Abstract
Alternative splicing contributes to the majority of protein diversity in higher eukaryotes by allowing one gene to generate multiple distinct protein isoforms. It adds another regulation layer of gene expression. Up to 95% of human multi-exon genes undergo alternative splicing to encode proteins with different functions. Moreover, around 15% of human hereditary diseases and cancers are associated with alternative splicing. Regulation of alternative splicing is attributed to a set of delicate machineries interacting with each other in aid of important biological processes such as cell development and differentiation. Given the importance of alternative splicing events, their accurate mapping and quantification are paramount for downstream analysis, especially for associating disease with alternative splicing. However, deriving accurate isoform expression from high-throughput RNA-seq data remains a challenging task. In this mini-review, we aim to illustrate I) mechanisms and regulation of alternative splicing, II) alternative splicing associated human disease, III) computational tools for the quantification of isoforms and alternative splicing from RNA-seq.
Keywords: Alternative splicing, Human disease, Isoform quantification, RNA-Seq
1. Introduction
Upon the completion of the Human Genome Project in 2003, the discrepancy between the number of annotated protein-coding genes and the number of observed human polypeptides reveals the widespread violation of the “one gene–one polypeptide” hypothesis. It is now commonly accepted that in higher eukaryotes, alternative splicing plays a remarkably important role in increasing protein diversity by allowing one gene to generate distinct protein isoforms and increasing the complexity of gene expression regulation [1]. In humans, up to 95% of multi-exon genes undergo alternative splicing to encode proteins with different functions in distinct cellular processes [2]. Furthermore, around 15% of human hereditary diseases and cancers are reported to be associated with alternative splicing [3], [4]. The analyses and studies of alternative splicing will fundamentally advance our understanding of mRNA complexity and its regulation, provide valuable insights to understand disease etiology, and assist the development of therapeutic interventions for splicing-related diseases.
2. Mechanisms and regulation of alternative splicing
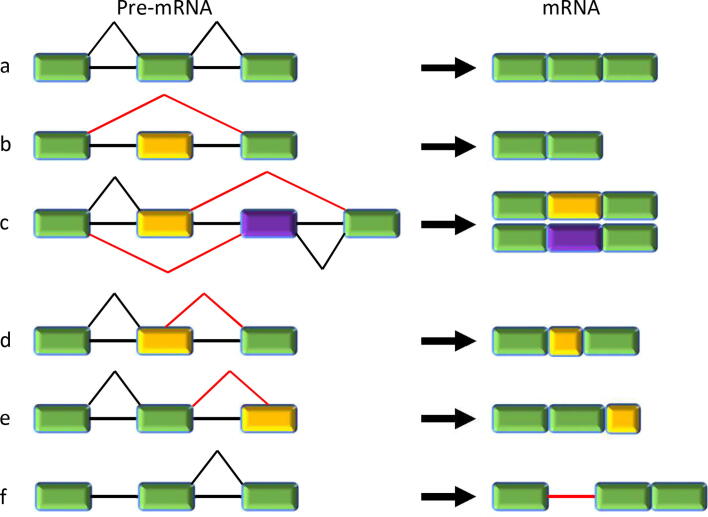
Constitutive splicing is the process that mRNA is spliced identically producing the same isoforms, while alternative splicing generates different isoforms through using different sets of exons. Typically, there are five major subtypes of alternative splicing [5] (Fig. 1). 1) Exon skipping (also known as cassette exons) is reported to be the most common alternative splicing event in mammalian cells, which results in complete skipping of one or more exons [6], [7]. 2) Mutually exclusive exons are a rare subtype where two or more splicing events are no longer independent. They are executed or disabled in a coordinated manner [8]. 3) Alternative 5′ splice sites (alternative donors): the usage of an alternative 5′ donor site, which changes the 3′ boundary of the upstream exon. 4) Alternative 3′ splice sites (alternative acceptors): opposite to alternative 5′ splice sites, it is the usage of an alternative 3′ splice junction site causing the change of the 5′ boundary of the downstream exon. 5) Intron retention (IR) is the process that one or more introns remain unspliced in the mRNA. The fate of those intron-retaining mRNA can be different [9], [10]. Some of them are degraded by the nonsense-mediated decay pathway, while others could generate new protein isoforms [11]. Often, malfunctional proteins could be produced from IR and lead to diseases. In addition to the five major subtypes, alternative polyadenylation sites and alternative promoters are often discussed under this topic. Although these two also increase the coding potential of genomes, they have very different mechanisms and are not directly related to splicing. We will not discuss them further in this mini-review.
Fig. 1.
Constitutive and five major types of alternative splicing. a: Constitutive splicing; b: exon skipping (cassette exons); c: mutually exclusive exons; d: alternative 5′ splice sites (alternative donors); e: alternative 3′ splice sites (alternative acceptors); f: intron retention.
2.1. General mechanisms of pre-mRNA splicing
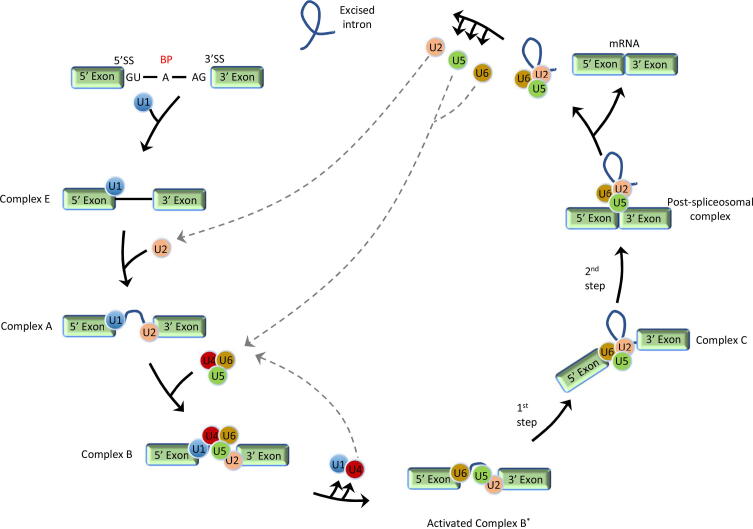
Pre-mRNA splicing occurs in a large ribonucleoprotein complex (RNP) known as spliceosome [12], [13], [14]. The spliceosome is a dynamic complex mainly consisting of five small nuclear ribonucleoproteins (snRNPs) (U1, U2, U4/U6, U5) that recognize and assemble on each intron to ultimately remove introns from a transcribed pre-mRNA (Fig. 2) [15], [16], [17], [18], [19]. During the assembly, U1 binds to the 5′ splice site with the assistance of the U2 auxiliary factor protein (U2AF) forming base pairing between the U1 snRNA and the splice site. This earliest formed complex (complex E or commitment complex) then binds to U2 to form the complex A (pre spliceosome). Formation of the complex A ensures the intron to be spliced out and the last exon to be retained during the final step. The complex B (precatalytic spliceosome) is formed by the complex A joining U4, U5 and U6. Then a series of intricate reorganization events occur in order to form the complex C (catalytic step 1 spliceosome). Firstly, the U1 interaction at the donor site is replaced by the U6 snRNP, followed by U1 and U4 leaving the complex B. This lastly formed complex C catalyzes two transesterification reactions of the splicing. During the first transesterification, the phosphate at the 5′ splice site is attacked by the 2′- hydroxyl group at the branch point which results in the 5′ end of the intron being cleaved from the upstream exon, and then joining to the branch point by a phosphodiester bond. In the second transesterification, the phosphate at the 3′ splice site of the intron is attacked by the 3′- hydroxyl of the downstream exon. This step finally releases the intron as well ligates the two exons by a phosphodiester bond [6].
Fig. 2.
Stepwise schematic presentation of general pre-mRNA splicing. Abbreviations: BP: brunch point; SS: splice site.
2.2. Alternative splicing mechanisms
Alternative splicing is regulated by the interaction between cis-acting regulatory sequences and corresponding trans-acting regulatory proteins. There are two major types of cis-acting elements that either promote (enhancers) or inhibit (silencers) splicing activity of nearby splice sites. Splicing enhancers can be located either in the exon (exonic splicing enhancers, ESEs) or in the intron (intronic splicing enhancers, ISEs). They bind to splicing activator proteins such as serine/arginine-rich family of nuclear phosphoproteins (SR protein family) to increase the chance of an adjacent site being spliced. Splicing silencers include exonic splicing silencers harbored in the exon (ESSs) and intronic splicing silencers harbored in the neighboring intron (ISSs). They bind to splicing repressor proteins such as heterogeneous nuclear ribonucleoproteins (hnRNPs) to inhibit the splicing of nearby sites or stimulate exon skipping.
Occasionally, the presence or absence of one single regulator is adequate to alternatively splice a pre-mRNA [20], [21], [22], [23]. But more often, more than one cis-acting regulatory sequences have to work together with splicing activators or repressors to enhance or inhibit the spliceosome activity at the splice site to determine an alternative spicing pathway [10], [20], [24], [25]. Interestingly, hnRNP proteins do not always act as splicing repressors nor does SR protein always act as splicing activators. HnRNPs is a well-conserved RNA-binding protein family in mammals [26]. In humans, hnRNPA1 usually binds to ESS or ISS to repress exon inclusion by steric actions, or binds to both sides of a cassette exon forming a loop to skip the exon [27], [28]. However, during the interaction with Fas pre-mRNA, hnRNPA1 promotes exon 6 inclusion through interrupting 5′ splice donor site selection of exon 5 [29]. Another member of the family hnRNPL also has the ability to both activate or suppress exon 5 of CD45 gene [30]. More importantly, this characteristic of splicing factors often appears to be strongly position-dependent [31]. For example, a splicing factor would serve as a splicing suppressor or an activator in different pre-mRNAs depending on whether it is bound to exons or introns. A well-studied splicing factor, neuron-specific RNA-binding protein Nova-1 is one of such splicing factors. CLIP Analysis has revealed that over 91% of Nova-dependent exon inclusion events occurred near either alternative 5′ splice sites or constitutive 3′ splice sites, while 74% of Nova-dependent exon skipping occurs near constitutive 5′ splice sites [32].
