Summary
Understanding the biological processes that determine the entry of three germ layers of human pluripotent stem cells (hPSCs) is a central question in developmental and stem cell biology. Here, we genetically engineered hPSCs with the germ layer reporter and inducible CRISPR/Cas9 knockout system, and a genome-scale screening was performed to define pathways restricting germ layer specification. Genes clustered in the key biological processes, including embryonic development, mRNA processing, metabolism, and epigenetic regulation, were centered in the governance of pluripotency and lineage development. Other than typical pluripotent transcription factors and signaling molecules, loss of function of mesendodermal specifiers resulted in advanced neuroectodermal differentiation, given their inter-germ layer antagonizing effect. Regarding the epigenetic superfamily, microRNAs enriched in hPSCs showed clear germ layer-targeting specificity. The cholesterol synthesis pathway maintained hPSCs via retardation of neuroectoderm specification. Thus, in this study, we identified a full landscape of genetic wiring and biological processes that control hPSC self-renewal and trilineage specification.
Subject areas: Biological Sciences, Cell Biology, Stem Cells Research, Developmental Biology
Graphical abstract
Highlights
-
•
Lineage reporter and CRISPR screening are powerful tools for studying cell fates
-
•
Lineage-specification preventing genes (LPGs) are identified in hPSCs
-
•
LPGs maintain pluripotency via targeting one or multiple germ layers
-
•
LPGs are clustered into distinct functional modules
Biological Sciences; Cell Biology; Stem Cells Research; Developmental Biology
Introduction
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), can self-renew and differentiate into all three germ layers (Thomson et al., 1998; Takahashi et al., 2007). Thus, hPSCs can be considered as highly advantageous cells for studying human development, modeling inherited disorders, and replenishing degenerated tissues and cells (Soldner and Jaenisch, 2018). Sophisticated protocols have been developed to target hPSCs into the ectoderm (Ec), mesoderm (Me), or endoderm (En) lineages, primarily via cocktailing developmentally related signaling cues. Activin/Nodal and fibroblast growth factor (FGF) signaling are two dominant pathways that maintain hPSCs in an undifferentiated state, and interference with the signaling molecules of these two pathways will lead to spontaneous differentiation (Vallier et al., 2005; Xu et al., 2005). Once leaving the culture conditions favors pluripotency maintenance, hPSCs adopt a neuroectoderm (nEc) fate in the absence of extra signaling activators, and dual inhibition of bone morphogenetic protein (BMP) and transforming growth factor β (TGF-β) signaling further strengthens this nEc fate (Chambers et al., 2009; Xu et al., 2005). On the other hand, activation of the BMP/Wnt/Activin/Nodal/FGF signaling pathways is major driving forces for generating primitive streak-like cells and Me or En lineage (D'Amour et al., 2005; Tam and Loebel, 2007; Zhang et al., 2008; Martyn et al., 2018; Wang and Chen, 2016).
Extracellular signaling pathways govern hPSC fates largely through their convergence on regulating fate-determining transcription factors (TFs) (Xu et al., 2008). OCT4 (POU5F1), SOX2, and NANOG are well-characterized core pluripotent genes that maintain hPSCs in pluripotency (Boyer et al., 2005). We previously showed that PAX6 is necessary and sufficient to generate a human nEc cell fate (Zhang et al., 2010; Chen et al., 2018). Both T (Brachury) and MIXL1 TFs are tightly associated with Me, whereas GATA4 and GATA6 are essential for the initiation of an En fate (Tam and Loebel, 2007; Faial et al., 2015).
Epigenetic machineries and metabolic states also offer proper genetic and cellular contexts to define specific cell fates (Wu et al., 2016; Xie et al., 2013; Gifford et al., 2013). Polycomb group, SWI/SNF, and MI-2/NURD proteins interact with pluripotent genes and play important roles in maintaining pluripotency (Zhang et al., 2014; Lee et al., 2006; Hu and Wade, 2012). Metabolites also have critical roles as substrates for epigenetic regulation, including acetylation and methylation (Moussaieff et al., 2015). While hPSCs are thought to depend primarily on glycolytic flux, several studies have suggested that the mitochondrial tricarboxylic acid (TCA) cycle and oxidative phosphorylation (OXPHOS) are active in hPSCs, and hPSCs consume more oxygen than the nEc cells (Birket et al., 2011; Lees et al., 2018).
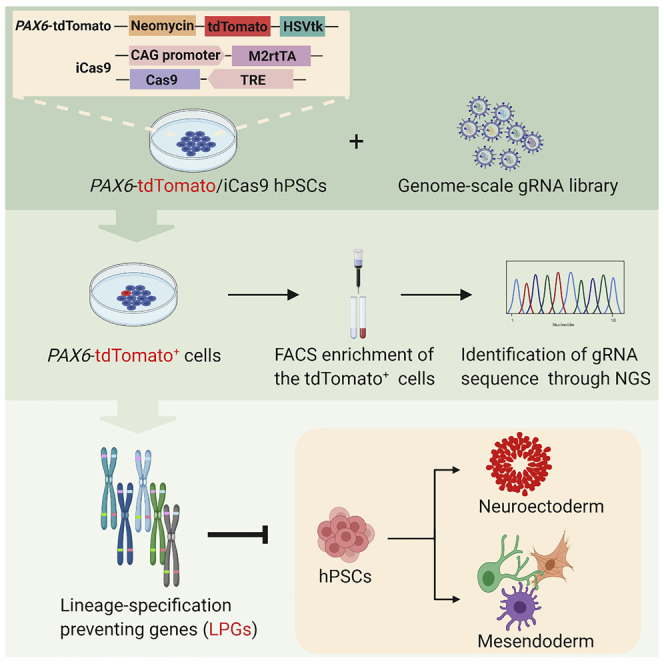
Though genetic, epigenetic, and metabolic processes are implicated in defining pluripotency, a systematic understanding of the transition from pluripotency to trilineage differentiation is still lacking (Li et al., 2019; Shparberg et al., 2019; Du et al., 2017). Due to advanced genome-wide screening technologies using either RNAi or CRISPR/Cas9 gRNA libraries, we now have abundant knowledge on essential genes (EGs) for survival and proliferation and pivotal pluripotent genes for self-renewal of hPSCs by monitoring growth behavior or using OCT4-GFP as a readout reporter (Yilmaz et al., 2018; Ihry et al., 2019; Chia et al., 2010; Mair et al., 2019). To date, genome-wide screening to elucidate the genetic wiring or biological processes underlying the commitment of trilineage from hPSCs is not yet to be performed. Here, we engineered hESCs with the PAX6-tdTomato reporter and inducible CRISPR/Cas9 knockout system. Genome-wide CRISPR loss-of-function screening was then conducted, and in-depth views of the genetic wiring and biological processes governing the dynamic transition from hPSCs to trilineage were provided.
