Abstract
Background:
Today, the role of microRNAs in the pathogenesis of breast cancer has been established. Genetic mutations play a significant role in determining the risk factors of cancer. The polymorphism of these two genes can alter their expression. This study has been performed to investigate the relationship between polymorphisms of rs6715345 of miR-375 gene and rs4939827 of the SMAD7 gene and development of breast cancer in a population in southeastern Iran.
Methods:
This case-control study was performed on the blood sample of 205 patients with breast cancer and 200 healthy individuals for investigating the rs34917480 and rs4939827 polymorphisms using the PCR-RFLP method. The data were analyzed by t-test, χ2, and logistic regression. The SPSS v18.0 used for data analysis.
Results:
The findings of this study indicated that the risk of developing breast cancer does not have a significant relationship with rs6715345 polymorphism of miR-375 gene (p>0.05). However, the rs4939827 polymorphism of the SMAD7 gene was significantly linked to the risk of developing breast cancer in the southeastern population in Iran (p<0.05).
Conclusion:
The results suggest that the rs4939827 polymorphism of the SMAD7 gene can lead to an increased risk of incidence of breast cancer in the southeastern population in Iran.
Key Words: Genetic polymorphisms, SMAD7, MIR-375, breast neoplasm
Introduction
Breast cancer is the most common cancer and the main cause of cancer-induced mortality among women worldwide whether in general or in less developed countries (DeSantis et al., 2019). According to the latest statistics of disease burden studies in 2019, it caused the death of more than 626,679 patients. It is also projected that by 2040, its prevalence would reach more than 46% (Pilevarzadeh et al., 2019). The risk of factors of breast cancer includes genetic, reproductive, and hormonal factors, the use of oral contraceptives, and having no children (Takalkar et al., 2014). Also, breast cancer during younger ages increases its chance of mortality (Zehentmayr et al., 2016). Clinically, breast cancer is considered a heterogeneous disease; although all of them have similar manifestations, they differ molecularly and in terms of expression of genes, response to conventional treatments, and prognosis. A conventional classification that has important clinical applications is based on the expression of HER2 (ErbB-2), estrogen receptor (ER), and progesterone receptor (PR) (Perou et al., 2000). The overexpression of the ErbB-2 gene, which encodes HER2 oncoprotein exists in 20-25% of breast cancers, which is associated with poor prognosis, the higher chance of metastasis, and relapse (Venturutti et al., 2016). Accordingly, the human anti-HER2 antibody, trastuzumab (Herceptin) has been used for treating HER2-positive cells (Slamon et al., 2001). Nevertheless, therapeutic response to monotherapy with trastuzumab in this type of breast cancer is less than 35%, and most patients with this type of treatment become resistant to this drug after initial response (Ye et al., 2014). Despite recent advances in adjuvant therapies such as hormone therapy, chemotherapy, and biological treatment, the prognosis of breast cancer is still poor (Pickle et al., 2007). For this reason, molecular properties and genetic susceptibility are very important factors that determine the prognosis of breast cancer, the extent of invasion and tumor relapse, as well as response to treatment (Lazaridis et al., 2014). For example, 70% of invasive cases of breast cancer show overexpression of ER- α receptor, which is known as an early process in tumorigenesis and progression of breast cancer (Simonini et al., 2010). Accordingly, in recent years, numerous studies have focused on identifying biomarkers for early diagnosis, prediction of response to treatment, and prognosis, where microRNAs have especially attracted attention (Nguyen-Dien et al., 2014). MicroRNAs (miRNAs) are a group of endogenous noncoding RNAs which are 18-23 nucleotides long and regulate the gene expression across the surface following transcription (Ambros, 2004). Thus, they can affect many biological processes and activities of cells including cell cycle, differentiation, growth, and metabolism (Kong et al., 2012). It has been found that miRNAs play a significant role in many human diseases especially cancers – whether as an oncogene or as a tumor inhibitor (Bartel, 2009). miRNAs control an extensive part of the human genome (Bartel, 2009), and it has been estimated that so far around 1,000 human miRNAs have been found(Friedman et al., 2009). Biogenesis of miRNAs begins from the nucleus and continues in the cytoplasm. Most miRNAs have been synthesized out of an initial transcript, including one head and one polyA tail transcribed by RNA polymerase II from the miRNA gene. The initial miRNA is broken by RNAse III Drosha–DGCR8 in the nucleus and Dicer in the cytoplasm to mature 22-nucleotide miRNAs (Babashah and Soleimani, 2011)(28). These single-stranded molecules regulate major biological processes across the surface following transcription through binding to the sequence of the target mRNA complement or inhibiting its translation (given the extent of mRNA and miRNA complement sequence) (Zhang et al., 2014).