Splicing factors are imperative in regulating splicing events. Being able to pinpoint the binding sites between RNA and splicing factors provides us valuable information about splicing regulation. The predominant methods for investigating RNA-protein binding sites are based on cross-linking followed by immunoprecipitation (CLIP) [22], [33]. The CLIP method coupling with high-throughput sequencing permits the transcriptome-wide investigation of RNAs that interact with the protein of interest [34]. A variety of CLIP-based methods have been developed. Among them, high-throughput sequencing-CLIP (HITS-CLIP or CLIP-Seq) [34], photoactivatable-ribonucleoside enhanced CLIP (PAR-CLIP) [35], and individual CLIP (iCLIP) [36] are the three major ones. HITS-CLIP was originally applied to identify genome-wide protein-RNA interactions for the Nova protein family [32]. It is a well-established and effective method, though the false negative rate is high as a result of low cross-linking efficiency [34]. Comparing to HITS-CLIP, PAR-CLIP has improved the efficiency due to the incorporation of photoreactive ribonucleoside analogs. This protocol has effectively increased the resolution and signal-to-noise ratio [35], [37]. The potential drawback of PAR-CLIP is that the treatment could be toxic [38], [39], but some studies suggest otherwise [35], [37]. iCLIP has achieved an even higher resolution and efficiency compared to the first two. However, the experimental set-up and computational analysis are complicated.
The three CLIP-based experiments along with other variations have extended the scope of the genome-wide map of protein-RNA interactions. Choices of CLIP-based methods, experimental design, and the selection of proper bioinformatic pipelines have been reviewed in detail [38], [40], [41]. Such high-resolution maps of protein-RNA interactions advance the understanding of splicing regulation, and how protein-RNA interactions play a role in human physiological and pathological process. In addition, the detection of RNA-protein interaction can reveal biomarkers, more importantly it could identify potential therapeutic targets [34].
The protein-RNA interaction is not the only player in regulating alternative splicing. RNA structures also play a significant role [42]. They could either promote splicing or inhibit splicing. In general, RNA structures aid splicing by bringing splicing signals close to each other. For example, in the adenoviruses ADML gene, a stem-loop is formed in order to bring the 3′ splice site into the proximity of three possible branch points [43]. Another astonishing example of promoting alternative splicing through RNA structures is through a docking and selector site in the Drosophila melanogaster DSCAM gene. DSCAM encodes over 38,000 distinct mature mRNA isoforms by mutually exclusive splicing of 95 alternative exons [44]. The majority of its diversity is generated by the exon 6 cluster, which contains 48 alternative exons. A conserved selector sequence is complementary to a portion of the docking site. This RNA-RNA base pairing between the docking site and selector sequence ensures that only one of the 48 alternative exons is used to produce mature mRNA [45]. This pattern illustrates that these competing RNA secondary structures are the key element to maintain and assist the formation of protein isoforms. As for RNA structure suppressing splicing, native RNA structures often sequester important sequences required for splicing, thus repress the usage of splice sites. In some other cases, structures close to exons or splice sites may have negative impact on the recruitment of U1 or U2 snRNPs [46]. To conclude, these delicate machineries integrate and interact with each other to regulate alternative splicing in distinct cellular machines.
3. Alternative splicing associated human disease
Considering the important role of pre-mRNA splicing in composing protein diversity and maintaining organism functionality, it is no surprise that disruption of normal splicing patterns can cause gene dysfunction and even disease. There are around 20,000 human protein-coding genes but almost 150,000 transcript isoforms. Thus, on average each human gene has about seven transcript isoforms. Meanwhile, a recent study finds that over 30% of tissue-dependent transcript variations are constituted by local splicing variations [47]. Given that such high level of human genome complexity is exemplified by the flexibility of each gene with alternative splicing, it is natural to consider such flexibility as a risk factor. Indeed, a great number of human diseases have been reported to be linked to defective splicing.
Mutation disrupting either trans-acting regulatory proteins or cis-acting regulatory sequences could lead to aberrant splicing. In general, trans-acting mutations are rarer than the other. It is very likely due to the fact that disruptions in basal factors of the splicing machinery are generally more lethal comparing to mutations altering splicing of a single gene in cis [48]. We will discuss some well-characterized diseases caused by mutations in trans-acting splicing factors as well as mutations in cis-acting regulatory sequences leading to different types of aberrant alternative splicing.
3.1. Cerebro‐Costo‐Mandibular syndrome caused by disruption in the core splicing machinery
Mutations that have impact on spliceosome components or regulatory factors of mRNA processing have been established as the basis of some craniofacial disorders. Cerebro-Costo-Mandibular Syndrome (CCMS) is one of rare craniofacial disorders characterized by the Pierre Robin sequence (severe micrognathia, glossoptosis, soft cleft palate, and upper airway obstruction) and posterior rib defects. Other symptoms including intellectual disability and microcephaly have been reported [49], [50]. Although most of cases appear to be de novo, familial examples have been reported as well with both autosomal dominant and recessive inheritance [51]. Both Lynch et al. and Bacrot et al. identified the mutations in SNRPB as a cause of CCMS [52], [53]. As mentioned above, the spliceosome consists of U1, U2, U4/U6, and U5 snRNPs. Each snRNP protein includes seven cores/Sm proteins. SNRPB-encoded small nuclear ribonucleoprotein polypeptides B and B1 belong to the Sm factors. Most of the SNRPB mutations are located on an alternative exon containing a premature termination codon in regions that serve as exonic splicing silencers. The inclusion of this exon increases the nonsense-mediated mRNA decay activity that eventually leads to CCMS. Other spliceosome-related craniofacial disorders have been reported. For example, mutations in EFTUD2 lead to mandibulofacial dysostosis, Guion-Almeida type (MFDGA) [54]; mutations in SF3B4 for Nager syndrome [55]; TXNL4A being identified as the cause of Burn-McKeown syndrome (BMKS) [56]; and EIF4A3 in Richieri-Costa-Pereira syndrome [57]. These disorders have been reviewed thoroughly by Lehalle et al. and Krausová et al. [58], [59].
3.2. Mutations in trans-splicing factors resulting in tumorigenesis
Alternative splicing possesses ubiquitous and flexible gene regulation in humans. Therefore, cancer cells often exploit this characteristic to expand and survive. Myelodysplastic syndromes (MDS) are a group of diverse cancers in the bone marrow caused by poorly formed immature blood cells [60]. The splicing factor SF3B1 encoding subunit 1 of the splicing factor 3b protein complex, which is a major component of U2 snRNP, is the most frequently mutated gene in MDS patients [61], [62], [63], [64]. The splicing factor 3b together with 3a bind the branchpoint sequence in pre-mRNA. The stable binding is indispensable to recruit and anchor U2 snRNP to the pre-mRNA [65]. SF3B1 also serves as a component of the minor U12-type spliceosome [66] and has a role in the commitment complex [67]. SF3B1 knockdown leads to growth inhibition and deregulation of numerous genes and pathways [61]. Due to the important role of SF3B1 in spliceosome machinery, it is no surprise that RNA-seq analysis of tumor tissues with SF3B1 mutations has revealed global splicing defects caused by the perturbed branch region fidelity [68], [69], [70], [71], [72]. Other splicing factors involving 3′-splice site recognition such as U2AF1 and SRSF2 have also been reported to harbor somatic mutations associated with MDS [73], [74]. The detailed mechanism and pathways of aberrant splicing in cancer progression have been reported and discussed extensively in those referred reviews [75], [76], [77], [78], [79]. More importantly, the identification of mutations in those splicing factors suggests that spliceosome machinery could be a therapeutic target for certain cancers [61], [62].
3.3. Spliceostatin A, a potent antitumor compound inhibiting splicing
As we discussed above, defects in spliceosome machinery lead to major dysfunctions and disorders. Interestingly, the inhibition of pre-mRNA splicing in cancer cells using Spliceostatin A (SSA) can suppress their proliferation [80], [81], [82]. SSA is a methylated derivative of a natural product FR901464 that inhibits pre-mRNA splicing in vitro and in vivo by binding to SF3b, a protein subcomplex of U2 snRNP which is indispensable for the recognition of pre-mRNA branchpoint. The interaction between SF3b 155-kDa subunit and mRNA is disrupted by SSA, which leads to nonproductive recruitment of U2 snRNP to 5′ branchpoint. Furthermore, down-regulation of genes that are crucial for cell division was observed, explaining the anti-proliferative effects of SSA [82]. SSA is not only a valuable target in cancer treatment, it can also suppress viral replication since splicing is vital for certain class of viruses to infect hosts [82], [83], [84].
3.4. Disease associated with changes in ratios of protein isoforms caused by dysregulation in alternative splicing
Tauopathies describe a class of neurodegenerative disorders characterized by neuronal and/or glial inclusions composed of the microtubule associated protein, tau [85]. The tau protein family consists of a group of six highly soluble protein isoforms produced by alternative splicing from a single gene MAPT (microtubule associated protein tau) [86]. The functionality of the tau protein family is mostly in maintaining the stability of microtubules in axons where it binds to microtubules via microtubule repeat regions. One of these microtubule binding regions is encoded by the alternatively spliced exon 10. The longest tau isoform (4R isoform) has four microtubules repeat-regions (R1, R2, R3, R4) at the C-terminal caused by exon 10 inclusion while the shortest isoform (3R isoform) has three repeats (R1, R3 and R4) due to the exon 10 skipping event. These splicing events are spatially and temporally regulated, and literatures documented that mutations in exon 10 alter its normal ratio of inclusion and exclusion. This fraction change has been confirmed to be the cause of frontotemporal dementia with parkinsonism linked to chromosome 17 [87], [88]. Growing evidence also shows that several other neuron degenerative diseases such as Alzheimer's disease, Parkinson's disease and Huntington’s disease can be associated with microtubule dysfunction caused by imbalanced ratio of tau isoforms [89], [90], [91], [92], [93], [94].
