Abstract
Hsp90 is often overexpressed with activated form in cancer cells, and many key cellular proteins are dependent upon the Hsp90 machinery (these proteins are called “client protein”). Nowadays, more client proteins and more inhibitors of Hsp90 are being discovered. Chaetocin has been identified as an inhibitor of histone methyl transferase SUV39H1. Herein, we find that Chaetocin is an inhibitor of Hsp90 which binds to the C-terminal of Hsp90α. Chaetocin inhibited a variety of Hsp90 client proteins including AMl1-ETO and BCL-ABL, the mutant fusion-protein in the K562 and HL-60 cells. SUV39H1 mediates epigenetic events in the pathophysiology of hematopoietic disorders. We found that inhibition of Hsp90 by Chaetocin and 17-AAG had ability to induce degradation of SUV39H1 through proteasome pathway. In addition, SUV39H1 interacted with Hsp90 through co-chaperone HOP. These results suggest that SUV39H1 belongs to a client protein of Hsp90. Moreover, Chaetocin was able to induce cell differentiation in the two cells in the concentration range of Hsp90 inhibition. Altogether, our results demonstrate that SUV39H1 is a new client protein of Hsp90 degradated by Chaetocin as a novel C-terminal inhibitor of Hsp90. The study establishes a new relationship of Chaetocin and SUV39H1, and paves an avenue for exploring a new strategy to target SUV39H1 by inhibition of Hsp90 in leukemia.
Keywords: Hsp90, Chaetocin, SUV39H1, Client protein, Cancer
INTRODUCTION
Hsp90 is an ubiquitous and conserved molecular chaperone molecule which plays an essential role in many cellular processes including cell cycle control, cell survival, hormone and other signaling pathways (Makhnevych and Houry, 2012; Mellatyar et al., 2018). Hsp90 is required for the correct maturation and activation of a number of key cellular proteins and protein complexes. These protein called “client protein” and many of them play important roles in signal transduction pathways (Mclaughlin et al., 2002; Li and Buchner, 2013). The first Hsp90 client protein discovered was the viral kinase v-Src (viral sarcoma) (Röhl et al., 2013). Later, Hsp90 was found to facilitate the maturation, stability, activation of more than 300 client proteins, and more client proteins are being discovered (Penkler et al., 2018).
Hsp90 is often overexpressed with activated form (Jackson, 2012), and many proteins in tumor cells are dependent upon the Hsp90 protein folding machinery, especially those overexpressed, or activated by mutation or translocation (Howes et al., 2014) Inhibition of Hsp90 targets client proteins associated with multiple hallmarks of cancer (Donnelly and Blagg, 2008; Khandelwal et al., 2016). Thus, Hsp90 has been a promising target for the treatment of cancer.
Leukemia is characterized by the presence of blasts in the bone marrow and deficient hematopoiesis (Orkin and Zon, 2008). There is increasing evidence that, in addition to genetic mutations, epigenetic events play a critical role in the pathophysiology of hematopoietic disorders (Plass et al., 2008). Human SUV39H1, the main methyl transferase responsible for the trimethylation of Lys9 in histone H3, is the main determinant of heterochromatin formation and also plays an important role in other processes, such as the inhibition of myod1-stimulated differentiation and regulation of the cell cycle, cell differentiation, and telomere length (Melcher et al., 2000; Rao et al., 2017). SUV39H1 interacts with oncogenes involved in acute myeloid leukemia (AML) and acts as a transcriptional repressor in hematopoietic differentiation and immortalization (Yang et al., 2010; Albacker et al., 2013). SUV39H1 interacts with the proto-oncogenes AML1, EVI-1 and PML-RARα, which play critical roles in the development of AML by influencing the transcriptional repression of target genes involved in hematopoietic differentiation and bone marrow immortalization (Carbone et al., 2006; Lakshmikuttyamma et al., 2009; Goyama et al., 2010).
Chaetocin, which is produced by Chaetomium sp., is an antibiotic having the thiodioxopiperazine structure (a disulfide-bridged piperazine) (Saito et al., 1988; Rombo et al., 2016). Chaetocin displayed anticancer properties against AML via oxidative stress induction (Chaib et al., 2012). Chaetocin was also previously identified as a specific inhibitor of histone methyltransferase SUV39H1 (Zhang et al., 2018). However, another report shows that Chaetocin was a nonspecific SUV39H1 inhibitor (Cherblanc et al., 2013). In our efforts to search new type inhibitor of Hsp90, we found that Chaetocin may be a new inhibitor. Herein, our results demonstrate that Chaetocin is a novel C-terminal inhibitor of Hsp90, and SUV39H1 is a new client protein of Hsp90 with HOP as a cruiter co-chaperone. SUV39H1 is induced to degradate by Chaetocin.
MATERIALS AND METHODS
Reagents
Roswell park memorial institute (RPMI) 1640 medium, fetal bovine serum (FBS) and trypsin were obtained from Gibco Invitrogen (Grand Island, NY, USA). Antibodies used to detect C-ABL, AML1-ETO, c-Raf, Akt, phosphorylated-Akt, phosphorylated-Erk, Erk, BCR-ABL, heat shock protein 90 (Hsp90), HOP, pp5, p23, Hsp70, SUV39H1, HOP, and me-H3K9 were purchased from Cell Signaling Technology (Boston, MA, USA). Primary antibodies (against Tubulin, Actin and GAPDH) and secondary antibodies were purchased from HuaBio (Hangzhou, China). Anti-Hsp90 antibody and protein A/G agarose were purchased from Santa Cruz Biotechnology, Inc (Dallas, TX, USA). Anti-Hsp90 (AC88) antibody, anti-p23 antibody and full-length Hsp90α (FL-Hsp90α) were purchased from Abcam (Cambridge, UK). 17-Allyl-17-demethoxygeldanamycin (17-AAG) was purchased from Apollo Scientific Limited (Stockport, UK). 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT), TPCK-treated trypsin, adenosine 5′-triphosphate (ATP)-agarose and alcohol dehydrogenase equine (ADH) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Cell lysis buffer for Western blotting (WB) and immunoprecipitation (IP), PMSF, MG132, a BCA Protein Assay Kit and IgG antibody were purchased from Beyotime Institute of Biotechnology (Shanghai, China).
Cell culture
The HL-60 and K562 cell lines were purchased from the Shanghai Cell Bank (Shanghai, China) and cultured in RPMI 1640 medium. All cell lines were maintained in medium with 10% FBS under 5% CO2 at 37°C.
Cell activity assay
MTT assays were used to measure the inhibition of cancer cell proliferation. For the MTT assays, suspended cells were seeded in a 96-well plate at a density of 6,000 cells per well. The cells were incubated with 17-AAG or Chaetocin (dissolved in 1% DMSO) at the indicated concentrations for 72 h. Twenty microliters of MTT solution was added to each well and incubated for 4 h at 37°C. Then, DMSO was added to the wells and incubated overnight at 37°C. The absorbance at 570 nm was measured using a microplate reader (BioTek, Winooski, VT, USA).
