Abstract
Rhizosphere bacteria, whether phytopathogenic or phytobeneficial, are thought to be perceived by the plant as a threat. Plant Growth-Promoting Rhizobacteria (PGPR), such as many strains of the Azospirillum genus known as the main phytostimulator of cereals, cooperate with host plants and favorably affect their growth and health. An earlier study of rice root transcriptome, undertaken with two rice cultivars and two Azospirillum strains, revealed a strain-dependent response during the rice-Azospirillum association and showed that only a few genes, including some implicated in plant defense, were commonly regulated in all tested conditions. Here, a set of genes was selected from previous studies and their expression was monitored by qRT-PCR in rice roots inoculated with ten PGPR strains isolated from various plants and belonging to various genera (Azospirillum, Herbaspirillum, Paraburkholderia). A common expression pattern was highlighted for four genes that are proposed to be markers of the rice-PGPR interaction: two genes involved in diterpenoid phytoalexin biosynthesis (OsDXS3 and OsDTC2) and one coding for an uncharacterized protein (Os02g0582900) were significantly induced by PGPR whereas one defense-related gene encoding a pathogenesis-related protein (PR1b, Os01g0382000) was significantly repressed. Interestingly, exposure to a rice bacterial pathogen also triggered the expression of OsDXS3 while the expression of Os02g0582900 and PR1b was down-regulated, suggesting that these genes might play a key role in rice-bacteria interactions. Integration of these results with previous data led us to propose that the jasmonic acid signaling pathway might be triggered in rice roots upon inoculation with PGPR.
Electronic supplementary material
The online version of this article (10.1007/s12298-020-00911-1) contains supplementary material, which is available to authorized users.
Keywords: Azospirillum, Genetic markers, Oryza sativa, PGPR, qRT-PCR, Root gene expression
Introduction
The rhizosphere, i.e. soil surrounding plant roots, is characterized by incredible diversity and abundance of microorganisms. Whether phytopathogenic or plant-beneficial, a microorganism is perceived by plants, as a threat and the success of the interaction is conditioned by the microbe’s ability to bypass plant immune reactions (Soto et al. 2009). Plant perception is based on cell surface-localized immune receptors that recognize specific microbe-associated molecular patterns (MAMP) leading to the activation of the defense response. In most cases, this plant basal immunity is believed to be sufficient to prevent the spreading of pathogens and apparition of symptoms; however well-adapted microbes can suppress or bypass defense mechanisms. Plant defense mechanisms occur as gene expression modifications that reflect a shift in the investment of resources into defense leading to reduced growth (Buscaill and Rivas 2014). The cross-talk among multiple plant hormone signaling has been proposed to regulate the growth-defense balance in plants (Wasternack and Hause 2013; Stringlis et al. 2018; Yang et al. 2020).
Some bacteria living in the rhizosphere and inside roots cooperate with host plants and favorably affect their growth and health. These, named Plant-Growth Promoting Rhizobacteria (PGPR), feed on root exudates, and induce plant beneficial effects through various mechanisms. PGPR can directly stimulate plant nutrition and development mainly through phytohormone production, phosphate solubilization, or nitrogen fixation. Besides, they can protect the plant against pathogens, by direct antagonism through the production of antimicrobial compounds or by inducing plant systemic immunity termed as Induced Systemic Resistance (ISR) (Spaepen et al. 2009). The ISR, well documented for dicots primed with PGPR of the genus Pseudomonas, allows the plant to display an accelerated defense response upon subsequent pathogen attack, and relies on Jasmonic acid (JA) and ethylene signalling (Pieterse et al. 2014). Recently, ISR has been reported in rice, with the involvement of the same signaling pathways (Laborda et al. 2020).
Members of the genus Azospirillum can improve the yield of many plant species including cereals (Pereg et al. 2016). One of the hallmarks of Azospirillum strains is the ability to fix nitrogen but only a fraction of the fixed nitrogen is thought to be retrieved by the plant. Azospirillum strains produce diverse phytohormones, among which indole-3-acetic acid (IAA) seems to play a major role in plant growth promotion (Spaepen et al. 2007). Induced resistance does not seem to be a common feature of Azospirillum, with only a few reports for an endophytic strain and the involvement of ethylene signalling was proposed (Kusajima et al. 2020). Azospirillum cooperation leads to enhancement of root development, which ultimately leads to increased yields (Dobbelaere et al. 2003). As described with other PGPR, an Azospirillum strain performed better with specific plant cultivars (Pereg et al. 2016). The affinity of strains for certain plant genotypes (together with other field parameters) leads to difficulties to obtain reliable and reproducible plant growth-promoting effect of inoculum for field application. Therefore, it is worth understanding molecular mechanisms ruling plant-Azospirillum “compatibility” and to decipher how this PGPR is perceived by the plant. Previously, a rice transcriptomic study was performed using two rice cultivars of the japonica group (Nipponbare and Cigalon) inoculated with two Azospirillum strains initially isolated from each of the cultivar (Drogue et al. 2014). Gene expression changes in rice roots were found to be dependent on the cultivar-strain combination with 83% of the differentially expressed genes appearing as combination-specific. A reduced number of genes involved in response to stress and plant defense appeared to be differentially expressed when a strain was inoculated onto its original host cultivar (Drogue et al. 2014). Besides, only 34 genes were found to be commonly regulated in all combinations and have been defined as potential “marker genes” of the Azospirillum-rice interaction. Interestingly, some of these marker genes have been found to be also regulated in rice roots following infection by a fungal pathogen (Drogue et al. 2014; Marcel et al. 2010). The present study is based on the hypothesis that some transcriptional changes induced in two rice cultivars by different Azospirillum strains may constitute a signature of plant perception to PGPR. Moreover, transcriptional changes specifically induced by a strain able to colonize inner root tissues might reflect the perception of an endophyte. Thus, we investigated by qRT-PCR the expression of a set of genes in rice roots inoculated with ten PGPR strains isolated from various plants and with a rice bacterial pathogen. Four genes are proposed to be markers of the rice-PGPR interaction. Besides, data suggests the activation of the JA signaling pathway in response to all PGPR strains.
Materials and methods
Bacterial strains and culture conditions
Ten gfp-tagged PGPR (Table 1) and one rice pathogenic strain B. glumae AU6208 were used throughout this study. Azospirillum strains were grown at 28°C onto nutrient agar (Difco) supplemented with 0.0005% (w/v) bromothymol blue (NAB medium) whereas other strains were grown on LBm medium, i.e. Luria–Bertani medium containing NaCl at only 5 g L−1. Liquid cultures for rice inoculation were performed at 28°C in NFb broth (Nelson and Knowles 1978) supplemented with 1/40 (v/v) LBm (i.e. NFb*), except for Herbaspirillum seropedicae SmR1 and Burkholderia glumae AU6208 that were grown in liquid LBm. When required, antibiotics were added at the following final concentrations µg mL−1: Gentamicin (Gm) 25; Kanamycin (Kan) 50.
Table 1.