The Wilms' tumor gene WT1 is another example that alteration of its isoform expression causes human disease: Frasier syndrome. WT1 encodes a zinc finger protein that binds to DNA. Its exon 5 and 9 are alternatively spliced leading to the formation of four isoforms [95], [96]. An intronic point mutation on WT1 causes the disruption of alternative splicing at the splice donor site of exon 9, which further prevents the synthesis of WT1 + KTS isoform. Studies suggest that the gonadal development may be particularly sensitive to the imbalance of WT1 + KTS and WT1 − KTS which eventually leads to Frasier syndrome [97].
The above examples illustrate that abnormal changes in isoform ratios can cause human diseases. Moreover, in such cases it is imperative to accurately estimate isoform abundance in order to shed light on the relative contribution of each isoform to the different physiologic states. We will discuss isoform quantification in detail in part III.
3.5. Distinct disease severity caused by point mutations in mutually exclusive exons
Timothy syndrome is a rare congenital disorder that primarily affects the heart but can also affect many other tissues including teeth, nervous systems, and immune systems. There are two documented types of Timothy syndrome: classical (type-1) and atypical (type-2). They are both caused by de novo point mutations in CACNA1C, a gene encodes an alpha-1 subunit of a voltage-dependent calcium channel [98]. At least 19 out of 55 exons of CACNA1C are subject to alternative splicing [99], [100], [101]. Its transmembrane segment IS6 is encoded by inclusion or exclusion of the mutually exclusive 8 and 8a exons [102]. Point mutations on those exons lead to a mutated channel that shows a much slower voltage-dependent inactivation leading to a larger influx of calcium ions. Since the mutations only occur in one of the mutually exclusive exons, usually the ion channel encoded by the unaffected exon will function normally. Exon 8 is expressed in the smooth muscle, while exon 8a is expressed in the cardiac muscle [100], [102]. Thus, mutations on CACNA1C cause different level of severity in patients with Timothy syndrome. The most fatal mutation takes places in the cardiac exon 8a, which often leads to cardiac arrhythmia, while mutations found in the smooth-muscle-related exon results in a less severe outcome [98], [103].
3.6. Familial dysautonomia caused by a mutation in the 5′ splice site leading to an exon skipping event
Familial dysautonomia (FD) is a rare, recessive genetic disorder that affects the development and survival of sensory sympathetic and parasympathetic nerve cells in the autonomic nervous system. The autonomic nervous system controls involuntary actions such as digestion, breathing, and the regulation of blood pressure and body temperature. Therefore, patients with FD show various symptoms including insensitivity to pain, difficulty swallowing, poor growth, pneumonia, labile blood pressure and gastrointestinal dysmotility. FD is the result of loss-of-function of an inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein (IKAP, also known as elongator complex protein 1, ELP1) [104]. Most FD patients have a point mutation in the 5′ splice donor site of exon 20. It is a T > C transition that weakens the intronic part of the 5′ splice site and results in a skipping event of exon 20. Translation of this mRNA produces a truncated IKAP protein, which misses all amino acids encoded from exon 20. Thus, the decreased level of functional IKAP protein expression causes many FD symptoms. In addition, studies also indicate that IKAP deficiency results in down regulation of genes involved in oligodendrocyte differentiation and myelination which could be the cause of demyelination symptoms of FD patients [48], [105]. This example not only demonstrates the complexity of mutations in splice sites, it also suggests that such a mutation may have impact on multiple physiological process.
3.7. Disease caused by intron retention
Intron retention (IR) has been the least described subtype of alternative splicing until recently. Intron-retaining transcripts were previously thought to be non-functional since they would be degraded by nonsense-mediated decay, a surveillance pathway that prevents such aberrant mRNA from being translated into potentially harmful abnormal proteins [11], [106], [107], [108], [109]. However, recent studies have confirmed that IR is not only a conserved form of alternative splicing, but also plays an essential role in controlling and enhancing the complexity of gene expression [9], [110]. Here, we will discuss some consequences of abnormal IR and the emergent role they play in diverse diseases.
IR is a widespread event occurring in many diseases. Point mutations occurring at critical splice control points such as donor or acceptor sites can result in partial or full retention of specific introns. Autoimmune polyendocrine syndrome type 1 (APS-1) is a rare and complex recessively inherited disorder of immune-cell dysfunction with multiple auto immunities [111], [112]. Loss-of-function mutations of the autoimmune regulator gene (AIRE) have been reported to cause APS-1. AIRE is a transcription factor expressed in the thymus medulla and lymph nodes. It controls the local transcription of tissue-specific proteins typically expressed in peripheral tissues, thus allowing the negative selection of self-reactive T cells [113], [114]. The c.463G > A transition in AIRE generates an aberrant transcript retaining intron 3. This aberrant intron 3-retaining transcript generates a truncated protein with a premature stop codon. The truncated protein contains only the first normal functional 154 amino acids followed by 48 aberrant amino acids [115].
Recent studies also revealed that IR events are very common across cancer patients. Partial retention of intron 6 of GSTP1 has been confirmed as one cause of the head and neck cancer [116]. Truncated cyclin D1b is produced by CCND1 with partially retained intron 4, which leads to the prostate and esophageal cancer [117], [118]. Moreover, in a recent large‐scale transcriptome profiling study, IR events are observed in solid cancers originating from bladder, breast, colon, colon, head and neck, kidney, liver, lung, prostate, rectum, stomach, thyroid, and endometrium, as well as in acute myeloid leukemia [119].
3.8. Transcriptome-wide alternative splicing analysis in disease
Aberrant splicing greatly contributes to the pathologies of many diseases as mentioned. Indeed, the detection of differential splicing not only serves as predictive, diagnostic, or prognostic biomarkers but also stratifies patients with different conditions. Here we briefly discuss examples of transcriptome-wide sequencing unveiling differential splicing events in diseases. Lin et al. identified 593 differential splicing events between Huntington’s disease (HD) and control brains, and four splicing factors with significantly altered expression [120]. The follow-up study verified the impact of the splicing factor PTBP1 on disease-associated splicing patterns in HD patients [120]. Similar study identified hundreds of aberrant splicing events in Alzheimer’s disease [121]. For cancers, differential alternative splicing is more than a diagnostic biomarker. Increasing evidence indicated that it could elucidate the progression and serve as prognostic biomarkers [122], [123].
Many studies employed univariate and multivariate Cox regression analyses to identify survival-associated alternative spicing events and to compute risk scores [124], [125], [126], [127], [128], [129], [130], [131], [132]. More recently, a novel statistical model SURVIV (Survival analysis of mRNA isoform variation) has shown its ability to outperforms the conventional Cox regression model [133]. SURVIV uses a survival-measurement-error model to estimate uncertainty of mRNA isoform ratios and assesses the association between isoform ratios and survival time of cancer patients. SURVIV outperforms Cox regression model under many different settings especially with low-depth or moderate-depth RNA-seq data. This makes SURVIV extra practical since many clinical studies have not yet reached deep sequencing depth.
Beyond the significant role of alternative splicing in disease diagnosis and prognosis, it is more than informative to deep dive into disease pathological mechanisms to investigate how dysfunctions of alternative splicing lead to distinct diseases and to connect phenotypes with genotypes. Despite the profound genetic understanding for many human genetic diseases, causative genetic variations or functional roles of highly correlated variations of many diseases still consist of a big piece of puzzle. To facilitate the understanding from genetic variants to human pathologies, it is necessary to accurately quantify gene expression at the individual transcript isoform level since isoforms are the ultimate units to execute gene functions. In addition to the expression quantification of annotated genes, the discovery of novel transcript isoforms and novel splicing events is also crucial for deciphering the role of alternative splicing in human physiology and pathologies.
4. Computational tools for isoform quantification from RNA-seq
Over the past few years, massively parallel RNA sequencing (RNA-seq) has become a powerful tool for comprehensive transcriptomic analysis. Furthermore, with the cost of deep sequencing dropping drastically, large scale studies for expression and alternative splicing have become plausible. Such studies will expedite the discovery of more splicing-related diseases and the gained knowledge has the potential to develop preventive and therapeutic interventions for these diseases.
However, precise quantification of alternative splicing is still hindered by the technological limitation, mainly the limited read length. During an RNA-seq experiment, mRNA is extracted from the tissue, then fragmented and reverse transcribed into cDNA, which is further amplified and sequenced by high-throughput, short-read sequencing methods. Ideally transcriptome assembly can be performed to reconstruct genome regions being transcribed. However, considering the typical read length of RNA-seq ranging from 50 bp to 150 bp, and the fact that transcript isoforms of the same genes are usually difficult to distinguish, the expression quantification of transcript isoforms remain challenging.
Moreover, the discovery of novel transcripts using short reads is one of the most challenging tasks in RNA-seq [134]. Short reads hardly ever spanning across splice junctions complicate the inference of full-length transcripts especially for lowly expressed transcripts. The identification of transcription start and end sites also remains a difficult mission [135]. Recently, another computation difficulty emerges as sequencing depth further increases due to the affordable sequencing. The massive sequence data may not only overwhelm hardware resources, but also confound heuristic algorithms that may not be scalable to that gigantic size of data [136].
Various computational tools have been developed in order to tackle these major complications in the last decade. In general, methods for alternative splicing analysis can be divided into two major categories: isoform-centric approaches and exon-centric approaches. Isoform-centric approaches quantify full-length transcript isoforms, and then the alternative splicing ratio can be estimated based on the ratio between isoforms including and isoforms excluding an alternative exon of interest. Exon-centric approaches directly focus on the quantification of exon inclusion ratios.
