator, and grants manager), and a Steering Committee (authors JAK, CK, CAC, DMC, KEN) that provides overall study guidance, including data use issues. The names of the sites and the governance overview are shown in Figure 1. The 45 site principal investigators are supported by a full‐time program manager (KLP) at the Indiana University School of Medicine who, with input from the overall principal investigator, answers questions about data entry and ensures each site has supporting documents including the detailed guidance and FAQ document.
The protocol for the registry was reviewed by the Institutional Review Boards at all participating sites, and with the exception of one site, the Institutional Review Boards deemed the protocol exempted from human subjects research. One Institutional Review Board deemed the 4‐digit zip code to represent protected health information and approved the protocol under waiver of authorization for participation in research as well as informed consent; therefore the zip code was reduced to 3 digits.
2.4. Patient selection
Figure 2 provides an overview of patient identification and flow of data. Patients selected for the registry included ED patients who had any diagnostic test for SARS‐CoV‐2, located by electronic query for charge procedure code 87635, which is unique for SARS‐CoV‐2 testing. All SARs‐CoV‐2 tests were based on molecular testing for SARS‐CoV‐2 RNA as opposed to antigen testing. Because many patients may have had a prior ED visit without testing, the “index visit” was defined as any ED visit that occurred within 14 days before the swab testing for SARS‐CoV‐2 (Figure 3), that did not meet specific exclusions. Exclusions to a prior ED visit within the previous 14 days being coded as the “index visit” included predefined circumstances where that ED visit lacked a reasonable probability of being related to possible COVID‐19 symptoms: (1) trauma, (2) alcohol or drug intoxication, (3) poisoning, (4) suicidality, (5) suspected rape or other domestic violence, (6) involuntary commitment, (7) other isolated chief complaints clearly not related to COVID‐19 (eg, suture removal), and (8) testing done purely for policy (eg, any admitted patient), rather than testing based on clinical suspicion. Patients were enrolled from March–September 2020 with intent to enroll eligible patients consecutively.
FIGURE 2.
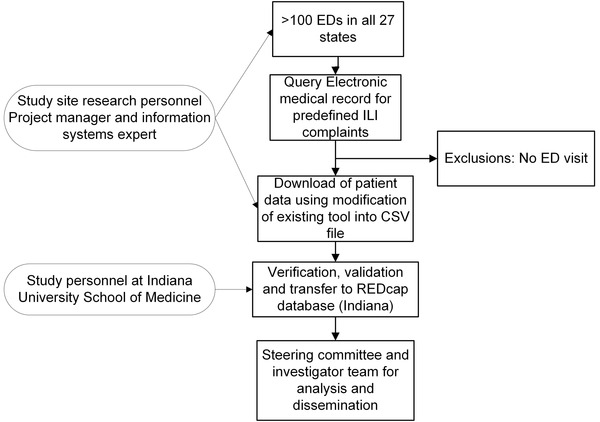
Diagram of data flow for the RECOVER registry
FIGURE 3.