Extensive evidence suggests that miRNAs are mal-expressed in a large number of human cancers (Farazi et al., 2013). Some of them act as an oncogene (oncomiR), while others as a tumor inhibitor, given which gene or pathway they affect (Zhang et al., 2014). Also, it has been found that miRNAs are also involved in metastasis (Edmonds et al., 2009). Recently, the relationship between miRNAs and hormonal receptors has attracted a great deal of attention (Di Leva et al., 2013). The Discovery of miRNAs has opened a novel way for diagnosis and treatment of cancer. miRNAs play a significant role in the regulation of tumor proliferation, metastasis, and invasion, and can be a potential target for cancer treatment and interventions (Lee and Dutta, 2009). In particular, they can be used as a non-invasive serum biomarker for the diagnosis of cancers (Chen et al., 2008). However, considering the population differences in the genome sequence and the presence of gene polymorphisms, further studies are required in this regard.
Single-nucleotide polymorphism (SNP) refers to single-nucleotide variations in the DNA sequence, causing the development of different genotypes. This can result in the development of population discrepancies with regards to the pathogenesis of diseases such as cancer with a special genotype (Wang et al., 1998). Thus, a better understanding of polymorphisms can support better treatment of breast cancer. This study was performed to investigate the relationship between rs6715345 and rs4939827 polymorphisms of miR-375 and SMAD-7 genes respectively in women with breast cancer as well as in healthy women.
Methods and Materials
Study Design
This case-control study was performed on women with breast cancer and on healthy women, who referred to an educational hospital affiliated with Zahedan University of Medical Sciences from 1 March 2019 to 30 August 2019. In this study, 205 samples belonging to breast cancer patients were investigated along with 200 samples of the control group who were healthy individuals. This disease of breast cancer patients was confirmed by pathology, while the healthy subjects had no history of the breast-associated disease. The samples were chosen thought a simple and available sampling method. Written and oral consent was taken from all participants. This study has been confirmed in the ethics committee of Zahedan University of Medical Sciences (IR.ZAUMS.REC.1396.325). STROBE checklist was used for reporting of the paper (Von Elm et al., 2008).
Data collection
To investigate the gene polymorphism, blood samples were taken from participants (5 ml) and collected in EDTA-containing tubes, and kept at -20ᵒC. The DNA was purified via standard salting out procedure (Hashemi et al., 2010). Specific primers for determining the gene polymorphism were designed via Restriction Fragment Length Polymorphism-PCR (RFLP-PCR) method for detecting single-nucleotide polymorphisms of rs4939827 and rs6715345 belonging to SMAD-7 and miR-375 genes, respectively (Table 1).
Table 1.
