Abstract
Regeneration of skeletal muscle after volumetric injury is thought to be impaired by a dysregulated immune microenvironment that hinders endogenous repair mechanisms. Such defects result in fatty infiltration, tissue scarring, chronic inflammation, and debilitating functional deficits. Here, we evaluated the key cellular processes driving dysregulation in the injury niche through localized modulation of sphingosine-1-phosphate (S1P) receptor signaling. We employ dimensionality reduction and pseudotime analysis on single cell cytometry data to reveal heterogeneous immune cell subsets infiltrating pre-clinical muscle defects due to S1P receptor inhibition. We show that global knockout of S1P receptor 3 (S1PR3) is marked by an increase of muscle stem cells within injured tissue, a reduction in classically-activated relative to alternatively-activated macrophages, and increased bridging of regenerating myofibers across the defect. We found that local S1PR3 antagonism via nanofiber delivery of VPC01091 replicated key features of pseudotime immune cell recruitment dynamics and enhanced regeneration characteristic of global S1PR3 knockout. Our results indicate that local S1P receptor modulation may provide an effective immunotherapy for promoting a pro-reparative environment leading to improved regeneration following muscle injury.
Keywords: immunomodulation, tissue regeneration, sphingolipid, inflammation, wound healing
Introduction
Volumetric muscle loss (VML) injuries commonly occur as the result of extremity trauma injuries and represent a significant clinical challenge among both military and civilian populations (1–5). VML injuries are characterized by traumatic or surgical loss of tissue which will not endogenously regenerate, resulting in chronic loss of function and permanent disability (6, 7). Autologous free or rotational muscle flap transfer is the current gold standard of treatment for VML injuries, and while these are successful procedures for limb salvage, flap transfer can lead to donor-site complications, morbidity and persistent functional strength deficits (4, 5). We have previously developed and utilized mouse skeletal muscle trauma models of full thickness VML injury in both the spinotrapezius (8, 9) and quadriceps muscles (10). Both injury models result in traumatic injury, similar to the clinical scenario, characterized by a chronic invasion of immune cells followed by significant fibrosis, fatty infiltration, and no bridging of new myofibers across the defect without intervention (11–14).
Tissue macrophages are important players in the immune response following VML, and persistent, unresolved inflammation is associated with pathological tissue remodeling in many injury contexts (9–12, 15). Tissue macrophages are a heterogeneous population whose function in injured tissues is largely dictated by microenvironmental cues (16, 17). Macrophages can be broadly classified into two phenotypes, classically-activated M1-like macrophages or alternatively-activated M2-like macrophages. M2-like macrophages in particular are believed to be the mediators of both pro-regenerative and pro-fibrotic signaling responses. M2-like macrophages often populate fibrotic tissue and their secretion of profibrotic factors, such as TGF-β1 and arginase-1, mediate activation of myofibroblasts and tissue scarring (18). However, M2-like macrophages are also a primary source of pro-angiogenic factors, including VEGF, in the post-injury microenvironment and support muscle stem cell (MuSC) proliferation and subsequent differentiation into myofibers (19–22). While VML injuries are populated with macrophages that express multiple M2 phenotypic markers, the environment is rich in both pro- and anti-inflammatory cues, eventually leading to pathological fibrosis (12–14, 23). Therefore, designing new strategies to stabilize pro-regenerative functions of tissue macrophages and achieve timely resolution of inflammation is critical to outpace pro-fibrotic signaling and restore function following VML injury.
Sphingosine-1-phosphate (S1P) is a bioactive lipid that is produced by and acts directly upon macrophages in response to extracellular cues (8, 24–31). Production of S1P and differential expression of the five G protein-coupled S1P receptors (S1PR1–5) in macrophages have potent effects on macrophage function and modulating the transition from pro-inflammatory to pro-reparative functions. We showed previously that differentiation and M2-polarization is associated with increased surface expression of S1PR3 (8, 32–35). Activation of S1PR3 also induces a switch toward pro-inflammatory macrophage responses (24, 36). Inflammatory cytokines such as TGFβ also redirect the S1P signaling outcome from being pro-myogenic to profibrotic by shifting the dominant receptor expression from S1PR2 to S1PR3 (37). TGFβ plays a crucial role in the promotion of chronic degenerative fibrosis of dystrophic muscles likely via S1PR3 (37). These observations suggest that local delivery of S1P receptor modulators, especially those which antagonize S1PR3 receptor activation, may be effective for enhancing muscle healing.
To understand the functional role of cellular progressions responsible for muscle healing, we assessed the high variability between individual cells throughout temporal regenerative processes. While the inflammatory cascade after injury is largely a function of time, we traditionally lack the analytical tools required to distinguish dynamic transitional states existing within bulk cell types. Variability in cell-to-cell expression levels of particular markers is often masked into averaged measurements defining a group of data, losing the trends existing between individual cells (38). Single-cell flow cytometry combined with dimensionality reduction can overcome such challenges to reconstruct cellular hierarchies and transitional phenotypes inferred through time- a concept known as ‘pseudotime’(38, 39). In this study, we utilize Uniform Manifold Approximation and Projection (UMAP), a dimensionality reduction algorithm that clusters cells into 2D projections based on marker similarities to reveal heterogeneity underlying immune cells taken at a snapshot in time (40). Furthermore, we leverage pseudotime trajectory analysis via Spanning-tree Progression Analysis of Density-normalized Events (SPADE) which organizes clusters of similar cells into distinct nodes along a 2D dendrogram, organized to track cellular lineages and progressive transitions (39). By harnessing such tools, we can reconstruct temporally regulated inflammatory and regenerative processes to reveal the trends in individual cellular expression levels influenced by S1P receptor modulation. Our studies suggest that modulation of S1P receptor signaling in VML defects may be effective for reducing pro-fibrotic macrophage functions and creating a pro-regenerative environment.
This study investigates the role of S1PR3 on the inflammatory cell response and phenotype within VML injuries in the quadriceps and spinotrapezius muscles. Full thickness VML injuries in WT mice or those with a global knockout of S1PR3 (S1PR3 KO) (Fig. 1A-I, II, III) were used to quantify frequency changes in M1-like and M2-like macrophages, MuSC frequency within the defect, diameter of regenerating fibers, and subsequent increased bridging across the void area (Fig. 1B). We explored cellular heterogeneity of the inflammatory response within VML defects by flow cytometry immunophenotyping and generation of pseudotime trajectories to visualize the interaction of diverse immune cell subsets with resident MuSCs (Figure 1C). We also investigated the ability of VPC01091 (a local S1PR3 antagonist) immunotherapy delivery from nanofiber-based meshes to recapitulate key features of the pseudotime trajectories observed in S1PR3 KO mice that are associated with muscle healing (Fig. 1A-II). We hypothesized that local modulation of S1PR signaling within pre-clinical VML defect models would result in timely resolution of inflammation, leading to enhanced muscle healing and regeneration.
Figure 1. Graphical workflow for targeting S1PR3 signaling and characterizing the regenerative capacity of immune modulation in muscle injuries.
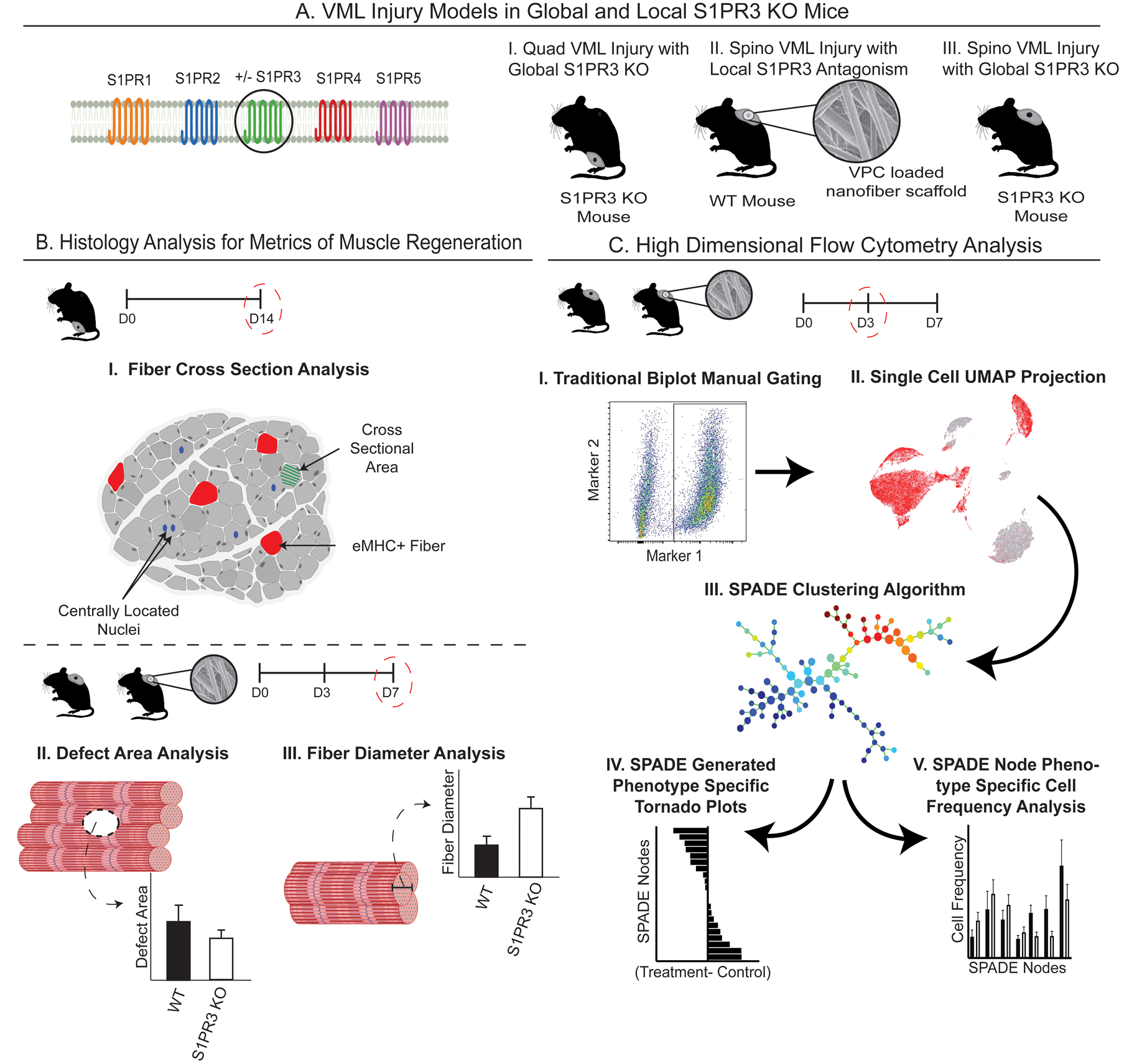
Graphical representation of the innovative methods employed in targeting of S1PR3 signaling for treatment of muscle injuries and the subsequent high dimensional analysis utilized to capture unique, infiltrating cellular subsets. (A) Introduction to the various mouse VML models used in these studies, including (A-I) quadriceps (quad) VML model in S1PR3 KO mice, (A-II) local inhibition of S1PR3 in the spinotrapezius (spino) with S1PR3 antagonist VPC01091-loaded (VPC) nanofibers in WT mice, and (A-III) a spino VML defect model in S1PR3 KO mice. (B) Metrics of muscle regeneration analyzed in these studies including histological analysis of quadricep cross sections (B-I) along with defect area and fiber diameter analysis via whole mount immunohistochemistry in the spino (B-II, III). (C) Workflow for analyzing single-cell high dimensional flow cytometry data after muscle injury. Our approach involves manually gating immune cell subsets (C-I), visualizing underlying heterogeneity via 2D projections using UMAP (C-II), high dimensional clustering algorithms with pseudotime trajectories in SPADE (C-III), and resulting tornado plots (C-IV) and SPADE node frequency quantifications (C-V). Created with BioRender.com.
Methods
Nanofiber scaffold fabrication:
Polycaprolactone (PCL, Sigma) and PLGA were combined in a 1:1 weight/weight ratio at 18% polymer concentration for VPC01091 (Avanti Polar Lipids) loaded fibers (41). Polymers were dissolved in a 1:3 volume ratio solution of methanol to chloroform and vortexed for 2+ hours prior to spinning. For VPC01091-loaded fibers, drug was added to the polymer solution at a 1:200 drug:polymer weight ratio. 2 mL of either polymer solution was loaded into a 3 mL syringe with diameter of 10mm. Electrospinning was performed at an applied voltage of 19 kV, and flow rate of 1 mL/hr for both blank and VPC01091 fibers. Working distance was set at 10 cm for blank fibers and 12 cm for VPC01091 fibers. After 2 mL of polymer was spun, fibers were wrapped in low-binding plastic folders and stored at −20°C.
VPC01091 release from nanofiber scaffolds:
3mm circular punches were made from electrospun nanofiber sheets. Discs were placed in 100μL of stimulated body fluid containing 4% bovine serum albumin and incubated at 37°C with agitation. Releasate was collected at predetermined timepoints and replaced with fresh release medium.
