Abstract
Rice grain yield is drastically reduced under low light especially in kharif (wet) season due to cloudy weather during most part of crop growth. Therefore, 50–60% of yield penalty was observed. To overcome this problem, identification of low light tolerant rice genotypes with a high buffering capacity trait such as photosynthetic rate has to be developed. Sedoheptulose-1,7 bisphosphatase, a light-regulated enzyme, plays pivotal role in the Calvin cycle by regenerating the substrate (RuBP) for RuBisCo and therefore, indirectly regulates the influx of CO2 for this crucial process. We found a potential role of SBPase expression and activity in low light tolerant and susceptible rice genotypes by analyzing its influence on net photosynthetic rate and biomass. We observed a significant relationship of yield with photosynthesis, SBPase expression and activity especially under low light conditions. Two tolerant and two susceptible rice genotypes were used for the present study. Tolerant genotypes exhibited significant but least reduction compared to susceptible genotypes in the expression and activity of SBPase, which was also manifested in its photosynthetic rate and finally in the grain yield under low light. However, susceptible genotypes showed significant reduction in SBPase activity along with photosynthesis and grain yield suggesting that tracking the expression and activity of SBPase could form a simple and reliable method to identify the low light tolerant rice cultivars. The data were analyzed using the Indostat 7.5, Tukey–Kramer method through Microsoft Excel 2019 and PAST4.0 software. The significant association of SBPase activity with the grain yield, net assimilation rate, electron transfer rate, biomass and grain weight were observed under low light stress. These traits should be considered while selecting and breeding for low light tolerant cultivars. Thus, SBPase plays a major role in the low light tolerance mechanism in rice.
Electronic supplementary material
The online version of this article (10.1007/s12298-020-00905-z) contains supplementary material, which is available to authorized users.
Keywords: Low light stress; Photosynthesis; Rice; Sedoheptulose-1,7-bisphosphatase; Yield
Introduction
Rice, a major wet season (July–October) crop is cultivated over an area of 156.1 m ha with a production of 476 million tonnes (International Grains Council, IGC 2013–14). However, the grain yield is significantly reduced during this season averaging around 1.2 t ha−1 in India due to the overcast skies that cause a low light (LL) stress (Praba et al. 2004; Panda et al. 2019). Additionally, in recent times, as a consequence of shrinkage in the cultivated areas, increasing planting density has been a potential practice of enhancing crop yields per unit land area (Duvick 1997). Additionally, plants are grown in a close planting system experiencing a low red to far‐red light ratio (R/FR), known as LL stress, which inevitably results in a decrement of the leaf CO2 assimilation rate (Marchiori et al. 2014). Consequently, this triggers an array of physiological changes collectively called shade‐avoidance syndrome (SAS) that results in the phenotypic readjustments of the plants while neglecting the harvestable organs, which would negatively influence the economic yield (Franklin et al. 2005). The grain yield and biomass accumulation are reduced by 50% whereas spikelet sterility is increased by 1.5 times (Singh 1988). This could be primarily attributed to a reduction in CO2 assimilating potency of plants that acts as a fundamental process behind regulating the yield capacity and chiefly depends on light (Kumar et al. 2019). Low light affects both the light and dark reactions of photosynthesis, which is highly sensitive to any type of environmental alterations. Photo system II and other Calvin cycle-related enzymes behave differently under normal light (NL) and LL conditions (Mathur et al. 2018). Therefore, identification of rice cultivars having better LL use efficiency through a minimum reduction in photosynthesis and grain yield during the kharif season has been a great challenge for the rice physiologists and breeders.
The CO2 fixation phase of photosynthesis comprises 11 different enzymes that catalyze 13 biochemical reactions, utilizing the products (ATP and NADPH) of the light reaction. In this cycle, triose phosphates are key intermediates which have two primary regeneration functions of the RuBP, the substrate for ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo), and biosynthesis of starch or sucrose (Marcus et al. 2011). Consequently, it is essential to maintain a balance between their participation in the process of carbohydrate biosynthesis and RuBP generation. To achieve this, the catalytic activity of certain enzymes such as seudoheptulose-1,7-bisphosphatase (SBPase, E.C. 3.1.3.37) needs to be strictly regulated. This is achieved by the redox potential generated through the ferredoxin/thioredoxin system, which modulates the activity of SBPase in response to light or dark conditions (Buchanan 1991). The C3 pathway of CO2 fixation in the stroma of chloroplasts is regulated by light dependant redox reactions that target specific enzymes including SBPase (Desiree et al. 2016). Light-driven reactions lead to electron transfer from water to ferredoxin, ultimately reducing the latter which regulates certain cardinal enzymes of the Calvin cycle. One key protein in these regulatory processes is thioredoxin. In chloroplasts, oxidized thioredoxin is reduced by ferredoxin in a reaction catalysed by ferredoxin-thioredoxin reductase. Reduced thioredoxin activates certain Calvin cycle enzymes including SBPase, by clearing regulatory disulphide bonds. Like several other chloroplast enzymes, such as RuBisCo, fructose 1,6-bisphosphatase, glyceraldehyde -3-phosphate dehydrogenase and phosphoribulo kinase, the SBPase are also light regulated (Buchanan 1980), especially by the light-dependent change in stromal pH and Mg2+-concentration (Woodrow et al. 1984). Therefore, it is expected that SBPase activity would decrease under LL stress, negatively influencing the rate of photosynthesis and the economic yield in comparison to NL condition. Alteration in SBPase activity influences the photosynthetic capacity, growth and yield in several crops such as tomato (Ding et al. 2016) and tobacco (Lefebvre et al. 2005). At the same time, overexpression of SBPase in tobacco was found to improve photosynthetic carbon gain and yield under fully open-air CO2 fumigation (FACE) (Rosenthal et al. 2011). Recent reports have revealed that a decrease in SBPase activity leads to dramatic reduction in carbon assimilation, growth rates and plant yield. At the same time, an overproduction of SBPase was reported to significantly increase CO2 assimilation (Driever et al. 2017). SBPase is encoded by nuclear DNA and synthesized as a precursor protein with a transit peptide, which helps it to enter chloroplast. The functional form of SBPase is a homodimer comprising two identical subunits of about 35 kDa each (Cadet and Meunier 1988). Due to the enormous significance of photosynthesis, SBPase is a potential target for photosynthesis research.
So far, the activity and expression of SBPase to track LL tolerance propensity of rice are not employed. At the same time, available literature lacks reports that could explain the direct role of SBPase on photosynthetic efficiency, growth, biomass and yield in rice plants under LL stress. We, therefore, studied SBPase expression and activity in LL tolerant and susceptible rice genotypes, with emphasis on its effect on the net photosynthetic rate, biomass and yield under LL and NL conditions.
Materials and methods
Plant materials and experimental design
Field experiments were conducted in the experimental plots of ICAR-National Rice Research Institute (NRRI), Cuttack, Odisha, India (20.4625° N, 85.8830° E) during the kharif and rabi seasons of 2017 and 2018 (Supplementary Fig. 1). Two tolerant genotypes, i.e., Purnendu and Swarnaprabha, one moderately susceptible genotype, Sasarang and one susceptible genotype, IR8 were used for study of expression and activity of SBPase, and its overall impact on the net photosynthetic rate during kharif and rabi seasons of 2017 and 2018. 25 days seedlings were transplanted to the main field. Low light stress was imposed after 15 days of transplantation up to maturity (active tillering stage to maturity) by using agro shade nets matted on wooden frame (50% and 25% interception of photosynthetically active photon flux density during rabi and kharif seasons, respectively), while no agro shade net was used for normal light condition. The spacing of plants was kept line to line 20 cm and plant to plant 15 cm. Readings were taken after 45 days of shade treatment from the flag leaves during the 50% flowering stage.
Measurement of photosynthetic active radiation (PAR)
Photosynthetic active radiation (PAR) above the canopy of plants under LL and NL conditions were recorded using a radiometer (LI-1500 LICOR, USA) at different times of the day (9.00 am, 12.00 pm and 4.00 pm of Indian Standard Time (IST), UTC + 05:30). Six replicas under each condition were maintained for accurate PAR measurement.
Physiological traits measurement
The net photosynthetic rate (NAR), stomatal conductance (SC), rate of transpiration (TR) and internal carbon dioxide to assimilated carbon dioxide ratio (Ci/Ca) were estimated from the five flag leaves of each genotype during 50% flowering stage using an open gas exchange system (Li-Cor 6400; Li-Cor Inc., Lincoln, NE, USA). CO2 concentration was maintained at 385 μmol mol−1 for NL-grown plants and LL-grown plants. We have measured 5 flag leaves per hill and for 6 hills per genotype, and then an average was made. The PAR was set at 1200 mol m−2s−1 for NL and 900 mol m−2s−1 for LL (LL) conditions which were measured by a 6400-2B LED light source. Chlorophyll a fluorescence from the ventral side of the flag leaves of different plants were measured with an open gas exchange system (Li-Cor 6400; Li-Cor Inc., Lincoln, NE, USA) and an integrated fluorescence chamber head (Li-Cor 6400-40; Li-Cor Inc., Lincoln, NE, USA) at LL and NL conditions as described by Dutta et al. (2009). Before each measurement, the leaf samples were dark-adapted for 20 min (Demmig et al. 1987). Variable fluorescence by maximum fluorescence ratio (Fv/Fm), electron transfer rate (ETR), photochemical quenching (qP) and nonphotochemical quenching (NPQ) was calculated as described by Schreiber (2004).
Total soluble protein estimation
Protein quantification was done as per Lowry et al. (1951) method. Leaf samples (1 g) were ground in 10 ml of Na-phosphate buffer in a mortar with a pestle and allowed to centrifuge for 15 min at 10,000 rpm. The supernatant was used for protein estimation after colour development and the absorbance was measured at 660 nm. The amount of protein was estimated from the standard curve made with bovine serum albumin and expressed in mg/g fresh weight.
Total soluble sugar estimation
Total soluble sugar content was determined as per Yemm and Willis (1954) method. Fresh three flag leaf samples were collected from individual plant/hill and for six plants/hills, and for each genotype at 3 days before and 3 days after flowering. Individual leaf sample (100 mg) was ground in liquid nitrogen using mortar and pestle and extracted with 10 ml of boiling 80% ethanol (v/v) in a 15 ml polypropylene tube for 30 min. The extract was centrifuged at 10,000 × g for 15 min to get clear supernatant. The above step was repeated twice. The collected supernatants from three flag leaves of same plant/hill were pooled together in a conical flask and dried in a boiling water bath at 100 °C. The contents of the flask were dissolved in 50 ml distilled water. An aliquot of 0.5 ml was used to determine total sugar content using the anthrone method (Yemm and Willis 1954) and expressed in µg glucose equivalent/g fresh weight. The estimation was carried out for six different plants/hills separately; data were pooled together, and mean of data was used for analysis.
Grain starch estimation
Grain starch content was estimated using α-amylase, amyloglucosidase, glucose oxidase plus peroxidase (GOPOD) and 4-aminoantipyrine reagents obtained from Megazyme (Total starch assay kit K-TSTA-100A, Megazyme International, Ireland Limited, Bray Business Park, Bray, Co. Wicklow, Ireland) as per Kumar et al. (2018). The grain amylose and amylopectin content were also determined as described by Kumar et al. (2018).
Antioxidant enzyme analysis
The activities of superoxide dismutase (SOD), catalase and peroxidase enzymes were determined spectrophotometrically as described by Dhindsa et al. (1981) and Aebi (1984), respectively. For these assays, 100 mg of leaf sample (flag leaf) was ground with 4 ml of extraction buffer (0.1 M phosphate buffer, pH 7.5, containing 0.5 mM EDTA) and filtered through 4 layers of cheesecloth. The filtrate was transferred to centrifuge tubes and centrifuged at 15,000 rcf for 20 min. The supernatant was used as the enzyme extract and 100 μl of this extract was used for each enzyme activity assay. Briefly, SOD activity was determined by measuring the decrease in the absorbance of blue coloured formazone and O·−2 at 560 nm. For determining catalase activity, the reaction was started by adding H2O2 (12.5 mM) to the enzyme extract and decrease in the absorbance was measured at 240 nm for 1 min. Peroxidase activity was determined by an increase in the optical density due to oxidation of guaiacol to tetra-guaiacol in the reaction mixture for 2 min, measured at 470 nm.
SBPase enzyme assay
Three flag leaves (1 g) were collected at 50% flowering stage per individual plant/hill for six hills and for each genotype. Leaves were ground to a fine powder using liquid nitrogen. 2 ml of extraction buffer containing 50 mM Tris-HCl, pH 7.5, 15% (v/v) glycerol, and 1 mM β-mercaptoethanol was added to the ground material. After vigorous vortexing, the extracts were clarified by centrifugation (10,000 rpm for 30 min). The supernatant was collected and incubated at 37 °C (Incubator, model: CI-10S, make: Remi instruments, India). The activity of SBPase was determined using the coupled assay method (Seuter et al. 2002; Chen et al. 2004) by adding 50 µl crude extract to assay mixture (50 mM Tris-HCl pH 8.3, 15 mM Mgcl2, 0.5 mM NADP+, glucose-6-Phosphate dehydrogenase-1U, phosphoglucose isomerase-2U, 0.5 mM EGTA and 1 mM FBP) and made volume up to 1 ml. Absorbance was measured at 340 nm with UV-vis spectrophotometer (Model Specord 210 Plus, Analytik Jena AG, Germany).