Results
Genetic engineering of lineage reporter for high-throughput screening in hPSCs
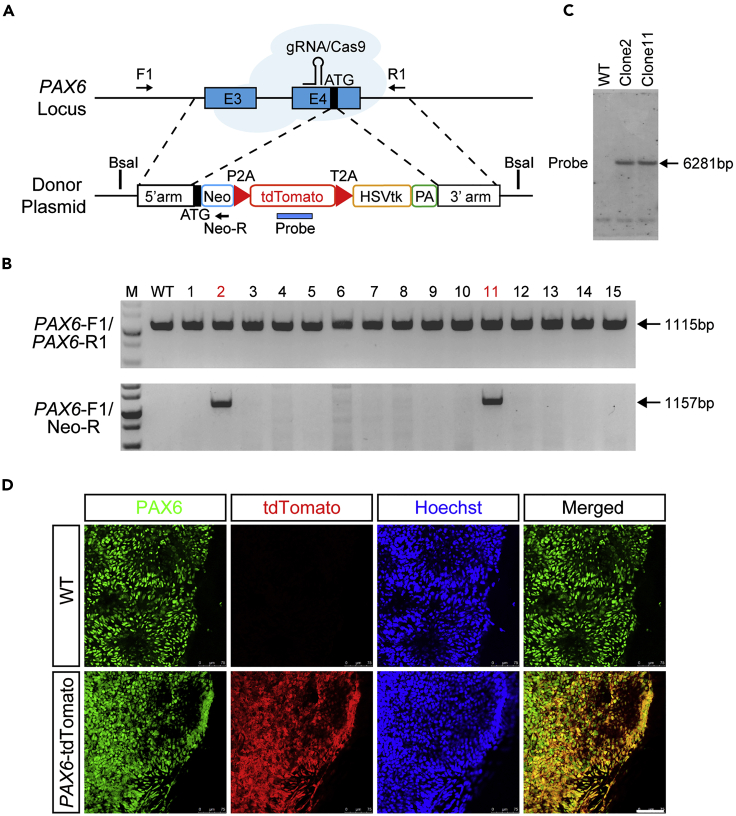
Within the three germ layers, nEc specification is independent of ectopic signaling pathways activation, referred to as a “default” cell fate induction from pluripotent stem cells (Munoz-Sanjuan and Brivanlou, 2002). We have revealed that the PAX6 TF is a human nEc cell fate determinant and thus can serve as a human nEc hallmark gene (Zhang et al., 2010; Chen et al., 2018). Moreover, nEc can be generated directly from hPSCs via overexpression of PAX6, even under strict hPSC culture conditions (Zhang et al., 2010). During early embryonic development, the expression of PAX6 is intricately regulated with cis-regulatory DNA machineries encompassing large genomic regions (Kleinjan et al., 2006; Tyas et al., 2006). We, therefore, engineered hESCs with a PAX6 reporter via CRISPR/Cas9-mediated homologous recombination (HR) to indicate PAX6 expression profiles and screen for intrinsic pathways that either drive hPSCs to three germ layer differentiations or to a nEc fate without changing culture conditions.
We first constructed a donor plasmid harboring an in-frame coding cassette of Neo-P2A-tdTomato-T2A-HSVtk flanked by 5′ AND-3′ homology arms of PAX6 (Figure 1A). A single guide RNA (gRNA) targeting exon 4 of PAX6 DNA near the ATG start codon was designed, and the cleavage efficacy was verified in HEK293 cells after co-transfection with Cas9 expression plasmids (Figure S1A). Sanger sequencing results showed that all 5 predicted loci which showed high similarity with the target sequences were identified intact with no mutations, suggesting no off-target effects of the designed gRNA (Figures S1B and S1C). After correctly HR, the putative integrated hESCs would express Neo-P2A-tdTomato-T2A-HSVtk protein under the tight control of endogenous PAX6 gene expression regulatory machineries. After protease cleavage, tdTomato mirrors an endogenous PAX6 expression and nEc fate, whereas neomycin (Neo) and HSVtk could serve as positive selection and negative depletion tools, respectively, when neomycin or ganciclovir are supplied to the cells. Genomic polymerase chain reaction (PCR) revealed that clones 2 and 11 were two monoallelic HR lines (Figures 1A and 1B). Southern blot analysis confirmed precise HR with no off-target recombination (Figure 1C). Both clone 2 and clone 11 retained typical hESC morphology and growth behavior compared with wild-type hESCs. After guiding the lines to a nEc fate with our standard differentiation protocol (Zhang and Zhang, 2010; Chen et al., 2018; Zhang et al., 2001; Li et al., 2009; Liu et al., 2019; Chi et al., 2016), both lines were normally specified to columnar neuroepithelial cells arranged in neural rosette-like structures, and cells started to express tdTomato from day 6 (Figure S1D). Immunostaining further revealed that tdTomato was co-labeled with endogenous PAX6 in the differentiated nEc cells (Figure 1D). Therefore, we successfully generated a nEc reporter line in hPSCs (referred to herein as PAX6-tdTomato reporter line), which can be used for lineage specification studies.
Figure 1.
Establishment and characterization of the PAX6-tdTomato reporter line
(A) Schematic view of the targeting strategy through gRNA-guided CRISPR/Cas9 system. The black box indicates the start codon (ATG) of the PAX6 gene. Arrows indicate primer sets used in genomic DNA PCR. Probe for Southern blot analysis is marked with blue bar at indicated genetic regions after homologous recombination (HR). E3, exon3; E4, exon4; PA, poly(A) signal; Neo, neomycin resistance gene.
(B) Genomic DNA PCR results of individual colonies after gene targeting. Primer sets of PAX6-F1 and PAX6-R1 were used for detecting the wild-type allele, and primer sets of PAX6-F1 and Neo-R were used for detecting the HR allele. Colonies #2 and #11 were monoallelically targeted.
(C) Southern blot analysis showed precise but no off-target recombination.
(D) PAX6-tdTomato hESCs were differentiated toward a neuroectoderm (nEc) fate. Immunostaining studies at day 10 showed that tdTomato was co-labeled with endogenous PAX6. Scale bar, 75 μm.
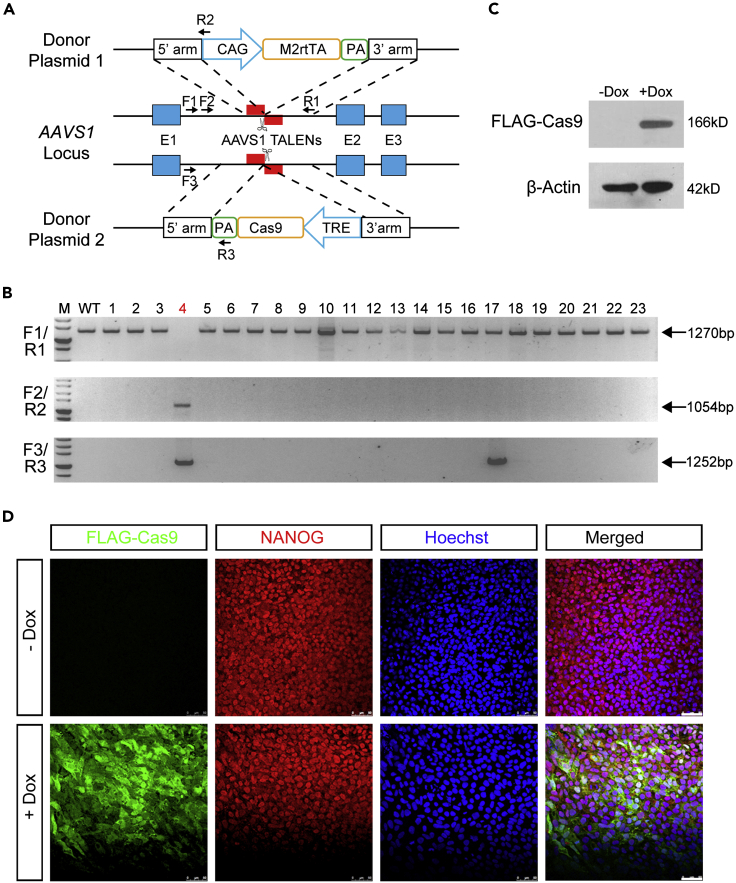
Construction of doxycycline-inducible Cas9 expression cassettes in PAX6-Tdtomato line
We modified the two-component inducible Cas9 (iCas9) system (Gonzalez et al., 2014) in the PAX6-tdTomato line. One donor plasmid containing a doxycycline-inducible Cas9 expression cassette (3×TRE-FLAG-Cas9) and another carrying a constitutive reverse tetracycline transactivator (M2rtTA) expression cassette (CAG-M2rtTA) were constructed (Figure 2A). Both donor plasmids were electroporated into the clone 11 PAX6-tdTomato reporter line together with left and right TALEN plasmids targeting AAVS1 (also known as PPP1R12C) loci. After genomic DNA PCR, we identified that clone 4 had both transgenes integrated in the AASV1 loci biallelically (Figure 2B). Western blotting and immunostaining confirmed the robust induction of Cas9 expression upon doxycycline treatment (Figures 2C and 2D). In addition, the constructed PAX6-tdTomato/iCas9 line showed typical hPSCs morphology and OCT4 and NANOG expression, as expected (Figures 2D and S2A). After doxycycline treatment, over 90% cells showed both NANOG and Cas9 staining, suggesting high induction efficiency (Figures S2B and S2C). To determine the capacity of the iCas9 hPSCs for DNA cleavage, we designed gRNA pairs targeting the human NF1 gene (Liu et al., 2016). The PAX6-tdTomato/iCas9 hESCs were then electroporated with the double gRNAs of NF1. After treatment with doxycycline, we observed ∼70% (5/7) knockout efficiency based on genomic DNA PCR (Figure S2D). We also designed a gRNA targeting P53 gene (Liu et al., 2016). The gRNA was packaged into lentivirus, which was then used to infect the PAX6-tdTomato/iCas9 cells. We observed prominent occurrence of indels surrounding the gRNA-guided cleavage site when doxycycline was supplied (Figure S2E). These results indicate that the iCas9 system constructed through HR at the genetically open AAVS1 loci is tight and functional and thus is suitable for large-scale CRISPR screening.
