Abstract
Labeling of biomolecules in live eukaryotic cells has been limited by low component stability and slow reaction rates. We show that genetically encoded tetrazine amino acids in proteins reach reaction rates of 8×104 M−1s−1 with sTCO reagents, making them the fastest site-specific bioorthogonal labels in eukaryotic systems. We demonstrate that tetrazine amino aids are stable on proteins and are capable of quantitative labeling with sTCO reagents. The exceptionally high reaction rate of this ligation minimizes label concentration, allowing for sub-stochiometric in vivo eukaryotic protein labeling where the concentration of the label is less than the concentration of the protein. This approach offers unprecedented control over the composition and stability of the protein tag. We anticipate that this system will have a broad impact on labeling and imaging studies because it can be used where all generally orthogonal PylRS/tRNA pairs are employed.
Graphical Abstract
Efficient labeling of proteins in living eukaryotic systems with fast bioorthogonal reactions is an essential tool for studying fundamental cell biology. The biological processes that we can monitor are currently limited by the rate of labeling reactions. The problem of slow labeling rates has been identified as the most critical factor in determining the labeling efficiency and specificity of proteins in cells.1–3 Reactions between strained alkynes and tetrazines can reach rate constants of 107 M−1s−1, however, optimizing the encoding of these functional groups in live cells while minimizing off-target reactions is an open challenge (Figure 1).1, 4–5 In recent years, two non-canonical amino acids (ncAAs) - lysine-derived bicyclononyne (BCNK) and trans-cyclooctenes (TCOK) - have been widely used in exploring biological processes because they are encoded by pyrrolysyl-tRNA synthetase PylRS/tRNACUA pairs from Methanosarcina species, which enables their use in many eukaryotic cell types and organisms.6 The fast reaction rates when encoded BCNK and TCOK are paired with tetrazine labels have enabled exciting live cell imaging, been used to probe receptors function, and been used in super resolution microscopy.2, 7–10
Figure 1.
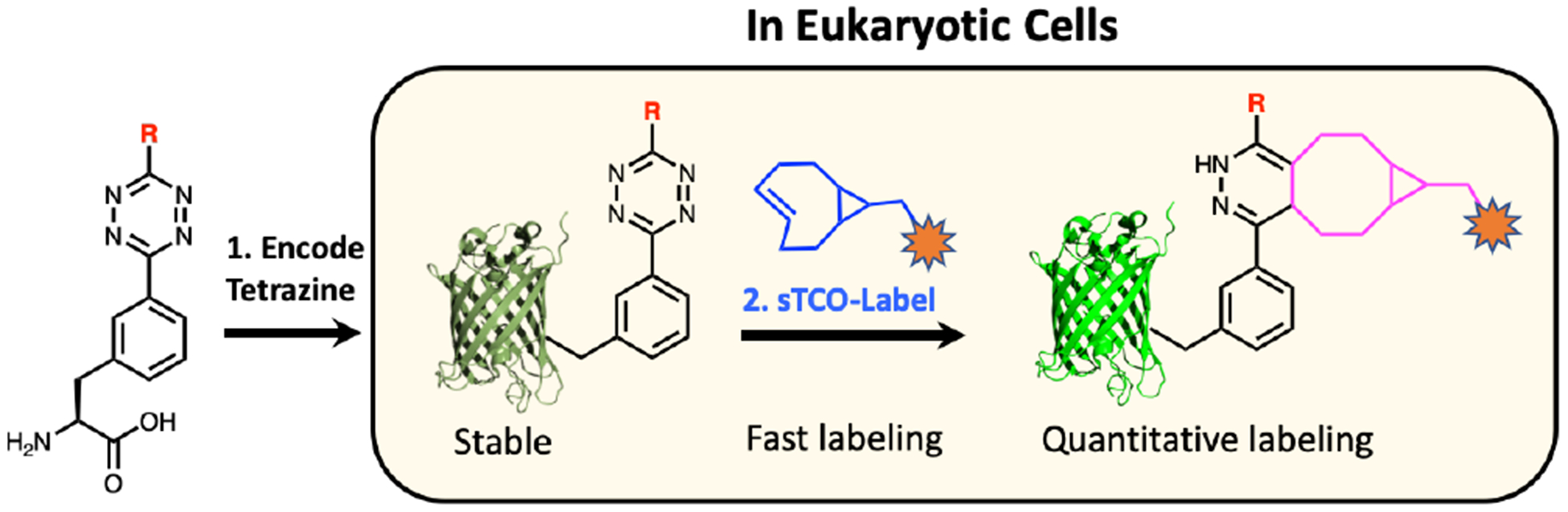
Eukaryotic encoded tetrazine amino acids provide quantitative labeling with increased labeling reaction rates and improve reagent stability in live cells.