Mass spectrometry quantification of VPC01091 release samples:
A long chain base sphingolipid extraction protocol was used to extract VPC01091 from release samples (42). 100μL of release media was placed in glass culture tubes with screw caps and 1.5 mL of 2:1 methanol to dichloromethane extraction solvent was added to each sample. For internal standard, 50 picomoles of C17 sphingosine was added to every tube. Tubes were capped, sonicated for 1 min and incubated overnight at 48°C. Tubes were allowed to cool to room temperature the following day and then were centrifuged at 3000rpm and supernatant was collected in separate open top glass tubes. 0.5 mL of extraction solvent was added to the pellet remaining in each original tube, and then sonicated for 1 min and centrifuged again at 3000rpm. The second supernatant collection was added to the first supernatant and then extraction solvent was evaporated off by speed vac for 4 hours. The following day, 300μL of mass spectrometry analysis solvent was added to each tube before sonicating for 1 minute. Samples were transferred to microcentrifuge tubes and centrifuged for 10 minutes at 14,000×g. 200μL of supernatant was transferred to mass spectrometry sample vials and all samples were analyzed on a Micromass Quattro LC mass spectrometer.
Force mapping of S1PR modulating nanofibers with Atomic Force Microscopy (AFM):
All AFM measurements were performed with a MFP-3D Bio-AFM (Asylum Research, Santa Barbara, CA, USA). Nanofiber scaffolds were prepared as described and fixed on glass slides with double-sided tape. For stiffness measurements, 9.6μm spherical PMMA particles (modulus ~ 3 GPa) were attached to tipless silicon cantilevers (NSC35-C, Mikromasch, Sofia, Bulgaria) using a two-part epoxy and dried for at least 24 hours. We use the thermal method (43) to calibrate the cantilever spring constant immediately prior to use by indenting a glass substrate and performing a Lorentzian fit to the thermal spectrum. Topological images and accompanying stiffness characterizations of nanofibers were obtained from regions of 80×80 μm2 force maps with 96×96 force curves per image. All the force-distance measurements were performed with 2 um/sec approach velocity using a 500 nN force trigger. Force-distance plots were transformed into force-indentation depth plots, then 10–100% indentation depth was used to evaluate Young’s modulus using a Hertzian contact model.
Spinotrapezius volumetric muscle loss injury:
Male C57BL/6J (The Jackson Laboratory) or S1PR3 KO (MMRRC) mice of age 6–8 months old were used for all animal studies. A 1mm full thickness defect in the spinotrapezius muscle was created as previously described (36). Briefly, a longitudinal 1-inch incision was made just after the bony prominence of the shoulder blade. The overlying fascia was dissected away and the spinotrapezius muscle identified. Using flat-tipped tweezers, the edge of the spinotrapezius was reflected and positioned against a sterile piece of wood. A 1mm biopsy punch was made through the muscle, using the wooden piece as support. The muscle was replaced and 4mm nanofiber scaffold was sutured over the punch defect if applicable to treatment. The incision was closed with mouse wound clips.
Quadriceps volumetric muscle loss injury:
Surgical procedure performed as previously reported [8]. Briefly, the left hindlimb was prepped and sterilized. A single incision was made above the quadriceps and a 3mm biopsy punch (VWR, 21909–136) was used to make a full-thickness muscle defect. Skin was closed and animals were allowed to heal without intervention for 14 days before euthanasia by CO2 inhalation.
Tissue harvest and flow cytometry:
Samples were prepared for flow cytometry analysis on a FACS AriaIII flow cytometer (BD Biosciences). Blood was collected from the jugular vein and red blood cells were lysed in ammonium chloride prior to immunostaining. For analysis of cell composition in spinotrapezius muscle, 6mm biopsy punches centered around the defect were harvested and digested with 5,500U/ml collagenase II and 2.5U/ml Dispase II for 1.5 hours in a shaking 37°C water bath. The digested muscles were filtered through a cell strainer to obtain a single cell suspension. Single-cell suspensions were stained for live cells using either Zombie Green or Zombie NIR (BioLegend) dyes in cell-culture grade PBS per manufacturer instructions. Cells were then stained with cell phenotyping antibodies in a 1:1 volume ratio of 3% FBS and Brilliant Stain Buffer (BD Biosciences) according to standard procedures. The following antibodies were used for cell phenotyping: BV605-conjugated anti-CD4 (BioLegend), BV785-conjugated anti-CD8 (BioLegend), PE-Cy7-conjugated anti-CD3ε (BioLegend), BV510-conjugated anti-CD11b (BioLegend), BV510-conjugated anti-Ly6C (BioLegend), APC-conjugated anti-Ly6C (BioLegend), FITC-conjugated anti-Ly6G/Ly6C (GR-1) (BioLegend), BV711-conjugated anti-CD64 (BioLegend), PE-conjugated anti-MerTK (BioLegend), PE-Cy7 conjugated anti-CD206 (BioLegend), FITC-conjugated anti-CD206 (BioLegend), PE-Cy5-conjugated anti-CD29 (BioLegend), PerCP-Cy5.5-conjugated anti-CXCR4 (BioLegend), and APC-conjugated Lineage antibody cocktail containing antibodies for mouse CD3e, CD11b, CD45R/B220, Ly-76, Ly6G and Ly6C (BD Pharmingen). For immunophenotyping, single, live cells were selected in FlowJo software for all subsequent analysis. Myeloid cells were gated as CD11b+ and lymphoid cells as CD3+. GR1 (Ly6C/Ly6G) was used to distinguish neutrophils (Ly6G+ or GR1+) from monocytes (Ly6G− or GR1lo) in homeostatic blood flow analysis (Fig. 2). Unless indicated otherwise, for all other flow analyses, macrophages were gated as CD11b+CD64+MerTK+ cells, where M1-like macrophages highly expressed Ly6C and M2-like macrophages highly expressed CD206. Monocytes were gated as CD11b+CD64+MerTK− cells, with inflammatory monocytes defined as Ly6Chi monocytes and anti-inflammatory as Ly6Clo monocytes. CD3+ T cells were gated as CD4+ or CD8+ T cells based on their high expression of CD4 and CD8, respectively. MuSCs were gated as Lineage negative (Lin−) CD29+ CXCR4+ cells. Where specified, whole blood cell composition was analyzed by Hemavet (Drew Scientific) veterinary hematology analyzer according to manufacturer’s protocol. The machine compares 20μL of whole blood samples to a standard and calculates total cells of the given types. Samples were calibrated to Drew Scientific whole mouse blood control.
Figure 2. Reduced circulation of immune cell subsets in homeostatic blood of 6–8 month mice lacking S1PR3.

SPADE trees consisting of live, single immune cell subsets from the peripheral blood of 6–8 month mice without injury (A, C). (A) SPADE tree generated from CD11b+ cells (A-I, II) with a manually gated overlay of monocytes (A-I) and neutrophils (A-II) annotated in the greater CD11b+ tree. The SPADE nodes are visualized in grayscale whereas colored nodes represent a heatmap form of normalized cell frequency within each node of the respective phenotypic manual overlay. All nodes corresponding to a single phenotypic subset are grouped into a single colored annotation, as denoted with a phenotype key. (B) Heatmap containing all nodes from the CD11b+ SPADE tree in (A) in which each phenotypic subset is denoted with a colored bar including all nodes that make up that phenotype. The colored bars on the bottom of the heatmap correspond to the colored annotations in (A). (C) SPADE tree generated from CD3+ T cells (C-I, II) where CD4+ (C-I) and CD8+ (C-II) T cells are manually gated and overlaid onto the T cell SPADE tree. (D) Heatmap containing all nodes from the CD3+ SPADE tree (C) in which CD4+ and CD8+ T cells are denoted with purple and green colored bars, respectively, accompanying their annotations in (C). (E) Hemavet concentrations of various immune cell subsets circulating the peripheral blood of mice lacking S1PR3 compared to their WT controls. Notable differences are seen within the 6–8 month age range. Statistical analyses conducted using two-way ANOVA (E-I) with Tukey’s multiple comparisons and two-tailed t-tests (E-II-E-V). Data presented as mean ± S.E.M, *p< 0.05. n=3 samples (A, B) or 7 samples (C, D) per genotype for SPADE analysis. n=4 WT samples and 6 S1PR3 KO samples for Hemavet analysis (E). WBCs: white blood cells.
High dimensional analysis of flow cytometry data
Uniform Manifold Approximation and Projection (UMAP):
UMAP is a nonlinear dimensionality reduction algorithm. UMAP is able to embed high-dimensional data into a space of two or three dimensions, and cells are visualized in a scatter plot, where similarity is demonstrated via proximity to other points. Prior to UMAP dimensional reduction, each flow cytometry sample was pre-gated to select cellular subsets of interest (i.e. monocytes, neutrophils, CD4+ and CD8+ T cells) and then imported into Python 3.7 using fcsparser (https://github.com/eyurtsev/fcsparser) and Pandas 2.5. Each was normalized by applying arcsinh/150. A composite UMAP projection that utilized data points from all desired samples was generated using Matplotlib. Each cell was then phenotyped by overlaying the pre-gated cell subset onto the UMAP projection. (https://github.com/lmcinnes/umap)
Spanning-tree Progression Analysis of Density-normalized Events (SPADE):
SPADE is a visualization tool creating a 2D minimum spanning tree organized into nodes representing clusters of cells similar in their surface marker expressions. The size and color of each node are relative to the number of cells present and the median marker expression. SPADE was performed through MATLAB and the source code is available at http://pengqiu.gatech.edu/software/SPADE/. MATLAB-based SPADE automatically generates the tree by performing density-dependent down-sampling, agglomerative clustering, linking clusters with a minimum spanning-tree algorithm and up-sampling based on user input. The SPADE tree was generated by exporting uncompensated pre-gated live, single cells or select pre-gated cellular subsets (i.e. CD3+ T cells). The following SPADE parameters were used: Apply compensation matrix in FCS header, Arcsinh transformation with cofactor 150, neighborhood size 5, local density approximation factor 1.5, max allowable cells in pooled downsampled data 50000, target density 20000 cells remaining, and number of desired clusters 100.
Whole mount immunofluorescence of spinotrapezius tissues:
Mice were euthanized 7 days after surgery via CO2 asphyxiation. Post-euthanasia, mouse vasculature was perfused with warm saline followed by 4% PFA until tissues were fixed. The entire spinotrapezius muscle was explanted and permeabilized overnight with 0.2% saponin, then blocked overnight in 10% mouse serum. For immunofluorescence, tissues were incubated at 4°C overnight in a solution containing 0.1% saponin, 5% mouse serum, 0.5% bovine serum albumin, conjugated fluorescent antibody Alexa Fluor 488 anti-desmin (1:200 dilution, Abcam), and Alexa Fluor 647 anti-CD68 (AbD Serotec) for macrophage visualization. Following immunostaining, tissues were washed 4 times for 30 minutes each in 0.2% saponin for the first two washes, 0.1% saponin for the third wash, and PBS for the final wash, and then mounted in 50/50 glycerol/PBS.
Imaging and quantification of whole-mount immunofluorescence:
Tissues were imaged on a Zeiss LSM 710 NLO confocal with acquisition parameters kept identical across all animals. Max intensity projections were generated from 3D confocal images for 2D analysis in Zen software (Zeiss). Quantification of defect area percentage and fiber diameter were done with FIJI and Zen software (Zeiss). The defect area was identified as the area that was desmin-negative. A 1mm circle (measurement of initial defect punch) was centered over each defect and the image was thresholded to display only areas of positive fluorescence signal within the region of interest. The number of pixels positive for desmin was normalized to the total area of the region of interest and subtracted from 100% to determine the area fraction that is desmin-negative. 2D fiber diameter was determined by measuring the widest portion of each regenerated muscle fiber (determined based on the morphology of desmin-positive regions) within the defect region from the day 7 2D maximum intensity projections. For both defect area and fiber diameter analysis, n=4 samples for blank NF and VPC01091 NF groups and n=6 for WT and S1PR3 KO groups.
Quadriceps histology and immunostaining:
Tissue processing and histology done as previously reported [8]. Briefly, muscle was dissected, weighed, and snap frozen in liquid nitrogen cooled isopentane. 10 μm cryosections (CryoStar NX70 Cryostat) were taken. Samples were blocked and permeabilized before staining with dystrophin (Abcam, ab15277, 1:200) and embryonic myosin heavy chain (eMHC) (DSHB, F1.652, 1:10) for 1-hour incubation at room temperature. Secondary antibodies conjugated to Alexa Fluor 555 (Thermo Fisher, Rb: A-21429, 1:250) and Alex Fluor 647 (Thermo Fisher, Ms: A-21236, 1:250) were incubated for 30 mins at room temperature. Slides were mounted with Fluoroshield Mounting Medium with DAPI (Abcam, ab104139) and stored at 4°C.