Specific Primers for Determining Gene Polymorphism via RFLP-PCR Method for Detecting Single-Nucleotide Polymorphisms of rs4939827 and rs6715345 Belonging to SMAD-7 and miR-375 Genes, Respectively
| Gene polymorphism | Primer sequence (5`->3`) 4 | Method | Restriction Enzyme | Fragment (bp) |
|---|---|---|---|---|
| SMAD-7 rs4939827 | F: ACAAGCCTAAGATAAAAGGGGACT | PCR-RFLP | TaaI | T allele=242 |
| R: GAGTCTGAGGGAGCTCTGGGGTAC | C allele=218+24 | |||
| miR-375 rs6715345 | F: CCCGTATTACGACGCAGAA | PCR-RFLP | NaeI | C allele=405 |
| R: ACGTGTCAGCCGCAGAT | G allele=226+179 |
Next, the genotyping of these two polymorphisms was done by PCR. Here, 0.20 of the reaction solution containing 1 mcL of DNA, 1 mcL of forwarding and reverse primers, and 10 mcL of Taq Premix, as well as 7 mcL of sterilized distilled water with no DNAse, was used. PCR conditions were optimized as follows: 1) preheating for 5 min at 95ᵒC for the primary denaturation step, 2) Thirty 30-sec cycles at 95ᵒC, 3) 59ᵒC (the selected annealing temperature) for 30 s and 72ᵒC for 30 s, and 4) the extension step for 5 min at 72ᵒC. In the next step, the PCR product was used via the limiting enzymes for each polymorphism (Table 1-3) so that the sequences of interest would be created (Burtis et al., 2012). The tubes were placed in Bain Marie at the activity temperature specified for every enzyme and within the duration predetermined in the protocol, to provide the conditions for the activity of enzymes. Eventually, the digestion product of DNA pieces was electrophoresed on agarose gel 2%. Finally, the alleles of the intended polymorphisms were observed through ethidium bromide staining.
Table 3.
The Frequency of Allele and Genotypes of rs4939827 Polymorphism of SMAD-7 Gene and Their Comparison in the Case and Control Groups
| SMAD-7 rs4939827 T>C | Case (breast cancer) N (%) | Control (healthy women) N (%) | OR (95%CI) | P-value |
|---|---|---|---|---|
| Codominant | ||||
| TT | 48 (23.4) | 71 (35.5) | 1 | - |
| CT | 121 (59.0) | 105 (52.5) | 1.71 (1.10-2.70) | 0.023 |
| CC | 36 (17.6) | 24 (12.0) | 2.22 (1.20-4.26) | 0.017 |
| Dominant | ||||
| TT | 48 (23.4) | 71 (35.5) | 1 | - |
| CT+CC | 157 (76.6) | 129 (64.5) | 1.80 (1.17-2.76) | 0.009 |
| Recessive | ||||
| CT+TT | 169 (82.4 | 176 (88.0) | 1 | - |
| CC | 36 (17.6) | 24 (12.0) | 1.56 (0.90-2.73) | 0.125 |
| Allele | ||||
| T | 217 (52.9) | 247 (62.7) | 1 | - |
| C | 193 (47.1) | 153 (38.3) | 1.44 (1.08-1.89) | 0.013 |
Data analysis
For data description, descriptive tests (percentage, frequency, mean) were used. The disease characteristics were compared using χ2 and independent t-test. To determine the relationship between polymorphisms and breast cancer, logistic regression was used along with odds ratio (OR) calculation and confidence interval (CI) 95%. SPSS Version 18.0 for Windows (SPSS Inc., Chicago, IL, USA) was used to analyze the data. The confidence interval of 95% and a significance level of P-value less than 0.05 was considered significant.