SBPase expression analysis
Total RNA was extracted from the flag leaves at 50% flowering stage using RNEasy Plant Mini Kit (Qiagen, USA) following the manufacturer’s protocol. First-strand cDNA synthesis was conducted using 5 mg of total DNase-treated RNA using primer script 1st strand cDNA synthesis kit (Takara Clontech, Japan). In order to estimate SBPase transcript levels in flag leaves, a real-time polymerization chain reaction was performed in QuantStudio®5 Real-Time PCR instrument (Applied Biosystems, Thermo Fisher Scientific, USA) using SYBR Green master mix (Takara Clontech) to monitor double-stranded DNA synthesis. The qRT-PCR reaction was performed by using gene-specific primer pair (FP: 50 AGTAGTGCGAGGGCCATAGA30) and (RP:50 TCTTGCAGGTGGTTTCAGTG 30), which were designed using PrimerQuest tool (IDT). The β-tubulin gene primer pair (FP: 50 ATGCGTGAGATTCTTCACATCC 30) and (RP:50 TGGGTACTCTTC ACGGATCTTAG 30) was used for its amplification and was treated as an internal control. Thermal cycling condition was as follows: initial denaturation at 94 °C for 4 min followed by 40 cycles of denaturation at 94 °C for 30 s; annealing at 60 °C for 30 s and extension at 72 °C for 1 min. The calculation was done in relation to the respective control samples as calibrator using equation 2−ΔΔct. The relative differences in expression for each sample were determined by normalizing the cycle threshold (ct) value of target gene against the ct value of the β-tubulin gene. Data analysis was done using the instrument’s software (QuantStudio™ Design & Analysis Software v1.4.3) followed by the manufacturer’s protocol. Dissociation curve analysis was also performed at the end of the assay to check for any non-specific amplification and/or contamination.
Grain yield and related traits
Four genotypes under study were evaluated for grain yield and related traits under LL stress in kharif and rabi seasons of 2017 and 2018 following randomized complete block design (RCBD) with three replications of each in a plot size of 4 × 4 m2 (spacing 15 cm × 20 cm). At maximum tillering stage, one set of each cultivar was subjected to LL regime (75% of natural available light, i.e., 900 µ mol quant m−2 s−1) using Agroshade nets matted on a wooden frame, while the other set was grown under open condition (under 100% natural light intensity, 1200 µ mol quant m−2 s−1). The recommended dose of nitrogen, P2O5 and K2O (80:40:40) fertilizers were applied. Recommended agronomic practices and plant protection measures were followed. Panicle emergence rate (%) was calculated from the first day to the fifth day as described by Panigrahy et al. (2019). The expression and activity of SBPase were studied on these genotypes and its overall impact on the net photosynthetic rate that ultimately influences the agronomic performance of rice under LL stress. Five hills were randomly selected from the middle of each plot and data for 6 traits, tiller/plant, panicle/plant, spikelet fertility %, 1000-grain weight, biomass/plant and grain yield/plant traits were recorded.
Statistical analysis
The experiments were carried out in three biological as well as three technical replicates. The statistical analysis on the mean values of five randomly selected plants from each of the three replications for four rice cultivars (both under NL and LL conditions) for the years 2017 and 2018 (kharif and rabi) was carried out on individual traits. The data of mean value for all the traits were analyzed for their variance following simple factorial RBD. Analysis of variance was done using the Indostat 7.5, along with the Tukey–Kramer method through Microsoft Excel 2019. In the Tukey–Kramer method, the minimum significant difference (MSD) is calculated for each pair of means. It depends on the sample size in each group, the average variation within the groups, and the total number of groups. It can be used to find means that are significantly different from each other. The significance was tested by referring to the table given by Fisher (1936). Range, phenotypic coefficient of variation (PCV), genotypic coefficient of variation (GCV), heritability (h2) and genetic advance (GA) were calculated comparing mean values of cultivar using the Indostat 7.5. The phenotypic correlations and principal component analysis (PCA) were estimated using PAST4.0 software (Hammer et al. 2001). The PCV, GCV, h2 and GA interpretation were based on Arp and Johnson (1955) and followed by Sanghamitra et al. (2018) for rice grain quality traits.
Results
Photosynthetic active radiation
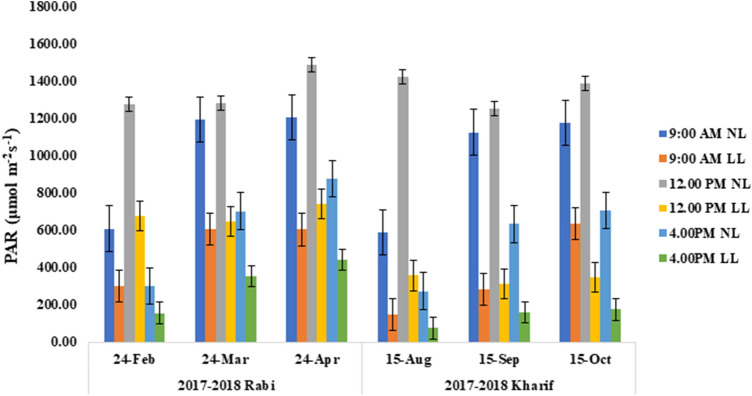
Under the agro shade mediated LL treatment, PAR reduced approximately to 50% and 25% in rabi and kharif seasons, respectively (Fig. 1 and Supplementary Table 1) than NL treatment, confirming the generation of LL stress for the grown set of plants.
Fig. 1.
Spatiotemporal distribution of photosynthetic active radiation (PAR) above the canopy of tolerant (Purnendu and Swarnaprabha) and susceptible (Sasarang and IR8) genotypes under normal (NL) and low light (LL) conditions during rabi and kharif seasons of 2017 and 2018. Each data point is the average of six replicates and the error bars represent standard error (SE)
Photosynthetic rate and internal carbon content
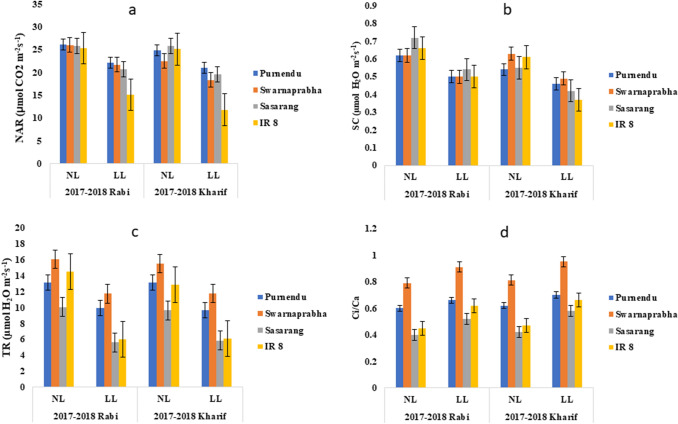
Gas exchange measurements were made to investigate the carbon assimilation rates in the tested cultivars under field conditions. Under LL, the net CO2 assimilation rate (NAR), stomatal conductance (SC) and transpiration rate (TR) were significantly (P < 0.05) decreased, whereas the ratio of internal CO2 to ambient CO2 (Ci/Ca) was significantly (P < 0.05) increased in all the genotypes during both rabi and kharif seasons of 2017 and 2018 (Supplementary Table 2). However, the decrease of NAR, SC, TR and increase in Ci/Ca during kharif was more than the rabi for all the genotypes under LL stress. Further, across the seasons, tolerant genotypes (Purnendu and Swarnaprabha) showed lesser reduction in NAR, SC and TR, while lesser increase in Ci/Ca ratio compared to susceptible cultivars (Sasarang and IR8) under LL stress (Supplementary Table 2). The reduction of PN (P < 0.05) under LL in comparison to NL was found to be 15.7% (Purnendu), 18.31% (Swarnaprabha), 24.28% (Sasarang) and 52.94% (IR8) in kharif seasons of 2017 and 2018, while 15.25% (Purnendu), 16.53% (Swarnaprabha), 19.79% (Sasarang) and 40.47% (IR8) in rabi seasons of 2017 and 2018 (Fig. 2a). Similarly, reduction in SC (P < 0.05) under LL in comparison to NL was found to be 14.72%, 23.02%, 22.63% and 39.51% in kharif seasons of 2017 and 2018, while 19.35%, 19.35%, 25% and 24.24% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8. The reduction in GS was manifested in significantly reduced TR (mol H2O m−2 s−1) in all rice genotypes (P < 0.05) under LL stress (Fig. 2b). Reduction in TR was found to be 26.49%, 24.03%, 39.46% and 52.80% in kharif seasons of 2017 and 2018, while 24.33%, 26.89%, 44.07% and 58.64% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 under LL compared to NL (Fig. 2c). The Ci/Ca ratio was significantly increased (P < 0.05) in all the tested rice cultivars under LL stress resulting in decrease in the photosynthetic rate (Fig. 2d). The increment (P < 0.05) in Ci/Ca ratio under LL compared to NL was found to be 12.9%, 17.28%, 38.09% and 40.42% in kharif seasons of 2017 and 2018, while 10%, 15.18%, 30% and 37.77% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 2 and Supplementary Table 2).
Fig. 2.
Net assimilation rate (a), stomatal conductance (b), transpiration rate (c), and Ci/Ca (d) in tolerant (Purnendu and Swarnaprabha) and susceptible (Sasarang and IR8) genotypes, monitored by infrared gas analyzer (IRGA (Licor 6400-XT portable photosynthetic system) under normal light (NL) and low light (LL) conditions in ambient CO2 during rabi and kharif seasons of 2017 and 2018. Each data point is the average of six replicates and the error bars represent standard error (SE)
Chlorophyll a fluorescence
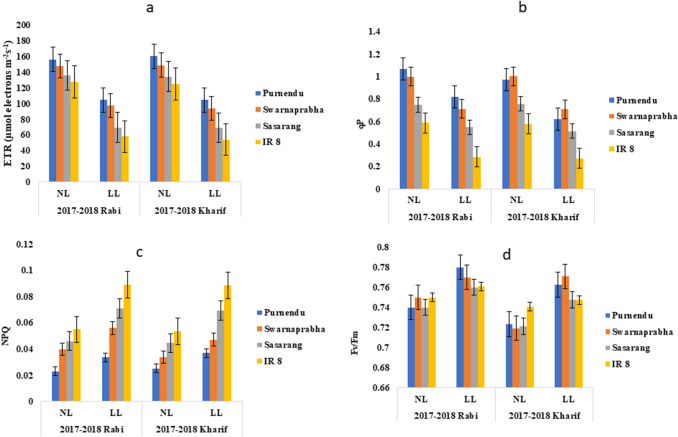
Chlorophyll a fluorescence measurement is used as a non-destructive and non-invasive signature of photosynthesis. Fv/Fm, ETR, qP were decreased, whereas NPQ was increased during kharif compared to rabi seasons for all the cultivars under LL stress (Fig. 3a–d). The maximum primary photochemical efficiency of PSII, which was measured as Fv/Fm, where Fv = Fm–F0, was significantly higher for all cultivars under LL in all the seasons (Fig. 3d). The significant increment (P < 0.05) in Fv/Fm ratio under LL in comparison to NL was found to be 5.43%, 7.211%, 3.63% and 0.922% in kharif seasons of 2017 and 2018, while 5.40%, 2.66%, 2.70% and 1.47% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 3d). Chlorophyll a fluorescence measurement revealed that the electron transport rate (ETR) (μmole electrons m−2 s−1) of PSII was decreased in response to LL significantly (P < 0.05) in all genotypes for all growing seasons (Fig. 3a). However, the decrease was greater in the susceptible genotypes than the tolerant ones. The decrease (P < 0.05) in ETR under LL in comparison to NL were observed as 34.67%, 36.98%, 48.34% and 56.70% in kharif seasons of 2017 and 2018, while 32.97%, 33.88%, 48.68% and 54.65% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 3a). The photochemical quenching (qP) was significantly lower for all the genotypes under LL in all the seasons. However, the decrease was greater in the susceptible genotypes than the tolerant ones. The decrement (P < 0.05) in qP under LL compared NL was found to be 36.20%, 29.25%, 32.21% and 52.92% in kharif 2017–2018, while 23.27%, 28.89%, 26.56% and 51.44% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 3b and Supplementary Table 3). Non-photochemical quenching (NPQ) increased in response to LL significantly (P < 0.05) in all genotypes for all grow seasons (Fig. 3c). However, the increase was greater in the susceptible genotypes than the tolerant ones. The increment (P < 0.05) in NPQ under LL in comparison to NL was found to be 47.01%, 38.72%, 56.17% and 65.73% in kharif seasons of 2017 and 2018, while 46.37%, 40%, 54.34% and 61.81% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 3c and Supplementary Table 3).
Fig. 3.
