Abstract
Hypoxia plays important roles in cancer progression by inducing angiogenesis, metastasis, and drug resistance. However, the effects of hypoxia on long noncoding RNA (lncRNA) expression have not been clarified. Herein, we evaluated alterations in lncRNA expression in lung cancer cells under hypoxic conditions using lncRNA microarray analyses. Among 40,173 lncRNAs, 211 and 113 lncRNAs were up- and downregulated, respectively, in both A549 and NCI-H460 cells. Uroplakin 1A (UPK1A) and UPK1A-antisense RNA 1 (AS1), which showed the highest upregulation under hypoxic conditions, were selected to investigate the effects of UPK1A-AS1 on the expression of UPK1A and the mechanisms of hypoxia-inducible expression. Following transfection of cells with small interfering RNA (siRNA) targeting hypoxia-inducible factor 1α (HIF-1α), the hypoxia-induced expression of UPK1A and UPK1A-AS1 was significantly reduced, indicating that HIF-1α played important roles in the hypoxia-induced expression of these targets. After transfection of cells with UPK1A siRNA, UPK1A and UPK1A-AS1 levels were reduced. Moreover, transfection of cells with UPK1A-AS1 siRNA downregulated both UPK1A-AS1 and UPK1A. RNase protection assays demonstrated that UPK1A and UPK1A-AS1 formed a duplex; thus, transfection with UPK1A-AS1 siRNA decreased the RNA stability of UPK1A. Overall, these results indicated that UPK1A and UPK1A-AS1 expression increased under hypoxic conditions in a HIF-1α-dependent manner and that formation of a UPK1A/UPK1A-AS1 duplex affected RNA stability, enabling each molecule to regulate the expression of the other.
Keywords: hypoxia, hypoxia-inducible factor 1α , microarray, uroplakin 1A, uroplakin 1A antisense RNA 1
INTRODUCTION
Hypoxia-inducible factor 1α (HIF-1α) is a key transcription factor that regulates the transcription of target genes under hypoxic conditions, thereby inducing hypoxic responses (Ke and Costa, 2006). Under normoxic conditions, HIF-1α protein is rapidly degraded by the ubiquitin-proteasome system (Maxwell et al., 1999). When oxygen levels are low, HIF-1α is stabilized and binds to the hypoxia response element (HRE) within the promoter to induce the transcription of downstream target genes (Jiang et al., 1996). Because HIF-1α is a key regulator of hypoxia-induced cellular processes, HIF-1α is a promising therapeutic target in the treatment of cancer (Hu et al., 2013; Semenza, 2003; Yu et al., 2017).
Long noncoding RNAs (lncRNAs) are endogenous, non-protein-coding RNAs greater than 200 nucleotides in length (Mercer et al., 2009). LncRNAs play roles in various biological processes (Autuoro et al., 2014; Kanduri, 2016; Penny et al., 1996; Pollex and Heard, 2012) and have been implicated in pathological processes, such as carcinogenesis (Faghihi et al., 2008; Scheuermann and Boyer, 2013; Schmitt and Chang, 2016; Schonrock et al., 2012). In cancer, dysregulation of lncRNAs affects the expression of oncogenes and tumor suppressors via epigenetic silencing, transcriptional regulation, and post-transcriptional processing (Tang et al., 2017). In tumor hypoxia, the expression of many genes and microRNAs is altered to adapt to the low oxygen environment via hypoxia-specific cellular processes (Elvidge et al., 2006; Hong et al., 2004; Kulshreshtha et al., 2007). However, the identities, biological functions, and mechanisms of action of lncRNAs with altered expression under hypoxic conditions have not been thoroughly studied.
Genome-wide expression profiling approaches such as microarray analysis and RNA sequencing (RNA-seq), have been used to identify differentially expressed lncRNAs under hypoxic conditions (Fiedler et al., 2015; Lin et al., 2015; Mimura et al., 2017; Voellenkle et al., 2016; Zhu et al., 2017). Recently, lncRNA microarray analysis of oral squamous cell carcinoma (Zhu et al., 2017) showed that hyaluronan synthase 2 antisense 1 (HAS2-AS1) contains an HRE within its promoter and is upregulated under hypoxic conditions in a HIF-1α-dependent manner. Under hypoxic conditions, HAS2-AS1 plays roles in the upregulation of HAS2, its sense counterpart, and induction of invasion in oral squamous cell carcinoma. In another study in human kidney-2 cells and renal proximal tubular cells (Mimura et al., 2017), RNA-seq revealed that aspartyl-tRNA synthetase anti-sense 1 was upregulated under hypoxic conditions in a HIF-1α-dependent manner. In addition, recent studies have investigated hypoxia-regulated lncRNAs in endothelial cells at the genome-wide scale (Fiedler et al., 2015; Voellenkle et al., 2016). Notably, many hypoxia-regulated lncRNAs are upregulated under hypoxic conditions by HIF-1α and have roles in hypoxia-induced cellular processes, including angiogenesis and metastasis (Chang et al., 2016; Gómez-Maldonado et al., 2015; Shih and Kung, 2017; Tee et al., 2016; Wang et al., 2014).
Uroplakins (UPKs) are urothelial-specific transmembrane proteins that form plaques on the asymmetric unit membrane in the umbrella cells of the urothelium (Wu et al., 2009). The plaques consist of four distinct UPKs, i.e., UPK1A, UPK1B, UPKII, and UPKIII, which are highly expressed in the urothelium. These proteins regulate the membrane permeability of umbrella cells and strengthen the urothelium through interaction between the asymmetric unit membrane and cytoskeleton structure (Hall et al., 2005). Because of their specific expression in the urothelium, UPKs have not been studied in other cell types. However, UPK1A has been shown to be downregulated in several types of tumor tissues compared with their adjacent normal tissues (He et al., 2014; Kong et al., 2010; Zheng et al., 2014). In esophageal squamous cell carcinoma (ESCC) cells, overexpression of UPK1A inhibits cell proliferation and tumor formation in nude mice, suggesting that UPK1A is a tumor-suppressor gene (Kong et al., 2010). In contrast, transfection of T24 human bladder carcinoma cells with an antisense nucleotide of UPK1A inhibits cell proliferation and enhances apoptosis (Zhu et al., 2015). It appears that the low expression of UPK1A in various cell types has led to conflicting results.
In this study, we evaluated the roles of lncRNAs during hypoxia in lung carcinoma cells using an lncRNA microarray assay. Our results provided important insights into the altered expression of lncRNAs in lung cancer cells at a genome-wide scale and the mechanisms underlying hypoxia-induced expression of UPK1A and UPK1A-AS1.
MATERIALS AND METHODS
RNA oligonucleotides
Small interfering RNA (siRNA) against UPK1A antisense RNA 1 (UPK1A-AS1) was obtained from Dharmacon (USA) (Lincode Smartpool siRNA). SiRNAs targeting HIF-1α, UPK1A, and a negative control (NC) were purchased from Shanghai GenePharma Company (China). Antisense LNA GapmeRs targeting UPK1A-AS1 and a NC (NC-A) were obtained from Exiqon (Denmark). The 19-mer siRNA target sequences are shown in Supplementary Table S1.
Cell lines and transfection
A549 (human epithelial, lung carcinoma-derived; KCLB 10185), NCI-H460 (human epithelial, lung carcinoma-derived; KCLB 30177), and T24 (human epithelial, bladder carcinoma; KCLB 30004) cell lines were obtained from the Korean Cell Line Bank (Korea). Cells were cultured in Roswell Park Memorial Institute 1640 medium supplemented with 10% fetal bovine serum in a humidified atmosphere of 5% CO2 at 37°C. To mimic hypoxia, HIF-1α protein was induced by treatment with 200 μM cobalt chloride (CoCl2) for 24 h at 21% oxygen. Cell culture under hypoxic conditions was achieved by incubation in a hypoxia chamber (MIC-101; Billups-Rothenberg, USA) containing 1% O2, 5% CO2, and 94% N2 at 37°C. For transfection, Lipofectamine 2000 and RNAiMAX (Invitrogen, USA) were used following the manufacturer’s protocol, as previously described (Kim et al., 2008).
Western blot analysis
Total protein was isolated using Pro-Prep protein extraction reagent (iNtRON Biotechnology, Korea). Western blotting was performed using standard procedures. Briefly, total proteins (25 μg) were separated on 4% to 12% precast protein gels (Koma Biotech, Korea) and then transferred to nitrocellulose membranes. Membranes were briefly washed with Tris-buffered saline containing 0.05% Tween 20 (TTBS), blocked for 1 h in TTBS containing 5% nonfat dry milk, and incubated with antibodies overnight at 4°C. Primary antibodies specific for HIF-1α (cat. No. 610958) and β-actin (C-4) were purchased from BD Biosciences (USA) and Santa Cruz Biotechnology (USA), respectively. Blots were then washed, incubated with secondary antibodies, and visualized using the Enhanced Chemiluminescence Plus western blotting reagent (Amersham Biosciences, USA).
