Abstract
The current coronavirus disease 2019 (COVID-19) pandemic has caused significant challenges throughout the world and a rapid, reliable diagnostic test is in high demand. Real-time reverse transcription polymerase chain reaction (RT-PCR) was one of the most quickly established methods of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection and is considered to be the gold standard. In this report, we share our experience of using two different testing platforms: the cobas 6800 SARS-CoV-2 test, an automated system that was recently granted Emergency Use Authorization by the FDA, and a laboratory-developed test based on the protocol from the Taiwan Centers for Disease Control (CDC). There was an overall 96.2% agreement between the two platforms. However, the positive agreement between the two platforms was only 80.0%. We found 3 instances of discordance between the two systems and this emphasized the need for timely diagnosis with a reliable testing platform.
Keywords: Coronavirus, Molecular diagnostics, Viral load, RT-PCR, RNA extraction
In December 2019, a cluster of patients with atypical pneumonia of unknown etiology emerged in Wuhan, China [1]. A novel beta-coronavirus, defined as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified as the cause of these cases [2]. According to the Johns Hopkins University COVID-19 Dashboard, as of May 29th 2020, there were over 5.8 million confirmed cases of COVID-19 and more than 360,000 deaths worldwide [3]. Testing capabilities are absolutely essential for managing a pandemic. Reverse transcription polymerase chain reaction (RT-PCR) was considered the primary mode of diagnosis for infection with SARS-CoV-2. Different viral targets were proposed for detection of the virus, including RNA-dependent RNA polymerase (RdRp), envelope (E), open reading frame (ORF) 1a and nucleocapsid (N) [4]. To confront the evolving healthcare crisis, the Taiwan Centers for Disease Control (CDC) organized COVID-19 testing and established a protocol for clinical laboratories to follow [5].
During the pandemic, timely and effective RT-PCR assay design was possible because of the availability of significant SARS-CoV-2 sequence data [6]. However, there is a need for a sensitive, accessible, and rapid diagnostic test for COVID-19 [7]. The aim of this manufacturer-independent study was to evaluate the clinical performance of a laboratory-developed test based on the Taiwan CDC protocol and the automated Roche cobas 6800 SARS-CoV-2 test (Roche, Pleasanton, CA, USA) that has recently received emergency use authorization (EUA) by the Food and Drug Administration (FDA) for use as a detection assay.
Methods
A total of 79 respiratory samples (nasopharyngeal or throat swabs) were obtained from 72 symptomatic patients or contact persons. After collection the swabs were placed in 1 ml viral transport medium and then the specimen volume specified by each assay was transferred as appropriate. All specimens were tested by both testing platforms. The protocol recommended by the Taiwan CDC was as follows: viral RNA was extracted from 300 μL of the sample using the LabTurbo automated purification system (TAIGEN, Taipei, Taiwan). Quantification RT-PCR (qRT-PCR) was performed using the TaqMan® Fast virus 1-Step Master Mix reagent (Thermo Fisher Scientific, NY, USA) with the previously published primer/probe [8]. Briefly, the 25 μL qRT-PCR mixture contained TaqMan® Fast virus 1-step Master Mix, 400 nM each of forward and reverse primer, 200 nM TaqMan probe, and 5 μL extracted RNA, or water for the no template controls. We purchased the AccuPlex™ SARS-CoV-2 reference material kit as the positive control (SeraCare, MA, USA). The samples were also performed and analyzed using the Roche cobas Z480 analyzer (Roche Molecular Systems, Rotkreuz, Switzerland) under the following conditions: 20 min at 50 °C and 20 s at 95 °C, followed by 45 cycles of 3 s at 95 °C and 30 s at 60 °C. The limit of detection for this assay was determined to be 3.8–5.2 RNA copies per reaction [8]. Positive detection of SARS-CoV-2 by the Taiwan CDC protocol was considered when a sample tested positive for the E, RdRp and N genes. Samples were considered SARS-Cov-2 negative if they tested negative for the E and RdRp genes, or negative for the RdRp gene but positive for the E gene [5].
Due to safety considerations, for the automated system, all of the samples were heat-inactivated (56 °C for 10 min) and handled inside a class BSC II biosafety cabinet before being loaded onto the cobas 6800 instrument (Roche Molecular Systems, Rotkreuz, Switzerland). The cobas SARS-CoV-2 kit was used. The cycle threshold (Ct) values reported by the cobas SARS-CoV-2 test were either “detected” (ORF1 gene and E gene detected), “presumptive positive” (ORF1 gene not detected; E gene detected), or “not detected” (ORF1 gene and E gene not detected). The limit of detection for this assay was determined to be 4.4 copies per reaction (package insert, AccuPlex™ SARS-CoV-2 reference material kit). All protocols were performed according to the manufacturer's instructions.