WB
HL-60 and K562 cells were incubated with 17-AAG or Chaetocin or other reagents at the indicated concentrations, washed twice with PBS, disrupted on ice for 45 min in loading buffer, and boiled for 10 min. The protein concentration was determined with a BCA Protein Assay Kit (Beyotime Institute of Biotechnology). Cell lysates containing equal amounts of protein were separated by SDS-PAGE, transferred to membranes, immunoblotted with the indicated primary and secondary antibodies, and detected by chemiluminescence with enhanced chemiluminescence (ECL) detection reagents. D means DMSO, NC means Negative control.
IP assays
HL-60 and K562 cells were cultured with 17-AAG, Chaetocin, CHX, MG132 or siRNA, washed twice with PBS, and disrupted on ice for 40 min. The lysates were centrifuged at 10,000×g for 15 min, and the supernatants were then harvested. Next, the lysates were incubated with control IgG or other corresponding antibodies overnight at 4°C. The mixtures were incubated with a 40 μL of protein A/G agarose slurry for 2-3 h at 4°C. The immunoprecipitates were harvested by centrifugation and then washed six times with wash buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Triton, pH 7.5). Finally, loading buffer was added, and the immunoprecipitates were boiled. Protein levels were assessed by WB.
Surface plasmon resonance spectroscopy (SPR)-based biomolecular interaction analysis
With Arrayit, the SpotBot3 Microarrayer control software in the SpotBot3 pinpoint platform (Kx5, Plexera, WA, USA) applied the compound to the surface of the biochip Graft-to-PCL and cross-linked UV light for 15 min to crosslink the compound onto the surface of the chip. The initial concentration of the protein was 10 mmol•L-1. Dilute with PBS buffer according to a certain ratio, and configure different concentrations of protein injection test. The injection flow rate was 1 μL/min, the injection time was 180 s, the dissociation flow rate was 1 μL•s-1, the injection time was 200 s, the regeneration flow rate was 2 μL•s-1, and the regeneration time was 200 s. The obtained data were analyzed and fitted according to the PLEXERA SPR Date Analysis Module (DAM) analysis software (Plexera) to obtain a binding kinetic constant Ka.
Proteolytic fingerprinting assay
A proteolytic fingerprinting assay was conducted as described previously (Li et al., 2009). Briefly, CTD-Hsp90α (0.6 μg) was incubated with novobiocin (20 μM) or Cheatocin (0.5-2 mM) in assay buffer (10 mM Tris-HCl, 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, pH 7.4) on ice for 1 h. The samples were digested on ice with TPCK-treated trypsin at concentrations of 10 and 30 μg/mL for 6 min. The reactions were terminated by the addition of SDS sample buffer, followed by boiling for 3-5 min. The digested products were analyzed by WB with the indicated antibody.
Plasmid constructs and protein expression and purification
GST-NTD-Hsp90α (9-236) was purchased from Addgene (Watertown, MA, USA). GST-CTD-Hsp90α construct containing CTD of Hsp90α encodes amino acids 535-732. Each insert was ligated into the BamH1 and Xho1 sites of pGEX-4 T3-GST. The inserts were transfected into BL21 cells. CTD-Hsp90α fusion protein expressions were induced by IPTG. Proteins were affinity purified with BeaverBeads GSH (Sangon Biotech, Shanghai, China). Protein aliquots were made and stored at −80°C. The primers for CTD-Hsp90α (535-732) were 5′-CGCGGATC CTTTGAGGGGAAGACTTTAGTGTC-3′ and 5′-CCGCTCGAGTTAGTCTA CTTCTTCCATGCGTG-3′.
ADH aggregation assays
ADH aggregation assays were conducted under the following conditions: ADH (6.2 μM) was incubated with GST-CTD-Hsp90α (1 μM), to which Chaetocin at different concentrations was added. Aggregation was induced at 55°C while the absorbance was measured at 360 nm every minute using a microplate reader for 60 min.
Hsp90α fragment vectors construction
FLAG-tagged Hsp90α N-, M-, and C-domains (encodes amino acids 9-236, 237-520, 535-732, respectively) were cloned from pGEX-4T3-GST-Hsp90α expression vector (Addgene) and inserted into the XhoI and BamHI sites of PLVX-IRES-ZsGreen1 vector. Then, these constructs were transformed into DH5α, amplified and extracted using Endo-free Plasmid Mini Kit I (Omega Bio-Tek, Inc., Norcross, GA, USA). Hsp90α domain-specific plasmids were amplified using the primer sequences used in this study (Table 1).
Table 1.
Primer sequence
| Plasmid name | Primer name | Primer sequence |
|---|---|---|
| Flag-C-Hsp90 | F1 | GGACGACGATGACAAGTTTGAGGGGAAGACTTTAGTGTC |
| F2 | CCGCTCGAGATGGATTACAAGGACGACGATGACAAG | |
| R | CGCGGATCCTTAGTCTACTTCTTCCATGCGTG | |
| Flag-N-Hsp90 | F1 | GGACGACGATGACAAG GACCAACCGATGGAGGAGGAG |
| F2 | CCGCTCGAGATGGATTACAAGGACGACGATGACAAG | |
| R | CGCGGATCCTCATTCAGCCTCATCATCGCTTA | |
| Flag-M-Hsp90 | F1 | GGACGACGATGACAAGAAGGAAGACAAAGAAGAAGAAAAAG |
| F2 | CCGCTCGAGATG GATTACAAGGACGACGATGACAAG | |
| R | CGCGGATCCTCACATATAGATCACTTCTAAGCC | |
| Flag- Hsp90 | F1 | GGACGACGATGACAAGCCTGAGGAAACCCAGACCCAA |
| F2 | CCGCTCGAGATGGATTACAAGGACGACGATGACAAG | |
| R | CGCGGATCCTTAGTCTACTTCTTCCATGCGTG |
HEK 293T cells were transfected with these plasmids using Lipofectamine 3000 transfection reagents (Thermo Scientific, MA, USA) according to the manufacturer’s instructions. After 48 h, cells with transfection efficiency >90% by detecting green fluorescence under the microscope (OLYMPUS, Tokyo, Japan) were utilized for further study.
Nitroblue tetrazolium (NBT) assays
HL-60 and K562 cells were incubated with Chaetocin at different concentrations for 72 h, collected, centrifuged at 1,000 r/min and 4°C for 5 min and washed with PBS twice, following which 1×106 cells from each group were reserved. One milliliter of NBT dye (plus 1 μL of 0.5 mg/mL PMA) was added to each group and dyed for 1 h at 37°C without light. The cells were collected after centrifugation and washed twice with PBS, following which 1 mL of 10% SDS (containing 0.04 M HCl) was added. The cells were allowed to stand for 12 h at 37°C, and the absorbance at a wavelength of 540 nm was detected.