Results
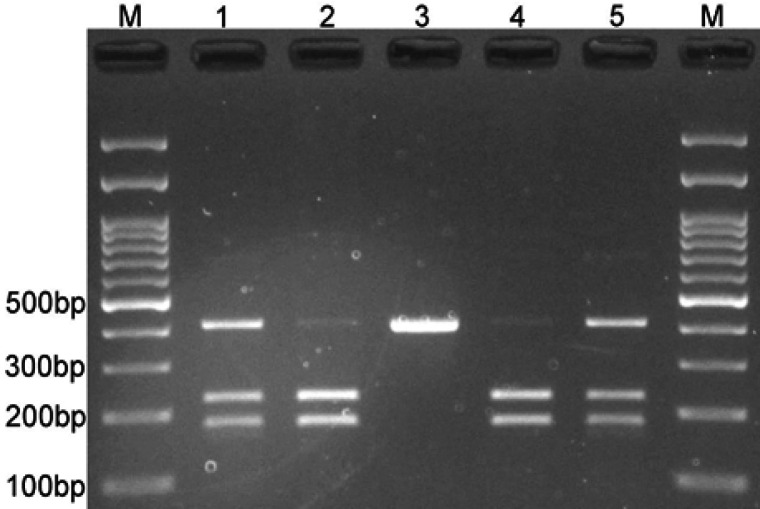
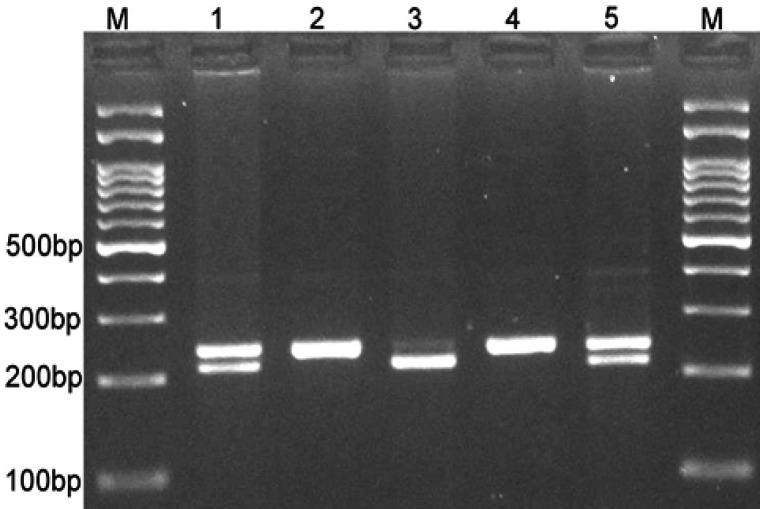
The mean age of women with breast cancer and healthy women was 48.56±10.67 and 48.41±11.29, respectively. The two groups did not differ significantly from each other statistically (p=0.894). The frequency of rs6715345 polymorphism of the miR-375 gene was examined in 205 patients with breast cancer and 225 healthy women, and the genotypes were compared. The results indicated that there is no relationship between hereditary models as well as alleles of rs6715375 polymorphism and the risk of developing breast cancer (p>0.05) (Table 2). The pattern of electrophoresis of rs6715345 polymorphism on agarose gel with a band length of 226 and 179 base pairs for GG genotype and band length of 405 base pairs for CC genotypes have been shown in Figure 1.
Table 2.
The Frequency of Alleles and Genotypes of rs6715375 Polymorphism of miR-375 Gene and Their Comparison in the Case and Control Groups
| miR-375 rs6715345 G>C | Breast cancer group N (%) | Healthy women N (%) | OR (95%CI) | P-value |
|---|---|---|---|---|
| Codominant | ||||
| GG | 164 (80.0) | 164 (82.0) | 1 | - |
| CG | 29 (14.1) | 32 (16.0) | 0.91 (0.52-1.57) | 0.781 |
| CC | 12 (5.9) | 4 (2.0) | 3.00 (0.95-8.63) | 0.071 |
| Dominant | ||||
| GG | 164 (80.0) | 164 (82.0) | 1 | - |
| CG+CC | 41 (20.0) | 36 (18.0) | 1.14 (0.70-1.86) | 0.615 |
| Recessive | ||||
| CG+GG | 193 (94.1) | 196 (98.0) | 1 | - |
| CC | 12 (5.9) | 4 (2.0) | 3.05 (0.97-8.75) | 0.071 |
| Allele | ||||
| G | 357 (87.1) | 360 (90.0) | 1 | - |
| C | 53 (12.9) | 40 (10.0) | 1.34 (0.86-2.07) | 0.225 |
Figure 1.