Chlorophyll-fluorescence measurements. Electron transfer rate (ETR) (a), photochemical quenching (qP) (b), non-photochemical quenching (NPQ) (c), Fv/Fm ratio (d) in tolerant (Purnendu and Swarnaprabha) and susceptible (Sasarang and IR8) genotypes, monitored by infrared gas analyzer (IRGA (Licor 6400-XT portable photosynthetic system) under normal light (NL) and low light (LL) conditions during rabi and kharif seasons of 2017 and 2018. Each data point is the average of six replicates and the error bars represent standard error (SE)
Soluble protein content in flag leaf
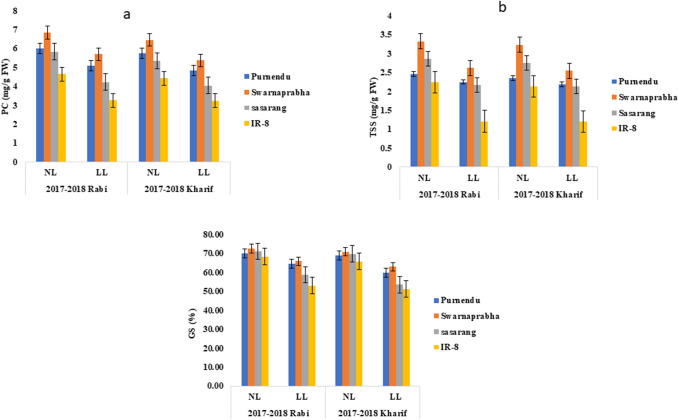
The soluble protein content of the flag leaf was estimated to find the possible relationship between photosynthetic rate and protein content. Flag leaf soluble protein decreased during kharif than rabi seasons in all genotypes. Under LL stress, there was a significant reduction (P < 0.05) in protein content of all the four genotypes. Tolerant genotypes Purnendu and Swarnaprabha showed lower reduction as compared to moderately susceptible genotype Sasarang and susceptible genotype, IR8 under LL stress as compared to NL condition for all the growth seasons. Reduction in protein content was observed as 16.08%,13.63%, 23.82% and 22.79% in kharif seasons of 2017 and 2018, while 15%, 16.67%, 27.62% and 29.96% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 4a and Supplementary Table 4a).
Fig. 4.
Total protein content (a), total soluble sugar of flag leaves (b) and grain starch (%) (c) in tolerant (Purnendu and Swarnaprabha) and susceptible (Sasarang and IR8)) genotypes. Plants were grown under normal light (NL) and low light (LL) conditions during rabi and kharif seasons of 2017 and 2018. Each data point is the average of six replicates and the error bars represent standard error (SE)
Total soluble sugar content in the flag leaf
The total soluble sugar content was estimated to compare the photosynthetic efficiency of the tested cultivars. Flag leaf total soluble sugar decreased during kharif than rabi seasons in all cultivars. LL stress caused reduction in the amount of TSS (P < 0.05) in the flag leaf irrespective of the cultivars for all the growth seasons. Reduction in protein content was found to be 7.02%, 21.05%, 22.4% and 43.79% in kharif seasons of 2017 and 2018, while 8.53%, 21.08%, 24.12% and 45.98% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 4b and Supplementary Table 4b).
Grain starch content
Grain starch content was estimated after the harvesting. Grain starch content was decreased during kharif compared to rabi seasons in all the genotypes. LL stress caused reduction in the amount of grain starch (P < 0.05) irrespective of the cultivars for all the growth seasons. Reduction in grain starch was pronounced in the Sasarang and IR8 than other tolerant genotypes. The reduction in starch content was found to be 13.33%, 11.14%, 23.30% and 22.25% in kharif seasons of 2017 and 2018, while 7.96%, 9.10%, 17.54% and 22.34% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8, respectively (Fig. 4c, Supplementary Table 4c).
Antioxidant enzymes
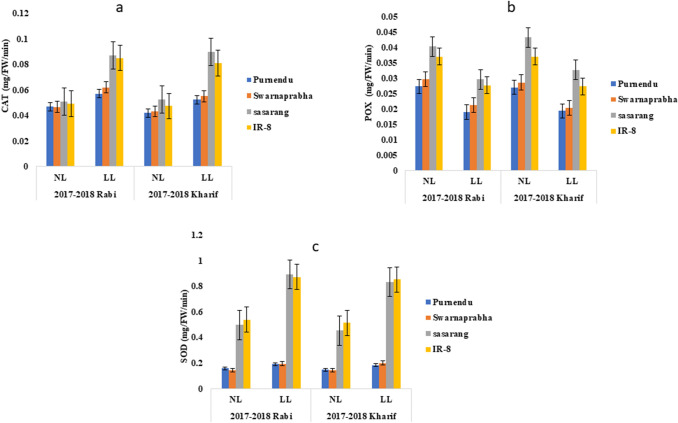
To understand the enzyme activity for detoxification of the reactive oxygen species (ROS) generated under LL stress, assays of superoxide dismutase (SOD), catalase (CAT) and peroxidase (POX) were performed. CAT activity showed negligible increase under LL in Purnendu and Swarnaprabha, whereas a sharp increase was observed in Sasarang and IR8. Increment in CAT was found to be 24.7%, 27%, 70.29% and 71.12% in kharif seasons of 2017 and 2018, while 21.27%, 32.83%, 70.60% and 72.29% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 5a and Supplementary Table 5). POX activity was decreased in all the genotypes tested under LL stress, however, the percent decrease was significant in Purnendu and Swarnaprabha than Sasarang and IR8. The decrease in POX was found to be 28.40%, 28.86%, 24.59% and 26.12% in kharif seasons of 2017 and 2018, while 30.46%, 27.99%, 26.42% and 25.16% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 5b and Supplementary Table 5). The SOD activity in Purnendu and Swarnaprabha under NL and LL conditions were significantly less than Sasarang and IR8. Increment in SOD was found to be 24.78%, 38.83%, 83.94% and 72.04% in kharif seasons of 2017 and 2018, while 21.02%, 35.64%, 79.94% and 61.80% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Fig. 5c and Supplementary Table 5).
Fig. 5.
Antioxidant enzyme activities. Catalase activity (a), peroxidase activity (b), superoxide dismutase activity (c) estimated from the flag leaf of tolerant (Purnendu and Swarnaprabha) and susceptible (Sasarang and IR8)) genotypes during 50% flowering stage from NL and LL conditions during rabi and kharif seasons of 2017 and 2018. Each data point is the average of six replicates and the error bars represent standard error (SE)
SBPase activity in rice flag leaf
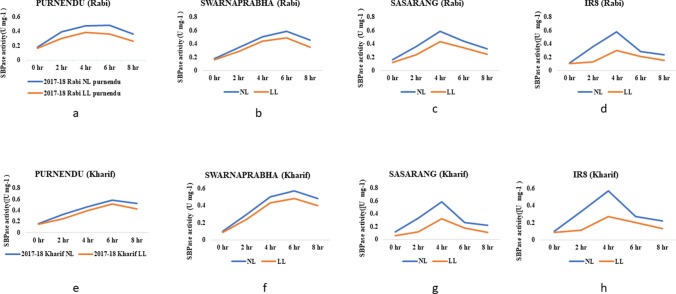
As SBPase is the principal enzyme for RuBP regeneration during the Calvin cycle, we measured its activity in flag leaves of LL tolerant genotypes, Purnendu and Swarnaprahbha, moderately susceptible genotype, Sasrang and susceptible genotype, IR8 under NL and LL stress conditions during rabi and kharif seasons of 2017 and 2018. The SBPase activity was reduced in all the four genotypes at the flowering stage under LL stress as compared to NL in all seasons (Fig. 6 and Supplementary Table 6). In tolerant genotypes (Purnendu and Swarnaprabha), SBPase activity was constantly increased up to the 6th hour of incubation and then non-significantly decreases irrespective of light condition. But, in case of susceptible genotypes (Sasarang and IR8), the SBPS activity was increased up to 4th hours of incubation and then significantly decreased (P < 0.05). The SBPase activity was maximum under NL than LL condition in all the genotypes. The SBPase activity was significantly reduced under LL compared to NL at 0th (5.45%), 2nd (22.36%), 4th (15.21%), 6th (11.24%) and 8th (19.13%) hour of incubations in in kharif 2017–2018 and similarly at 0th (8.45%), 2nd (24.01%), 4th (18.37%), 6th (24.58%) and 8th (27.63%) hour of incubations in rabi 2017–2018 in Purnendu (Fig. 6). In Swarnaprabha, under LL stress, the reduction was observed at 0th (7.22%), 2nd (21.59%), 4th (13.8%), 6th (15.67%) and 8th (16.84%) hour of incubations in kharif seasons of 2017 and 2018, and similarly at 0th (10.86%), 2nd (16.39%), 4th (13.61%), 6th (16.43%) and 8th (22.75%) hour of incubations during rabi seasons of 2017 and 2018 (Fig. 6). In Sasarang, under LL stress, the reduction was found to be at 0th (47.83%), 2nd (63.23%), 4th (45.5%), 6th (33.33%) and 8th (48.49%) hour of incubations in kharif seasons of 2017 and 2018, and similarly at 0th (22.5%), 2nd (33.23%), 4th (25.96%), 6th (21.86%) and 8th (24.49%) hour of incubations in rabi seasons of 2017 and 2018 (Fig. 6). In IR8, under LL stress, the reduction was found to be at 0th (11.94%), 2nd (68.02%), 4th (52.67%), 6th (24.58%) and 8th (37.88%) hour of incubations in kharif seasons of 2017 and 2018, and similarly at 0th (11.84%), 2nd (63.43%), 4th (48.70%), 6th (24.69%) and 8th (34.10%) hour of incubations in rabi seasons of 2017 and 2018 (Fig. 6).The critical observation was considered at 4th and 6th hour, where the difference in SBPase activity was highest. The SBPase activity in tolerant and susceptible genotypes at 4th and 6th hour was statistically tested (T-test). We observed significant differences in both types of genotypes for SBPase activity, especially under the LL. In case of tolerant genotypes, the SBPase activity at the 4th hour was reduced up to 15.21% in kharif and 18.37% in rabi, whereas the reduction was up to 52.67% in kharif and 48.70% in rabi under LL condition for susceptible genotypes. There was an increase in enzyme activity both under NL and LL conditions after 4th hour of incubation in tolerant genotypes (Purnendu and Swarnaprabha). In contrast, the enzyme activity was drastically reduced after the 4th hour, both under NL and LL conditions in susceptible genotypes (Sasarang and IR8). Even at the 6th hour, SBPase activity in tolerant genotypes (Purnendu and Swarnaprabha) was effectively maintained under NL and LL conditions, whereas it was significantly reduced in susceptible genotypes (Sasarang and IR8) (Supplementary Table 6).
Fig. 6.
Sedoheptulose 1,7 bisphosphatase(SBPase) activity under NL and LL conditions. Purnendu (a), Swarnaprabha (b), Sasarang (c), IR8 (d). The assay of SBPase activity was carried out at 0, 4, 2, 6, and 8 h from the flag leaf collected from NL and LL conditions during the flowering stage in rabi and kharif seasons of 2017 and 2018. Each data point is the average of six replicates and the error bars represent standard error (SE)
Real time expression of SBPase gene in rice flag leaf
To further check if SBPase activity is influenced by LL stress, we investigated the expression pattern of SBPase transcript in rice plants through quantitative real-time PCR analysis. The data suggested that the primary process of biosynthesis of SBPase protein was optimum during 50% flowering, when net photosynthetic rate is expected to be high. The LL stress reduced the expression of the SBPase in all genotypes as compared to NL (Fig. 7). Tolerant genotypes, Purnendu (0.55 time; 44.8%) and Swarnaprabha (0.40 time; 52.61%) showed comparatively lower down regulation in SBPase expression as compared to susceptible genotypes, Sasarang (P < 0.05) (0.36 time; 63.33%) and IR-8 (0.35 time; 64.33%) under LL stress.
Fig. 7.
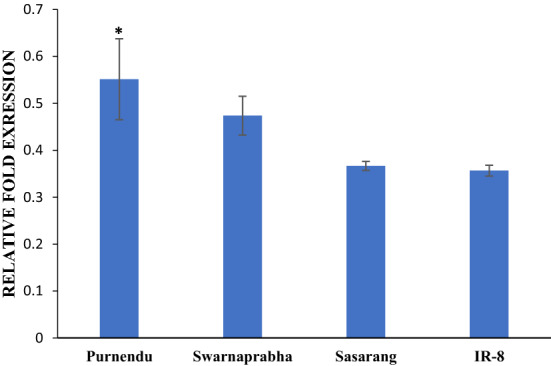
Expression pattern of SBPase gene at flowering stage in tolerant (Purnendu and Swarnaprabha) and susceptible (Sasarang and IR8)) genotypes under normal light (NL) as compared to low light (LL) condition. The expression level was determined by qRTPCR. β-tubulin was used as the internal standard and leaf SBPase was used for expression value normalization at the flowering stage. Each value represents the mean standard error (SE) (n6)
Grain Yield and related traits
Grain yield and yield components for the growth seasons of rabi and kharif seasons of 2017 and 2018 under NL and LL are shown in Supplementary Table 7 (a–f). Reduction in grain yield was significant (P < 0.05) between two tolerant (Purnendu, Swarnaprabha) and two susceptible (Sasarang, IR8) genotypes under LL stress in kharif and rabi seasons, however, the percent reduction was pronounced in Sasarang and IR8. Decrement in grain yield was observed as 13.02%, 17.86%, 42.17% and 53.09% in kharif seasons of 2017 and 2018, while 10.44%,14%, 37.90% and 45.92% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8 (Supplementary Table 7a). The significant reduction (P < 0.05) was found in panicle/plant (Supplementary Table 7f), tiller/plant (Supplementary Table 7e), total biomass/plant (Supplementary Table 7b), spikelet fertility percent (Supplementary Table 7d) and 1000-grain weight (Supplementary Table 7c) under LL stress. Reduction in total biomass/plant (P < 0.05) was observed as 7.85%, 16.98%, 39.64% and 51.74% in kharif seasons of 2017 and 2018, while 7.5%, 14%, 37.99% and 46.02% in rabi seasons of 2017 and 2018, respectively in Purnendu, Swarnaprabha, Sasarang and IR8. Rate of panicle emergence (RPE) was higher in rabi than kharif seasons. In all the growth seasons, RPE was significantly reduced in all the cultivars under low light but was pronounced in Sasarang and IR8 (Supplementary Table 8).