Base on whether the reference genome or the reference transcriptome is utilized, transcript quantification can be categorized as reference-based approaches or de novo assembly-based ones [137]. Reference-based approaches first align reads to a reference genome. Then those mapped reads are distributed to known annotated isoforms computationally. Or a splicing graph representing all possible alternative splicing events can be built upon the mapped reads and individual isoforms are finally assembled by traversing the graph. The high sensitivity of reference-based approach permits the discovery of novel transcript as well [138]. For example, several popular methods, such as StringTie [139] and Cufflink [138] apply genome-aligned reads and take advantage of existing annotations when assembling transcripts including novel ones. Transcript isoform quantification is performed simultaneously or afterwards. Although the majority of reference-based quantification methods align reads to the genome, recently there are methods that developed as alignment-free approaches. Such alignment-free methods pre-index reference transcripts, break sequence reads into k-mers, and then preform fast matches to the pre-indexed transcripts as “pseudo”-alignments.
In general, referenced-based approaches are computationally economic and more suitable for isoform quantification especially when the high-quality reference is available. But for non-model organisms de novo assembly-based method is indispensable. De novo assembly-based approaches usually rely on a De Bruijn graph to assemble isoforms, therefore the required computational resources are enormous. Regarding to the alternative splicing analysis, even though some studies have demonstrated that de novo assembly-based method is capable of detecting novel splicing events, the application on human datasets remain sparse [140], [141].
In this review, we will mainly focus on reference-based isoform-centric or exon-centric methods. Representative packages for the detection and quantification of isoforms or splicing events with or without the discovery of novel isoforms or splicing events will be discussed. To facilitate users’ selection, we summarized a collection of benchmark characteristics for these representative tools based on simulated studies, literature reviews [142], [143], [144], and software documentation (Table 1). We will briefly mention de novo assembly-based methods afterwards.
Table 1.
Methods of isoform or splicing analysis from RNA-seq. We summarized a collection of benchmark characteristics from simulated studies, literature reviews [142], [143], [144], and software documentation. Abbreviations: ICA: isoform-centric approaches; ECA: exon-centric approaches; EM: expectation maximization algorithm; VB: variational Bayes inference algorithm, MCMC: Markov chain Monte Carlo.
| Methods | Speed | Alignment-free | Novel transcript/splicing event discovery | Major algorithm | Input format | Memory Usage | Multi-threading | |
|---|---|---|---|---|---|---|---|---|
| Cufflinks | ICA | Relative slow | No | Yes | EM | SAM/BAM | Medium | Yes |
| StringTie | ICA | Fast | No | Yes | Network flow | SAM/BAM | Medium | No |
| RSEM | ICA | Relative slow | No | No | EM | SAM/BAM/FASTQ | Medium | No |
| WemIQ | ICA | Fast | No | No | EM | SAM | Medium | No |
| eXpress | ICA | Fast | No | No | EM | SAM/BAM | Small | Yes |
| Sailfish | ICA | Extremely fast | Yes | No | EM, VB | FASTA/FASTQ | Small | Yes |
| Kallisto | ICA | Very fast | Yes | No | EM | FASTA/FASTQ | Small | Yes |
| Salmon | ICA | Very fast | Yes | No | EM, VB | SAM/BAM/FASTQ | Small | Yes |
| MISO | ECA | Fast | No | No | MCMC | SAM | Small | Yes |
| SUPPA | ECA | Fast | No | Yes (with de novo assembler) | Density-based clustering algorithm | Expression in TPM | Small | Yes |
| SplAdder | ECA | Fast | No | Yes | Splicing graph | BAM, GTF/GFF | Small | No |
4.1. Isoform-centric analysis tools
4.1.1. Cufflinks
Although developed nearly a decade ago, Cufflinks is perhaps one of the most popular approach for expression quantification, especially for simultaneous novel transcript discovery and abundance estimation [138]. Cufflinks adopts and extends the ideas from expressed sequence tag (EST) assemblers such as PASA, which collapse alignments to transcripts on the basis of splicing compatibility [145]; as well as Dilworth’s theorem which was originally applied to assemble a parsimonious set of haplotypes from sequencing reads of a mixed virus population [146]. The approach reduces the transcript assembly problem into searching a maximum matching in a weighted bipartite graph that represents compatibilities among fragments. Their validation results suggest that Cufflinks is capable of not only improving transcriptome-based genome annotation but also discovery of novel transcripts.
4.1.2. StringTie
StringTie is another highly efficient transcript quantification tool which has the ability to discover novel transcripts [139]. It uses a novel network flow algorithm combined with an optional de novo assembly step to discover and quantify transcripts concurrently. StringTie has assembled 53% more transcripts than Cufflinks in a benchmark of 90 million reads from human blood. Moreover, it runs faster and consumes less memory. StringTie belongs to the updated Tuxedo protocol (HISAT, StringTie, Ballgown) which achieves faster speed, substantially memory saving, and more accurate overall results than the previous Tuxedo protocol (Tophat, Cufflinks, Cuffdiff) [147]. The output of StringTie is also compatible with downstream specialized tools such as Cuffdiff, DESeq2, edgeR, and so on.
4.1.3. RSEM
RSEM is also a popular tool for quantifying gene and isoform abundances from single-end or paired-end RNA-Seq data [142], [143], [148], [149]. RSEM takes original FASTQ sequence files as well as BAM/SAM alignment files. It is equipped with Bowtie2, STAR and HISAT aligners internally for read alignment. Or users can choose their preferred alternative aligner by providing aligned BAM/SAM files. RSEM is an Expectation-Maximization (EM) algorithm-based method, which assigns mapped reads to transcript isoforms and estimates the maximum likelihood (ML) of relative abundances of transcript isoforms. However, since the assignment of reads to isoforms are resulted from iterations of the EM method, the performance of RSEM is relatively inefficient [142], [149]. Given the recent massive sequence data size, the speed of RSEM has become a major drawback.
4.1.4. WemIQ
Precise transcriptome quantification is also hindered by non-uninform short-read sampling. The severity of overdispersion can be exemplified by non-uniform read distribution along single-isoform genes. The hidden bias is multifactorial with many unknown causes, which varies between sequencing platforms and protocols. To address this fundamental problem and fully harness the power of transcriptomics data, Chen’s group has developed a series of statistical models to tackle the overdispersion issue in bulk RNA-seq and single-cell RNA-seq (scRNA-seq) [150], [151], [152]. The RNA-seq bias in these models is estimated by a generalized-Poisson model (GPSeq) in a data-adaptive and assumption-free manner [150]. GPSeq estimates the bias directly from read-count distribution and does not need to specify the (often unknown) bias sources. GPSeq outperforms commonly used bias correction methods and still efficiently removes bias in cases that traditional methods fail [151].
For isoform-level quantification, they developed WemIQ [151]. The heterogeneity of read counts along a multi-isoform gene is caused by both RNA-seq bias and annotation heterogeneity (i.e., exonic positions are shared by different isoforms). The challenge is to separate bias from signals during deconvolution of isoform expression. WemIQ addresses this challenge by assigning different weights to reads at different positions. Weights are derived from bias-correction factors using GPSeq. Then an EM algorithm is used to distribute reads among different isoforms and maximize the weighted loglikelihood. Both simulation and empirical analyses showed that WemIQ significantly improves the accuracy of isoform quantification and estimation of exon inclusion rates.
4.1.5. eXpress
eXpress is a more recent tool that takes advantage of an online-EM inference procedure that permits accurate inference of transcript abundance after a single pass over the read alignments [153]. In eXpress, Roberts and Pachter modified the online EM algorithm that resolves the fragment-assignment problem into one that works directly with estimated counts instead of relative abundances. In the algorithm, each incoming fragment can be mapped to any number of target sequences and assigned to its mapped target based on the previously estimated counts. As fragments being processed, their assignment allows the algorithm to update and improve parameter estimates. This dynamic scheme greatly enhances the convergence speed and improves software performance. However, since eXpress is still an alignment-based method, it requires input alignments in the SAM/BAM format. Thus, although the procedure itself is fast, the alignment step is still inevitable and time consuming.
4.1.6. Sailfish
Alignment-based quantification tools are usually relatively slow, below we will introduce three alignment-free methods developed in the past few years. Sailfish is a k-mer-based approach, which completely avoids the time-consuming step of mapping reads to a reference genome [154]. Expression quantification is performed by extraction of k-mers from reads followed by exact matching of the k-mers using a hash table. This approach provides much faster quantification estimates than other approaches (typically 20 times faster). Although it claimed there was no trade-off of accuracy loss, fragmenting sequence reads into k-mers indeed loses valuable information [142], [143]. K-mers may align to more transcripts than the read itself since they are shorter, thereby leading to loss of accuracy.
4.1.7. Kallisto
Kallisto is another alignment-free k-mer-based method developed to tackle the inadequate accuracy mentioned in the previous method [143]. It is based on pseudoalignment of reads and uses fast hashing of k-mers together with the transcriptome de Bruijn graph, which has been proven to be crucial for DNA and RNA assembly [155]. Clustering of pseudoalignments originated from the same transcripts into equivalence classes permits simpler likelihood function and more efficient algorithm convergence. The accuracy of Kallisto is similar to those of alignment-based RNA-seq quantification approaches since its pseudoalignments explicitly maintain the information provided by k-mers across reads, yet the speed of Kallisto is two orders of magnitude faster.