Figure 2.
Construction of inducible Cas9 expression systems in PAX6-tdTomato reporter hESCs
(A) Schematic diagram for constructing inducible Cas9 (iCas9) expression cassette in PAX6-tdTomato line through TALEN (marked in red bars)-mediated gene targeting at the AAVS1 loci. Genotyping PCR primer sets are labeled with arrows. E1, exon1; E2, exon2; E3, exon3; CAG, CMV enhancer/chicken β-actin promoter; M2rtTA, reverse tetracycline transactivator; TRE, tetracycline response element; PA, poly(A) signal.
(B) Genomic DNA PCR results showed colony #4 was biallelically targeted and had both CAG-M2rtTA and 3×TRE-Flag-Cas9 expression cassettes integrated at the AAVS1 loci. Primer sets of F1 and R1 were used for detecting the wild-type allele, primer sets of F2 and R2 were used for detecting the recombinated CAG-M2rtTA allele, and primer sets of F3 and R3 were used for detecting the recombinated 3×TRE-FLAG-Cas9 allele, respectively.
(C) Western blot analysis of Cas9 expression in PAX6-tdTomato/iCas9 hESCs treated with or without doxycycline.
(D) Immunostaining results of FLAG-Cas9 and NANOG expression in PAX6-tdTomato/iCas9 hESCs treated with or without doxycycline. Scale bar, 75 μm.
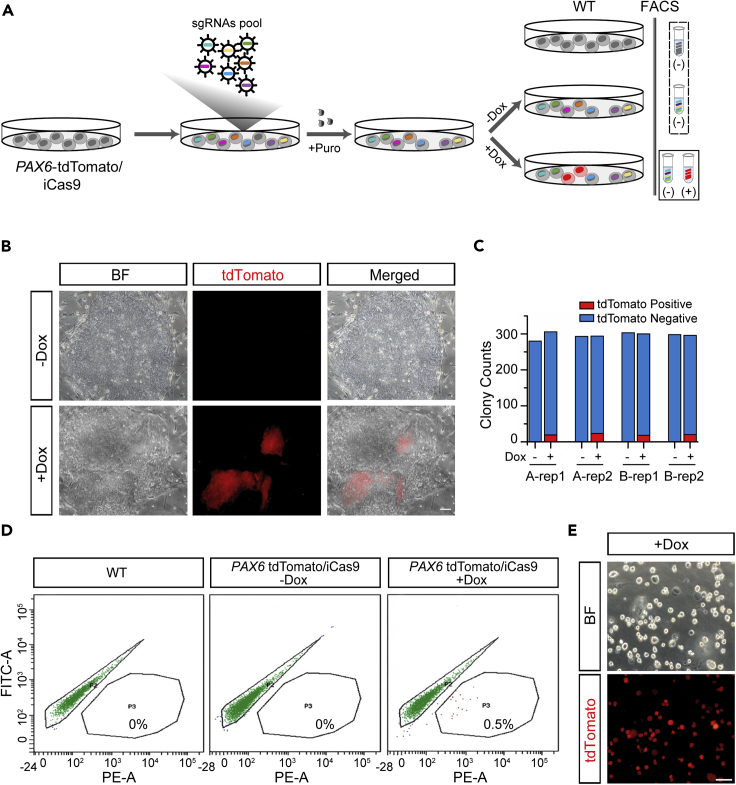
FACS-based forward genome-wide CRISPR screening
We hypothesized that genetic loss of function in the PAX6-tdTomato line which caused tdTomato expression would indicate the role of this gene in preventing either trilineage or nEc specification. Therefore, we performed genome-wide loss-of-function screening using the high-throughput CRISPR/Cas9 platform in the PAX6-tdTomato/iCas9 line (Figure 3A). Lentiviral libraries expressing gRNAs (GeCKO) targeting all coding genes and microRNAs were prepared (Shalem et al., 2014). To avoid spontaneous differentiation, the PAX6-tdTomato/iCas9 hESCs were digested into small clusters (not single cells) and infected with lentiviral libraries at a multiplicity of infection (MOI) of 0.3 to ensure high enrichment in the proportion of cells infected with only one viral particle and thus target a unique gene. As the gRNA lentiviral libraries had a puromycin drug-resistant gene, we removed non-infected cells by 4 days puromycin treatment. The PAX6-tdTomato/iCas9 hESCs infected with the gRNA lentiviral libraries were subsequently treated with doxycycline for another 5 days. Without doxycycline treatment, we did not see tdTomato-positive cells containing colonies, suggesting high quality hESC maintenance under strict culture conditions. Furthermore, doxycycline treatment repetitively yielded tdTomato-positive clusters within the hESC colonies at a frequency of 4%–5% (Figures 3B and 3C). Notably, the tdTomato-expressing cells showed columnar neuroepithelial morphology, forming clusters of similar size and containing distinct boarders with surrounding tdTomato-negative hPSCs (Figure 3B). These tdTomato-positive cells completely lost pluripotent marker OCT4 but were positive for SOX2, a known TF also for facilitating nEc fate decision (Wang et al., 2012) (Figure S3A). These results indicate that the tdTomato-expressed clusters were guided to a nEc fate and their uniform clonal appearance implied a single-cell origin likely caused by a loss-of-function mutation of a gene. Fluorescence-activated cell sorting (FACS) experiment showed that approximately 0.5% of entire hESC population was tdTomato positive after doxycycline treatment (Figure 3D). We then enriched the tdTomato-positive cells of interest using FACS and validated their tdTomato expression under the microscope immediately and their nEc characteristics by analyzing the expression of nEc marker genes including PAX6, SOX1, DLK1, MEIS2, and ZIC1 at the mRNA level (Figures 3E and S3B). After validation, the total genomic DNA of these tdTomato-positive cells was extracted, the fragments containing the integrated gRNA were amplified via PCR, and target gRNAs were profiled through sequencing.
Figure 3.
Genome-scale CRISPR screening in PAX6-tdTomato/iCas9 hESCs
(A) Schematic illustration of the genome-scale CRISPR screening strategy. PAX6-tdTomato/iCas9 hESCs were transduced with lentiviral particles of either A pool or B pool gRNA libraries at an MOI of 0.3. Puromycin was applied for 4 days to eliminate non-infected cells. Doxycycline treatment for another 5 days led to target gene loss of functions, and loss of function of germ layer specification preventing genes therefore caused lineage differentiation and aberrant expression of tdTomato under hPSC culture conditions. Wild-type (WT) cells and PAX6-tdTomato cells with no doxycycline treatment did not show tdTomato expression, and they served as negative controls. tdTomato-positive cells induced by doxycycline were purified by FACS. DNA isolation, two-step PCR amplification, and next-generation sequencing were subsequently used for elucidating gRNA enrichment in tdTomato-positive cells.