Analysis of variances (ANOVA)
Analysis of variance (ANOVA) refers to the observable differences for a particular trait. ANOVA was performed using Tukey-Kramer method. To study the ANOVA, total seven treatments were prepared by taking three factors into consideration, i.e., season, light and genotypes. Average season data was taken as treatment one irrespective of light and genotypes. For treatment two, average light conditions data was taken. Similarly, season x light data as taken as treatment three, average genotype data was taken as treatment four, season x variety and light x variety data were taken as treatment five and six, respectively. Season x light x variety data were taken as treatment seven. To know the extent of variations of observed characters, all the seven treatments were taken for analysis, and analysis of variance was performed. The results of ANOVA revealed highly significant mean sum of squares for all the 22 traits, suggesting the presence of sufficient variations among genotypes for these traits. Analysis of variance for treatment one indicated significant variation for 20 out of 22 traits except for non-photochemical quenching and total soluble sugar. Except for Ci/Ca, all the 21 traits showed significant variation for treatment two. For treatment three, a total of 13 traits showed significant variation (Net assimilation rate, transpiration rate, Ci/Ca, Electron transfer rate, non-photochemical quenching, SBPase at 4th hr, SBPase at 6th hr, catalase, peroxidase, grain starch (%), 1000-grain weight, spikelet fertility percentage and tiller number). For treatment four, a total of 20 traits revealed significant variation except for stomatal conductance and Fv/Fm. Except for stomatal conductance, Ci/Ca and Fv/Fm, rest of 19 traits showed significant variation for treatment six. A total of eleven traits, i.e., net assimilation rate, electron transfer rate, photochemical quenching, SBPase at 4th hr, SBPase at 6th hr, catalase, peroxidase, grain starch, 1000-grain weight, spikelet fertility percentage and panicle number revealed significant variation for treatment seven (Table 1).
Table 1.
Analysis of variance (ANOVA) for physiological, biochemical and yield-related traits in tolerant and susceptible rice genotypes grown under normal light and low light conditions during rabi and kharif seasons of 2017 and 2018
| DF | NAR | SC | TR | Ci/Ca | ETR | qP | NPQ | FV/FM | SBP4hr | SBP6hr | PC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Season Treatment | 1.000 | 0.000 | 0.008 | 0.046 | 0.001 | 0.052 | 0.000 | 0.155 | 0.010 | 0.000 | 0.000 | 0.000 |
| Light *Treatment | 1.000 | 0.000 | 0.000 | 0.000 | 0.542 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Season * Light | 1.000 | 0.004 | 0.946 | 0.041 | 0.000 | 0.011 | 0.509 | 0.049 | 0.543 | 0.011 | 0.000 | 0.265 |
| Variety *Treatment | 3.000 | 0.000 | 0.801 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.431 | 0.000 | 0.000 | 0.000 |
| Season * Variety | 3.000 | 0.000 | 0.225 | 0.422 | 0.148 | 0.001 | 0.000 | 0.019 | 0.856 | 0.000 | 0.000 | 0.531 |
| Light * Variety | 3.000 | 0.000 | 0.567 | 0.000 | 0.979 | 0.000 | 0.001 | 0.000 | 0.232 | 0.000 | 0.000 | 0.026 |
| Season * Light * Variety | 3.000 | 0.001 | 0.754 | 0.186 | 0.265 | 0.048 | 0.007 | 0.378 | 0.436 | 0.000 | 0.000 | 0.812 |
| SE | 32.000 | 0.271 | 0.007 | 0.287 | 0.003 | 2.106 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.048 |
| Total | 47.000 | 17.036 | 0.013 | 11.736 | 0.036 | 1208.67 | 0.066 | 0.000 | 0.001 | 0.009 | 0.020 | 1.089 |
| Catalase | Peroxidase | SOD | TSS | GS | GYLD | BIOM | TGW | SFP | TN | PN | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Season Treatment | 0.000 | 0.000 | 0.000 | 0.102 | 0.000 | 0.000 | 0.000 | 0.023 | 0.000 | 0.000 | 0.000 |
| Light *Treatment | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Season * Light | 0.021 | 0.000 | 0.216 | 0.508 | 0.001 | 0.816 | 0.431 | 0.001 | 0.001 | 0.042 | 0.352 |
| Variety *Treatment | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Season * Variety | 0.000 | 0.000 | 0.838 | 0.000 | 0.460 | 0.000 | 0.000 | 0.012 | 0.000 | 0.006 | 0.279 |
| Light * Variety | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Season * Light * Variety | 0.000 | 0.000 | 0.054 | 0.554 | 0.033 | 0.377 | 0.452 | 0.000 | 0.042 | 0.081 | 0.000 |
| SE | 0.000 | 0.000 | 0.000 | 0.023 | 0.992 | 0.164 | 0.589 | 0.014 | 0.206 | 0.058 | 0.023 |
| Total | 0.026 | 0.001 | 0.089 | 0.545 | 47.665 | 13.668 | 53.593 | 8.321 | 178.352 | 2.874 | 3.344 |
DF degree of freedom, SE standard error, NAR net assimilation rate (µmol CO2 m−2 s−1), SC stomatal conductance (µmol H2O m−2 s−1), TR transpiration rate (µmol H2O m−2 s−1), Ci/Ca ratio of internal to atmospheric CO2 concentration, ETR electron transfer rate (µmol electrons m−2 s−1), qP photochemical quenching, NPQ non-photochemical quenching, FV/FM variance fluorescence/maximal fluorescence, SBP4hr SBPase activity at 4th hour (U mg−1), SBP6hr SBPase activity at 6th hour (U mg−1), PC protein content (mg/gFW), SOD superoxide dismutase (mg/FW/min), TSS total soluble sugar(mg/gFW), GS grain starch (%), GYLD grain yield/plant (g), BIOM biomass/plant (g), SFP spikelet fertility percentage, TN tiller number/plant, PN panicle number/plant
T test: *Significant at 0.05 level, ** Significant at 0.01 level, *** Significant at 0.001 level
Trait association and genetic variability
The association analysis was estimated to establish a relationship between grain yield and SBPS activity under different light situations. In the case of tolerant genotypes, the grain yield exhibited significantly positive association with SBPase activity at both 4th hour and 6th hour under LL, but it was non-significant for susceptible genotypes. Under NL, the tolerant genotypes showed non-significant association with SBPase activity at both hours, whereas SBPase activity at 4th hour and 6th hour showed a significant correlation with grain yield in susceptible genotypes (Table 2).
Table 2.
Phenotypic correlation for physiological, biochemical and yield-related traits in tolerant and susceptible rice genotypes grown under normal light and low light conditions
| Normal light (tolerant) | Normal light (susceptible) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAR | SC | TR | Ci/Ca | ETR | qP | NPQ | FV/FM | SBP4hr | S SBP6hr | PC | Catalase | Peroxidase | SOD | TSS | GS | GYLD | BIOM | TGW | SFP | TN | PN | |
| NAR | 0.286 | 0.174 | 0.777 | 0.083 | 0.077 | 0.003 | 0.370 | 0.240 | 0.417 | 0.079 | 0.176 | 0.740 | 0.163 | 0.770 | 0.078 | 0.499 | 0.429 | 0.057 | 0.093 | 0.116 | 0.053 | |
| SC | 0.955 | 0.688 | 0.214 | 0.995 | 0.610 | 0.560 | 0.260 | 0.844 | 0.074 | 0.649 | 0.460 | 0.551 | 0.990 | 0.006 | 0.302 | 0.107 | 0.133 | 0.931 | 0.318 | 0.298 | 0.670 | |
| TR | 0.550 | 0.140 | 0.127 | 0.002 | 0.003 | 0.001 | 0.133 | 0.419 | 0.358 | 0.003 | 0.366 | 0.133 | 0.003 | 0.843 | 0.042 | 0.014 | 0.004 | 0.001 | 0.061 | 0.268 | 0.006 | |
| Ci/Ca | 0.322 | 0.149 | 0.002 | 0.939 | 0.979 | 0.214 | 0.024 | 0.593 | 0.033 | 0.897 | 0.823 | 0.446 | 0.244 | 0.104 | 0.511 | 0.000 | 0.003 | 0.417 | 0.292 | 0.089 | 0.878 | |
| ETR | 0.549 | 0.138 | 0.000 | 0.001 | 0.001 | 0.005 | 0.256 | 0.161 | 0.063 | 0.000 | 0.261 | 0.026 | 0.004 | 0.916 | 0.001 | 0.209 | 0.121 | 0.002 | 0.000 | 0.002 | 0.000 | |
| qP | 0.077 | 0.427 | 0.124 | 0.002 | 0.067 | 0.003 | 0.406 | 0.232 | 0.021 | 0.000 | 0.209 | 0.016 | 0.006 | 0.448 | 0.001 | 0.266 | 0.163 | 0.003 | 0.000 | 0.016 | 0.000 | |
| NPQ | 0.694 | 0.865 | 0.003 | 0.001 | 0.006 | 0.041 | 0.185 | 0.429 | 0.402 | 0.006 | 0.161 | 0.322 | 0.002 | 0.980 | 0.017 | 0.035 | 0.024 | 0.001 | 0.047 | 0.129 | 0.004 | |
| FV/FM | 0.026 | 0.968 | 0.597 | 0.202 | 0.485 | 0.065 | 0.634 | 0.916 | 0.272 | 0.566 | 0.831 | 0.688 | 0.068 | 0.034 | 0.523 | 0.021 | 0.021 | 0.127 | 0.601 | 0.929 | 0.297 | |
| SBP4hr | 0.799 | 0.849 | 0.010 | 0.058 | 0.023 | 0.284 | 0.016 | 0.698 | 0.128 | 0.158 | 0.423 | 0.549 | 0.584 | 0.987 | 0.360 | 0.048 | 0.603 | 0.359 | 0.196 | 0.075 | 0.229 | |
| SBP6hr | 0.184 | 0.947 | 0.093 | 0.026 | 0.204 | 0.048 | 0.006 | 0.183 | 0.093 | 0.015 | 0.686 | 0.091 | 0.457 | 0.017 | 0.026 | 0.013 | 0.265 | 0.195 | 0.004 | 0.005 | 0.040 | |
| PC | 0.995 | 0.667 | 0.012 | 0.014 | 0.005 | 0.071 | 0.003 | 0.222 | 0.004 | 0.123 | 0.164 | 0.035 | 0.012 | 0.552 | 0.002 | 0.338 | 0.230 | 0.002 | 0.000 | 0.004 | 0.000 | |
| Catalase | 0.069 | 0.768 | 0.090 | 0.649 | 0.286 | 0.866 | 0.124 | 0.528 | 0.336 | 0.140 | 0.342 | 0.849 | 0.771 | 0.692 | 0.046 | 0.570 | 0.444 | 0.198 | 0.276 | 0.430 | 0.197 | |
| Peroxidase | 0.783 | 0.170 | 0.102 | 0.020 | 0.006 | 0.070 | 0.184 | 0.352 | 0.125 | 0.931 | 0.055 | 0.134 | 0.039 | 0.774 | 0.082 | 0.547 | 0.419 | 0.052 | 0.026 | 0.058 | 0.012 | |
| SOD | 0.022 | 0.861 | 0.073 | 0.977 | 0.335 | 0.309 | 0.437 | 0.562 | 0.363 | 0.514 | 0.470 | 0.003 | 0.447 | 0.622 | 0.047 | 0.027 | 0.017 | 0.001 | 0.030 | 0.101 | 0.003 | |
| TSS | 0.073 | 0.151 | 0.001 | 0.048 | 0.001 | 0.495 | 0.047 | 0.782 | 0.048 | 0.655 | 0.035 | 0.131 | 0.064 | 0.050 | 0.638 | 0.092 | 0.095 | 0.823 | 0.462 | 0.664 | 0.869 | |