The Band of Genotypes CG (1), GG (2), and CC (3) in rs6715375 of miR-375 Gene on Agarose Gel Following Electrophoresis
Also, regarding the frequency of rs4939827 polymorphism of SMAD-7 gene and its genotypes in women patients with breast cancer and healthy women, it was found that the codominant hereditary models (OR=1.71, 95%CI=1.10-2.70, p=0.023) in comparison with TT (OR=2.22, 95%CI=1.20-4.26, p=0.017), CC in comparison with TT and dominant (OR=1.80, 95%CI=1.17-2.76, p=0.009, CT+CC in comparison with TT) and allele C (OR=1.44, 95%CI=1.08-1.89, P-value=0.013, allele C in comparison with allele T) were significantly related to the risk of developing breast cancer. The electrophoresis pattern of rs4939827 polymorphism on agarose gel with the band length of 242 base pairs for genotype TT and band length of 28 and 218 base pairs for genotype CC have been shown in Figure 2.
Figure 2.
The Band of Genotypes of TC (1), TT (2), and CC (3) in rs4939827 of SMAD-7 Gene on Agarose Gel Following Electrophoresis
Discussion
Today, considering the progressive prevalence of breast cancer despite the medical advances in the early treatment of breast cancer and increasing the survival of patients with this cancer, the causes of breast cancer should still be more profoundly understood as a complex issue (Shieh et al., 2020). Recognizing the risk factors of breast cancer in different dimensions can help us in better understanding the methods of prevention and early treatment of breast cancer (McPherson et al., 2000). In recent decades, molecular and cellular technological advances have provided a suitable opportunity to obtain a better understanding of the molecular and cellular aspects of breast cancer and the role of genetic factors in it (Network, 2012). On the other hand, further, understanding of the human genome and its regulating factors have resulted in the development of new areas for examining human diseases including cancer at the gene level and the factors affecting the expression or suppression of genes. Meanwhile, the role of microRNAs is of interest in the development of cancers (Hannafon et al., 2016). These noncoding RNAs and their polymorphisms have a significant impact on the regulation of gene expression in humans and can be used as biomarkers in the early diagnosis of human diseases (Bertoli et al., 2015). The results of the present study indicated that there is no significant relationship between rs6715345 polymorphism of the MIR375 gene and breast cancer in the studied population. To the best of the authors’ knowledge, no similar study has been found with this polymorphism. Unlike the present study, the study by Zhang et al., (2018), who had examined the variation of the MIR275 gene in patients with breast cancer, showed that this gene is associated with diminished risk of breast cancer. This difference can be due to the difference in the type of gene polymorphism in the two studies. Also, another study on populations suffering esophageal squamous cell carcinoma in Kazakh patients (Zhu et al., 2015) indicated that the variation in rs6715345 polymorphism of MIR375 gene leads to diminished risk of developing breast cancer. Based on our findings, rs4939827 polymorphism of the SMAD-7 gene is significantly linked to the risk of developing breast cancer. SMAD proteins are considered as the main mediators in TGF-β signaling pathway, playing a significant role in tumorigenesis-associated processes. Various studies have shown the role of SMAD-7 in tumorigenesis of different types of cancer (Greenwood and Bruna, 2019). Unlike the results of the present study, research on Caucasian English and Scottish populations suggest that there is no relationship between rs4939827 polymorphism of SMAD-7 gene and developing breast cancer (Gibson et al., 2009; Scollen et al., 2011). This can be due to the differences in the study populations as well as the type of polymorphisms examined in the two studies. Also, the study performed by Xin Lin testing the relationship between three polymorphisms (rs12953717, rs4939827, and rs4464148) showed that rs4939827 polymorphism was not associated with breast cancer, and the only rs1295371 was linked to this cancer (Li et al., 2011). However, concerning other cancers, in line with the present study, research on colorectal cancer in Caucasian and Asian populations (Zhang et al., 2018), stomach and lung cancers in a western population (Li et al., 2011) has also shown that SMAD-7 gene is associated with developing cancer. The study by Kirac showed that rs4939827 polymorphism of the SMAD-7 gene is associated with developing colorectal cancer in a Croatian population (Kirac et al., 2013). Meta-analysis studies also show a relationship between rs4939827 polymorphism and the risk of developing different types of cancer including colorectal and esophageal cancers (Hong et al., 2016; Huang et al., 2016; Zhao et al., 2017). The most important limitations of the present study were: low sample size which may affect the study power regarding the relationship. Another limitation was the identical race of participants, all being Iranian and chosen from one single province in the east of the country.
