Abstract
The asexual taxon Phialolunulospora vermisporagen. et sp. nov., collected from submerged dicotyledonous leaves in Hainan, China, is described and illustrated herein. Phialolunulosporagen. nov. is characterized by macronematous, semimacronematous, septate and pigmented conidiophores and acrogenous, long lunate, vermiform to sigmoid, hyaline conidia with an eccentric basal appendage. Complete sequences of internal transcribed spacer (ITS) and partial sequences of nuclear large subunits ribosomal DNA (LSU) genes are provided. Phylogenetic analyses of combined ITS and LSU sequences revealed its placement in the Chaetosphaeriaceae. The new fungus is compared with morphologically similar genera.
Keywords: Biodiversity, Chaetosphaeriales , phylogeny, taxonomy
Introduction
China is considered an important Asian reservoir of biodiversity. The southern area of China ranks 34th in biodiversity hotspots (Myers et al. 2000; Williams et al. 2001). Hainan Island, located in the south of China, harbors an incredibly high diversity of fungi. Its humid, subtropical climate, with an average annual temperature of 22 to 27 °C and an average annual precipitation of 1000–2600 mm, favors development of fungi. Our group has conducted investigations of freshwater fungi to increase knowledge of this important ecological group in China (Qiao et al. 2017a, b, 2018a, b, 2019a, b, 2020).
During our present investigation of freshwater fungi in Hainan Island, South China, an interesting species was collected on dead leaves of an unidentified dicotyledonous tree. This species is characterized by unbranched and septate conidiophores, phialidic conidiogenous cells and vermiform to sigmoid and aseptate conidia with an eccentric basal appendage. Based on preliminary analysis of morphological data, we place this unknown fungus in Chaetosphaeriaceae, but a literature search found that it did not belong to any known genus. To further confirm the position of the species, phylogenetic analyses with related taxa within Chaetosphaeriaceae were carried out based on complete sequences of internal transcribed spacer (ITS) and partial sequences of nuclear large subunits ribosomal DNA (LSU) genes.
Materials and methods
Isolation and morphological study
Submerged dicotyledonous leaves were collected from Limu Mountain Nature Reserve in Hainan Province. Samples were preserved in zip-lock plastic bags, labelled, and transported to the laboratory. The decomposed leaves were cut into several 2–4 × 2–4 cm sized fragments and then spread on to the surface of corn meal agar (CMA, 20 g cornmeal, 18 g agar, 40 mg streptomycin, 30 mg ampicillin, 1000 ml distilled water) medium for 10 days; single conidium was isolated with a sterilized needle and transferred to CMA plates while viewing with an Olympus BX51 microscope. The pure strain was further transferred to potato dextrose agar (PDA, 200 g potato, 20 g dextrose, 18 g agar, 40 mg streptomycin, 30 mg ampicillin, 1000 ml distilled water) medium. Colony morphology and microscopic characteristics were examined, and photographs were taken with an Olympus BX51 microscope connected to a DP controller digital camera. Measurement data were based on 30 random conidia and 10 conidiophores.
Pure cultures were deposited in the Herbarium of the Laboratory for Conservation and Utilization of Bio resources, Yunnan University, Kunming, Yunnan, China (YMF, formerly Key Laboratory of Industrial Microbiology and Fermentation Technology of Yunnan) and at the China General Microbiological Culture Collection Center (CGMCC).
DNA extraction, PCR amplification, and sequencing
Pure cultures were grown on PDA medium for 5 days at 25 °C. Actively growing mycelium was scraped off from the surface of the culture and transferred to 2 ml Eppendorf micro-centrifuge tubes. Total genomic DNA was extracted according to the procedures in Turner et al. (1997). Primers used for PCR amplification and sequencing of the nuclear large subunits ribosomal DNA (LSU) and the internal transcribed spacer (ITS) were LROR-LR7 and ITS1-ITS4, respectively (Vilgalys and Hester 1990; White et al. 1990). PCR products were purified and stored at -20 °C until sequencing. The same pairs of primers were used to obtain sequences, which was performed by Macrogen Europe (Macrogen Inc. Amsterdam, The Netherlands). Finally, the sequences were assembled and edited using SeqMan v. 7.0.0 (DNAStar Lasergene, Madison, WI, USA) to obtain the consensus sequences. The newly obtained sequences were submitted to GenBank nucleotide database (Table 1).
Table 1.
List of strains analyzed in this study, with GenBank accession numbers.