| GS | 0.603 | 0.243 | 0.001 | 0.000 | 0.001 | 0.066 | 0.000 | 0.346 | 0.009 | 0.017 | 0.012 | 0.545 | 0.023 | 0.596 | 0.025 | 0.688 | 0.447 | 0.006 | 0.000 | 0.001 | 0.000 | |
| GYLD | 0.571 | 0.355 | 0.000 | 0.001 | 0.000 | 0.077 | 0.001 | 0.456 | 0.063 | 0.154 | 0.001 | 0.190 | 0.005 | 0.225 | 0.002 | 0.000 | 0.000 | 0.041 | 0.985 | 0.535 | 0.294 | |
| BIOM | 0.367 | 0.345 | 0.001 | 0.005 | 0.000 | 0.139 | 0.001 | 0.601 | 0.005 | 0.218 | 0.002 | 0.176 | 0.006 | 0.176 | 0.001 | 0.000 | 0.000 | 0.020 | 0.731 | 0.763 | 0.178 | |
| TGW | 0.179 | 0.461 | 0.002 | 0.013 | 0.000 | 0.218 | 0.001 | 0.871 | 0.014 | 0.199 | 0.006 | 0.099 | 0.158 | 0.074 | 0.001 | 0.004 | 0.002 | 0.001 | 0.003 | 0.054 | 0.001 | |
| SPF | 0.057 | 0.500 | 0.002 | 0.071 | 0.003 | 0.492 | 0.019 | 0.845 | 0.019 | 0.335 | 0.016 | 0.005 | 0.182 | 0.004 | 0.000 | 0.038 | 0.002 | 0.003 | 0.002 | 0.000 | 0.000 | |
| TN | 0.000 | 0.738 | 0.790 | 0.120 | 0.929 | 0.012 | 0.494 | 0.067 | 0.951 | 0.270 | 0.712 | 0.046 | 0.252 | 0.004 | 0.251 | 0.407 | 0.989 | 0.784 | 0.444 | 0.149 | 0.001 | |
| PN | 0.004 | 0.740 | 0.644 | 0.289 | 0.843 | 0.083 | 0.978 | 0.022 | 0.968 | 0.997 | 0.654 | 0.004 | 0.047 | 0.000 | 0.371 | 0.549 | 0.953 | 0.869 | 0.465 | 0.159 | 0.004 | |
| Low light (tolerant) | Low light (susceptible) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAR | SC | TR | Ci/Ca | ETR | qP | NPQ | FV/FM | SBP4hr | S SBP6hr | PC | Catalase | Peroxidase | SOD | TSS | GS | GYLD | BIOM | TGW | SFP | TN | PN | |
| NAR | 0.000 | 0.006 | 0.004 | 0.000 | 0.001 | 0.000 | 0.015 | 0.000 | 0.011 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.001 | 0.054 | 0.000 | 0.000 | 0.007 | |
| SC | 0.002 | 0.006 | 0.001 | 0.000 | 0.003 | 0.000 | 0.019 | 0.001 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.002 | 0.063 | 0.000 | 0.000 | 0.020 | |
| TR | 0.002 | 0.001 | 0.000 | 0.002 | 0.296 | 0.004 | 0.022 | 0.002 | 0.179 | 0.085 | 0.001 | 0.018 | 0.004 | 0.123 | 0.004 | 0.000 | 0.000 | 0.000 | 0.028 | 0.024 | 0.601 | |
| Ci/Ca | 0.262 | 0.161 | 0.002 | 0.001 | 0.110 | 0.001 | 0.048 | 0.002 | 0.032 | 0.014 | 0.001 | 0.007 | 0.002 | 0.043 | 0.001 | 0.001 | 0.001 | 0.002 | 0.015 | 0.008 | 0.242 | |
| ETR | 0.000 | 0.001 | 0.008 | 0.381 | 0.010 | 0.000 | 0.010 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.001 | 0.014 | 0.000 | 0.000 | 0.054 | |
| qP | 0.002 | 0.001 | 0.001 | 0.056 | 0.002 | 0.002 | 0.115 | 0.045 | 0.004 | 0.000 | 0.032 | 0.000 | 0.026 | 0.000 | 0.002 | 0.045 | 0.042 | 0.820 | 0.000 | 0.000 | 0.000 | |
| NPQ | 0.054 | 0.073 | 0.913 | 0.115 | 0.026 | 0.217 | 0.013 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.002 | 0.046 | 0.000 | 0.000 | 0.022 | |
| FV/FM | 0.070 | 0.153 | 0.173 | 0.400 | 0.084 | 0.138 | 0.610 | 0.013 | 0.069 | 0.051 | 0.011 | 0.018 | 0.015 | 0.104 | 0.011 | 0.015 | 0.019 | 0.059 | 0.061 | 0.077 | 0.289 | |
| SBP4hr | 0.003 | 0.001 | 0.000 | 0.067 | 0.003 | 0.000 | 0.193 | 0.137 | 0.001 | 0.001 | 0.001 | 0.002 | 0.000 | 0.002 | 0.000 | 0.081 | 0.001 | 0.002 | 0.001 | 0.000 | 0.113 | |
| SBP6hr | 0.002 | 0.002 | 0.000 | 0.087 | 0.003 | 0.000 | 0.182 | 0.117 | 0.000 | 0.000 | 0.005 | 0.000 | 0.000 | 0.006 | 0.001 | 0.067 | 0.013 | 0.456 | 0.016 | 0.006 | 0.016 | |
| PC | 0.006 | 0.001 | 0.000 | 0.001 | 0.016 | 0.000 | 0.986 | 0.227 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.003 | 0.004 | 0.368 | 0.000 | 0.000 | 0.001 | |
| Catalase | 0.868 | 0.626 | 0.047 | 0.000 | 0.768 | 0.398 | 0.012 | 0.846 | 0.493 | 0.543 | 0.034 | 0.001 | 0.002 | 0.007 | 0.001 | 0.001 | 0.000 | 0.004 | 0.001 | 0.001 | 0.101 | |
| Peroxidase | 0.012 | 0.003 | 0.000 | 0.001 | 0.028 | 0.001 | 0.976 | 0.392 | 0.001 | 0.002 | 0.000 | 0.043 | 0.000 | 0.000 | 0.000 | 0.002 | 0.003 | 0.126 | 0.000 | 0.000 | 0.009 | |
| SOD | 0.000 | 0.002 | 0.006 | 0.258 | 0.001 | 0.001 | 0.078 | 0.349 | 0.000 | 0.000 | 0.011 | 0.985 | 0.009 | 0.000 | 0.000 | 0.002 | 0.002 | 0.006 | 0.000 | 0.000 | 0.073 | |
| TSS | 0.105 | 0.079 | 0.000 | 0.000 | 0.158 | 0.009 | 0.366 | 0.393 | 0.014 | 0.016 | 0.000 | 0.000 | 0.001 | 0.050 | 0.000 | 0.016 | 0.013 | 0.484 | 0.000 | 0.000 | 0.000 | |
| GS | 0.003 | 0.001 | 0.000 | 0.021 | 0.002 | 0.001 | 0.408 | 0.246 | 0.001 | 0.001 | 0.000 | 0.226 | 0.001 | 0.002 | 0.005 | 0.001 | 0.001 | 0.049 | 0.000 | 0.000 | 0.013 | |
| GYLD | 0.044 | 0.123 | 0.000 | 0.000 | 0.232 | 0.031 | 0.245 | 0.497 | 0.030 | 0.036 | 0.000 | 0.001 | 0.002 | 0.080 | 0.000 | 0.016 | 0.000 | 0.000 | 0.001 | 0.001 | 0.158 | |
| BIOM | 0.147 | 0.126 | 0.001 | 0.000 | 0.237 | 0.028 | 0.238 | 0.510 | 0.030 | 0.036 | 0.000 | 0.001 | 0.001 | 0.083 | 0.000 | 0.014 | 0.000 | 0.000 | 0.001 | 0.000 | 0.159 | |
| TGW | 0.049 | 0.040 | 0.000 | 0.000 | 0.084 | 0.008 | 0.407 | 0.324 | 0.009 | 0.014 | 0.000 | 0.000 | 0.000 | 0.076 | 0.000 | 0.002 | 0.000 | 0.000 | 0.201 | 0.150 | 0.738 | |
| SPF | 0.093 | 0.173 | 0.711 | 0.039 | 0.065 | 0.488 | 0.000 | 0.409 | 0.375 | 0.379 | 0.577 | 0.002 | 0.557 | 0.199 | 0.138 | 0.674 | 0.081 | 0.082 | 0.163 | 0.000 | 0.001 | |
| TN | 0.018 | 0.022 | 0.295 | 0.803 | 0.010 | 0.030 | 0.006 | 0.696 | 0.034 | 0.036 | 0.406 | 0.146 | 0.359 | 0.003 | 0.705 | 0.077 | 0.844 | 0.839 | 0.829 | 0.012 | 0.002 | |
| PN | 0.005 | 0.003 | 0.049 | 0.537 | 0.003 | 0.028 | 0.086 | 0.217 | 0.018 | 0.042 | 0.062 | 0.796 | 0.120 | 0.115 | 0.406 | 0.019 | 0.475 | 0.465 | 0.159 | 0.176 | 0.225 | |
NAR net assimilation rate (µmol CO2 m−2 s−1), SC stomatal conductance (µmol H2O m−2 s−1), TR transpiration rate (µmol H2O m−2 s−1), Ci/Ca ratio of internal to atmospheric CO2 concentration, ETR electron transfer rate (µmol electrons m−2 s−1), qP photochemical quenching, NPQ non-photochemical quenching, FV/FM variance fluorescence/maximal fluorescence, SBP4hr SBPase activity at 4th hour (U mg−1), SBP6hr SBPase activity at 6th hour (U mg−1), PC protein content (mg/gFW), SOD superoxide dismutase (mg/FW/min), TSS total soluble sugar (mg/gFW), GS grain starch (%), GYLD grain yield/plant (g), BIOM biomass/plant (g), SFP spikelet fertility percentage, TN tiller number/plant, PN Panicle number/plant
T test: * Significant at 0.05 level,** Significant at 0.01 level, *** Significant at 0.001 level
Besides, under LL, SBPase activity, both at 4th hour and 6th hour has significant positive correlation with net assimilation rate, stomatal conductance, transpiration rate, ETR, qP, protein content, peroxidase, SOD, total soluble sugar, grain starch, total biomass, TGW, TN and PN in tolerant and susceptible genotypes under both the conditions. Grain yield showed significant positive correlation with net assimilation rate, transpiration rate, Ci/Ca, qP, protein content, catalase, peroxidase, total soluble sugar, grain starch, biomass, TGW and panicle number under LL condition in tolerant genotypes (Table 2). Along with standard statistical measures, heritable and non-heritable components of variance in the form of genotypic coefficient of variation (GCV), phenotypic coefficient of variation (PCV), heritability (broad sense, h2) and genetic advance as a percentage of mean (GA at 5%) was estimated. The highest estimate of GCV (89.05, 88.06) and PCV (89.08, 88.27) in kharif seasons, while GCV (137.13, 93.20) and PCV (137.19, 93.19) in rabi seasons were observed for catalase under NL and LL, respectively. Similarly, the lowest estimate of GCV (1.31, 1.40) in kharif seasons, while GCV (1.01, 1.80) in rabi seasons were observed for FV/FM under NL and LL conditions, respectively. For PCV, FV/FM also showed the lowest value (1.34, 1.55) in kharif seasons, while 0.67 and 1.24) in rabi seasons under NL and LL, respectively. The reliability of the phenotypic values depend on the estimates of heritability for a particular character. Therefore, high heritability helps in the effective selection of a particular character. The highest broad-sense heritability for TGW was observed (0.99, 0.99) under NL and LL in kharif seasons, while TGW, ETR showed highest value (i.e. 0.99, 0.99) under NL and LL, respectively in rabi seasons. The lowest heritability was -observed for stomatal conductance (0.14, 0.22) both under NL and LL in kharif seasons. In rabi seasons, the lowest heritability was observed for Fv/FM (-2.25, -2.24) under NL and LL, respectivelly. In kharif seasons, the highest estimate of GA (5%) was observed for peroxidase (183.39,180.99) under NL and LL. Similarly, in rabi seasons, while the highest estimate of GA(5%) was observed for catalase, i.e., 282.37, 191.81 under NL and LL conditions, respectively. The lowest GA (5%) was observed for FV/FM under NL and LL in both the seasons (Table 3). The SBPase activity inheritability (h2) and genetic advance is the major criteria for trait selection. In the present experiment, we observed h2 coupled with GA (5%) ≥ 0.30 for most of the traits including SBPase activity at 4th and 6th hours. This implies an indication for the tolerance activity in genotypes in relation to SBPase activity. High h2 and GA with SBPase activity, enables us for the easy selection of traits while breeding LL tolerance in rice through recombination breeding.
Table 3.