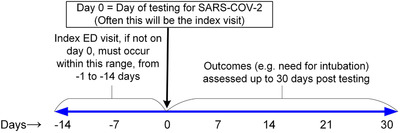
Timeline of eligibility and follow‐up. The index visit refers to the first ED visit that occurred within the previous 14 days of SARS‐CoV‐2 testing. In many cases, the index visit is the same as the day of testing (day 0). All patients are followed for outcomes up to 30 days after the day of SARS‐CoV‐2 testing
2.5. Outcomes—COVID‐19+ and severe COVID‐19
The primary outcomes were COVID‐19 diagnosis and severe COVID‐19. The criterion standard for COVID‐19+ was a positive swab for SARS‐CoV‐2 nucleic acid by a molecular detection (rtPCR or other amplification technique) or positive convalescent sera for anti‐SARS‐CoV‐2 IgM or IgG. 15 , 16 We recognize the imperfect test sensitivity of the swab‐based rtPCR and also serum antibody testing. 17 The most pragmatic method to categorize patients as “clinically negative” for COVID‐19 disease is the absence of return visit for 30 days, or consistently negative testing. Sites were encouraged in the course of investigating the index visit and 30‐day outcomes to review any and all available data from clinical care data networks that allowed access to medical records from other hospitals. For example, in Indianapolis, the investigators examined the CareWeb medical record of the Indiana Health Information Exchange (https://www.ihie.org/) which includes results from over 100 EDs in Indiana. In the case of serial testing, any positive test within 30 days would be considered COVID‐19+, unless the record clearly indicates a negative test at the index visit, followed by >7 days of no symptoms, then new symptoms, or exposure and then a new positive test. These patients were excluded from CORC rule derivation because their COVID‐19 status at the index visit was considered ambiguous. The definition of “severe COVID‐19” was death, or the decision to perform endotracheal intubation followed by mechanical ventilation for >24 hours within 30 days of a positive SARS‐CoV‐2 test.
2.6. Role of funding
Funding was derived from unrestricted internal monies from the Department of Emergency Medicine at Indiana University School of Medicine under the direction of the senior principal investigator (JAK).
2.7. Data analysis and sample size
The first defined aim of the study was to develop pragmatic criteria for a COVID‐19 Rule Out Criteria‐CORC rule that predicts an acceptably low probability of SARS‐CoV‐2 infection to forego testing. The general analytic strategy will encompass the use of logistic regression with positive SARS‐CoV‐2 testing as the primary outcome, and demographic, symptomatic, risk factors, vital signs, and past medical history as the predictor variables. The logistic regression coefficients will be used to create a pretest probability scoring system. To make the CORC rule widely applicable, the predictor variables will only include variables available in almost all cases at the bedside, including demographics, patient‐reported exposure, vital signs, history of present illness, medications, past medical history, and findings from physical examination. The sample size estimate for the first aim was based on preliminary work done in March 2020, including a survey of 20 participating sites. We anticipated a SARS‐CoV‐2 positive rate between 8% to 66%, with an average of 31% (ie, 69% of testing negative). We also anticipated that ∼15% of the COVID‐19 patients would have atypical presentations and ≈20% would go on to require mechanical ventilation or experience death, defining severe disease. Thus, a minimum sample size of 20,000 patients was anticipated to produce ≈6000 positive cases, including 600 with atypical features and 800 with severe disease. These samples would allow sufficient patients to derive pretest probability on a random sample of 10,000 patients using bivariate‐multivariable approach and assuming a traditional 10:1 rule for the ratio of outcomes to predictors in a logistic regression equation; thus, this sample will allow inclusion of at least 25 predictor variables without significant overfit for atypical presentations. 18 The derived rule will be tested in the remaining independent sample.
2.8. Ancillary studies and data sharing
Each site is asked to enroll at least 500 patients with a complete REDCap form before request for data access. Requests for data access require submission of a REDCap survey that asks 5 questions: (1) investigator name and contact, (2) title of work, (3) brief description, (4) study objective(s), and (5) method of analysis. As of October 12, 2020, site investigators, or their local collaborators, have submitted 39 proposals for ancillary studies. The Steering Committee reviews these submissions on an ad‐hoc basis. Data will be embargoed from the general public until approximately June 1, 2021 and thereafter will be freely, publicly available with a written request (including contact information, title and main objective of the work, and brief analytic plan), approved by the Steering Committee. The RECOVER data are devoid of protected health information.
2.9. Progress and timeline
The goal was to complete 20,000 forms within 6 months of study initiation (ie, September 2020). As of October 12, 2020, over 31,500 forms have been uploaded. The end of data collection is planned for October 16, 2020.