Statistical analysis
Data are presented as the mean values ± standard deviations. Statistical comparisons among groups were performed by Student’s t-test. A p-value <0.05 was considered statistically significant.
RESULTS
Chaetocin inhibits Hsp90
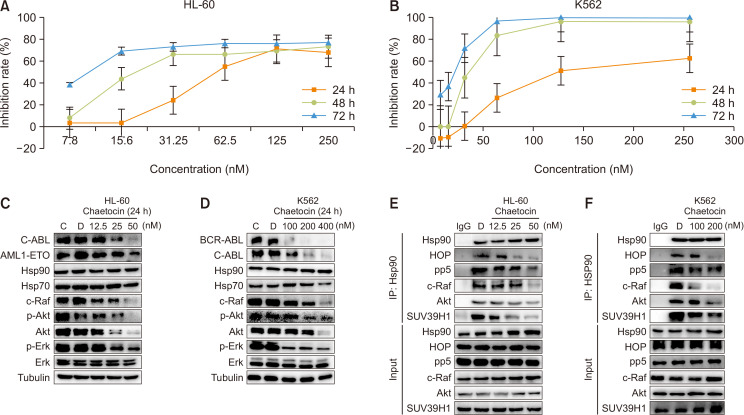
To determine if Chaetocin is a new inhibitor of Hsp90, an MTT assay was first used to measure the inhibitory effects of Chaetocin on the viability of HL-60 and K562 cells. As shown in Fig. 1A and 1B, Chaetocin inhibited the two cell lines with IC50 values of 25, 48, and 125 nM (K562 cells) and 7.8, 16.4, and 62.4 nM (HL-60 cells) after 24, 48, and 72 h of treatment, respectively.
Fig. 1.
Chaetocin inhibits the activities of cancer cells and displays potent Hsp90 inhibition effects. (A, B) Chaetocin inhibits activities of K562 and HL-60 cells. Cells were treated with Chaetocin at the indicated concentrations for 24 h, 48 h, and 72 h. Cell vitality was determined by a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay. Data are presented as the mean ± SD from three independent experiments. (C, D) Chaetocin inhibits the expression and activation of some client proteins in K562 and HL-60 cells. Cells were treated with Chaetocin at the indicated concentration for 24 h. Protein levels were analyzed by WB. (E, F) Chaetocin at different concentrations was incubated with K562 and HL-60 cells for 24 h, following which the interaction of Hsp90 with its co-chaperones and client proteins were tested with IP by Hsp90 antibody. Results are representative of at least three independent experiments.
The Hsp90 chaperone complex regulates many client oncoproteins that play key roles in tumor growth and progression (Yin et al., 2005; Song et al., 2015) and inhibition of Hsp90 causes client proteins to subsequent degradation by the proteasome (Ochel and Gademann, 2002; Banerji, 2015). Therefore, we next examined whether Chaetocin induced degradation of Hsp90 client oncoproteins. As shown in Fig. 1C and 1D, study with western blot analysis showed that Chaetocin inhibited the content and phosphorylation level of a variety of Hsp90 client proteins in the K562 and HL-60 cells including C-ABL, c-Raf, Akt, p-Akt, Erk and p-ERK, which are all client protein of Hsp90. We then treated HL-60 and K562 cells with Chaetocin for 24 h and detected the association of clients and co-chaperones with Hsp90 by IP assays. As shown in Fig. 1E and 1F, cell lysates were co-immunoprecipitated by Hsp90 antibody and detected with indicated antibody by western blot, Chaetocin was revealed to significantly inhibit the interaction of Hsp90 with the client proteins c-Raf, Akt in HL-60 and K562 cells. It also noted that the interaction of Hsp90 with the co-chaperones HOP, pp5 were dose-dependently inhibited by Chaetocin in the two cell lines. Altogether, these results show that Chaetocin is a new kind of Hsp90 inhibitor.
Chaetocin interacts with the C-terminal of Hsp90α
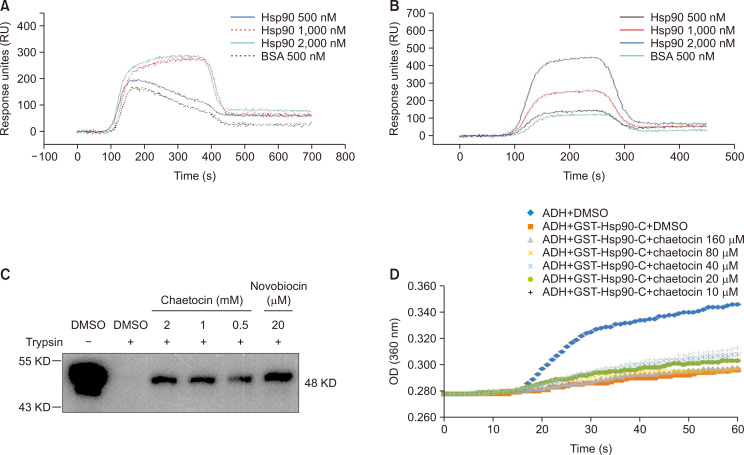
The chemical structure of Chaetocin is similar to that of HDN-1, which targets the C-terminal of Hsp90α (Song et al., 2015). Analogically, we speculate that the Chaetocin binds to C-terminal of Hsp90α. We first determined whether Chaetocin interacted directly with the C-terminal of Hsp90α by surface plasmon resonance (SPR) technology with immobilized purified FL-Hsp90α and CTD-Hsp90α chips. The obtained sensor grams showing the Chaetocin-FL-Hsp90α and Chaetocin-CTD-Hsp90α interactions were enhanced in a concentration-dependent manner, with comparative dissociation constants (KD values) of 8.96 uM (Chaetocin-CTD-Hsp90α) and 2.82 uM (Chaetocin-FL-Hsp90α), respectively (Fig. 2A, 2B), indicating that Chaetocin binds to the C-terminal of Hsp90. In addition, our studies showed that Chaetocin did not cause an increase in Hsp70 expression detected by western blot (As show in Fig. 1C and 1D). To further make sure the Chaetocin-binding domain in Hsp90, we performed a proteolytic fingerprinting assay to detect the interaction of Chaetocin with recombinant GST-CTD-Hsp90. The trypsin cuts specifically at arginine and lysine sites within prot which supported that Chaetocin bound to the C-terminal of Hsp90. Proein in the presence of TPCK, whereas these sites may be resistant to hydrolysis of trypsin due to protein conformation change by a small molecule binds to (Yu et al., 2016). After the GST-CTD-Hsp90 was incubated with Chaetocin, it was treated with TPCK-processed trypsin, and finally analyzed by WB for protein fingerprint detection. We used novobiocin, which inhibits the C-terminal of Hsp90, as a control. As shown in Fig. 2C, similar to novobiocin , Chaetocin protected the C-terminus of the Hsp90 protein from trypsin hydrolysis. The results further suggested that Chaetocin bound to the C-terminal of Hsp90. We then used ADH polymerization experiment to observe effect of Cheatocin on chaperone-like activity CTD of the Hsp90 to confirm whether Chaetocin interacted with the C-terminus of Hsp90. As shown in Fig. 2D, it was found that ADH proteins aggregated when the temperature rose and the aggregation could be measured using light absorption at 360 nm. The aggregation of ADH proteins was substantially reduced in the presence of CTD-Hsp90, which was reversed after treatment with Chaetocin in a dose-dependent manner.