LncRNA microarray experiments
Total RNA was isolated from A549 and NCI-H460 cells cultured under normoxic or hypoxic conditions using an RNeasy Mini kit (Qiagen, Germany). Each group consisted of three replicates. Microarray experiments were performed using Arraystar Human LncRNA Microarray v4.0, which contained 40,173 lncRNA and 20,730 mRNA probes (Arraystar, USA). Sample labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies, USA), with minor modifications. Briefly, mRNA was purified from total RNA after removal of rRNA (mRNA-ONLY Eukaryotic mRNA Isolation Kit; Epicentre, USA). Next, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method (Arraystar Flash RNA Labeling Kit; Arraystar). The labeled cRNAs were purified using an RNeasy Mini Kit (Qiagen). One microgram of each labeled cRNA was fragmented by adding 5 μl of 10× Blocking Agent and 1 μl of 25× Fragmentation Buffer. The mixture was then heated at 60°C for 30 min, and 25 μl of 2× GE Hybridization buffer was added to dilute the labeled cRNA. Next, 50 μl hybridization solution was dispensed into the gasket slide and assembled to produce the lncRNA expression microarray slide. The slides were incubated for 17 h at 65°C in an Agilent Hybridization Oven. The hybridized arrays were washed, fixed, and scanned using an Agilent DNA Microarray Scanner (part No. G2505C).
Data analysis
For data analysis, Agilent Feature Extraction software (ver. 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using GeneSpring GX v12.1 software (Agilent Technologies). Differentially expressed lncRNAs and mRNAs with a fold-change of at least 1.5 and statistical significance were identified through Volcano Plot filtering between the two groups. Hierarchical clustering was performed using R software. Gene ontology (GO) analysis was performed using the topGO package in the R environment for statistical computing and graphics, and pathway analysis was performed using Fisher’s exact test. The microarray data is available in the Gene Expression Omnibus (GEO)/NCBI repository (accession No. GSE151120).
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis
Total RNA was isolated using an RNeasy Mini kit (Qiagen). Total RNA from human bladder tissues was obtained from BioChain (USA). For cDNA synthesis, 1 μg total RNA was reverse transcribed using an iScript cDNA synthesis Kit (Bio-Rad, USA). To determine relative mRNA levels, qRT-PCR was performed in triplicate in 384-well plates on an ABI Prism 7900 Sequence Detection System (Applied Biosystems, USA) using 2× SYBR Green PCR Master Mix (Applied Biosystems). The thermal cycling conditions consisted of an initial 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The expression of each cDNA was normalized to that of β-actin, and the comparative Ct method was used to obtain relative expression levels. The primers used for qRT-PCR are shown in Supplementary Table S2.
Nuclear and cytoplasmic RNA fractionation
Nuclear and cytoplasmic RNA fractions were isolated using a Cytoplasmic & Nuclear RNA purification kit (Norgen Biotek, Canada) according to the manufacturer’s instructions. After incubating NCI-H460 cells at 37°C for 24 h in a hypoxia chamber (MIC-101) under conditions of 1% O2, 5% CO2, and 94% N2, 2 × 106 cells were harvested. The process was carried out according to the manufacturer’s protocol, and lysis buffer J was diluted to 15% before use. The nuclear RNA and cytoplasmic RNA concentrations were measured using a NanoDrop ND-1000, after which, qRT-PCR was performed. The primer sequences used in qRT-PCR are as shown in Supplementary Table S2.
Ribonuclease protection assay
NCI-H460 cells were harvested after incubating in a hypoxia chamber at 37°C for 24 h. Subsequent to RNA extraction using an RNeasy Mini kit (Qiagen), 90 μl RNA was incubated at 37°C for 1 h. After adding 10 μl of 10× RPA buffer (100 mM Tris-HCl, 3 M NaCl, and 50 mM ethylenediaminetetraacetic acid), the mixture was divided into two tubes (50 μl each). In one tube, 1 μl RNase A + T1 (Thermo Fisher Scientific, USA) was added, and both tubes were incubated at 37°C for 30 min. Following treatment with 200 μg/ml proteinase K (Biosesang, Korea), the tubes were incubated at 50°C for 1 h. RNA was isolated using an RNeasy Mini kit (Qiagen), and the concentration was measured. Subsequently, cDNA was synthesized by adding UPK-ss-primer and UPK-ds-primer using an iScript Select cDNA Synthesis Kit (Bio-Rad). After using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) to perform qRT-PCR on 1.5 μl cDNA, the product was identified by agarose gel electrophoresis. The thermal cycling conditions were as follows: 95°C for 10 min; and 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The primers used were as follows: UPK1A forward/UPK1A reverse and UPK-ds forward/UPK-ds reverse.
RNA stability
After incubating NCI-H460 cells for 24 h under hypoxic conditions, the cells were treated with 4 μg/ml actinomycin D (Thermo Fisher Scientific) and transfected with 20 nM NC, UPK1A siRNA-916, and UPK1A-AS1 siRNA (Dharmacon). After incubation for 12 h and 24 h under hypoxic conditions, the cells were harvested, and RNA was isolated. During synthesis of cDNA from 1 μg RNA using an iScript cDNA Synthesis Kit (Bio-Rad), 2 μl TATAA Universal RNA Spike I (TATAA Biocenter, Sweden) was added. After cDNA synthesis, SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) was used to perform qRT-PCR with 1.5 μl cDNA. The thermal cycling conditions were as follows: 95°C for 10 min; and 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The primers used in this assay were as follows: UPK1A forward/UPK1A reverse, UPK1A-AS1 forward/UPK1A-AS1 reverse, and Spike I assay primer forward/reverse. TATAA Universal RNA Spike I serves as a normalization control. For transfection with plasmids, NCI-H460 cells were transfected with pcDNA3.1, pcDNA3.1-UPK1A-AS1-Wild, and pcDNA-UPK1A-AS1-Mutant at a concentration of 70 ng/well on 6-well plate using TransIT-X2 transfection reagent (Mirus Bio, USA). After 6 h, transfected cells were incubated under hypoxia. The following day, cells were treated with 4 μg/ml actinomycin D. After 0 h and 24 h, RNA was isolated and subjected to qRT-PCR as described above. For construction of plasmids, full-length and deleted UPK1A-AS1 transcripts were cloned into pcDNA3.1 to yield plasmids pcDNA3.1-UPK1A-AS1-Wild and pcDNA-UPK1A-AS1-Mutant, respectively (GenScript, USA). In the pcDNA-UPK1A-AS1-Mutant, 6-300 nt from the 5′ end containing overlapping region between UPK1A and UPK1A-AS1 was deleted from the UPK1A-AS1 transcript.
DNA methylation analysis
Genomic DNAs were isolated from A549, NCI-H460, and T24 cells using a Wizard Genomic DNA purification kit (Promega, USA). Genomic DNAs from human lung, liver, and bladder tissues were obtained from BioChain. Genomic DNA was treated with bisulfite using an EZ DNA Methylation-Gold kit (Zymo Research, USA). Bisulfite-modified genomic DNA was then amplified by PCR using EpiMark Hot Start Taq DNA Polymerase (New England BioLabs, USA). PCR was performed in a 25 μl reaction volume at 95°C for 60 s, followed by 40 cycles of 95°C for 25 s, 55°C for 45 s, and 68°C for 45 s. The primer sequences were as follows: UPK-296, 5′-GGTTTTGGGTTATTATTTTTGTATG-3′; UPK-479, 5′-CTTAACTTATAAATTTACCCATCTAC-3′ (bisulfite sequencing for a 184-bp fragment within a CpG island located approximately 1100 bp upstream of exon 1); UPK-40, 5′-GGGTGTTTTTTTGTGTAAAATGTTT-3′; UPK-287, 5′-ATATTACCCACAACTAACAAACCCA-3′ (bisulfite sequencing for a 248-bp fragment containing an ATG initiation codon within exon 2); UPKAS-202, 5′-TATTTTTTTAGTGATGTTTTTTTGA-3′; UPKAS-402, 5′-CCTCCCATACAAATCAAACC-3′ (bisulfite sequencing for a 201-bp fragment containing exon 1 of UPK1A-AS1). After purification using a DNA Clean and Concentrator kit (Zymo Research, USA), PCR products were cloned into the pGEM-T easy vector (Promega), and 10 to 20 independent clones were sequenced for each sample.
Statistical analysis
Data are shown as mean ± SD. Differences between groups were analyzed by two-tailed Student’s t-tests using Microsoft Excel software (Microsoft, USA). Results with P values of less than 0.05 were considered statistically significant.
