Abstract
The important achievements in kidney physiological and pathophysiological mechanisms can largely be ascribed to progress in the technology of microscopy. Much of what we know about the architecture of the kidney is based on the fundamental descriptions of anatomic microscopists using light microscopy and later by ultrastructural analysis provided by electron microscopy. These two techniques were used for the first classification systems of kidney diseases and for their constant updates. More recently, a series of novel imaging techniques added the analysis in further dimensions of time and space. Confocal microscopy allowed us to sequentially visualize optical sections along the z-axis and the availability of specific analysis software provided a three-dimensional rendering of thicker tissue specimens. Multiphoton microscopy permitted us to simultaneously investigate kidney function and structure in real time. Fluorescence-lifetime imaging microscopy allowed to study the spatial distribution of metabolites. Super-resolution microscopy increased sensitivity and resolution up to nanoscale levels. With cryo-electron microscopy, researchers could visualize the individual biomolecules at atomic levels directly in the tissues and understand their interaction at subcellular levels. Finally, matrix-assisted laser desorption/ionization imaging mass spectrometry permitted the measuring of hundreds of different molecules at the same time on tissue sections at high resolution. This review provides an overview of available kidney imaging strategies, with a focus on the possible impact of the most recent technical improvements.
Keywords: immunohistochemistry, kidney biopsy, podocytes, proximal tubule, stem cells
INTRODUCTION
The kidney’s fascinating structural complexity is required to fulfil its diverse physiological functions. At the beginning, studying the kidney was limited to macroscopic observations and their contribution to the diagnosis of kidney diseases was undeniably limited. Clinical manifestations were described but the underlying pathogenetic mechanisms were unknown. The achievements in understanding kidney physiology and pathophysiology can be largely ascribed to continuing progress in the microscopy field. Attempts at kidney imaging started a long time ago [1]. In 1666, the anatomist Malpighi examined kidney tissue with a light microscope that had a power estimated to be ×25–30 and described for the first time the glomerulus as ‘a gland in which urine was separated from blood’[2]. This contribution was ignored until 1842, when nephron structures were unravelled with a 10 times more powerful light microscope by Sir William Bowman. Bowman discovered the periglomerular capsule (later named the Bowman capsule) as anatomically connected to the first part of the tubule [3]. In 1862, Jacob Henle described the loop-like tubule segment that took his name, connecting the cortical tubule and renal papilla.
Thanks to the introduction of light microscopy, a histomorphological classification of kidney diseases became possible [1]. The chance to observe what was occurring in diseased kidneys allowed one to understand that pathological changes varied considerably between patients with similar clinical manifestations. However, the first studies on pathological changes in renal tissue were performed on autopsy samples, leading mostly to the description of chronic diseases. The lack of information about the evolution of kidney pathologies and the technical problem of autolytic phenomena, correlated to the use of post-mortem tissues, limited the knowledge of human diseases. In 1951, the introduction of kidney biopsy permitted direct observation and study not only of chronic, but also of acute kidney diseases. Meanwhile, microscopy gradually became more sophisticated with progressively increased sensitivity and resolution up to nanoscale levels. In this review we will provide an overview of fundamental kidney discoveries made possible by advancements in microscopy technology, with a focus on the possibilities evolving from the most recent technical developments.
VISUALIZING IMMUNOPATHOLOGY OF THE KIDNEY
Immunolabelling and fluorescence microscopy
The possibility to obtain a representative fragment of living tissue permitted application of emerging techniques that required maximal tissue preservation, such as the immunohistochemical methods (fluorescent first, then enzymatic). In fact, in 1950, Coons developed the method of labelling antibodies with fluorescein isocyanate, that is, immunofluorescence, without destroying the capacity to react specifically with their tissue antigens. Since then immunofluorescence has been incorporated in the diagnostic procedure for assessment of frozen renal biopsy specimens. For the first time, these techniques permitted the observation of intrarenal immunoglobulins (Igs) and complement deposits in patients affected by glomerulonephritis, contributing to the discrimination of different pathophysiologies underlying certain unspecific tissue lesions, for example, in crescentic glomerulonephritis [4]. In addition, the possibility to observe a characteristic staining pattern and the coupling of fluorescein to antibodies other than Immunoglobulin G (IgG) led to the recognition of Immunoglobulin A (IgA) deposits in IgA nephropathy, as well as complement deposits in post-infectious glomerulonephritis and membranoproliferative glomerulonephritis [4].
Confocal microscopy
Fluorescence microscopy underwent considerable technical improvement with the introduction of confocal microscopy in the late 1980s. Key elements in confocal microscopy are point illumination focused and scanned over the specimen and a variable confocal aperture (pinhole) in front of the detector that allows the collection of light from only a thin section around the focal plane (called the optical section). Confocal microscopy enabled the first in vivo studies of the kidney in experimental models and observation of the uptake and transport of fluorescent molecules through tubules in intact kidneys after micropuncture. This allowed studies of tubular function in physiological and pathological conditions [5].
The possibility to sequentially image optical sections along the z-axis and the availability of specific analysis software permitted a three-dimensional (3D) rendering of thicker tissue specimens, showing the frequent inconsistency and misrepresentations provided by 2D analyses [6, 7] (Figure 1A and a’). In addition, the introduction of genetically encoded fluorescent proteins in transgenic mice permitted the labelling of specific cells and tracing them directly within tissues without immunostaining (lineage tracing strategy) [6, 7, 8]. However, confocal microscopy still did not resolve details >200 nm, due to the diffraction limit of the visible light, photobleaching and phototoxicity.