Fig. 2.
Chaetocin interacts with the C-terminal of Hsp90α. (A, B) FL-Hsp90 (A) and CTD-Hsp90α (B) at various concentrations (500-2,000 nM) was injected onto an Cheatocin-immobilized chip, and RU values were recorded. (C) Proteolytic fingerprinting assay assessed the interaction of Chaetocin on CTD-Hsp90α. Hsp90α (0.6 μg) was incubated with Chaetocin (2, 1, and 0.5 mM) or novobiocin (20 μM), digested with TPCK-treated trypsin and analyzed by WB with anti-GST antibody. (D) Effect of Chaetocin on the anti-aggregation function of CTD-Hsp90α. Equine ADH (ADH, 6.2 μM) was incubated with GST-CTD-Hsp90α (1 μM) and Chaetocin at the indicated concentration. Aggregation was induced at 55°C while the absorbance at 360 nm was measured every minute for 60 min. Results are representative of at least three independent experiments.
SUV39H1 is a client protein of Hsp90
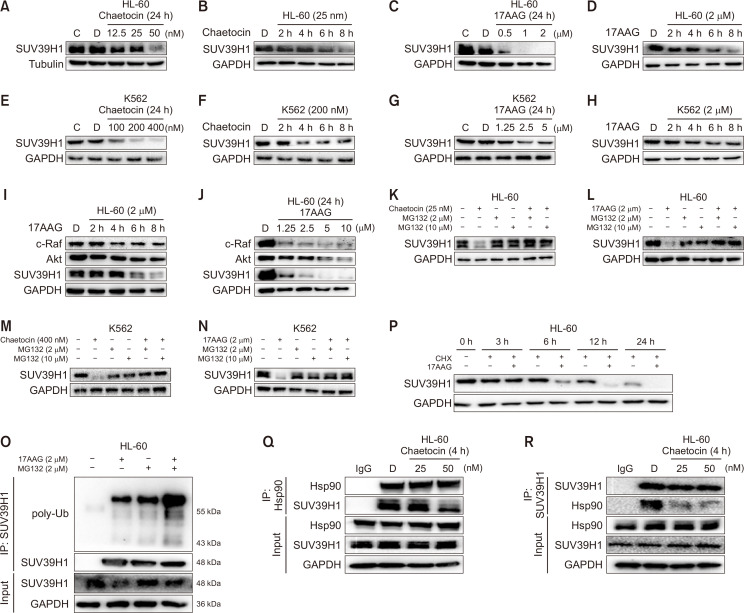
As referred above, Chaetocin is a new kind of Hsp90 C-terminal inhibitor. We wondered if its target molecule SUV39H1 was a client protein of Hsp90. With Hsp90 inhibitors 17-AAG and Chaetocin, we found that the protein level of SUV39H1 was reduced in a concentration- and time-dependent manner (Fig. 3A-3H), and SUV39H1 was more sensitive than c-Raf and Akt to concentration- and time-dependent inhibition by Chaetocin (Fig. 3I, 3J). Inhibition of Hsp90 blocks Hsp90 client proteins to mature, eventually leading to their degradation by the proteasome pathway. To ensure the SUV39H1 was degradation via proteasome pathway, the Hsp90 inhibitors Chaetocin and 17AAG were incubated with HL-60 cells for 8 h, and the cells were treated with the proteasome inhibitor MG132 for 2 h, then amount of SUV39H1 was determined by western blot. As shown in Fig. 3K and 3L, after HL-60 cells were exposed to Chaetocin and 17AAG for 8 h, the protein levels of SUV39H1 were significantly reduced, while in the presence of MG132, the protein levels of SUV39H1 recovered to the same level as control group. The same results were obtained in K562 cells (Fig. 3M, 3N). Furthermore, we detected SUV39H1 ubiquitination level by SUV39H1 antibody pulldown assay after treatment of Hsp90 inhibitor. As shown in Fig. 3O, compared with the group treated with only MG132, the ubiquitination of SUV39H1 significantly increased in the presence of MG132 and 17AAG. This finding supported that SUV39H1 was degraded through the proteasome pathway when Hsp90 was inhibited. To confirm that the decrease in the SUV39H1 protein level after Hsp90 inhibition was not the result of reduced SUV39H1 protein synthesis, we conducted further experiments with cycloheximide (CHX) to inhibit protein synthesis. Compared with the group treated with CHX alone, in HL-60 cells incubated with both CHX and 17AAG, the protein level of SUV39H1 was further decreased (Fig. 3P), suggesting that the decrease in SUV39H1 content was independent of gene transcription and translation.
Fig. 3.
SUV39H1 is a client protein of Hsp90. (A, C, E, G) The protein level changes of SUV39H1 with concentrations of Chaetocin or 17AAG. After the incubation of HL-60 or K562 cells with Chaetocin or 17AAG at different concentrations for 24 h, the amount of SUV39H1 was detected by WB. (B, D, F, H) The protein level changes of SUV39H1 with time in the presence of Chaetocin or 17AAG. After the incubation of HL-60 or K562 cells with Chaetocin or 17AAG at indicated time, the amount of SUV39H1 was detected by WB assay. (I, J) HL-60 cells were treated with 17AAG (2 μM) for different times (I) and different concentrations (J), the amount of different client proteins of Hsp90 and the SUV39H1 protein were detected using WB assay. (K-N) The decreases of SUV39H1 were reversed by MG132. The expression of SUV39H1 was detected in HL-60 or K562 cells treated by Chaetocin or 17AAG for 8 h after preincubated with MG132 (2 μM and 10 μM) for 2 h, and the levels of different client proteins SUV39H1 were detected using WB assay. (O) HL-60 cells were treated by 17AAG (2 μM) for 8 h with or without MG132 (2 μM). The ubiquitination of SUV39H1 was detected by immunoprecipitation assay. (P) The decrease of SUV39H1 protein level by 17-AAG was not due to reduction of SUV39H1 protein synthesis 17AAG (2 μM) were incubated with HL-60 for different time with or without CHX (2 μM) , WB assay was used to observe the amount of SUV39H1. (Q, R). HL-60 cells were treated with Chaetocin at different concentrations for 4 h, following which the interaction of Hsp90 with SUV39H1 was detected by Hsp90 antibody (Q) or SUV39H1 antibody (R). Results are representative of at least three independent experiments.