RESULTS
Identification of hypoxia-induced lncRNAs by microarray
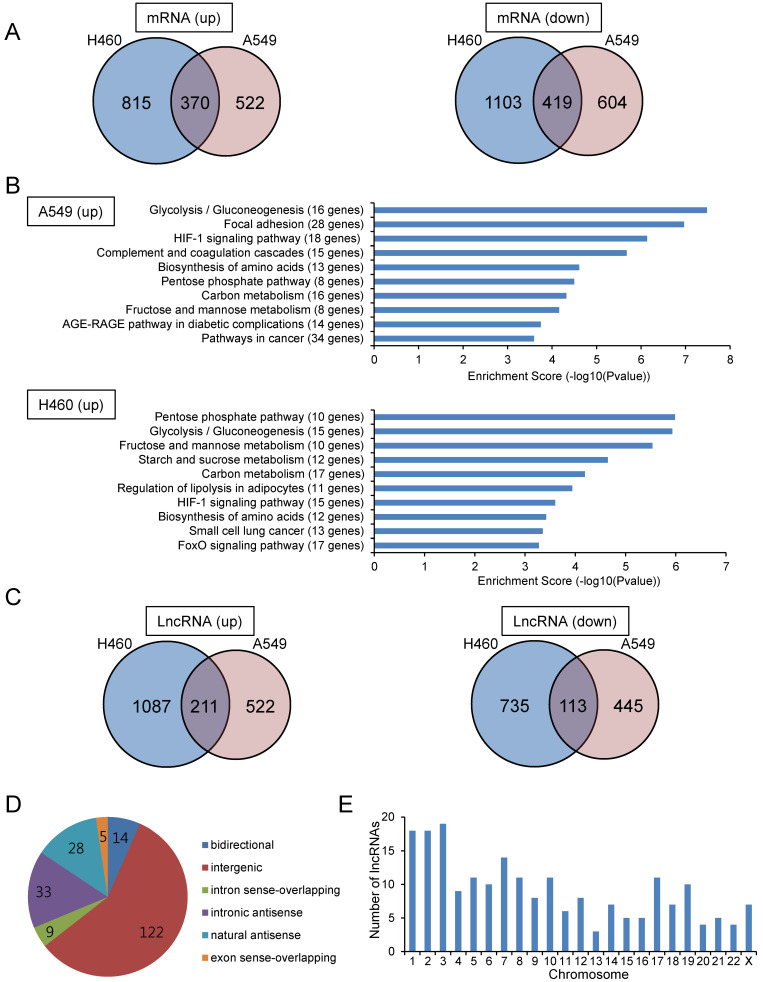
To identify lncRNAs whose expression was altered under hypoxic conditions, we performed microarray experiments using total RNA isolated from A549 and NCI-H460 lung carcinoma cells. The human microarray slides used in the experiment contained 40,173 lncRNA and 20,730 mRNA probes. Among 40,173 lncRNAs, 7,506 lncRNAs were labeled as “Gold Standard LncRNAs”; these lncRNAs are well annotated, have been functionally studied, and have been experimentally identified as full-length lncRNAs. The cells were cultured under hypoxic or normoxic conditions, and total RNA was subjected to microarray analysis. Figs. 1A and 1C show mRNAs and lncRNAs that were up- or downregulated by greater than 1.5-fold under hypoxic conditions compared with those under normoxic conditions in A549 and NCI-H460 cells. For mRNAs, 892 and 1,185 mRNAs were upregulated in A549 and NCI-H460 cells, respectively. As expected, the gene showing the greatest upregulation in both A549 and NCI-H460 cells was CA9, a known marker of hypoxia. Pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database showed that genes upregulated in hypoxia were involved in the HIF-1 signaling pathway, glycolytic pathway, and pentose phosphate pathway (Fig. 1B); these genes were also known downstream target genes of HIF-1α, including SLC2A1, PGK1, ALDOA, and GAPDH. For lncRNAs, 211 and 113 lncRNAs were up- and downregulated, respectively, in both A549 and NCI-H460 cells. Among the 211 upregulated lncRNAs, more than half were classified as intergenic lncRNAs, followed by intronic and natural antisense lncRNAs (Fig. 1D). The upregulated lncRNAs were located on all chromosomes, with more lncRNAs being located on chromosomes 1, 2, and 3 (Fig. 1E). The top 20 lncRNAs upregulated under hypoxic conditions in both A549 and NCI-H460 cells are shown in Table 1.
Fig. 1. Microarray analysis identified mRNAs and lncRNAs with altered expression under hypoxic conditions.
(A) Venn diagram of up- and downregulated mRNAs under hypoxic conditions compared with that under normoxic conditions in A549 and NCI-H460 cells. Cells were incubated under hypoxic conditions for 24 h, and total RNA was subjected to microarray analysis. (B) Pathway analysis based on the KEGG database for upregulated mRNAs under hypoxic conditions in A549 and NCI-H460 cells. The top 10 pathways with significant enrichment of differentially expressed mRNAs are shown. The P value denotes the significance of the pathway (cutoff: P < 0.05). (C) Venn diagram of up- and downregulated lncRNAs under hypoxic conditions compared with that under normoxic conditions in A549 and NCI-H460 cells. (D) LncRNA subgroup analysis based on the relationship between lncRNAs and their associated protein-coding genes. LncRNAs upregulated under hypoxic conditions in both A549 and NCI-H460 cells were classified. (E) Chromosomal distribution of lncRNAs with hypoxia-induced upregulation in both A549 and NCI-H460 cells.
Table 1.
Top 20 upregulated lncRNAs under hypoxic condition in both A549 and NCI-H460 cells
| Sequence name | Gene symbol | Fold change A549 | Fold change H460 | RNA length | Chromosome | Strand | Start | End | Source |
|---|---|---|---|---|---|---|---|---|---|
| NR_046420 | UPK1A-AS1 | 21.5191323 | 38.5241975 | 813 | chr19 | – | 36158849 | 36164193 | RefSeq |
| ENST00000522547 | RP11-14I17.2 | 10.6497245 | 38.3353098 | 508 | chr8 | + | 26280108 | 26281445 | GENCODE |
| ENST00000417355 | AC114803.3 | 8.5930288 | 46.8519287 | 311 | chr2 | + | 220163723 | 220168852 | GENCODE |
| ENST00000546523 | RP5-1057I20.2 | 8.384609 | 8.9300753 | 353 | chr12 | + | 48223276 | 48228337 | GENCODE |
| T054705 | G012608 | 7.9549952 | 26.5872803 | 1448 | chr11 | + | 10382151 | 10383599 | RNA-seq: Iyer et al., 2015 |
| T203015 | G046886 | 6.8663933 | 5.958497 | 1343 | chr2 | – | 173461092 | 173462435 | RNA-seq: Iyer et al., 2015 |
| T036689 | G008310 | 6.8583296 | 4.7583565 | 773 | chr10 | + | 6428518 | 6429558 | RNA-seq: Iyer et al., 2015 |
| ENST00000458314 | AC078883.3 | 6.1392631 | 5.030675 | 512 | chr2 | – | 173328989 | 173330750 | GENCODE |
| TCONS_00000467 | XLOC_000695 | 5.9628345 | 10.5276413 | 410 | chr1 | – | 8066073 | 8066784 | RNA-seq: Cabili et al., 2011 |
| ENST00000564127 | RP11-480I12.10 | 5.6380716 | 5.7666391 | 592 | chr1 | – | 202779365 | 202779957 | GENCODE |
| NR_015421 | LOC154761 | 5.626012 | 3.2081902 | 2579 | chr7 | – | 143509060 | 143533810 | RefSeq |
| T085710 | G019879 | 4.5601655 | 9.3665869 | 998 | chr12 | – | 106740693 | 106743112 | RNA-seq: Iyer et al., 2015 |
| NR_040079 | LOC399715 | 4.3338854 | 5.1864333 | 2967 | chr10 | + | 6368506 | 6377943 | RefSeq |
| NR_026804 | KLF3-AS1 | 4.2354962 | 6.9351289 | 2368 | chr4 | – | 38614321 | 38666249 | RefSeq |
| T336396 | G078889 | 4.1126362 | 9.7690188 | 3072 | chr7 | + | 153646863 | 153654276 | RNA-seq: Iyer et al., 2015 |
| NR_024006 | LINC00950 | 3.8685642 | 7.478096 | 5245 | chr9 | + | 35860270 | 35865515 | RefSeq |
| T036645 | G008294 | 3.8119865 | 3.0235726 | 590 | chr10 | + | 6296150 | 6296740 | RNA-seq: Iyer et al., 2015 |
| NR_120598 | GACAT2 | 3.7435975 | 2.183135 | 835 | chr18 | – | 8695853 | 8707619 | RefSeq |