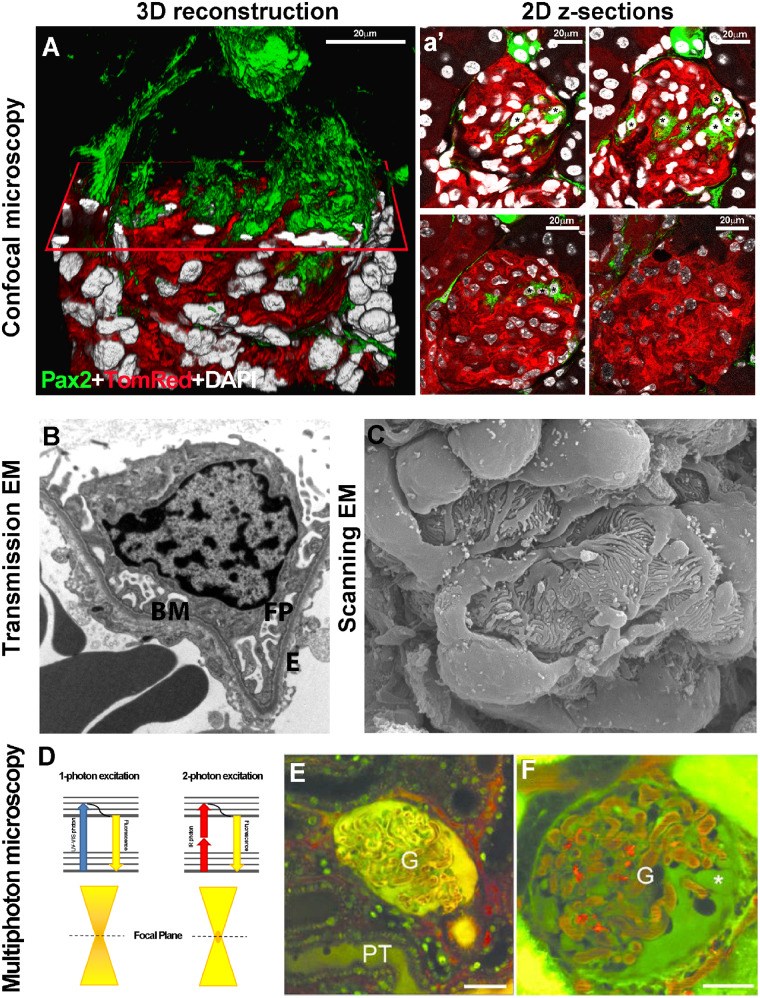
FIGURE 1.
(A) 3D reconstruction of a glomerulus from a 50-μm-thick kidney section, showing de novo podocyte regeneration in a transgenic mouse model of adriamycin-induced nephropathy. The fluorescent reporter GFP (green) labels Pax2+ renal progenitors, whereas Tomato Red labels all other cell types. An x-plane (red) has been added to virtually dissect the glomerulus in two parts. In the upper part, only green signal is shown; in the lower part, all three colours are shown. (a’) Representative 2D z-section stacks of the 3D reconstruction shown in (A). Asterisks indicate the different number of Pax2+ progenitor-derived podocytes counted inside the glomerular tuft in each 2D image. The z-stack analysis used to perform the 3D reconstruction of the glomerulus showed that the detection of regenerated podocytes was highly dependent on which section was used for 2D analysis and its thickness. (Adapted from Romoli et al. [6]. This article is licenced under a Creative Common Attribution 4.0 International License. http://creativecommons.org/licenses/by/4.0/.) (B) Representative image of a glomerular capillary loop as seen with TEM. BM, basement membrane; FP, foot processes; E, endothelial cell (TEM ×5000) (Adapted from Liapis [10] by permission of the publisher Taylor & Francis, http://www.tandfonline.com.) (C) SEM of a healthy glomerulus reveals octopus-like podocytes wrapping the capillary loops with interdigitating foot processes capillary loops (SEM ×140 000). (Adapted from Liapis [10] by permission of the publisher Taylor & Francis, http://www.tandfonline.com.) (D) The Jablonski diagram shows a comparison between one- and two-photon absorption. Conventional one-photon excitation uses ultraviolet or visible light to excite fluorescent molecules, whereas two-photon excitation depends on the simultaneous absorption of two photons with double λ (infrared light). Compared with one-photon excitation, the fluorescence excitation (orange) of two-photon excitation is spatially restricted to a small point within the focal plane, because the fluorescence excitation occurs only where the density of illuminating photons is highest. This reduces tissue photobleaching compared with the confocal approach. Representative MPM images of glomeruli (G) in vivo in (E) control or (F) PAN-treated Munich–Wistar–Fromter rat kidneys. The freely filtered dye, Lucifer Yellow, injected intravenously, labelled the Bowman’s space and early proximal tubule (PT) and allowed negative labelling of podocytes and parietal cells (which do not take up the dye). Cell nuclei were labelled using Hoechst33342 (green). The intravascular space (plasma) was labelled red by 70-kDa dextran rhodamine B injected intravenously. Unlike a normal glomerular structure (E), podocytes develop numerous pseudocysts (asterisk) after PAN treatment (F). Scale bars: 20 μm. (Adapted from Peti-Peterdi [17] with permission from Elsevier.)
UNDERSTANDING THE GLOMERULAR FILTRATION BARRIER: ELECTRON MICROSCOPY
The first attempt to overcome the resolution limit of light and fluorescence microscopes was in 1931 when Ernst Ruska and Max Knoll invented the electron microscope (EM), which uses a beam of accelerated electrons, instead of a beam of light, as an excitation source, and has a higher resolving power than light microscopes (up to 0.2 nm). For this innovation, in 1986 Ernst Ruska was awarded the Nobel Prize in Physics. Thanks to the increased resolution power, it soon became evident that the glomerulus was not simply a mechanical filter, but an extremely complex organ composed of three layers: endothelial cells, the glomerular basement membrane (GBM) and podocytes [9]. Transmission EM (TEM) permitted the visualization of glomerular cells from inside [10] and revealed that endothelial cells had thin fenestrated cell bodies, whereas podocytes had large cell bodies, and extending primary and secondary ‘foot’ processes anchored to the GBM and interconnected through linear junctions (the slight diaphragm) (Figure 1B). Furthermore, scanning EM (SEM), capturing backscattered electrons, permitted the visualization of glomerular cells from their outer surface [10] and revealed that foot processes of neighbouring podocytes interdigitated and covered the GBM (Figure 1C). EM has remained a powerful instrument not only for the comprehension of normal glomerular ultrastructure, but also for understanding kidney physiology and pathophysiology [11]. Numerous studies performed on human samples or in experimental models have revealed that different disease processes are associated with a set of distinctive patterns of ultrastructural alterations, which implies further updates on an evolving classification system of kidney diseases. The technical ability to visualize a further complexity of the kidney’s ultrastructure endorsed the specialization of ‘nephro’pathologists.