Genetic variability parameters for physiological, biochemical and yield-related traits in tolerant and susceptible rice genotypes grown under normal light and low light conditions during kharif and rabi seasons of 2017 and 2018
| NAR | SC | TR | Ci/Ca | ETR | qP | NPQ | FV/FM | SBP 4 h | SBP 6 h | PC | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic parameters (summary) | |||||||||||
| Rabi | |||||||||||
| GCV | |||||||||||
| NL | 5.719 | 2.837 | 18.504 | 29.116 | 10.999 | 23.936 | 31.386 | 1.316 | 14.926 | 31.081 | 3.088 |
| LL | 22.642 | 5.162 | 34.181 | 22.612 | 28.516 | 35.632 | 27.735 | 1.4 | 20.917 | 48.809 | 9.575 |
| PCV | |||||||||||
| NL | 5.839 | 7.547 | 18.756 | 30.036 | 11.008 | 23.95 | 31.396 | 1.348 | 15.382 | 31.214 | 3.161 |
| LL | 22.791 | 11.058 | 34.528 | 23.541 | 28.543 | 35.636 | 27.747 | 1.553 | 21.174 | 48.902 | 9.611 |
| h2 (broad sense) | |||||||||||
| NL | 0.96 | 0.141 | 0.973 | 0.94 | 0.998 | 0.999 | 0.999 | 0.952 | 0.942 | 0.992 | 0.954 |
| LL | 0.987 | 0.218 | 0.98 | 0.923 | 0.998 | 1 | 0.999 | 0.812 | 0.976 | 0.996 | 0.993 |
| GA (5%) | |||||||||||
| NL | 11.541 | 2.197 | 37.606 | 58.144 | 22.638 | 49.278 | 64.637 | 2.645 | 29.837 | 63.754 | 6.214 |
| LL | 46.339 | 4.964 | 69.706 | 44.74 | 58.686 | 73.396 | 57.111 | 2.599 | 42.567 | 100.355 | 19.652 |
| Genetic parameters (summary) | |||||||||||
| Kharif | |||||||||||
| GCV | |||||||||||
| NL | 1.097 | 7.056 | 18.968 | 25.863 | 8.83 | 28.553 | 29.387 | 1.01 | 10.143 | 27.851 | 15.533 |
| LL | 16.44 | 3.565 | 36.118 | 34.08 | 26.979 | 39.044 | 30.08 | 1.868 | 2.618 | 8.87 | 23.351 |
| PCV | |||||||||||
| NL | 1.416 | 7.215 | 18.974 | 25.903 | 8.831 | 28.829 | 29.979 | 0.673 | 10.251 | 27.914 | 15.555 |
| LL | 16.48 | 3.922 | 36.126 | 34.279 | 26.981 | 39.073 | 30.38 | 1.247 | 2.805 | 11.292 | 23.385 |
| h2 (broad sense) | |||||||||||
| NL | 0.599 | 0.956 | 0.999 | 0.997 | 1 | 0.981 | 0.961 | -2.25 | 0.979 | 0.995 | 0.997 |
| LL | 0.995 | 0.826 | 1 | 0.988 | 1 | 0.999 | 0.98 | -2.242 | 0.871 | 0.617 | 0.997 |
| GA (5%) | |||||||||||
| NL | 1.749 | 14.216 | 39.06 | 53.195 | 18.187 | 58.257 | 59.342 | -3.121 | 20.673 | 57.242 | 31.952 |
| LL | 33.785 | 6.676 | 74.387 | 69.798 | 55.574 | 80.371 | 61.352 | -5.763 | 5.031 | 14.353 | 48.034 |
| Catalase | Peroxidase | SOD | TSS | GS | GYLD | Biomass | TGW | SFP | TN | PN | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic parameters (summary) | |||||||||||
| Rabi | |||||||||||
| GCV | |||||||||||
| NL | 0 | 89.055 | 63.752 | 10.356 | 45.399 | 13.441 | 22.435 | 17.998 | 11.19 | 7.992 | 16.955 |
| LL | 62.297 | 88.069 | 73.296 | 20.158 | 62.59 | 32.463 | 35.246 | 33.281 | 13.448 | 29.729 | 33.889 |
| PCV | |||||||||||
| NL | 6.491 | 89.087 | 63.808 | 10.451 | 45.485 | 13.471 | 22.459 | 17.999 | 11.194 | 7.995 | 16.982 |
| LL | 62.354 | 88.279 | 73.328 | 20.278 | 62.612 | 32.487 | 35.254 | 33.284 | 13.458 | 29.73 | 33.901 |
| h2 (broad sense) | |||||||||||
| NL | 0 | 0.999 | 0.998 | 0.982 | 0.996 | 0.995 | 0.998 | 1 | 0.999 | 0.999 | 0.997 |
| LL | 0.998 | 0.995 | 0.999 | 0.988 | 0.999 | 0.999 | 1 | 1 | 0.999 | 1 | 0.999 |
| GA (5%) | |||||||||||
| NL | 0 | 183.388 | 131.215 | 21.141 | 93.346 | 27.625 | 46.166 | 37.072 | 23.041 | 16.46 | 34.87 |
| LL | 128.213 | 180.993 | 150.922 | 41.278 | 128.893 | 66.825 | 72.589 | 68.552 | 27.683 | 61.242 | 69.789 |
| Genetic parameters (summary) | |||||||||||
| Kharif | |||||||||||
| GCV | |||||||||||
| NL | 137.131 | 20.126 | 59.889 | 17.468 | 2.451 | 12.519 | 12.518 | 10.666 | 7.378 | 7.678 | 21.183 |
| LL | 93.206 | 18.738 | 73.209 | 29.051 | 9.678 | 23.66 | 23.768 | 14.697 | 25.742 | 24.959 | 28.514 |
| PCV | |||||||||||
| NL | 137.19 | 20.333 | 59.908 | 17.473 | 2.552 | 12.65 | 12.649 | 10.667 | 7.391 | 7.883 | 21.234 |
| LL | 93.299 | 18.887 | 73.214 | 29.148 | 9.775 | 23.853 | 23.956 | 14.698 | 25.747 | 25.166 | 28.543 |
| h2 (broad sense) | |||||||||||
| NL | 0.999 | 0.98 | 0.999 | 0.999 | 0.923 | 0.979 | 0.979 | 1 | 0.996 | 0.949 | 0.995 |
| LL | 0.998 | 0.984 | 1 | 0.993 | 0.98 | 0.984 | 0.984 | 1 | 1 | 0.984 | 0.998 |
| GA (5%) | |||||||||||
| NL | 282.367 | 41.037 | 123.333 | 35.975 | 4.85 | 25.522 | 25.52 | 21.971 | 15.171 | 15.405 | 43.534 |
| LL | 191.812 | 38.295 | 150.801 | 59.644 | 19.74 | 48.344 | 48.579 | 30.274 | 53.021 | 50.992 | 58.68 |
NAR net assimilation rate (µmol CO2 m−2 s−1), SC stomatal conductance (µmol H2O m−2 s−1), TR transpiration rate (µmol H2O m−2 s−1), Ci/Ca ratio of internal to atmospheric CO2 concentration, ETR electron transfer rate (µmol electrons m−2 s−1), qP photochemical quenching, NPQ non-photochemical quenching, FV/FM variance fluorescence/maximal fluorescence, SBP4hr SBPase activity at 4th hour (U mg−1), SBP6hr SBPase activity at 6th hour (U mg−1), PC protein content (mg/gFW), SOD superoxide dismutase (mg/FW/min), TSS total soluble sugar (mg/gFW), GS grain starch (%), GYLD grain yield/plant (g), BIOM biomass/plant (g), SFP spikelet fertility percentage, TN tiller number/plant, PN panicle number/plant, GCV genotypic coefficient of variation, PCV phenotypic coefficient of variation, h2 (broad sense) heritability, GA genetic advance as a percentage of mean
Principal component analysis
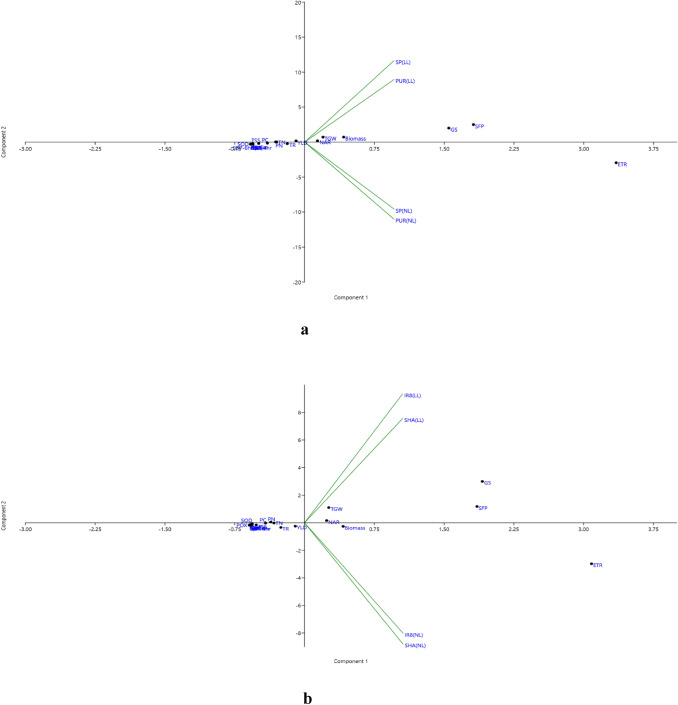
The principal component analysis was used to establish the patterns and interrelationships existing between the four genotypes with all the 22 traits (Table 4). In PCA biplot, twenty-two parameters were distributed in the four quadrants for tolerant and susceptible genotypes (Fig. 8). In first biplot, traits distributed in 3rd and 4th quadrant were associated with all the tolerant genotypes, but traits on 1st and 4th quadrant failed to associate with any genotype. Among all the traits, net assimilation rate, 1000-grain weight, biomass, grain starch (%) and spikelet fertility percentage were positively associated with tolerant genotypes under LL condition, whereas they were negatively associated with tolerant genotypes under NL condition. Only electron transfer rate was positively associated with tolerant genotypes under NL. Similarly, the second biplot is for susceptible genotypes, distributed only in 2nd and 3rd quadrant. Here, both the susceptible genotypes under LL came under 2nd quadrant, while under NL in 3rd quadrant. Traits like net assimilation rate, 1000-grain weight, grain starch(%) and spikelet fertility percentage were positively associated with tolerant genotypes under LL condition, whereas, they were negatively associated with tolerant genotypes under NL condition. Here, biomass and ETR were positively associated with genotypes under NL condition. Similarly, for tolerant genotypes, the first three principal components explained a total of 99.99% variability in all traits. For PC1, NAR, ETR, GS, biomass, TGW and SFP showed a positive association with tolerant genotypes, while for PC2, NAR, GS, GYLD, biomass, TGW, SFP, TN and PN displayed a positive association with tolerant genotypes. For susceptible genotypes, first three PC explained a total of 99.93% variability in all the traits. For PC1, NAR, ETR, GS, biomass, TGW and SFP showed a positive association with susceptible genotypes, while in PC2, NAR, GS, TGW, SFP and PN displayed positive association. Overall both tolerant and susceptible genotypes displayed a positive association with NAR, ETR, GS, TGW and SFP. The analysis of eigenvectors gave the information of qualitative traits for a percentage of variation to the first three principal components, which were 98.84%, 0.86%, and 0.28%, respectively for tolerant genotypes, while 97.82%, 1.53%, and 0.57% for susceptible genotypes (Table 4).
Table 4.
Principal component analysis (PCA) among the tolerant and susceptible rice genotypes for all the considered traits under the normal and low light conditions during kharif and rabi seasons of 2017 and 2018
| Tolerant | Susceptible | |||||
|---|---|---|---|---|---|---|
| PC 1 | PC 2 | PC 3 | PC 1 | PC 2 | PC 3 | |
| NAR | 0.38772 | 0.051786 | − 0.056502 | 0.66435 | 0.051273 | − 0.16558 |
| SC | − 1.5889 | − 0.075919 | − 0.045206 | − 1.6018 | − 0.056033 | − 0.0056285 |
| TR | − 0.52147 | − 0.059236 | 0.090385 | − 0.71035 | − 0.11776 | 0.10645 |
| Ci/Ca | − 1.5641 | − 0.06596 | − 0.036343 | − 1.6005 | − 0.046106 | 0.0017683 |
| ETR | 9.4127 | − 0.77689 | − 0.015718 | 8.6317 | − 1.0413 | 0.16704 |
| qP | − 1.5582 | − 0.079419 | − 0.042142 | − 1.6025 | − 0.062955 | − 0.018679 |
| NPQ | − 1.6331 | − 7.66E−02 | − 0.04693 | − 1.6546 | − 0.058681 | − 0.0062294 |
| FV/FM | − 1.5693 | − 6.64E−02 | − 4.67E−02 | − 1.5751 | − 0.037714 | 0.00023915 |
| SBp− 4h | − 1.598 | − 0.076059 | − 0.045562 | − 1.6137 | − 0.063213 | − 0.005336 |
| SBP 6h | − 1.6001 | − 0.080767 | − 0.046426 | − 1.6378 | − 0.062126 | − 0.0058663 |
| PC | − 1.1246 | − 0.040708 | − 0.004171 | − 1.1725 | − 0.008559 | − 0.012654 |
| Catalase | − 1.622 | − 0.074326 | − 0.057099 | − 1.6478 | − 0.051958 | − 0.0057213 |
| Peroxidase | − 1.6326 | − 0.079098 | − 0.050158 | − 1.6591 | − 0.062671 | − 0.0076727 |
| SOD | − 1.6211 | − 0.073977 | − 0.04756 | − 1.5749 | − 0.017511 | 0.0013418 |
| TSS | − 1.3872 | − 0.053324 | 0.0014164 | − 1.4516 | − 0.052218 | − 0.0022526 |
| GS | 4.3577 | 0.52804 | 0.20269 | 5.3469 | 1.0558 | 0.39428 |
| GYLD | − 0.25758 | 0.048947 | 0.19099 | − 0.27816 | − 0.083992 | 0.072351 |
| BIOM | 1.1817 | 0.1953 | 0.439 | 1.159 | − 0.087551 | 0.15735 |
| TGW | 0.55654 | 0.18187 | 0.10174 | 0.73183 | 0.38427 | 0.29239 |
| SFP | 5.1021 | 0.65458 | − 0.39507 | 5.1807 | 0.40683 | − 0.77601 |
| TN | − 0.84522 | 0.010242 | − 0.058798 | − 0.91992 | − 0.005776 | − 0.072849 |
| PN | − 0.87523 | 0.007921 | − 0.031882 | − 1.0142 | 0.018009 | − 0.10873 |
| Eigenvalue | 7.90719 | 0.0691232 | 0.0226003 | 7.82584 | 0.122638 | 0.0456028 |
| % variance | 98.84 | 0.86404 | 0.2825 | 97.823 | 1.533 | 0.57003 |
| Cumulitive Variance | 98.84 | 99.704 | 99.99 | 97.823 | 99.36 | 99.93 |
NAR net assimilation rate (µmol CO2 m−2 s−1), SC stomatal conductance (µmol H2O m−2 s−1), TR transpiration rate (µmol H2O m−2 s−1), Ci/Ca ratio of internal to atmospheric CO2 concentration, ETR electron transfer rate (µmol electrons m−2 s−1), qP photochemical quenching, NPQ non-photochemical quenching, FV/FM variance fluorescence/maximal fluorescence, SBP4hr SBPase activity at 4th hour (U mg−1), SBP6hr SBPase activity at 6th hour (U mg−1), PC protein content (mg/gFW), SOD superoxide dismutase (mg/FW/min), TSS total soluble sugar (mg/gFW), GS grain starch (%), GYLD grain yield/plant (g), BIOM biomass/plant (g), SFP spikelet fertility percentage, TN tiller number/plant, PN panicle number/plant
Fig. 8.