| Species | Strain | ITS | LSU |
|---|---|---|---|
| Adautomilanezia caesalpiniae | LAMIC 010212 | NR_153560 | NG_058594 |
| Anacacumisporium appendiculatum | HMAS 245593T | KT001555 | KT001553 |
| Anacacumisporium appendiculatum | HMAS 245602 | KT001556 | KT001554 |
| Bahusutrabeeja dwaya | CBS 261.77T | MH861059 | MH872829 |
| Brunneodinemasporium brasiliense | CBS 112007T | JQ889272 | JQ889288 |
| Brunneodinemasporium jonesii | GZCC 16–0050T | KY026058 | KY026055 |
| Cacumisporium capitulatum | FMR 11339 | HF677176 | HF677190 |
| Cacumisporium capitulatum | SMH 3766 | – | AY017374 |
| Calvolachnella guaviyunis | CBS 134695 | NR_153892 | NG_058879 |
| Chaetosphaeria ciliata | CBS 122131T | MH863180 | MH874726 |
| Chaetosphaeria ciliata | ICMP 18253 | – | GU180637 |
| Chloridium chloroconium | FMR 11940 | KY853435 | KY853495 |
| Chloridium sp. | HGUP 1806 | MK372070 | MK372068 |
| Codinaea lambertiae | CBS 143419T | NR_156389 | NG_059053 |
| Codinaea pini | CBS 138866T | NR_137943 | NG_058902 |
| Conicomyces pseudotransvaalensis | HHUF 29956T | NR_138015 | LC001708 |
| Cryptophiale hamulata | MFLUCC 180098 | – | MG386756 |
| Cryptophiale udagawae | MFLUCC 180422 | MH758198 | MH758211 |
| Cryptophialoidea fasciculata | MFLUCC 172119 | MH758195 | MH758208 |
| Dendrophoma cytisporoides | CBS 223.95T | JQ889273 | JQ889289 |
| Dictyochaeta ellipsoidea | MFLUCC 181574T | MK828628 | MK835828 |
| Dictyochaeta lignicola | DLUCC 0899T | MK828630 | MK835830 |
| Dictyochaeta assamica | CBS 242.66 | MH858788 | MH870426 |
| Dictyochaetopsis gonytrichoides | CBS 593.93 | AF178556 | AF178556 |
| Dinemasporium morbidum | CBS 129.66T | JQ889280 | JQ889296 |
| Dinemasporium polygonum | CBS 516.95T | NR_137786 | NG_059109 |
| Echinosphaeria canescens | SMH 4791 | – | AY436403 |
| Eucalyptostroma eucalypti | CBS 142074T | NR_154027 | NG_059257 |
| Eucalyptostroma eucalyptorum | CPC 31800T | NR_159834 | MH327838 |
| Exserticlava vasiformis | TAMA 450 | – | AB753846 |
| Gelasinospora tetrasperma | CBS 178.33 | NR_077163 | DQ470980 |
| Helminthosphaeria clavariarum | SMH 4609T | – | AY346283 |
| Infundibulomyces cupulata | BCC 11929T | EF113976 | EF113979 |
| Infundibulomyces oblongisporus | BCC 13400T | EF113977 | EF113980 |
| Kionochaeta castaneae | GZCC 18–0025T | MN104610 | MN104621 |
| Kionochaeta microspora | GZCC 18–0036T | MN104607 | MN104618 |
| Lasiosphaeria ovina | SMH 4605 | AY587923 | AY436413 |
| Lecythothecium duriligni | CBS 101317 | – | AF261071 |
| Leptosporella arengae | MFLUCC 150330T | MG272255 | MG272246 |
| Leptosporella gregaria | SMH 4290T | – | AY346290 |
| Linocarpon arengae | MFLUCC 150331T | – | MG272247 |
| Linocarpon cocois | MFLUCC 150812T | MG272257 | MG272248 |
| Menispora glauca | FMR 12089 | HF678528 | HF678538 |
| Menispora tortuosa | DAOM 231154 | KT225527 | AY544682 |
| Menispora tortuosa | CBS 214.56 | AF178558 | AF178558 |
| Menisporopsis breviseta | GZCC 18–0071T | MN104612 | MN104623 |
| Menisporopsis dushanensis | GZCC 18–0084T | MN104615 | MN104626 |
| Morrisiella indica | HKUCC 10827 | – | DQ408578 |
| Multiguttulispora sympodialis | MFLUCC 180153T | MN104606 | MN104617 |
| Nawawia filiformis | MFLUCC 160853 | – | MH758206 |
| Nawawia filiformis | MFLUCC 172394 | MH758196 | MH758209 |
| Neonawawia malaysiana | CBS 125544T | GU229886 | GU229887 |
| Paliphora intermedia | CBS 896.97T | NR_160203 | NG_057766 |
| Paliphora intermedia | CBS 199.95 | – | EF204500 |
| Phaeostalagmus cyclosporus | CBS 663.70 | MH859892 | MH871680 |
| Phaeostalagmus cyclosporus | CBS 312.75 | – | MH872661 |
| Phialolunulospora vermispora | YMF 1.04260T | MK165444 | MK165442 |
| Phialosporostilbe scutiformis | MFLUCC 170227T | MH758194 | MH758207 |
| Phialosporostilbe scutiformis | MFLUCC 181288 | MH758199 | MH758212 |
| Pseudodinemasporium fabiforme | MAFF 244361T | AB934068 | AB934044 |
| Pseudolachnea fraxini | CBS 113701T | JQ889287 | JQ889301 |
| Pseudolachnea hispidula | MAFF 244364 | AB934071 | AB934047 |
| Pseudolachnella longiciliata | HHUF 29962 | AB934081 | AB934057 |
| Pseudolachnella yakushimensis | HHUF 29683T | AB934087 | AB934063 |
| Pseudolachnella pachyderma | HHUF 29955 | AB934085 | AB934061 |
| Pyrigemmula aurantiaca | CBS 126743T | HM241692 | HM241692 |
| Pyrigemmula aurantiaca | CBS 126744 | HM241693 | HM241693 |
| Rattania setulifera | GUFCC 15501 | GU191794 | HM171322 |
| Ruzenia spermoides | SMH 4606 | – | AY436422 |
| Sordaria fimicola | CBS 508.50 | MH856730 | MH868251 |
| Sporoschisma hemipsilum | SMH 2125 | – | AF466083 |
| Sporoschisma hemipsilum | SMH 3251 | – | AF466084 |
| Stanjehughesia vermiculata | HKUCC 10840 | – | DQ408570 |
| Striatosphaeria codinaeaphora | MR 1230 | AF178546 | AF178546 |
| Striatosphaeria codinaeaphora | SMH 1524 | – | AF466088 |
| Synaptospora plumbea | SMH 3962 | – | KF765621 |
| Tainosphaeria jonesii | GZCC 16–0053 | KY026059 | KY026056 |
| Tainosphaeria jonesii | GZCC 16–0065 | KY026060 | KY026057 |
| Tainosphaeria monophialidica | MFLUCC 180146T | – | MN104616 |
| Thozetella pandanicola | MFLUCC 160253T | MH388366 | MH376740 |
| Thozetella tocklaiensis | CBS 378.58T | MH857817 | MH869349 |
| Verhulstia trisororum | CBS 143234T | MG022181 | MG022160 |
| Zanclospora iberica | CBS 130426T | KY853480 | KY853544 |
| Zanclospora iberica | FMR 12186 | KY853481 | KY853545 |
*Sequences generated in this study are emphasized in bold face. Tex-type cultures.