Confocal imaging and quantification of quadriceps fiber cross-sectional area, eMHC+ fibers, and centrally located nuclei:
Imaging and quantification done as previously reported [8]. Briefly, Immunofluorescence images were taken on Zeiss 710 Laser Scanning Confocal microscopes at 20X and stitched together with Zen Black software (Zeiss). The dystrophin images were analyzed using ImageJ by thresholding and using the Analyze Particle function for particles of 0.25–1.0 circularity and 150–6000 μm2 in area to measure muscle fiber cross sectional area (n=4 for WT VML, n=8 for S1PR3 KO VML). Area histograms were created in GraphPad Prism 7. Quantification of eMHC+ myofibers was done on images containing all channels (eMHC, dystrophin, DAPI). The brightest fibers of entire stitched sections were counted using the ImageJ multipoint tool (n=4). Centrally located nuclei were analyzed by taking 5 representative regions of each section, 3 replicates per animal (n=4 per group) and counted using the Image J multipoint tool.
SPADE node heatmap:
SPADE dendrogram heatmaps were constructed with calculated z-scores of fluorescence marker expression intensities for each surface marker used to immunophenotype a specific cell type across all nodes to visualize each node by marker expression intensity. Each row of the heatmap corresponds to a specific surface marker while each column represents an individual SPADE node.
Tornado plot generation:
For each manually gated cell population overlaid onto generated SPADE trees, normalized cell frequencies were averaged across replicates for each treatment for every SPADE node containing the specified cellular phenotype. All nodes of a particular SPADE tree were ordered largest to smallest in average cell frequency difference between treatment groups and input into Graphpad Prism version 8.0 for tornado plot generation.
Statistical analysis:
All statistical analyses were performed using Graphpad Prism version 8.0. Data is presented as mean ± standard error of the mean (S.E.M.) unless otherwise indicated. For pairwise comparisons, unpaired two-tailed t-tests were used. For multiple comparisons, one-way and two-way ANOVA was used with Tukey’s multiple comparisons tests.
Results
Heterogeneity in immune subsets of WT and S1PR3 KO mice.
To gain an understanding of the heterogeneity in immune cell subsets in the blood upon knockout of S1PR3, we first characterized immune cell populations circulating the peripheral blood of naïve wild-type (WT) control or S1PR3 KO mice. CD11b+ myeloid blood cells were identified by traditional biplot gating strategies and then fed into our SPADE workflow to generate a pooled CD11b+ SPADE tree representing both WT and KO mice (Fig. 2A). This SPADE dendrogram was overlaid with annotation of manually-gated monocytes (Fig. 2A-I, yellow) and neutrophils (Fig. 2A-II, light green or orange) to visualize where each respective phenotype was located, with GR1 (Ly6C/Ly6G) being used to distinguish neutrophils (Ly6G+ or GR1+) from monocytes (Ly6G− or GR1lo). Monocytes make up approximately 2/3 of the tree with several distinct branched structures, reflecting the known heterogeneity within the monocyte population (Fig. 2A-I, circled in yellow) (44). Neutrophils are located in two distinct pools on either end of the tree (Fig. 2A-II), suggesting phenotypic heterogeneity due to the distant tree locations (45). Further surface marker analysis revealed that one neutrophil subset (light green annotation) is GR1hi while the other subset (orange annotation) is GR1int as visualized in the heatmap of relative marker expression for each SPADE node (Figure 2B).
T cells were identified by manual gating of CD3+ cells and a SPADE tree was constructed from pooled WT and S1PR3 KO blood (Fig. 2C). Populations of manually-gated CD4+ helper T cells (Fig. 2B-I, purple annotation) and CD8+ cytotoxic T cells (Fig. 2B-II, dark green annotation) were overlaid onto the dendrogram and were found to reside on opposite ends of the tree. This is further visualized and validated in the CD3+ SPADE node heatmap (Fig. 2D) where undefined nodes containing low expression of CD4 and CD8 cluster together. Heterogeneity of monocytes, neutrophils, and T cells in circulation is further observed within separate WT and S1PR3 KO generated UMAP analysis, where cell subgroups are spread amongst several islands, and corresponding SPADE trees, where subgroups are spread among many nodes and trajectories (Supplementary Fig. 1).
Reduced circulating WBCs in the peripheral blood of young, mature mice lacking S1PR3
Peripheral blood taken from WT and S1PR3 KO mice of an immature age (2–3 months) showed no differences in total white blood cell (WBC) count per mL of blood by Hemavet hematology analyzer, while WBC counts were drastically reduced in the blood of mature adult mice (6–8 months) lacking S1PR3 compared to their WT counterparts (Fig. 2E-I). Within these WBCs, we analyzed neutrophils (Fig. 2E-II), monocytes (Fig. 2E-III), CD4+ T cells (Fig. 2E-IV), and CD8+ T cells (Fig. 2E-V); each cell population was significantly reduced in S1PR3 KO mice compared to WT mice. The lack of S1PR3 caused a distinct change in the circulation of WBCs and therefore leads us to ask how the loss of S1PR3 affects local modulation of the immune microenvironment after severe tissue injury.
Loss of S1PR3 stimulates endogenous regeneration and repair of injured myofibers
To evaluate the effects of S1PR3 deletion on tissue regeneration, we first studied a 3mm full-thickness VML injury to the murine quadriceps which was previously defined as a critical, non-healing VML injury defect (10). We evaluated metrics of regenerating muscle fibers of S1PR3 KO compared to WT control mice. Representative cross sections of quadriceps 14 days post VML injury were stained for dystrophin to identify muscle fiber borders, embryonic myosin heavy chain (eMHC) to identify regenerating fibers, and DAPI to mark nuclei (Fig. 3A, B). These sections from both genotypes revealed the location of the defect (Fig. 3A and B, dotted white box) while contralateral quadricep cross sections showed no sign of injury (Fig. 3C, D). Centrally-located nuclei (Fig. 3I–J, white arrows), a hallmark of myofiber regeneration (46), were enhanced at the interface of healthy myofibers and the defect itself in both genotypes (Fig. 3I, J, M) relative to contralateral controls (Fig. 3K–M). Although centrally located nuclei were found within injured WT muscle fibers, the increased cellular infiltration observed within the void area of WT fibers (Fig. 3I) compared to S1PR3 KO fibers is indicative of chronic inflammation that delays the onset of endogenous regenerative mechanisms (47). Furthermore, loss of S1PR3 leads to significantly more fibers expressing the early regenerative marker, eMHC (48) (Fig. 3E, F white arrows), around the defect compared to WT (Fig. 3N), suggesting more regeneration within S1PR3 deficient muscle. Contralateral muscle cross sections from both genotypes did not show eMHC+ fibers, indicating no newly regenerating fibers in uninjured tissue, as expected (Fig. 3G, H, N). While both WT and S1PR3 KO mice exhibited some level of regeneration with increases in centrally located nuclei, a much higher eMHC expression profile indicates a continued regenerative response in mice lacking S1PR3. No differences were noted between cross sectional area of myofibers from VML injured WT and S1PR3 KO muscle (Fig. 3O).
Figure 3. Lack of S1PR3 enhances regenerative metrics of muscle healing.
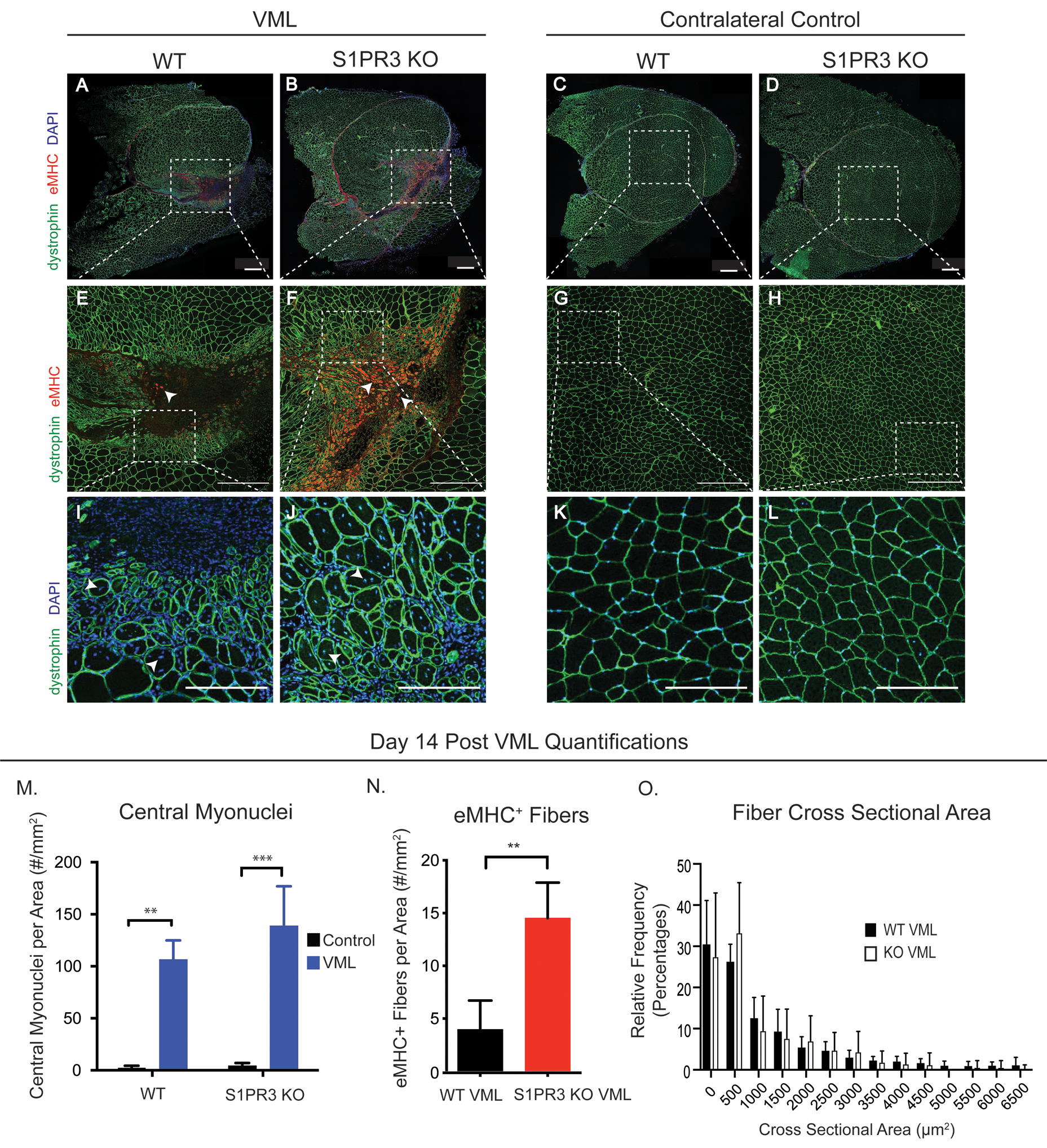
Representative immunofluorescence images of quadricep cross sections taken 14 days post 3mm full thickness VML injury. Muscle cross sections are stained for dystrophin (green), eMHC (red), and DAPI (blue). Quadriceps with a VML defect are seen in the left panel while uninjured contralateral control quadriceps are seen in the right panel, each including WT and S1PR3 KO mice. The first row of images (A-D) are full cross quadricep cross sections with all 3 stains depicted in the images. Dotted white boxes in injured sections (A, B) or uninjured sections (C, D) illustrate the defect area, or lack thereof, within the cross section that is zoomed in on in the second row of images (E-H) where white arrows point to eMHC+ fibers. Dotted white boxes in the second row (E-H) represent the ROI for a third row of images in which we further zoom in to display myonuclei location within regenerating and naïve muscle fibers (I-L). White arrows point to the centrally located myonuclei in regenerating fibers (I, J) while DAPI+ nuclei are located in the periphery of the fibers of contralateral quadriceps (K, L). (M) Centrally located nuclei in WT and S1PR3 KO mice with VML injuries compared to their contralateral controls. (N) eMHC+ fibers in VML defects from WT and S1PR3 KO mice normalized to cross sectional area. (O) Histogram of fiber cross sectional area between VML defects in WT and S1PR3 KO mice. Scale bars of 500μm (A-H) and 200 μm (I-L). Statistical tests conducted include two-way ANOVA with Tukey’s multiple comparisons (M) and two-tailed t-tests (N, O) with error bars representing standard deviation. n=4–8 samples per experimental group. **p< 0.01, ***p< 0.001
S1PR3 KO increases M2-like macrophages and reduces T cell infiltration into injured muscle
To evaluate the S1PR3-modulated immune response that may play a role in the promising regenerative metrics emerging from our quadriceps VML study in S1PR3 KO mice, we generated SPADE dendrograms from flow cytometry data from cells extracted from the muscle tissue 3 days after a 1mm full thickness VML defect to the murine spinotrapezius muscle, a model more sensitive to visualization of cell populations and tissue microstructures critical to injury repair. Figure 4 shows SPADE trees containing live, single cell events in both S1PR3 KO and WT control groups. For selected manually gated populations, including Ly6Clo anti-inflammatory monocytes (Fig. 4A), Ly6Chi classical inflammatory monocytes (Fig. 4B), CD206+ M2-like macrophages (Fig. 4C), Ly6Chi M1-like macrophages (Fig. 4D), CD4+ T cells (Fig. 4E), or CD8+ (Fig. 4F) T cells, we annotated the SPADE dendrogram with each respective immune subset. Nodes were colored by overall cell frequency. For each selected manually gated population, we generated vertically ordered bar graphs, or “tornado plots,” to visualize differences in average cell frequency percentage recruited to the muscle tissue between genotypes per annotated SPADE node. Bars of the tornado plots within the blue annotation represent nodes where the S1PR3 KO animals had fewer cells compared to WT, while the red annotation represents more cells in the KO versus the WT. These tornado plots revealed a reduction in the overall infiltration of CD4+ and CD8+ T cells and monocyte subsets and an enrichment of CD206+ M2-like macrophages within S1PR3 KO muscle defects. Statistical differences between genotypes by node are represented in Supplementary Figure 2. While we did not observe statistical differences in the heterogeneity of monocyte populations within the wound, there were significant increases in CD206+ M2-like macrophages from S1PR3 KO muscle within several of the annotated nodes compared to WT controls (Supplementary Fig. 2C) and a significant decrease in a Ly6Chi M1-like subset in annotated node 34 in S1PR3 KO injury versus WT control (Supplementary Fig. 2D). These results indicate that modulation of S1PR signaling in muscle injuries through loss of S1PR3 may reduce inflammatory cell infiltration while simultaneously encouraging the recruitment or polarization of pro-resolving immune subsets such as CD206+ M2-like macrophages which act as key players in immune modulation and myoblast differentiation (21).