However, BCNK and TCOK have reached their limit of reactivity and biocompatibility in vivo with rate constants of 2×104 M−1s−1.1, 4–5 Efforts to reach faster rates with more strained TCO functional groups using ligases or direct encoding into proteins have proven unsuccessful.1, 4 Our lab took a different approach and directly encoded tetrazine (tet) amino acids (Tet-v2.0) into proteins. These Tet-v2.0-proteins can be efficiently labeled with cyclopropane-fused transcyclooctene (sTCO) in E. coli at a reaction rate of 8×104 M−1s−1 with no detectable off-target reactivity within the timescale of the experiment (Figure 2A).11
Figure 2.
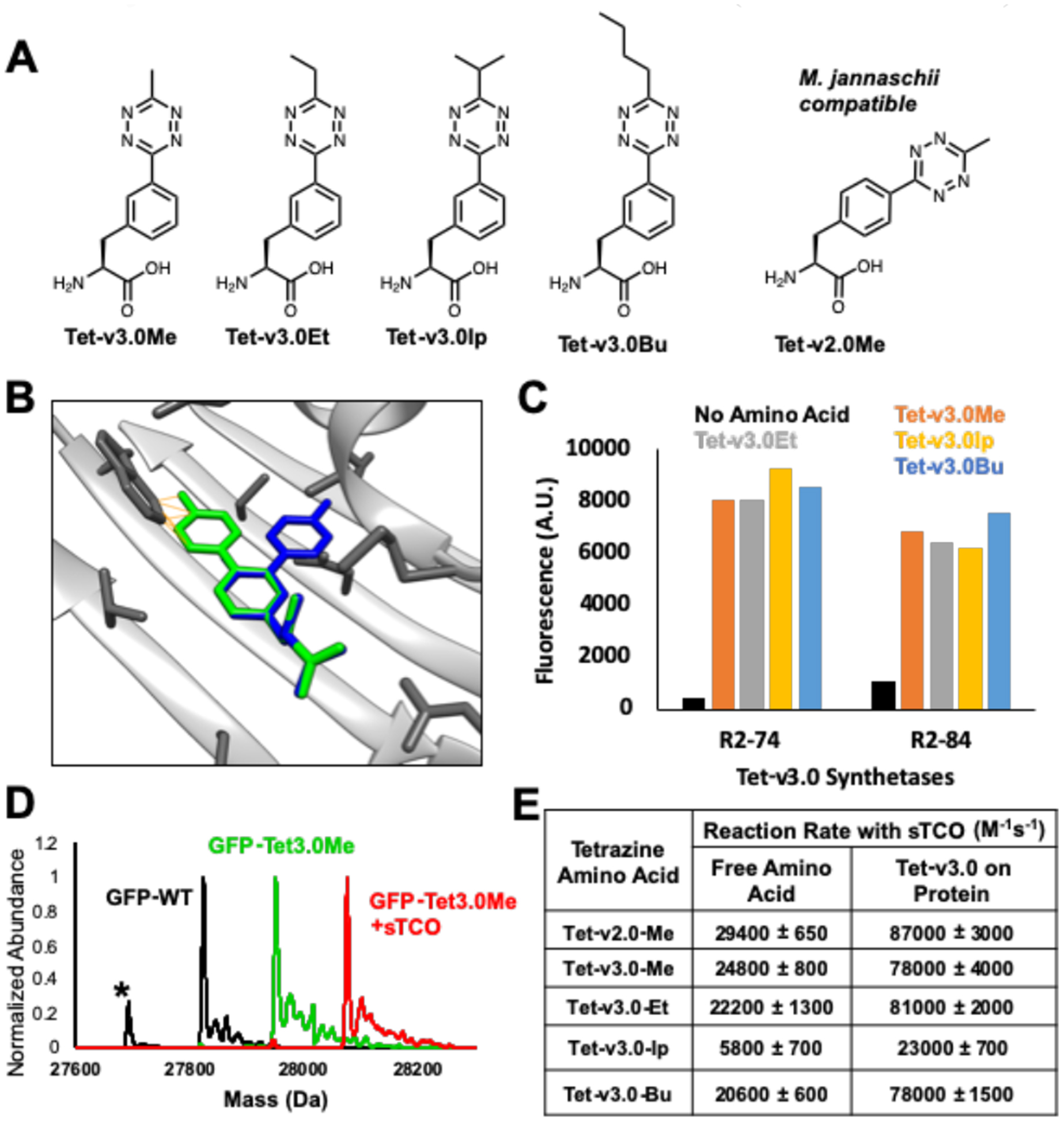
Selection of Tet-v3.0 RSs and characterization of Tet-v3.0 reactivity. (A) Tet-v3.0 amino acids with similar reactivity to Tet-v2.0 (B) Tet-v2.0Me (green) clashes with RS backbone whereas Tet-v3.0Me is eliminates backbone clash. (C) Measuring fluorescence of expressed GFP-Tet-v3.0 showed high efficiency and broad permissivity when other Tet-v3.0 amino acids were supplemented at 0.4 mM. (D) ESI-Q-TOF mass spectrometry confirmed Tet-v3.0Me incorporation into GFP and quantitative labeling with sTCO. (GFP-wt predicted: 27827.02 Da avg, observed 27827.49 Da avg; GFP-Tet-v3.0Me predicted: 27954.17 Da avg, observed: 27954.54 Da avg; GFP-Tet-v3.0Me+sTCO predicted: 28078.39 Da avg, observed: 28078.61 Da avg). The lower mass peak labeled with * is a loss of n-terminal methionine and higher mass peaks are salt sodium and potassium adducts. (E) Measured second order rate constants of free Tet-v3.0 and GFP-Tet-v3.0 with sTCO in PBS, pH 7.4 (Fig S3 and S7).