(B) Epithelial cell clusters expressing tdTomato within the hESCs colony appeared after doxycycline treatment. Scale bar, 100 μm.
(C) Counts of colonies comprising at least one tdTomato-positive cluster of a 6-well plate with or without doxycycline treatment.
(D) FACS of tdTomato-positive cells after doxycycline treatment.
(E) Fluorescent images showed that FACS-sorted cells had uniform tdTomato expression. Scale bar, 100 μm.
Recovered gRNAs are enriched in biological processes essential for hPSC maintenance and lineage specification
To evaluate the CRISPR/Cas9 screening performance, we first verified the integrality of the gRNA pools (A pool and B pool as designed) transduced in the PAX6-tdTomato/iCas9 hESCs without doxycycline treatment, with results showing more than 90% coverage (Figure S4A). The Gini index was then calculated to reflect disparities in the distribution of gRNAs. Both the A and B pool in the doxycycline-untreated cells showed a low Gini index value, thus representing highly uniform transduction with no bias (Figure S4B). In contrast, gRNAs retrieved from doxycycline-induced tdTomato-positive cells showed a high Gini index value, indicating target gene selectivity and enrichment (Figure S4B). Furthermore, high repeatability between independent screens reinforced the reliability of the forward screen system based on lineage reporter and inducible gene knockout (Figure S4C).
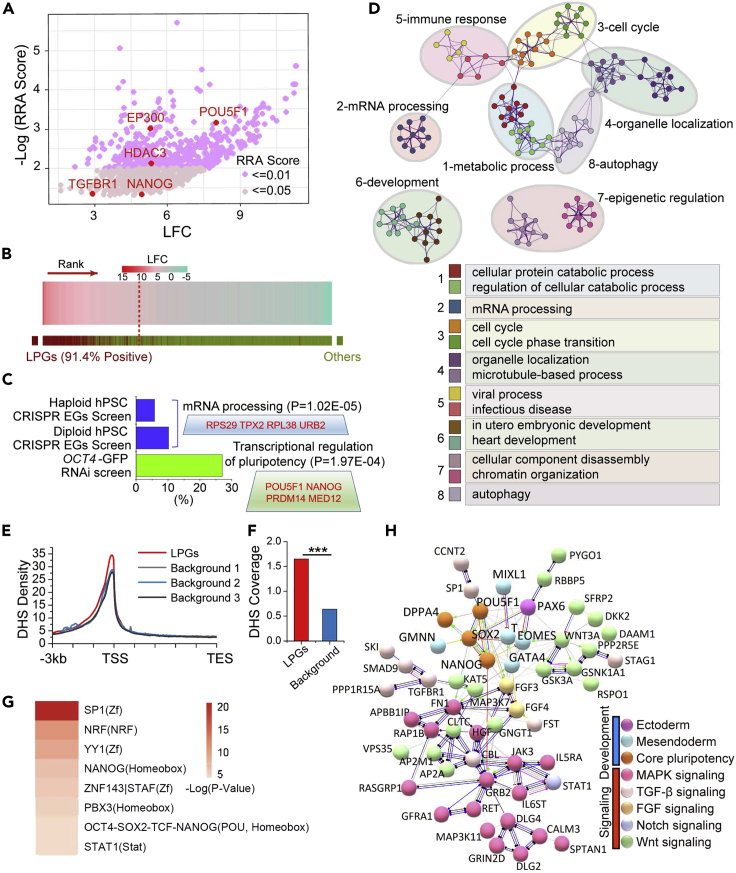
The MAGeCK algorithm (Li et al., 2014) was used to score the statistical significance (using a negative binomial test) of enrichment for individual gRNAs in tdTomato-positive cells by comparison with the overall gRNA pool. Significantly, enriched gRNAs were defined with an over 2-fold change and false discovery rate (FDR) below 0.05 (Figure S5A). Genes considered relevant were identified by their targeting gRNAs shown in the screen ranked consistently higher (by significance) using robust rank aggregation (RRA). Based on these criteria, we identified ∼4% (829/19,062) of overall genes in the coding genome as essential and their loss of function in hPSCs triggered lineage specification (Figure 4A). We therefore termed these genes as lineage-specification preventing genes (LPGs). To further validate these LPGs, we designed a sub-pool library, which included 182 non-targeting control gRNAs, and 7,324 gRNAs for the 829 LPGs and for those genes which were identified for at least one screen but showed no statistical significance. The sub-pool gRNAs were then verified by next-generation sequencing and showed a Gini index value at 0.029, suggesting uniform distribution (Figure S5B). Two sub-pool validation screenings were then performed, and more than 90% of previously identified LPGs were preferentially enriched with over 2-fold changes and FDRs below 0.05 (Figure 4B). The top 100 LPGs were listed in Table S1.
Figure 4.
Identification of lineage-specification prevention genes in hPSCs
(A) MAGeCK algorithm was used to estimate the statistical significance of enriched gRNA in tdTomato-positive cells after doxycycline induction. gRNA-associated genes were ranked through robust rank aggregation (RRA) and were termed lineage preventing genes (LPGs). Log2 fold change (LFC) was used to calculate the enrichment index as compared with the library. Each dot of the scatterplot represented one single LPG and genes known for regulating self-renewal of hPSCs were labeled in red.
(B) Over 90% of LPGs were efficiently enriched in the second sub-pool library screening.
(C) Percentage of overlapped genes of LPGs and reported essential genes (EGs) related to hPSC proliferation and survival or hPSC self-renewal-required genes. Gene Ontology (GO) analyses showed these overlapped genes were mostly related to mRNA processing and transcriptional regulation of pluripotency, with representative genes of each GO term been boxed and marked in red.
(D) LPGs clustered into functional modules as analyzed by the Metascape tool. Enriched terms retrieved from GO, KEGG pathway, and Reactome Gene Sets of all LPGs were assigned to modules based on Kappa-statistical similarities among their gene memberships. Eight modules were defined and each module represented a group of similar functional categories, and edges were connected where terms with similarity above 0.3. Terms with the best p values were depicted as network nodes, and the size of nodes represented number of enrichment of genes.
(E) LPGs and randomly selected background genes at equal size were subjected to DNase hypersensitive sites (DHSs) analysis. As compared with all 3 background controls, LPGs showed significantly higher DHS density around transcriptional start sites (TSS), indicating their active or poised transcriptional state.
(F) Statistics of DHS coverage of LPGs and background controls. Chi-square (x2) test was used to examine the significant difference (x2 = 892.2; df = 1; p = 0).
(G) DHSs of LPGs were enriched in binding sites of typical pluripotent transcription factors as revealed by HOMER.
(H) Protein-protein interaction (PPI) network analysis generated by STRING revealed that LPGs within the embryonic development module comprised core pluripotent transcription factors and mesendoderm specifiers, both heavily interacted with the nEc determinant PAX6. Signaling molecules related to Wnt, TGF-β, FGF/MAPK, and Notch pathways are early embryonic developmental cues, which showed a tight link with pluripotent genes and lineage specifiers. This PPI analysis pointed to an antagonistic effect within early lineage specifiers in hPSCs, given loss of function of mesendoderm specifiers led to spontaneous nEc differentiation.
Both OCT4 and NANOG were among the LPGs, coinciding with their pivotal role in pluripotency (Boyer et al., 2005). TGFBR1, receptor of the Activin/Nodal signaling pathway, which is essential for hPSC maintenance, was also among the identified LPGs (Vallier et al., 2005; Xu et al., 2008). In addition, HDAC3 and EP300, which are known to facilitate self-renewal while inhibiting lineage specification of hPSCs, were also detected within the LPGs (Qiao et al., 2015). Importantly, the identified LPGs were distributed across all chromosomes without enrichment in specific chromosomal regions (Figure S5C). These results suggest that the genome-wide CRISPR screening tool revealed a plethora of candidate LPGs and the mutation of which caused hPSC pluripotency exit and germ layer entry.