Figure 4. Pseudotime visualization of immune cell recruitment following injury in S1PR3 KO and WT control mice.
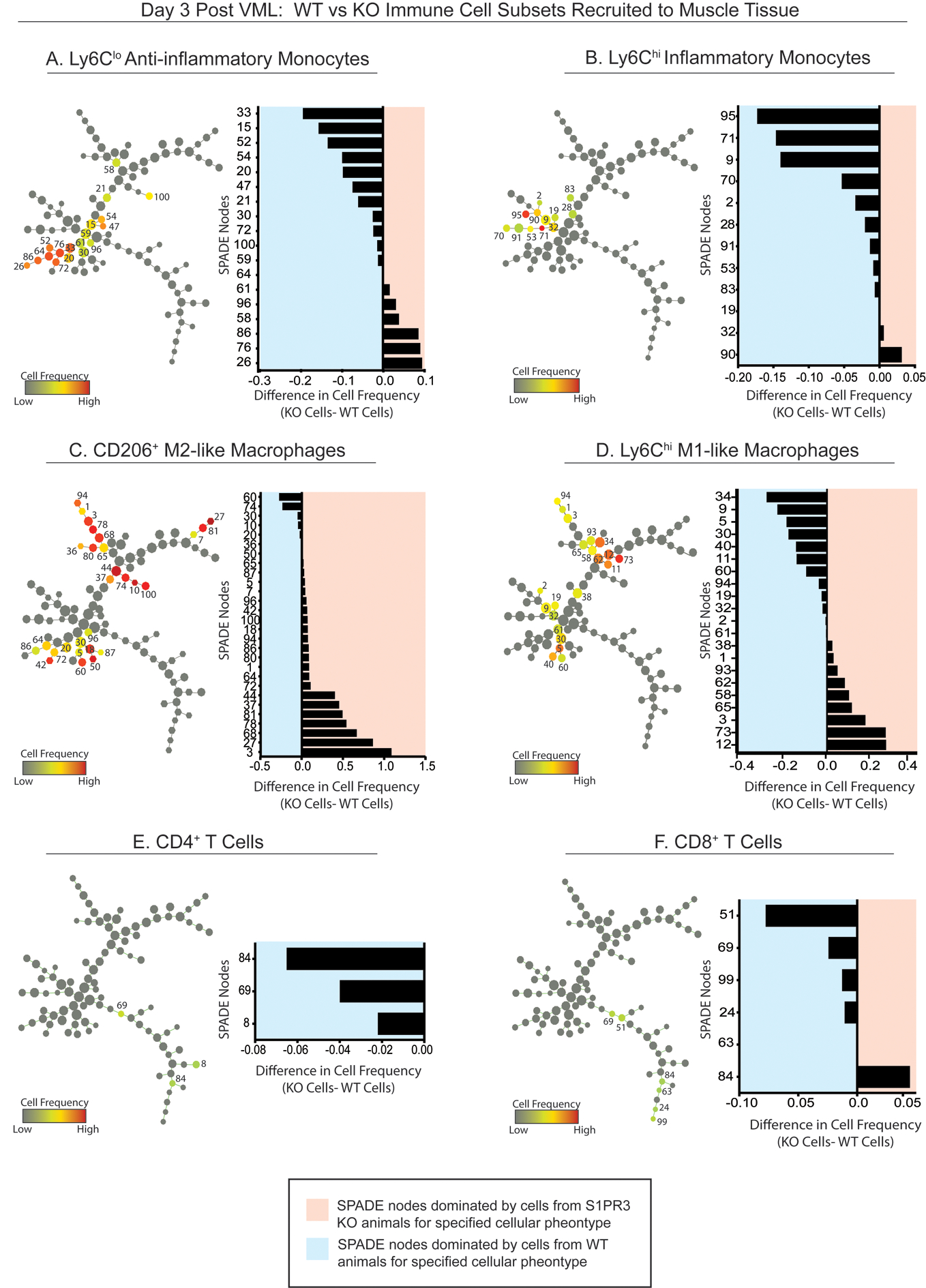
SPADE tree consisting of live, single cellular events extracted from the spinotrapezius muscle of S1PR3 KO and WT mice 3 days after volumetric muscle injury. Tree dendrograms are annotated to distinguish the different immune cell subpopulations that emerge through pseudotime trajectory projections after initial sequential biplot manual gating analysis. All SPADE nodes are visualized in grayscale so that color annotation overlay in each panel permits visualization of the normalized cell frequency within the node for each manually gated cell type (A-F) in the form of a heatmap. From SPADE tree dendrograms, corresponding tornado plots can be created to represent the difference in average cell frequency percentage between S1PR3 KO (KO) mice and WT controls for each cellular phenotype. SPADE node number assignments annotated on the dendrogram trees correlate to the node assignment on the y-axis of the accompanying tornado plot. n=7 samples per genotype.
Local pharmacological inhibition of S1PR3 mimics changes in immune infiltration in S1PR3 KO mice
To test whether localized delivery of a pharmacological S1PR3 antagonist could recapitulate the immune response of S1PR3 KO mice following injury, we made a 1mm full thickness defect in the spinotrapezius of WT mice and overlaid the defect with PLGA-PCL nanofibers releasing VPC01091. Nanofibers exhibited sustained release of VPC01091 over several days in vitro (Supplementary Fig. 3). Atomic force microscopy (AFM) was used to confirm similar topography between blank and VPC01091 nanofibers as well as to assess their mechanical properties. Nanofiber scaffolds loaded with VPC01091 were found to have an average Young’s modulus value around 11 MPa, consistent with the modulus values of blank nanofiber scaffolds (Supplementary Fig. 3). SPADE tree dendrograms were generated from flow cytometry data of live, single cell events of spinotrapezius muscle tissue treated with either blank control or VPC01091-releasing nanofibers 3 days after injury (Fig. 5). Selected manually gated populations, Ly6Clo anti-inflammatory and Ly6Chi classical inflammatory monocytes (Fig. 5A, B, respectively), CD206+ M2-like and Ly6Chi M1-like macrophages (Fig. 5C, D, respectively), and CD4+ and CD8+ T cells (Fig. 5E, F, respectively), were overlaid onto the SPADE tree. The tornado plots show that there was an overall reduction in inflammatory mononuclear subsets with a concurrent increase in anti-inflammatory phenotypes as well as a reduction in CD4+ T cells with VPC01091-mediated antagonism of S1PR3. Although VPC01091 delivery caused a bias towards CD8+ versus CD4+ T cell infiltration compared to controls, overall frequency of T cells in the muscle was low in both groups. We observed significant increases in the frequency of Ly6Clo anti-inflammatory monocytes (Fig. 5A) and CD206+ M2-like macrophages (Fig. 5C) along with significant decreases in the infiltration of Ly6Chi classical inflammatory monocytes (Fig. 5B), Ly6Chi M1-like macrophages (Fig. 5D), and CD4+ T cells (Fig. 5E) within at least one SPADE node (Supplementary Fig. 4). Within the blood, CD4 and CD8 lymphocyte numbers showed no significant differences between S1PR3 KO and WT mice or between animals treated with VPC01091-loaded scaffolds and blank control scaffolds 3 days after injury (Supplementary Fig. 5). Our results showed that local acute delivery of VPC01091 from a nanofiber scaffold mimicked key elements of our pseudotime analyses observed in S1PR3 KO mice, including increased CD206+ M2-like macrophages and decreased T cell invasion, demonstrating its effectiveness as an immunomodulatory therapy for targeting S1P receptor signaling after injury.
Figure 5. Pseudotime visualization of immune cell recruitment following injury in mice treated with VPC01091-loaded nanofibers and nanofiber vehicle controls.
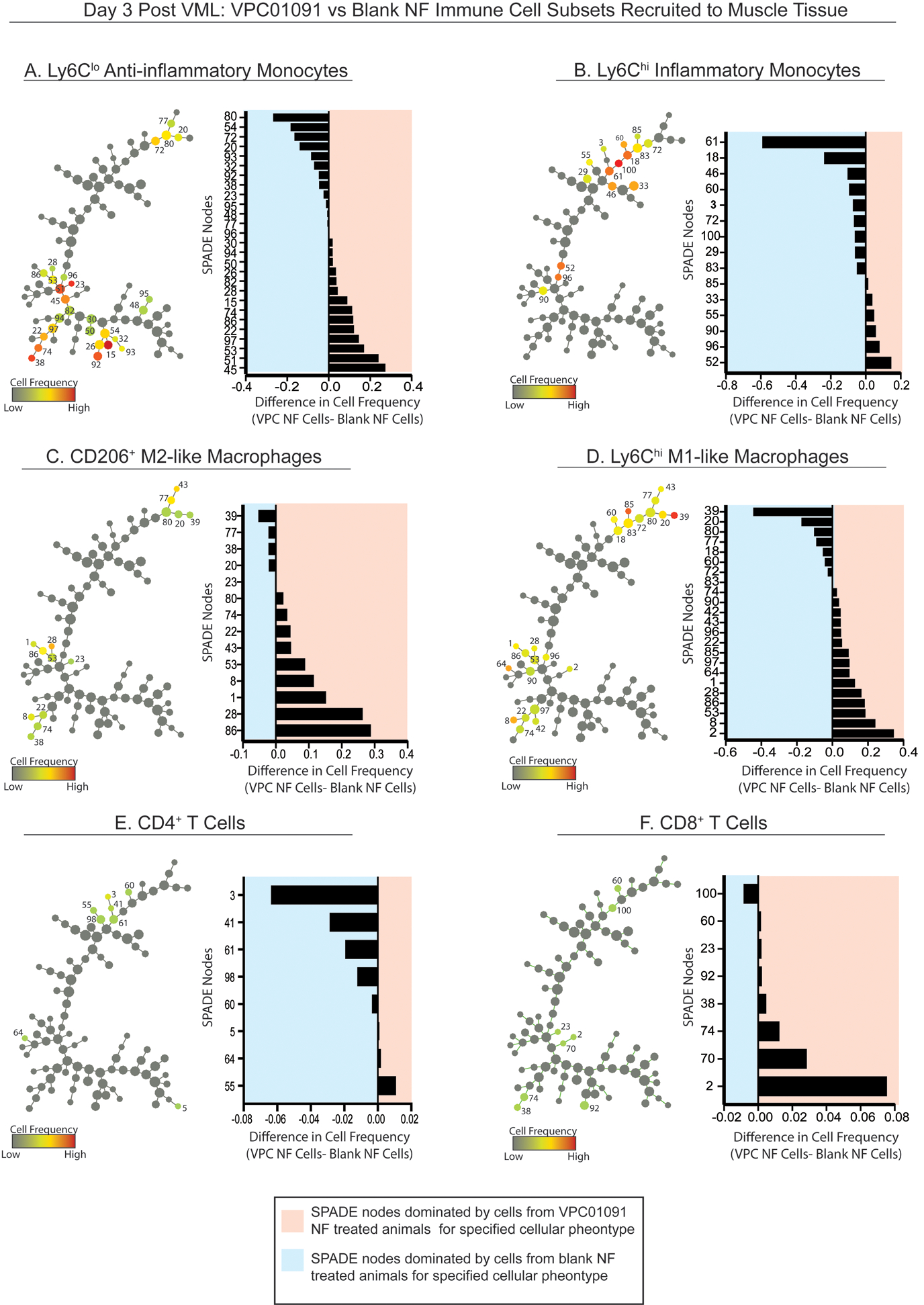
SPADE tree consisting of live, single cellular events extracted from the spinotrapezius muscle 3 days after volumetric muscle injury from WT mice treated with blank or VPC01091 nanofibers. Tree dendrograms are annotated to distinguish the different immune cell subpopulations that emerge through pseudotime trajectory projections after initial sequential biplot manual gating analysis. All SPADE nodes are visualized in grayscale so that color annotation overlay in each panel permits visualization of the normalized cell frequency within the node for each manually gated cell type (A-F) in the form of a heatmap. From SPADE tree dendrograms, corresponding tornado plots can be created to represent the difference in average cell frequency percentage between muscle defects treated with PGLA-PCL nanofibers (NF) releasing VPC01091 (VPC) and blank NF controls. SPADE node number assignments annotated on the dendrogram trees correlate to the node assignment on the y-axis of the accompanying tornado plot. n=8 samples per treatment group.