Sequence alignment and phylogenetic analysis
Preliminary BLAST searches with the ITS and LSU sequences of our strain against the GenBank nucleotide database determined the closely related species (Altschul et al. 1990). BLAST search showed that our strain has homology to species in Chaetosphaeriaceae. Based on this information, related sequences of the two marker loci, which include 72 representatives belonging to Chaetosphaeriaceae, 4 representatives of Helminthosphaeriaceae, 2 representatives of Linocarpaceae and 2 representatives of Leptosporellaceae, were downloaded according to recent studies (Yang et al. 2016, 2018; Wei et al. 2018; Lin et al. 2019). Sordaria fimicola (Roberge ex Desm.) Ces. & De Not, Gelasinospora tetrasperma Dowding and Lasiosphaeria ovina (Pers.) Ces. & De Not were used as the outgroup. These, together with the newly generated sequences, were aligned with ClustalX 1.83 (Thompson et al. 1997) with default parameters, and the consensus sequences were manually adjusted and linked through BioEdit v.7.0 (Hall 1999). Manual gap adjustments were done to improve the alignment and ambiguously aligned regions were excluded. Then, the combined alignment was converted to a NEXUS file using the program MEGA6 (Tamura et al. 2013) and a PHY files using the program ClustalX 1.83. The resulting combined sequence matrix included 1475 nucleotide positions (with alignment gaps) from two regions (607 from ITS, 868 from LSU). GenBank accession numbers of downloaded sequences are given in Table 1.
Maximum-likelihood (ML) analysis was computed with RAxML (Stamatakis 2006) with the PHY files generated with CLUSTAL_X version 1.83, using the GTR-GAMMA model. ML bootstrap proportions (MLBPs) were computed with 1000 replicates. Bayesian inference (BI) analysis was conducted with MrBayes version 3.2.2 (Ronquist and Huelsenbeck 2003). The Akaike information criterion (AIC) implemented in jModelTest version 2.0 was used to select the best fit models after likelihood score calculations were done (Posada 2008). The base tree for likelihood calculations was ML-optimized. HKY+I+G was estimated as the best-fit model under the output strategy of the AIC. Metropolis-coupled Markov chain Monte Carlo (MCMCMC) searches were run for 5 000 000 generations, sampling every 500th generation. Two independent analyses with four chains each (one cold and three heated) were run until the average standard deviation of the split frequencies dropped below
0.01. The initial 25% of the generations of MCMC sampling were discarded as burn-in. The refinement of the phylogenetic tree was used for estimating BI posterior probability (BIPP) values. The tree was viewed in FigTree version 1.4 (Rambaut 2012).