4.1.8. Salmon
Salmon is introduced by the same developers of Sailfish; it is also an ultra-fast alignment-free method [156]. Interestingly the developers no longer utilize k-mer-based algorithm. Salmon applies a two-phase parallel inference procedure consisting of a reduced data representation, and a novel lightweight read alignment algorithm. The first online phase uses a variant of stochastic, collapsed variational Bayesian inference (SCVB0) to estimate initial expression levels, model parameters, and to construct equivalence classes over the input fragments. During the second offline phase, a variational Bayesian EM algorithm is applied over a reduced representation of the data to refine the initial transcript abundance estimates until a data-dependent convergence criterion is achieved. This method achieves both significantly faster speed and state-of-art accuracy, and it is now wildly applied in RNA-seq data analysis because of those merits. One convenience Salmon provides is that, unlike other two alignment-free methods, it also takes SAM/BAM files as input. Therefore, for analyses requiring better quality control, Salmon may be the choice.
4.2. Exon-centric analysis tools
4.2.1. MISO
As mentioned above, event-based approaches are the emerging direction for alternative splicing analysis. MISO (Mixture-of-Isoforms) is a statistical model that quantifies expression of alternatively spliced exons and isoforms and identifies differentially regulated ones across samples [157]. The MISO model uses probabilistic method (Bayesian inference) to compute the probability of the reads originated from a certain isoform. MISO enables both exon-centric analysis and isoform-centric analysis with improved accuracy for quantification and differential detection across samples.
4.2.2. SUPPA
SUPPA is another powerful tool to investigate alternative splicing events [158]. SUPPA generates alternative splicing events from an input annotation file. It calculates the percentage or “proportion spliced-in” (PSI) to describe relative abundance of splicing events or transcript isoforms, exploiting the fast transcript quantification. Differences of these relative abundances (ΔPSI) across different samples are also reported to quantify differential splicing. SUPPA achieves a much faster computational speed, while its accuracy is on a par with most standard methods based on analysis of experimentally validated events. SUPPA would be an effective and affordable choice for splicing analysis of big datasets. While SUPPA itself is restricted to annotated splicing events, coupling with novel transcript reconstruction methods such as StringTie makes it a powerful tool to identify novel splicing events.
4.2.3. SplAdder
While most of event-based methods do not support the identification of novel splicing events, SplAdder is a powerful tool to tackle the task. To avoid the computational burden posed by the identification of complete transcripts, SplAdder takes an innovative approach which treats individual splicing events as proxy for transcriptome characteristics [159]. SplAdder takes a given annotation and summarizes all transcripts of a gene into a splicing graph. The splicing graph is further augmented with new information extracted from RNA-seq (i.e. new introns and exon segments detected in the alignment). Alternative splicing events can be identified through the augmented annotation graph, and eventually be quantified with RNA-seq data. SplAdder is able to detect all main types of alternative splicing as well as multiple (coordinated) exon skips. In terms of accuracy, SplAdder has achieved an overall high accuracy in the benchmark comparing to various other state-of-the-art methods, while remaining computationally efficient.
4.3. De novo assembly-based and reference-free tools
Discovery of novel transcripts has been challenging ever since the question was posed. Although tools such as Cufflinks [138], StringTie [139], SLIDE [160], [158], and IsoLasso [161] take advantage of different algorithms (EM, Lasso, network flow algorithm, and so on) and incorporate existing annotations to perform transcript discovery, the results of accurate transcript reconstruction exhibit certain disagreements and remain unsatisfactory [135]. More importantly, those genome-guided novel transcript detection may be biased by the process itself [134], [162]. One way to mitigate the bias from the reference genome is to apply de novo transcript assembly through packages such as Trinity [163], Trans-AbySS [164], and Oases [165]. Trinity is by far the most widely applied de novo assembler with over 10, 000 citations. It is named Trinity as three software modules were involved. Inchworm first assembles the RNA-seq data into linear contigs; Chrysalis then groups related contigs and constructs de-Bruijn graphs; finally Butterfly examines the reads and reports full-length transcripts through dynamic programming. In splicing analysis, de novo assembly-based approaches usually first reconstruct and quantify isoforms, and then quantify alternative splicing events. One exception, KisSplice [166], de novo assembles alternative splicing events directly, and achieves a higher accuracy than other de novo full-length transcript assemblers.
4.4. Comparison of different analysis tools
Most of the isoform quantification with known transcripts exploit either EM algorithms or Bayesian inference to perform the read-count deconvolution. The difference mainly lies in the algorithm convergence speed [142]. One exception is WemIQ which also handles the bias correction in RNA-seq to improve the quantification accuracy. Generally, alignment-free methods, such as Salmon and Kallisto are extremely fast. One should take advantage of those tools when processing tremendously large-scale data. Alignment-based methods sometimes have slightly better performance in accuracy than alignment-free methods according to previous evaluation [142].
The accuracy of novel transcript or splicing event discovery still remains questionable, due to the lack of agreement of different methods, one should consider combine two or more approaches to validate with each other if high accuracy is desired. As mentioned before, the quantification of novel isoforms suffers from bias when deploying methods incorporating existing annotation. Using de novo assembly-based methods such as Trinity followed by downstream quantification packages such as RESM, eXpress, or Salmon may mitigate the issue. A recent study shows that in terms of conducting alternative splicing annotation and differential analysis, the results of de novo assembly-based methods and reference-based methods overlap by only 70% with noticeable differences [141]. Since assembly-based methods are capable of detecting more novel events and reference-based methods perform better for lowly expressed transcripts, Benoit-Pilven et al. suggest that the combination of those would be a better alternative [141].
Isoform-centric methods and exon-centric methods complement with each other. The choice should be decided by the raised biological questions. Each of the different approaches could shed light on different discovery. While isoform-centric methods rely on quantification of full-length transcripts, exon-centric approaches have bypassed this step by directly exploiting the quantification of alternative splicing events. Some studies concluded that exon-centric methods are superior to isoform-centric methods [167], [168], [169]. Another study showed that some exon-centric methods may not handle multiple replicates well [170]. We also argue that although exon-centric methods are more sensitive with known transcripts, isoform-centric ones provide more flexibility in novel and complicated alternative splicing events. Current isoform-centric methods are mainly hindered by the computational accuracy issue resulting from short reads. Recent advances in third-generation sequencing (long-read technology) such as single molecule real time sequencing (SMRT) from Pacific Biosciences, or the Nanopore sequencing from Oxford can produce substantially longer reads. With reads longer than a typical full-length transcript, in the near future, the accuracy of isoform quantification will allow the direct quantification of splicing events.
5. Conclusion
It has been more than four decades since Walter Gilbert first proposed the concept of alternative splicing: special combination of exons being spliced together to make special differentiation products [171]. Our understanding about alternative splicing is advancing rapidly with the help of molecular research, high-throughput sequencing and bioinformatic tools, yet there are still more to explore how alternative splicing functions at cellular level.
As discussed above, quantification of full-length transcripts and lowly expressed genes are the two major stumbling blocks for alternative splicing studies based on RNA-seq. We envision advances in two types of technologies may resolve those issues in the near future. 1) Long-read sequencing, SMRT and Nanopore sequencing technologies, has the potential to sequence the entire transcript bypassing the reconstruction issue. Those technologies have been proven applicable in revealing full-length transcripts and alternative splicing events [172], [173], [174], [175], [176]. 2) Single-cell RNA-seq (scRNA-seq) examines gene expression on a cellular resolution with optimized next-generation sequence technologies. With proper protocols and bioinformatic pipeline, scRNA-seq has the potential to better unravel lowly expressed transcripts and splicing events [177], [178], [179], [180]. No doubt there are still limitations in both technologies. Long-read sequencing suffers from high error rates, thus de novo transcript detection and quantification is still unsatisfied [181]; scRNA-seq usually yields fewer expressed genes caused by limited sequence depth and suffers from little distinguishment between technical and biological noise [180]. But with the combination of those newer technologies with traditional ones, more and more powerful computing tools, complete understanding of alternative splicing will be fulfilled in the near future.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work has been supported by the National Institutes of Health [R01GM137428].