Local modulation of S1P receptor signaling leads to an enhanced resident MuSC population
The interplay between immune cell subsets, namely macrophages, and MuSCs’ ability to proliferate and differentiate into myofibers is a vital cellular mechanism underlying muscle regeneration. The injury niche and its precise transition from inflammatory M1-like macrophage dominance to M2-like macrophage recruitment can thus significantly influence MuSC function and subsequent repair (21). To first understand the resting population of MuSCs in uninjured muscle, we explanted both spinotrapezius and quadricep muscle from S1PR3 KO and WT controls for MuSC frequency analysis. Our results showed that there were no significant differences in homeostatic MuSC frequency within the spinotrapezius muscle between groups, yet there were significantly more MuSCs in the uninjured quadriceps of S1PR3 KO mice compared to WT controls (Supplementary Fig. 6).
To understand how the MuSC population changes due to local modulation of S1PR signaling after injury, we evaluated the early effects of VPC01091 nanofiber treatment on local MuSCs within a 1mm spinotrapezius muscle defect. The manually pre-gated Lineage (Lin)− CD29+ CXCR4+ MuSC population was overlaid onto a SPADE tree of Lin− cells (Fig. 6A) and revealed to cluster into a particular branching of the tree with clear double expression of CD29 and CXCR4 (Fig. 6B). We generated a heatmap from the Lin− SPADE tree in which all 100 nodes were visualized for expression levels of CD29, CXCR4, and Lin, normalized with z-scores calculated for each marker (Fig. 6C). A distinct Lin−CD29+CXCR4+ grouping of nodes, the nodes containing MuSCs, is denoted with a black bracket, and the nodes within this bracketed section of the heatmap correspond exactly to the node assignments of the MuSC manual overlay. While all MuSC nodes were dominated by cells from VPC01091 treated muscle, nodes 1, 15, and 62 specifically showed significant increases in MuSC frequency in response to this local S1PR3 inhibition (Fig. 6D-I). A tornado plot generated from MuSC containing nodes shows that MuSC numbers are extensively expanded from pharmacological S1PR3 antagonism (Fig. 6D-II). Finally, overall MuSC frequency as a percentage of all events shows that the pool of MuSCs within the spinotrapezius muscle is drastically enriched with VPC01091 delivery (Fig. 6D-III). These results suggest that local immunomodulation can not only modulate the inflammatory cues within the muscle injury, but it can also expand the MuSC pool within the injured muscle, two promising feats for driving a regenerative muscle microenvironment.
Figure 6. SPADE analysis reveals enhanced pool of MuSCs with local immune modulation.
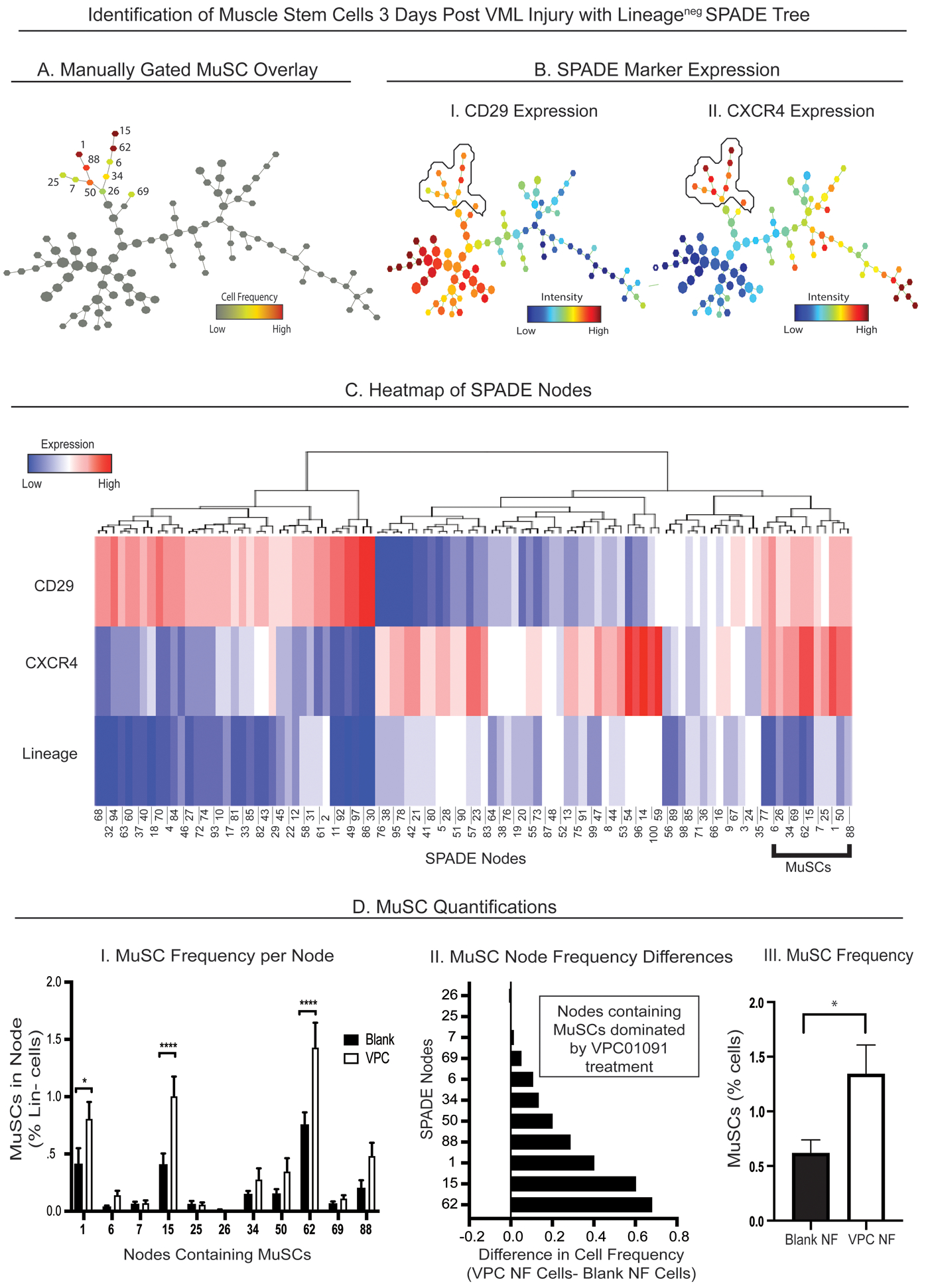
SPADE tree dendrogram consisting of live, single Lin− cells (negatively selected for hematopoietic lineage) extracted from the muscle tissue 3 days after injury from animals treated with blank vehicle nanofibers (NFs) and NFs releasing VPC01091 (A-B). (A) MuSCs were manually gated and then overlaid onto the SPADE tree as a unique subpopulation of all Lin− cells represented in the tree. Each node containing MuSCs is given a node assignment number as shown besides each colored node, where the color of the node corresponds to normalized cell frequency of MuSCs in heatmap form. (B) Lin− SPADE trees overlaid with CD29 (B-I) and CXCR4 (B-II) fluorescence marker intensities with the manual MuSC annotation circled to represent the double CD29+CXCR4+ expression defining the MuSC subpopulation. (C) Heatmap representation of each SPADE node from the Lin− tree generated with z-scores calculated per surface marker used to identify MuSCs. Black bracket defines the CD29+CXCR4+ nodes containing MuSCs. (D) MuSC quantifications from muscle tissue. (D-I) Frequency of MuSCs per annotated node from animals treated with NFs releasing VPC01091 (VPC) and blank NFs. (D-II) Tornado plot representing the difference in average cell frequency percentage between the 2 treatment groups, and (D-III) overall MuSC frequency between blank and VPC01091-loaded NF treatments. Data presented as mean ± S.E.M. Statistical analysis performed include two-way ANOVA with Tukey’s multiple comparisons (D-I) and two-tailed t-test (D-III), *p< 0.05, ****p< 0.0001. n=8 samples per treatment group.
Improved skeletal muscle regeneration with localized S1PR3 targeted immunotherapy
To determine whether accumulation of pro-regenerative immune cells and increased MuSCs resulting from loss of S1PR3 signaling translates to improved wound healing after injury, we assessed parameters of muscle regeneration 7 days after VML injury to the spinotrapezius muscle in both the S1PR3 KO and the VPC01091 nanofiber treated animals. Representative images of the muscle tissue with 1mm full thickness defects across 3 different treatment groups were stained with desmin (green) to identify muscle fibers and CD68 (blue) to visualize infiltrating macrophages (Figure 7A–C). Notably, treatment with blank nanofiber scaffolds resulted in large void defect areas (Fig. 7A) while newly developed fibers attempting to bridge the defect space were observed in groups lacking S1PR3 signaling through pharmacologic or genetic means (Fig. 7B, C). At the interface between the defect space and the muscle fibers surrounding the area (interface denoted with white dotted box), there was an overwhelming infiltration of macrophages (denoted with white arrows) into the defect area of muscle treated with blank nanofibers, likely impairing subsequent regeneration due to chronic inflammatory conditions (Fig. 7D). This observation is further supported by the significantly increased infiltration of macrophages in muscle tissue treated with blank nanofibers at day 3 post injury compared to WT mice that received no treatment (Supplementary Fig. 7). However, VPC01091 treated muscle as well as S1PR3 KO derived muscle showed less macrophage infiltration and more regenerating myofibers bridging across the injury site (Fig. 7E, F). The percentage of the defect area remaining 7 days post injury is significantly decreased in S1PR3 KO defects as well as in defects treated with VPC01091 delivery compared to WT and blank scaffold controls, respectively (Fig. 7G). Furthermore, we measured the fiber diameter of all regenerating fibers growing into the defect space (example measurement denoted with red bracket) across animals of all treatment groups and found that fiber diameter was significantly greater in S1PR3 KO and VPC01091 treated groups compared to both untreated WT and blank scaffold controls (Fig. 7H–J). Taken together, this data indicates that muscle defects from groups modulating S1PR3 signaling exhibit improved regeneration following injury, and local immunotherapy via VPC01091 delivery results in muscle healing just as effective as with global S1PR3 knockout.
Figure 7. Local S1PR3 inhibition and global S1PR3 knockout have comparable regenerative effects after muscle injury.