Results
Phylogenetic analyses
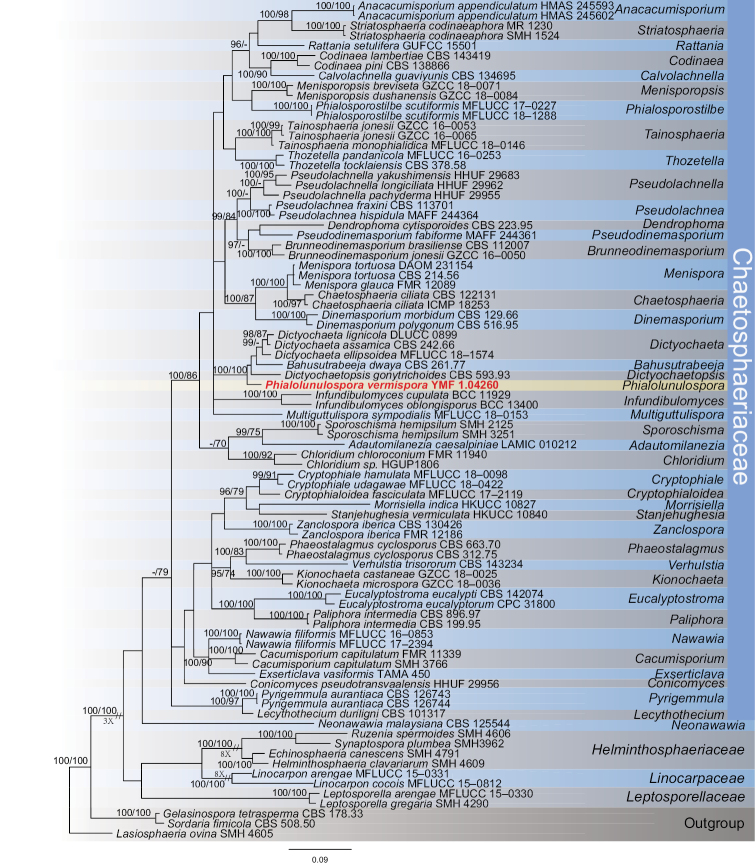
The combined dataset comprised 71 taxa (including our strain) representing 52 genera, which include 60 species in the family Chaetosphaeriaceae, 4 species in Helminthosphaeriaceae, 2 species in Linocarpaceae and 2 species in Leptosporellaceae, with Gelasinospora tetrasperma CBS 178.33, Sordaria fimicola CBS 508.50 and Lasiosphaeria ovina SMH 4605 as the outgroup. The final alignment comprised a total of 1475 base pairs, containing the ITS and LSU sequences, and were analyzed by BI and ML method. The topology of the tree is shown in Fig. 1, with the Bayesian posterior probabilities above 95% and ML bootstrap support greater than 70% indicated for respective clades. In this tree, our strain occurred on an isolated clade within Chaetosphaeriaceae, and clustered together with Dictyochaetopsis Aramb. & Cabello, Bahusutrabeeja Subram. & Bhat and Dictyochaeta Speg. with good Bayesian posterior probabilities (100%) and ML bootstrap proportions (100%). Considering distinct morphological characters with these three genera, we propose to describe our unknown isolate as a new genus and species, Phialolunulospora vermispora.
Figure 1.
Phylogenetic tree derived from Bayesian analysis based on ITS and LSU sequences, depicting the relationships of the new taxon Phialolunulospora vermispora with closely related taxa. The numbers above branches represent BIPP (left) and MLBPs (right). BIPP over 95% and MLBPs greater than 70% are shown on the respective branches, and the bar represents the substitutions per nucleotide position. Gelasinospora tetrasperma CBS 178.33, Sordaria fimicola CBS 508.50 and Lasiosphaeria ovina SMH 4605 were used as outgroup.
Taxonomy
Phialolunulospora
Z. F. Yu & R. F. Castaneda gen. nov.
C8C22876-17B4-5984-B075-7703241266A8
828716
Type species.
Phialolunulospora vermispora Z. F, Yu & R. F. Castañeda
Etymology.
Phialo-Prefix, Phia. lis N.L fem. S. Phialide referring to the phialidic conidiogenous cells, and lunulospora, (lu.nu.la.tus N.L. adj. mean crescent-shaped + spo.ra N.L. fem. S. spora, referred to the conidia), referring to the genus Lunulospora.
Description.
Asexual fungus. Conidiophores macronematous, semimacronematous, mononematous, septate, prostrate or erect, straight or flexuous, pigmented. Conidiogenous cells integrated, terminal, cylindrical to subulate, pale brown to brown, monophialidic or polyphialidic, enteroblastic. Conidial secession schizolytic. Conidia solitary, acrogenous, long lunate, vermiform to sigmoid, unicellular, hyaline, truncate at the conspicuous or inconspicuous basal frill, with a cellular, unbranched, eccentric basal appendage.
Phialolunulospora vermispora
Z. F. Yu & R. F. Castaneda sp. nov.
6A1EB2AB-F23B-54E4-B808-E0AEF505B12D
828717
Figure 2.
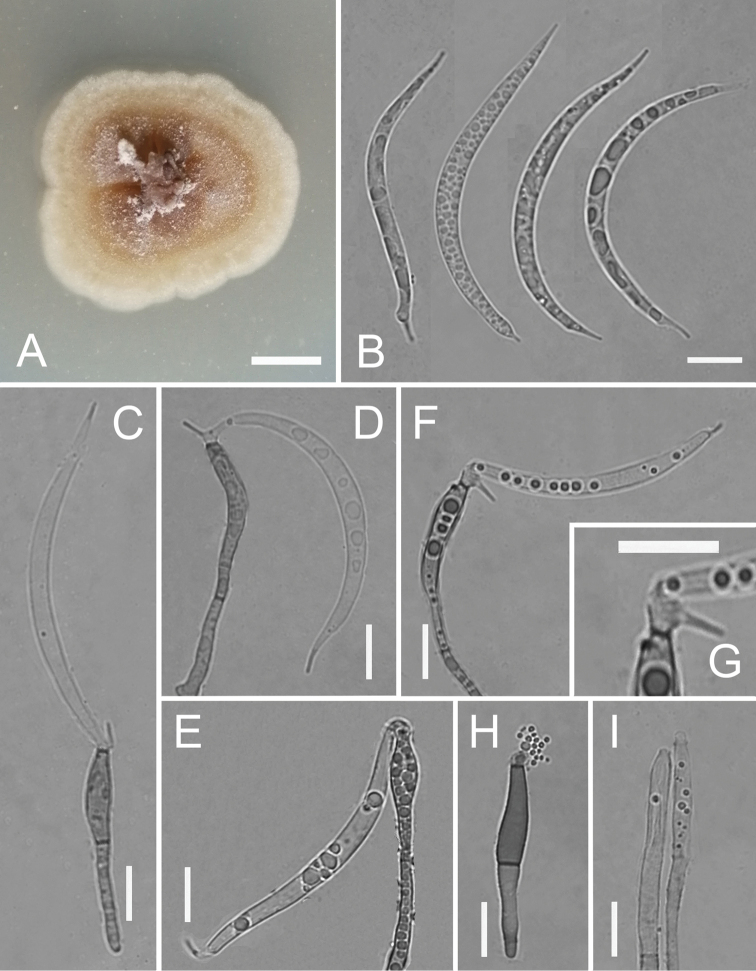
Phialolunulospora vermispora (YMF 1.04260) A colony on PDA at day 10 B conidia C–F conidiophores, conidiogenous cells and conidia G conidiogenous cells H, I conidiophores and conidiogenous cells. Scale bars: 10 mm (A); 10 μm (B–I).