A recent SEM technique, based on a highly sensitive detector, allowed the study of the deepest portion of the slit diaphragm and revealed that it was characterized by pores with varying sizes [12]. In rats, the onset of proteinuria was associated with large pores, raising the possibility that ultrastructural changes of the slit diaphragm could be causative for the changes in glomerular permselectivity [12]. As a further technical advancement, block face SEM permitted passage from a 2D to a 3D view of the glomerular filtration barrier with a resolution sufficient to follow the nanostructure of the thinnest cellular processes in healthy mice, diseased mice and during kidney development [13, 14].
Finally, the coupling of EM to an X-ray microanalysis system (X-ray microscopy) allowed an elemental analysis of cells or parts of cells at an ultrastructural level. X-ray microscopy uses a scanning TEM with a narrow electron beam to excite the sample. The impact of the high-energy electrons from the beam with the atoms in the specimens produces energy in the form of X-ray photons. This X-ray signal, collected with an energy-dispersive semiconductor detector, is used to identify the elements present in the specimen, with a resolution of, at best, 30 nm. In the kidney field, the X-ray microanalysis was successfully utilized by Beck and Thurau to study transepithelial tubule ion transport [15] and the intracellular elemental concentration in tubular cells [16].
VISUALIZING RENAL PHYSIOLOGY WITH MULTIPHOTON MICROSCOPY
Multiphoton microscopy (MPM) provided a further advancement in kidney research, that is, the possibility to simultaneously investigate renal function and structure relationships in real time [17]. Different from conventional light microscopes, two-photon excitation depends on the simultaneous absorption of two photons with double the excitation wavelength that is necessary to excite the fluorophore in classic one-photon excitation (Figure 1D). The use of commercially available pulsed infrared, low-energy excitation lasers increased deep tissue penetration and minimized phototoxicity (Figure 1D). This enabled researchers to visualize dynamic cellular processes directly in live animals, a feature not previously achievable with any other technique. MPM permits the quantification of basic renal functions and pathological alterations such as glomerular permeability, vascular blood flow, single-nephron glomerular filtration rate and tubular flow [17] (Figure 1E and F). In addition, MPM permits the study of intracellular processes in response to injury, including cell death and shedding, leukocyte rolling and recruitment and disruptions of the GBM [17]. Further technical improvements have permitted prolonged intravital imaging over days to weeks [18] that, in combination with transgenic mice expressing fluorescent proteins, have been used to track the fate of single cells directly in vivo in healthy and injured kidney [19] and to better understand the mechanisms of renal cell turnover and regeneration.
Label-free imaging techniques were also possible with MPM, exploiting the natural autofluorescence of nicotinamide adenine dinucleotide hydrate (NADH) to study the redox state of tubular cells after injury [20]. In addition, second-harmonic generation from fibrillar collagen was used to quantify collagen deposition, a marker of renal fibrosis [21]. Second-harmonic generation is a non-linear optical process in which two excitation photons with the same wavelength interact with non-centrosymmetric structures in biological tissues, such as collagen or microtubules, and generate a new photon with half the wavelength.
A recent innovation of intravital microscopy was the introduction of a miniaturized multiphoton endoscope successfully used to image kidneys in living anesthetized mice [22]. This minimally invasive MPM may pave the way for real-time in vivo diagnosis, without tissue removal and in a very short time.
THE KIDNEY IN 3D: TISSUE-CLEARING TECHNIQUES AND EXPANSION MICROSCOPY
Despite the technological improvements, one of the principal limits for deeper volumetric imaging is renal tissue opacity, which makes the kidney one of the most optically challenging organs. This is the reason why tissue-clearing techniques have raised a lot of interest [23]. The aim of these approaches is to convert an opaque tissue into a transparent one, preserving proteins and eventually fluorescent markers, to image thick tissue slices. The combination of optical clearing with state-of-the-art microscopy provides the possibility of morphometric analysis, such as the quantification of glomerular volume and number, in thick tissues or even in the intact kidney [24]. Other applications include the analysis of distinct tubule segments [25], their functions [26] and quantifying podocyte loss [27].
To image large volumes of tissue and, at the same time, resolve fine structural details, a new technical approach was recently introduced, that is, expansion microscopy (ExM). The idea is to physically expand an entire organ 4- to 5-fold while preserving the overall architecture and the 3D proteome content. Using an expanding polymer, directly synthesized within the specimen, molecules closer than the diffraction limit of light are isotropically separated in space to greater distances and therefore can be optically resolved even by a conventional fluorescent microscope. In addition, the ExM provides greater optical transparency and reduces light scattering, allowing one to visualize a larger volume of tissue. Using this approach, the localization of multiple proteins in the slit diaphragm and the GBM can be dissected even with a normal confocal microscope [28]. For example, it can reveal foot process effacement in proteinuric mice, both in 2D and 3D, at a nanometre resolution never reached before [29]. Recently ExM has been used for clinical histopathologic assessment of paraffin-embedded specimens already stained with haematoxylin and eosin [30]. This technique can visualize foot process effacement at a nanoscale level that was previously possible only with EM. Advances in volumetric imaging permit high-resolution 3D analysis of the entire organ in a way that the main limitation to deep imaging is now represented by the capacity for antibodies to penetrate in the tissue.