In conclusion, this study indicated that single-nucleotide rs4939827 polymorphism of the SMAD-7 gene is associated with developing breast cancer. However, single-nucleotide rs6715345 polymorphism of the miR-375 gene was not significantly correlated with the risk of developing breast cancer. The results of the present study suggest attention to genetic dimensions of disease for the population suffering breast cancer in earlier diagnosis and better treatment of diseases. For better detection of the existing relationship, studies with a larger sample size are suggested.
Disclosure of potential conflicts of interest
All authors declare that they have no competing interests.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were under the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Compliance with Ethical Standards
All procedures performed in studies involving human participants were under the ethical standards of The Cancer Genome Atlas Human Subjects Protection and Data Access Policies, adopted by the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI). The study approved by the ethics committee of the Zahedan University of Medical Science.
Funding
This study was funded by the Zahedan University of Medical Science and Technology Plan Project (Grant number: 8632).
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Babashah S, Soleimani M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer. 2011;47:1127–37. doi: 10.1016/j.ejca.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438–51. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- Di Leva G, Piovan C, Gasparini P, et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013;9:e1003311. doi: 10.1371/journal.pgen.1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK-H, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson L, Johnson N, Fraser A, et al. No breast cancer association for transforming growth factor-β pathway colorectal cancer single nucleotide polymorphisms. Cancer Epidemiol Biomarkers Prev. 2009;18:1934–6. doi: 10.1158/1055-9965.EPI-09-0214. [DOI] [PubMed] [Google Scholar]
- Hannafon BN, Trigoso YD, Calloway CL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi M, Moazeni-Roodi A, Fazaeli A, et al. Lack of association between paraoxonase-1 Q192R polymorphism and rheumatoid arthritis in southeast Iran. Genet Mol Res. 2010;9:333–9. doi: 10.4238/vol9-1gmr728. [DOI] [PubMed] [Google Scholar]
- Hong Y, Wu G, Li W, et al. A comprehensive meta-analysis of genetic associations between five key SNPs and colorectal cancer risk. Oncotarget. 2016;7:73945. doi: 10.18632/oncotarget.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wu W, Nie M, et al. SMAD7 polymorphisms and colorectal cancer risk: a meta-analysis of case-control studies. Oncotarget. 2016;7:75561. doi: 10.18632/oncotarget.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirac I, Matošević P, Augustin G, et al. SMAD7 variant rs4939827 is associated with colorectal cancer risk in Croatian population. PLoS One. 2013;8:e74042. doi: 10.1371/journal.pone.0074042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YW, Ferland-McCollough D, Jackson TJ, et al. microRNAs in cancer management. Lancet Oncol. 2012;13:249–58. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- Lazaridis G, Lambaki S, Karayannopoulou G, et al. Prognostic and predictive value of p-Akt, EGFR, and p-mTOR in early breast cancer. Strahlentherapie und Onkologie. 2014;190:636–45. doi: 10.1007/s00066-014-0620-6. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang X-x, Hu N-y, et al. A risk-associated single nucleotide polymorphism of SMAD7 is common to colorectal, gastric, and lung cancers in a Han Chinese population. Mol Biol Rep. 2011;38:5093–7. doi: 10.1007/s11033-010-0656-3. [DOI] [PubMed] [Google Scholar]