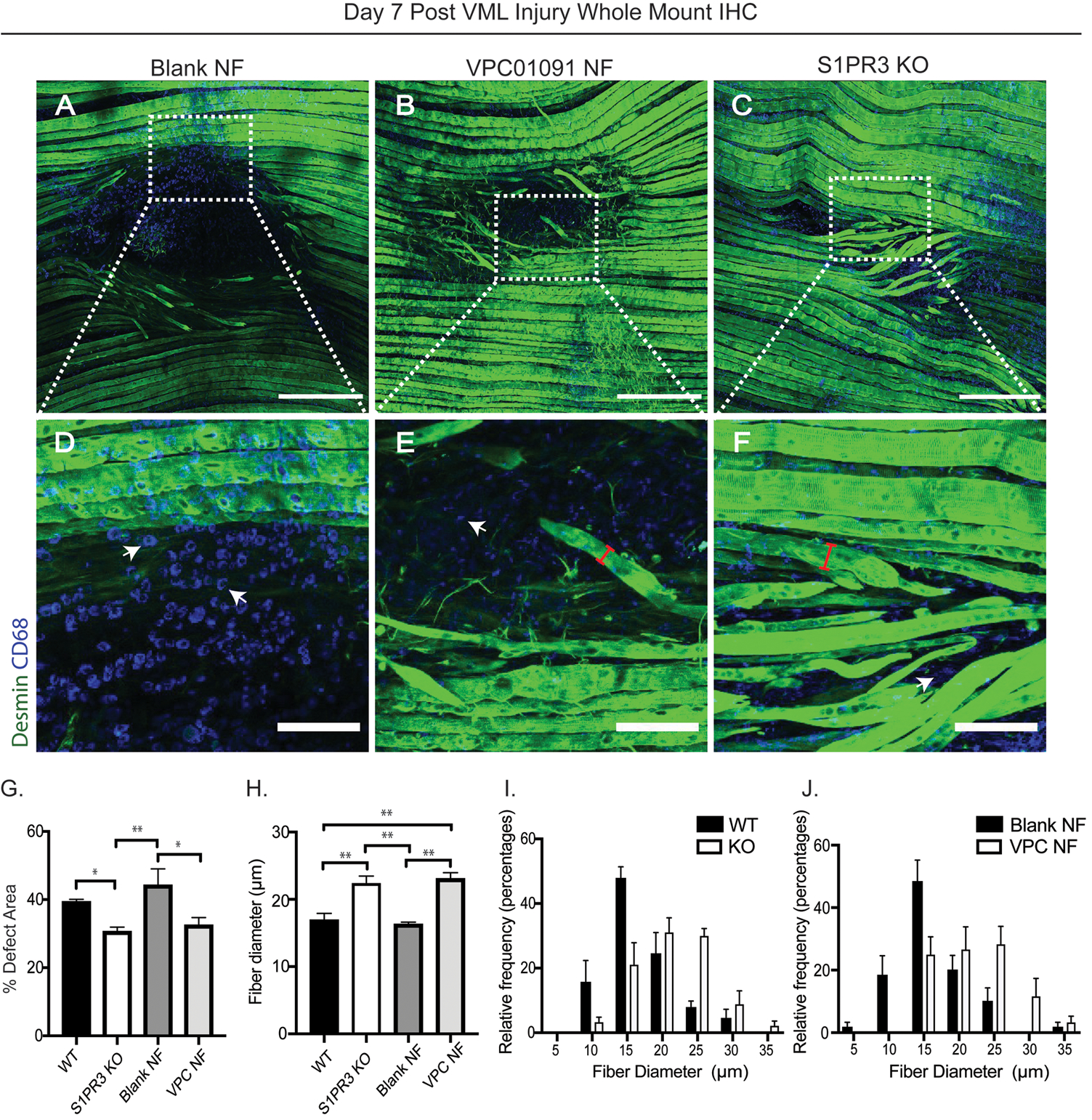
Whole mount immunohistochemistry (IHC) of explanted spinotrapezius muscle 7 days post injury (A-F) stained with desmin (green) and CD68 (blue). Full image of spinotrapezius defects are shown in the first row (A-C) while ROI sections of the full image, as denoted with white boxes, are displayed in the second row (D-F) directly below its respective full image. Representative images from animals treated with blank vehicle nanofibers (NF) (A), VPC01091 loaded NFs (B), and S1PR3 KO mice (C) display the bridging of myofibers across the defect void or lack thereof, and further, the interface between the fibers and defect area can be visualized in (D-F) for each treatment. White arrows point to CD68+ macrophages within the defect space (D-F) while red brackets indicate example diameter measurements across regenerating myofibers (E, F) bridging across the defect. (G) Percentage of defect area remaining 7 days post injury across treatments. (H) Fiber diameter measurements from animals across all treatment groups with corresponding fiber diameter histograms displaying the relative distributions of fibers from WT vs KO mice (I) and VPC01091 vs blank control NFs (J) in diameter measurements. Scale bars of 500 μm (A-C) and 100 μm (D-F). Data presented as mean ± S.E.M. Statistical tests performed include a one-way ANOVA with Tukey’s multiple comparisons (G, H) with n= 4–6 samples for experimental groups. *p< 0.05, **p< 0.01
Discussion
Regenerative immunotherapies, in which local cues are provided in situ to influence the mobilization, homing, phenotype, and function of key inflammatory endogenous host cells, represent an attractive new approach for the treatment of volumetric muscle injuries. However, these approaches require an understanding of immune responses to injury and the ability to target multiple signaling pathways in a wide range of specialized host cell types (49). Our team developed a series of severe rodent trauma models of full thickness VML defects wherein we utilize dimensionality reduction and pseudotime analysis to characterize the chronically dysregulated immune response following injury as a function of S1P receptor function. With severe defects showing high levels of both Ly6Chi M1-like and CD206+ M2-like macrophage subsets as early as 3 days post injury, controlling the invasion and polarization of immunologically defined monocyte and macrophage populations in vivo represents an important step in harnessing their role in the intricate muscle injury repair.
In this study, we investigated novel immunotherapies which increase recruitment of Ly6Clo non-classical monocytes and other pro-regenerative immune cells while also stabilizing pro-regenerative macrophage functions. Our findings show that S1PR3 KO drastically increased the number of infiltrating CD206+ M2-like macrophages into the injured muscle. Furthermore, early invasion of CD4+ and CD8+ T cells was reduced at day 3 along with decreased infiltration of pro-inflammatory monocytes. S1PR3 KO in a VML quadriceps model also resulted in increased expression of eMHC 14 days post injury along with increasing numbers of centrally located nuclei compared to WT controls. As eMHC is transiently expressed in the beginning stages of muscle regeneration (48), loss of function of S1PR3 is directly linked to enhanced myofiber regeneration. Moreover, the presence of centrally located nuclei within S1PR3 KO mice reveals that myoblasts are fusing together to form new, regenerating myofibers as myonuclei accumulate at the center during the early stages of myofiber development before ultimately migrating to the periphery. These results demonstrate the potential of developing immunotherapies which modulate S1P receptor activation to improve skeletal muscle regeneration following severe trauma. With this in mind, we aimed to take a deeper look into the cellular players driving injury repair.
To test whether delivery of a pharmacological inhibitor of S1PR3 could replicate changes in the immune infiltration of S1PR3 KO mice, PLGA-PCL nanofibers releasing VPC01091 were sutured over the defect of WT mice. VPC01091 is a local inhibitor of endogenous S1P production and inhibits S1PR3 signaling in circulating monocytes (33). We showed that local delivery of VPC01091 recapitulates the immune infiltration of S1PR3 KO mice, including relative increases in CD206+ M2-like macrophages. Within injured spinotrapezius tissue, CD4+ T lymphocyte invasion was decreased at day 3 with delivery of VPC01091 and overall frequency of T cells was quite low likely due to removal of surface S1PR1 by functional antagonism. Pseudotime dimensionality reduction of flow cytometry profiles using SPADE enabled novel evaluation of the various intermediate states characteristic of temporally regulated inflammation cascades, and we visualized the innate cellular heterogeneity contained within immune subsets. Manually overlaying selected gates such as M2-like macrophages allowed us to elucidate important transitions in macrophage marker expression profiles, primarily involving Ly6C and CD206, and to evaluate the expression profile changes of these markers as a response to S1PR modulation. The increases in CD206+ M2-like macrophages shown at day 3 with both global and localized loss of S1PR3 are likely key contributors to the improved healing and regeneration observed at day 7 within the defect area, as these M2-like macrophages support extracellular matrix deposition and wound closure via their high expression of arginase-1 allowing them to generate collagen precursors (50). Future studies are necessary to further assess the carefully coordinated balance between recruitment of anti-inflammatory M2-like macrophages for the promotion of wound healing with a prolonged M2-like macrophage presence leading to undesired fibrosis in injured tissue.
Determining direct functional links between alternatively-activated M2-like macrophages and other pro-regenerative changes to the inflammatory microenvironment on MuSCs remains a challenge due to the marked heterogeneity that M2-like macrophages exhibit both in in vitro polarization paradigms, and in in vivo injury niches. Since macrophages are known to impact MuSC behavior, we evaluated the effect of VPC01091-releasing nanofibers on MuSC numbers. Unbiased pseudotimes of Lin− cellular events generated using SPADE as well as traditional biplot gating showed that MuSC frequency was significantly increased by local immunotherapy 3 days after VML injury. These compelling results are supported by previous studies in which the overexpression of S1PR3 led to suppressed myoblast proliferation and reduced myotube fusion (51). While the increased population of M2-like macrophages likely supports the proliferation and differentiation of MuSCs (21, 52), further studies are necessary to elucidate the interaction between this macrophage subset and MuSC progressions due to S1PR modulation. IL-4-polarized macrophages have been shown in vitro to suppress proliferation and promote differentiation of myoblasts (53), whereas macrophages polarized via IL-10 exposure promoted C2C12 myoblast proliferation but had no effects on myoblast differentiation as evaluated by myoD and myogenin protein expression (54). An intriguing hypothesis is that the decrease in CD4+ (which produce IL-4) T-lymphocytes due to VPC01091 delivery and local manipulation of S1P receptor signaling may shift the injury cytokine milieu towards favoring IL-10-mediated M2 polarization. Further studies that probe the muscle cytokine profile in response to VPC01091 are necessary in order to further elucidate the connection between lymphoid and myeloid cell populations within skeletal muscle injury.
Critically sized VML defects are also characterized by fatty infiltration, significant fibrosis, and no new bridging of myofibers across the defect area. We evaluated local S1P inhibition in a spinotrapezius model of VML and saw significant increases in fiber diameter from both S1PR3 KO mice as well as those treated with VPC01091 in comparison to their respective controls. Moreover, we show that both S1PR3 KO mice and VPC01091 delivery greatly reduces the total defect area, as the regenerating myofibers bridge the void to repair the defect as they align with healthy fibers. This work demonstrates the effectiveness of local immunomodulatory approaches for remodeling S1P receptor function to promote muscle regeneration and repair. These findings suggest that the combination of pro-reparative immune cell subset recruitment and the expanded MuSC population in response to local antagonism of S1PR3 has a direct influence on functional regeneration of skeletal muscle.
Overall, our results demonstrate that local immunomodulation of innate and adaptive immune responses via VPC01091 delivery and modulation of S1P receptor signaling coincide with increased MuSC numbers after volumetric muscle injury along with pro-regenerative immune cell infiltration and enhanced muscle defect repair. Our work highlights the promise of leveraging advanced pseudotime analytics to identify patterns of pro-resolving immune subset infiltration that can be modulated with novel engineered immunotherapies to promote the regeneration of skeletal muscle.
Supplementary Material
Supplemental Figure 1. Visualization of circulating immune cell subsets with single cell 2D projections and pseudotime trajectories reveals innate leukocyte heterogeneity. High dimensional flow cytometry data of peripheral blood taken from uninjured animals represented as 2D projections using UMAP and SPADE dimensionality reduction techniques. (A) SPADE trees and accompanying UMAPs generated with manually gated CD11b+ cells circulating homeostatic blood from WT mice (A-I, III) as well as S1PR3 KO (KO) mice (A-II, IV). Within the CD11b+ SPADE trees, neutrophils (A-I, II) and monocytes (A-III, IV) were manually gated and overlaid onto their respective SPADE trees, where the node color represents normalized cell frequency of its specified phenotype in heatmap form. A red overlay on the corresponding UMAPs represent cells identified as a neutrophil or monocyte, respectively. (B) SPADE trees and UMAPS generated with manually gated CD3+ T cells from the blood of uninjured WT mice (B-I, III) and S1PR3 KO mice (B-II, IV). CD4+ T cells (B-I, II) and CD8+ T cells (B-III, IV) were manually gated and overlaid onto the SPADE trees, as well as onto the single cell UMAP projections. UMAP and SPADE representations of high dimensional flow data reveal discrete heterogeneities as observed through the multiple islands depicted on the UMAPs and many nodes clustered via SPADE per phenotypic characterization. n=3 samples (A) and 7 samples (B) per genotype.
Supplemental Figure 2. Cell frequency quantifications between WT and S1PR3 KO mice within each SPADE node containing cells of specified phenotype. Bar graph representation and statistical analysis applied to the tornado plots presented in Figure 4. For each specified cellular phenotype, every node containing cells of this particular subset are analyzed for phenotype-specific cell frequency between experimental groups to evaluate which nodes are significantly dominated by subsets from a particular genotype. Nodes containing cells of the respective phenotype are denoted along the x-axis while the phenotype-specific cell frequency is quantified as a percentage of all live cells. Two-way ANOVAs with Tukey's multiple comparisons were performed for statistical analyses with n=7 samples per genotype. Data is presented as mean ± S.E.M. *p< 0.05, **p< 0.01, ****p< 0.0001
Supplemental Figure 3. Characterization of VPC01091-loaded nanofiber scaffold. Release of VPC01091 from PLGA-PCL nanofiber scaffolds and biomaterial characterization by atomic force microscopy (AFM). (A) Experimental setup for electrospinning VPC01091-loaded PLGA-PCL nanofibers. (B) VPC01091 in-vitro release profile exhibits sustained delivery. (C) Average Young’s modulus values obtained via AFM for blank control nanofiber scaffold (NF) and NF loaded with VPC01091 (VPC). (D, G) AFM generated topography (height) map of blank (D) and VPC01091-loaded (G) PLGA-PCL nanofiber scaffold. Height values throughout nanofiber range from low (black) to high (white) regions. (E, H) AFM generated stiffness map of blank (E) and VPC01091-loaded (H) PLGA-PCL nanofiber scaffold in which white regions represent areas of greater stiffness. (F, I) Histogram of Young's modulus values (MPa) obtained from AFM over multiple contact points on blank (F) and VPC01091-loaded (I) PLGA-PCL nanofiber scaffold. Unpaired two-sided t-test was performed for statistical analysis (C) and data is presented as mean ± S.E.M.
Supplemental Figure 4. Cell frequency quantifications between mice treated with control and VPC01091-loaded nanofibers within each SPADE node containing cells of specified phenotype. Bar graph representation and statistical analysis applied to the tornado plots presented in Figure 5. For each specified cellular phenotype, every node containing cells of this particular subset are analyzed for phenotype-specific cell frequency between experimental groups to evaluate which nodes are significantly dominated by subsets from a particular treatment. Nodes containing cells of the respective phenotype are denoted along the x-axis while the phenotype-specific cell frequency is quantified as a percentage of all live cells. Two-way ANOVAs with Tukey’s multiple comparisons were performed for statistical analysis with n=8 samples per treatment. Data is presented as mean ± S.E.M. *p< 0.05, **p< 0.01, ***p< 0.001, ****p< 0.000.