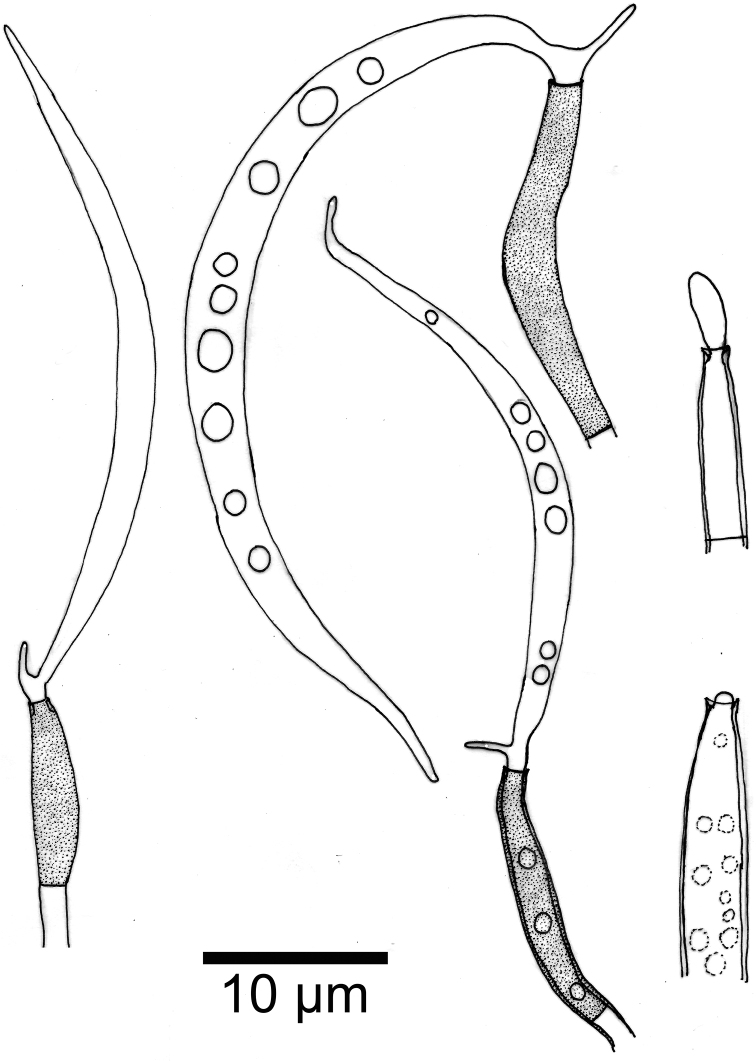
Figure 3.
Phialolunulospora vermispora conidiogenous cells and conidia.
Figure 4.
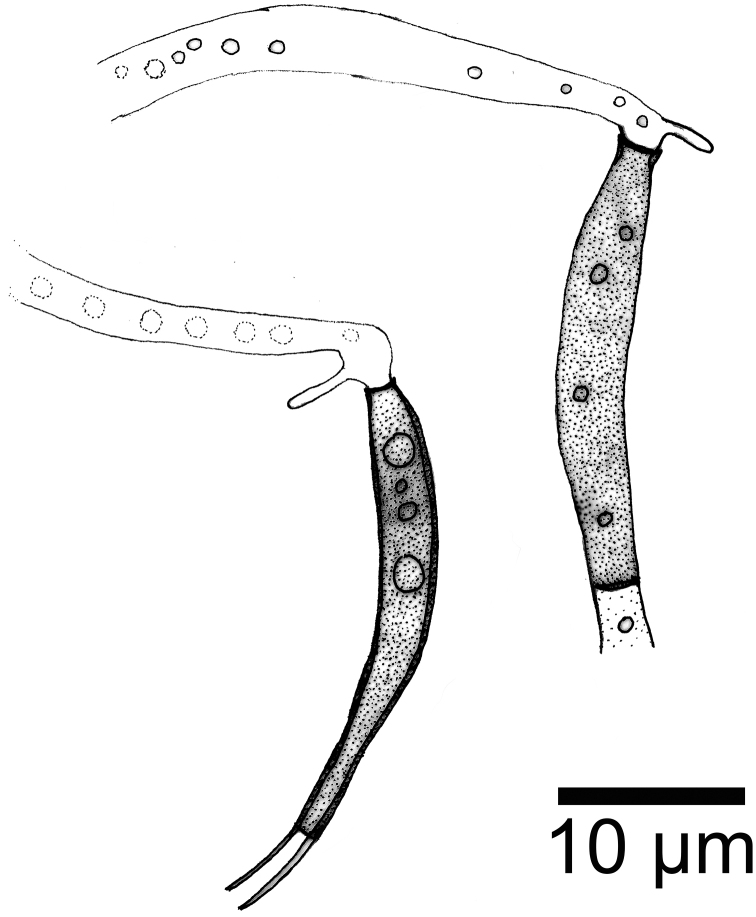
Phialolunulospora vermispora conidiogenous cells.