References
- 1.Graveley B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17(2):100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 2.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 3.Marquez Y., Brown J.W., Simpson C., Barta A., Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012;22(6):1184–1195. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Y., Cai M., Stanley H.E. Comparative analysis and classification of cassette exons and constitutive exons. Biomed Res Int. 2017;2017 doi: 10.1155/2017/7323508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sammeth M., Foissac S., Guigó R. A general definition and nomenclature for alternative splicing events. PLoS Comput Biol. 2008;4(8) doi: 10.1371/journal.pcbi.1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72(1):291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 7.Sugnet C.W., Kent W.J., Ares M., Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac Symp Biocomput. 2004;66–77 doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- 8.Sammeth M. Complete alternative splicing events are bubbles in splicing graphs. J Comput Biol. 2009;16(8):1117–1140. doi: 10.1089/cmb.2009.0108. [DOI] [PubMed] [Google Scholar]
- 9.Vanichkina D.P., Schmitz U., Wong J.J.-L., Rasko J.E., editors. Challenges in defining the role of intron retention in normal biology and disease. Seminars in cell & developmental biology. Elsevier; 2018. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z., Xiao X., Van Nostrand E., Burge C.B. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23(1):61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaillon O., Bouhouche K., Gout J.-F., Aury J.-M., Noel B., Saudemont B. Translational control of intron splicing in eukaryotes. Nature. 2008;451(7176):359–362. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- 12.Wahl M.C., Will C.L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Will C.L., Lührmann R. Spliceosome structure and function. Cold Spring Harbor Perspect Biol. 2011;3(7) doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staley J.P., Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92(3):315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 15.Wolf E., Kastner B., Deckert J., Merz C., Stark H., Luehrmann R. Exon, intron and splice site locations in the spliceosomal B complex. EMBO J. 2009;28(15):2283–2292. doi: 10.1038/emboj.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander B., Golas M.M., Makarov E.M., Brahms H., Kastner B., Lührmann R. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6. U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol Cell. 2006;24(2):267–278. doi: 10.1016/j.molcel.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Boehringer D., Makarov E.M., Sander B., Makarova O.V., Kastner B., Lührmann R. Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat Struct Mol Biol. 2004;11(5):463–468. doi: 10.1038/nsmb761. [DOI] [PubMed] [Google Scholar]
- 18.Jurica M.S., Sousa D., Moore M.J., Grigorieff N. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat Struct Mol Biol. 2004;11(3):265–269. doi: 10.1038/nsmb728. [DOI] [PubMed] [Google Scholar]
- 19.Behzadnia N., Golas M.M., Hartmuth K., Sander B., Kastner B., Deckert J. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26(6):1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matlin A.J., Clark F., Smith C.W. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6(5):386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 21.Jensen K.B., Dredge B.K., Stefani G., Zhong R., Buckanovich R.J., Okano H.J. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25(2):359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 22.Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 23.Polydorides A.D., Okano H.J., Yang Y.Y., Stefani G., Darnell R.B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci. 2000;97(12):6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., Burge C.B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14(5):802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Liu J., Huang B., Xu Y.M., Li J., Huang L.F. Mechanism of alternative splicing and its regulation. Biomed Rep. 2015;3(2):152–158. doi: 10.3892/br.2014.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eversole A., Maizels N. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol Cell Biol. 2000;20(15):5425–5432. doi: 10.1128/mcb.20.15.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 28.Blanchette M., Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18(7):1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.kyung Oh H, Lee E, Jang HN, Lee J, Moon H, Sheng Z, et al. hnRNP A1 contacts exon 5 to promote exon 6 inclusion of apoptotic Fas gene. Apoptosis. 2013;18(7):825-35. [DOI] [PMC free article] [PubMed]
- 30.Motta-Mena L.B., Heyd F., Lynch K.W. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol Cell. 2010;37(2):223–234. doi: 10.1016/j.molcel.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim K.H., Ferraris L., Filloux M.E., Raphael B.J., Fairbrother W.G. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci. 2011;108(27):11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ule J., Jensen K., Mele A., Darnell R.B. CLIP: a method for identifying protein–RNA interaction sites in living cells. Methods. 2005;37(4):376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22(5):535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 35.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B. iCLIP-transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. JoVE (Journal of Visualized Experiments) 2011;50 doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spitzer J, Hafner M, Landthaler M, Ascano M, Farazi T, Wardle G, et al. PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation): a step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods in enzymology. 539: Elsevier; 2014. p. 113-61. [DOI] [PMC free article] [PubMed]
- 38.Wang T., Xiao G., Chu Y., Zhang M.Q., Corey D.R., Xie Y. Design and bioinformatics analysis of genome-wide CLIP experiments. Nucleic Acids Res. 2015;43(11):5263–5274. doi: 10.1093/nar/gkv439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garzia A., Meyer C., Morozov P., Sajek M., Tuschl T. Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods. 2017;118:24–40. doi: 10.1016/j.ymeth.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishore S., Jaskiewicz L., Burger L., Hausser J., Khorshid M., Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8(7):559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 41.Uhl M., Houwaart T., Corrado G., Wright P.R., Backofen R. Computational analysis of CLIP-seq data. Methods. 2017;118:60–72. doi: 10.1016/j.ymeth.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 42.Buratti E., Baralle F.E. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24(24):10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989;9(11):4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmucker D., Clemens J.C., Shu H., Worby C.A., Xiao J., Muda M. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101(6):671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 45.Graveley B.R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123(1):65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warf M.B., Berglund J.A. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35(3):169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaquero-Garcia J, Barrera A, Gazzara MR, Gonzalez-Vallinas J, Lahens NF, Hogenesch JB, et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. elife. 2016;5:e11752. [DOI] [PMC free article] [PubMed]
- 48.Tazi J., Bakkour N., Stamm S. Alternative splicing and disease. Biochim Biophys Acta (BBA)-Mol Basis Dis. 2009;1792(1):14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plötz F.B., van Essen A.J., Bosschaart A.N., Bos A.P. Cerebro-costo-mandibular syndrome. Am J Med Genet. 1996;62(3):286–292. doi: 10.1002/(SICI)1096-8628(19960329)62:3<286::AID-AJMG16>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 50.James P.A., Aftimos S. Familial cerebro-costo-mandibular syndrome: a case with unusual prenatal findings and review. Clin Dysmorphol. 2003;12(1):63–68. doi: 10.1097/00019605-200301000-00012. [DOI] [PubMed] [Google Scholar]
- 51.Su P.-H., Chen J.-Y., Chiang C.-L., Ng Y.-Y., Chen S.-J. Exclusion of MYF5, GSC, RUNX2, and TCOF1 mutation in a case of cerebro-costo-mandibular syndrome. Clin Dysmorphol. 2010;19(2):51–55. doi: 10.1097/MCD.0b013e328335c133. [DOI] [PubMed] [Google Scholar]
- 52.Bacrot S., Doyard M., Huber C., Alibeu O., Feldhahn N., Lehalle D. Mutations in SNRPB, encoding components of the core splicing machinery, cause cerebro-costo-mandibular syndrome. Hum Mutat. 2015;36(2):187–190. doi: 10.1002/humu.22729. [DOI] [PubMed] [Google Scholar]
- 53.Lynch D.C., Revil T., Schwartzentruber J., Bhoj E.J., Innes A.M., Lamont R.E. Disrupted auto-regulation of the spliceosomal gene SNRPB causes cerebro–costo–mandibular syndrome. Nat Commun. 2014;5(1):1–6. doi: 10.1038/ncomms5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lines M.A., Huang L., Schwartzentruber J., Douglas S.L., Lynch D.C., Beaulieu C. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet. 2012;90(2):369–377. doi: 10.1016/j.ajhg.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernier F.P., Caluseriu O., Ng S., Schwartzentruber J., Buckingham K.J., Innes A.M. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet. 2012;90(5):925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wieczorek D., Newman W.G., Wieland T., Berulava T., Kaffe M., Falkenstein D. Compound heterozygosity of low-frequency promoter deletions and rare loss-of-function mutations in TXNL4A causes Burn-McKeown syndrome. Am J Hum Genet. 2014;95(6):698–707. doi: 10.1016/j.ajhg.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Favaro F.P., Alvizi L., Zechi-Ceide R.M., Bertola D., Felix T.M., de Souza J. A noncoding expansion in EIF4A3 causes Richieri-Costa-Pereira syndrome, a craniofacial disorder associated with limb defects. Am J Hum Genet. 2014;94(1):120–128. doi: 10.1016/j.ajhg.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krausova M, Staněk D, editors. snRNP proteins in health and disease. Seminars in cell & developmental biology; 2018: Elsevier. [DOI] [PubMed]
- 59.Lehalle D., Wieczorek D., Zechi-Ceide R., Passos-Bueno M.R., Lyonnet S., Amiel J. A review of craniofacial disorders caused by spliceosomal defects. Clin Genet. 2015;88(5):405–415. doi: 10.1111/cge.12596. [DOI] [PubMed] [Google Scholar]
- 60.Board PATE. Myelodysplastic Syndromes Treatment (PDQ®): Patient Version. PDQ Cancer Information Summaries [Internet]. 2002.