STUDYING THE METABOLIC PROFILE: FREQUENCY DOMAIN FLUORESCENCE-LIFETIME IMAGING MICROSCOPY
The kidney is a metabolic organ with a high content of mitochondria. The reduced form of NADH and other metabolites are naturally fluorescent and this autofluorescence can be used as an index of the redox state [20]. However, the emission spectra of these endogenous fluorophores are overlapping and identifying them separately had been impossible. But the fluorescence lifetimes (i.e. the time decay of fluorescence) were found to be significantly different. Hence fluorescence-lifetime imaging microscopy (FLIM) allows the study of metabolic profiles [31]. Recently, using a combination of MPM and FLIM, it has become possible to characterize cell-specific metabolic signatures and to monitor metabolic changes in vivo during kidney disease [32]. FLIM is expected to provide further insights into kidney metabolism in vivo in health and disease.
PUSHING RESOLUTION BEYOND LIMITS: SUPER-RESOLUTION IMAGING OF THE KIDNEY
Standard light microscopy faces a limit of optical resolution beyond 200 nm. New technologies, referred to as super-resolution imaging, now allow fluorescent microscopy to image beyond its diffraction limit as a starting point of the era of nanoscopy [33]. These approaches combine nanoscopic resolution with the advantages of multicolour fluorescence labelling. In recognition of the potential impact of these innovations, the 2014 Nobel Prize in Chemistry was awarded to Eric Betzig, Stefan Hell and William Moerner ‘for the development of super-resolved fluorescence microscopy’. Different strategies of super-resolution imaging can be applied to address different biological questions. These approaches can be grouped into two main categories, depending on whether the super-resolution is exploited at the level of single molecules or at ensembles [33].
Ensemble-based techniques
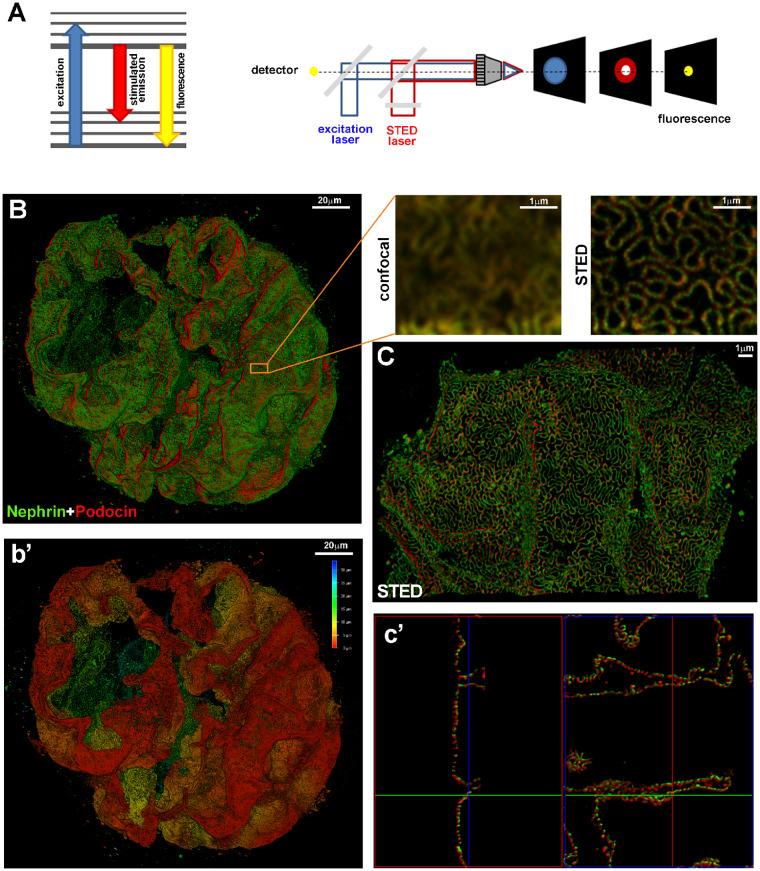
Ensemble-based techniques brake the diffraction limit by temporally and spatially modulating the excitation light beam. Among them, super-resolution stimulated emission depletion (STED) imaging uses two lasers, an excitation laser and a super-imposed, red-shifted, depletion laser (STED laser), with a doughnut shape, to suppress the fluorescence emission from the fluorophores located off the centre of the excitation. This suppression is achieved through stimulated emission and allows collecting only the signal originating from a focal spot with a dimension below the diffraction limit (Figure 2A). STED imaging was used to image the slit diaphragm in optically cleared kidney tissues (Figure 2B–c’) and allowed localizing of the spatial distribution of podocin and nephrin at the nanometre scale up to at least 30 μm of depth [34]. In addition, the possibility to observe and quantify foot process effacement in an experimental model can make STED imaging useful for studies of the glomerular filtration barrier [34].
FIGURE 2.
(A) The Jablonski diagram (left) shows the process of stimulated emission in STED microscopy. In normal fluorescence, a fluorophore can absorb a photon from the excitation light (blue arrow) and jump from the ground state to the excited state. Spontaneous fluorescence emission (with longer wavelength) (yellow arrow) brings the fluorophore back to the ground state. Stimulated emission of the excited molecules (red arrow) causes the emitted light to be of sufficiently longer wavelength and shorter fluorescent lifetime so that it can be separated from normal fluorescence. Schematic drawing (right) of a STED microscope: the excitation laser (blue) and STED laser (red) are focused into the sample through the objective. A phase mask is placed in the light path of the STED laser to create a specific doughnut-shaped pattern at the objective focal point. With this configuration, a diffraction-limited spot is excited (blue spot) while a superimposed, red-shifted STED laser with a doughnut shape (red spot) depletes all emission laterally, leaving only a central focal spot with a dimension less than the diffraction limit (yellow spot). Only emitted photons from the centre of the doughnut are collected. (B) A z-stack and 3D rendering of a glomerulus in a cleared tick slice (500 μm thick) of kidney tissue acquired with confocal microscopy. The optical transparency and antibody penetration depth were sufficient for imaging thick samples with confocal microscopy, enabling a global view of protein expression in the whole glomerulus. Magnifications of the boxed area show the comparison between confocal and STED acquisitions. The sample was stained for nephrin (green) and podocin (red). Localization of podocin and nephrin can clearly be resolved at the nanometre scale with super-resolution STED imaging. (b’) Depth coding profile of the same glomerulus in (B). (C) Volumetric representation of a 3D STED z-stack. The combination of optical clearing, immunostaining and higher-resolution imaging permitted visualization of the spatial distribution of proteins in the slit diaphragm (in c’, 3D projections of C). All images were deconvolved with SVI Huygens software. STED and confocal images were acquired with a Leica TCS SP8 STED 3X.