| uc001gwt.1 | AX747377 | 3.7160007 | 2.6729957 | 2162 | chr1 | – | 201604354 | 201606516 | UCSC_knowngene |
| ENST00000608142 | RP11-1399P15.1 | 3.6876088 | 19.1472439 | 540 | chr2 | – | 87777013 | 87777553 | GENCODE |
Real-time PCR validation of mRNAs and associated antisense lncRNAs
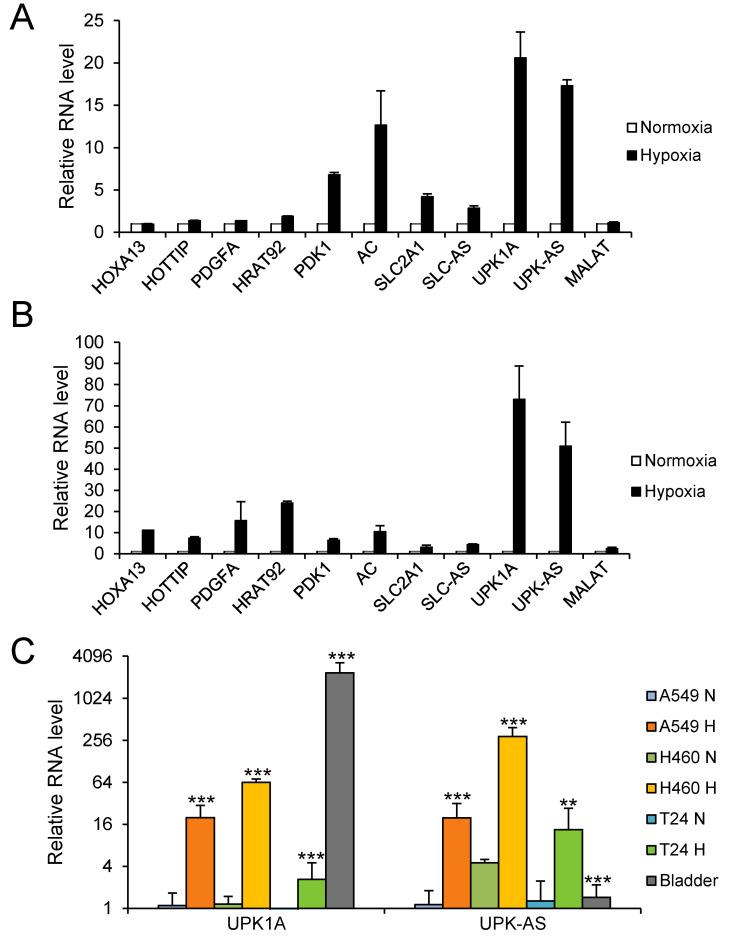
Antisense lncRNAs are a subgroup of lncRNAs that are transcribed in the opposite direction of an associated protein-coding gene. Increasing evidence indicates that antisense lncRNAs regulate the expression of nearby sense coding genes in cis, often via epigenetic modulation (Su et al., 2017; Yap et al., 2010; Yu et al., 2008). Therefore, we next focused on antisense lncRNAs and their nearby coding genes. Microarray data analysis yielded a table of differentially expressed antisense lncRNAs and their associated coding gene pairs under hypoxic conditions. Among them, we selected the following five pairs based on their functions and fold changes in gene expression under hypoxic conditions compared with those under normoxic conditions: homeobox A13 (HOXA13) and HOXA distal transcript antisense RNA (HOTTIP: Gold standard lncRNA; an oncogenic lncRNA regulating HOXA genes; both HOTTIP and HOXA13 are upregulated in hypoxia and HOTTIP might promote glycolysis under hypoxia), platelet derived growth factor subunit A (PDGFA) and heart tissue-associated transcript 92 (HRAT92: Gold standard lncRNA; not yet characterized), pyruvate dehydrogenase kinase 1 (PDK1: hypoxia-inducible metabolic switch; suppresses the glucose metabolism through the tricarboxylic acid cycle (TCA) by inactivating the TCA enzyme, pyruvate dehydrogenase) and AC093818.1, solute carrier family 2 member 1 (SLC2A1: HIF-1α-regulated glucose transporter GLUT1; switch from oxygen mitochondrial process to glycolysis by upregulating glycolytic enzymes including GLUT1—the Warburg effect), SLC2A1 antisense RNA 1 (SLC2A1-AS1), and UPK1A and UPK1A-AS1 (highly upregulated genes under hypoxia; their role has not yet been well characterized in cells other than bladder cells). Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which is upregulated in hypoxia (Lelli et al., 2015), was included as a positive control. As shown in Fig. 2A, PDK1 and AC093818.1, SLC2A1 and SLC2A1-AS1, and UPK1A and UPK1A-AS1 pairs were substantially upregulated under hypoxic conditions, whereas HOXA13, HOTTIP, PDGFA, and MALAT1 were not appreciably upregulated under hypoxic conditions in A549 cells. In NCI-H460 cells, all examined pairs, including MALAT1, were substantially upregulated under hypoxic conditions (Fig. 2B). Notably, UPK1A and UPK1A-AS1 were the most upregulated genes under hypoxic conditions in both A549 and NCI-H460 cells. Consistent with qRT-PCR results, microarray data revealed that UPK1A and UPK1A-AS1 genes were highly upregulated under hypoxic conditions in both A549 and NCI-H460 cells. Therefore, we selected the UPK1A and UPK1A-AS1 pair for further analyses.
Fig. 2. Validation of five lncRNAs and their associated coding genes by qRT-PCR.
(A and B) A549 (A) and NCI-H460 (B) cells were incubated under hypoxic conditions for 24 h, and total RNA was subjected to qRT-PCR. For analysis of each gene, the RNA level under normoxic conditions was set at 1. MALAT1 was used as a positive control. (C) Hypoxia-induced upregulation of UPK1A and UPK1A-AS1 in A549, NCI-H460, and T24 cells. The RNA level in A549 cells under normoxic conditions was set as 1. RNA from human bladder tissue was included as a positive control. **P < 0.01; ***P < 0.001. SLC-AS, SLC2A1-AS1; UPK-AS, UPK1A-AS1; MALAT, MALAT1.
HIF-1α-dependent upregulation of UPK1A and UPK1A-AS1 under hypoxic conditions
In addition to A549 and NCI-H460 cells, we examined the expression of UPK1A and UPK1A-AS1 under hypoxic conditions in T24 bladder carcinoma cells, using human bladder tissues as a positive control. As shown in Fig. 2C, both UPK1A and UPK1A-AS1 were upregulated under hypoxic conditions in all three cell lines; upregulation was greatest in NCI-H460 cells, followed by A549 cells and then T24 cells. Although UPK1A was expressed at a very high level in bladder tissue, the expression level of UPK1A-AS1 in bladder tissue was very low, suggesting that UPK1A-AS1 may have no substantial role in the expression of UPK1A in the bladder.
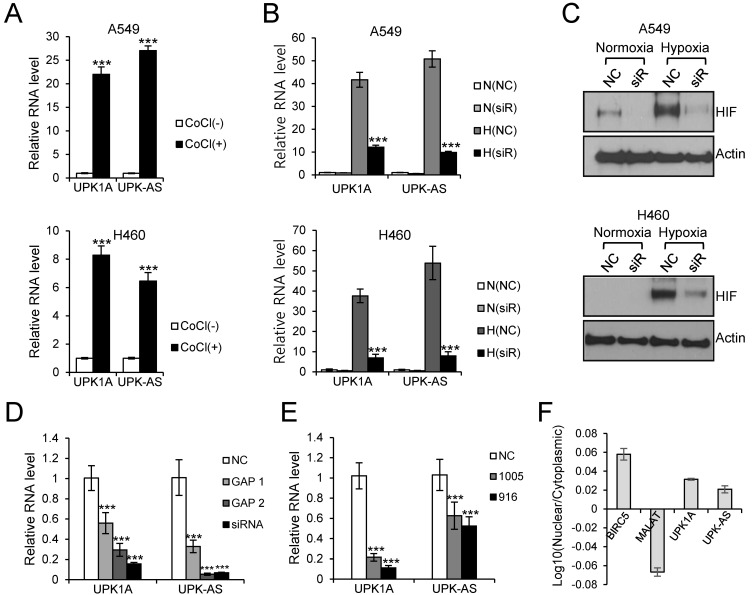
To examine whether HIF-1α was involved in hypoxia-induced expression of UPK1A and UPK1A-AS1, A549 and NCI-H460 cells were treated with the hypoxia mimetic CoCl2, which stabilizes and upregulates HIF-1α, for 24 h. As shown in Fig. 3A, both UPK1A and UPK1A-AS1 were upregulated after cells were treated with CoCl2. To further demonstrate HIF-1α-mediated upregulation of UPK1A and UPK1A-AS1 under hypoxic conditions, cells were transfected with siRNA targeting HIF-1α (siRNA-2210), and RNA levels were measured by qRT-PCR. As shown in Fig. 3B, hypoxia-induced upregulation of UPK1A and UPK1A-AS1 was greatly decreased following treatment with siRNA-2210, indicating a key role for HIF-1α in upregulation of UPK1A and UPK1A-AS1 under hypoxic conditions. The efficient knockdown of HIF-1α by siRNA-2210 under hypoxic conditions, as determined by western blot analysis, is shown in Fig. 3C.