These results suggest that inhibitor of Hsp90 has ability to induce degradation of SUV39H1 through proteasome pathway. To confirm that SUV39H1 is a client protein of Hsp90, we further observed the effect of Cheatocin on interaction of SUV39H1 through IP assay with Hsp90 antibody. Immunobloting analyses of Hsp90 immunoprecipitates showed that Hsp90 were associated with SUV39H1, and Cheatocin obviously inhibited the Hsp90-SUV39H1 interaction compared with the control group (Fig. 3Q, 3R). Reciprocal IP with an anti- SUV39H1 antibody confirmed SUV39H1 were associated with Hsp90, and effect of Cheatocin on the two molecules interaction. Overall, the above results indicate that SUV39H1 is not properly folded when the binding of it to Hsp90 impaired by Hsp90 inhibitor, leading to ubiquitinated of SUV39H1 that is eventually degraded through the proteasome pathway. Thus, we demonstrate that SUV39H1 is a client protein of Hsp90.
SUV39H1 interacts with Hsp90 through co-chaperone HOP
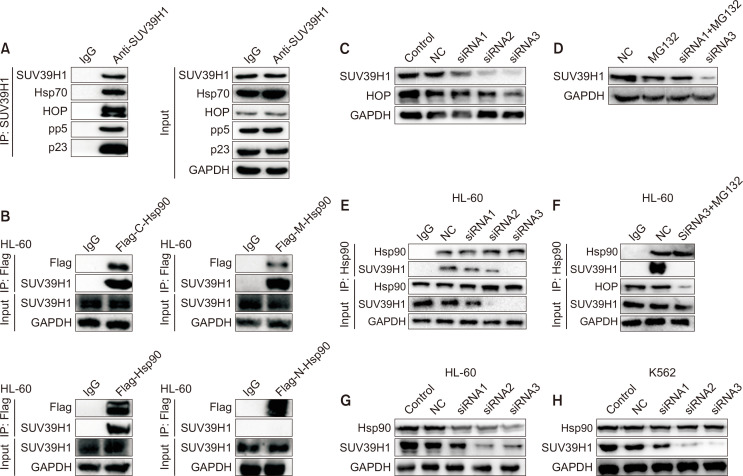
It has been known that Hsp90 requires co-chaperones to transport, fold and stability of its client proteins (Condelli et al., 2019). To study how SUV39H1 interacts with Hsp90, co-chaperones involved in the interaction of SUV39H1 with Hsp90 were investigated. We first conducted IP assay with SUV39H1 antibody, and found that co-chaperones Hsp70, HOP, pp5 and p23 bound to SUV39H1 (Fig. 4A). Meanwhile, recombinant Flag-CTD, -NTD and -MD of Hsp90 fragment plasmids were separately transfected into 293T cells to explore which domain of Hsp90 that SUV39H1 binds to. As shown in Fig. 4B, immunoblotting analyses of immunoprecipitates with flag antibody showed SUV39H1 was found to bind to the CTD and MD of Hsp90, rather than to NTD of Hsp90. Given that Hop was reported to deliver client protein to Hsp90 and bind with C-terminal and M-terminal of Hsp90 (Onuoha et al., 2008). Therefore, we speculate that SUV39H1 interacts with Hsp90 through HOP. SiRNA-mediated Hop depletion was further performed to verify this assumption. As shown in Fig. 4C, the expression levels of HOP and SUV39H1 were significantly reduced after HOP was knock-downed, suggesting that the SUV39H1 binding to Hsp90 was probably impaired and degradated subsequently due to be lack of HOP transport. To further confirm that the interaction of Hsp90 with SUV39H1 is mediated by HOP, we further carry out co-immunoprecipitation assay with SUV39H1 antibody. Compared with the control group, after the knockout of HOP, the interaction of SUV39H1 with Hsp90 was significantly reduced (Fig. 4E), while MG132 did not reverse the reduction in the interaction of SUV39H1 with Hsp90 (Fig. 4F) although degradation of SUV39H1 was inhibited by MG132 (Fig. 4D). These results indicate that SUV39H1 interacts with Hsp90 mediated by HOP, further confirm that SUV39H1 is a client protein of Hsp90. To make sure SUV39H1 is a new client of Hsp90, we detect SUV39H1 after Hsp90 was knock-downed, as shown in Fig. 4G and 4H, we could find that the expression levels of Hsp90 and SUV39H1 were significantly reduced.
Fig. 4.
SUV39H1 interacts with Hsp90 through co-chaperone HOP. (A) HL-60 cells were collected, and the interaction of SUV39H1 with Hsp90 co-chaperones was detected by SUV39H1 antibody. (B) SUV39H1 was found to bind to the CTD and MD of Hsp90. The Plasmids of Hsp90 CTD, NTD, and MD and the FL-Hsp90α protein were constructed and transfected to HEK293 cells, and their interactions with SUV39H1 were detected by IP assay. (C) The siRNA1, 2, 3 were used to deplete the HOP, and protein levels of HOP and SUV39H1 were then detected by WB assay. (D) The decrease of SUV39H1 by HOP depletion was reversed by MG132. HOP was blocked by siRNA3 with or without MG132 for 2 h, and protein expression was detected by WB assay. (E) After HOP was knocked out with siRNA 1, 2, 3, the interaction of Hsp90 with SUV39H1 was detected by IP. (F) After HOP was depleted by siRNA3 with or without MG132 for 2 h, and then collected to assess the interaction of Hsp90 with SUV39H1 by Hsp90 antibody. It is under the same conditions as Fig. 4D. Results are representative of at least three independent experiments. (G, H) The siRNA1, 2, 3 were used to deplete the Hsp90, and protein levels of Hsp90 and SUV39H1 were then detected by WB assay.
Chaetocin induces HL-60 and K562 cell differentiation
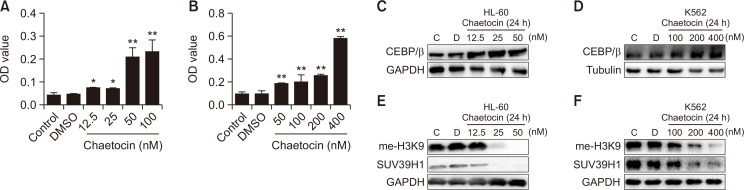
To investigate if Chaetocin induces HL-60 and K562 cell differentiation in indicated concentration of Hsp90 inhibition. NBT reduction assay was further performed. Differentiation was measured by induction of superoxide production as monitored by percentage of cells able to reduce NBT (Kumagai et al., 2005). As shown in Fig. 5A, 5B, compared with the control group, NBT-reducing activity was significantly increased after Chaetocin treatment, and the amount of NBT reduction by K562 and HL-60 cells was found to be directly proportional to the concentration of Chaetocin, suggesting Chaetocin was able to induce cell differentiation in two cells in the concentrations with Hsp90 inhibition.
Fig. 5.