The bacterial strains used in the study
| Straina | Isolated from | Referencesb |
|---|---|---|
| Azospirillum | ||
| Azospirillum sp. B510 | Disinfected stem of O. sativa subsp. japonica (cv Nipponbare), Japan | Elbeltagy et al. (2001), Chamam et al. (2013) |
| A. brasilense Sp245 | Surface-disinfected root of Triticum aestivum, Brazil | Baldani et al. (1986a), Valette et al. (2020) |
| A. lipoferum 4B | Rhizosphere of Oryza sativa subsp. japonica (cv Cigalon), France | Thomas-Bauzon et al. (1982), Chamam et al. (2013) |
| A. lipoferum B518 | Disinfected stem of O. sativa subsp. indica rice (cv Kasalath), Japan | Elbeltagy et al. (2001), Valette et al. (2020) |
| A. lipoferum TVV3 | Rhizosphere of O. sativa subsp. indica rice (cv IR64), Vietnam | Trân Vân et al. (1997), Valette et al. (2020) |
| A. lipoferum Sp59b | Undisinfected root of Triticum aestivum, Brazil | Tarrand et al. (1978), Valette et al. (2020) |
| A. lipoferum Br17 | Undisinfected root of Zea mays, Brazil | Tarrand et al. (1978), Valette et al. (2020) |
| A. zeae N7 | Rhizosphere of Zea mays, Canada | Mehnaz and Lazarovits (2006), Valette et al. (2020) |
| Other genera | ||
| Herbaspirillum seropedicae SmR1 | Surface sterilized root of Sorghum bicolor | Baldani et al. (1986b) |
| Paraburkholderia phytofirmans PsJN | Surface sterilized root of Allium cepa | Sessitsch et al. (2005), Compant et al. (2005) |
| Burkholderia glumae AU6208 | Clinical isolate, pathogenic on rice | Devescovi et al. (2007) |
aAll strains used (except B. glumae AU6208) were labelled previously, either with egfp via introduction of pMP2444 (All Azospirillum strains) or by gfp via transposition (Herbaspirillum seropedicae SmR1 and Paraburkholderia phytofirmans PsJN)
bThe last reference refers to the labelling
Acetylene-reduction assay
The acetylene-reduction assay (ARA) was performed on free-living cells grown in a semi-solid medium conferring the favourable microaerobic atmosphere required for N2 fixation. Ethylene production was measured using a gas chromatograph (Girdel Série 30 F) equipped with a flame ionization detector. Cultures were grown overnight in Nfb broth supplemented with 5 mM NH4Cl. Cultures were then washed 3 times in NFb broth. Five mL of washed cultures were then mixed with 45 mL of lukewarm semi-solid NFb medium to obtain a final concentration of 2 × 107 cells mL−1 and a final agar concentration of 1.8 g L−1 in 150 mL plasma flasks (Verre équipement, France). For each strain, three flasks were prepared as experimental replicates. Cultures were then incubated at 28 °C for 60 h before sealing them with a rubber septum and replacing 10% of the atmosphere with acetylene (bottles without acetylene were used as control). The exponential phase of nitrogenase activity started around 2 h post-acetylene injection. From that point, 400 µL gas samples were taken from each bottle every 2 h (for 10 h) and injected into the gas chromatograph for ethylene quantification, to obtain the kinetic of ethylene production. Quantification of ethylene was achieved using external standard calibration with serially diluted ethylene-containing bottles. Since cell count is not feasible in a semi-solid medium, total protein quantification was performed in order to normalize nitrogenase activity between strains. The cultures were boiled and NaOH was added to a final concentration of 0.1 M before a 15 min-incubation in an ultrasonic bath for cell lysis. Then, total protein content was measured using a fluorometer with the Qubit Protein Assay Kit following the manufacturer’s instructions (Invitrogen). Nitrogenase activity was calculated and expressed in nmol ethylene produced over 1 hour, normalised by protein content.
In vitro production of bacterial phytohormones
Strains were grown for 48 h (28°C, 150 rpm) in NFb* broth supplemented or not with 250 mM of the auxin precursor tryptophan or with 0.1 mM of the cytokinin precursor adenine. 750 µL of bacterial cultures were lyophilized before being extracted in 750 µL of 100% methanol followed by sonication for 20 min. The extraction was performed twice successively and extracts were pooled. Solutions were filtered, dried, and resuspended in 50 µL 100% methanol. Extracts were subjected to UHPLC separation on an Agilent 1290 Series instrument using a 100 mm × 3 mm reverse phase column (Agilent Poroshell 120 EC-C18, 2.7 µm particle size) with a diode array detector as previously described (Vacheron et al. 2016). The following 16 compounds were searched for: abscisic acid (sesquiterpenoides), jasmonic acid (jasmonates), gibberellic acid and gibberellin A1 (gibberellins), kinetin, 6-benzylaminopurine, trans-zeatin, trans-zeatin riboside, and isopentenyl adenosine (cytokinins), indole acetic acid, indole butyric acid, indole lactic acid, indole carboxylic acid, indole pyruvic acid, indole propionic acid and tryptophol (auxins and auxinic compounds). These compounds were detected with an Agilent 6530 Quadrupole Time-of-Flight (Q-TOF) mass spectrometer in positive and negative electrospray ionization, based on comparison with commercial standards on both mass and UV (280 nm) chromatograms. Absolute quantification of indole acetic acid, indole lactic acid, indole carboxylic acid was achieved using an external standard calibration by DAD analysis with commercial standards (Sigma-Aldrich, St. Louis, USA) serially diluted in methanol.
Rice seed disinfection and inoculation
For all experiments, rice seeds of cultivar Nipponbare were chosen based upon the availability of genetic resources and previous work. Seeds were surface sterilized using the previously described method (Drogue et al. 2014; Valette et al. 2020) and germinated on water agar plates (8 g L−1) for 2 or 3 days in the dark at 28 °C. Fresh bacterial cultures were mixed with water agar at a final bacterial concentration of 2 × 107 cells mL−1. The whole experiment was performed independently three times for the transcriptional analysis.
Assessing endophytic colonisation
For determining the bacterial ability to colonize plant tissues, 7 days-old plants were harvested and three plantlets were transferred in a tube containing 40 mL of a disinfecting solution (1% chloramine T with 3 drops of Tween-20 for 100 mL). Tubes were mixed by inversion for 10 min (for Azospirillum strains) or 15 min (for Paraburkholderia and Herbaspirillum strains). The plantlets were rinsed four times with 0.85% NaCl. Roots were aseptically removed from plantlets and crushed in 1 mL MgSO4 (10 mM) using sterile mortar and pestle. The solution of crushed roots was then plated onto NAB medium. To test the efficiency of the surface-sterilization procedure, 150 µL of the last wash plated on LBm, as well as a fragment of the surface-disinfected root. After 3 days of incubation at 28 °C and if no any colony appeared in control plates, we recorded the growth of any individual colonies appared in experimental plate, the fluorescence of such colonies was checked. At least two independent biological repeats were performed for each strain.
Preparation of samples for qRT-PCR and qRT-PCR
Twenty-five root systems from each experimental conditions (i.e. 5 root systems × 5 plates) were collected after 7 days of inoculations. Roots on each plate were collected separately into a 2 mL tube to avoid the inter-plate variability. RNA extraction was performed with RNAzol RT reagent (Sigma, St. Louis, USA), according to previously described methods (Breitler et al. 2016; Valette et al. 2020). Primers used in this study (Table S1) were either taken from previous reports or designed using Primer3plus software (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi) with the following criteria: product size ranges 80–180 bp, primer size comprises between 18 and 24 bases, optimal primer Tm 60 °C. All qRT-PCR reactions were performed in a CFX96 Touch Real Time PCR Detection System (BioRad, Hercules, USA) in a final volume of 20 µL, using the method previously described (Valette et al. 2020). To normalize the data, two reference genes, Os03g0177400 encoding the elongation factor 1A (Blanvillain-Baufumé et al. 2017) and Os06g0215200 encoding a Zinc finger domain containing protein (Narsai et al. 2010) were selected. The election was based on their stable expression in our experimental conditions (data not shown). The level of gene expression of each inoculated condition was compared to that of the uninoculated condition using the Pfaffl method 2−ΔΔCt (Livak and Schmittgen 2001), and corrected by each primer pair efficiency.
Results
Colonization behaviour and potential for plant growth promotion of studied PGPR strains
We selected a panel of PGPR comprising Azospirillum strains isolated from japonica rice cultivars (A. lipoferum 4B and Azospirillum sp. B510), from indica rice cultivars (A. lipoferum B518 TVV3), from wheat (A. brasilense Sp245 and A. lipoferum Sp59b), and from maize (A. lipoferum Br17 and A. zeae N7) (Table 1). Two non-Azospirillum PGPR strains, Paraburkholderia phytofirmans PsJN and Herbaspirillum seropedicae SmR1 were also isolated from onion and sorghum respectively (Table 1). Since some of our selected strains have been found within plant roots (Olivares et al. 1997; Schloter and Hartmann 1998; Elbeltagy et al. 2001; Compant et al. 2005) and since the transcriptional response of rice roots is more affected by an endophytic strain than by a rhizospheric one (Drogue et al. 2014), we choose to characterize the root colonization behaviour of each selected strain onto Nipponbare rice. Normally confocal microscopy is used to study gfp-labelled strains however, this technique failed to answer our question, likely due to the very low endophytic colonization level, and the bacterial settlement occurring mainly in superficial root cell layers. Thus, a surface-disinfection procedure with chloramine-T was used, followed by root crushing and plating. The main difficulty encountered with surface sterilization was to remove all the rhizoplane-colonizing bacteria (thus avoiding false positive) without harming the potential endophytes colonizing in the superficial root cell layers. In agreement with previous observations (Chamam et al. 2013; Elbeltagy et al. 2001), Azospirillum sp. B510 has been recovered from all the surface-disinfected roots confirming its rice endophytic colonization. A. zeae N7, initially isolated from maize and for which no endophytic behaviour has so far been described, has been repeatedly recovered from Nipponbare disinfected roots (Table 2). For strains A. lipoferum B518, Sp59b, Br17, A. brasilense Sp245 and H. seropedicae SmR1, colonies could be recovered from surface-disinfected roots on some occasions but not systematically; this may indicate that their endophytic colonization rate is very low and not easily detectable. Finally, A. lipoferum 4B and TVV3, both isolated from rice, were found to colonize exclusively the rhizoplane, a feature already described for strain 4B (Chamam et al. 2013). Finally, our data seem to indicate that P. phytofirmans PsJN is not able to colonize rice as an endophyte under our experimental conditions, which is rather surprising, as this strain has been described as an endophyte of various plants like grapevine (Ait Barka et al. 2002), maize and lupin (Kost et al. 2013).