Supplemental Figure 5. Circulating lymphocytes after muscle injury unaffected by S1PR3 deficiency. SPADE trees generated with CD3+ T cells from the peripheral blood of WT and global S1PR3 KO mice (A-C) and mice treated with blank control nanofibers (NF) and those releasing VPC01091 (D-F) 3 days post VML injury. (A, D) CD4+ (I) and CD8+ (II) T cells were manually gated and overlaid onto their respective SPADE trees where colored node annotation represents normalized cell frequency in the form of a heatmap, illustrating the phenotypic differences in these T cell subpopulations as they are located on opposing sides of the tree. (B, E) CD4 (I) and CD8 (II) fluorescence marker intensities overlaid onto the SPADE trees with the manually gated annotation circled in black to further characterize their expression as displayed by SPADE clustering. (C, F) SPADE quantifications of total CD4+ and CD8+ T cells circulating the blood of WT vs S1PR3 KO mice (C) and mice treated with blank control or VPC01091-loaded (VPC) nanofibers (F). Data represented as mean ± S.E.M. using a two-way ANOVA with Tukey’s multiple comparisons test. n=7 samples per experimental group.
Supplemental Figure 6. Lack of S1PR3 leads to increased numbers of muscle resident stem cells. SPADE trees generated from Lin− cell events taken from uninjured spinotrapezius and hindlimb muscles. (A) MuSCs from the spinotrapezius (A-I) and hindlimb (A-II) are manually gated and overlaid onto their respective trees, annotated with colored nodes representing normalized cell frequency and displayed as a heatmap on the tree. Each node containing MuSCs is given a node assignment number as given next to each colored node. (B) MuSC frequency within each node annotated on spinotrapezius and hindlimb derived SPADE trees. Nodes from the SPADE tree generated with Lin− spinotrapezius derived cells are quantified for MuSC frequency (B-I) while those derived from the hindlimb are quantified by node in (B-II). (C) Tornado plot representation of the difference in average MuSC frequency percentage between S1PR3 deficient mice and their WT counterparts shows that nodes containing MuSCs largely contain those derived from mice lacking S1PR3 (KO) in both the spinotrapezius and hindlimb (C-I, II). Two-way ANOVAs with Tukey’s multiple comparisons were conducted for statistical analyses and data is presented as mean ± S.E.M. with n=3 per genotype for hindlimb samples and an n=4 per genotype for spinotrapezius samples. *p< 0.05
Supplemental Figure 7. Treatment with blank nanofiber scaffold increases macrophage infiltration to defect area 3 days post injury. Flow cytometry analysis of CD11b+CD64+MerTK+ macrophages in injured spinotrapezius muscle 3 days post injury between WT mice receiving no treatment (injury control, black bar) and WT mice treated with blank nanofiber scaffolds (white bar). Statistical analysis performed with two-tailed t-test. Data presented as mean ± S.E.M. n=7 for WT injury control and n=8 for mice treated with blank nanofibers.
Acknowledgements
General: We thank the core facilities at the Parker H. Petit Institute for Bioengineering and Bioscience at the Georgia Institute of Technology for the use of their shared equipment, services, and expertise.
Funding: This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1650044 as well as by the NIH under award numbers R01AR071708 and R56AR071708, and the NIH Cell and Tissue Engineering training grant under award number GM008433.
References
- 1.Banerjee M, Bouillon B, Shafizadeh S, Paffrath T, Lefering R, Wafaisade A, German Trauma Registry G. Epidemiology of extremity injuries in multiple trauma patients. Injury. 2013;44(8):1015–21. Epub 2013/01/05. doi: 10.1016/j.injury.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Dougherty AL, Mohrle CR, Galarneau MR, Woodruff SI, Dye JL, Quinn KH. Battlefield extremity injuries in Operation Iraqi Freedom. Injury. 2009;40(7):772–7. Epub 2009/05/20. doi: 10.1016/j.injury.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Owens BD, Kragh JF Jr., Wenke JC, Macaitis J, Wade CE, Holcomb JB. Combat wounds in operation Iraqi Freedom and operation Enduring Freedom. J Trauma. 2008;64(2):295–9. Epub 2008/02/28. doi: 10.1097/TA.0b013e318163b875. [DOI] [PubMed] [Google Scholar]
- 4.Kyba M Skeletal Muscle Regeneration in the Mouse Methods and Protocols Preface. Methods Mol Biol. 2016;1460:V–V. doi: 10.1007/978-1-4939-3810-0. [DOI] [Google Scholar]
- 5.Garg K, Ward CL, Hurtgen BJ, Wilken JM, Stinner DJ, Wenke JC, Owens JG, Corona BT. Volumetric muscle loss: persistent functional deficits beyond frank loss of tissue. J Orthop Res. 2015;33(1):40–6. Epub 2014/09/19. doi: 10.1002/jor.22730. [DOI] [PubMed] [Google Scholar]
- 6.Corona BT, Rivera JC, Owens JG, Wenke JC, Rathbone CR. Volumetric muscle loss leads to permanent disability following extremity trauma. J Rehabil Res Dev. 2015;52(7):785–92. Epub 2016/01/09. doi: 10.1682/JRRD.2014.07.0165. [DOI] [PubMed] [Google Scholar]
- 7.Hsu JR, Grogan BF. Volumetric Muscle Loss. Skeletal Trauma Research Consortium [Internet]. 2011; 19:[S35–S7 pp.]. [DOI] [PubMed] [Google Scholar]
- 8.Sefcik LS, Aronin CEP, Awojoodu AO, Shin SJ, Gabhann FM, MacDonald TL, Wamhoff BR, Lynch KR, Peirce SM, Botchwey EA. Selective Activation of Sphingosine 1-Phosphate Receptors 1 and 3 Promotes Local Microvascular Network Growth. Tissue Eng Pt A. 2011;17(5–6):617–29. doi: 10.1089/ten.tea.2010.0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emeterio CLS, Olingy CE, Chu Y, Botchwey EA. Selective recruitment of non-classical monocytes promotes skeletal muscle repair. Biomaterials. 2017;117(Physiol. Rev. 93 1 2013):32–43. doi: 10.1016/j.biomaterials.2016.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson SE, Han WM, Srinivasa V, Mohiuddin M, Ruehle MA, Moon JY, Shin E, San Emeterio CL, Ogle ME, Botchwey EA, Willett NJ, Jang YC. Determination of a Critical Size Threshold for Volumetric Muscle Loss in the Mouse Quadriceps. Tissue Eng Part C Methods. 2019;25(2):59–70. Epub 2019/01/17. doi: 10.1089/ten.TEC.2018.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy D, Besnard A, Latil M, Jouvion G, Briand D, Thepenier C, Pascal Q, Guguin A, Gayraud-Morel B, Cavaillon JM, Tajbakhsh S, Rocheteau P, Chretien F. Comparative Study of Injury Models for Studying Muscle Regeneration in Mice. PLoS One. 2016;11(1):e0147198. Epub 2016/01/26. doi: 10.1371/journal.pone.0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao WH, Liu Y, Chen PJ. Macrophage Depletion Impairs Skeletal Muscle Regeneration: the Roles of Pro-fibrotic Factors, Inflammation, and Oxidative Stress. Inflammation. 2016;39(6):2016–28. doi: 10.1007/s10753-016-0438-8. [DOI] [PubMed] [Google Scholar]
- 13.Shen W, Li Y, Zhu JH, Schwendener R, Huard J. Interaction between macrophages, TGF-beta 1, and the COX-2 pathway during the inflammatory phase of skeletal muscle healing after injury. J Cell Physiol. 2008;214(2):405–12. doi: 10.1002/jcp.21212. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Wehling-Henricks M, Samengo G, Tidball JG. Increases of M2a macrophages and fibrosis in aging muscle are influenced by bone marrow aging and negatively regulated by muscle-derived nitric oxide. Aging Cell. 2015;14(4):678–88. Epub 2015/05/27. doi: 10.1111/acel.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown BN, Ratner BD, Goodman SB, Amar S, Badylak SF. Macrophage polarization: an opportunity for improved outcomes in biomaterials and regenerative medicine. Biomaterials. 2012;33(15):3792–802. Epub 2012/03/06. doi: 10.1016/j.biomaterials.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donati C, Cencetti F, Bruni P. Sphingosine 1-phosphate axis: a new leader actor in skeletal muscle biology. Front Physiol. 2013;4:338. Epub 2013/12/11. doi: 10.3389/fphys.2013.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159(6):1327–40. Epub 2014/12/07. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbons MA, MacKinnon AC, Ramachandran P, Dhaliwal K, Duffin R, Phythian-Adams AT, van Rooijen N, Haslett C, Howie SE, Simpson AJ, Hirani N, Gauldie J, Iredale JP, Sethi T, Forbes SJ. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am J Respir Crit Care Med. 2011;184(5):569–81. Epub 2011/06/18. doi: 10.1164/rccm.201010-1719OC. [DOI] [PubMed] [Google Scholar]
- 19.Tonkin J, Temmerman L, Sampson RD, Gallego-Colon E, Barberi L, Bilbao D, Schneider MD, Musaro A, Rosenthal N. Monocyte/Macrophage-derived IGF-1 Orchestrates Murine Skeletal Muscle Regeneration and Modulates Autocrine Polarization. Mol Ther. 2015;23(7):1189–200. doi: 10.1038/mt.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng B, Wehling-Henricks M, Villalta SA, Wang Y, Tidball JG. IL-10 Triggers Changes in Macrophage Phenotype That Promote Muscle Growth and Regeneration. Journal of immunology. 2012;189(7):3669–80. doi: 10.4049/jimmunol.1103180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wosczyna MN, Rando TA. A Muscle Stem Cell Support Group: Coordinated Cellular Responses in Muscle Regeneration. Dev Cell. 2018;46(2):135–43. Epub 2018/07/18. doi: 10.1016/j.devcel.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spiller KL, Anfang RR, Spiller KJ, Ng J, Nakazawa KR, Daulton JW, Vunjak-Novakovic G. The role of macrophage phenotype in vascularization of tissue engineering scaffolds. Biomaterials. 2014;35(15):4477–88. Epub 2014/03/05. doi: 10.1016/j.biomaterials.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sommerfeld SD, Cherry C, Schwab RM, Chung L, Maestas DR Jr., Laffont P, Stein JE, Tam A, Ganguly S, Housseau F, Taube JM, Pardoll DM, Cahan P, Elisseeff JH. Interleukin-36gamma-producing macrophages drive IL-17-mediated fibrosis. Sci Immunol. 2019;4(40). Epub 2019/10/13. doi: 10.1126/sciimmunol.aax4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olingy CE, San Emeterio CL, Ogle ME, Krieger JR, Bruce AC, Pfau DD, Jordan BT, Peirce SM, Botchwey EA. Non-classical monocytes are biased progenitors of wound healing macrophages during soft tissue injury. Scientific reports. 2017;7(1):447. Epub 2017/03/30. doi: 10.1038/s41598-017-00477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes JE, Srinivasan S, Lynch KR, Proia RL, Ferdek P, Hedrick CC. Sphingosine-1-phosphate induces an antiinflammatory phenotype in macrophages. Circ Res. 2008;102(8):950–8. Epub 2008/03/08. doi: 10.1161/CIRCRESAHA.107.170779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo B, Gan W, Liu Z, Shen Z, Wang J, Shi R, Liu Y, Liu Y, Jiang M, Zhang Z, Wu Y. Erythropoeitin Signaling in Macrophages Promotes Dying Cell Clearance and Immune Tolerance. Immunity. 2016;44(2):287–302. Epub 2016/02/14. doi: 10.1016/j.immuni.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Yang L, Tian L, Ji X, Yang L, Li L. Sphingosine 1-Phosphate (S1P)/S1P Receptor2/3 Axis Promotes Inflammatory M1 Polarization of Bone Marrow-Derived Monocyte/Macrophage via G(alpha)i/o/PI3K/JNK Pathway. Cell Physiol Biochem. 2018;49(5):1677–93. Epub 2018/09/20. doi: 10.1159/000493611. [DOI] [PubMed] [Google Scholar]
- 28.Weigert A, Weis N, Brune B. Regulation of macrophage function by sphingosine-1-phosphate. Immunobiology. 2009;214(9–10):748–60. Epub 2009/07/25. doi: 10.1016/j.imbio.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Wynn TA, Vannella KM. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity. 2016;44(3):450–62. doi: 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nature Reviews Immunology. 2010;10(6):427–39. doi: 10.1038/nri2779 [DOI] [PubMed] [Google Scholar]