EGs critical for pluripotent stem cell survival and proliferation have been defined in both diploid and haploid hPSCs through CRISPR loss-of-function screening (Yilmaz et al., 2018; Mair et al., 2019). We made an intersection between LPGs and EGs and found that less than 10% of LPGs overlapped with the EGs, and these overlapped genes were enriched in the biological function of RNA processing (Figure 4C). With the OCT4-GFP reporter, genes required for hPSC self-renewal were also identified through RNAi library screening (Chia et al., 2010). LPGs shared far more genes with self-renewal-required genes than with the common genes of LPGs and EGs (Figures 4C and S5D). In addition, the shared LPGs and self-renewal-required genes were mostly clustered in the transcriptional regulation of pluripotency (Figures 4C and S5D). These results suggested that our CRISPR screening successfully retrieved a group of genes essential for hPSC self-renewal. However, given the advanced lineage reporter design and forward screening system, we also discovered those previously undefined genes essential for other biological processes, which shape trilineage specification.
We further annotated the LPGs to determine their biological function using the Panther Classification System (Mi et al., 2005). Biological process analysis showed strong enrichment in metabolic processes, biological regulation, and cellular component organization pathways (Figure S5E). Protein classification analysis showed enrichment in nucleic acid binding, transcription factors, enzyme modulators, and signaling molecules (Figure S5F). Additionally, we assigned various functional categories to hierarchical clusters based on Kappa-statistical similarities among their gene memberships using Metascape (Tripathi et al., 2015). This revealed that the LPGs were mostly categorized into eight functional modules, among which embryonic development, epigenetic regulation, metabolic regulation, and mRNA processing were the most prominent (Figures 4D and S5G).
To further delineate the regulatory networks controlling LPG expression, we profiled the DNase hypersensitive sites (DHSs) for these genes. DHSs are regions of open chromatin that make DNA accessible, and these accessible chromatin zones are functionally related to transcriptional activity via recruitment of dominant TFs. Here, compared with random or background genes, LPGs harbored significantly higher DHS coverage through chi-square tests (Figures 4E and 4F), suggesting their open chromatin states and active or poised transcription. The DNA sequences around the DHSs of all LPGs were subsequently subjected to TF binding motif analysis. We found that the top motifs were well-characterized TF binding sites associated with pluripotency, such as SP1 of the Z-finger domain, OCT4/NANOG/SOX2 core pluripotency factors, PBX3 of homeobox domain, NF-κB repressing factor (NRF), and Yin Yang-1 (YY1) (Figure 4G). These results suggest that LPGs are largely transcriptionally active genes in hPSCs and are tightly regulated by core pluripotency factors, reinforcing the fidelity and biological relevance of the identified LPGs. Although we found specific motif enrichment in modules such as metabolism, autophagy, and immune response, the SP1, OCT4/NANOG/SOX2, and YY1 binding motifs appeared in multiple modules (Figure S5H), reflecting their broad effect in regulating pluripotency and lineage specification.
Mesendodermal lineage specifiers antagonize nEc cell fate
Those LPGs within the embryonic development module could be clearly separated into a large-sized and a small-sized group. The large-sized group contained core pluripotency TFs, such as OCT4 and NANOG, and key molecules related to TGF-β, FGF/MAPK, and Notch signaling, which largely overlapped with pluripotency genes retrieved from previous RNAi screening using the OCT4-GFP reporter (Chia et al., 2010). Strikingly, the small-sized group was centered by typical lineage specifiers, such as T and MIXL1 for Me and GATA4 and EOMES for En. Protein-protein interaction (PPI) network analysis showed the interplay of these pluripotency genes and Me or En specifiers (Figure 4H). Deng and colleagues revealed that a combination of nEc, Me, and En lineage specifiers can efficiently reprogram fibroblasts into iPSCs (Shu et al., 2013). It has been reported that germ layer genes and pluripotent genes are co-expressed in a small population of hPSCs, and these cells still preserved their clonogenicity and potency to differentiate into all three germ layers (Allison et al., 2018; Han et al., 2010; Hough et al., 2014). Thus, expression of trilineage specifiers in hPSCs could act against each other to maintain a state of equilibrium in hPSCs, and upsetting such a balance will cause the differentiation of hPSCs toward antagonistic lineage.
Mutations of epigenetic machineries lead to hPSC pluripotency exit or lineage differentiation
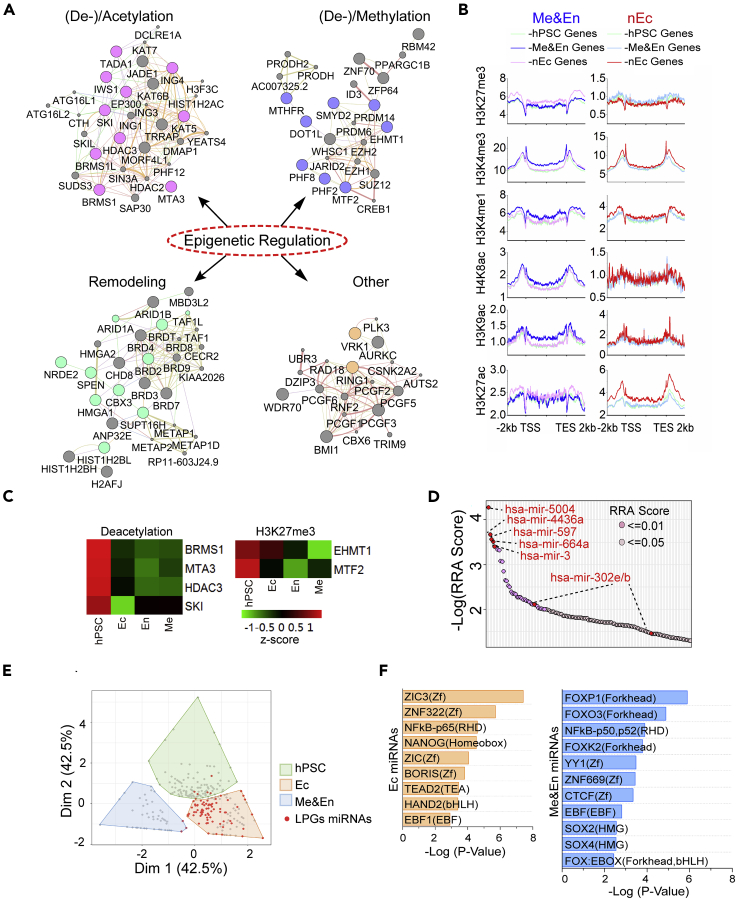
Epigenetic regulation, especially chromatin modification and remodeling, plays an important role in hPSC maintenance and differentiation (Gifford et al., 2013; Xie et al., 2013). We identified a functional module for epigenetic regulation in LPGs, mostly categorized into families of histone acetylation or de-acetylation, methylation or de-methylation, chromatin remodelers, and others related to ubiquitination or phosphorylation (Figure 5A). Many of these genes were downregulated during lineage specification (Figure S6A). To understand the roles of these epigenetic regulations in trilineage specification, we profiled the histone modifications (HMs) in gene categories of hPSCs-, mesendoderm-, and neural lineage-specific genes. Results showed that during mesendoderm and nEc lineage specification, the levels of histone acetylation/H3K4me3 in promoter regions and H3K4me1/H3K27ac in enhancer regions of lineage-specific genes were significantly increased, whereas those of H3K27me3 were decreased (Figures 5B and S6B). Heatmap analysis confirmed an obvious decrease in histone deacetylases, such as BRMS1, MTA3, HDAC3, and SKI, and methyl transferases for H3K27, including EHMT1 and MTF2 along with trilineage differentiation of hPSCs (Figure 5C). These results suggest that active epigenetic machineries are repressed, while repressive epigenetic machineries are highly active in lineage genes in hPSCs. Thus, loss of function of genes centered in these epigenetic machineries caused aberrant activation of lineage genes in hPSCs and therefore led to spontaneous lineage differentiation (Figure S6C).