PCA Biplot (two-dimensional diagram) analysis for 22 traits under two light conditions (a) for low light tolerant genotypes (Purnendu and Swarnaprabha) (b) for low light susceptible genotypes (IR8 and Sasarang) resulting from principal component analysis (PCA)
Discussion
The agronomic performance of crops is negatively influenced by a reduction in R/FR ratio of the incident light (LL stress), which is the characteristic of canopy. Under LL stress, rice plants are subjected to SAS along with a depleted photosynthetic performance, which consequently results in the various phenotypic readjustments in the plants while neglecting the harvestable organs. In this study, we have tracked the expression and activity patterns of the light-sensitive Calvin cycle enzyme, SBPase, along with various associated physiological, biochemical and agronomic traits under simulated LL stress conditions to understand the role of SBPase in modulating yield performance in rice under light depleted environment. Factorial ANOVA for all the 22 traits including seasonal and light variations was captured. This seasonal variation was significant for 19 traits (including SBPase activity and grain yield) indicating both the seasons had different total light availability which influenced the expression of traits. The variations raised due to light were significant indicating that LL imposed during the experimentation was significantly different compared to NL condition. This difference in the light significantly influenced the 21 traits including SBPase activity and grain yield. Further, the interaction between the season x light and season x genotype was significant for most of the traits, which suggested a greater relationship between the performance of genotypes and light availability in different seasons. Such significant interaction effects under stress situation and seasonal variations for different traits in rice and other cereals are also reported by Subudhi et al. (2020).
The rate of photosynthesis is comparatively higher at the flowering stage in the flag leaf compared to the second leaf at the active tillering stage of rice (He et al. 2014). Therefore, we assessed the CO2 fixation efficiency of rice grown under LL stress at this stage. Our data suggested a significant reduction in NAR, GS and TR under LL stress than NL condition, which was significantly higher in the susceptible (Sasarang and IR8) than tolerant genotypes (Swarnaprabha and Purnendu). A similar pattern in the reduction of net assimilation rate under LL was previously reported by Dai et al. (2009). The use of chlorophyll fluorescence and photosynthesis measurement systems has provided a non-destructive means for assessing the shade tolerance of plants (Zhang et al. 2010). We found a significant (p ≤ 0.001) positive correlation between NAR and ETR irrespective of the type of genotype (susceptible and tolerant), which suggested that CO2 fixation process is dependent on photon availability that regulates the efficiency of light reaction in photosynthesis. In a similar study by Kumagai et al. (2009), the sustainability of photosynthesis in the flag leaves was reported to be dependent upon PSII photochemistry and electron transport. Additionally, in our previous work (Sudhanshu et al. 2019), we reported down regulation of the expression of the oxygen-evolving complex genes, OEEP1 and OEEP2, along with various other proteins of PSII such as PSIIPSB27-H1 and PSII 10kd proteins that play an essential role in the electron transport chain of the light reaction; thus further impairs the electron transfer rate (ETR). Therefore, a decrease in the ETR under LL stress might be attributed to the depletion inefficiency of excitation capture in all the tested genotypes, which was more in susceptible compared to tolerant cultivars.
Additionally, in this study, the maximum efficiency of PSII photochemistry under dark adaption (Fv/Fm) significantly increased in tolerant genotypes under NL, however in case of LL, this association was positive but up to the significant level. Such kind of positive association of Fv/Fm with net assimilation rate was observed by Mauro et al. (2011). The above relation may have significant changes if the genotype would be chosen from a different genetic background. Our data suggested that Swarnaprabha and Purnendu enhanced their light adaptive capacity under LL stress by optimizing the light-harvesting potential, remodeling photosynthetic characteristics, and chlorophyll fluorescence traits (such as arising Fv/Fm, ETR, qP but reducing NPQ) to maintain a sustainable light-use efficiency and limit the dissipation of light energy. In our study, we found Ci/Ca ratio to be negatively correlated with NAR in all the genotypes. This suggested that inefficiency in the CO2 utilization potency by the plants under LL stress might be contributing to a hampered NAR output. A similar observation was previously reported by Tooulakou et al. (2016).
As SBPase plays a significant role in CO2 fixation process, we further tracked its expression and activity in tolerant and susceptible genotypes under LL stress. We observed a significant down regulation of SBPase transcript under LL stress as compared to NL condition in all the genotypes under study, which was pronounced in susceptible genotypes compared to tolerant genotypes under low light. Bilgin et al. (2010) reported light-regulated GATA motifs in the upstream sequence of SBPase gene in Arabidopsis thaliana. Additionally, WF1, a transcription regulatory protein in wheat nuclei was reported to interact with various Calvin cycle genes, including SBPase (Gutle et al. 2016). Recently, the binding site for WF1 is found to be localized at ACGT motifs within the promoter of SBPase gene which is a core sequence for the binding of bZIP class of transcription factors whose activity is light-regulated (Driever et al. 2017; Hao et al. 2019). These studies along with our observation of the behavior of SBPase expression under LL suggest their dependence on light signal for optimal regulation, which has a direct influence on CO2 fixation efficiency and finally on economic yield. Among all the four genotypes tested, tolerant genotypes, Purnendu and Swarnaprabha showed a lower reduction, whereas moderately susceptible (Sasarang) and susceptible (IR8) genotypes demonstrated a higher reduction in SBPase expression under stress. It is evident from mean % reduction, correlation and PCA. In general, while selecting a trait, we consider a single statistics, ignoring other sources of variation, which may lead to the false positive section of the trait. Thus, in the present experiment, we used multiple statistical selection criteria (correlation, heritability. genetic advance and PCA) for efficient selection of the traits which are net assimilation, ETR, biomass and 1000-grain weight. These traits are highly related to the SBPase activity and grain yield under LL stress, and thus could be attributed to LL tolerance mechanism in rice. This pattern of differences in the expression efficiency of all four rice genotypes could possibly be attributed to their intrinsic genetic capacity, which greatly contributes to the entire process of the involvement of various classes of transcription factors, for example, bZIP class of transcription factors, that regulates the expression level of SBPase transcript. Previous studies have established a correlation between SBPase expression, accumulation of total biomass and net assimilation rate (Driever et al. 2017), which is also mirrored in our work.
In the present study, SBPase activity significantly declined in all genotypes under LL stress. We have observed a correlation between the SBPase activity and photosynthetic rate under LL conditions that is ultimately reflected in the total soluble sugar and starch accumulation in the rice flag leaf and grains, respectively. Recent studies have demonstrated that low SBPase activity is associated with reduced net photosynthetic rate and carbohydrate level in the leaf (Harrison et al. 1998; Ding et al. 2016). Overall (4th and 6th hour), we observed that tolerant genotypes Purnendu (15.21%, 11.24% in kharif; 18.37%, 24.58% in rabi) and Swarnaprabha (13.8%, 15.67% in kharif; 13.61%, 16.44% in rabi) had 3.81 times lesser reduction in SBPase activity than the susceptible genotypes (Sasarang (45.5%, 33.33% in kharif; 25.96%, 21.87% in rabi and IR8 (52.67%, 24.58% in Kharif; 48.70%, 24.69% in rabi). SBPase is activated via ferredoxin-thioredoxin pathway (FTP) that derives electrons through the electron transfer chain of the light reaction (Buchanan 2016). Therefore, a possible reduction in ETR might further discourage electron supply to the FTP ultimately reducing the activation of SBPase activity under LL stress. Additionally, SBPase activity is also dependent on the RuBisCo (Parry et al. 2013). We observed a significant down regulation of soluble protein content in the flag leaf in all the tested rice genotypes under LL stress. As RuBisCo is the most abundant protein of the rice flag leaf (Evans 1989) and its expression is light-regulated (Panda et al. 2014), reduction in its amount under LL stress would in all likelihood affect the activity of SBPase and ultimately the net assimilation rate.
Among antioxidant enzymes, CAT and SOD increased, whereas POX decreased under LL stress. In a previous study by Moradi and Abdelbagi Ismail (2007), up-regulation of the anti-oxidant system was found to play a regulatory role in stress tolerance of rice, which helped the plants to maintain a supportable photosynthetic function during vegetative and reproductive stages. In our work, we found a lesser increase in SOD activity under LL (T-test value 0.055) in Swarnaprabha and Purnendu compared to NL (T-test value 0.059) treatment, which might play a role in meeting the expected photosynthetic requirements of these particular genotypes, imparting them shade tolerance over Sasarang and IR8. Our data suggested that only SOD activity is correlated with high yield under NL and LL conditions. A similar result was previously reported by Panigrahy et al. (2019). We hypothesize that tolerant genotypes might have maintained an optimal gradation of their enzyme activity associated with CO2 fixation under LL stress through a systematic regulation of antioxidant enzymes that possibly contributed to their final photosynthetic output.
One of the major challenges for eastern India is to sustain grain yield in the prevailing LL stress conditions. To address such an issue, 22 traits were measured to find out the relationship with the grain yield. In our case, grain yield was significantly and positively correlated with SBPase activity under LL but not under NL condition. Further, tolerant genotypes had a strong and positive correlation with SBPase activity compared to the susceptible genotypes under LL (Table 2). The above findings justify that the SBPase activity is a significant and important parameter to be considered for improving the grain yield in the genotypes while breeding for LL tolerance. Such type of evidential information was also reported by Suzuki et al. (2019) where there was no correlation between SBPase activity and photosynthetic rate under NL condition. In contrast to this, our data suggested that depletion in the light quality under LL reduces SBPase activity that could possibly be generating a photosynthetic loophole ultimately hampering rice yield. This is also evidenced by the trait association performed in the current experiment separately for NL and LL conditions (Table 2). Similarly, grain yield has a significant positive correlation with transpiration rate, Ci/Ca, protein content, total soluble sugar, tiller number, panicle number, biomass and thousand-grain weight-under both conditions. Similar results were obtained by Oetting et al. (2003) and Akinwale et al. (2011). Purnendu and Swarnaprabha exhibited less down regulation of SBPase transcript under LL stress as evident from only less reduction in total biomass and yield. Conversely, an apparent reduction in the SBPase expression of Sasarang and IR8 under LL stress was manifested in a dramatic reduction in total biomass-accumulation and grain yield. Moreover, the significant down regulation of soluble protein content in the flag leaf in all the four rice genotypes suggested that the protein synthesis is reduced, in general, under LL condition. This finally resulted in a lesser reduced spikelet filling and a comparative trifle reduction in economic yield. This observation is bolstered by the findings of Wang et al. (2015) who observed LL tolerance in rice to be associated with a better light-harvesting efficiency during the grain-filling period. Additionally, our results suggested a positive correlation between the soluble sugar in leaves with the grain starch content in all the genotypes, which is also evidenced from the report of Luo and Qiufeng (2011). This could be attributed to the hampering of the source-sink communication under LL stress which could be resulted as a result of the inadequacy of photosynthesis in the source organs accompanied by a retarded starch biosynthesis rate in the sinks (Panda et al. 2020). Any kind of stress leads to loss and gain in the phenotypic changes in plants which are mostly related to the biochemical mechanism involved in the tolerance or susceptibility of the genotype. These changes decide the quantity of yield penalty in a genotype through an increase or decrease in the buffering capacity of the individual trait under selection. Thus, a significant relation was established in LL stress-tolerant genotypes in relation to SBPase activity and grain yield.
Conclusion
The present work has critically analyzed the impact of low light stress on the expression and activity of SBPase enzyme, and its possible influences on the photosynthetic performance and grain yield under photon depleted condition in low light tolerant and susceptible rice genotypes. It provided a comprehensive insight into the involvement of this particular enzyme, differentially, in LL tolerant and susceptible rice genotypes. The study indicated that by tracking the expression and activity of SBPase, we can screen for the LL tolerant rice genotypes. It, therefore, could be used as a marker enzyme for the screening process. However, it is not clear how exactly the expression and activity of SBPase are maintained under LL tolerant genotypes, and what is the exact physiological basis for the regulation of SBPase. Since photosynthesis is highly sensitive to the available light intensity, we proposed that SBPase could enhance LL tolerance in rice plants by precisely maintaining a delicate balance between its expression and activity by efficiently utilizing the available photons. The enhanced tolerance to LL stress could be due to overexpression of SBPase in vivo. A further study on other light-regulated Calvin cycle genes and their interaction with SBPase could provide information about the role of other enzymes for a better understanding of this complex process.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The financial assistance received from the Indian Council of Agricultural Research, New Delhi, India and logistic support provided by the Director, ICAR-NRRI, Cuttack is gratefully acknowledged.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Awadhesh Kumar, Email: awadh_iari@yahoo.com.
Darshan Panda, Email: darshan.panda216@gmail.com.
Soumya Mohanty, Email: smmohanty.mohanty9@gmail.com.
Monalisha Biswal, Email: monalisha.horti@gmail.com.
Prajjal Dey, Email: ahaan.agri@gmail.com.
Manaswini Dash, Email: manaswini.crri@gmail.com.
Rameswar Prasad Sah, Email: ramesh.pbg@gmail.com.
Sudhir Kumar, Email: sudhirnpf@gmail.com.
Mirza Jaynul Baig, Email: mjbaigcrri@gmail.com.
Lambodar Behera, Email: lambodarjamujhadi@gmail.com, Email: lbehera.publi2018@gmail.com.