Chaetocin has ability to induce cell differentiation with decrease of H3K9 methylation in the concentration of Hsp90 inhibition. (A, B) After the incubation of HL60 cells (A) and K562 cells (B) with Chaetocin at different concentrations for 72 h, cells were collected, centrifuged at 1,000 r/min and 4°C for 5 min and washed with PBS for twice. 1×106 cells in each well was added 1ml NBT dye (add 0.5 mg/ml PMA 1 μL) for 1 h at 37°C without light. Cells were collected, and 1 mL 10% SDS (containing 0.04 M HCL) was added for 12 h at 37°C, then the absorbance was detected at the wavelength of 540 nm. Data are presented as the mean ± SD from three independent experiments. (*p<0.05,**p<0.01 vs. DMSO). (C, D) After the incubation of HL60 cells and K562 cells with Chaetocin at different concentrations for 24 h, cells were collected. The protein levels of CEBP/β were analyzed by WB. Results are representative of at least three independent experiments. (E, F) HL-60 and K562 cells were treated with Chaetocin for 24 h. The protein levels of me-H3K9 and SUV39H1 were analyzed by WB. Results are representative of at least three independent experiments.
We also detected the C/EBPβ, the family of C/EBP has shown to be involved in different stages of myeloid development, and C/EBPβ are important in the late process of granulocytic differentiation. As shown in Fig. 5C and 5D.
We detected the methylation level of H3K9 after Chaetocin treatment, the result showed that me-H3K9 was obviously inhibited accompanied by decrease of SUV39H1 (Fig. 5E, 5F). These results indicate that Chaetocin binds to Hsp90 and impaired the function of Hsp90, thereby induces the degradation of SUV39H1, resulting in cell differentiation of K562 and HL-60 cells.
DISCUSSION
The data presented here establish in detailed Chaetocin as an inhibitor of Hsp90, and SUV39H1 as a strong Hsp90 client. Our findings provide a new context for understanding Chaetocin biological activity, and for treating SUV39H1-related disease. Histone lysine methyl transferases (HKMTs) are important epigenetic enzymes and exciting new drug gable targets for drug discovery (Copeland et al., 2009; Cherblanc et al., 2012). The fungal metabolite Chaetocin has identified as the first inhibitor of a lysine-specific histone methyltransferase SUV39H1 (Greiner et al., 2005). Chaetocin was found to have inhibitory activity with IC50 value at 0.11-0.8 μM on purified human SUV39H1 in vitro and obviously inhibitory effect at 0.1 μM in cells, although specificity on inhibition of Chaetocin is still in dispute (Cherblanc et al., 2013). Moreover, the level of AMl1-ETO and BCL-ABL, which is mutant fusion-protein recognized as a client protein of Hsp90 (Wang et al., 2009; Li et al., 2013; Roh et al., 2016), were also inhibited by Chaetocin on HL-60 and K562 cells, respectively. These results imply Chaetocin may be an inhibitor of Hsp90. Hsp90 inhibitor affects the binding of Hsp90 to its client protein and co-chaperones to some extent. In our study, we demonstrate that Chaetocin is an inhibitor of Hsp90 associating with the C-domain. We found that Chaetocin significantly inhibited the amount or phosphorylation levels of a variety of Hsp90 client proteins, including Akt, c-Raf, C-ABL, BCR-ABL and AML1-ETO, and even SUV39H1 at very low concentrations (with lowest concentration at 12.5 nM). SUV39H1 was verified by us to be a new client protein of Hsp90. Based on these reports and our findings, we think that Chaetocin inhibits Hsp90 displays stronger inhibitory activity on Hsp90 than on SUV39H1, indicating that Chaetocin inhibits Hsp90. Our results also supported that Chaetocin bound to the C-terminal of Hsp90, as inhibition of Hsp90 by targeting its N-terminus but not its C-terminus is often compensated by increased expression of Hsp70 (Mcconnell et al., 2015). Previous studies showed that C-terminal inhibitors inhibit the anti-aggregation function of C-terminus of Hsp90α (Allan et al., 2006). Determining anti-aggregation function is a standard method used to evaluate the chaperone-like activity of a protein (Gliniewicz et al., 2019). Our results indicate that Chaetocin has the ability to inhibit the chaperone function of Hsp90, consolidating that Chaetocin binds to C-terminus of Hsp90, and is a new kind of C-terminal inhibitor of Hsp90.
Hsp90 is involved in initial protein folding as well as in stabilizing proteins and its constitutively active with unstable domains (Makhnevych and Houry, 2012). Many Hsp90-stabilized proteins are oncogenic, which has led to the development of Hsp90 inhibitors for cancer therapy (Kim et al., 2009). Hundreds of client proteins in the eukaryotic cytosol has identified, and new clients are still being discovered (see http://www.picard.ch/downloads/Hsp90 inter actors.pdf). In the client proteins, AKT and c-RAF was recognized to be “strong client” which is more depend on Hsp90 constitutively for their stability (Xu et al., 2005; Theodoraki et al., 2007). In the presence of paper, we ensure that SUV39H1 is a new strong client protein. SUV39H1 was revealed to bind to Hsp90, and degrade after treatment of 17-AAG and Chaetocin by proteasome pathway. SUV39H1 as a histone H3K9 methyltransferase is responsible for the trimethylation of Lys9 in histone H3, and interacts with proto-oncogenes which play critical roles in the development of AML (Yang et al., 2010; Albacker et al., 2013). The degradation of SUV39H1 is more sensitive to Hsp90 inhibitor than that of AKT and c-RAF. SUV39H1 is the main HMT responsible for the accumulation of histone H3 containing a tri-methyl group at its lysine 9 position (H3K9me3) in heterochromatin (Chaib et al., 2012). SUV39H1 interacts with proto-oncogenes which play critical roles in the development of leukemia, influencing aberrant gene silencing of target genes related to hematopoietic differentiation and survival (Goyama et al., 2010). Consistently, our data show that Chaetocin decreases the amount of SUV39H1 accompanied by reduction of H3K9 methylation, in turn inducing the differentiation and apoptosis of HL-60 and K562.
In the eukaryotic cytosol, Hsp90 operates with the help of numerous co-chaperones (Sahasrabudhe et al., 2017). Considering their influence on Hsp90 and client maturation, co-chaperones can be divided into three categories: client recruiters, remodelers of Hsp90, and late-acting co-chaperones. One of common client recruiters is the TPR protein Hop (stress inducible 1/Hsp70-Hsp90 organizing protein), which delivers substrates from Hsp70 to Hsp90 (Scheufler et al., 2000). Cdc37 (cell division cycle 37 homolog) is another common Hsp90 ATPase-inhibiting co-chaperone that has been implicated in client recruiting, but clients recruited by Cdc37 seem to be limited to kinases (Taipale et al., 2012). In line with these references, we demonstrate that SUV39H1 binding to Hsp70, Hsp90 and Hop, and Hop is a client recruiter of SUV39H1, which was responsible for transferring SUV39H1 from Hsp70 to Hsp90.