To generate a eukaryotic tetrazine-based bioorthogonal conjugation system, we focused on evolving the generally orthogonal Methanosarcina barkeri (Mb) PylRS/tRNACUA pair to incorporate Tet-v2.0 in response to an amber codon. Failure to identify any functional tRNA/aaRS pairs for Tet-v2.0 from among 107 RS active site variants prompted us to evaluate the crystal structure of the M. mazei PylRS in regard to its ability to incorporate Tet-v2.0 (Figure S1).12 Analyzing the active site of the Mb-PylRS with Tet-v2.0 superimposed with the substrate revealed clashes between the tetrazine ring, the beta sheet backbones, and residues sidechains (Figure 2B. Figure S2).
To avoid significant active-site engineering, the structure of the tetrazine amino acid was altered to minimize overlap with the RS peptide backbone. We found the best option for maintaining tetrazine stability and the reaction rate constant was to shift the para-substituted tetrazine ring on the phenylalanine to the meta-position, positioning the tetrazine ring deeper into the binding pocket of the PylRS active site. The 3-(6-alkyl-s-tetrazin-3-yl) phenylalanine amino acids (Tet-v3.0) (Figure 2A) were synthesized in two steps using standard coupling conditions with final yields between 60–80% (Scheme S1).13
The reaction rate constants sTCO with the Tet-v3.0 substituted with methyl, ethyl, isopropyl and butyl, named Tet-v3.0Me, Tet-v3.0Et, Tet-v3.0Ip, Tet-v3.0Bu, respectively, was determined by stopped flow absorbance spectroscopy (Figure 2E, S3). When reacted with sTCO, the Tet-v3.0 variants containing methyl, ethyl and butyl substituents all have similar 2nd order rate constants as Tetv2.0Me, whereas the sterically hindered Tet-v3.0Ip had an 80% reduction in reactivity.14
Standard selection methods for Tet-v3.0Me with a 3.2 million-member aaRS library resulted in six unique members that have high efficiency in the presence of Tet-v3.0Me and high fidelity in the absence of Tet-v3.0Me (Figure S4).15 The top Tet-v3.0-RS/tRNA variants were evaluated for permissivity toward the Tet-v3.0 ncAAs and found to robustly incorporate substituted Tet-v3.0 amino acids (Figure 2C). The top two Tet-v3.0 tRNA/RS pairs, R2–74 and R2–84 RSs, are highly permissive and both contain mutations to the N346 and C348 residues. These mutations likely open the active site pocket, allowing it to fit different tetrazine substituents and thus enabling tuning of the sTCO coupling rate constant (Figure 2B, S2).16
To verify Tet-v3.0-RS/tRNA efficiency and fidelity, GFP was expressed with Tet-v3.0 incorporated at site 150 using pDule vectors containing the top two Tet-v3.0 RSs and then purified by metal affinity chromatography. Protein expression resulted in yields of up to 80 mg/L in the presence of Tet-v3.0 amino acids with minimal GFP150-TAG suppression in the absence of amino acid (Figure S5). ESI-Q-TOF mass spectrometry was used to confirm the site-specific incorporation of Tet-v3.0 amino acids into GFP and their reactivity with sTCO (Figure 2D, S6). All GFP-Tet-v3.0 proteins show the expected mass increase from GFP-wt corresponding to single Tet-v3.0 incorporation. Purified GFP150-Tet-v3.0 proteins were exposed to a 10-fold molar excess of sTCO for 5 minutes and each Tet-v3.0 protein showed the expected 124.2 Da increase in mass corresponding to the addition of sTCO and loss of N2 (Figure 2D, S6). No unreacted GFP150-Tet-v3.0 was detected, verifying the reaction of genetically encoded Tet3.0 amino acids with sTCO was quantitative. In contrast, wt-GFP undergoes no mass change when exposed to sTCO (Figure S6).