Table 2.
Characteristic features of Plant Growth-Promoting Rhizobacteria (PGPR) used in the study
| Species | Strain (host plant) | Colonisation level of roots (Log10 UFC mg−1 fresh root)a | Detection in surface-sterilized rootsb | Nitrogenase activityc (nmol C2H4 mg proteins−1 h−1) | Indole acetic acidd (µg mL−1 A−1600) | Indole lactic acidd (µg mL−1 A−1600) | Indole carboxylic acidd (µg mL−1 A−1600) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Trp− | Trp+ | Trp− | Trp+ | Trp− | Trp+ | |||||
| A. lipoferum | 4B (rice) | 3–5 | – | 1975 ± 93 | < 1 | 1.48 | ND | 13.16 | ND | 40.66 |
| B518 (rice) | 3–5 | (+) | 11,074 ± 1920 | ND | 1.50 | ND | ND | ND | 2.37 | |
| TVV3 (rice) | 3–5 | – | 3074 ± 58 | ND | 1.76 | ND | ND | ND | 21.89 | |
| Sp59b (wheat) | 3–5 | (+) | 824 ± 102 | ND | < 1 | ND | 158.09 | ND | ND | |
| Br17 (maize) | 3–5 | (+) | 3849 ± 70 | < 1 | 7.96 | ND | 5.94 | ND | 30.90 | |
| Azospirillum sp. | B510 (rice) | 3–5 | + | 10,763 ± 877 | ND | ND | ND | ND | ND | 1.64 |
| A. brasilense | Sp245 (wheat) | 3–5 | (+) | 2035 ± 160 | < 1 | 3.73 | ND | 2.46 | ND | 6.01 |
| A. zeae | N7 (maize) | 3–5 | + | 4397 ± 272 | ND | ND | ND | 1.69 | ND | < 1 |
| P. phytofirmans | PsJN (onion) | > 6 | – | NT | ND | ND | ND | ND | ND | 32.50 |
| H. seropedicae | SmR1 (sorghum) | > 6 | (+) | 575 ± 60 | ND | 1.48 | ND | 498.11 | ND | 1.21 |
aData retrieved from (Valette et al. 2020)
b−, a strain not retrieved inside roots in any of the experiments; (+), a strain retrieved from root tissue in at least one experiment, but not in all; +, a strain retrieved from root tissues in all experiments
cValues of nitrogenase activity represent the means of three independent repetitions with standard deviation. NT: not tested as PsJN lacks nitrogenase genes
dUsing standard curves, the quantity of these compounds was determined per mL of culture and for inter-strain comparison the quantity was expressed per unit of absorbance at 600 nm. Values represent the means of two independent repetitions. ND: not detected
Nitrogen fixation is one of the bacterial mechanisms contributing to plant-growth promotion. Nitrogenase activity of the eight Azospirillum strains and the Herbaspirillum strain was measured via the acetylene reduction assay method in semi-solid culture medium and data were normalized by total proteins content for inter-strain comparison. Under our experimental conditions, a strong variability was observed between the strains. A. lipoferum B518 and Azospirillum sp. B510 appeared to be the best nitrogen fixers by far with more than 10,000 nmol C2H4 mg proteins−1 h−1 produced (Table 2). Conversely, A. lipoferum Sp59b and H. seropedicae SmR1 displayed the lowest nitrogenase activity with around 800 and 500 nmol C2H4 mg proteins−1 h−1 respectively. Thus, the intensity of the nitrogenase activity observed in vitro is not related to the phylogeny of the strains nor with the original host plant used for bacterial isolation, since large variations between strains of the same species (for instance A. lipoferum) were observed.
One of the main mechanisms used by PGPR to stimulate growth is to interfere with plant hormones homeostasis and this is achieved by bacterial biosynthesis of phytohormones, an ability described for various PGPR (Egamberdieva et al. 2017). The potential to synthesize hormones has been analysed for all strains grown in liquid medium supplemented or not with tryptophan or adenine, by searching 16 compounds using UHPLC-DAD-MS. Strains were not able to synthesize abscisic acid, jasmonic acid, gibberellic acid, gibberellin A1 and the cytokinins, even if the medium was supplemented with adenine, a cytokinin precursor. Out of the the seven auxinic compounds studied, four auxinic compounds were detected with variable levels (Table 2). The production of the indole acetic acid (IAA) assessed in the presence of tryptophan has been detected in H. seropedicae and into six Azospirillum strains out of the eight used in the study. The highest level of IAA production was detected in A. lipoferum Br17. The variability in IAA production from one strain to another might be linked to the presence or absence of ipdC, the genetic determinant encoding the main pathway for tryptophan-dependent IAA biosynthesis. This gene is present in A. brasilense Sp245 genome (that produces moderate levels of IAA) whereas it is absent from Azospirillum sp. B510 and A. lipoferum 4B genomes which produce respectively no and low IAA level (Table 2; Wisniewski-Dyé et al. 2012). The three Azospirillum strains produced IAA in absence of exogenous tryptophan, a property already shown for A. brasilense Sp245 (Ona et al. 2005), indicating that they can probably use a tryptophan-independent IAA biosynthesis pathway (Spaepen et al. 2007). Indole lactic acid and indole carboxylic acid have been detected in six and nine strains grown in the medium supplimented with tryptophan respectively, Among all the Azospirillum strains A. lipoferum Sp59b was found producing the highest quantity of indole lactic acid (Table 2). Finally, synthesis of indole pyruvic acid was observed only for A. lipoferum TVV3 but at a low level (data not shown).
Altogether, the panel of selected PGPR strains displays contrasted features (colonization behaviour, nitrogenase activity, phytohormone production) relevant for plant growth promotion. Moreover, among Azospirillum strains, none of these traits appeared to be correlated with species or to the host plant.
Choice of genes for qRT-PCR analysis
Differential gene expression had been reported in roots of two rice cultivars (Cigalon and Nipponbare) 7 days post-inoculation with two Azospirillum strains isolated from rice (A. lipoferum 4B isolated from Cigalon and Azospirillum sp. B510 isolated from Nipponbare) (Drogue et al. 2014). A strain-specific response of rice, with 83% of the differentially expressed genes being specific to the cultivar-strain combination, was reported. Only a small set of genes (i.e. 34) showed a common induction in the four different combinations and had been defined as “plant marker genes” for the Azospirillum-rice interaction. In order to characterize the transcriptional changes of these potential marker genes in response to a larger set of PGPR, we first selected genes following these criteria: (1) genes showed a strong regulation in the previous transcriptomic study (Drogue et al. 2014); (2) when possible, genes involved in signaling pathways linked to plant response to biotic interactions. Then, up to four qRT-PCR primer pairs were designed for each gene, targeting all known splicing variants for one gene. Out of the 34 genes initially identified, functional primer pairs have been finally obtained for ten potential marker genes of the Azospirillum-rice association (Drogue et al. 2014; Table S1).
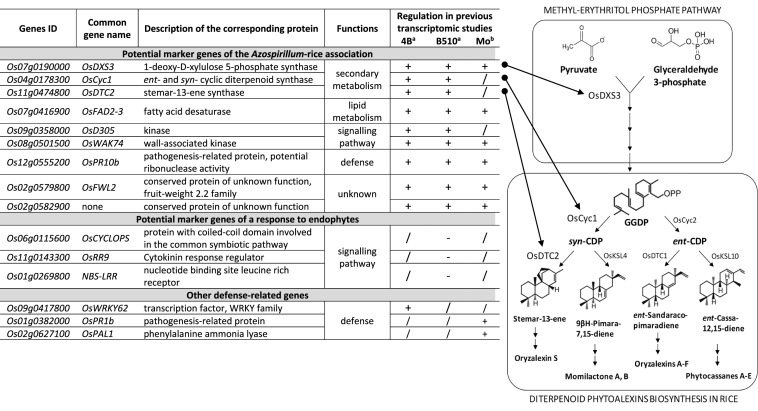
The expression profiles of these genes were analysed after 48 h, 72 h, 7 days and 10 days of inoculation with A. lipoferum 4B to observe the transcriptional changes. The strongest transcriptional changes have been observed 7 days post-inoculation (data not shown), which was in agreement of previous studies (Drogue et al. 2014; Valette et al. 2020). However, one of the 10 selected genes showed no transcriptional changes following bacterial inoculation, a result contrasting to previous data (Drogue et al. 2014), so this gene (Os03g0237100) was further omitted from the study. Among the remaining nine genes (Table 3), three code for proteins involved in secondary metabolism: the product of OsDXS3 (Os07g0190000) was involved in the first step of the Methyl-Erythitol Phosphate pathway (Okada et al. 2007) whereas the products encoded by OsCyc1 (Os04g0178300) and OsDTC2 (Os11g0474800) were involved in the biosynthesis of diterpenoid phytoalexins, especially Oryzalexin S (Toyomasu et al. 2008) (Table 3). This group also contains OsFAD2-3 (Os07g0416900), involved in lipid metabolism (Zaplin et al. 2013). Two genes, OsD305 (Os09g0358000) and OsWAK74 (Os08g0501500), encode proteins containing kinase domain that might be involved in signalling pathways. A defense-related gene, OsPR10b (Os12g0555200) encoding a ribonuclease from the pathogenesis-related protein family (Mcgee et al. 2001) was also selected. Finally, two genes Os02g0579800 (also named OsFWL2) and Os02g0582900 encode uncharacterized products for which a strong up-regulation in Nipponbare roots in response to both Azospirillum strains was observed (Drogue et al. 2014) (Table 3).