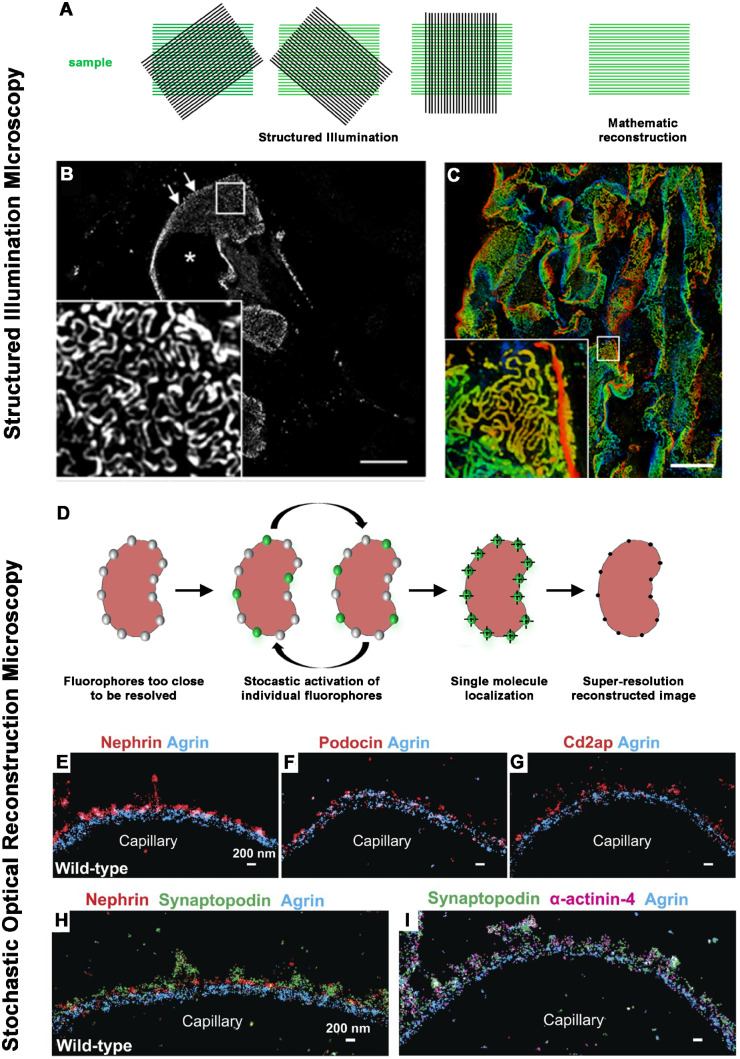
STED microscopy requires specific protocols for sample preparation, special fluorophores and technical training. For this reason, over the last few years more attention has been given to another super-resolution technique, structured illumination microscopy (SIM), which can extend the diffraction limit by a factor of two both laterally and axially. SIM uses a wide-field microscope setup with a fine-striped illumination pattern, allowing the capture of high-frequency information (corresponding to fine details in the sample) at lower spatial frequencies. By acquiring multiple images with illumination patterns of different phases and orientations, a high-resolution image can be reconstructed (Figure 3A). SIM is compatible with standard fluorophores, does not require specific sample preparation and permits 3D visualization. Given all these advantages, different studies were performed to evaluate the possibility of using SIM as a rapid diagnostic tool [35], for example, with a potential shorter turnaround time than EM. Recently a 3D SIM and automated image processing have been used as a rapid diagnostic tool of foot process effacement using routine paraffin sections from biopsies of patients with minimal change disease [36] (Figure 3B and C). This capacity to successfully identify alterations in diseases known to exhibit nanoscale pathology confirmed SIM as a promising tool for routine diagnostics.
FIGURE 3.
(A) A schematic drawing shows the basic principle of SIM. SIM uses a wide-field microscope setup with a fine-striped illumination pattern. The interaction between illumination patterns and structures in the sample produces moiré fringes, allowing the capture of high-frequency information (corresponding to fine details in the sample) at lower spatial frequencies. By acquiring multiple images with illumination patterns of different phases and orientations, a high-resolution image can be reconstructed. Because the illumination pattern itself is also limited by the diffraction of light, SIM is only capable of doubling the spatial resolution. (B, C) Representative images of nephrin-stained glomeruli after SIM reconstruction. The asterisk indicates the glomerular capillary lumen and arrows indicate the plan view areas on the glomerular capillary. The magnification of the boxed area in (B) shows a regular nephrin staining pattern with a nanometric resolution of the slit diaphragm. Scale bar = 10 µm. Depth coding profile of a 4.5 µm z-stack acquisition (C). (Adapted from Siegerist [36]. This article is licenced under a Creative Common Attribution 4.0 International License. http://creativecommons.org/licenses/by/4.0/.) (D) A schematic drawing shows the basic principle of localization-based microscopy techniques, such as STORM. All these techniques are based on the possibility to cyclically switch on and off individual fluorescent molecules that are too close to be resolved. In these approaches, molecules within a diffraction-limited region can be activated at different time points so that they can be individually imaged and subsequently localized by computationally finding their centres. (E–I) STORM imaging of slit diaphragm proteins to orient the localization of them relative to the GBM (Adapted from Suleiman [39] with permission). (E–G) Double-colour imaging of the GBM protein agrin (blue) and podocyte proteins nephrin (E, red), podocin (F, red) and Cd2ap (G, red) shows that these proteins cluster adjacent to the GBM. (H) Triple-colour imaging of agrin (blue), nephrin (red) and synaptopodin (green) shows that synaptopodin clusters are located between the nephrin clusters. (I) Triple-colour imaging of agrin (blue), synaptopodin (green) and the cytoskeletal protein α-actinin-4 (magenta) shows that synaptopodin and α-actinin-4 clusters have a similar pattern. Scale bars: 200 nm.