GFP fluorescence is quenched 4–6 fold when Tet-v3.0 is incorporated at site 150 but fluorescence returns when reacted with sTCO (Figure S7a).17 By measuring this return in fluorescence, we determined the second order rate constants for each Tet-v3.0 protein. The reaction rate of the methyl, ethyl and butyl forms of GFP-Tet-v3.0 with sTCO were all ~80,000 M−1s−1 (Figure 2E and S7b). As expected, this is significantly faster than the rate observed for the reaction between the free amino acid and sTCO due to the relative polarity between the protein surface and buffer.11, 17 Once again, the rate constant of the reaction between Tet-v3.0-isopropyl and sTCO is reduced by 80% compared to the other Tet-v3.0 amino acids, likely due to steric bulk.14
We next assessed the substrate concentration dependence of the Tet-RSs for each Tet-v3.0 amino acid in order to find the optimal balance between efficient expression of Tet-v3.0 containing protein and potential HEK293T cell toxicity (Figure S8 and S9). The R2–84-RS functioned at maximum efficiency at 100 μM Tet-v3.0 and 60–70% efficiency at 20 μM, while the R2–74-RS required greater than 400 μM to reach maximum efficiency for most Tet-v3.0 amino acids. Overall Tet-3.0Bu with R2–84-RS was the most efficient combination for expressing Tet-v3.0-protein with the lowest amount of Tet-v3.0.
To site-specifically incorporate Tet-v3.0 in a eukaryotic system, we cloned codon-optimized R2–84 RS into the pAcBac1 plasmid (Figure S10).18 The resulting pAcBac1-R2–84 plasmid was transfected into HEK293T cells with the reporter pAcBac1-sfGFP-TAG150 plasmid. While GFP-Tet-v3.0 could be selectively expressed when all Tet-v3.0 amino acids were supplemented at 100 μM (Figure S11), only Tet-v3.0Bu functioned efficiently at 10–30 μM (Figure 3A). Further optimization of the system by the addition of a nuclear export sequence (NES) to the aaRS resulted in a 40% increase in Tet-v3.0-butyl-GFP expression compared to the absence of the NES (Figure 3B, Figure S12).19 With 10-fold less ncAA, this Tet3.0 system matched the eukaryotic suppression efficiency of the gold-standard Boc-lysine-tRNA/RS pair (Figure S13).20
Figure 3.
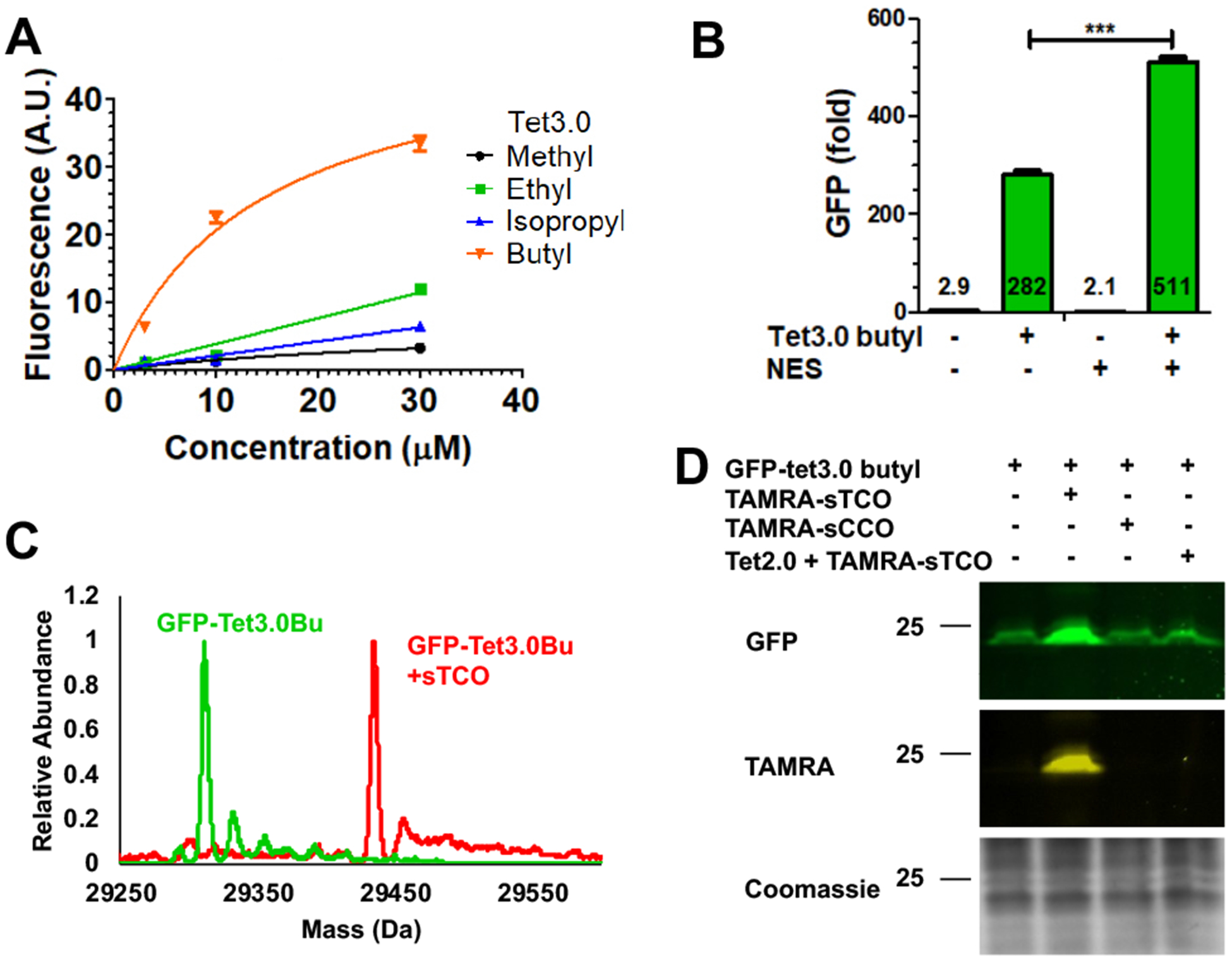