Table 3.
Description of the 15 genes selected for gene expression analysis in rice roots
The right panel displays secondary metabolism pathways in which three selected genes are involved: the Methyl-Erythritol Phosphate pathway followed by the diterpenoid phytoalexins biosynthesis in rice. GGDP geranylgeranyl diphosphate, CDP copalyl diphosphate
aTranscriptional regulation based on microarray analysis of Nipponbare roots 7 days post-inoculation with A. lipoferum 4B or Azospirillum sp. B510 (Drogue et al. 2014)
bTranscriptional regulation based on microarray analysis of Nipponbare roots 6 days post-infection with the fungal pathogen Magnaporthe oryzae (Marcel et al. 2010)
For a and b: + means up-regulation, − means down-regulation, /, means no significant regulation
The previous transcriptomic study also allowed the identification of genes regulated specifically in response to the endophytic strain Azospirillum sp. B510 in both cultivars (Drogue et al. 2014). Among them, a selection had been done following the same criteria as above and functional primers were obtained for three genes that corresponded to: (1) OsCYCLOPS (Os06g0115600) involved in the common symbiotic pathway and required for arbuscular mycorrhization in rice roots (Gutjahr et al. 2008; Yano et al. 2008); (2) OsRR9 (Os11g0143300) a type-A response regulator controlling the expression of cytokinin-responsive genes (Ito and Kurata 2006); and (3) a NBS-LRR gene (Os01g0269800) its product contains a Nucleotide-Binding Site and a Leucine-Rich Repeat domain, and belong to a protein family involved in the detection of diverse plant pathogens (Marone et al. 2013).
The three additional genes related to immunity included in our analysis were not previously defined as marker genes. First, the choice was put on OsWRKY62 (Os09g0417800), a gene coding for a transcription factor involved in rice immune response and induced by JA (Peng et al. 2008), and shown to be up-regulated in Nipponbare roots in response to A. lipoferum 4B (Drogue et al. 2014). Second, OsPAL1 (Os02g0627100) encoding Phenylalanine Ammonia-Lyase, an enzyme that catalyses the first step of the phenylpropanoid pathway by converting phenylalanine into trans-cinnamic acid (PAL, EC4.3.1.5) (MacDonald and D’Cunha 2007), has been chosen because PGPR strains belonging to Azospirillum genus can significantly affect secondary metabolites content in Nipponbare roots and particularly flavonoids and hydroxycinnamic acid derivatives that are outcome of the phenylpropanoid pathway (Chamam et al. 2013; Valette et al. 2020). Finally, OsPR1b (Os01g0382000) encoding a pathogenesis-related protein that responds positively to fungal and bacterial pathogens in Nipponbare leaves was selected (Mitsuhara et al. 2008; Table 3).
Expression of rice genes previously qualified as potential markers of the Azospirillum-rice association
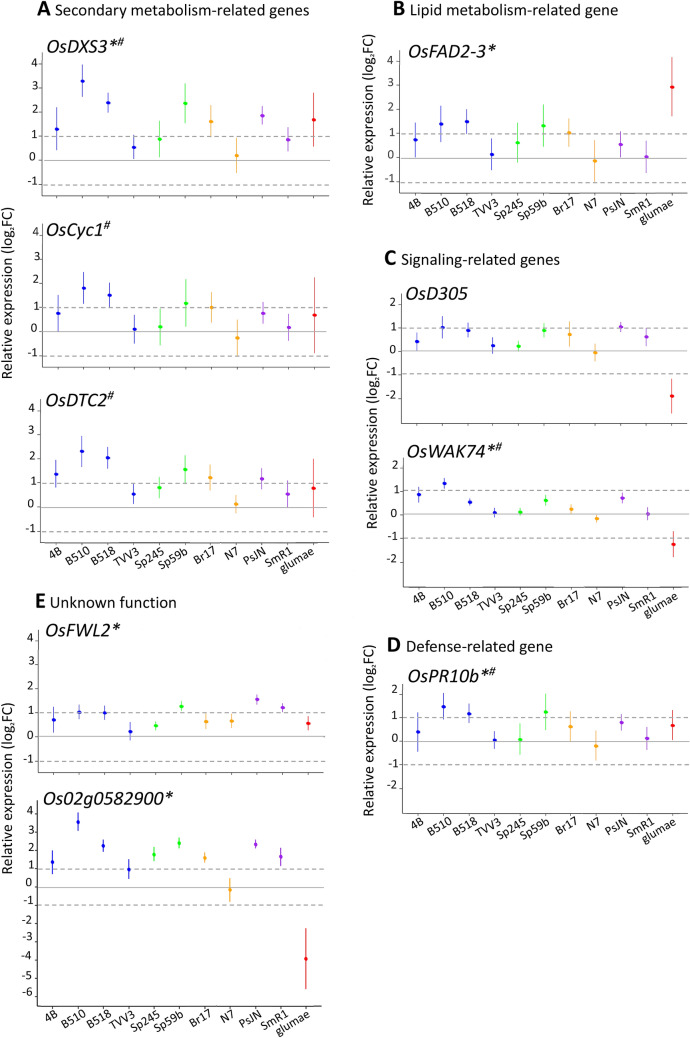
Expression of the selected genes was analysed in Nipponbare roots inoculated with the selected strains (Table 1). The relative expression (expressed in log2 Fold Change) of the nine potential marker genes of the Azospirillum-rice association was determined by qRT-PCR and compared with to non-inoculated controls, the values displayed by 95% confidence interval of the mean (Fig. 1). Two genes related to secondary metabolism, OsDXS3 and OsDTC2, showed significant up-regulation upon inoculation of all the tested PGPRs except A. zeae N7 (Fig. 1a). The expression of OsCyc1 was significantly induced with six PGPR strains (4B, B510, B518, Sp59b, Br17 and PsJN) (Fig. 1a). OsFAD2-3, a gene involved in lipid metabolism, appeared to be significantly up-regulated in rice roots in response to a few PGPR strains however the expression level was low (Fig. 1b). For all these metabolism-related genes, higher fold changes were observed with Azospirillum sp. B510 and A. lipoferum B518 inoculation. Regarding the two signaling-related genes, OsD305 and OsWAK74, up-regulation was observed for several PGPR strains (4B, B510, B518, Sp59b, Br17 and PsJN), albeit with low fold-changes (Fig. 1c). The defense-related gene OsPR10b was shown to be significantly induced by at least twofold (log2FC > 1) by only three PGPR strains (B510, B518 and Sp59b); an up-regulation was also observed with PsJN and Br17, although at lower levels (Fig. 1c).
Fig. 1.