- McPherson K, Steel C, Dixon J. ABC of breast diseases: breast cancer-epidemiology, risk factors, and genetics. Br Med J. 2000;321:624. doi: 10.1136/bmj.321.7261.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Dien GT, Smith RA, Haupt LM, et al. Genetic polymorphisms in miRNAs targeting the estrogen receptor and their effect on breast cancer risk. Meta Gene. 2014;2:226–36. doi: 10.1016/j.mgene.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pickle LW, Hao Y, Jemal A, et al. A new method of estimating United States and state-level cancer incidence counts for the current calendar year. CA Cancer J Clin. 2007;57:30–42. doi: 10.3322/canjclin.57.1.30. [DOI] [PubMed] [Google Scholar]
- Pilevarzadeh M, Amirshahi M, Afsargharehbagh R, et al. Global prevalence of depression among breast cancer patients: a systematic review and meta-analysis. Breast Cancer Res Tr. 2019;176:519–33. doi: 10.1007/s10549-019-05271-3. [DOI] [PubMed] [Google Scholar]
- Scollen S, Luccarini C, Baynes C, et al. TGF-β signaling pathway and breast cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2011;20:1112–9. doi: 10.1158/1055-9965.EPI-11-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh Y, Ziv E, Kerlikowske K. Interval breast cancers-insights into a complex phenotype. Nat Rev Clin Oncol. 2020;17:138–9. doi: 10.1038/s41571-020-0327-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini PdSR, Breiling A, Gupta N, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor α in breast cancer cells. Cancer Res. 2010;70:9175–84. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Takalkar U, Asegaonkar S, Kodlikeri P, et al. Hormone related risk factors and breast cancer: hospital based case control study from India. Research in Endocrinology. 2014 [Google Scholar]
- Venturutti L, Romero LV, Urtreger AJ, et al. Stat3 regulates ErbB-2 expression and co-opts ErbB-2 nuclear function to induce miR-21 expression, PDCD4 downregulation and breast cancer metastasis. Oncogene. 2016;35:2208–22. doi: 10.1038/onc.2015.281. [DOI] [PubMed] [Google Scholar]
- Von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. 2008;61:344–9. doi: 10.1016/j.jclinepi.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan J-B, Siao C-J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–82. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Ye X-M, Zhu H-Y, Bai W-D, et al. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer. 2014;14:134. doi: 10.1186/1471-2407-14-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehentmayr F, Hauser-Kronberger C, Zellinger B, et al. Hsa-miR-375 is a predictor of local control in early stage breast cancer. Clin Epigenetics. 2016;8:28. doi: 10.1186/s13148-016-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wei B, Hu H, et al. The association between differentially expressed micro RNAs in breast cancer cell lines and the micro RNA205 gene polymorphism in breast cancer tissue. Oncol Lett. 2018;15:2139–46. doi: 10.3892/ol.2017.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu J, Wang G. The role of microRNAs in human breast cancer progression. Tumour Biol. 2014;35:6235–44. doi: 10.1007/s13277-014-2202-8. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yao F, Tang W, et al. S100A14 rs11548103 G> A polymorphism is associated with a decreased risk of esophageal cancer in a Chinese population. Oncotarget. 2017;8:86917. doi: 10.18632/oncotarget.20868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yang L, You W, et al. Genetic variation in miR-100 rs1834306 is associated with decreased risk for esophageal squamous cell carcinoma in Kazakh patients in northwest China. Int J Clin Exp Pathol. 2015;8:7332. [PMC free article] [PubMed] [Google Scholar]