Type.
China, Hainan province, Limu Mountain, 19°29'40"N, 107°80'45"E, ca. 350 m alt., from leaves of an unidentified dicotyledonous plant submerged in a stream, Apr 2015, Zefen Yu, YMF 1.04260 – holotype; CGMCC 3.19632 – culture ex-type.
Etymology.
ver.mi- (from vermiformis), NL fem. adj mean worm-shaped + spo.ra N.L. fem. S. spora, referred to worm-shaped conidia.
Description.
Mycelium partly superficial and partly immersed, composed of septate, branched, smooth, hyaline, 1–2 μm wide hyphae. Conidiophores solitary, macronematous, semimacronematous, erect or prostrate, straight or flexuous, unbranched, up to 4-septate, cylindrical, up to 150 μm long, 3–4 μm wide, pale brown to brown, smooth, sometimes reduced to conidiogenous cells. Conidiogenous cells integrated, terminal, cylindrical to subulate, sometimes lageniform, determinate, smooth, pale brown to brown, mostly darker than conidiophores, phialidic, after secession leaving an inconspicuous basal frill, 12–47 × 2.6–3 μm. Conidia solitary, acrogenous, long lunate, vermiform to sigmoid, unicellular, guttulate, hyaline, smooth-walled, 31–55 × 2.5–3.5 μm, acute at the apex and narrow truncate at the base bearing minute marginal frills and a cellular, single, unbranched, somewhat attenuated or acuminate, eccentric basal appendage, 1.5–4.6 μm long.
Culture characteristics.
Colonies attain 2.4 cm diameter on PDA and 2.8 cm diameter on CMA after 10 days at 25 °C. On PDA, colonies flat to slightly raised, aerial mycelium abundant, margin entire to undulate, surface white initially, then become buff and grey with age, reverse same color. Colonies on CMA, center with aerial mycelium cottony, periphery with scarce aerial mycelium, olivaceous grey, dark green exudate and soluble pigment produced, reverse same color.
Distribution and ecology.
The species occurs on submerged leaves in stream. This species is currently known only from the type locality.
Discussion
The family Chaetosphaeriaceae was firstly introduced by Réblová et al. (1999) to accommodate Chaetosphaeria and its allies. Réblová et al. (1999) also suggested that Chaetosphaeriaceae should be placed in the Sordariales. However, based on the nuclear large subunit ribosomal RNA gene (LSU) sequence, Huhndorf et al. (2004) placed Chaetosphaeriaceae in order Chaetosphaeriales. In a recent review of the family Chaetosphaeriaceae based on morphology and phylogenetic analysis, Lin et al. (2019) accepted 49 genera (including three uncertain genera) within the family, among which 44 were asexual genera.
The asexual morph of the Chaetosphaeriaceae is hyphomycetous taxa. It is characterized by septate, branched or unbranched conidiophores with the conidiogenous cell monophialidic or polyphialidic, holoblastic or enteroblastic, smoothwalled (Réblová et al. 1999, 2011). Our new fungus, Phialolunulospora vermispora, fits the general description of asexual hyphomycetous Chaetosphaeriaceae well. Phialolunulospora is mainly distinguished from other species in the Chaetosphaeriaceae in having vermiform to sigmoid conidia. Conidia of typical members of the family, including Dictyochaeta and Codinaea Maire (Réblová 2000; Whitton et al. 2000; Cruz et al. 2008; Crous et al. 2014), are aseptate or 1-septate; they may be setulose or not. In this study, the phylogenetic analyses combining ITS and LSU sequences showed that P. vermispora is close to three asexual genera in Chaetosphaeriaceae (Fig. 1), Dictyochaetopsis, Bahusutrabeeja and Dictyochaeta. Morphologically, Bahusutrabeeja and Dictyochaeta are superficially similar to P. vermispora in septate and cylindrical conidiophores, but can be distinguished from the new genus in having globose conidia without appendages and long fusiform conidia with long appendage (Subramanian and Bhat 1977; Li et al. 2014; Liu et al. 2016; Lin et al. 2019), respectively. P. vermispora is clearly different from Dictyochaetopsis species in morphology, such as smooth and pale brown or brown conidiophores and long lunate, vermiform to sigmoid conidia (Arambarri and Cabello 1990; Whitton et al. 2000; Castañeda-Ruíz et al. 2008).
Phialolunulospora is morphologically similar to some other genera species of Chaetosphaeriaceae in hyaline conidia with basal eccentric cellular appendages, including Neopseudolachnella A. Hashim. & Kaz. Tanaka, Pseudolachnea Ranoj., Pseudolachnella Teng and Rattania Prabhug. & Bhat. Of these, species of Neopseudolachnella, Pseudolachnea and Pseudolachnella are different from Phialolunulospora in acervular, setose and stromatic conidiomata (Ranojevic 1910; Teng and Ling 1936; Hashimoto et al. 2015). The genus Rattania is distinguished from Phialolunulospora in having seta and smaller septate conidia (Prabhugaonkar and Bhat 2009). In addition, the type species of Lunulospora Ingold, L. curvula Ingold (Sordariomycetes, Sordariales incertae sedis), also has resemblance to Phialolunulospora in conidial shape (Ingold 1942; Seifert et al. 2011), but it has obviously bigger size of conidia, 70–90 × 4–5 μm vs. 12–47 × 2.6–3 μm, in Lunulospora.