Relative expression of potential “Azospirillum-rice interaction marker genes” in Nipponbare roots. Expression values (from three independent experiments) were normalised by the non-inoculated condition and error bars represent 95% confidence interval of the mean. The color code is as follows: blue, for strains originally isolated from rice rhizosphere or tissues; green, for strains isolated from wheat; orange, for strains isolated from maize; purple, for strains isolated from other plants (Table 1). An asterisk indicates that the corresponding gene was also induced following infection with the fungal pathogen Magnaporthe Oryzae (Marcel et al. 2010; Table 3). A sharp sign (#) indicates that the gene was induced by a jasmonate treatment in rice roots (TENOR database). Dashed lines represent log2FC of −1 and 1 cut off (color figure online)
Among the nine potential marker genes for the Azospirillum-rice association selected, six have been reported to be induced in Nipponbare roots in response to the rice fungal pathogen Magnaporthe oryzae (Marcel et al. 2010; Table 3), this suggests the common regulation mechanisms for perception of phytobeneficial and pathogenic microorganisms. This interested us to investigate how the inoculation of another rice bacterial pathogen, Burkholderia glumae AU6208, would affect the expression of these genes in Nipponbare roots. The expression level of two metabolism-related genes (OsDXS3 and OsFAD2-3) appeared to be also induced in response to B. glumae infection, especially for OsFAD2-3 (log2FC 2.94) (Fig. 1). The two genes involved in the diterpenoid phytoalexins biosynthesis (OsCyc1 and OsDTC2) were found to be upregulated though it was not statistically significant due to the high variability between the replicates. Interestingly, whereas PGPR strains triggered a weak induction of the two signalling-related genes (OsD305 and OsWAK74), B. glumae significantly repressed both genes (OsD305 and OsWAK74) compared to the non-inoculated condition (Fig. 1c). For the defense-related gene OsPR10b, a weak induction was observed in response to the bacterial pathogen (Fig. 1d). One of the gene encoding an uncharacterized product (Os02g0582900) found to be strongly repressed by B. glumae (log2FC − 3.92) whereas PGPR mostly led to an up-regulation (Fig. 1e).
Altogether, these results demonstrated that genes previously considered as potential markers of the Azospirillum-rice interaction were significantly induced by most of the PGPR strains, albeit at different levels. Based on the number of PGPR strains triggering the expression (9 out of 10) and on the values of fold-change, three genes (OsDXS3, OsDTC2 and Os02g0582900) could be considered as markers of a PGPR-rice interaction. Interestingly, two of these genes also responded to pathogens, OsDXS3 was up-regulated by B. glumae and M. oryzae, while Os02g0582900 was inversely regulated in response to the pathogens (Marcel et al. 2010).
Expression patterns of other selected rice genes
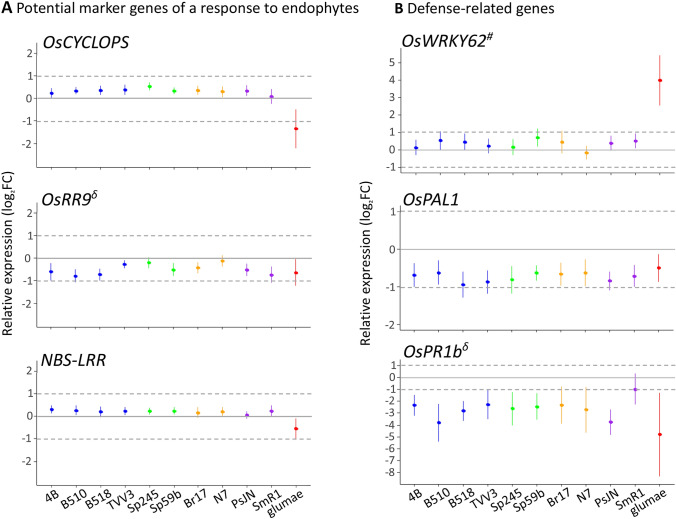
It was shown previously that expression of a set of genes was specifically affected by the endophytic strain Azospirillum sp. B510, with many genes being down-regulated (Drogue et al. 2014). Given the different behaviours of the PGPR strains used in our study, we analysed the transcriptional response of three genes OsCYCLOPS, OsRR9 and NBS-LRR that might be relevant for plant-bacteria interaction. OsRR9 indeed found to be slightly (log2FC ≤ 1) down-regulated following the inoculations of nearly all the PGPRs except the two strains A. brasilense Sp245 and A. zeae N7, and the pathogenic strain (Fig. 2a). OsCYCLOPS was found significantly up-regulated by 9 strains out of 10 PGPRs and down-regulated by B. glumae (log2FC of − 1.34). Surprisingly, by using the same threshold value of |log2FC| ≥ 1 (Drogue et al. 2014), none of these three genes appeared to be differentially regulated upon inoculation with all the 10 PGPR strains, irrespective of their colonization behaviour (Drogue et al. 2014) (Fig. 2a).
Fig. 2.
Relative expression of the potential endophyte-regulated markers and of defense-related genes in Nipponbare roots. Expression values (from three independent experiments) were normalised by the non-inoculated condition and error bars represent 95% confidence interval of the mean. The color code is as follows: blue, for strains originally isolated from rice rhizosphere or tissues; green, for strains isolated from wheat; orange, for strains isolated from maize; purple, for strains isolated from other plants (Table 1). The sharp sign (#) and the delta sign (δ) indicate that the corresponding gene was described as being respectively induced or repressed by a jasmonate treatment in rice roots (TENOR database) (color figure online)
The last three genes were selected on the basis of their involvment in plant-defense mechanisms; expression of OsPAL1 was attenuated in response to all strains tested including B. glumae, however, at a value lower than twofold and with a high variability between replicates. The expression of OsWRKY62 was barely affected by PGPR, and this gene unveiled to be strongly induced in response to the bacterial pathogen (log2FC of 3.99) (Fig. 2b). Finally, only OsPR1b was found significantly and strongly down-regulated in response to all PGPR except H. seropedicae SmR1, with log2FC values ranging from − 2.27 (A. lipoferum TVV3) to − 3.79 (Azospirillum sp. B510) (Fig. 2b). Interestingly, infection by B. glumae led to a similar response for this gene (log2FC of − 3.79).
Thus, OsPR1b, encoding a pathogenesis-related protein, could be considered as a marker of rice-bacteria interactions.
Discussion
Several studies demonstrated that PGPR inoculation leads to large plant transcriptional reprogramming associated with the activation of the plant immune system (Spaepen et al. 2014, Rekha et al. 2018; van de Mortel et al. 2012). The majority of documented transcriptional plant responses to PGPR appeared to be specific to the bacterial strain, with for instance the inoculation of A. lipoferum 4B induced more than 1600 genes in roots. On the contrary, the inoculation of the endophyte Azospirillum sp. B510 inhibited more than 1600 other genes, mainly related to plant defense, in the same rice cultivar Nipponbare (Drogue et al. 2014). Only a small part of the root transcriptional changes has been found to be commonly regulated in two rice cultivars in response to two different Azospirillum strains (Drogue et al. 2014). The transcriptional changes of 15 rice genes measured in Nipponbare roots inoculated with ten various PGPR strains and revealed significant activation of three genes (OsDXS3, OsDTC2 and Os02g0582900) while a defense-related gene OsPR1b was inhibited. Thus, these four genes could be considered as markers of a PGPR-rice interaction.
Phytoalexins are plant defense molecules usually accumulated during plant-microorganism interactions (Paxton 1981). Interestingly, the expression of two phytoalexin biosynthetic genes OsDXS3 and OsDTC2 (also OsCyc1 to a lesser extent) were induced by most of the PGPR strains. The enzyme encoded by OsDXS3 was involved in the first step of the methyl-erythritol phosphate pathway which acted upstream of both products of OsCyc1 and OsDTC2 genes involved in the diterpenoid phytoalexins biosynthesis, especially the oryzalexin S (Table 3, right panel). Attempts were failed to detect oryzalexin S by mass-spectrometry either in roots extracts or in extracts recovered from the gelified medium surrounding roots (data not shown). However, the accumulation of oryzalexin S might not be that easy to observe since previous attempts at detecting this phytoalexin in experimental conditions where the expression of biosynthetic genes was induced have also been unsuccessful (Liang et al. 2017).
OsDXS3 expression also revealed to be induced upon M. oryzae or B. glumae infections while none of the two phytoalexin biosynthetic genes appeared to be regulated by these two pathogens. Some bacterial phytopathogens alter specific host components in order to facilitate bacterial colonisation through the injection of effectors into plant cells by the type III secretion system. Although this secretion system is known to be required for virulence of B. glumae (Kang et al. 2008), no bacterial effector has been characterized so far; it is likely that one of the B. glumae effectors secreted into rice root cells could block the activation of the diterpenoid phytoalexins biosynthetic pathway. Regarding the fungal pathogen, M. oryzae is able to affect the rice immune system by suppressing JA-signaling (Zhang et al. 2018). Rice diterpenoid phytoalexins are not only known to accumulate in response to biotic or abiotic stresses but also to jasmonate (Yamane 2013), it is possible that M. Oryzae prevents the activation of the diterpenoid phytoalexins biosynthesis to promote its colonisation by suppressing the JA-signaling pathway in rice roots. An antifungal activity has been reported for oryzalexins A, B, C, and D against M. oryzae (Akatsuka et al. 1985; Sekido and Akatsuka 1987) while no antibacterial property has been reported so far. Whether PGPR growth is impacted by oryzalexins remains an open question. Altogether, these results suggest that the activation of phytoalexins biosynthesis is part of the common rice root in responses to various PGPRs. Besides, by being induced by both beneficial and pathogenic microorganisms, OsDXS3 might be considered as a common marker of biotic interactions.