Optimization and reactivity of Tet-v3.0 in eukaryotic cells using R2–84-RS/tRNA. (A) Expression of GFP-vTet-3.0 with different Tet-v3.0 amino acids. (B) Addition of NES improved GFP-Tet-v3.0 expression. (C) ESI-Q-TOF mass spectrometry confirms eukaryoticTet-3.0 incorporation and quantitative labeling. (D) Labeling of GFP-Tet-v3.0 in eukaryotic cells with sTCO and TAMRA-sTCO labels.
Using this optimized expression system, we set out to characterize the stability and reactivity of eukaryotic proteins containing Tet-v3.0Bu. The mass of purified GFP-Tet-v3.0Bu was compared to GFP-wt using ESI-Q-TOF mass spectrometry. GFP-Tet-v3.0Bu exhibits the expected mass increase to 29311.10 Da avg (predicted 29310.71 Da), verifying that Tet-v3.0Bu is stably incorporated at a single site (Figure 3C, S14).
To evaluate the selective reactivity of Tet-v3.0-butyl-protein with sTCO-labels in eukaryotic cells, HEK293T cells expressing Tet-v3.0-butyl-GFP were treated with 100 nM TAMRA-sTCO and TAMRA-sCCO for 30 minutes. The reaction was quenched with free Tet2.0 amino acid prior to analysis with fluorescent SDS-PAGE. As expected, only GFP-Tet-v3.0Bu containing cells treated with TAMRA-sTCO showed TAMRA labeled GFP protein (Figure 3D, S15). Prereaction of TAMRA-sTCO with Tet2.0, or isomerized and therefore unreactive cis-cyclooctene-label, TAMRA-sCCO, showed no GFP-Tet-v3.0Bu labeling. To verify complete reactivity, GFP-Tet-v3.0-butyl protein expressed in eukaryotic cells was exposed to sTCO and the reaction product was characterized by ESI-Q-TOF mass spectrometry. Mass spectra showed complete labeling with the expected mass increase to 29434.44 Da avg (predicted 29434.93 Da avg) (Figure 3C, S14). These results demonstrate that Tet-v3.0-butyl is incorporated into eukaryotic proteins with high fidelity using the pAcBac1-NES-R2–84 plasmid and the expressed protein is stable and reacts quantitatively with sTCO-labels.
Next, we demonstrated that this exceptional reaction rate can be used to label Tet-v3.0-proteins in live cells using short reaction times and low concentrations of sTCO-label in three ways: with sTCO, TAMRA-sTCO, and in-cell protein dimerization reagents. First, we monitored labeling of Tet-protein by sTCO in HEK293T cells by measuring the extent of fluorescence return from quenched GFPTet-v3.0Bu. When HEK293T cells expressing GFP-Tet-v3.0Bu are exposed to increasing sTCO concentrations for 30 minutes at 37°C in media, there is a clear fluorescence increase for cells containing GFP-Tet-v3.0Bu but no increase for GFP-wt (Figure 4A, 4B, S17).
Figure 4.