Table 1.
Imaging techniques and their contribution to the comprehension of renal physiology and pathology so far
| Imaging technique | Contribution to renal research |
|---|---|
| Light microscopy | Understanding of the nephron structure [1–3] |
| Visualization of morphological alterations in renal diseases [1] | |
| First classification of renal diseases [1] | |
| Fluorescence microscopy | Application of immunofluorescence as a diagnostic procedure for assessment of renal biopsy [1] |
| Visualization of aetiologic and pathogenic factors of renal diseases [4] | |
| Confocal microscopy | 3D rendering of thicker tissue specimens [6, 7] |
| Possibility to acquire multiple fluorophores simultaneously [6, 7] | |
| Lineage tracing strategy for the comprehension of renal pathology and kidney regeneration [6–8] | |
| Electron microscopy | Identification of the ultrastructural bases for glomerular filtration [9, 10] |
| Understanding of ultrastructural changes in renal diseases [11] | |
| Essential component for the diagnosis on renal human biopsies [10] | |
| MPM | Visualization of renal function and structure in vivo in healthy and diseased kidney [17, 18] |
| Tracking of the fate of single cells in vivo [19] | |
| Label-free imaging techniques to study the redox state of cells and to quantify renal fibrosis [20, 21] | |
| Tissue clearing techniques and ExM | Imaging of thick tissue slices or intact organs with confocal microscopy [24–30] |
| Antibody penetration sufficient for imaging thick samples [23] | |
| Frequency domain FLIM | Study of metabolism in vivo in healthy and disease states [31, 32] |
| Super-resolution microscopy STED, STORM and SIM | Nanoscale localization of GBM proteins in mouse and human [34, 37] |
| Identification the of actin cytoskeleton organization in podocytes in healthy and pathological conditions with nanoscopic resolution [39] | |
| Localization of podocin and nephrin spatial distribution [34] | |
| Identification of foot process effacement in biopsies from patients with minimal change disease [36] | |
| Cryo-electron microscopy | High-resolution structure determination of biomolecules in solution [41] |
| Structural characterization of proteins whose mutations are involved in human disease to reveal the molecular basis of pathogenic mutations [42–44] | |
| MALDI-IMS | Label-free map of the total spectrum of molecules in kidney tissues [45] |
| Identification of molecular markers in renal cell carcinoma [46] | |
| Identification of molecular markers to establish the margin between cancerous and normal tissue | |
| Study of renal drug toxicity [47] | |
| Study of lipids and their role in renal pathological processes [50, 51] | |
| Molecular profiles of different renal pathologies [51–54] |
Single-molecule localization-based imaging methods
These techniques rely on the possibility to cyclically switch on and off individual fluorescent molecules that are too close to be resolved. In these approaches, molecules within a diffraction-limited region can be activated at different time points so that they can be individually imaged and subsequently localized with nanometre precision by computationally finding their centres (Figure 3D). Among these approaches, stochastic optical reconstruction microscopy (STORM) was first introduced in kidney research in 2013, with a study that revealed the intricate molecular organization of the GBM at nanometre precision, including the orientation of molecules within [37]. In a mouse model of a rare congenital nephrotic syndrome, STORM revealed the integration and correct orientation of laminin 521 within the GBM following intravenous injection, opening the way for advanced therapeutic options for patients [38]. Another study described the organization of actin cytoskeleton in podocytes at a molecular scale resolution [39] (Figure 3E–I). As a new finding, podocytes contain contractile actin bundles connected with non-contractile actin bundles in foot processes. In podocyte injury models, the actin structure in the foot processes disassembled, leading to collapse of the contractile structure in the main body [39].
VISUALIZING 3D STRUCTURES OF FUNDAMENTAL BIOMOLECULES AT THE ATOMIC LEVEL: CRYO-ELECTRON MICROSCOPY
Until recently, the possibility of studying the structure of fundamental biomolecules was limited to electron microscopy, with a resolution of few nanometres. Technical hurdles, such as sample damage by intense electron beam, low-image contrast and molecule movements after interaction with electrons or difficulties to preserve water in biological samples in the vacuum applied inside the EM chamber, made it impossible to resolve molecules below this size. Cooling the specimen can reduce water evaporation and electron-induced damage [40]. From this idea cryo-EM was introduced in the 1950s. A few years ago, the introduction of a new single-electron counting detector finally pushed resolution beyond the previous limits. In 2017, the Nobel Prize in Chemistry was awarded to Jacques Dubochet, Joachim Frank and Richard Henderson for ‘developing cryo-electron microscopy for the high-resolution structure determination of biomolecules in solution’. These advances now allow structural determination of biomolecules in solution [41].
In kidney research, the structures of fundamental proteins were determined at the atomic level [42, 43]. Among them, the structure of human polycystin-2, whose mutations are responsible for autosomal dominant polycystic kidney disease [44], and the structure of the transient receptor potential canonical 6 ion channels, whose mutations cause FSGS in humans [43], were determined with a resolution of a few angstroms [44].
UNDERSTANDING THE MOLECULAR BASIS OF KIDNEY DISEASES: INTEGRATIVE KIDNEY MICROSCOPY
The last few years have seen increased interest in label-free approaches able to specifically and simultaneously map the total spectrum of molecules in tissues [45]. Among them, matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI-IMS) allows the measurement of hundreds of different molecules at the same time directly on tissue sections at high resolution. In a MALDI-IMS experiment, a thin tissue section, frozen or paraffin embedded, is coated with a matrix that absorbs the laser energy and promotes the ionization of tissue analytes. The analysis in the mass spectrometer generates a spectrum for each x/y coordinate. Subsequent to MALDI measurement, the same tissue section can be stained with traditional histochemical and immunohistochemical procedures to integrate the molecular pattern with histological details. In renal research, MALDI-IMS is used to study drugs, metabolites, lipids, peptides and proteins in normal and diseased tissues [45].