Figure 5.
Comprehensive epigenome contributes to hPSC maintenance
(A) Four epigenetic regulation clusters as labeled were highlighted in LPGs. Networks were constructed by using GeneMANIA integrated with Cytoscape visualization. Within each tightly inter-connected cluster, colored nodes represented the genes identified in LPGs.
(B) Profiling histone modifications (HMs) of hESC-, mesendoderm (Me&En)-, and nEc-specific genes in hPSCs differentiated Me&En or nEc lineages. Histone acetylation, H3K4me1, and H3K4me3 of lineage-specific genes were significantly increased, and H3K27me3 were decreased during corresponding lineage specification.
(C) Heatmap showing a decreased expression of core regulators for histone deacetylation and H3K27me3 along with trilineage specification of hPSCs. The expression level was normalized to RPKM and z-scored.
(D) miRNAs within the scope of LPGs were identified by MAGeCK algorithm. The significance of each miRNA was ranked through RRA. Each dot of the scatterplot represented one single miRNA and top 5 miRNA, as well as hsa-mir-302e/b known for regulating self-renewal of hPSCs were labeled in red.
(E) Among all miRNAs in miRBae, hPSCs-, Ec-, or Me&En-related miRNAs were defined by using PCA classifier according to the enrichment score of all 3 categories based on GO annotation of its target genes. Red dots represented miRNAs shown in defined LPGs, and they were highly enriched in the Ec-related group.
(F) Motif analyses with HOMER for the promoter regions of Ec- or Me&En-related miRNAs.
hPSCs-expressed miRNAs are clustered and maintain pluripotency by targeting trilineage-specific genes
We identified a cluster of miRNAs (85/1864) within the screened LPGs (Figure 5D). Among these miRNAs, the hsa-mir-302 family is reported to play a role in hPSC maintenance by targeting COUP-TFII, a TF expressed in the nEc (Rosa and Brivanlou, 2011). To profile the function of miRNAs in LPGs, we annotated the top five miRNAs through miRBase (Griffiths-Jones et al., 2006) and performed Gene Ontology (GO) analysis for their target genes. We found that most of the target genes were highly enriched in the nEc lineage and were associated with nEc development (Figures S6D and S6E). Indeed, when we sub-grouped the miRNA superfamily into hPSC-, mesendoderm-, and ectoderm-related groups according to their target genes, we found that most miRNAs in our defined LPGs were in the Ec-related group (Figure 5E). These data suggested that hPSC-expressed miRNAs act as differentiation barriers, which specifically inhibit differentiation of an individual lineage. In line with this hypothesis, the promoter regions of Ec-related miRNAs primarily exhibited binding sites for Zinc finger family TFs and NANOG, whereas the mesendoderm-related miRNAs were enriched with distinct binding motifs of TFs, such as SOX2/4 and forkhead family TFs (Figure 5F).
Metabolic pathways profoundly regulate hPSC maintenance and lineage specification
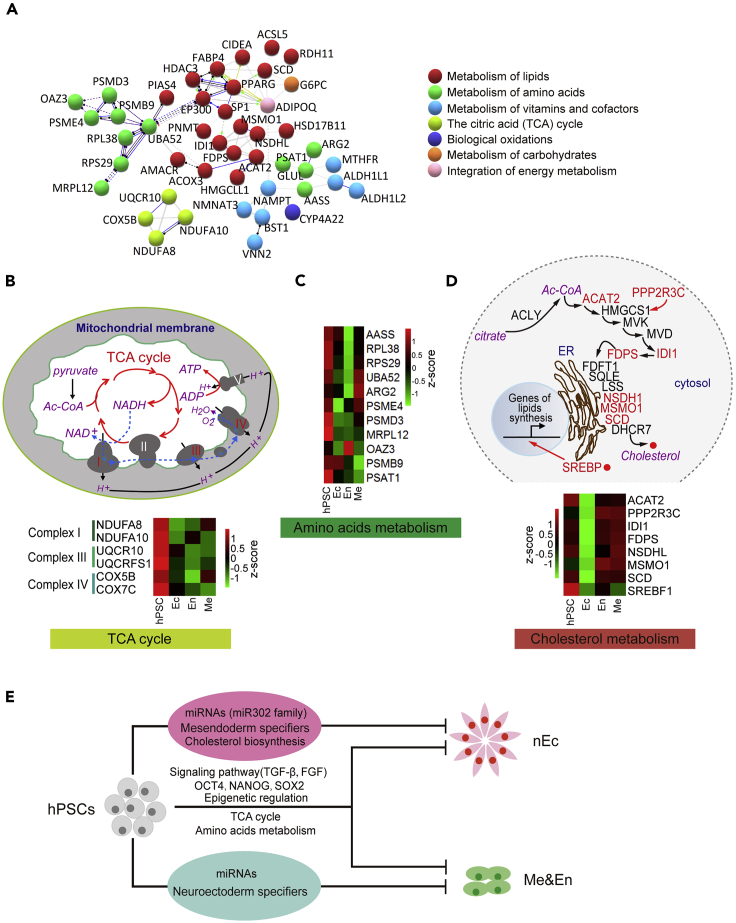
An accumulation of evidence has demonstrated that metabolic pathways play important roles in hPSC maintenance (Wu et al., 2016). Here, genes belonging to the metabolic pathways were the largest superfamily in the identified LPGs. GO analysis showed that these metabolism-related genes were mostly clustered in functional modules of the TCA cycle, energy production, and lipid or amino acid metabolism (Figure 6A).
Figure 6.
Cholesterol synthesis pathway restricts nEc specification from hPSCs
(A) PPI network analysis generated by STRING revealed that LPGs within the metabolism module comprised key molecules function in multiple metabolic pathways.
(B) Genes related to TCA cycle and oxidative phosphorylation were enriched in LPGs, and heatmap showed decreased expression of these LPGs along trilineage differentiation of hPSCs. The expression level was normalized to RPKM and z-scored.
(C) Heatmap showed decreased expression of LPGs related to amino acid metabolism along trilineage differentiation of hPSCs. The expression level was normalized to RPKM and z-scored.
(D) Genes of cholesterol synthesis pathway that were enriched in LPGs were labeled in red. Heatmap showed that the expression of these cholesterol synthesis pathway-related LPGs was specifically downregulated when the hPSCs were specified toward a nEc fate. The expression level was normalized to RPKM and z-scored.
(E) A schematic model for hPSC self-renewal and trilineage differentiation. Extracellular signaling, epigenetic regulation, transcriptional regulation, and metabolic pathways play a synergistic role in regulating hPSC self-renewal and trilineage differentiation. Histone modifications share common mechanisms in leveraging the transcriptional activity of genes belonging to all three lineages. The TCA cycle, the respiratory chain, and the amino acid metabolism are all important for maintaining an undifferentiated state of hPSCs, though glycolytic flux is highly active. A mutual antagonistic effect occurs in Me&En and nEc lineage specifiers, and a balance of these lineage specifiers is required for the self-renewal of hPSCs. hPSC-enriched miRNAs showed clear lineage gene targeting preference. The cholesterol synthesis pathway plays a pivotal role in maintaining hPSCs via restraining the nEc differentiation.
While hPSCs generally use glycolysis for energy production, a switch from glycolysis to OXPHOS is observed during their differentiation (Zhang et al., 2016). Within the LPGs, we were surprised to find a cluster of mitochondrial electron transport complex genes, considered as the endpoint of the TCA cycle. More importantly, these mitochondrial TCA-related genes were highly expressed in undifferentiated hPSCs but exhibited decreased expression upon differentiation (Figure 6B), implying their important role in hPSCs. They may act like c-Myc, an important modulator for mitochondrial biogenesis and is key for hPSC pluripotency maintenance and hiPSC reprogramming (Takahashi et al., 2007; Cliff et al., 2017). Although systematic research is still required to fully elucidate the impact of bioenergetic pathways on the fate of hPSCs, our data suggest that the mitochondrial TCA cycle and OXPHOS are absolutely required for the self-renewal of hPSCs, as loss of function of genes in these pathways caused spontaneous lineage differentiation.