Many freshwater species occur in the family Chaetosphaeriaceae. So far, approximately 16 genera in this family have been reported from fresh water, such as Codinaea (Luo et al. 2019). In this study, Phialolunulospora vermispora was also collected from freshwater habitats.
Supplementary Material
Acknowledgements
This work was financed by the National Natural Science Foundation Program of PR China (31770026, 31970013). We are grateful to two reviewers for critically reviewing the manuscript and for providing helpful suggestions to improve this paper.
Citation
Zheng H, Wan Y, Li J, Castañeda-Ruiz RF, Yu Z (2020) Phialolunulospora vermispora (Chaetosphaeriaceae, Sordariomycetes), a novel asexual genus and species from freshwater in southern China. MycoKeys 76: 17–30. https://doi.org/10.3897/mycokeys.76.57410
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Arambarri AM, Cabello MN. (1990) Considerations about Dictyochaeta, Codinaeopsis and a new genus Dictyochaetopsis. Mycotaxon 38: 11–14. 10.1007/BF02277306 [DOI] [Google Scholar]
- Castañeda-Ruíz RF, Gusmão LFP, Guarro J, Stchigel AM, Stadler M, Saikawa M, Leão-Ferreira SM. (2008) Two new anamorphic fungi from Brazil: Dictyochaetopsis polysetosa and Myrothecium compactum. Mycotaxon 103: 1–8. 10.1007/BF02277306 [DOI] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, Summerell BA, Giraldo A, Gené J, Guarro J, Wanasinghe DN, Hyde KD, Camporesi E, Garethjones EB, Thambugala KM, Malysheva EF, Malysheva VF, Acharya K, Álvarez J, Alvarado P, Assefa A, Barnes CW, Bartlett JS, Blanchette RA, Burgess TI, Carlavilla JR, Coetzee MPA, Damm U, Decock CA, Denbreeÿen A, Devries B, Dutta AK, Holdom DG, Rooney-Latham S, Manjón JL, Marincowitz S, Mirabolfathy M, Moreno G, Nakashima C, Papizadeh M, Shahzadehfazeli SA, Amoozegar MA, Romberg MK, Shivas RG, Stalpers JA, Stielow B, Stukely MJC, Swart WJ, Tan YP, Vanderbank M, Wood AR, Zhang Y, Groenewald JZ. (2014) Fungal Planet description sheets: 281–319. Persoonia 33: 212–289. 10.3767/003158514X685680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz ACR, Leáo-ferreira SM, Barbosa FR, Gusmão LFP. (2008) Conidial fungi from semi-arid Caatinga biome of Brazil. New and interesting Dictyochaeta species. Mycotaxon 106: 15–27. [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. 10.1021/bk-1999-0734.ch008 [DOI] [Google Scholar]
- Hashimoto A, Sato G, Matsuda T, Matsumura M, Hatakeyama S, Harada Y, Ikeda H, Tanaka K. (2015) Taxonomic revision of Pseudolachnea and Pseudolachnella and establishment of Neopseudolachnella and Pseudodinemasporium gen. nov. Mycologia 107(2): 383–408. 10.3852/14-171 [DOI] [PubMed] [Google Scholar]
- Huhndorf SM, Miller AN, Fernandez FA. (2004) Molecular systematics of the Sordariales: the order and the family Lasiosphaeriaceae redefined. Mycologia 96: 368–387. 10.2307/3762068 [DOI] [PubMed] [Google Scholar]
- Ingold CT. (1942) Aquatic hyphomycetes of decaying alder leaves. Transactions of the British Mycological Society 25(4): 339–417. [IN1–IN6] 10.1016/S0007-1536(42)80001-7 [DOI] [Google Scholar]
- Li XX, Xia JW, Ma LG, Castaneda-Ruiz RF, Zhang XG. (2014) A new species of Bahusutrabeeja from Guangxi, China. Mycotaxon 126: 227–230. 10.5248/126.227 [DOI] [Google Scholar]
- Lin CG, McKenzie EHC, Liu JK, Jones EBG, Hyde KD. (2019) Hyaline-spored chaetosphaeriaceous hyphomycetes from Thailand and China, with a review of the family Chaetosphaeriaceae. Mycosphere 10(1): 655–700. 10.5943/mycosphere/10/1/14 [DOI] [Google Scholar]
- Liu J, Yang J, Maharachchikumbura SS, Mckenzie EH, Jones EB, Hyde KD, Liu ZY. (2016) Novel chaetosphaeriaceous hyphomycetes from aquatic habitats. Mycological Progress 15(10–11): 1157–1167. 10.1007/s11557-016-1237-1 [DOI] [Google Scholar]
- Luo Z, Hyde KD, Liu J, Maharachchikumbura SSN, Jeewon R, Bao D-F, Bhat DJ, Lin C-G, Li W-L, Yang J, Liu N-G, Lu Y-Z, Jayawardena RS, Li J-F, Su H-Y. (2019) Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. 10.1007/s13225-019-00438-1 [DOI] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. (2000) Biodiversity hotspots for conservation priorities. Nature 403: 853–858. 10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- Posada D. (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Prabhugaonkar A, Bhat DJ. (2009) Rattania setulifera, an undescribed endophytic hyphomycete on rattans from Western Ghats, India. Mycotaxon 108: 217–222. 10.5248/108.217 [DOI] [Google Scholar]