Fig. 3. Involvement of HIF-1α in the upregulation of UPK1A and UPK1A-AS1 under hypoxic conditions.
(A) A549 and NCI-H460 cells were treated with CoCl2 (200 μM) for 24 h, and total RNA was subjected to qRT-PCR. The RNA level under normoxic conditions was set as 1. CoCl, CoCl2. ***P < 0.001. (B) A549 and NCI-H460 cells were transfected with NC or siRNA-2210 (20 nM). The next day, cells were incubated under normoxic or hypoxic conditions. After 24 h, total RNA was isolated and subjected to qRT-PCR. The RNA level from NC-transfected cells cultured under normoxic conditions was set as 1. N, normoxia; H, hypoxia; siR, siRNA-2210. ***P < 0.001. (C) Western blot analysis of HIF-1α in lysates isolated from NC- or siRNA-2210-transfected cells. After transfection and cultivation under normoxic or hypoxic conditions, as stated in (B), total protein was subjected to western blotting. Actin was used as a loading control. siR, siRNA-2210. (D) NCI-H460 cells were transfected with NC, GapmeR 1 UPK1A-AS1, GapmeR 2 UPK1A-AS1, or siRNA targeting UPK1A-AS1. After 24 h of incubation under hypoxic conditions, total RNA was isolated, and the expression levels of UPK1A and UPK1A-AS1 were analyzed by qRT-PCR. ***P < 0.001. (E) NCI-H460 cells were transfected with NC, UPK1A siRNA-916, or UPK1A siRNA-1005. qRT-PCR was performed using total RNA after incubation for 24 h under hypoxic conditions. ***P < 0.001. (F) NCI-H460 cells were cultured under hypoxic conditions for 24 h. Nuclear and cytoplasmic RNAs were isolated. BIRC5 and MALAT1 served as cytoplasmic and nuclear controls, respectively. UPK-AS, UPK1A-AS1; MALAT, MALAT1.
Knockdown of UPK1A or UPK1A-AS1 downregulated both UPK1A and UPK1A-AS1
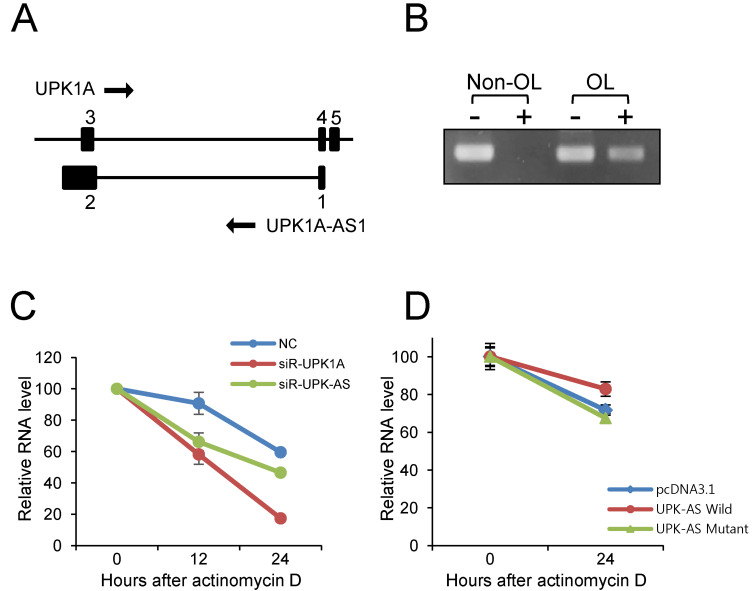
To investigate the interactions among UPK1A and UPK1A-AS1 expression, NCI-H460 cells were transfected with siRNA targeting UPK1A or UPK1A-AS1. Notably, knockdown of UPK1A-AS1 under hypoxic conditions resulted in downregulation of both UPK1A-AS1 and UPK1A (Fig. 3D). Moreover, knockdown of UPK1A resulted in downregulation of both UPK1A and UPK1A-AS1 (Fig. 3E). Thus, the expression levels of UPK1A and UPK1A-AS1 were mutually regulated under hypoxic conditions. One possible explanation for this mutual dependence of UPK1A and UPK1A-AS1 expression is that UPK1A and UPK1A-AS1 may form a duplex, resulting in increased stability of UPK1A and UPK1A-AS1. Indeed, we found that both UPK1A and UPK1A-AS1 were present in the cytoplasm under hypoxic conditions (Fig. 3F), and we identified a sequence of approximately 260 bp that was complementary between UPK1A and UPK1A-AS1, supporting the possibility of duplex formation (Fig. 4A).
Fig. 4. UPK1A formed a duplex with UPK1A-AS1.
(A) Schematic diagram of UPK1A and UPK1A-AS1 genes. The number above the black box represents the exon number. (B) Duplex formation between UPK1A and UPK1A-AS1. NCI-H460 cells were cultured under hypoxic conditions for 24 h. Total RNA was isolated and treated (+) or not (–) with RNase. Nonoverlapping (Non-OL) and overlapping (OL) sequences were analyzed by qRT-PCR. (C) NCI-H460 cells were treated with actinomycin D (4 μg/ml) and transfected with 20 nM NC, UPK1A siRNA-916, or UPK1A-AS1 siRNA. After cultivation under hypoxic conditions for 12 or 24 h, total RNA was isolated and subjected to qRT-PCR. (D) NCI-H460 cells were transfected with pcDNA3.1, pcDNA3.1-UPK1A-AS1-Wild, and pcDNA-UPK1A-AS1-Mutant plasmids. At 0 h and 24 h after treatment of actinomycin D, RNA was isolated and subjected to qRT-PCR. Relative levels of UPK1A mRNA are shown.
RNase protection assays revealed duplex formation between UPK1A and UPK1A-AS1
Next, to investigate the potential of duplex formation between UPK1A and UPK1A-AS1, we performed RNase protection assays using total RNA isolated from NCI-H460 cells incubated under hypoxic conditions. As shown in Fig. 4B, after digestion with RNase, no PCR product was generated for the nonoverlapping parts, whereas a PCR product was generated for the overlapping parts. These results indicated that UPK1A and UPK1A-AS1 formed duplexes in a complementary manner.
siRNA against UPK1A-AS1 negatively affected the RNA stability of UPK1A
To investigate whether UPK1A-AS1 affected the stability of UPK1A mRNA, the RNA remaining at 12 h and 24 h after treatment with actinomycin D was quantified by qRT-PCR. As shown in Fig. 4C, there was a greater decrease in the amount UPK1A mRNA after treatment with siRNA against UPK1A-AS1, as compared with that after treatment with NC. Thus, we expected that UPK1A-AS1 affected the stability of UPK1A mRNA, which could be attributed to duplex formation between UPK1A and UPK1A-AS1, thereby contributing to increased stability. To examine whether the overlapping region of UPK1A-AS1 is important in modulating the stability of UPK1A, we constructed two plasmids, pcDNA3.1-UPK1A-AS1-Wild and pcDNA-UPK1A-AS1-Mutant which express a full-length UPK1A-AS1 transcript and a shorter UPK1A-AS1 transcript, respectively, in which the overlapping region was deleted. The plasmids were transfected into NCI-H460 cells together with pcDNA3.1 plasmid as a control. Transfected cells were treated with actinomycin D for 24 h. As shown in Fig. 4D, transfection of pcDNA3.1-UPK1A-AS1-Wild resulted in an increase in the UPK1A mRNA level compared with the control vector pcDNA3.1. However, UPK1A mRNA level was not increased when transfected with the pcDNA-UPK1A-AS1-Mutant plasmid. Thus, the results indicated that the overlapping region of UPK1A-AS1 is important for the stability of the UPK1A mRNA.