Diterpenoid phytoalexins biosynthesis activated by PGPR may illustrate a moderate defense reaction. JA is an important component of the plant immune system during biotic interactions (Yang et al. 2013). Interestingly, we noticed that among the genes induced by most of the PGPR, many of them are also strongly induced by a JA treatment on rice roots (based on the TENOR: Transcriptome ENcyclopedia Of Rice database: https://tenor.dna.affrc.go.jp) including OsDXS3, OsCyc1, OsDTC2, OsPR10b, OsWAK74, and OsWRKY62. In addition, the expression of both genes OsRR9 and OsPR1b were found to be significantly down-regulated (although at a low level for OsRR9) in response to all PGPR inoculations and they both are repressed by the JA treatment in rice roots (TENOR). These observations suggest that PGPR inoculations lead to an activation of the JA signalling pathway which could be associated with an enhanced JA level. However, several attempts to quantify JA levels failed so far. Interestingly, a recent study performed on rice inoculated with two beneficial Burkholderia strains exhibiting contrasted colonizing patterns revealed a co-expression JA network with opposite regulation in response to each strain (King et al. 2019); a temporal shift in the JA systemic response was also observed throughout the establishment of the interaction, indicating the complexity of the plant response. Further confirmation of the interference with JA to the strains used in this study would be interesting to investigate if ISR can be triggered by these strains, since JA and ethylene signalling have been shown to play crucial roles in ISR in crops including rice (Laborda et al. 2020; Pieterse et al. 2014).
Os02g0582900 encodes an uncharacterized small protein of 12 kDa containing a putative N-terminal transmembrane domain. Interestingly, this gene displays the strongest expression changes and the most contrasted expression patterns, being strongly induced by all PGPR inoculation and by M. Oryzae infection (Marcel et al. 2010), and strongly inhibited in response to B. glumae infection. This protein may play a key role in rice roots during biotic interactions. The availability of two mutant accessions affecting this gene in French and Chinese rice mutant banks, can open a new opportunity for further functional analysis.
Even though most of the “potential markers” display commonly induced expressions, variation in intensity between strains were observed and these variations neither correlated to bacterial colonization rates nor to their ability to colonize inner roots. Indeed, strain A. zeae N7 induced the weakest transcriptional responses even though it colonised Nipponbare roots to the same extent as strain Azospirillum sp. B510 that triggered the strongest transcriptional changes. Moreover, strains P. phytofirmans PsJN and H. seropedicae SmR1 did not induce exarcebated transcriptional changes even though they strongly colonised Nipponbare roots (Valette et al. 2020). A. zeae N7 may be able to escape recognition by rice. Bacteria, like Azospirillum, can undergo frequent phenotypic changes through the adaptive process of phase variation, resulting in differentially expressed surface molecules (Vial et al. 2006). Whether A. zeae N7 could generate variants and minimize the activation of the plant immune system is not known. P. phytofirmans PsJN was recently shown to generate in vitro a variant form displaying reduced expression of several MAMP including flagellin, and consequently failing to induce common plant defense markers in grapevine (Rondeau et al. 2019).
Variations in plant gene expression cannot either be attributed to specific features of PGPR, and especially to phytohormone production, the main trait by which PGPR can stimulate plant growth and development and that is intimately linked to plant defense (Yang et al. 2020). Indeed, the two more potent IAA producers, A. lipoferum Br17 and A. brasilense Sp245, and the more potent indole lactic acid producer, H. seropedicae SmR1, did not trigger the more intense transcriptional response of selected genes. However, it is noteworthy that strain A. zeae N7 appeared, in the tested conditions and for the hormones that could be quantified, the poorest phytohormone producer. Moreover, phytohormone production might be different when bacteria interact with plants compared to in vitro. It was recently suggested that IAA production in H. seropedicae SmR1 might be induced by the plant (Pankievicz et al. 2016); such a feature could explain why despite harbouring the genetic determinants of the four IAA biosynthetic pathways (Pedrosa et al. 2011), only a low level of IAA was detected in vitro. Tryptophan accumulation was shown to be reduced in Nipponbare root extracts in response to inoculation of PGPR (Valette et al. 2020), thus PGPR might use plant-derived tryptophan as a substrate for IAA biosynthesis. Assessing phytohormone production and nitrogenase activity on bacteria associated with plant roots would constitute an interesting perspective in order to attest how the plant influence such phytobeneficial features.
Previously, some rice genes have been found to be regulated only by the endophytic strain Azospirillum sp. B510 in two rice cultivars (Nipponbare and Cigalon) and some of these genes have consequently been proposed as potential “endophyte-responsive marker genes” (Drogue et al. 2014). To explore this possibility, we analyzed the expression of three of these “markers” with strains differing in their endophytic potential, including Azospirillum sp. B510 and A. zeae N7 that are able to colonise internal Nipponbare root tissues (Table 1). None of these genes (i.e. OsCYCLOPS, OsRR9 and NBS-LRR) displayed a specific regulation in response to the putative Nipponbare endophytes (B510 and N7) nor to any other strains. All PGPR triggered a significant but weak induction of OsCYCLOPS and NBS-LRR expression and a reduction of OsRR9 expression. OsCYCLOPS belongs to the common symbiotic pathway and its product is required for establishment of rice mycorrhizal symbiosis (Chen et al. 2008). A previous study revealed that OsCYCLOPS expression in rice roots was not affected by the inoculation of the endophytic PGPR strain Azoarcus sp. BHGN3.1 (Chen et al. 2015). However, it is possible that OsCYCLOPS expression is triggered only in a few root cells.
The experimental design of this work allowed the detection of statistically significant transcriptional changes. Nevertheless, several significant transcriptional changes appeared to be relatively low (|log2FC ≤ 1|), thus inevitably questioning whether these significant but weak gene expression changes were biologically relevant. As the root system contains a high proportion of differentiated cell layers, sampling of the whole root system may dilute the signal of transcripts displaying a strong but localized expression pattern (Cartieaux et al. 2003). This was indeed supported experimentally: by using a transcriptional fusion, the AtTLP1 gene of A. thaliana was shown to be specifically induced in the root vascular bundle in response to colonization of a non-pathogenic Pseudomonas fluorescens strain (Léon-Kloosterziel et al. 2005) while no differential expression was detected when total root RNA was used for microarray analysis (Verhagen et al. 2004). Thus, exploring gene expression patterns in situ could be relevant to visualise the localized expression pattern of some specific genes, like OsCYCLOPS.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We wish to thank Emmanuel Guiderdoni (AGAP laboratory, CIRAD Montpellier) for gift of Nipponbare seeds and Lisa Sanchez for providing the GFP-tagged P. phytofirmans PsJN strain. We are grateful to Jonathan Gervaix (AME platform of UMR CNRS-5557) for his invaluable help in acetylene reduction assay, to Danis Abrouk (iBio platform of UMR CNRS-5557) and Samuel Barreto for their great help in statistical analyses. The platform “Serre” of FR41 (University Lyon 1) was used to carry out this work. MV receives a fellowship from «Ministère de l’enseignement supérieur et de la recherche».
Abbreviations
- IAA
Indole acetic acid
- ISR
Induced systemic resistance
- JA
Jasmonic acid
- MAMP
Microbe-associated molecular patterns
- PGPR
Plant Growth-Promoting Rhizobacteria
Authors’ contribution
M.V. conducted experiments and wrote the paper; M.R., J.D. and F.G. conducted experiments; F.W.D. designed experiments and wrote the paper; all authors discussed the data, and read and approved the contents of this manuscript.
Funding
This study was supported by reccurent funding from CNRS and University Lyon 1. MV receives a fellowship from « Ministère de l’enseignement supérieur et de la recherche » .
Availability of data and material
All used data are reported in the manuscript.
Code availability
Not applicable.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interest.
Ethics approval
Not applicable.
Consent to participate
All authors agreed to participate.