Altogether, our results demonstrate that SUV39H1 is a new client protein of Hsp90 degradated by Chaetocin as a novel C-terminal inhibitor of Hsp90. The study establishes new relationship of Chaetocin and SUV39H1, and paves an avenue for exploring a new strategy to target SUV39H1 by inhibition of Hsp90 in leukemia.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 81673450), the NSFC Shandong Joint Fund (U1606403), the Scientific and Technological Innovation Project financially supported by Qingdao National Laboratory for Marine Science and Technology (No. 2015ASKJ02), Fundamental Research Funds for the Central Universities (No. 201762002) and the National Science and Technology Major Project for Significant New Drugs Development (2018ZX09735-004).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- Albacker C. E., Storer N. Y., Langdon E. M., Anthony D. B., Yi Z., Langenau D. M., Zhou Y., Langenau D. M., Zon L. I. The histone methyltransferase suv39h1 suppresses embryonal rhabdomyosarcoma formation in zebrafish. PLoS ONE. 2013;8:e64969- doi: 10.1371/journal.pone.0064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan R. K., Mok D., Ward B. K., Ratajczak T. Modulation of chaperone function and cochaperone interaction by novobiocin in the c-terminal domain of Hsp90. J. Biol. Chem. 2006;281:7161–7171. doi: 10.1074/jbc.M512406200. [DOI] [PubMed] [Google Scholar]
- Banerji U. O7.6 - HSP90 inhibitors in the clinic: what have we learnt? Ann. Oncol. 2015;26:ii10. doi: 10.1093/annonc/mdv085.6. [DOI] [Google Scholar]
- Carbone R., Botrugno O. A., Ronzoni S., Insinga A., Di Croce L., Pelicci P. G., Minucci S. Recruitment of the histone methyltransferase suv39h1 and its role in the oncogenic properties of the leukemia-associated pml-retinoic acid receptor fusion protein. Mol. Cell. Biol. 2006;26:1288–1296. doi: 10.1128/MCB.26.4.1288-1296.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaib H., Nebbioso A., Prebet T., Castellano R., Garbit S., Restouin A., Vey N., Altucci L., Collette Y. Anti-leukemia activity of chaetocin via death receptor-dependent apoptosis and dual modulation of the histone methyl-transferase suv39h1. Leukemia. 2012;26:662–674. doi: 10.1038/leu.2011.271. [DOI] [PubMed] [Google Scholar]
- Cherblanc F. L., Chapman K. L., Brown R., Fuchter M. J. Chaetocin is a nonspecific inhibitor of histone lysine methyltransferases. Nat. Chem. Biol. 2013;9:136–137. doi: 10.1038/nchembio.1187. [DOI] [PubMed] [Google Scholar]
- Cherblanc F. L., Chapman-Rothe N., Brown R., Fuchter M. Current limitations and future opportunities for epigenetic therapies. Future Med. Chem. 2012;4:425–446. doi: 10.4155/fmc.12.7. [DOI] [PubMed] [Google Scholar]
- Condelli V., Crispo F., Pietrafesa M., Lettini G., Matassa D. S., Esposito F., Landriscina M., Maddalena F. HSP90 molecular chaperones, metabolic rewiring, and epigenetics: impact on tumor progression and perspective for anticancer therapy. Cells. 2019;8:532. doi: 10.3390/cells8060532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland R. A., Solomon M. E., Richon V. M. Protein methyltransferases as a target class for drug discovery. Nat. Rev. Drug Discov. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- Donnelly A., Blagg B. S. Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket. Curr. Med. Chem. 2008;15:2702–2717. doi: 10.2174/092986708786242895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliniewicz E. F., Chambers K. M., De Leon E. R., Sibai D., Campbell H. C., McMenimen K. A. Chaperone-like activity of the N-terminal region of a human small heat shock protein and chaperone-functionalized nanoparticles. Proteins. 2019;87:401–415. doi: 10.1002/prot.25662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S., Nitta E., Yoshino T., Kako S., Watanabe-Okochi N., Shimabe M., Imai Y., Takahashi K., Kurokawa M. EVI-1 interacts with histone methyltransferases suv39h1 and g9a for transcriptional repression and bone marrow immortalization. Leukemia. 2010;24:81–88. doi: 10.1038/leu.2009.202. [DOI] [PubMed] [Google Scholar]
- Greiner D., Bonaldi T., Eskeland R., Roemer E., Imhof A. Identification of a specific inhibitor of the histone methyltransferase su(var)3-9. Nat. Chem. Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- Howes J., Lu B. F., Powers M., Mitsopoulos C., Al-Lazikani B., Linardopoulos S., Clarke P., Workman P. Abstract 2730: RNAi knockdown or chemical inhibition of anaphase-promoting complex components is synthetic lethal with HSP90 inhibition. Cancer Res. 2014;74:2730. doi: 10.1158/1538-7445.AM2014-2730. [DOI] [Google Scholar]
- Jackson S. E. Hsp90: structure and function. Top. Curr. Chem. 2012;328:155–240. doi: 10.1007/128_2012_356. [DOI] [PubMed] [Google Scholar]
- Khandelwal A., Crowley V. M., Blagg B. S. J. Natural product inspired N-terminal Hsp90 inhibitors: from bench to bedside? Med. Res. Rev. 2016;36:92–118. doi: 10.1002/med.21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Alarcon S., Lee S., Lee M. J., Giaccone G., Neckers L., Trepel J. B. Update on Hsp90 inhibitors in clinical trial. Curr. Top. Med. Chem. 2009;9:1479–1492. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T., Shih L. Y., Hughes S. V., Desmond J. C., O'Kelly J., Hewison M., Koeffler H. P. 19-Nor-1,25(OH)2D2 (a novel, noncalcemic vitamin D analogue), combined with arsenic trioxide, has potent antitumor activity against myeloid leukemia. Cancer Res. 2005;65:2488–2497. doi: 10.1158/0008-5472.CAN-04-2800. [DOI] [PubMed] [Google Scholar]
- Lakshmikuttyamma A., Scott S. A., Decoteau J. F., Geyer C. R. Reexpression of epigenetically silenced AML tumor suppressor genes by SUV39H1 inhibition. Oncogene. 2009;29:576–588. doi: 10.1038/onc.2009.361. [DOI] [PubMed] [Google Scholar]
- Li J., Buchner J. Structure, function and regulation of the Hsp90 machinery. Biomed. J. 2013;36:106–117. doi: 10.4103/2319-4170.113230. [DOI] [PubMed] [Google Scholar]
