Abstract
Circulating tumor cell (CTC) clusters have an enhanced capacity to initiate metastases, compared with single cells, but it is unclear how they gain such advantage. A recent study by Gkountela et al. (Cell 2019;176:98–112) links the physical state of clusters with increased accessibility of stem cellrelated transcription factors, providing novel insights into the epigenetic regulation of ‘stemness’ in CTC clusters.
Circulating tumor cells (CTCs) refer to the population of tumor cells in blood circulation, shed from primary or metastatic tumors [1]. They contain metastatic precursors that can eventually initiate new metastases. CTCs can exist as single cells or clusters, ranging from two cells to dozens of cells. It has long been speculated that CTC clusters are more likely to initiate metastasis, possibly because they can be easily trapped in small blood vessels in various organs. The significance of this old hypothesis has newly been demonstrated in CTCs from patients. In an earlier report by Aceto et al. [2], CTC clusters are found to be more metastatic than single CTCs. CTC clusters are more likely to resist initial cell death after lodging in distant organs, thereby promoting metastasis. However, exactly how CTC clusters enhance their metastatic potential is unclear.
To follow up on this question, Gkountela et al. [3] performed an elegant study to analyze the epigenomes of CTC clusters and single CTCs. They showed that CTC clusters have stem cell signatures that contribute to their metastatic initiating properties. In this study, they employed a newly developed single cell whole genome bisulfite sequencing (WGBS) method [4] to analyze DNA methylation differences between single CTCs and cluster CTCs isolated from breast cancer patients and xenograft mouse models generated using ex vivo expanded CTC lines [5]. This analysis revealed a significant amount of differentially methylated regions, enriching for different sets of transcription factor (TF) binding sites. The hypomethylated regions (typically correlated with more accessibility to transcriptional regulation) in CTC clusters were enriched with binding sites for several key TFs that play important roles in embryonic stem cells, such as OCT4, NANOG, and SOX2, whereas hypomethylated regions in single CTCs had enrichment of different sets of TFs. RNA-sequencing analysis of CTC clusters and single CTCs confirmed an enrichment of downstream pathways that are regulated by these stem cell TFs in the clusters. The authors then employed a high-content image-based screen with a large panel of FDA approved drugs and identified a small list of compounds that can disrupt CTC clusters, including several cardiac glycosides that are Na+/K+-ATPases, such as digitoxin. Intriguingly, disrupting the clusters into single cells, via either Na+/K+-ATPases inhibitors or knockout of cell–cell junction receptors, led to gain of DNA methylation in the cluster-specific hypomethylated regions and loss of accessibility of stem cell TF binding. Furthermore, xenograft assays showed that digitoxin significantly inhibited the ability of CTC clusters to generate metastases.
Cancers often hijack stem cell programs to gain so-called ‘sternness’ features that promote tumor initiation or progression. The majority of studies have focused on the genetic and epigenetic regulation of the stemness of tumor cells at the primary or metastatic sites, while the epigenetic landscapes of CTCs remain largely unexplored. This study provides exciting novel insights into the stemness of metastasis-prone CTC clusters, linking the epigenetic regulation of this trait with the phenotypic properties of CTCs (Figure 1). The finding that disrupting cell–cell connections could lead to the rewiring of DNA methylation and block stem cell TF binding is quite intriguing and opens up a new area of research. For example, what is the minimum number of cells that can establish or maintain the epigenetic feature that promotes stemness? It is known that the CTC clusters could be quite heterogeneous in size and cell types. CTC clusters sometimes also contain other cell types, such as tumor-associated macrophages, fibroblasts, or leukocytes. Do these heterotypic cell types influence epigenetic status? Furthermore, it remains unknown whether the epigenetic states of the CTC clusters are established prior to shedding or during their journey through circulation? Are these epigenetic states maintained in the metastases initiated by CTC clusters? From a mechanistic point of view, it remains to be discovered how cell–cell clustering mediates the establishment and maintenance of low DNA methylation status in cluster-specific regions. Is it by influencing DNA methyltransferases (DNMTs) or demethylases (TETs)? It is known that some embryonic stem cell TFs are also pioneer factors, such as OCT4 [6], that participate in modulating the accessibility of chromatin during reprogramming. Therefore, it is unknown whether these stem cell TFs facilitate the establishment and maintenance of the DNA hypomethylation status of their targets. Understanding the detailed mechanisms behind such dynamic regulation may provide precise therapeutic designs to target clusters.
Figure 1. Epigenetic Regulation of Stemness in Circulating Tumor Cell (CTC) Clusters and Single CTCs.