We also found that most LPGs related to amino acid metabolism were downregulated during the process of hPSC differentiation (Figure 6C), implying an important role of amino acid metabolism in maintaining hPSCs (Shiraki et al., 2014). Surprisingly, genes related to cholesterol synthesis were highly enriched in LPGs (8 genes). These cholesterol synthesis genes were highly expressed in undifferentiated hPSCs and Me or En lineages but were downregulated when a nEc cell fate was adopted (Figure 6D). This implies that, different from other metabolic pathways, the cholesterol synthesis pathway is a preset barrier and maintains hPSCs via retardation of nEc specification.
Discussion
With a nEc-specific reporter and inducible CRISPR/Cas9 expression system, we performed genome-scale loss-of-function screening in hPSCs to identify dominant genes that determine pluripotency and lineage specification. We discovered various LPGs for hPSCs and loss of function of which led to either pluripotency exit or specific lineage commitment. Our study demonstrated that lineage-based reporters combined with CRISPR/Cas9 whole-genome screening offer a powerful strategy for revealing the biological processes and gene networks underlying specific cell fate determination, which is a key question in developmental biology and the basis for regenerative medicine. The identified LPGs could also serve as an invaluable resource for future studies on cellular and molecular events regarding hPSC self-renewal and trilineage development.
Our study re-emphasized the pivotal role of Notch/TGF-β/FGF signaling and OCT4/NANOG core transcriptional networks in regulating pluripotency of hPSCs (Vallier et al., 2005; Xu et al., 2008; Boyer et al., 2005). Our screening also revealed that common cellular biological processes, such as cellular organization, cell cycle regulation, and RNA processing, also play universal and important roles in hPSCs. Some of these functional modules overlap with EGs crucial for hPSC survival and proliferation. As demonstrated by our screening, there was a specific module for immunological responses within the LPGs. Eggenberger et al. showed that ectopic activation of the canonical type I interferon antiviral pathway drives hPSCs away from pluripotency and induces aberrant trilineage differentiation, supporting the conclusion that immune responses are also tightly associated with pluripotency and lineage development (Eggenberger et al., 2019).
Epigenetic modifiers establish a fundamental genomic environment for precise gene expression control and cellular context homeostasis. We identified that HMs and chromatin remodelers had profound effects on hPSC self-renewal and lineage differentiation, based on the following observations: (1) HM-related machineries and chromatin remodelers in LPGs were largely downregulated during lineage specification; (2) active HMs in the promoter and enhancer regions of lineage specific genes were enriched, whereas repressive HMs were de-enriched during corresponding lineage commitment; thus we concluded that HMs and chromatin remodelers maintained hPSCs in an undifferentiated state by establishing transcriptional barriers to restrict lineage gene expression. Notably, those miRNAs highly expressed in hPSCs retained pluripotency by targeting lineage genes. However, these miRNAs could be clearly sub-grouped into specific lineage-related clusters. Moreover, the promoter regions of the ectoderm-related miRNAs were enriched in binding sites of NANOG, the core pluripotency factor that antagonizes Ec but favors definitive En development. With our analysis, we also found that the mesendoderm-related miRNAs were enriched with binding motifs for SOX2, a known TF for facilitating nEc but inhibiting Me or En specification (Wang et al., 2012).
A small but intriguing cluster within the identified LPGs was the key mesendoderm specifiers, i.e., GATA4, EOMES, MIXL1, and T. Deng and his colleagues revealed that GATA3 and DLX3 can substitute OCT4 and SOX2 to reprogram mouse fibroblasts into iPSCs. Furthermore, they established a “seesaw” model, which demonstrated the balance between pluripotency factors and/or counteracting lineage specifiers (Shu et al., 2013). The same model also applies in humans, with lineage-specific genes able to efficiently reprogram human fibroblasts into hiPSCs (Mai et al., 2018; Shu et al., 2015). With the advent of single-cell RNA-seq technology, pluripotent stem cells are considered relatively heterogeneous, and lineage-specific genes are also expressed in some subpopulations of pluripotent stem cells responsible for clonal expansion (Allison et al., 2018; Hough et al., 2014). Thus, we hypothesized that, at least, in some populations of pluripotent stem cells, lineage specifiers counteract with each other and therefore maintain their pluripotency. In the future, the same strategy by applying mesendoderm-specific reporters could be used to identify Ec including nEc specifiers, LPGs antagonizing mesendoderm differentiation.
Here, genes within the metabolic pathway accounted for the largest family among identified LPGs. Although glycolytic flux has been shown to be crucial for the maintenance of hPSCs (Folmes et al., 2011; Panopoulos et al., 2012), we identified LPGs that were clustered in the functional module of mitochondrial TCA cycle and OXPHOS. Multiple studies have shown that proteins function in the TCA cycle and OXPHOS are higher in hPSCs than in their differentiated derivatives (Varum et al., 2011). Thus, the mitochondrial TCA cycle and OXPHOS and are also indispensable for hPSC pluripotency. Surprisingly, loss of function of genes within the cholesterol synthesis pathway led to nEc differentiation of hPSCs. It has been reported that the expression of pluripotent gene OCT4 was decreased when the cholesterol biosynthesis pathway was inhibited (Matsuzaki et al., 2018). Moreover, the cholesterol synthesis pathway is highly active in hPSCs and Me and En lineages but low in nEc. Thus, both glycolysis and OXPHOS bioenergetic pathways are crucial for the maintenance of hPSCs, and cholesterol synthesis process is required for human pluripotency via retardation of nEc specification.
In summary, this study provides a full landscape of biological processes underlying hPSC self-renewal and trilineage differentiation (Figure 6E). Pathways involved in bioenergetic production, HMs and chromatin remodelers, immune responses, RNA processing, as well as core pluripotency-related signaling and transcriptional networks, function synergistically to maintain a pluripotent state in hPSCs via inhibition of trilineage differentiation. However, hPSC-enriched miRNAs, lineage specifiers, and the cholesterol synthesis pathway exhibit germ layer selectivity, and these biological processes promote or antagonize specific germ layers along the maintenance of hPSCs or early lineage differentiation.
Limitations of the study
In this paper, we performed CRISPR/Cas9 screen in hPSCs to identify those genes that prevent lineage specification. However, when taking the trilineage cell fate decision in count, we only used the PAX6 reporter and lacked parallel screen with the mesendodermal reporters. The uncovered library of lineage specification preventing genes is a valuable source for studying mechanisms underlying hPSC self-renewal and trilineage specification. However, specific validation is apparently required when comes to individual genes.
Resource availability
Lead contact
Xiaoqing Zhang (xqzhang@tongji.edu.cn)
Materials availability
Requests for materials and reagents should be directed to the Lead contact.
Data availability
All relevant data are available from the correspondence authors upon reasonable request. The accession number for the CRISPR/Cas9 screening data reported in this paper is GEO: GSE132309.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (Grant no. 2018YFA0108000 and 2019YFA0110300), the National Natural Science Foundation of China (Grant No. 82025020, 31400934, 31771132, 31872760, 31801204, and 31800858), the Science and Technology Commission of Shanghai Municipality (19JC1415100), the Shanghai Municipal Education Commission (C120114), China Postdoctoral Science Foundation (Grant No. 2017M621526), the Fundamental Research Funds for the Central Universities, and the Major Program of Development Fund for Shanghai Zhangjiang National Innovation Demonstration Zone (Stem Cell Strategic Biobank and Clinical Translation Platform of Stem Cell Technology, ZJ2018-ZD-004). Graphical abstract of this study was created with BioRender.com.