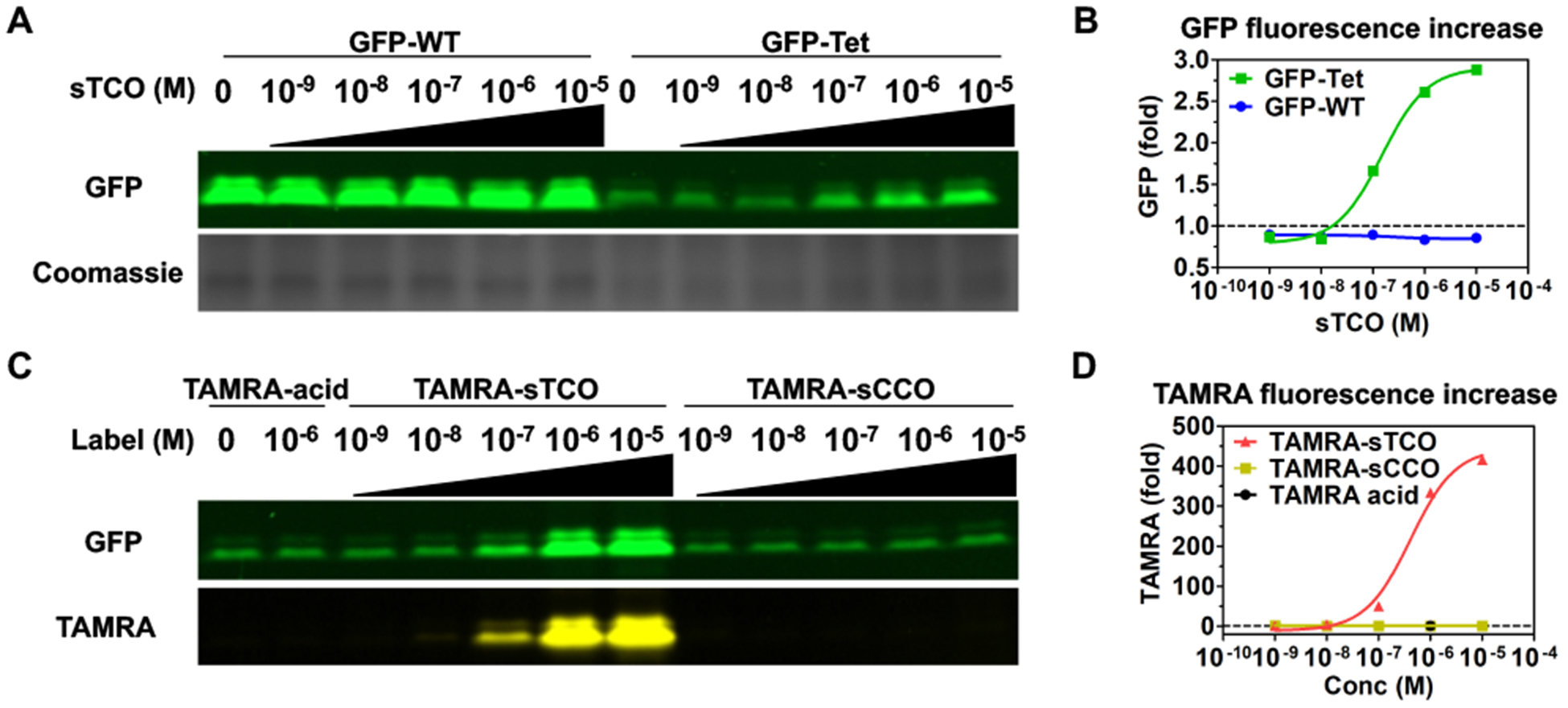
Concentration dependent labeling of Tet3.0 protein in eukaryotic cells. Superfolder-GFP fluorescent can be quantified in SDS-PAGE if the sample is not heat denatured during sample preparation (A, B) sTCO labeling (C, D) TAMRA-sTCO and TAMRA-sCCO labeling.
Second, we verified sub-stoichiometric labeling by repeating this experiment using low levels of TAMRA-sTCO and TAMRA-sCCO and tracking the fluorescence of the conjugated dye (Figure 4C, 4D, S15, S16). TAMRA-sTCO and TAMRA-sCCO did not show detectable background labeling of proteins at these label concentrations (Figure S17, S18). This demonstrates the ability of this bioorthogonal reaction to label a portion of Tet-v3.0-protein in HEK293T cells using low concentrations of label and short reaction times.
To verify that the cytosolic Tet-protein is undergoing sub-stoichiometric labeling, we employed small, peg linked-double-headed sTCO molecules (dh-sTCO) (11) that pass rapidly into the cell and dimerize Tet-protein. Reactions between dh-sTCO and GFP150-Tet-v3.0Bu only create protein dimers if the dh-sTCO is the limiting reagent, because an excessive concentration of dh-sTCO results in each Tet-protein with a singly reacted dh-sTCO molecule (Figure 5A).
Figure 5.