- 61.Dolatshad H., Pellagatti A., Fernandez-Mercado M., Yip B.H., Malcovati L., Attwood M. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 2015;29(5):1092–1103. doi: 10.1038/leu.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malcovati L., Papaemmanuil E., Bowen D.T., Boultwood J., Della Porta M.G., Pascutto C. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood, J Am Soc Hematol. 2011;118(24):6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thol F., Kade S., Schlarmann C., Löffeld P., Morgan M., Krauter J. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood, J Am Soc Hematol. 2012;119(15):3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 65.Will C.L., Urlaub H., Achsel T., Gentzel M., Wilm M., Lührmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21(18):4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Will C.L., Schneider C., Hossbach M., Urlaub H., Rauhut R., Elbashir S. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004;10(6):929–941. doi: 10.1261/rna.7320604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das R., Zhou Z., Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell. 2000;5(5):779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- 68.DeBoever C., Ghia E.M., Shepard P.J., Rassenti L., Barrett C.L., Jepsen K. Transcriptome sequencing reveals potential mechanism of cryptic 3’splice site selection in SF3B1-mutated cancers. PLoS Comput Biol. 2015;11(3) doi: 10.1371/journal.pcbi.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alsafadi S., Houy A., Battistella A., Popova T., Wassef M., Henry E. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7(1):1–12. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darman R.B., Seiler M., Agrawal A.A., Lim K.H., Peng S., Aird D. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 2015;13(5):1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 71.Wang L., Brooks A.N., Fan J., Wan Y., Gambe R., Li S. Transcriptomic characterization of SF3B1 mutation reveals its pleiotropic effects in chronic lymphocytic leukemia. Cancer Cell. 2016;30(5):750–763. doi: 10.1016/j.ccell.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kesarwani A.K., Ramirez O., Gupta A.K., Yang X., Murthy T., Minella A.C. Cancer-associated SF3B1 mutants recognize otherwise inaccessible cryptic 3′ splice sites within RNA secondary structures. Oncogene. 2017;36(8):1123–1133. doi: 10.1038/onc.2016.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Je E.M., Yoo N.J., Kim Y.J., Kim M.S., Lee S.H. Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors. Int J Cancer. 2013;133(1):260–265. doi: 10.1002/ijc.28011. [DOI] [PubMed] [Google Scholar]
- 74.Haferlach T., Nagata Y., Grossmann V., Okuno Y., Bacher U., Nagae G. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.David C.J., Manley J.L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24(21):2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Venables J.P. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64(21):7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 77.Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33(46):5311-8. [DOI] [PubMed]
- 78.Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre‐mRNA splicing in cancer. Genes, Chromosomes and Cancer. 2005;42(4):342-57. [DOI] [PubMed]
- 79.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. Journal of cell science. 2006;119(13):2635-41. [DOI] [PubMed]
- 80.Kaida D., Motoyoshi H., Tashiro E., Nojima T., Hagiwara M., Ishigami K. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3(9):576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 81.Roybal G.A., Jurica M.S. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res. 2010;38(19):6664–6672. doi: 10.1093/nar/gkq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corrionero A., Miñana B., Valcárcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25(5):445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heaton N.S., Moshkina N., Fenouil R., Gardner T.J., Aguirre S., Shah P.S. Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity. 2016;44(1):46–58. doi: 10.1016/j.immuni.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang N., Gibbs J.S., Hickman H.S., Reynoso G.V., Ghosh A.K., Bennink J.R. Defining Viral DRiPs: standard and alternative translation initiation events generate a common peptide from influenza A Virus M2 and M1 mRNAs. J Immunol (Baltimore, Md 1950) 2016;196(9):3608. doi: 10.4049/jimmunol.1502303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Irwin D.J. Tauopathies as clinicopathological entities. Parkinsonism Related Disorders. 2016;22:S29–S33. doi: 10.1016/j.parkreldis.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sergeant N., Delacourte A., Buée L. Tau protein as a differential biomarker of tauopathies. Biochim Biophys Acta (BBA)-Mol Basis Dis. 2005;1739(2–3):179–197. doi: 10.1016/j.bbadis.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 87.Gallo J.-M., Noble W., Martin T.R. RNA and protein-dependent mechanisms in tauopathies: consequences for therapeutic strategies. Cell Mol Life Sci. 2007;64(13):1701–1714. doi: 10.1007/s00018-007-6513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andreadis A. Springer; 2006. Misregulation of tau alternative splicing in neurodegeneration and dementia. Alternative Splicing and Disease; pp. 89–107. [DOI] [PubMed] [Google Scholar]
- 89.Spillantini M.G., Goedert M. Tau protein pathology in neurodegenerative diseases. Trends Neurosci. 1998;21(10):428–433. doi: 10.1016/s0166-2236(98)01337-x. [DOI] [PubMed] [Google Scholar]
- 90.Mudher A., Lovestone S. Alzheimer's disease–do tauists and baptists finally shake hands? Trends Neurosci. 2002;25(1):22–26. doi: 10.1016/s0166-2236(00)02031-2. [DOI] [PubMed] [Google Scholar]
- 91.Fernández-Nogales M., Lucas J.J. Altered levels and isoforms of tau and nuclear membrane invaginations in Huntington’s disease. Front Cell Neurosci. 2019;13 doi: 10.3389/fncel.2019.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wolfe M.S. The role of tau in neurodegenerative diseases and its potential as a therapeutic target. Scientifica. 2012;2012 doi: 10.6064/2012/796024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cisbani G., Maxan A., Kordower J.H., Planel E., Freeman T.B., Cicchetti F. Presence of tau pathology within foetal neural allografts in patients with Huntington’s and Parkinson’s disease. Brain. 2017;140(11):2982–2992. doi: 10.1093/brain/awx255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fernández-Nogales M., Cabrera J.R., Santos-Galindo M., Hoozemans J.J., Ferrer I., Rozemuller A.J. Huntington's disease is a four-repeat tauopathy with tau nuclear rods. Nat Med. 2014;20(8):881–885. doi: 10.1038/nm.3617. [DOI] [PubMed] [Google Scholar]
- 95.Haber D.A., Sohn R.L., Buckler A.J., Pelletier J., Call K.M., Housman D.E. Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc Natl Acad Sci. 1991:9618–9622. doi: 10.1073/pnas.88.21.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gessler M., König A., Bruns G. The genomic organization and expression of the WT1 gene. Genomics. 1992;12(4):807–813. doi: 10.1016/0888-7543(92)90313-h. [DOI] [PubMed] [Google Scholar]
- 97.Klamt B., Koziell A., Poulat F., Wieacker P., Scambler P., Berta P. Frasier syndrome is caused by defective alternative splicing of WT1 leading to an altered ratio of WT1+/− KTS splice isoforms. Hum Mol Genet. 1998;7(4):709–714. doi: 10.1093/hmg/7.4.709. [DOI] [PubMed] [Google Scholar]
- 98.Splawski I., Timothy K.W., Decher N., Kumar P., Sachse F.B., Beggs A.H. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci. 2005;102(23):8089–8096. doi: 10.1073/pnas.0502506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abernethy D.R., Soldatov N.M. Structure-functional diversity of human L-type Ca2+ channel: perspectives for new pharmacological targets. J Pharmacol Exp Ther. 2002;300(3):724–728. doi: 10.1124/jpet.300.3.724. [DOI] [PubMed] [Google Scholar]
- 100.Liao P., Yong T.F., Liang M.C., Yue D.T., Soong T.W. Splicing for alternative structures of Cav1. 2 Ca2+ channels in cardiac and smooth muscles. Cardiovasc Res. 2005;68(2):197–203. doi: 10.1016/j.cardiores.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 101.Tang ZZ, Liang MC, Lu S, Yu D, Yu CY, Yue DT, et al. Transcript scanning reveals novel and extensive splice variations in human L-type voltage-gated calcium channel, Cav1. 2 α1 subunit. Journal of Biological Chemistry. 2004;279(43):44335-43. [DOI] [PubMed]
- 102.Welling A., Ludwig A., Zimmer S., Klugbauer N., Flockerzi V., Hofmann F. Alternatively spliced IS6 segments of the α1C gene determine the tissue-specific dihydropyridine sensitivity of cardiac and vascular smooth muscle L-type Ca2+ channels. Circ Res. 1997;81(4):526–532. doi: 10.1161/01.res.81.4.526. [DOI] [PubMed] [Google Scholar]
- 103.Splawski I., Timothy K.W., Sharpe L.M., Decher N., Kumar P., Bloise R. CaV1. 2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 104.Anderson S.L., Coli R., Daly I.W., Kichula E.A., Rork M.J., Volpi S.A. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet. 2001;68(3):753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheishvili D., Maayan C., Smith Y., Ast G., Razin A. IKAP/hELP1 deficiency in the cerebrum of familial dysautonomia patients results in down regulation of genes involved in oligodendrocyte differentiation and in myelination. Hum Mol Genet. 2007;16(17):2097–2104. doi: 10.1093/hmg/ddm157. [DOI] [PubMed] [Google Scholar]
- 106.Gudipati R.K., Xu Z., Lebreton A., Séraphin B., Steinmetz L.M., Jacquier A. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell. 2012;48(3):409–421. doi: 10.1016/j.molcel.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ni T., Yang W., Han M., Zhang Y., Shen T., Nie H. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res. 2016;44(14):6817–6829. doi: 10.1093/nar/gkw591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mauger O., Lemoine F., Scheiffele P. Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron. 2016;92(6):1266–1278. doi: 10.1016/j.neuron.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 109.Gontijo A.M., Miguela V., Whiting M.F., Woodruff R., Dominguez M. Intron retention in the Drosophila melanogaster Rieske Iron Sulphur Protein gene generated a new protein. Nat Commun. 2011;2(1):1–12. doi: 10.1038/ncomms1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wong J.J.L., Au A.Y., Ritchie W., Rasko J.E. Intron retention in mRNA: No longer nonsense: known and putative roles of intron retention in normal and disease biology. BioEssays. 2016;38(1):41–49. doi: 10.1002/bies.201500117. [DOI] [PubMed] [Google Scholar]
- 111.Perheentupa J. Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J Clin Endocrinol Metab. 2006;91(8):2843–2850. doi: 10.1210/jc.2005-2611. [DOI] [PubMed] [Google Scholar]
- 112.Buzi F., Badolato R., Mazza C., Giliani S., Notarangelo L.D., Radetti G. Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome: time to review diagnostic criteria? J Clin Endocrinol Metab. 2003;88(7):3146–3148. doi: 10.1210/jc.2002-021495. [DOI] [PubMed] [Google Scholar]
- 113.Consortium F-GA. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nature genetics. 1997;17(4):399. [DOI] [PubMed]
- 114.Conteduca G., Indiveri F., Filaci G., Negrini S. Beyond APECED: an update on the role of the autoimmune regulator gene (AIRE) in physiology and disease. Autoimmun Rev. 2018;17(4):325–330. doi: 10.1016/j.autrev.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 115.Zhang J., Liu H., Liu Z., Liao Y., Guo L., Wang H. A functional alternative splicing mutation in AIRE gene causes autoimmune polyendocrine syndrome type 1. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0053981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Masood N., Malik F.A., Kayani M.A. Unusual intronic variant in GSTP1 in head and neck cancer in Pakistan. Asian Pac J Cancer Prev. 2012;13(4):1683–1686. doi: 10.7314/apjcp.2012.13.4.1683. [DOI] [PubMed] [Google Scholar]