Consent for publication
All authors approved the content of this manuscript and provided their consent for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ait Barka E, Gognies S, Nowak J, Audran J-C, Belarbi A. Inhibitory effect of endophyte bacteria on Botrytis cinerea and its influence to promote the grapevine growth. Biol Control. 2002;24:135–142. doi: 10.1016/S1049-9644(02)00034-8. [DOI] [Google Scholar]
- Akatsuka T, Sekido H, Takeuchi S, Kono Y, Kodama O. Novel phytoalexins (oryzalexins a, b and c) isolated from rice blast leaves infected with Pyricularia oryzae. Agric Biol Chem. 1985;49:1689–1701. [Google Scholar]
- Baldani VLD, de Alvarez MAB, Baldani JI, Döbereiner J. Establishment of inoculated Azospirillum spp. in the rhizosphere and in roots of field grown wheat and sorghum. Plant Soil. 1986;90:37–40. doi: 10.1007/BF02277385. [DOI] [Google Scholar]
- Baldani JI, Baldani VLD, Seldin L, Dobereiner J. Characterization of Herbaspirillurn seropedicae gen. nov. sp. nov. a root-associated nitrogen-fixing bacterium. Int J Syst Bacteriol. 1986;36:86–93. doi: 10.1099/00207713-36-1-86. [DOI] [Google Scholar]
- Blanvillain-Baufumé S, Reschke M, Solé M, Auguy F, Doucoure H, Szurek B, Meynard D, Portefaix M, Cunnac S, Guiderdoni E, Boch J, Koebnik R. Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol J. 2017;15:306–317. doi: 10.1111/pbi.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitler J, Campa C, Georget F, Bertrand B, Etienne H. A single-step method for RNA isolation from tropical crops in the field. Sci Rep. 2016;6:38368. doi: 10.1038/srep38368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill P, Rivas S. Transcriptional control of plant defence responses. Curr Opin Plant Biol. 2014;20:35–46. doi: 10.1016/j.pbi.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Cartieaux F, Thibaud MC, Zimmerli L, Lessard P, Sarrobert C, David P, Gerbaud A, Robaglia C, Somerville S, Nussaume L. Transcriptome analysis of Arabidopsis colonized by a plant-growth promoting rhizobacterium reveals a general effect on disease resistance. Plant J. 2003;36:177–188. doi: 10.1046/j.1365-313X.2003.01867.x. [DOI] [PubMed] [Google Scholar]
- Chamam A, Sanguin H, Bellvert F, Meiffren G, Comte G, Wisniewski-Dyé F, Bertrand C, Prigent-Combaret C. Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum-Oryza sativa association. Phytochemistry. 2013;87:65–77. doi: 10.1016/j.phytochem.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Chen C, Ané JM, Zhu H. OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol. 2008;180:311–315. doi: 10.1111/j.1469-8137.2008.02612.x. [DOI] [PubMed] [Google Scholar]
- Chen X, Miché L, Sachs S, Wang Q, Buschart A, Yang H, Vera Cruz CM, Hurek T, Reinhold-Hurek B. Rice responds to endophytic colonization which is independent of the common symbiotic signaling pathway. New Phytol. 2015;208:531–543. doi: 10.1111/nph.13458. [DOI] [PubMed] [Google Scholar]
- Compant S, Reiter B, Sessitsch A, Nowak J, Clément C, Barka EA. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl Environ Microbiol. 2005;71:1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devescovi G, Bigirimana J, Degrassi G, Cabrio L, LiPuma JJ, Kim J, Hwang I, Venturi V. Involvement of a quorum-sensing-regulated lipase secreted by a clinical isolate of Burkholderia glumae in severe disease symptoms in rice. Appl Environ Microbiol. 2007;73:4950–4958. doi: 10.1128/AEM.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere S, Vanderleyden J, Okon Y. Plant growth-promoting effects of diazotrophs in the rhizosphere. CRC Crit Rev Plant Sci. 2003;22:107–149. doi: 10.1080/713610853. [DOI] [Google Scholar]
- Drogue B, Sanguin H, Chamam A, Mozar M, Llauro C, Panaud O, Prigent-Combaret C, Picault N, Wisniewski-Dyé F. Plant root transcriptome profiling reveals a strain-dependent response during Azospirillum-rice cooperation. Front Plant Sci. 2014;5:1–14. doi: 10.3389/fpls.2014.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egamberdieva D, Wirth SJ, Alqarawi AA, Abd-Allah EF, Hashem A. Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front Microbiol. 2017;8:2104. doi: 10.3389/fmicb.2017.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbeltagy A, Nishioka K, Sato T, Suzuki H, Ye B, Hamada T, Isawa T, Mitsui H, Minamisawa K. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. Isolated from wild rice species. Appl Environ Microbiol. 2001;67:5285–5293. doi: 10.1128/AEM.67.11.5285-5293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, Hirochika H, Imaizumi-Anraku H, Paszkowski U. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell. 2008;20:2989–3005. doi: 10.1105/tpc.108.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Kurata N. Identification and characterization of cytokinin-signalling gene families in rice. Gene. 2006;382:57–65. doi: 10.1016/j.gene.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Kang Y, Kim J, Kim S, Kim H, Lim JY, Kim M, Kwak J, Moon JS, Hwang I. Proteomic analysis of the proteins regulated by HrpB from the plant pathogenic bacterium Burkholderia glumae. Proteomics. 2008;8:106–121. doi: 10.1002/pmic.200700244. [DOI] [PubMed] [Google Scholar]
- King E, Wallner A, Rimbault I, Barrachina A, Klonowska A, Moulin L, Czernic P. Monitoring of rice transcriptional responses to contrasted colonizing patterns of phytobeneficial Burkholderia s.l. reveals a temporal shift in JA systemic response. Front Plant Sci. 2019;10:1141. doi: 10.3389/fpls.2019.01141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost T, Stopnisek N, Agnoli K, Eberl L, Weisskopf L. Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofirmans. Front Microbiol. 2013;4:421. doi: 10.3389/fmicb.2013.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusajima M, Shima S, Fujita M, Minamisawa K, Che FS, Yamakawa H, Nakashita H. Involvement of ethylene signaling in Azospirillum sp. B510-unduced disease resistance in rice. Biosci Biotechnol Biochem. 2020;82:1522–1526. doi: 10.1080/09168451.2018.1480350. [DOI] [PubMed] [Google Scholar]
- Laborda P, Chen X, Wu GC, Wang SY, Lu XF, Ling J, Li KH, Liu FQ. Lysobacter gummosus OH17 induces systemic resistance in Oryza sativa ‘Nipponbare’. Plant Pathol. 2020;69:838–848. doi: 10.1111/ppa.13167. [DOI] [Google Scholar]
- Léon-Kloosterziel KM, Verhagen BWM, Keurentjes JJB, Vanpelt JA, Rep M, Vanloon LC, Pieterse CMJ. Colonization of the Arabidopsis rhizosphere by fluorescent Pseudomonas spp. activates a root-specific, ethylene-responsive PR-5 gene in the vascular bundle. Plant Mol Biol. 2005;57:731–748. doi: 10.1007/s11103-005-3097-y. [DOI] [PubMed] [Google Scholar]
- Liang X, Chen X, Li C, Fan J, Guo Z. Metabolic and transcriptional alternations for defense by interfering OsWRKY62 and OsWRKY76 transcriptions in rice. Sci Rep. 2017;7:1–15. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- MacDonald MJ, D’Cunha GB. A modern view of phenylalanine ammonia lyase. Biochem Cell Biol. 2007;85:759. doi: 10.1139/o07-147. [DOI] [PubMed] [Google Scholar]
- Marcel S, Sawers R, Oakeley E, Angliker H, Paszkowski U. Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell. 2010;22:3177–3187. doi: 10.1105/tpc.110.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone D, Russo MA, Laidò G, De Leonardis AM, Mastrangelo AM. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci. 2013;14:7302–7326. doi: 10.3390/ijms14047302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee JD, Hamer JE, Hodges TK. Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol Plant-Microbe Interact. 2001;14:877–886. doi: 10.1094/MPMI.2001.14.7.877. [DOI] [PubMed] [Google Scholar]
- Mehnaz S, Lazarovits G. Inoculation effects of Pseudomonas putida, Gluconacetobacter azotocaptans, and Azospirillum lipoferum on corn plant growth under greenhouse conditions. Microb Ecol. 2006;51:326–335. doi: 10.1007/s00248-006-9039-7. [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Iwai T, Seo S, Yanagawa Y, Kawahigasi H, Hirose S, Ohkawa Y, Ohashi Y. Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds. Mol Genet Genom. 2008;279:415–427. doi: 10.1007/s00438-008-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Ivanova A, Ng S, Whelan J. Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 2010;10:56. doi: 10.1186/1471-2229-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LM, Knowles R. Effect of oxygen and nitrate on nitrogen fixation and denitrification by Azospirillum brasilensegrown in continuous culture. Can J Microbiol. 1978;24:1395–1403. doi: 10.1139/m78-223. [DOI] [PubMed] [Google Scholar]