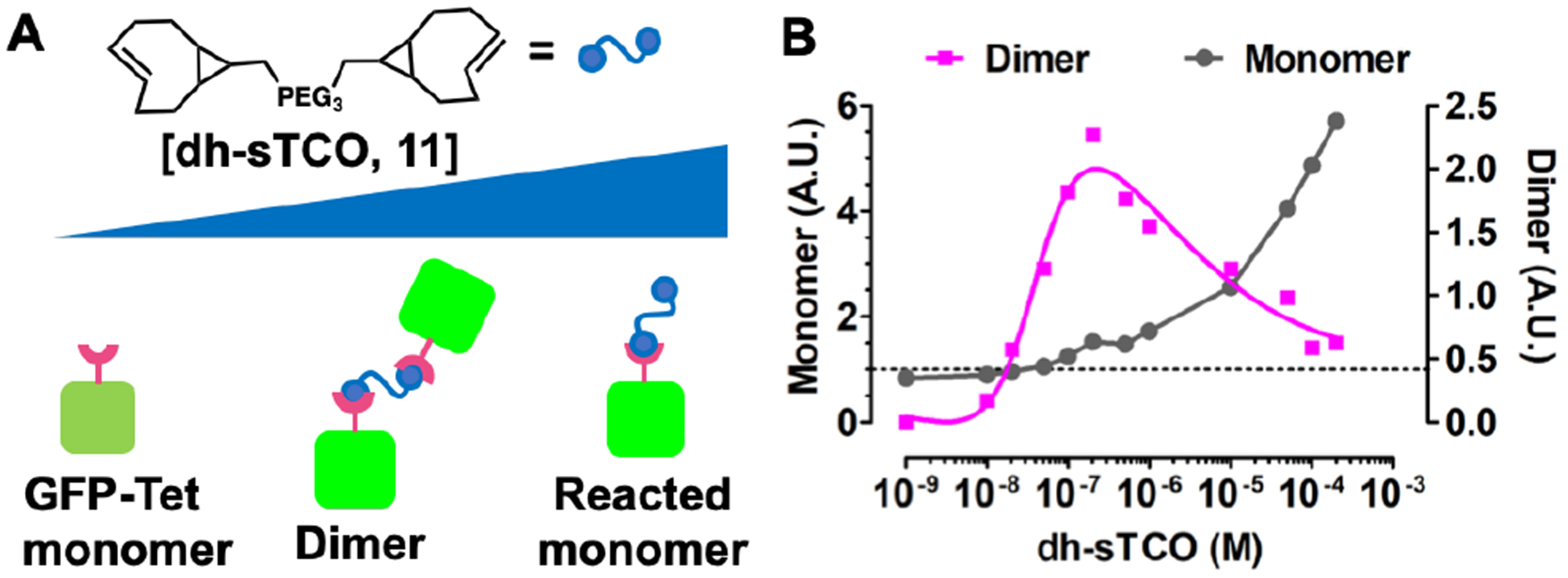
Concentration-dependent protein dimerization in eukaryotic cells. (A) Model for concentration-dependent dimerization with double headed linker. (B) At low concentration of dh-sTCO dimers form but at higher concentrations monomers dominate.
A dh-sTCO containing a short linker optimized for rapid cellular uptake was synthesized containing 3-PEG spacer between sTCO head groups (Scheme S3). Cells expressing GFP150-Tet-v3.0Bu were exposed to increasing concentrations of dh-sTCO for 30 min., quenched with Tetv2.0, and analyzed for mobility changes via SDS-PAGE. Fluorescent analysis shows that the dh-sTCO are capable of dimerizing GFP in a concentration dependent manner, confirming that this reaction is capable of sub-stoichio-metric protein labeling in eukaryotic cells (Figure 5B, Supplementary Fig 19). As expected, dh-sTCO shows the expected concentration dependent effect, dimerizing protein at low concentration of dh-sTCO but forming monomer at high concentrations (Supplementary S19).
In summary, we have developed a Tet-v3.0 GCE system with the broadly used PylRS/tRNA pairs that can function at low tetrazine amino acid concentrations to efficiently and site-specifically encode tetrazine amino acids in eukaryotic cells. The Tet-v3.0-protein exhibited ideal bioorthogonal labeling behavior, showing no detectable degradation products and sub-stoichiometric sTCO labeling with a reaction rate constant of 8×104 M−1s−1. To our knowledge, this is the fastest bioorthogonal labeling reaction yet reported for site-specific protein labeling in eukaryotic cells. This general approach should provide access to even faster eukaryotic bioorthogonal labeling rates by altering the tetrazine amino acid substitution, which will offer greater chemical control over the composition and stability of tag and reporter.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Joe Beckman and e-MSion for their mass spectrometry assistance and the Confocal Microscope Facility of the Center for Genome Research and Biocomputing at Oregon State University.
Funding Sources
This work was supported by NSF Grant MCB-1518265 and NSF MRI 1337774.
Footnotes
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website.
Materials and Methods, Schemes S1-S3, Figures S1-S42.