One of the most frequent applications of MALDI-IMS is the identification of molecular markers to classify renal cell carcinoma [46] and to establish the true margin between cancerous and normal tissue at the molecular as well as the histological level [47]. MALDI-IMS has also been successfully used to study renal drug toxicity to provide an early detection of toxic kidney mechanisms of damage mediated by pharmaceutical candidates [48]. Moreover, MALDI-IMS is used to study lipids and their role in renal pathologies, such as diabetic nephropathy [49], acute kidney injury [50] and polycystic kidney disease [51], and also to reveal endogenous metabolic profiles for a deeper understanding of disease-related mechanisms [52]. The identification of molecular profiles was also finalized for possible biomarkers that could be new potential diagnostic and prognostic biomarkers [53, 54].
CONCLUSIONS AND FUTURE PERSPECTIVES
Constant improvements in imaging techniques have progressively solved many critical technical barriers in kidney research and have allowed a dynamic portrayal of the structure and function in normal and diseased kidneys. Indeed, the advent of new microscopy technologies has enabled fundamental scientific discoveries and subsequently changed our view of kidney physiology and pathophysiology that can now be visualized in 3D and even filmed in videos, where time represents the fourth dimension. With the recent technical innovation of super-resolution microscopy and the advancement of molecular imaging techniques, researchers can now visualize individual biomolecules directly in the tissues and determine how they interact at cellular and subcellular levels.
The possibility to apply these tools to pathological kidney specimens, focusing on the molecular and subcellular details in clinical biopsies, should help with disease re-classifications. If and when this will create an impact on the diagnosis of kidney disease or even on patient management is currently unclear.
FUNDING
Funding was provided by the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement 648274).
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Weening JJ, Jennette JC.. Historical milestones in renal pathology. Virchows Arch 2012; 461: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Malpighi M. De Viscerum Structura exercitatio anatomica. Bologna1666
- 3. Bowman W. On the structure and use of the Malpighian bodies of the kidney, with observations on the circulation through that gland. Philos Trans R Soc A 1842; 132: 57–80 [Google Scholar]
- 4. D'Agati VD, Mengel M.. The rise of renal pathology in nephrology: structure illuminates function. Am J Kidney Dis 2013; 61: 1016–1025 [DOI] [PubMed] [Google Scholar]
- 5. Ohno Y, Birn H, Christensen EI.. In vivo confocal laser scanning microscopy and micropuncture in intact rat. Nephron Exp Nephrol 2005; 99: e17–e25 [DOI] [PubMed] [Google Scholar]
- 6. Romoli S, Angelotti ML, Antonelli G.. CXCL12 blockade preferentially regenerates lost podocytes in cortical nephrons by targeting an intrinsic podocyte-progenitor feedback mechanism. Kidney Int 2018; 94: 1111–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lazzeri E, Angelotti ML, Peired A. et al. Endocycle-related tubular cell hypertrophy and progenitor proliferation recover renal function after acute kidney injury. Nat Commun 2018; 9: 1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. LeHir M, Kriz W.. New insights into structural patterns encountered in glomerulosclerosis. Curr Opin Nephrol Hypertens 2007; 16: 184–191 [DOI] [PubMed] [Google Scholar]
- 9. Tisher CC. Functional anatomy of the kidney. Hosp Pract 1978; 13 [DOI] [PubMed] [Google Scholar]
- 10. Liapis H. Electron microscopy in kidney research: seeing is believing. Ultrastruct Pathol 2013; 37: 340–345 [DOI] [PubMed] [Google Scholar]
- 11. Strong ML, Evers P.. The third dimension in renal diagnosis. Scanning electron microscopy of normal and abnormal kidney. S Afr Med J 1979; 55: 174–177 [PubMed] [Google Scholar]
- 12. Gagliardini E, Conti S, Benigni A. et al. Imaging of the porous ultrastructure of the glomerular epithelial filtration slit. J Am Soc Nephrol 2010; 21: 2081–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lausecker F, Tian X, Inoue K. et al. Vinculin is required to maintain glomerular barrier integrity. Kidney Int 2018; 93: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ichimura K, Kakuta S, Kawasaki Y. et al. Morphological process of podocyte development revealed by block-face scanning electron microscopy. J Cell Sci 2017; 130: 132–142 [DOI] [PubMed] [Google Scholar]
- 15. Rick R, Dorge A, Beck FX. et al. Electron-probe X ray microanalysis of transepithelial ion transport. Ann N Y Acad Sci 1986; 483: 245–259 [DOI] [PubMed] [Google Scholar]
- 16. Thurau K, Dorge A, Mason J. et al. Intracellular elemental concentrations in renal tubular cells. An electron microprobe analysis. Klin Wochenschr 1979; 57: 993–999 [DOI] [PubMed] [Google Scholar]
- 17. Peti-Peterdi J, Kidokoro K, Riquier-Brison A.. Novel in vivo techniques to visualize kidney anatomy and function. Kidney Int 2015; 88: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schuh CD, Haenni D, Craigie E. et al. Long wavelength multiphoton excitation is advantageous for intravital kidney imaging. Kidney Int 2016; 89: 712–719 [DOI] [PubMed] [Google Scholar]
- 19. Hackl MJ, Burford JL, Villanueva K. et al. Tracking the fate of glomerular epithelial cells in vivo using serial multiphoton imaging in new mouse models with fluorescent lineage tags. Nat Med 2013; 19: 1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall AM, Unwin RJ, Parker N. et al. Multiphoton imaging reveals differences in mitochondrial function between nephron segments. J Am Soc Nephrol 2009; 20: 1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Strupler M, Hernest M, Fligny C. et al. Second harmonic microscopy to quantify renal interstitial fibrosis and arterial remodeling. J Biomed Opt 2008; 13: 054041. [DOI] [PubMed] [Google Scholar]
- 22. Ducourthial G, Leclerc P, Mansuryan T. et al. Development of a real-time flexible multiphoton microendoscope for label-free imaging in a live animal. Sci Rep 2015; 5: 18303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Puelles VG, Moeller MJ, Bertram JF.. We can see clearly now: optical clearing and kidney morphometrics. Curr Opin Nephrol Hypertens 2017; 26: 179–186 [DOI] [PubMed] [Google Scholar]