- 31.Sassoli C, Pierucci F, Zecchi-Orlandini S, Meacci E. Sphingosine 1-Phosphate (S1P)/S1P Receptor Signaling and Mechanotransduction: Implications for Intrinsic Tissue Repair/Regeneration. International journal of molecular sciences. 2019;20(22):5545. doi: 10.3390/ijms20225545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olingy CE, Emeterio CL, Ogle ME, Krieger JR, Bruce AC, Pfau DD, Jordan BT, Peirce SM, Botchwey EA. Non-classical monocytes are biased progenitors of wound healing macrophages during soft tissue injury. Scientific reports.7(1). doi: 10.1038/s41598-017-00477-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Awojoodu AO, Ogle ME, Sefcik LS, Bowers DT, Martin K, Brayman KL, Lynch KR, Peirce-Cottler SM, Botchwey E. Sphingosine 1-phosphate receptor 3 regulates recruitment of anti-inflammatory monocytes to microvessels during implant arteriogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(34):13785–90. doi: 10.1073/pnas.1221309110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sefcik LS, Petrie Aronin CE, Wieghaus KA, Botchwey EA. Sustained release of sphingosine 1-phosphate for therapeutic arteriogenesis and bone tissue engineering. Biomaterials. 2008;29(19):2869–77. Epub 2008/04/15. doi: 10.1016/j.biomaterials.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sefcik LS, Aronin CE, Awojoodu AO, Shin SJ, Mac Gabhann F, MacDonald TL, Wamhoff BR, Lynch KR, Peirce SM, Botchwey EA. Selective activation of sphingosine 1-phosphate receptors 1 and 3 promotes local microvascular network growth. Tissue Eng Part A. 2011;17(5–6):617–29. Epub 2010/09/30. doi: 10.1089/ten.TEA.2010.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.San Emeterio CL, Olingy CE, Chu Y, Botchwey EA. Selective recruitment of non-classical monocytes promotes skeletal muscle repair. Biomaterials. 2017;117:32–43. Epub 2016/12/09. doi: 10.1016/j.biomaterials.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cencetti F, Bernacchioni C, Nincheri P, Donati C, Bruni P. Transforming growth factor-beta1 induces transdifferentiation of myoblasts into myofibroblasts via up-regulation of sphingosine kinase-1/S1P3 axis. Mol Biol Cell. 2010;21(6):1111–24. Epub 2010/01/22. doi: 10.1091/mbc.E09-09-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–6. Epub 2014/03/25. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anchang B, Hart TD, Bendall SC, Qiu P, Bjornson Z, Linderman M, Nolan GP, Plevritis SK. Visualization and cellular hierarchy inference of single-cell data using SPADE. Nat Protoc. 2016;11(7):1264–79. Epub 2016/06/17. doi: 10.1038/nprot.2016.066. [DOI] [PubMed] [Google Scholar]
- 40.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018. Epub 2018/12/12. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 41.Das A, Segar CE, Hughley BB, Bowers DT, Botchwey EA. The promotion of mandibular defect healing by the targeting of S1P receptors and the recruitment of alternatively activated macrophages. Biomaterials. 2013;34(38):9853–62. Epub 2013/09/26. doi: 10.1016/j.biomaterials.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogle ME, Sefcik LS, Awojoodu AO, Chiappa NF, Lynch K, Peirce-Cottler S, Botchwey EA. Engineering in vivo gradients of sphingosine-1-phosphate receptor ligands for localized microvascular remodeling and inflammatory cell positioning. Acta Biomater. 2014;10(11):4704–14. Epub 2014/08/17. doi: 10.1016/j.actbio.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutter JL, Bechhoefer J. Calibration of atomic‐force microscope tips. Review of Scientific Instruments. 1993;64(7):1868–73. doi: 10.1063/1.1143970. [DOI] [Google Scholar]
- 44.Geissmann F, Jung S, Littman DR. Blood Monocytes Consist of Two Principal Subsets with Distinct Migratory Properties. Immunity. 2003;19(1):71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 45.Sok MCP, Tria MC, Olingy CE, San Emeterio CL, Botchwey EA. Aspirin-Triggered Resolvin D1-modified materials promote the accumulation of pro-regenerative immune cell subsets and enhance vascular remodeling. Acta Biomater. 2017;53:109–22. Epub 2017/02/19. doi: 10.1016/j.actbio.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roman W, Gomes ER. Nuclear positioning in skeletal muscle. Semin Cell Dev Biol. 2018;82:51–6. Epub 2017/12/16. doi: 10.1016/j.semcdb.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Hurtgen BJ, Ward CL, Garg K, Pollot BE, Goldman SM, McKinley TO, Wenke JC, Corona BT. Severe muscle trauma triggers heightened and prolonged local musculoskeletal inflammation and impairs adjacent tibia fracture healing. J Musculoskelet Neuronal Interact. 2016;16(2):122–34. Epub 2016/06/11. [PMC free article] [PubMed] [Google Scholar]
- 48.Schiaffino S, Rossi AC, Smerdu V, Leinwand LA, Reggiani C. Developmental myosins: expression patterns and functional significance. Skelet Muscle. 2015;5:22. Epub 2015/07/17. doi: 10.1186/s13395-015-0046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang T, Carbone EJ, Lo KWH, Laurencin CT. Electrospinning of polymer nanofibers for tissue regeneration. Progress in Polymer Science. 2015;46:1–24. doi: 10.1016/j.progpolymsci.2014.12.001. [DOI] [Google Scholar]
- 50.Ogle ME, Segar CE, Sridhar S, Botchwey EA. Monocytes and macrophages in tissue repair: Implications for immunoregenerative biomaterial design. Exp Biol Med (Maywood). 2016;241(10):1084–97. Epub 2016/05/28. doi: 10.1177/1535370216650293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fortier M, Figeac N, White RB, Knopp P, Zammit PS. Sphingosine-1-phosphate receptor 3 influences cell cycle progression in muscle satellite cells. Dev Biol. 2013;382(2):504–16. Epub 2013/08/06. doi: 10.1016/j.ydbio.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dziki JL, Sicari BM, Wolf MT, Cramer MC, Badylak SF. Immunomodulation and Mobilization of Progenitor Cells by Extracellular Matrix Bioscaffolds for Volumetric Muscle Loss Treatment. Tissue Eng Part A. 2016;22(19–20):1129–39. Epub 2016/10/18. doi: 10.1089/ten.TEA.2016.0340. [DOI] [PubMed] [Google Scholar]
- 53.Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. The Journal of experimental medicine. 2007;204(5):1057–69. Epub 2007/05/09. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng B, Wehling-Henricks M, Villalta SA, Wang Y, Tidball JG. IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J Immunol. 2012;189(7):3669–80. Epub 2012/08/31. doi: 10.4049/jimmunol.1103180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Visualization of circulating immune cell subsets with single cell 2D projections and pseudotime trajectories reveals innate leukocyte heterogeneity. High dimensional flow cytometry data of peripheral blood taken from uninjured animals represented as 2D projections using UMAP and SPADE dimensionality reduction techniques. (A) SPADE trees and accompanying UMAPs generated with manually gated CD11b+ cells circulating homeostatic blood from WT mice (A-I, III) as well as S1PR3 KO (KO) mice (A-II, IV). Within the CD11b+ SPADE trees, neutrophils (A-I, II) and monocytes (A-III, IV) were manually gated and overlaid onto their respective SPADE trees, where the node color represents normalized cell frequency of its specified phenotype in heatmap form. A red overlay on the corresponding UMAPs represent cells identified as a neutrophil or monocyte, respectively. (B) SPADE trees and UMAPS generated with manually gated CD3+ T cells from the blood of uninjured WT mice (B-I, III) and S1PR3 KO mice (B-II, IV). CD4+ T cells (B-I, II) and CD8+ T cells (B-III, IV) were manually gated and overlaid onto the SPADE trees, as well as onto the single cell UMAP projections. UMAP and SPADE representations of high dimensional flow data reveal discrete heterogeneities as observed through the multiple islands depicted on the UMAPs and many nodes clustered via SPADE per phenotypic characterization. n=3 samples (A) and 7 samples (B) per genotype.
Supplemental Figure 2. Cell frequency quantifications between WT and S1PR3 KO mice within each SPADE node containing cells of specified phenotype. Bar graph representation and statistical analysis applied to the tornado plots presented in Figure 4. For each specified cellular phenotype, every node containing cells of this particular subset are analyzed for phenotype-specific cell frequency between experimental groups to evaluate which nodes are significantly dominated by subsets from a particular genotype. Nodes containing cells of the respective phenotype are denoted along the x-axis while the phenotype-specific cell frequency is quantified as a percentage of all live cells. Two-way ANOVAs with Tukey's multiple comparisons were performed for statistical analyses with n=7 samples per genotype. Data is presented as mean ± S.E.M. *p< 0.05, **p< 0.01, ****p< 0.0001
Supplemental Figure 3. Characterization of VPC01091-loaded nanofiber scaffold. Release of VPC01091 from PLGA-PCL nanofiber scaffolds and biomaterial characterization by atomic force microscopy (AFM). (A) Experimental setup for electrospinning VPC01091-loaded PLGA-PCL nanofibers. (B) VPC01091 in-vitro release profile exhibits sustained delivery. (C) Average Young’s modulus values obtained via AFM for blank control nanofiber scaffold (NF) and NF loaded with VPC01091 (VPC). (D, G) AFM generated topography (height) map of blank (D) and VPC01091-loaded (G) PLGA-PCL nanofiber scaffold. Height values throughout nanofiber range from low (black) to high (white) regions. (E, H) AFM generated stiffness map of blank (E) and VPC01091-loaded (H) PLGA-PCL nanofiber scaffold in which white regions represent areas of greater stiffness. (F, I) Histogram of Young's modulus values (MPa) obtained from AFM over multiple contact points on blank (F) and VPC01091-loaded (I) PLGA-PCL nanofiber scaffold. Unpaired two-sided t-test was performed for statistical analysis (C) and data is presented as mean ± S.E.M.
Supplemental Figure 4. Cell frequency quantifications between mice treated with control and VPC01091-loaded nanofibers within each SPADE node containing cells of specified phenotype. Bar graph representation and statistical analysis applied to the tornado plots presented in Figure 5. For each specified cellular phenotype, every node containing cells of this particular subset are analyzed for phenotype-specific cell frequency between experimental groups to evaluate which nodes are significantly dominated by subsets from a particular treatment. Nodes containing cells of the respective phenotype are denoted along the x-axis while the phenotype-specific cell frequency is quantified as a percentage of all live cells. Two-way ANOVAs with Tukey’s multiple comparisons were performed for statistical analysis with n=8 samples per treatment. Data is presented as mean ± S.E.M. *p< 0.05, **p< 0.01, ***p< 0.001, ****p< 0.000.
Supplemental Figure 5. Circulating lymphocytes after muscle injury unaffected by S1PR3 deficiency. SPADE trees generated with CD3+ T cells from the peripheral blood of WT and global S1PR3 KO mice (A-C) and mice treated with blank control nanofibers (NF) and those releasing VPC01091 (D-F) 3 days post VML injury. (A, D) CD4+ (I) and CD8+ (II) T cells were manually gated and overlaid onto their respective SPADE trees where colored node annotation represents normalized cell frequency in the form of a heatmap, illustrating the phenotypic differences in these T cell subpopulations as they are located on opposing sides of the tree. (B, E) CD4 (I) and CD8 (II) fluorescence marker intensities overlaid onto the SPADE trees with the manually gated annotation circled in black to further characterize their expression as displayed by SPADE clustering. (C, F) SPADE quantifications of total CD4+ and CD8+ T cells circulating the blood of WT vs S1PR3 KO mice (C) and mice treated with blank control or VPC01091-loaded (VPC) nanofibers (F). Data represented as mean ± S.E.M. using a two-way ANOVA with Tukey’s multiple comparisons test. n=7 samples per experimental group.
Supplemental Figure 6. Lack of S1PR3 leads to increased numbers of muscle resident stem cells. SPADE trees generated from Lin− cell events taken from uninjured spinotrapezius and hindlimb muscles. (A) MuSCs from the spinotrapezius (A-I) and hindlimb (A-II) are manually gated and overlaid onto their respective trees, annotated with colored nodes representing normalized cell frequency and displayed as a heatmap on the tree. Each node containing MuSCs is given a node assignment number as given next to each colored node. (B) MuSC frequency within each node annotated on spinotrapezius and hindlimb derived SPADE trees. Nodes from the SPADE tree generated with Lin− spinotrapezius derived cells are quantified for MuSC frequency (B-I) while those derived from the hindlimb are quantified by node in (B-II). (C) Tornado plot representation of the difference in average MuSC frequency percentage between S1PR3 deficient mice and their WT counterparts shows that nodes containing MuSCs largely contain those derived from mice lacking S1PR3 (KO) in both the spinotrapezius and hindlimb (C-I, II). Two-way ANOVAs with Tukey’s multiple comparisons were conducted for statistical analyses and data is presented as mean ± S.E.M. with n=3 per genotype for hindlimb samples and an n=4 per genotype for spinotrapezius samples. *p< 0.05
Supplemental Figure 7. Treatment with blank nanofiber scaffold increases macrophage infiltration to defect area 3 days post injury. Flow cytometry analysis of CD11b+CD64+MerTK+ macrophages in injured spinotrapezius muscle 3 days post injury between WT mice receiving no treatment (injury control, black bar) and WT mice treated with blank nanofiber scaffolds (white bar). Statistical analysis performed with two-tailed t-test. Data presented as mean ± S.E.M. n=7 for WT injury control and n=8 for mice treated with blank nanofibers.