References
- Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/S0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- Akinwale MG, Gregorio G, Nwilene F, Akinyele BO, Ogunbayo SA, Odiyi AC. Heritability and correlation coefficient analysis for yield and its components in rice (Oryza sativa L.) Afr J Plant Sci. 2011;5:207–212. [Google Scholar]
- Arp HC, Johnson HL. The globular cluster M13. Astrophys J. 1955;122:171. doi: 10.1086/146065. [DOI] [Google Scholar]
- Bilgin DD, Zavala JA, Zhu JIN, Clough SJ, Ort DR, Delucia EH. Biotic stress globally down regulates photosynthesis genes. Plant Cell Environ. 2010;33:1597–1613. doi: 10.1111/j.1365-3040.2010.02167.x. [DOI] [PubMed] [Google Scholar]
- Buchanan BB. Role of light in the regulation of chloroplast enzymes. Ann Rev Plant Physiol. 1980;31:341–374. doi: 10.1146/annurev.pp.31.060180.002013. [DOI] [Google Scholar]
- Buchanan BB. Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Arch Biochem Biophys. 1991;288:1–9. doi: 10.1016/0003-9861(91)90157-E. [DOI] [PubMed] [Google Scholar]
- Buchanan BB. The path to thioredoxin and redox regulation in chloroplasts. Annul Rev Plant Biol. 2016;67:1–24. doi: 10.1146/annurev-arplant-043015-111949. [DOI] [PubMed] [Google Scholar]
- Cadet F, Meunier JC. pH and kinetic studies of chloroplast sedoheptulose-1,7-bisphosphatase from spinach (Spinacia oleracea) Biochem J. 1988;253:249–254. doi: 10.1042/bj2530249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xiong J, Yu T, Li X, Li S, Hua Y, Li Y, Zhu Y. Molecular cloning and characterization of rice sedoheptulose-1, 7-bisphosphatase gene that is regulated by environmental stresses. J Plant Biochem Biotechnol. 2004;13:93–99. doi: 10.1007/BF03263201. [DOI] [Google Scholar]
- Dai Y, Shen Z, Liu Y, Wang L, Hannaway D, Lu H. Effects of shade treatments on the photosynthetic capacity, chlorophyll fluorescence, and chlorophyll content of Tetrastigma hemsleyanum Diels et Gilg. Environ Exp Bot. 2009;65:177–182. doi: 10.1016/j.envexpbot.2008.12.008. [DOI] [Google Scholar]
- Demmig B, Winter K, Krüger A, Czygan FC. Photoinhibition and zeaxanthin formation in intact leaves: a possible role of the xanthophyll cycle in the dissipation of excess light energy. Plant Physiol. 1987;84:218–224. doi: 10.1104/pp.84.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiree D, Gutle TR, Stefanie J, Muller JC, Stephane D, Lemaire AH, Tiphaine D, Buchanan BB, Ralf R, Oliver E, Jean-Pierre J. Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc Natl Acad Sci USA. 2016;113:6779–6784. doi: 10.1073/pnas.1606241113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa RS, Pamela PD, Trevor AT. Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J Exp Bot. 1981;32:93–101. doi: 10.1093/jxb/32.1.93. [DOI] [Google Scholar]
- Ding F, Wang M, Zhang S, Ai X. Changes in SBPase activity influence photosynthetic capacity, growth, and tolerance to chilling stress in transgenic tomato plants. Sci Rep. 2016;6:32741. doi: 10.1038/srep32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever SM, Simkin AJ, Alotaibi S, Fisk SJ, Madgwick PJ, Sparks CA, Jones HD, Lawson T, Parry MA, Raines CA. Increased SBPase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Philos Trans R Soc Lond B Biol Sci B Biol Sci. 2017;372:20160384. doi: 10.1098/rstb.2016.0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Mohanty S, Tripathy BC. Role of temperature stress on chloroplast biogenesis and protein import in pea. Plant Physiol. 2009;150:1050–1106. doi: 10.1104/pp.109.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick DN (1997) What is yield? Developing drought-and low N-tolerant maize. In: Proceedings of a symposium; El Batan, Mex.(Mexico); 25–29 Mar 1996. Developing drought and low N-tolerant maize. Proceedings of a symposium; El Batan, Mex.(Mexico); 25–29 Mar, 1997
- Evans J. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia. 1989;78:9–19. doi: 10.1007/BF00377192. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The use of multiple measurements in taxonomic problems. Ann Eugen. 1936;7:179–188. doi: 10.1111/j.1469-1809.1936.tb02137.x. [DOI] [Google Scholar]
- Franklin K, Garry A, Whitelam C. Phytochromes and shade-avoidance responses in plants. Ann Bot. 2005;96:169–175. doi: 10.1093/aob/mci165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutle DD, Roret T, Müller SJ. Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc Natl Acad Sci USA. 2016;113:6779–6784. doi: 10.1073/pnas.1606241113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer O, Harper DA, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9. [Google Scholar]
- Hao X, Zhong Y, Nutzmann HW, Fu X, Yan T, Shen Q, Chen M, Ma Y, Zhao J, Osbourn A, Li L. Light-induced artemisinin biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua. Plant Cell Physiol. 2019;60:1747–1760. doi: 10.1093/pcp/pcz084. [DOI] [PubMed] [Google Scholar]
- Harrison EP, Willingham NM, Lloyd JC, Raines CA. Reduced seudoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate portioning. Planta. 1998;204:27–36. doi: 10.1007/s004250050226. [DOI] [Google Scholar]
- He H, Yang R, Jia B, Chen L, Fan H, Cui J, Yang D, Li M, Ma FY. Rice photosynthetic productivity and psii photochemistry under non-flooded irrigation. Sci World J. 2014;2014:839658. doi: 10.1155/2014/839658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai E, Takuya A, Fumitake K. Characteristics of gas exchange and chlorophyll fluorescence during senescence of flag leaf in different rice (Oryza sativa L.) cultivars grown under nitrogen-deficient condition. Plant Prod Sci. 2009;12:285–292. doi: 10.1626/pps.12.285. [DOI] [Google Scholar]
- Kumar A, Sahoo U, Baisakha B, Okpani OA, Ngangkham U, Parameswaran C, Basak N, Kumar G, Sharma SG. Resistant starch could be decisive in determining the glycemic index of rice cultivars. J Cereal Sci. 2018;79:348–353. doi: 10.1016/j.jcs.2017.11.013. [DOI] [Google Scholar]
- Kumar A, Panda D, Biswal M, Dey P, Behera L, Baig MJ, Nayak L, Ngangkham U, Sharma S. Low light stress influences resistant starch content and glycemic index of rice (O. sativa L) Starch-Starke. 2019;71:5–6. [Google Scholar]
- Lefebvre S, Lawson T, Zakhleniuk OV, Lloyd JC, Raines CA, Fryer M. Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol. 2005;138:451–460. doi: 10.1104/pp.104.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Bio Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Luo X, Qiufeng H. Relationships between leaf and stem soluble sugar content and tuberous root starch accumulation in cassava. J Agric Sci. 2011;3:64. [Google Scholar]
- Marchiori Paulo ER, Machado Eduardo C, Ribeiro Rafael V. Photosynthetic limitations imposed by self-shading in field-grown sugarcane varieties. Field Crops Res. 2014;155:30–37. doi: 10.1016/j.fcr.2013.09.025. [DOI] [Google Scholar]
- Marcus Y, Hagit AG, Yael W, Michael R. Rubisco mutagenesis provides new insight into limitations on photosynthesis and growth in Synechocystis PCC6803. J Exp Bot. 2011;62:4173–4182. doi: 10.1093/jxb/err116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S, Jain L, Jajoo A. Photosynthetic efficiency in sun and shade plants. Photosynthetica. 2018;56:354–365. doi: 10.1007/s11099-018-0767-y. [DOI] [Google Scholar]
- Mauro RP, Occhipinti A, Longo AMG, Mauromicale G. Effects of shading on chlorophyll content, chlorophyll fluorescence and photosynthesis of subterranean clover. J Agron Crop Sci. 2011;197:57–66. doi: 10.1111/j.1439-037X.2010.00436.x. [DOI] [Google Scholar]
- Moradi F, Abdelbagi Ismail M. Responses of photosynthesis, chlorophyll Fluorescence and ROS-Scavenging systems to salt stress during seedling and reproductive stages in rice. Ann Bot. 2007;99(6):1161–1173. doi: 10.1093/aob/mcm052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetting WS, Fryer JP, Shriram S, King RA. Oculocutaneous albinism type 1: the last 100 years. Pigment Cell Res. 2003;16:307–311. doi: 10.1034/j.1600-0749.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- Panda D, Baig MJ, Tripathy BC. Phytochrome mediated growth and development in rice: a review. Agrotechnology. 2014;2:4. [Google Scholar]
- Panda D, Biswal M, Behera L, Baig MJ, Dey P, Nayak L, Sharma S, Samantaray S, Ngangkham U, Kumar A. Impact of low light stress on physiological, biochemical and agronomic attributes of rice. J Pharmacogn Phytochem. 2019;8:1814–1821. [Google Scholar]
- Panda D, Biswal M, Mohanty S, Dey P, Swain A, Behera D, Baig MJ, Kumar A, Sah RP, Tripathy BC, Behera L. contribution of phytochrome a in the regulation of sink capacity, starch biosynthesis, grain quality, grain yield and related traits in rice. Plant Arch. 2020;20:1179–1194. [Google Scholar]
- Panigrahy M, Ranga A, Das J, Panigrahi KCS. Shade tolerance in Swarnaprabha rice is associated with higher rate of panicle emergence and positively regulated by genes of ethylene and cytokinin pathway. Sci Rep. 2019;9:1–17. doi: 10.1038/s41598-019-43096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MA, Andralojc PJ, Scales JC, Salvucci ME, Carmo-Silva AE, Alonso H, Whitney SM. Rubisco activity and regulation as targets for crop improvement. J Exp Bot. 2013;64:717–730. doi: 10.1093/jxb/ers336. [DOI] [PubMed] [Google Scholar]
- Praba ML, Vanangamudi M, Thandapani V. Effects of low light on yield and physiological attributes of rice. Int Rice Res Note. 2004;29:1. [Google Scholar]
- Rosenthal DM, Locke AM, Khozaei M, Raines CA, Long SP, Ort DR. Over-expressing the C3 photosynthesis cycle enzyme sedoheptulose-1-7 bisphosphatase improves photosynthetic carbon gain and yield under fully open-air CO2 fumigation (FACE) BMC Plant Biol. 2011;11:123. doi: 10.1186/1471-2229-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghamitra P, Sah RP, Bagchi TB, Sharma SG, Kumar A, Munda S, Sahu RK. Evaluation of variability and environmental stability of grain quality and agronomic parameters of pigmented rice (O. sativa L.) J Food Sci Technol. 2018;55:879–890. doi: 10.1007/s13197-017-2978-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U (2004) Pulse-amplitude-modulation (PAM) fluorometry and saturation pulse method: an overview. Chlorophyll a fluorescence. Springer, Dordrecht, pp 279–319
- Seuter A, Busch M, Ruediger DH. Overexpression of potential herbicide target seudoheptulose-1,7-bisphosphatase from Spinacia oleracea. Mol Breed. 2002;9:53–61. doi: 10.1023/A:1019297521424. [DOI] [Google Scholar]
- Singh VP. Effect of low light stress on growth and yield of rice. Ind J Plant Physiol. 1988;31:84–91. [Google Scholar]
- Subudhi HN, Prasad KVSV, Ramakrishna C, Rameswar PS, Pathak H, Ravi D, Khan AA, Padmakumar V, Blümmel M. Genetic variation for grain yield, straw yield and straw quality traits in 132 diverse rice varieties released for different ecologies such as upland, lowland, irrigated and salinity prone areas in India. Field Crops Res. 2020;245:107626. doi: 10.1016/j.fcr.2019.107626. [DOI] [Google Scholar]
- Sudhanshu S, Panda D, Kumar J, Mohanty N, Biswal M, Baig Kumar A, Umakanta N, Samantaray S, Pradhan SK, Shaw BP, Swain P, Behera L. Comparative transcriptome profiling of low light tolerant and sensitive rice varieties induced by low light stress at active tillering stage. Sci Rep. 2019;9:1–14. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Wada S, Kondo E, Yamori W, Makino A. Effects of co-overproduction of sedoheptulose-1,7-bisphosphatase and Rubisco on photosynthesis in rice. Soil Sci Plant Nutr. 2019;65:36–40. doi: 10.1080/00380768.2018.1530053. [DOI] [Google Scholar]
- Tooulakou G, Giannopoulos A, Nikolopoulos D, Bresta P, Dotsika E, Orkoula MG, Kontoyannis CG, Fasseas C, Liakopoulos G, Klapa MI, Karabourniotis G. Alarm photosynthesis: calcium oxalate crystals as an internal CO2 source in plants. Plant Physiol. 2016;171:2577–2585. doi: 10.1104/pp.16.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fei D, Wan-Jun R. Shading tolerance in rice is related to better light harvesting and use efficiency and grain filling rate during grain filling period. Field Crop Res. 2015;180:54–62. doi: 10.1016/j.fcr.2015.05.010. [DOI] [Google Scholar]
- Woodrow IE, Murphy DJ, Latzko E. Regulation of stromal sedoheptulose1,7-bisphosphatase activity by pH and Mg2+ concentration. BioChem. 1984;259:3791–3795. [PubMed] [Google Scholar]
- Yemm EW, Willis AJ. The estimation of carbohydrates in plant extracts by anthrone. Biochem J. 1954;57:508–514. doi: 10.1042/bj0570508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Pan X, Mu G, Wang J. Toxic effects of antimony on photosystem II of Synechocystis sp. as probed by in vivo chlorophyll fluorescence. J Appl Phycol. 2010;22:479–488. doi: 10.1007/s10811-009-9482-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.