Figure 4.
Subthreshold Contextual Selectivity in DG Granule Cells
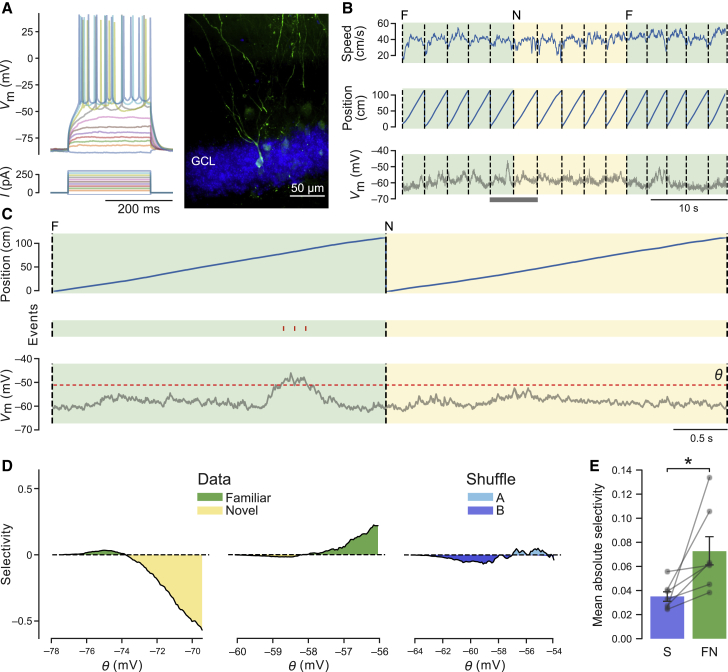
(A) Example recording. Left: sub- and suprathreshold membrane potential responses to current injections in a DG granule cell. Right: fluorescence image of a biocytin-filled granule cell. GCL, granule cell layer.
(B) Example recording from a “silent” neuron in F and N virtual environments. Traces show animal speed (top), position along the virtual corridor (center), and Vm (bottom).
(C) The same recording at higher magnification, as indicated by the gray bar at the bottom in (B). Membrane potential was thresholded (see bottom trace, threshold θ). The resulting predicted spikes are shown as a raster plot (red tick marks) in the middle.
(D) Selectivity for an environment was computed from the predicted spikes (STAR Methods) and plotted against the threshold θ. Selectivity was computed for 100 values of θ spanning a range of 2 standard deviations around the mean Vm of the recording. Examples show a recording with higher selectivity for the N than for the F environment (left), a recording with higher selectivity for the F environment (center), and a bootstrapped recording showing lack of clear selectivity for any two groups of S laps (A versus B, right).
(E) Bar graph summarizing mean absolute selectivity ± SEM across cells recorded with environment alternation (FN) versus bootstrap (S) (FN, 0.073 ± 0.012; S, 0.035 ± 0.004; n = 7 cells, Wilcoxon test, t = 0, p = 0.018).
∗p < 0.05. See also Figure S4.