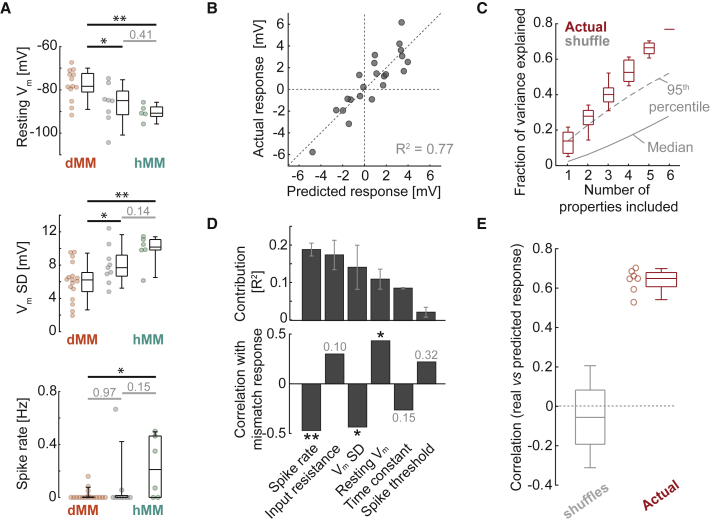
Figure 3.
Mismatch Responses Are Predictable from Electrophysiological Properties
(A) Boxplots to compare properties between dMM, hMM, and unclassified categories. From top to bottom: resting Vm (one-way ANOVA: p < 0.01, F-statistic = 6.5; dMM versus hMM: p < 0.01, dMM versus unclassified: p < 0.05, hMM versus unclassified: p = 0.41, t test), standard deviation (SD) of Vm during stationary periods (one-way ANOVA: p < 0.01, F-statistic = 6.5; dMM versus hMM: p < 0.01, dMM versus unclassified: p < 0.05, hMM versus unclassified: p = 0.14, t test), and baseline spike rate (one-way ANOVA: p = 0.06, F-statistic = 6.5; dMM versus hMM: p < 0.05, dMM versus unclassified: p = 0.97, hMM versus unclassified: p = 0.15, t test).
(B) Scatterplot between predicted mismatch response (from a multiple linear regression model including six electrophysiological properties) and actual mismatch response for each neuron.
(C) Fraction of variance in mismatch response explained as a function of the number of electrophysiological properties included in the model. Red boxplots show data for all model subsets. In gray are the median (solid line) and 95th percentile (dashed line) for shuffle controls, in which mismatch responses were randomly permuted with respect to electrophysiological properties.
(D) Top: bar plot of the average additional R2 when including each electrophysiological property in the linear model. Bottom: bar plot of correlation coefficients between each electrophysiological property and mismatch response. ∗p < 0.05; ∗∗p < 0.01.
(E) Results of leave-one-out analysis. Plotted in red are correlation coefficients between predicted and actual mismatch responses (for the neurons left out during model generation) for all seven model subsets including five or six properties. The gray boxplot shows the values for models generated with data in which mismatch responses were randomly permuted relative to electrophysiological properties.