- Qiao M, Du X, Bian ZH, Peng J, Yu ZF. (2017a) Ellisembia pseudokaradkensis sp. nov. from Hainan, China. Mycotaxon 132: 813–817. 10.5248/132.813 [DOI] [Google Scholar]
- Qiao M, Huang Y, Deng C, Yu ZF. (2017b) Tripospermum sinense sp. nov. from China. Mycotaxon 132(3): 513–517. 10.5248/132.513 [DOI] [Google Scholar]
- Qiao M, Guo JS, Tian WG, Yu ZF. (2018a) Ellisembia hainanensis sp. nov. from Hainan, China. Mycotaxon 133: 97–101. 10.5248/133.97 [DOI] [Google Scholar]
- Qiao M, Li WJ, Huang Y, Xu JP, Zhang L, Yu ZF. (2018b) Classiculasinensis, a new species of basidiomycetous aquatic hyphomycetes from southwest China. MycoKeys 40: 1–12. 10.3897/mycokeys.40.23828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M, Tian WG, Castañeda-Ruiz RF, Xu JP, Yu ZF. (2019a) Two new species of Verruconis from Hainan, China. MycoKeys 48: 41–53. 10.3897/mycokeys.48.32147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M, Zheng H, Zhang Z, Yu ZF. (2019b) Seychellomyces sinensis sp. nov. from China. Mycotaxon 134(2): 391–398. 10.5248/134.161 [DOI]
- Qiao M, Zheng H, Lv RL, Yu ZF. (2020) Neodactylariales, Neodactylariaceae (Dothideomycetes, Ascomycota): new order and family, with a new species from China. MycoKeys 73: 69–85. 10.3897/mycokeys.73.54054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2012) FigTree v1.4.2. http://tree.bio.ed.ac.uk/software/figtree/
- Ranojevic N. (1910) Zweiter Beitrag zur Pilzflora Serbiens. Annales Mycologici 8: 347–402. [Google Scholar]
- Réblová M. (2000) The genus Chaetosphaeria and its anamorphs. Studies in Mycology 143: 149–168. 10.1007/s005720050286 [DOI] [Google Scholar]
- Réblová M, Barr ME, Samuels GJ. (1999) Chaetosphaeriaceae, a new family for Chaetosphaeria and its relatives. Sydowia 51: 49–70. [Google Scholar]
- Réblová M, Gams W, Seifert K. (2011) Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Studies in Mycology 68: 163–191. 10.3114/sim.2011.68.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Seifert KA, Morgan-Jones G, Gams W, Kendrick B. (2011) The genera of Hyphomycetes. CBS Biodiversity Series 9, CBSKNAW Fungal Biodiversity Centre, Utrecht.
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Subramanian CV, Bhat DJ. (1977) Bahusutrabeeja, a new genus of the hyphomycetes. Botany 55(16): 2202–2206. 10.1139/b77-249 [DOI] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Ling L. (1936) Additional fungi from China IV. Sinensia 7: 752–823. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D, Kovacs W, Kuhls K, Lieckfeldt E, Peter B, Arisan-Atac I, Strauss J, Samuels GJ, Börner T, Kubicek CP. (1997) Biogeography and phenotypic variation in Trichoderma sect. Longibrachiatum and associated Hypocrea species. Mycological Research 101(4): 449–459. 10.1017/S0953756296002845 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Zhang H, Dong W, Boonmee S, Zhang D. (2018) Introducing Dictyochaeta aquatica sp. nov. and two new species of Chloridium (Chaetosphaeriaceae, Sordariomycetes) from aquatic habitats. Phytotaxa 362(2): 187–199. 10.11646/phytotaxa.362.2.5 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a guide to methods and applications 18(1): 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Whitton SR, Mckenzie EHC, Hyde KD. (2000) Dictyochaeta and Dictyochaetopsis species from the Pandanaceae. Fungal Diversity 4: 133–158. [Google Scholar]
- Williams J, Read C, Norton A, Dovers S, Burgman M, Proctor W, Anderson H. (2001) Biodiversity, Australia State of the Environment Report 2001 (Theme Report). CSIRO Publishing on behalf of the Department of the Environment and Heritage, Canberra, 217 pp. [Google Scholar]
- Yang J, Liu JK, Hyde KD, Bhat DJ, Jones EBG, Liu Z-Y. (2016) New species of Sporoschisma (Chaetosphaeriaceae) from aquatic habitats in Thailand. Phytotaxa 289(2): 147–157. 10.11646/phytotaxa.289.2.4 [DOI] [Google Scholar]
- Yang J, Liu NG, Liu J, Hyde KD, Jones EB, Liu ZY. (2018) Phylogenetic placement of Cryptophiale, Cryptophialoidea, Nawawia, Neonawawia gen. nov. and Phialosporostilbe. Mycosphere 9(6): 1132–1150. 10.5943/mycosphere/9/6/5 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.