- 24. Klingberg A, Hasenberg A, Ludwig-Portugall I. et al. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J Am Soc Nephrol 2017; 28: 452–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saritas T, Puelles VG, Su XT. et al. Optical clearing in the kidney reveals potassium-mediated tubule remodeling. Cell Rep 2018; 25: 2668–2675.e2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schuh CD, Polesel M, Platonova E. et al. Combined structural and functional imaging of the kidney reveals major axial differences in proximal tubule endocytosis. J Am Soc Nephrol 2018; 29: 2696–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Puelles VG, van der Wolde JW, Schulze KE. et al. Validation of a three-dimensional method for counting and sizing podocytes in whole glomeruli. J Am Soc Nephrol 2016; 27: 3093–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chozinski TJ, Mao C, Halpern AR. et al. Volumetric, nanoscale optical imaging of mouse and human kidney via expansion microscopy. Sci Rep 2018; 8: 10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Unnersjo-Jess D, Scott L, Sevilla SZ. et al. Confocal super-resolution imaging of the glomerular filtration barrier enabled by tissue expansion. Kidney Int 2018; 93: 1008–1013 [DOI] [PubMed] [Google Scholar]
- 30. Zhao Y, Bucur O, Irshad H. et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat Biotechnol 2017; 35: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blacker TS, Mann ZF, Gale JE. et al. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat Commun 2014; 5: 3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hato T, Winfree S, Day R. et al. Two-photon intravital fluorescence lifetime imaging of the kidney reveals cell-type specific metabolic signatures. J Am Soc Nephrol 2017; 28: 2420–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galbraith CG, Galbraith JA.. Super-resolution microscopy at a glance. J Cell Sci 2011; 124: 1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Unnersjo-Jess D, Scott L, Blom H. et al. Super-resolution stimulated emission depletion imaging of slit diaphragm proteins in optically cleared kidney tissue. Kidney Int 2016; 89: 243–247 [DOI] [PubMed] [Google Scholar]
- 35. Wang M, Tulman DB, Sholl AB. et al. Partial nephrectomy margin imaging using structured illumination microscopy. J Biophotonics 2018; 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Siegerist F, Ribback S, Dombrowski F. et al. Structured illumination microscopy and automatized image processing as a rapid diagnostic tool for podocyte effacement. Sci Rep 2017; 7: 11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suleiman H, Zhang L, Roth R. et al. Nanoscale protein architecture of the kidney glomerular basement membrane. Elife 2013; 2: e01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin MH, Miller JB, Kikkawa Y. et al. Laminin-521 protein therapy for glomerular basement membrane and podocyte abnormalities in a model of Pierson syndrome. J Am Soc Nephrol 2018; 29: 1426–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suleiman HY, Roth R, Jain S. et al. Injury-induced actin cytoskeleton reorganization in podocytes revealed by super-resolution microscopy. JCI Insight 2017; 2: e94137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fernandez-Moran H. Low-temperature preparation techniques for electron microscopy of biological specimens based on rapid freezing with liquid helium II. Ann N Y Acad Sci 1960; 85: 689–713 [DOI] [PubMed] [Google Scholar]
- 41. Fernandez-Leiro R, Scheres SH.. Unravelling biological macromolecules with cryo-electron microscopy. Nature 2016; 537: 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Su Q, Hu F, Liu Y. et al. Cryo-EM structure of the polycystic kidney disease-like channel PKD2L1. Nat Commun 2018; 9: 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Azumaya CM, Sierra-Valdez F, Cordero-Morales JF. et al. Cryo-EM structure of the cytoplasmic domain of murine transient receptor potential cation channel subfamily C member 6 (TRPC6). J Biol Chem 2018; 293: 10381–10391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen PS, Yang X, DeCaen PG. et al. The structure of the polycystic kidney disease channel PKD2 in lipid nanodiscs. Cell 2016; 167: 763–773.e711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Prentice BM, Caprioli RM, Vuiblet V.. Label-free molecular imaging of the kidney. Kidney Int 2017; 92: 580–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Na CH, Hong JH, Kim WS. et al. Identification of protein markers specific for papillary renal cell carcinoma using imaging mass spectrometry. Mol Cells 2015; 38: 624–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oppenheimer SR, Mi D, Sanders ME. et al. Molecular analysis of tumor margins by MALDI mass spectrometry in renal carcinoma. J Proteome Res 2010; 9: 2182–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sakurai T, Kamio K, Sasaki K. et al. Imaging mass microscopy of kidneys from azithromycin-treated rats with phospholipidosis. Am J Pathol 2018; 188: 1993–2003 [DOI] [PubMed] [Google Scholar]
- 49. Miyamoto S, Hsu CC, Hamm G. et al. Mass spectrometry imaging reveals elevated glomerular ATP/AMP in diabetes/obesity and identifies sphingomyelin as a possible mediator. EBioMedicine 2016; 7: 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rao S, Walters KB, Wilson L. et al. Early lipid changes in acute kidney injury using SWATH lipidomics coupled with MALDI tissue imaging. Am J Physiol Renal Physiol 2016; 310: F1136–F1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ruh H, Salonikios T, Fuchser J. et al. MALDI imaging MS reveals candidate lipid markers of polycystic kidney disease. J Lipid Res 2013; 54: 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu H, Li W, He Q. et al. Mass spectrometry imaging of kidney tissue sections of rat subjected to unilateral ureteral obstruction. Sci Rep 2017; 7: 41954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith A, L'Imperio V, Ajello E. et al. The putative role of MALDI-MSI in the study of membranous nephropathy. Biochim Biophys Acta 2017; 1865: 865–874 [DOI] [PubMed] [Google Scholar]
- 54. Smith A, L'Imperio V, De Sio G. et al. α-1-Antitrypsin detected by MALDI imaging in the study of glomerulonephritis: its relevance in chronic kidney disease progression. Proteomics 2016; 16: 1759–1766 [DOI] [PubMed] [Google Scholar]