- Okada A, Shimizu T, Okada K, Kuzuyama T, Koga J, Shibuya N, Nojiri H, Yamane H. Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol Biol. 2007;65:177–187. doi: 10.1007/s11103-007-9207-2. [DOI] [PubMed] [Google Scholar]
- Olivares FL, James EK, Baldani JI, Döbereiner J. Infection of mottled stripe disease-susceptible and resistant sugar cane varieties by the endophytic diazotroph Herbaspirillum. New Phytol. 1997;135:723–727. doi: 10.1046/j.1469-8137.1997.00684.x. [DOI] [Google Scholar]
- Ona O, Van Impe J, Prinsen E, Vanderleyden J. Growth and indole-3-acetic acid biosynthesis of Azospirillum brasilense Sp245 is environmentally controlled. FEMS Microbiol Lett. 2005;246:125–132. doi: 10.1016/j.femsle.2005.03.048. [DOI] [PubMed] [Google Scholar]
- Pankievicz VCS, Camilios-Neto D, Bonato P, Balsanelli E, Tadra-Sfeir MZ, Faoro H, Chubatsu LS, Donatti L, Wajnberg G, Passetti F, Monteiro RA, Pedrosa FO, Souza EM. RNA-seq transcriptional profiling of Herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant Mol Biol. 2016;90:589–603. doi: 10.1007/s11103-016-0430-6. [DOI] [PubMed] [Google Scholar]
- Paxton JD. Phytoalexins—a working redefinition. J Phytopathol. 1981;101:106–109. doi: 10.1111/j.1439-0434.1981.tb03327.x. [DOI] [Google Scholar]
- Pedrosa FO, Monteiro RA, Wassem R, Cruz LM, Ayub RA, Colauto NB, Fernandez MA, et al. Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet. 2011;7:e1002064. doi: 10.1371/journal.pgen.1002064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R. OSWRKY62 is a negative regulator of basal and XA21 -mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant. 2008;1:446–458. doi: 10.1093/mp/ssn024. [DOI] [PubMed] [Google Scholar]
- Pereg L, de-Bashan LE, Bashan Y. Assessment of affinity and specificity of Azospirillum for plants. Plant Soil. 2016;399:389–414. doi: 10.1007/s11104-015-2778-9. [DOI] [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- Rekha K, Kumar RM, Ilango K, Rex A, Usha B. Transcriptome profiling of rice roots in early response to Bacillus subtilis (RR4) colonization. Botany. 2018;96:749–765. doi: 10.1139/cjb-2018-0052. [DOI] [Google Scholar]
- Rondeau M, Esmaeel Q, Crouzet J, Blin P, Gosselin I, Sarazin C, Pernes M, Beaugrand J, Wisniewski-Dyé F, Vial L, Faure D, Clément C, Ait Barka E, Jacquard C, Sanchez L. Biofilm constructing variants of Paraburkholderia phytofirmans PsJN outcompete the wild-type form in free-living and static conditions but not in planta. Appl Environ Microbiol. 2019;85:e02670-18. doi: 10.1128/AEM.02670-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloter M, Hartmann A. Endophytic and surface colonization of wheat roots (Triticum aestivum) by different Azospirillum brasilense strains studied with strain-specific monoclonal antibodies. Symbiosis. 1998;25:159–179. [Google Scholar]
- Sekido H, Akatsuka T. Mode of action of oryzalexin D against Pyricularia oryzae. Agric Biol Chem. 1987;51:1967–1971. [Google Scholar]
- Sessitsch A, Coenye T, Sturz AV, Vandamme P, Barka EA, Salles JF, Van Elsas JD, Faure D, Reiter B, Glick BR, Wang-Pruski G, Nowak J. Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int J Syst Evol Microbiol. 2005;55:1187–1192. doi: 10.1099/ijs.0.63149-0. [DOI] [PubMed] [Google Scholar]
- Soto MJ, Domínguez-Ferreras A, Pérez-Mendoza D, Sanjuán J, Olivares J. Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell Microbiol. 2009;11:381–388. doi: 10.1111/j.1462-5822.2009.01282.x. [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev. 2007;31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Okon Y. Plant growth-promoting actions of rhizobacteria. In: Van Loon LC, editor. Plant innate immunity. Amsterdam: Elsevier; 2009. pp. 283–320. [Google Scholar]
- Spaepen S, Bossuyt S, Engelen K, Marchal K, Vanderleyden J. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 2014;201:850–861. doi: 10.1111/nph.12590. [DOI] [PubMed] [Google Scholar]
- Stringlis IA, Proietti S, Hickman R, Van Verk M, Zamioudis C, Pieterse CMJ. Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 2018;93:166–180. doi: 10.1111/tpj.13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrand JJ, Krieg NR, Döbereiner J. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol. 1978;24:967–980. doi: 10.1139/m78-160. [DOI] [PubMed] [Google Scholar]
- Thomas-Bauzon D, Weinhard P, Villecourt P, Balandreau J. The spermosphere model. I. Its use in growing, counting, and isolating N 2 -fixing bacteria from the rhizosphere of rice. Can J Microbiol. 1982;28:922–928. doi: 10.1139/m82-139. [DOI] [Google Scholar]
- Toyomasu T, Kagahara T, Okada K, Koga J, Hasegawa M, Mitsuhashi W, Sassa T, Yamane H. Diterpene phytoalexins are biosynthesized in and exuded from the roots of rice seedlings. Biosci Biotechnol Biochem. 2008;72:562–567. doi: 10.1271/bbb.70677. [DOI] [PubMed] [Google Scholar]
- Trân Vân V, Ngôkê S, Berge O, Hebbar P, Heulin T, Faure D, Bally R. Isolation of Azospirillum lipoferum from the rhizosphere of rice by a new, simple method. Can J Microbiol. 1997;43:486–490. doi: 10.1139/m97-068. [DOI] [Google Scholar]
- Vacheron J, Moënne-Loccoz Y, Dubost A, Gonçalves-Martins M, Muller D, Prigent-Combaret C. Fluorescent Pseudomonas strains with only few plant-beneficial properties are favored in the maize rhizosphere. Front Plant Sci. 2016;7:1–13. doi: 10.3389/fpls.2016.01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valette M, Rey M, Gerin F, Comte G, Wisniewski-Dyé F. A common metabolomic signature is observed upon inoculation of rice roots with various rhizobacteria. J Integr Plant Biol. 2020;62:228–246. doi: 10.1111/jipb.12810. [DOI] [PubMed] [Google Scholar]
- van de Mortel JE, de Vos RCH, Dekkers E, Pineda A, Guillod L, Bouwmeester K, van Loon JJA, Dicke M, Raaijmakers JM. Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 2012;160:2173–2188. doi: 10.1104/pp.112.207324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen BWM, Glazebrook J, Zhu T, Chang H-S, van Loon LC, Pieterse CMJ. The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant-Microbe Interact. 2004;17:895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- Vial L, Lavire C, Mavingui P, Blaha D, Haurat J, Moënne-Loccoz Y, Bally R, Wisniewski-Dyé F. Phase variation and genomic architecture changes in Azospirillum. J Bacteriol. 2006;188:5364–5373. doi: 10.1128/JB.00521-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski-Dyé F, Herrera AM, Drogue B, Acosta-Cruz E, Prigent-Combaret C, Lozano L, González V, Barbe V, Rouy Z, Mavingui P, Borland S. Genome sequence of Azospirillum brasilense CBG497 and comparative analyses of Azospirillum core and accessory genomes provide insight into niche adaptation. Genes. 2012;3:576–602. doi: 10.3390/genes3040576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H. Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Biosci Biotechnol Biochem. 2013;77:1141–1148. doi: 10.1271/bbb.130109. [DOI] [PubMed] [Google Scholar]
- Yang DL, Yang Y, He Z. Roles of plant hormones and their interplay in rice immunity. Mol Plant. 2013;6:675–685. doi: 10.1093/mp/sst056. [DOI] [PubMed] [Google Scholar]
- Yang J, Duan GH, Li CQ, Liu L, Han GY, Zhang YL, Wang CM. The crosstalks between jasmonic acid and other plant hormone signaling highlight the Involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front Plant Sci. 2020;10:1349. doi: 10.3389/fpls.2019.01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Satoko Y, Muller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, Asamizu E, Tabata S, Murooka Y, Perry J, Wang TL, Kawaguchi M, Imaizumi-Anraku H, Hayashi M, Parniske M. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci. 2008;105:20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaplin ES, Liu Q, Li Z, Butardo VM, Blanchard CL, Rahman S. Production of high oleic rice grains by suppressing the expression of the OsFAD2-1 gene. Funct Plant Biol. 2013;40:996–1004. doi: 10.1071/FP12301. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bao Y, Shan D, Wang Z, Song X, Wang Z, Wang J, He L, Wu L, Zhang Z, Niu D, Jin H, Zhao H. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 2018;177:352–368. doi: 10.1104/pp.17.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All used data are reported in the manuscript.
Not applicable.