REFERENCES
- 1.Murrey HE; Judkins JC; Am Ende CW; Ballard TE; Fang Y; Riccardi K; Di L; Guilmette ER; Schwartz JW; Fox JM; Johnson DS, Systematic Evaluation of Bioorthogonal Reactions in Live Cells with Clickable HaloTag Ligands: Implications for Intracellular Imaging. J Am Chem Soc 2015, 137 (35), 11461–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng T; Hang HC, Site-Specific Bioorthogonal Labeling for Fluorescence Imaging of Intracellular Proteins in Living Cells. J Am Chem Soc 2016, 138 (43), 14423–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang K; Chin JW, Bioorthogonal Reactions for Labeling Proteins. ACS Chemical Biology 2014, 9 (1), 16–20. [DOI] [PubMed] [Google Scholar]
- 4.Fang Y; Zhang H; Huang Z; Scinto SL; Yang JC; Am Ende CW; Dmitrenko O; Johnson DS; Fox JM, Photochemical syntheses, transformations, and bioorthogonal chemistry of trans-cycloheptene and sila trans-cycloheptene Ag(i) complexes. Chem Sci 2018, 9 (7), 1953–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darko A; Wallace S; Dmitrenko O; Machovina MM; Mehl RA; Chin JW; Fox JM, Conformationally Strained trans-Cyclooctene with Improved Stability and Excellent Reactivity in Tetrazine Ligation. Chem Sci 2014, 5 (10), 3770–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang K; Davis L; Wallace S; Mahesh M; Cox DJ; Blackman ML; Fox JM; Chin JW, Genetic Encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels-Alder reactions. J Am Chem Soc 2012, 134 (25), 10317–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumdick M; Gelleri M; Uttamapinant C; Beranek V; Chin JW; Bastiaens PIH, A conformational sensor based on genetic code expansion reveals an autocatalytic component in EGFR activation. Nat Commun 2018, 9 (1), 3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neubert F; Beliu G; Terpitz U; Werner C; Geis C; Sauer M; Doose S, Bioorthogonal Click Chemistry Enables Site-specific Fluorescence Labeling of Functional NMDA Receptors for Super-Resolution Imaging. Angew Chem Int Ed Engl 2018, 57 (50), 16364–16369. [DOI] [PubMed] [Google Scholar]
- 9.Uttamapinant C; Howe JD; Lang K; Beranek V; Davis L; Mahesh M; Barry NP; Chin JW, Genetic code expansion enables live-cell and super-resolution imaging of site-specifically labeled cellular proteins. J Am Chem Soc 2015, 137 (14), 4602–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beliu G; Kurz AJ; Kuhlemann AC; Behringer-Pliess L; Meub M; Wolf N; Seibel J; Shi ZD; Schnermann M; Grimm JB; Lavis LD; Doose S; Sauer M, Bioorthogonal labeling with tetrazine-dyes for super-resolution microscopy. Commun Biol 2019, 2, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blizzard RJ; Backus DR; Brown W; Bazewicz CG; Li Y; Mehl RA, Ideal Bioorthogonal Reactions Using A Site-Specifically Encoded Tetrazine Amino Acid. J Am Chem Soc 2015, 137 (32), 10044–7. [DOI] [PubMed] [Google Scholar]
- 12.Yanagisawa T; Ishii R; Fukunaga R; Kobayashi T; Sakamoto K; Yokoyama S, Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J Mol Biol 2008, 378 (3), 634–52. [DOI] [PubMed] [Google Scholar]
- 13.Yang J; Karver MR; Li W; Sahu S; Devaraj NK, Metal-catalyzed one-pot synthesis of tetrazines directly from aliphatic nitriles and hydrazine. Angew Chem Int Ed Engl 2012, 51 (21), 5222–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F; Liang Y; Houk KN, Bioorthogonal Cycloadditions: Computational Analysis with the Distortion/Interaction Model and Predictions of Reactivities. Acc Chem Res 2017, 50 (9), 2297–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young DD; Schultz PG, Playing with the Molecules of Life. ACS Chem Biol 2018, 13 (4), 854–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hohl A; Karan R; Akal A; Renn D; Liu X; Ghorpade S; Groll M; Rueping M; Eppinger J, Engineering a Polyspecific Pyrrolysyl-tRNA Synthetase by a High Throughput FACS Screen. Sci Rep 2019, 9 (1), 11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seitchik JL; Peeler JC; Taylor MT; Blackman ML; Rhoads TW; Cooley RB; Refakis C; Fox JM; Mehl RA, Genetically encoded tetrazine amino acid directs rapid site-specific in vivo bioorthogonal ligation with trans-cyclooctenes. J Am Chem Soc 2012, 134 (6), 2898–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee A; Xiao H; Bollong M; Ai HW; Schultz PG, Efficient viral delivery system for unnatural amino acid mutagenesis in mammalian cells. Proc Natl Acad Sci U S A 2013, 110 (29), 11803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikic I; Estrada Girona G; Kang JH; Paci G; Mikhaleva S; Koehler C; Shymanska NV; Ventura Santos C; Spitz D; Lemke EA, Debugging Eukaryotic Genetic Code Expansion for Site-Specific Click-PAINT Super-Resolution Microscopy. Angew Chem Int Ed Engl 2016, 55 (52), 16172–16176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willis JCW; Chin JW, Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat Chem 2018, 10 (8), 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