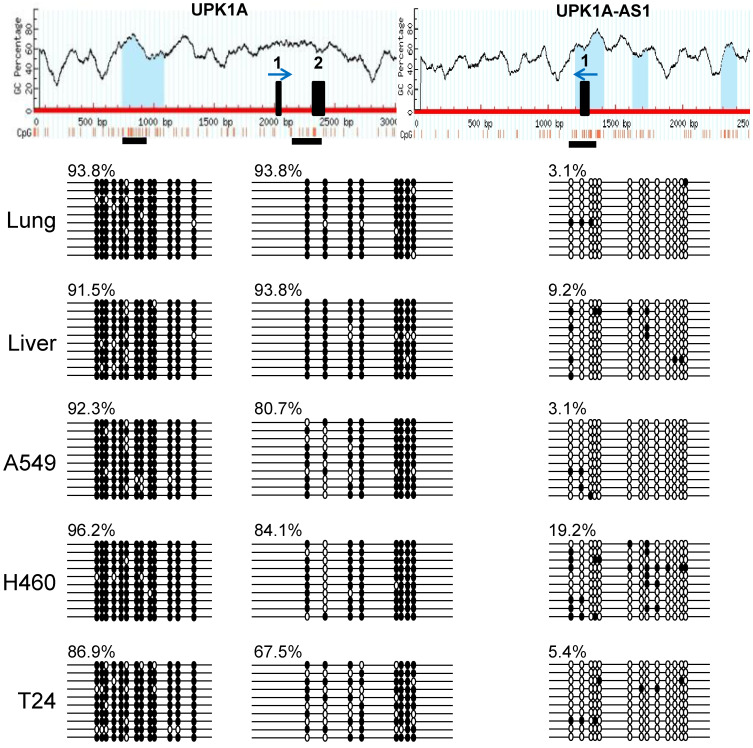
Methylation of UPK1A and UPK1A-AS1
Because antisense lncRNAs often play important roles in the expression of their sense counterparts via epigenetic regulation, we examined the methylation statuses of UPK1A and UPK1A-AS1 in several cell lines and human tissues. As shown in Fig. 5, a CpG island located upstream of exon 1 in UPK1A was heavily methylated in A549, NCI-H460, and T24 cells and in lung and liver tissues. In a region containing the ATG initiation codon of UPK1A, a high degree of methylation was also observed in all examined cell lines and tissues, although slightly lower levels of methylation were detected in the three cell lines. In contrast, a region comprising exon 1 of UPK1A-AS1 was not methylated in all cell lines and tissues examined. These results suggested that heavy methylation of UPK1A in several cell lines and tissues could create a closed chromatin structure in the UPK1A promoter region, which could be unfavorable for expression. In contrast, the lack of UPK1A-AS1 methylation could create a chromatin structure that was favorable for expression. Notably, methylation of the UPK1A promoter region was found in the bladder, where UPK1A was expressed (data not shown). UPK1A was expressed only in umbrella cells in the upper-most layer of the epithelium and not in most bladder cells in the lower layers; thus, the proportion of umbrella cells was relatively low in bladder samples, resulting in overall high levels of methylation.
Fig. 5. Methylation analysis of UPK1A and UPK1A-AS1.
Sodium bisulfite-modified genomic DNA was amplified by PCR. The resulting fragments were cloned, and 10 to 20 independent clones were sequenced. The top panel shows the MethPrimer results, including the CpG percentage, CpG distribution, and location of CpG islands (blue regions). The horizontal black boxes (two for UPK1A and one for UPK1A-AS1) below the map represent the regions in which the methylation status of CpG dinucleotides was determined by sequencing. The vertical black boxes on the map show exons 1 and 2 for UPK1A and exon 1 for UPK1A-AS1. The blue arrows above the black boxes indicate the direction of transcription. The methylation status of the CpG dinucleotide is indicated by filled (methylated) and open (not methylated) circles. Each line represents the methylation status of an individual clone. The degree of methylation is indicated by a percentage.
DISCUSSION
The expression of UPK1A is highly specific to urothelial cells, although low expression levels have been observed in several other tissues, including the cervix and esophagus (Hall et al., 2005; Genotype-Tissue Expression Portal [https://www.gtexportal.org]). Moreover, the expression of UPK1A-AS1 is low in bladder tissues and most other tissues, suggesting that UPK1A-AS1 may not play important roles in the expression of UPK1A under normoxic conditions. However, under hypoxic conditions, both UPK1A-AS1 and UPK1A showed increased expression in a HIF-1α-dependent manner, as revealed by RNA interference (RNAi) experiments. Several studies have shown that lncRNA expression levels are upregulated under hypoxic conditions in a HIF-1α-dependent manner (Lauer et al., 2020; Li et al., 2018; Xue et al., 2014; Zhou et al., 2015). In such cases, HIF-1α increases expression under hypoxia by binding to HREs (5′-RCGTG-3′) present on the promoter or enhancer regions of lncRNAs. Because HIF-1α also acts on hypoxia-induced expression of UPK1A and UPK1A-AS1, the promoter region containing the HRE site was cloned into the pGL4.10 luciferase vector to investigate the possibility of HIF-1α binding directly to the promoter of UPK1A and UPK1A-AS1 to modulate expression under hypoxic conditions. For UPK1A, a 2619-bp region containing four HREs was cloned, and for UPK1A-AS1, a 1212-bp region containing two HREs was cloned. In the case of UPK1A, four HREs are located 1001, 1220, 1513, and 2011 nt upstream of the first nucleotide of the UPK1A transcript. For UPK1A-AS1, two HREs are located 5 and 283 nt downstream of the first nucleotide of the UPK1A-AS1 transcript. In the case of UPK1A-AS1, several Alu sequences were continuously distributed outside the 900 bp containing exon 1; thus, the promoter region may have a small size of approximately 900 bp. When luciferase activity was measured after transfecting A549 cells with the pGL4.10 luciferase vector harboring the cloned promoter region, the results showed no significant increase under hypoxic conditions compared with that under normoxic conditions. Therefore, it may be necessary to repeat these assays after cloning a larger promoter region. Moreover, additional experiments may be required to examine the possibilities that (1) UPK1A and UPK1A-AS1 may be regulated by a single HIF-1α molecule rather than regulated independently by HIF-1α under hypoxic conditions and (2) UPK1A-AS1 molecule may be involved in the transcription of UPK1A.
LncRNAs regulate the expression of many genes at the transcriptional and post-transcriptional levels (Bach and Lee, 2018). One of the mechanisms involved in regulation at the post-transcriptional level is the formation of a duplex between antisense lncRNA and the neighboring sense gene RNA to regulate RNA stability (Huang et al., 2016; Kimura et al., 2013; Sun et al., 2016; Zhang et al., 2019). For example, the lncRNA MUC5B-AS1 forms a duplex with the sense transcript MUC5B, and the two RNAs then mutually regulate their expression levels (Yuan et al., 2018). Our results showed that UPK1A and UPK1A-AS1 also formed a duplex based on complementary sequences. When cells were transfected with UPK1A-AS1 siRNA, the stability of UPK1A mRNA was reduced, suggesting that the formation of a duplex between UPK1A and UPK1A-AS1 could affect the stability of UPK1A mRNA. It is notable that UPK1A-AS1 contains approximately 100 nt Alu J element at the 3′ end, potentially allowing UPK1A-AS1 to form a duplex with mRNAs and lncRNAs, which have an Alu sequence in an inverted position in their transcript. It is known that the Alu element plays a role in subcellular localization and alternative splicing (Hu et al., 2016; Lubelsky and Ulitsky, 2018). Transfection with an expression plasmid in which the Alu J sequence is deleted from UPK1A-AS1 and RNA pulldown experiments would be necessary in future studies to identify biological functions of the Alu J element of UPK1A-AS1.
DNA methylation regulates gene expression by affecting the chromatin structure (Jones and Takai, 2001; Klose and Bird, 2006). In this study, we found that the promoter region of UPK1A was methylated, whereas the promoter region of UPK1A-AS1 was not methylated in several cell and tissue types. Bladder tissues showed methylation of the promoter region of UPK1A; however, the cells that actually expressed UPK1A were umbrella cells. Therefore, it will be necessary to isolate only umbrella cells and investigate the methylation patterns in these cells. Moreover, UPK1A expression was higher under hypoxic conditions, although the promoter region of UPK1A was methylated, which would be expected to suppress expression. When the methylation pattern was investigated after incubation for 48 h under hypoxic conditions, the results showed that demethylation of the UPK1A promoter did not occur. Therefore, further studies are needed to evaluate the methylation patterns of the promoter region of UPK1A after incubation for a longer period under hypoxic conditions. In addition, it is noteworthy that there are increasing number of examples showing that DNA hypermethylation correlates with gene activation (Chatterjee et al., 2017; Guillaumet-Adkins et al., 2014; Smith et al., 2020). For instance, there is a correlation between DNA hypermethylation and increased expression of survivin in endometrial tumors, as revealed by methylation-specific PCR and pyrosequencing (Nabilsi et al., 2009). Methylation has been demonstrated to inhibit the binding of p53, a repressor of survivin expression. Another study showed that methylation of the promoter region of hTERT gene was positively correlated with gene expression (Guilleret et al., 2002; Lee et al., 2019). One possible mechanism of gene activation from the methylated promoter is the inhibition of the binding of a transcriptional repressor. Another possibility is the binding of a transcriptional enhancer to the methylated promoter. Interestingly, a consensus core motif (5′-A/GCGTG-3′) of the HIF-1α binding site contains a CG sequence which is the substrate for DNA methyltransferases. Electrophoretic mobility shift and chromatin immunoprecipitation assays may be necessary to examine the effects of DNA methylation on HIF-1α binding and to identify functional HRE for UPK1A and UPK1A-AS1 expression in lung cancer cells.