Results
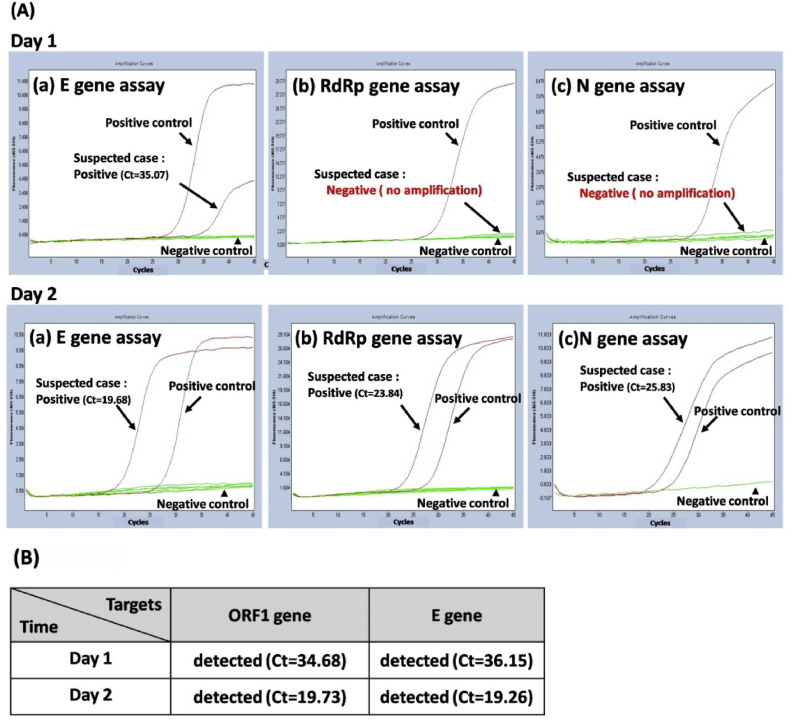
Of the 79 specimens tested, SARS-CoV-2 was detected by both methods in 12 specimens, while 64 specimens were negative by both assays [Table 1]. The overall agreement between the two platforms was 96.2%. However, the positive agreement between the two platforms was only 80.0%. A total of 3 discrepant results were noted, with samples testing SARS-CoV-2 positive by the automated system but negative by the laboratory-developed test based on the Taiwan CDC protocol. An example of a discordant case is shown in Fig. 1. A sample was reported as negative for SARS-CoV-2 virus on the first day by the Taiwan CDC protocol because the RdRp and N gene were undetected, but the E gene was detected. The respiratory sample was collected again after 24 h, and this new sample tested positive for SARS-CoV-2 as the three targets were all detected by the laboratory-developed test based on the Taiwan CDC protocol [Fig. 1A]. These two samples were examined by the cobas SARS-CoV-2 test and both tested positive for SARS-CoV-2 as the ORF1 and E gene targets were detected [Fig. 1B]. The second patient had the similar situation as mentioned above. The third one was a patient who diagnosed with COVID-19. After one month of treatment and quarantine, the throat swab showed a negative result by the Taiwan CDC protocol because only the E gene was detected. A positive result of CDC protocol was demonstrated on the next day. However, these two samples were positive results by the cobas SARS-CoV-2 test.
Table 1.
Results of SARS-CoV-2 testing by two molecular diagnosis methods.
| Method | Roche cobas 6800 systema |
Total | |
|---|---|---|---|
| Detected | Not-detected | ||
| Taiwan CDCb | |||
| Detected | 12 | 0 | 12 |
| Not-detected | 3 | 64 | 67 |
| Total | 15 | 64 | 79 |
Cohen's k coefficients: 0.866 (95% CI, 0.719–1.000).
Overall percent agreement = 96.2%.
Positive percent agreement = 80.0%.
A sample was considered positive for SARS-CoV-2 when both the ORF1 and E genes were detected, a sample was considered presumptive positive when the ORF1 gene was not detected but the E gene was detected, and a result was defined as negative if neither the ORF1 nor E gene were detected.
A sample was considered positive for SARS-CoV-2 when the E gene, RdRp gene and N gene were detected. A sample was considered negative for SARS-CoV-2 if the sample was negative for the E and RdRp genes, or negative for the RdRp gene but positive for the E gene.
Fig. 1.
Example of discordant cases between the two detection methods. (A) Amplification curves of the (a) E gene assay, (b) RdRp gene assay, and (c) N gene assay under the Taiwan CDC protocol for SARS-CoV-2 detection. (B) Results from the cobas 6800 SARS-CoV-2 test. The samples were collected from a suspected patient at day 1 and day 2.
Discussion
The continued need for increased testing capacity is because of the desire to test both asymptomatic and symptomatic individuals, particularly as countries begin to restart their economies. Detection of SARS-CoV-2 RNA from respiratory tract samples is an important component of outbreak management. Rapid and accurate identification of infected individuals is critical for enforcing self-isolation recommendations, community contact tracing and the use of appropriate enhanced personal protective equipment in healthcare settings.