3. LIMITATIONS
This registry is restricted to ED patients who received a SARS‐CoV‐2 diagnostic test. This leads to bias toward reporting characteristics of patients with overt symptoms, and the risk of missing patients with atypical or no symptoms. Furthermore, there was variation in national SARS‐CoV‐2 testing strategy over time driven in part by availability of the different tests and the frequently changing recommendations in testing strategy at most institutions. The diagnostic sensitivity of molecular tests on swab samples may result in misclassification error in terms of criterion standard; however, the 30‐day follow‐up data should help to clarify which patients suffered significant morbidity or mortality and help to mitigate this issue. The electronic surveillance methodology may miss important outcomes of patients who were discharged from the index visit (eg, death at home or presentation to another hospital out of state). Another limitation is the lack of consecutive enrollment and the assumption that data not charted were negative. Although registries can show associations, they have limited ability to establish causal inference. 19
4. DISCUSSION
The RECOVER registry will provide data describing the presenting clinical characteristics and 30‐day outcomes of over 30,000 US ED patients tested for SARS‐CoV‐2. This database will serve as a resource to create probabilistic tools for diagnosis and prognosis and to generate hypotheses about diverse aspects of the COVID‐19 pandemic. 19 To our knowledge, this will be the largest and most geographically diverse registry of patients with suspected COVID‐19 in emergency care. The Steering Committee and the site investigators debated, discussed, and pilot‐tested each question to ensure its scientific importance, clarity, or adequate explanation by “field notes,” so that persons with modest research experience could complete them with minimal training. This was essential, given the need for remote training and data entry. The fact that over 30,000 records from 45 sites have been uploaded into REDCap between April and September 2020 suggests functionality of this data collection form. Each question was designed to be scientifically important. In addition to producing the proposed CORC rule and prognostic criteria, the RECOVER registry will allow risk ratio calculations for patient demographics, COVID‐19 exposure patterns, social behaviors, medications, and medical conditions with respect to the outcomes of infection and severe illness over the subsequent 30 days. Moreover, the RECOVER registry will provide a resource to test the external validity of the many predictive and prognostic criteria that have been published for COVID‐19. 20 , 21
Although many prognostic criteria have been proposed, a recent systematic review of literature yielded an urgent need for high‐quality diagnostic and prognostic data for COVID‐19. 22 The authors note that “Of 541 papers that reported clinical characteristics, 295 were commentaries/expert opinions and 36 were case reports. There were no randomized clinical trials, 45 case series studies, 58 narrative reviews, 1 cohort study, and 5 systematic reviews.” 22 The RECOVER registry may be unique because it includes ED patients who are both SARS‐CoV‐2 positive, as well as those who had suspected COVID‐19, but who have negative diagnostic testing for SARS‐CoV‐2. This methodology specifically addresses the current critical need for a data set to derive and test–pretest probability and exclusionary rules for COVID‐19, as has been done with many other diseases, notably pulmonary embolism. 23 The proposed CORC rule can be used to guide the decision whether to test or not, and assuming that accurate likelihood ratios can be produced for diagnostic tests for SARS‐CoV‐2, the pretest probability criteria derived from this work will allow an estimate of post‐test probability. 6 , 24 With upcoming influenza, respiratory syncytial, and other viral challenges approaching, the need for clinical criteria to exclude or estimate the probability of COVID‐19 may increase. Therefore, evidence‐based pretest probability criteria, an exclusionary rule, and criteria to predict mild versus severe disease course, will be important to facilitate prudent decisions about the optimal and safe location of post ED care, timing of follow‐up, or need for inpatient monitoring for clinical deterioration.