Several studies have indicated UPK1A to be a tumor suppressor which is downregulated in several tumor tissues compared with nontumorous tissues. Recently, it was reported that UPK1A-AS1 is downregulated in ESCC and overexpression of UPK1A-AS1 suppressed the proliferation, migration, and invasion of ESCC cells (Du et al., 2020). UPK1A-AS1 functions as a tumor suppressor by sponging miR-1248 in ESCC cells. Herein, we showed that UPK1A-AS1 forms a duplex with UPK1A, thereby increasing the stability of UPK1A; thus, UPK1A-AS1 acts as a tumor suppressor by stabilizing UPK1A. Therefore, low levels of UPK1A and UPK1A-AS1 in tumor cells may be an indication of cancer and overexpression of UPK1A-AS1 may have potential in cancer therapy. Our study also showed that UPK1A and UPK1A-AS1 are upregulated under hypoxic conditions. As UPK1A-AS1 was the most upregulated gold standard lncRNA in both A549 and NCI-H460 cells, it has the potential to serve as a marker for hypoxia. Although tumor hypoxia provides a survival advantage to tumor cells by promoting angiogenesis, glycolysis, proliferation, and metastasis, the reason tumor suppressors UPK1A and UPK1A-AS1 are upregulated under hypoxia remains unclear. Recently, the existence of a positive feedback loop was reported between UPK1A and HIF-1α: Downregulation of UPK1A was found to decrease the expression levels of HIF-1α and the downstream target genes in hepatocellular carcinoma (Song et al., 2020). Thus, it is likely that the upregulation of UPK1A in hypoxia increases the expression of HIF-1α and the downstream target genes, which results in the promotion of glycolysis and proliferation under hypoxia in hepatocellular carcinoma.
Overall, in this study, we found that UPK1A and UPK1A-AS1 expression levels were induced under hypoxic conditions in lung and bladder carcinoma cells. The hypoxia-induced expression of UPK1A and UPK1A-AS1 was mediated by HIF-1α, as revealed by RNAi experiments. Moreover, we found that UPK1A and UPK1A-AS1 expression levels were mutually dependent and that duplex formation between UPK1A and UPK1A-AS1 may be one possible mechanism responsible for this interdependence. Further studies of hypoxia-induced UPK1A protein may provide a basis for understanding the role of UPK1A in cancer growth and metastasis under hypoxic conditions.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (grant No. 2015R1D1A1A01057433).
Footnotes
AUTHOR CONTRIBUTIONS
Y.C.C., J.Y., and K.B. conceived and designed the study. Y.B. and Y.C.C. performed the experiments. Y.J. analyzed the data. Y.C.C. and K.B. wrote and edited the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Autuoro J.M., Pirnie S.P., Carmichael G.G. Long noncoding RNAs in imprinting and X chromosome inactivation. Biomolecules. 2014;4:76–100. doi: 10.3390/biom4010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach D.H., Lee S.K. Long noncoding RNAs in cancer cells. Cancer Lett. 2018;419:152–166. doi: 10.1016/j.canlet.2018.01.053. [DOI] [PubMed] [Google Scholar]
- Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.N., Zhang K., Hu Z.M., Qi H.X., Shi Z.M., Han X.H., Han Y.W., Hong W. Hypoxia-regulated lncRNAs in cancer. Gene. 2016;575:1–8. doi: 10.1016/j.gene.2015.08.049. [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Stockwell P.A., Ahn A., Rodger E.J., Leichter A.L., Eccles M.R. Genome-wide methylation sequencing of paired primary and metastatic cell lines identifies common DNA methylation changes and a role for EBF3 as a candidate epigenetic driver of melanoma metastasis. Oncotarget. 2017;8:6085–6101. doi: 10.18632/oncotarget.14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F., Guo T., Cao C. Restoration of UPK1A-AS1 expression suppresses cell proliferation, migration, and invasion in esophageal squamous cell carcinoma cells partially by sponging microRNA-1248. Cancer Manag. Res. 2020;12:2653–2662. doi: 10.2147/CMAR.S239418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvidge G.P., Glenny L., Appelhoff R.J., Ratcliffe P.J., Ragoussis J., Gleadle J.M. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J. Biol. Chem. 2006;281:15215–15226. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent G., 3rd, Kenny P.J., Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler J., Breckwoldt K., Remmele C.W., Hartmann D., Dittrich M., Pfanne A., Just A., Xiao K., Kunz M., Müller T., et al. Development of long noncoding RNA-based strategies to modulate tissue vascularization. J. Am. Coll. Cardiol. 2015;66:2005–2015. doi: 10.1016/j.jacc.2015.07.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Maldonado L., Tiana M., Roche O., Prado-Cabrero A., Jensen L., Fernandez-Barral A., Guijarro-Muñoz I., Favaro E., Moreno-Bueno G., Sanz L., et al. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene. 2015;34:2609–2620. doi: 10.1038/onc.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumet-Adkins A., Richter J., Odero M.D., Sandoval J., Agirre X., Catala A., Esteller M., Prósper F., Calasanz M.J., Buño I., et al. Hypermethylation of the alternative AWT1 promoter in hematological malignancies is a highly specific marker for acute myeloid leukemias despite high expression levels. J. Hematol. Oncol. 2014;7:4. doi: 10.1186/1756-8722-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilleret I., Yan P., Grange F., Braunschweig R., Bosman F.T., Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int. J. Cancer. 2002;101:335–341. doi: 10.1002/ijc.10593. [DOI] [PubMed] [Google Scholar]
- Hall G.D., Weeks R.J., Olsburgh J., Southgate J., Knowles M.A., Selby P.J., Chester J.D. Transcriptional control of the human urothelial-specific gene, uroplakin Ia. Biochim. Biophys. Acta. 2005;1729:126–134. doi: 10.1016/j.bbaexp.2005.04.004. [DOI] [PubMed] [Google Scholar]
- He Y., Kong F., Du H., Wu M. Decreased expression of uroplakin Ia is associated with colorectal cancer progression and poor survival of patients. Int. J. Clin. Exp. Pathol. 2014;7:5031–5037. [PMC free article] [PubMed] [Google Scholar]
- Hong S.S., Lee H., Kim K.W. HIF-1alpha: a valid therapeutic target for tumor therapy. Cancer Res. Treat. 2004;36:343–353. doi: 10.4143/crt.2004.36.6.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Wang X., Shan G. Insertion of an Alu element in a lncRNA leads to primate-specific modulation of alternative splicing. Nat. Struct. Mol. Biol. 2016;23:1011–1019. doi: 10.1038/nsmb.3302. [DOI] [PubMed] [Google Scholar]
- Hu Y., Liu J., Huang H. Recent agents targeting HIF-1α for cancer therapy. J. Cell. Biochem. 2013;114:498–509. doi: 10.1002/jcb.24390. [DOI] [PubMed] [Google Scholar]
- Huang B., Song J.H., Cheng Y., Abraham J.M., Ibrahim S., Sun Z., Ke X., Meltzer S.J. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927–4936. doi: 10.1038/onc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S., et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B.H., Rue E., Wang G.L., Roe R., Semenza G.L. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kanduri C. Long noncoding RNAs: lessons from genomic imprinting. Biochim. Biophys. Acta. 2016;1859:102–111. doi: 10.1016/j.bbagrm.2015.05.006. [DOI] [PubMed] [Google Scholar]