- Li M., Zhang X., Zhou W. J., Chen Y. H., Liu H., Liu L., Yang C. M., Qan W. B. Hsp90 inhibitor BIIB021 enhances triptolide-induced apoptosis of human T-cell acute lymphoblastic leukemia cells in vitro mainly by disrupting p53-MDM2 balance. Acta Pharmacol. Sin. 2013;34:1545–1553. doi: 10.1038/aps.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang T., Jiang Y., Lee H. F., Schwartz S. J., Sun D. (-)-Epigallocatechin-3-gallate inhibits Hsp90 function by impairing Hsp90 association with cochaperones in pancreatic cancer cell line Mia Paca-2. Mol. Pharm. 2009;6:1152–1159. doi: 10.1021/mp900037p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T., Houry W. A. The role of Hsp90 in protein complex assembly. Biochim. Biophys. Acta. 2012;1823:674–682. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Mcconnell J., Wang Y., Mcalpine S. Heat Shock Protein Inhibitors. Springer International Publishing; 2015. Targeting the C-terminus of Hsp90 as a cancer therapy. [DOI] [Google Scholar]
- Mclaughlin S. H., Smith H. W., Jackson S. E. Stimulation of the weak ATPase activity of human Hsp90 by a client protein. J. Mol. Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- Melcher M., Schmid M., Aagaard L., Selenko P., Laible G., Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol. 2000;20:3728–3741. doi: 10.1128/MCB.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellatyar H., Talaei S., Pilehvar-Soltanahmadi Y., Barzegar A., Akbarzadeh A., Shahabi A., Barekati-Mowahed M., Zarghami N. Targeted cancer therapy through 17-DMAG as an Hsp90 inhibitor: overview and current state of the art. Biomed. Pharmacother. 2018;102:608–617. doi: 10.1016/j.biopha.2018.03.102. [DOI] [PubMed] [Google Scholar]
- Ochel H. J., Gademann G. Heat-shock protein 90: potential involvement in the pathogenesis of malignancy and pharmacological intervention. Onkologie. 2002;25:466–473. doi: 10.1159/000067442. [DOI] [PubMed] [Google Scholar]
- Onuoha S. C., Coulstock E. T., Grossmann J. G., Jackson S. E. Structural studies on the co-chaperone Hop and its complexes with Hsp90. J. Mol. Biol. 2008;379:732–744. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Zon L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkler D. L., Atilgan C., Tastan Bishop Ö. Allosteric modulation of human hsp90α conformational dynamics. J. Chem. Inf. Model. 2018;58:383–404. doi: 10.1021/acs.jcim.7b00630. [DOI] [PubMed] [Google Scholar]
- Plass C., Oakes C., Blum W., Marcucci G. Epigenetics in acute myeloid leukemia. Semin. Oncol. 2008;35:378–387. doi: 10.1053/j.seminoncol.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V. K., Pal A., Taneja R. A drive in SUVs: from development to disease. Epigenetics. 2017;12:177–186. doi: 10.1080/15592294.2017.1281502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh S. H., Kasembeli M., Galaz-Montoya J. G., Trnka M., Lau W. C., Burlingame A., Chiu W., Tweardy D. J. Chaperonin TRiC/CCT modulates the folding and activity of leukemogenic fusion oncoprotein AML1-ETO. J. Biol. Chem. 2016;291:4732–4741. doi: 10.1074/jbc.M115.684878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombo R., Weiher H., Schmidt-Wolf I. G. Effect of chaetocin on renal cell carcinoma cells and cytokine-induced killer cells. Ger. Med. Sci. 2016;14:Doc04. doi: 10.3205/000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhl A., Rohrberg J., Buchner J. The chaperone Hsp90: changing partners for demanding clients. Trends Biochem. Sci. 2013;38:253–262. doi: 10.1016/j.tibs.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Sahasrabudhe P., Rohrberg J., Biebl M. M., Rutz D. A., Buchner J. The plasticity of the Hsp90 co-chaperone system. Mol. Cell. 2017;67:947–961.e5. doi: 10.1016/j.molcel.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Saito T., Suzuki Y., Koyama K., Natori S., Iitaka Y., Kinosita T. Chetracin A and chaetocins B and C, three new epipolythiodioxopiperazines from Chaetomium spp. Chem. Pharm. Bull. 1988;36:1942–1956. doi: 10.1248/cpb.36.1942. [DOI] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Song X., Zhao Z., Qi X., Tang S., Wang Q., Zhu T., Gu Q., Liu M., Li J. Identification of epipolythiodioxopiperazines HDN-1 and chaetocin as novel inhibitor of heat shock protein 90. Oncotarget. 2015;6:5263–5274. doi: 10.18632/oncotarget.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K., Karras G. I., Lindquist S. Quantitative analysis of Hsp90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoraki M. A., Kunjappu M., Sternberg D. W., Caplan A. J. Akt shows variable sensitivity to an Hsp90 inhibitor depending on cell context. Exp. Cell Res. 2007;313:3851–3858. doi: 10.1016/j.yexcr.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fiskus W., Natarajan K., Yang Y., Rao R., Chen J., Joshi A., Koul S., Upadhyay S., Balusu R., Fernandez P., Buckley K., Jillella A., Quadt C., Atadja P., Levine R., Bhalla K. Abstract #3722: co-treatment with pan-HDAC inhibitor panobinostat or heat shock protein (hsp) 90 inhibitor AUY922 with JAK2 inhibitor TG101209 is highly active against bone marrow progenitor cells from myeloproliferative neoplasms (MPN) Cancer Res. 2009;69:3722. [Google Scholar]
- Xu W., Yuan X., Xiang Z., Mimnaugh E., Marcu M., Neckers L. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat. Struct. Mol. Biol. 2005;12:120–126. doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- Yang Y. J., Han J. W., Youn H. D., Cho E. J. The tumor suppressor, parafibromin, mediates histone H3 K9 methylation for cyclin D1 repression. Nucleic Acids Res. 2010;38:382–390. doi: 10.1093/nar/gkp991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Zhang H., Burrows F., Zhang L., Shores C. G. Potent activity of a novel dimeric heat shock protein 90 inhibitor against head and neck squamous cell carcinoma in vitro and in vivo. Clin. Cancer Res. 2005;11:3889–3896. doi: 10.1158/1078-0432.CCR-04-2272. [DOI] [PubMed] [Google Scholar]
- Yu H., Cai S., Gao J., Wang C., Qiao X., Wang H., Feng L., Wang Y. Express sequence tag analysis - identification of anseriformes trypsin genes from full-length cDNA library of the duck (Anas platyrhynchos) and characterization of their structure and function. Biochemistry Mosc. 2016;81:152–162. doi: 10.1134/S0006297916020097. [DOI] [PubMed] [Google Scholar]
- Zhang Y. M., Gao E. E., Wang Q. Q., Tian H., Hou J. Effects of histone methyltransferase inhibitor chaetocin on histone H3K9 methylation of cultured ovine somatic cells and development of preimplantation cloned embryos. Reprod. Toxicol. 2018;79:124–131. doi: 10.1016/j.reprotox.2018.06.006. [DOI] [PubMed] [Google Scholar]