Finally, in terms of ancillary (hypothesis‐generating) work, the RECOVER registry is a unique cooperative effort intended to be a public resource to address many other questions. As one example, the observation of increased risk of venous thromboembolism in critically ill patients with COVID‐19 patients has particular concern in emergency care, because early anticoagulation may improve outcomes. 25 , 26 , 27 Opinions conflict about the use of empiric full‐dose anticoagulation for patients with known or suspected COVID‐19. 28 , 29 , 30 This debate is at least partly fueled by the lack of data to estimate the risk ratio for the short‐term incidence of venous thromboembolism diagnosis after a positive SARS‐CoV‐2 test, compared with symptomatic patients who test negative for SARS‐CoV‐2 in the emergency care setting. 31 , 32 The RECOVER registry will directly address that knowledge gap. For ancillary proposals, contributing sites in Figure 1 have first rights to submit for approval by the Steering Committee. After publication of the initial set of manuscripts, in approximately June, 2021, we will open the RECOVER registry to proposals from the general scientific community. Data access will be free of monetary charge, but will be subject to approval by the Steering Committee.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
JAK conceived the study, wrote the initial protocol, drafted the case report form, organized sites, obtained funding, collected data, and wrote, edited, and approved this manuscript. KLP assisted with drafting and revising of the case report form, organized sites, collected data, maintained the database, and edited and approved this manuscript. CK assisted with drafting and revising of the case report form, collected data, maintained the database, and edited and approved this manuscript. DMC assisted with drafting and revising of the case report form, collected data, maintained the database, and edited and approved this manuscript. KEN assisted with study design, data collection, manuscript preparation, and editing. CAC assisted with drafting and revising of the case report form, organized sites, collected data, maintained the database, and edited and approved this manuscript. All authors approved the submitted version of this manuscript. JAK takes the final responsibility of the article.
ACKNOWLEDGMENTS
The authors present this work on behalf of all site principal investigators in the RECOVER network. This work was made possible by the administrative oversite of Patti Hogan and Amanda Klimeck.
Biography
Jeffrey A. Kline, MD, is Vice Chair of Research in emergency medicine and a professor of physiology at Indiana University School of Medicine.

Kline JA, Pettit KL, Kabrhel C, Courtney DM, Nordenholz KE, Camargo CA . Multicenter registry of United States emergency department patients tested for SARS‐CoV‐2. JACEP Open. 2020;1:1341–1348. 10.1002/emp2.12313
Funding and support: Unrestricted internal funds.
Supervising Editor: Henry E. Wang, MD, MS
REFERENCES
- 1. U.S. Department of Health & Human Services CfDC. Coronavirus Disease 2019 (COVID‐19). https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/cases-in-us.html. Published 2020. Accessed October 8, 2020.
- 2. Lipsitch M, Swerdlow DL, Finelli L. Defining the Epidemiology of Covid‐19 ‐ Studies Needed. N Engl J Med. 2020;382(13):1194–1196. [DOI] [PubMed] [Google Scholar]
- 3. Peirlinck M, Linka K, Sahli Costabal F, et al. Visualizing the invisible: the effect of asymptomatic transmission on the outbreak dynamics of COVID‐19. medRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grassly NC, Pons‐Salort M, Parker EPK, White PJ, Ferguson NM. Comparison of molecular testing strategies for COVID‐19 control: a mathematical modelling study. Lancet Infect Dis. 2020;S1473‐3099(20):30630–30637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grubaugh ND, Hanage WP, Rasmussen AL. Making sense of mutation: what D614G means for the COVID‐19 pandemic remains unclear. Cell. 2020;182(4):794–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carpenter CR, Mudd PA, West CP, Wilber E, Wilber ST. Diagnosing COVID‐19 in the emergency department: a scoping review of clinical examinations, laboratory tests, imaging accuracy, and biases. Acad Emerg Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dinnes J, Deeks JJ, Adriano A, et al. Rapid, point‐of‐care antigen and molecular‐based tests for diagnosis of SARS‐CoV‐2 infection. Cochrane Database Syst Rev. 2020;8:Cd013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu Z, Song C, Xu C, et al. Clinical characteristics of 24 asymptomatic infections with COVID‐19 screened among close contacts in Nanjing. China Sci China Life Sci. 202063(5):706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Young BE, Ong SWX, Kalimuddin S, et al. Epidemiologic features and clinical course of patients infected with SARS‐CoV‐2 in Singapore. JAMA:2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang W, Cao Q, Qin L, et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID‐19):a multi‐center study in Wenzhou city, Zhejiang, China. J Infect. 2020;80(4):388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Statistics UDoHaHSCfDCaPNCfH. National Hospital Ambulatory Medical Care Survey, 2017. https://www.cdc.gov/nchs/data/nhamcs/web_tables/2017_ed_web_tables-508.pdf2017.