- 117.Solomon D.A., Wang Y., Fox S.R., Lambeck T.C., Giesting S., Lan Z. Cyclin D1 splice variants Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278(32):30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- 118.Comstock C.E., Augello M.A., Benito R.P., Karch J., Tran T.H., Utama F.E. Cyclin D1 splice variants: polymorphism, risk, and isoform-specific regulation in prostate cancer. Clin Cancer Res. 2009;15(17):5338–5349. doi: 10.1158/1078-0432.CCR-08-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dvinge H., Bradley R.K. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7(1):45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lin L., Park J.W., Ramachandran S., Zhang Y., Tseng Y.T., Shen S. Transcriptome sequencing reveals aberrant alternative splicing in Huntington's disease. Hum Mol Genet. 2016;25(16):3454–3466. doi: 10.1093/hmg/ddw187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Raj T., Li Y.I., Wong G., Humphrey J., Wang M., Ramdhani S. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer's disease susceptibility. Nat Genet. 2018;50(11):1584–1592. doi: 10.1038/s41588-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brinkman B.M. Splice variants as cancer biomarkers. Clin Biochem. 2004;37(7):584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 123.Pajares M.J., Ezponda T., Catena R., Calvo A., Pio R., Montuenga L.M. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol. 2007;8(4):349–357. doi: 10.1016/S1470-2045(07)70104-3. [DOI] [PubMed] [Google Scholar]
- 124.Dales J.-P., Beaufils N., Silvy M., Picard C., Pauly V., Pradel V. Hypoxia inducible factor 1α gene (HIF-1α) splice variants: potential prognostic biomarkers in breast cancer. BMC Med. 2010;8(1):44. doi: 10.1186/1741-7015-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li Y., Sun N., Lu Z., Sun S., Huang J., Chen Z. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017;393:40–51. doi: 10.1016/j.canlet.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 126.He R-q, Zhou X-g, Yi Q-y, Deng C-w, Gao J-m, Chen G, et al. Prognostic signature of alternative splicing events in bladder urothelial carcinoma based on spliceseq data from 317 cases. Cellular Physiology and Biochemistry. 2018;48(3):1355-68. [DOI] [PubMed]
- 127.Song J., Liu Y.D., Su J., Yuan D., Sun F., Zhu J. Systematic analysis of alternative splicing signature unveils prognostic predictor for kidney renal clear cell carcinoma. J Cell Physiol. 2019;234(12):22753–22764. doi: 10.1002/jcp.28840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hu C., Wang Y., Liu C., Shen R., Chen B., Sun K. Systematic profiling of alternative splicing for sarcoma patients reveals novel prognostic biomarkers associated with tumor microenvironment and immune cells. Med Sci Monit. 2020;26:e924126–e924131. doi: 10.12659/MSM.924126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang D., Duan Y., Cun J., Yang Q. Identification of prognostic alternative splicing signature in breast carcinoma. Front Genet. 2019;10:278. doi: 10.3389/fgene.2019.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang X, Huang W-t, He R-q, Ma J, Lin P, Xie Z-c, et al. Determining the prognostic significance of alternative splicing events in soft tissue sarcoma using data from The Cancer Genome Atlas. Journal of translational medicine. 2019;17(1):1-21. [DOI] [PMC free article] [PubMed]
- 131.Zong Z., Li H., Yi C., Ying H., Zhu Z., Wang H. Genome-wide profiling of prognostic alternative splicing signature in colorectal cancer. Front Oncol. 2018;8:537. doi: 10.3389/fonc.2018.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li S., Hu Z., Zhao Y., Huang S., He X. Transcriptome-wide analysis reveals the landscape of aberrant alternative splicing events in liver cancer. Hepatology. 2019;69(1):359–375. doi: 10.1002/hep.30158. [DOI] [PubMed] [Google Scholar]
- 133.Shen S., Wang Y., Wang C., Wu Y.N., Xing Y. SURVIV for survival analysis of mRNA isoform variation. Nat Commun. 2016;7:11548. doi: 10.1038/ncomms11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17(1):13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Steijger T., Abril J.F., Engström P.G., Kokocinski F., Akerman M., Alioto T. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods. 2013;10(12):1177–1184. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Roberts A. Ambiguous fragment assignment for high-throughput sequencing experiments. UC Berkeley. 2013 [Google Scholar]
- 137.Martin J.A., Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12(10):671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 138.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dargahi D., Swayze R.D., Yee L., Bergqvist P.J., Hedberg B.J., Heravi-Moussavi A. A pan-cancer analysis of alternative splicing events reveals novel tumor-associated splice variants of matriptase. Cancer Inform. 2014;13:167–177. doi: 10.4137/CIN.S19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Benoit-Pilven C., Marchet C., Chautard E., Lima L., Lambert M.-P., Sacomoto G. Complementarity of assembly-first and mapping-first approaches for alternative splicing annotation and differential analysis from RNAseq data. Sci Rep. 2018;8(1):1–13. doi: 10.1038/s41598-018-21770-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang C., Zhang B., Vincent M., Zhao S. Bioinformatics tools for RNA-seq gene and isoform quantification. Next Generat Sequenc Appl. 2016;3:140. [Google Scholar]
- 143.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 144.Zhang C., Zhang B., Lin L.-L., Zhao S. Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genomics. 2017;18(1):583. doi: 10.1186/s12864-017-4002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Jr, Hannick L.I. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31(19):5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Eriksson N., Pachter L., Mitsuya Y., Rhee S., Wang C., Gharizadeh B. Viral population estimation using pyrosequencing. PLoS Comput Biol Public Library Sci. 2008;4 doi: 10.1371/journal.pcbi.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kanitz A., Gypas F., Gruber A.J., Gruber A.R., Martin G., Zavolan M. Comparative assessment of methods for the computational inference of transcript isoform abundance from RNA-seq data. Genome Biol. 2015;16(1):150. doi: 10.1186/s13059-015-0702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Srivastava S., Chen L. A two-parameter generalized Poisson model to improve the analysis of RNA-seq data. Nucleic Acids Res. 2010;38(17) doi: 10.1093/nar/gkq670. e170-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang J., Kuo C.-C.J., Chen L. WemIQ: an accurate and robust isoform quantification method for RNA-seq data. Bioinformatics. 2015;31(6):878–885. doi: 10.1093/bioinformatics/btu757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen L., Zheng S. BCseq: accurate single cell RNA-seq quantification with bias correction. Nucleic Acids Res. 2018;46(14) doi: 10.1093/nar/gky308. e82-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Roberts A., Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10(1):71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Patro R., Mount S.M., Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol. 2014;32(5):462–464. doi: 10.1038/nbt.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Compeau P.E., Pevzner P.A., Tesler G. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol. 2011;29(11):987–991. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7(12):1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alamancos G.P., Pagès A., Trincado J.L., Bellora N., Eyras E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA. 2015;21(9):1521–1531. doi: 10.1261/rna.051557.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kahles A., Ong C.S., Zhong Y., Rätsch G. SplAdder: identification, quantification and testing of alternative splicing events from RNA-Seq data. Bioinformatics. 2016;32(12):1840–1847. doi: 10.1093/bioinformatics/btw076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li J.J., Jiang C.-R., Brown J.B., Huang H., Bickel P.J. Sparse linear modeling of next-generation mRNA sequencing (RNA-Seq) data for isoform discovery and abundance estimation. Proc Natl Acad Sci. 2011;108(50):19867–19872. doi: 10.1073/pnas.1113972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li W., Feng J., Jiang T. IsoLasso: a LASSO regression approach to RNA-Seq based transcriptome assembly. J Comput Biol. 2011;18(11):1693–1707. doi: 10.1089/cmb.2011.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Engström P.G., Steijger T., Sipos B., Grant G.R., Kahles A., Alioto T. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods. 2013;10(12):1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Robertson G., Schein J., Chiu R., Corbett R., Field M., Jackman S.D. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7(11):909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 165.Schulz M.H., Zerbino D.R., Vingron M., Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28(8):1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sacomoto G.A., Kielbassa J., Chikhi R., Uricaru R., Antoniou P., Sagot M.F. KISSPLICE: de-novo calling alternative splicing events from RNA-seq data. BMC Bioinf. 2012;13(Suppl 6):S5. doi: 10.1186/1471-2105-13-S6-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mehmood A., Laiho A., Venäläinen M.S., McGlinchey A.J., Wang N., Elo L.L. Systematic evaluation of differential splicing tools for RNA-seq studies. Brief Bioinform. 2019 doi: 10.1093/bib/bbz126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu R., Loraine A.E., Dickerson J.A. Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems. BMC Bioinf. 2014;15:364. doi: 10.1186/s12859-014-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Soneson C., Matthes K.L., Nowicka M., Law C.W., Robinson M.D. Isoform prefiltering improves performance of count-based methods for analysis of differential transcript usage. Genome Biol. 2016;17:12. doi: 10.1186/s13059-015-0862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Alamancos G.P., Agirre E., Eyras E. Methods to study splicing from high-throughput RNA sequencing data. Methods Mol Biol. 2014;1126:357–397. doi: 10.1007/978-1-62703-980-2_26. [DOI] [PubMed] [Google Scholar]
- 171.Gilbert W. Why genes in pieces? Nature. 1978;271(5645):501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 172.Ma J., Xiang Y., Xiong Y., Lin Z., Xue Y., Mao M. SMRT sequencing analysis reveals the full-length transcripts and alternative splicing patterns in Ananas comosus var. bracteatus. PeerJ. 2019;7 doi: 10.7717/peerj.7062. e7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sharon D., Tilgner H., Grubert F., Snyder M. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013;31(11):1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Park E., Pan Z., Zhang Z., Lin L., Xing Y. The expanding landscape of alternative splicing variation in human populations. Am J Hum Genet. 2018;102(1):11–26. doi: 10.1016/j.ajhg.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Byrne A., Beaudin A.E., Olsen H.E., Jain M., Cole C., Palmer T. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun. 2017;8(1):1–11. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.de Jong L.C., Cree S., Lattimore V., Wiggins G.A., Spurdle A.B., Miller A. Nanopore sequencing of full-length BRCA1 mRNA transcripts reveals co-occurrence of known exon skipping events. Breast Cancer Res. 2017;19(1):1–9. doi: 10.1186/s13058-017-0919-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50(8):1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sekula M., Gaskins J., Datta S. Detection of differentially expressed genes in discrete single-cell RNA sequencing data using a hurdle model with correlated random effects. Biometrics. 2019;75(4):1051–1062. doi: 10.1111/biom.13074. [DOI] [PubMed] [Google Scholar]
- 179.Picelli S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10(11):1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 180.Ramsköld D., Luo S., Wang Y.-C., Li R., Deng Q., Faridani O.R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Au K.F., Underwood J.G., Lee L., Wong W.H. Improving PacBio long read accuracy by short read alignment. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046679. [DOI] [PMC free article] [PubMed] [Google Scholar]