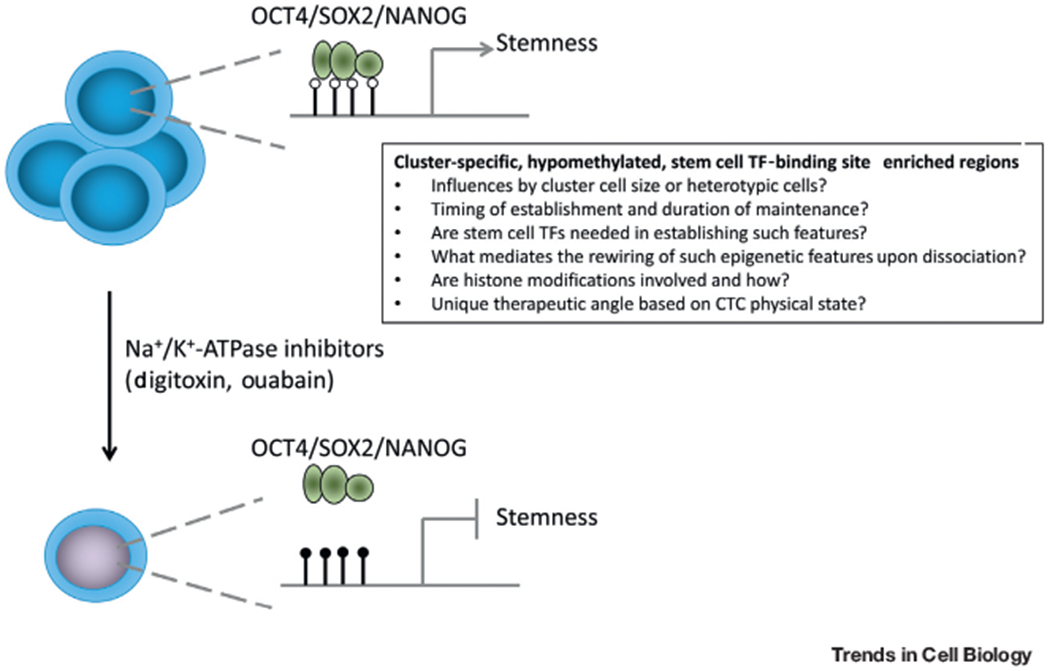
By comparing DNA methylation differences between CTC clusters and single CTCs, the authors discovered that hypomethylation regions in CTC clusters are enriched with embryonic stem cell transcription factor (TF) binding sites (e.g., OCT4, SOX2, NANOG), which correlated with stemness-related transcriptional pathways and metastatic phenotype. Upon dissociation of CTC clusters into single CTCs via treatment with Na+/K+-ATPase inhibitors, the cluster-specific hypomethylated regions gained methylation and reduced the accessibility of TFs, leading to suppression of stemness phenotype. This exciting finding has raised new questions related to dynamic epigenetic regulation and some of the examples are listed in the box.
Cancer stem cell properties have been linked with epithelial mesenchymal transition (EMT) [7], a developmental process frequently hijacked by cancer. EMT often leads to the loss of cell–cell connections and to mesenchymal-like single CTCs entering circulation. It seems that stemness may be regulated via distinct mechanisms in single versus clustered CTCs, calling for therapeutic strategies tailored according to the physical state of CTCs in circulation.
It is intriguing to see that the class of cardiac glycosides, traditionally used for heart diseases, can have a specific effect in disrupting CTC cluster formation. In recent years, studies have also reported the benefits of digitoxin and its analogs in cancer patients [8]. However, the exact mechanisms affecting cancer cells are not entirely clear. Disruption of cell signaling mediated by the signalosome and subsequent cell cycle genes has been proposed. It is tempting to speculate that the benefit of digitoxin is partly due to its effect in preventing cluster formation in circulation.
Single cell analysis has emerged as a collection of new tools to dissect the molecular properties of cells [9]. Many single cell epigenetic analyses still suffer from low coverage. For example, the WGBS only has coverage of a small percentage of the total CpGs. Despite this limitation, single cell epigenetic analyses have helped elucidate crucial biological properties of cells by increasing the number of cells analyzed with focused questions, such as in the case of single versus cluster states of CTCs. We expect that in the near future the advance of technologies will further improve our ability to explore the fascinating biology at single cell level. For example, in this context, it will be interesting to evaluate the roles of histone modifications in establishing and maintaining the DNA methylation states of single CTCs and clusters, due to the known interactions between these two epigenetic features [10].
In conclusion, CTC clusters are intriguing features of tumor cells in circulation. The study by Gkountela et al. [3] provided a fascinating finding, linking CTC clusters with epigenetics and stem cell factors, which provide the ‘stemness’ to promote metastasis.
References
- 1.Yu M et al. (2011) Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol. 192, 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aceto N et al. (2014) Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gkountela S et al. (2019) Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 176, 98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farlik M et al. (2015) Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 10, 1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu M et al. (2014) Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soufi A et al. (2015) Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mani SA et al. (2008) The epithelial-mesenchymal transition generates cels with properties of stem cels. Cell 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbaz HA et al. (2012) Digitoxin and its analogs as novel cancer therapeutics. Exp. Hematol. Oncol 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortiz V and , Yu M (2018) Analyzing circulating tumor cells one at a time. Trends Cell Biol. 28, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baylin SB and , Jones PA (2016) Epigenetic determinants of cancer. Cold Spring Harb. Perspect Biol. 8, a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]