Compared to a similar study [9], our study results revealed that the limit of detection for both systems was better than the laboratory-developed test based on the modified USA CDC protocol. The differences between the systems that affected performance may include the efficiency of the designed primer/probe gene targets, the amount of sample collected and the input volume of the initial specimen. Hundreds of genomic sequences for SARS-CoV-2 were uploaded from different research groups around the world. Different national CDC and biotechnology companies designed the primer and probe based on these sequences for accurately detected SARS-CoV-2 virus by qRT-PCR. The genomic sequence variation of SARS-CoV-2 is important for molecular diagnostic method development. The detection targets of USA CDC protocol only for N gene, but Taiwan CDC protocol target RdRp, E and N gene otherwise automated system target ORF1 and E gene. The finding is consistent with other study reported that tests targeting the N gene to have a higher limitation of detection than the E gene [9]. The laboratory-developed test based on the modified USA CDC protocol has a lower input volume for the initial specimen (120–140 μL) compared with our study (300 μL). In addition, most commercially available viral (universal) transport mediums have a volume of 3.0 mL but to enhance the sensitivity of the SARS-CoV-2 detection methods, we used 1.0 mL viral transport medium to achieve a concentrated specimen.
There were three samples in which SARS-CoV-2 was “not-detected” by the laboratory-developed test based on the Taiwan CDC protocol but was “detected” by the automated system. This may have happened because the amount of virus in the sample was below the detection limit for this method, thus leading to failure to detect a positive result. Conventional molecular diagnostic methods include several manual steps, such as nucleic acid extraction, master mix preparation and RT-PCR setup, as well as the interpretation of results. These steps are labor intensive and time consuming. A fully automated system allows for the handling/testing of large numbers of samples and also significantly reduces the hands-on time required by up to 50%. In addition, it is easier and quicker to train personnel who are not familiar with molecular diagnostic assays to work with automated tests. However, a limitation of automated tests is that the sputum specimen cannot be placed directly on the instrument; it must be pre-treated to avoid instruction malfunctions.
Taiwan successfully used big data analytics for efficient contact tracing management, and surveillance of those who require quarantine and isolation [10]. Therefore, the number of patients infected with COVID-19 in Taiwan was limited and decreased after mid-April [11]. A major limitation of the current study was the small number of clinical samples, and not included those asymptomatic cases, the number of positive samples in particular. However, the early detection or diagnosis is an important key to prevent transmission, especially for those asymptomatic detections. Because of this, the positive agreement by both platforms may have been under- or over-estimated by our laboratories. At present, there is no clinical or laboratory gold standard to serve as an absolute reference for comparison. Our experience revealed some differences between these two methods. The samples collected from suspected patients on day 1 could report a negative result for SARS-CoV-2 when tested by the Taiwan CDC protocol. As a result, repeat testing on suspected patients is needed even if the result is initially negative. The cobas 6800 SARS-CoV-2 test might provide a solution to this problem.
Conclusions
Our results suggest that the Roche cobas 6800 automated system might have a lower limit of detection compared with the laboratory-developed test based on the Taiwan CDC protocol. In addition to this lower limit of detection, the cobas 6800 SARS-CoV-2 test might shorten the diagnosis time for COVID-19. Further studies are needed in the future.
Conflicts of interest
The authors declare that they have no conflict of interest.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johns Hopkins Coronavirus Resource Center . 2020. COVID-19 map.https://coronavirus.jhu.edu/map.html [Google Scholar]
- 4.CDC . 2020. Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes.https://signagen.com/blog/2020/10/07/research-use-only-2019-novel-coronavirus-2019-ncov-real-time-rt-pcr-primers-and-probes/ [Google Scholar]
- 5.Taiwan Centers of Disease Control . 2020. 2019-nCoV virus nucleic acid test.https://www.cdc.gov.tw/En/Category/QAPage/LnqBFJsulw6fW3nswc04Yw [Google Scholar]
- 6.Decaro N., Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesion from animal coronaviruses. Vet Microbiol. 2020;244:10869. doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfefferle S., Reucher S., Norz D., Lutgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25:2000152. doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pujadas E., Ibeh N., Hernandez M.M., Waluszko A., Sidorenko T., Flores V. Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. J Med Virol. 2020;92:1695–1698. doi: 10.1002/jmv.25988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C.M., Jyan H.W., Chien S.C., Jen H.H., Hsu C.Y., Lee P.C. Containing COVID-19 among 627,386 persons in contact with the diamond princess cruise ship passengers who disembarked in Taiwan: big data analytics. J Med Internet Res. 2020;22 doi: 10.2196/19540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taiwan centers of disease control. 2020. https://www.cdc.gov.tw/En [Google Scholar]