- 14. West CP, Montori VM, Sampathkumar P. COVID‐19 testing: the threat of false‐negative results. Mayo Clin Proc. 2020;95(6):1127–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID‐19). Clin Infect Dis. 2020;71(15):778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019 [published online ahead of print March 28, 2020]. Clin Infect Dis. 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cassaniti I, Novazzi F, Giardina F, et al. Performance of VivaDiagTM COVID‐19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID‐19 in acute patients referring to emergency room department [published online ahead of print March 30, 2020]. J Med Virol. 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Concato J, Feinstein AR, Holford TR. The risk of determining risk with multivariable models. Ann Intern Med. 1993;118(3):201–210. [DOI] [PubMed] [Google Scholar]
- 19. Pop B, Fetica B, Blaga ML, et al. The role of medical registries, potential applications and limitations. Med Pharm Rep. 2019;92(1):7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid‐19 infection: systematic review and critical appraisal. Bmj. 2020;369:m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Struyf T, Deeks JJ, Dinnes J, et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID‐19 disease. Cochrane Database Syst Rev. 2020;7(7):Cd013665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang S, Li A, Eshaghpour A, et al. Quality of early evidence on the pathogenesis, diagnosis, prognosis and treatment of COVID‐19. BMJ Evid Based Med. 2020(111499). [DOI] [PubMed] [Google Scholar]
- 23. Tritschler T, Kraaijpoel N, Le Gal G, Wells PS. Venous Thromboembolism: advances in diagnosis and treatment. JAMA. 2018;320(15):1583–1594. [DOI] [PubMed] [Google Scholar]
- 24. Zehtabchi S, Kline JA. The art and science of probabilistic decision‐making in emergency medicine. Acad Emerg Med. 2010;17(5):521–523. [DOI] [PubMed] [Google Scholar]
- 25. Roncon L, Zuin M, Barco S, et al. Incidence of acute pulmonary embolism in COVID‐19 patients: systematic review and meta‐analysis [Epub ahead of print September 17, 2020]. Eur J Intern Med. 10.1016/j.ejim.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang C, Shen L, Le KJ, et al. Incidence of venous thromboembolism in hospitalized Coronavirus disease 2019 patients: a systematic review and meta‐analysis. Front Cardiovasc Med. 2020;7:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Birkeland K, Zimmer R, Kimchi A, Kedan I. Venous thromboembolism in hospitalized COVID‐19 patients: systematic review. Interact J Med Res. 2020;9(3):e22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parks AL, Auerbach AD, Schnipper JL, et al. COVID‐19 coagulopathy and thrombosis: analysis of hospital protocols in response to the rapidly evolving pandemic. Thromb Res. 2020;196:355–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moores LK, Tritschler T, Brosnahan S, et al. Prevention, diagnosis, and treatment of VTE in patients with coronavirus disease. CHEST Guideline and Expert Panel Report Chest. 2019;158(3):1143–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spyropoulos AC. The management of venous thromboembolism in hospitalized patients with COVID‐19. Blood Adv. 2020;4(16):4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miró Ò, Llorens P, Aguirre A, et al. Association between Covid‐19 and pulmonary embolism (AC‐19‐PE study). Thromb Res. 2020;196:322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Freund Y, Drogrey M, Miró Ò, et al. Association between pulmonary embolism and COVID‐19 in emergency department patients undergoing computed tomography pulmonary angiogram: the PEPCOV International Retrospective Study. Acad Emerg Med. 2020;27(9):811–820. [DOI] [PubMed] [Google Scholar]