- Ke Q., Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol. Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- Kim S., Lee U.J., Kim M.N., Lee E.J., Kim J.Y., Lee M.Y., Choung S., Kim Y.J., Choi Y.C. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J. Biol. Chem. 2008;283:18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- Kimura T., Jiang S., Nishizawa M., Yoshigai E., Hashimoto I., Nishikawa M., Okumura T., Yamada H. Stabilization of human interferon-α1 mRNA by its antisense RNA. Cell. Mol. Life Sci. 2013;70:1451–1467. doi: 10.1007/s00018-012-1216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Bird A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Kong K.L., Kwong D.L., Fu L., Chan T.H., Chen L., Liu H., Li Y., Zhu Y.H., Bi J., Qin Y.R., et al. Characterization of a candidate tumor suppressor gene uroplakin 1A in esophageal squamous cell carcinoma. Cancer Res. 2010;70:8832–8841. doi: 10.1158/0008-5472.CAN-10-0779. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R., Ferracin M., Wojcik S.E., Garzon R., Alder H., Agosto-Perez F.J., Davuluri R., Liu C.G., Croce C.M., Negrini M., et al. A microRNA signature of hypoxia. Mol. Cell. Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer V., Grampp S., Platt J., Lafleur V., Lombardi O., Choudhry H., Kranz F., Hartmann A., Wullich B., Yamamoto A., et al. Hypoxia drives glucose transporter 3 expression through hypoxia-inducible transcription factor (HIF)-mediated induction of the long noncoding RNA NICI. J. Biol. Chem. 2020;295:4065–4078. doi: 10.1074/jbc.RA119.009827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.D., Leão R., Komosa M., Gallo M., Zhang C.H., Lipman T., Remke M., Heidari A., Nunes N.M., Apolónio J.D., et al. DNA hypermethylation within TERT promoter upregulates TERT expression in cancer. J. Clin. Invest. 2019;129:223–229. doi: 10.1172/JCI128527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli A., Nolan K.A., Santambrogio S., Gonçalves A.F., Schönenberger M.J., Guinot A., Frew I.J., Marti H.H., Hoogewijs D., Wenger R.H. Induction of long noncoding RNA MALAT1 in hypoxic mice. Hypoxia (Auckl.) 2015;3:45–52. doi: 10.2147/HP.S90555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Xiao Y., Huang T. HIF-1α-induced upregulation of lncRNA UCA1 promotes cell growth in osteosarcoma by inactivating the PTEN/AKT signaling pathway. Oncol. Rep. 2018;39:1072–1080. doi: 10.3892/or.2018.6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Zhang X., Xue C., Zhang H., Shashaty M.G., Gosai S.J., Meyer N., Grazioli A., Hinkle C., Caughey J. The long noncoding RNA landscape in hypoxic and inflammatory renal epithelial injury. Am. J. Physiol. Renal Physiol. 2015;309:F901–F913. doi: 10.1152/ajprenal.00290.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y., Ulitsky I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018;555:107–111. doi: 10.1038/nature25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P.H., Wiesener M.S., Chang G.W., Clifford S.C., Vaux E.C., Cockman M.E., Wykoff C.C., Pugh C.W., Maher E.R., Ratcliffe P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mimura I., Hirakawa Y., Kanki Y., Kushida N., Nakaki R., Suzuki Y., Tanaka T., Aburatani H., Nangaku M. Novel lnc RNA regulated by HIF-1 inhibits apoptotic cell death in the renal tubular epithelial cells under hypoxia. Physiol. Rep. 2017;5:e13203. doi: 10.14814/phy2.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabilsi N.H., Broaddus R.R., Loose D.S. DNA methylation inhibits p53-mediated survivin repression. Oncogene. 2009;28:2046–2050. doi: 10.1038/onc.2009.62. [DOI] [PubMed] [Google Scholar]
- Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Pollex T., Heard E. Recent advances in X-chromosome inactivation research. Curr. Opin. Cell Biol. 2012;24:825–832. doi: 10.1016/j.ceb.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Scheuermann J.C., Boyer L.A. Getting to the heart of the matter: long non-coding RNAs in cardiac development and disease. EMBO J. 2013;32:1805–1816. doi: 10.1038/emboj.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonrock N., Harvey R.P., Mattick J.S. Long noncoding RNAs in cardiac development and pathophysiology. Circ. Res. 2012;111:1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- Semenza G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Shih J.W., Kung H.J. Long non-coding RNA and tumor hypoxia: new players ushered toward an old arena. J. Biomed. Sci. 2017;24:53. doi: 10.1186/s12929-017-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., Sen S., Weeks R.J., Eccles M.R., Chatterjee A. Promoter DNA hypermethylation and paradoxical gene activation. Trends Cancer. 2020;6:392–406. doi: 10.1016/j.trecan.2020.02.007. [DOI] [PubMed] [Google Scholar]
- Song Y., Wang H., Zou X.J., Zhang Y.X., Guo Z.Q., Liu L., Wu D.H., Zhang D.Y. Reciprocal regulation of HIF-1α and Uroplakin 1A promotes glycolysis and proliferation in Hepatocellular Carcinoma. J. Cancer. 2020;11:6737–6747. doi: 10.7150/jca.48132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Xu M., Chen X., Chen N., Gong J., Nie L., Li L., Li X., Zhang M., Zhou Q. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol. Cancer. 2017;16:142. doi: 10.1186/s12943-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Wang X., Fu C., Wang X., Zou J., Hua H., Bi Z. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol. Biol. Rep. 2016;43:427–436. doi: 10.1007/s11033-016-3975-1. [DOI] [PubMed] [Google Scholar]
- Tang Y., Cheung B.B., Atmadibrata B., Marshall G.M., Dinger M.E., Liu P.Y., Liu T. The regulatory role of long noncoding RNAs in cancer. Cancer Lett. 2017;391:12–19. doi: 10.1016/j.canlet.2017.01.010. [DOI] [PubMed] [Google Scholar]
- Tee A.E., Liu B., Song R., Li J., Pasquier E., Cheung B.B., Jiang C., Marshall G.M., Haber M., Norris M.D., et al. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget. 2016;7:8663–8675. doi: 10.18632/oncotarget.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellenkle C., Garcia-Manteiga J.M., Pedrotti S., Perfetti A., De Toma I., Da Silva D., Maimone B., Greco S., Fasanaro P., Creo P., et al. Implication of long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Sci. Rep. 2016;6:24141. doi: 10.1038/srep24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu X., Zhang H., Sun L., Zhou Y., Jin H., Zhang H., Zhang H., Liu J., Guo H., et al. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting γ-synuclein. Neoplasia. 2014;16:1094–1106. doi: 10.1016/j.neo.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.R., Kong X.P., Pellicer A., Kreibich G., Sun T.T. Uroplakins in urothelial biology, function, and disease. Kidney Int. 2009;75:1153–1165. doi: 10.1038/ki.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Li X., Li Z., Chen W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biol. 2014;35:6901–6912. doi: 10.1007/s13277-014-1925-x. [DOI] [PubMed] [Google Scholar]
- Yap K.L., Li S., Muñoz-Cabello A.M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M.J., Zhou M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Tang B., Sun X. Development of inhibitors targeting hypoxia-inducible factor 1 and 2 for cancer therapy. Yonsei Med. J. 2017;58:489–496. doi: 10.3349/ymj.2017.58.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Gius D., Onyango P., Muldoon-Jacobs K., Karp J., Feinberg A.P., Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Liu Q., Hu Z., Zhou Z., Wang G., Li C., Xie W., Meng G., Xiang Y., Wu N., et al. Long non-coding RNA MUC5B-AS1 promotes metastasis through mutually regulating MUC5B expression in lung adenocarcinoma. Cell Death Dis. 2018;9:450. doi: 10.1038/s41419-018-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Dong Q., Zhu H., Li S., Shi L., Chen X. Long non-coding antisense RNA GAS6-AS1 supports gastric cancer progression via increasing GAS6 expression. Gene. 2019;696:1–9. doi: 10.1016/j.gene.2018.12.079. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wang D.D., Wang W., Pan K., Huang C.Y., Li Y.F., Wang Q.J., Yuan S.Q., Jiang S.S., Qiu H.B., et al. Reduced expression of uroplakin 1A is associated with the poor prognosis of gastric adenocarcinoma patients. PLoS One. 2014;9:e93073. doi: 10.1371/journal.pone.0093073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Ye L., Jiang C., Bai J., Chi Y., Zhang H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1α activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol. 2015;36:9179–9188. doi: 10.1007/s13277-015-3453-8. [DOI] [PubMed] [Google Scholar]
- Zhu G., Wang S., Chen J., Wang Z., Liang X., Wang X., Jiang J., Lang J., Li L. Long noncoding RNA HAS2-AS1 mediates hypoxia-induced invasiveness of oral squamous cell carcinoma. Mol. Carcinog. 2017;56:2210–2222. doi: 10.1002/mc.22674. [DOI] [PubMed] [Google Scholar]
- Zhu H., Tang Y., Zhang X., Jiang X., Wang Y., Gan Y., Yang J. Downregulation of UPK1A suppresses proliferation and enhances apoptosis of bladder transitional cell carcinoma cells. Med. Oncol. 2015;32:84. doi: 10.1007/s12032-015-0541-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.