Author contributions
X.Z. and L.L. conceived and designed the project. X.X., Y.D., and L.M. performed most of the experiments and analyses. SW.Z., L.S., Z.C., Z.Z., Y.H., Y.L., Y.F., B.F., Z.L., N.L., SS.Z., and L.L., helped to set up the genetic engineering system and performed Western blot, immunostaining, and genomic DNA PCR experiments. C.J. helped with bioinformatics analyses. X.X., Y.D., and X.Z. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 22, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101926.
Contributor Information
Ling Liu, Email: lliu@tongji.edu.cn.
Xiaoqing Zhang, Email: xqzhang@tongji.edu.cn.
Supplemental information
References
- Allison T.F., Smith A.J.H., Anastassiadis K., Sloane-Stanley J., Biga V., Stavish D., Hackland J., Sabri S., Langerman J., Jones M. Identification and single-cell functional characterization of an Endodermally biased pluripotent substate in human embryonic stem cells. Stem Cell Reports. 2018;10:1895–1907. doi: 10.1016/j.stemcr.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birket M.J., Orr A.L., Gerencser A.A., Madden D.T., Vitelli C., Swistowski A., Brand M.D., Zeng X. A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. J. Cell Sci. 2011;124:348–358. doi: 10.1242/jcs.072272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.R., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Ren X., Xu X., Zhang X., Hui Y., Liu Z., Shi L., Fang Y., Ma L., Liu Y. Genetic engineering of human embryonic stem cells for precise cell fate tracing during human lineage development. Stem Cell Reports. 2018;11:1257–1271. doi: 10.1016/j.stemcr.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi L., Fan B., Zhang K., Du Y., Liu Z., Fang Y., Chen Z., Ren X., Xu X., Jiang C. Targeted differentiation of regional ventral neuroprogenitors and related neuronal subtypes from human pluripotent stem cells. Stem Cell Reports. 2016;7:941–954. doi: 10.1016/j.stemcr.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N.Y., Chan Y.S., Feng B., Lu X., Orlov Y.L., Moreau D., Kumar P., Yang L., Jiang J., Lau M.S. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Cliff T.S., Wu T., Boward B.R., Yin A., Yin H., Glushka J.N., Prestegaard J.H., Dalton S. MYC controls human pluripotent stem cell fate decisions through regulation of metabolic flux. Cell Stem Cell. 2017;21:502–516 e9. doi: 10.1016/j.stem.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Du Y., Liu Z., Cao X., Chen X., Chen Z., Zhang X., Zhang X., Jiang C. Nucleosome eviction along with H3K9ac deposition enhances Sox2 binding during human neuroectodermal commitment. Cell Death Differ. 2017;24:1121–1131. doi: 10.1038/cdd.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenberger J., Blanco-Melo D., Panis M., Brennand K.J., Tenoever B.R. Type I interferon response impairs differentiation potential of pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2019;116:1384–1393. doi: 10.1073/pnas.1812449116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faial T., Bernardo A.S., Mendjan S., Diamanti E., Ortmann D., Gentsch G.E., Mascetti V.L., Trotter M.W., Smith J.C., Pedersen R.A. Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development. 2015;142:2121–2135. doi: 10.1242/dev.117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes C.D.L., Nelson T.J., Martinez-Fernandez A., Arrell D.K., Lindor J.Z., Dzeja P.P., Ikeda Y., Perez-Terzic C., Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford C.A., Ziller M.J., Gu H., Trapnell C., Donaghey J., Tsankov A., Shalek A.K., Kelley D.R., Shishkin A.A., Issner R. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V., Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., Van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.W., Tapia N., Joo J.Y., Greber B., Arauzo-Bravo M.J., Bernemann C., Ko K., Wu G., Stehling M., Do J.T., Scholer H.R. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143:617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Hough S.R., Thornton M., Mason E., Mar J.C., Wells C.A., Pera M.F. Single-cell gene expression profiles define self-renewing, pluripotent, and lineage primed states of human pluripotent stem cells. Stem Cell Reports. 2014;2:881–895. doi: 10.1016/j.stemcr.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Wade P.A. NuRD and pluripotency: a complex balancing act. Cell Stem Cell. 2012;10:497–503. doi: 10.1016/j.stem.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihry R.J., Salick M.R., Ho D.J., Sondey M., Kommineni S., Paula S., Raymond J., Henry B., Frias E., Wang Q. Genome-scale CRISPR screens identify human pluripotency-specific genes. Cell Rep. 2019;27:616–630 e6. doi: 10.1016/j.celrep.2019.03.043. [DOI] [PubMed] [Google Scholar]
- Kleinjan D.A., Seawright A., Mella S., Carr C.B., Tyas D.A., Simpson T.I., Mason J.O., Price D.J., Van Heyningen V. Long-range downstream enhancers are essential for Pax6 expression. Dev. Biol. 2006;299:563–581. doi: 10.1016/j.ydbio.2006.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J.G., Gardner D.K., Harvey A.J. Mitochondrial and Glycolytic Remodeling during Nascent Neural Differentiation of Human Pluripotent Stem Cells. Development. 2018;145:dev168997. doi: 10.1242/dev.168997. [DOI] [PubMed] [Google Scholar]
- Li Q.V., Dixon G., Verma N., Rosen B.P., Gordillo M., Luo R., Xu C., Wang Q., Soh C.L., Yang D. Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat. Genet. 2019;51:999–1010. doi: 10.1038/s41588-019-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.J., Zhang X., Johnson M.A., Wang Z.B., Lavaute T., Zhang S.C. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136:4055–4063. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Chen Z., Zhang X., Li S., Hui Y., Feng H., Du Y., Jin G., Zhou X., Zhang X. Protection of ZIKV infection-induced neuropathy by abrogation of acute antiviral response in human neural progenitors. Cell Death Differ. 2019;26:2607–2621. doi: 10.1038/s41418-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Hui Y., Shi L., Chen Z., Xu X., Chi L., Fan B., Fang Y., Liu Y., Ma L. Efficient CRISPR/Cas9-Mediated versatile, predictable, and donor-free gene knockout in human pluripotent stem cells. Stem Cell Reports. 2016;7:496–507. doi: 10.1016/j.stemcr.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai T., Markov G.J., Brady J.J., Palla A., Zeng H., Sebastiano V., Blau H.M. NKX3-1 is required for induced pluripotent stem cell reprogramming and can replace OCT4 in mouse and human iPSC induction. Nat. Cell Biol. 2018;20:900. doi: 10.1038/s41556-018-0136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair B., Tomic J., Masud S.N., Tonge P., Weiss A., Usaj M., Tong A.H.Y., Kwan J.J., Brown K.R., Titus E. Essential gene profiles for human pluripotent stem cells identify uncharacterized genes and substrate dependencies. Cell Rep. 2019;27:599–615 e12. doi: 10.1016/j.celrep.2019.02.041. [DOI] [PubMed] [Google Scholar]
- Martyn I., Kanno T.Y., Ruzo A., Siggia E.D., Brivanlou A.H. Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature. 2018;558:132–135. doi: 10.1038/s41586-018-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki T., Matsumoto S., Kasai T., Yoshizawa E., Okamoto S., Yoshikawa H.Y., Taniguchi H., Takebe T. Defining lineage-specific membrane fluidity signatures that regulate adhesion kinetics. Stem Cell Reports. 2018;11:852–860. doi: 10.1016/j.stemcr.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A., Kogan N.M., Aberdam D. Concise review: energy metabolites: key mediators of the epigenetic state of pluripotency. Stem Cells. 2015;33:2374–2380. doi: 10.1002/stem.2041. [DOI] [PubMed] [Google Scholar]
- Munoz-Sanjuan I., Brivanlou A.H. Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 2002;3:271–280. doi: 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- Panopoulos A.D., Yanes O., Ruiz S., Kida Y.S., Diep D., Tautenhahn R., Herrerias A., Batchelder E.M., Plongthongkum N., Lutz M. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Wang R., Yang X., Tang K., Jing N. Dual roles of histone H3 lysine 9 acetylation in human embryonic stem cell pluripotency and neural differentiation. J. Biol. Chem. 2015;290:2508–2520. doi: 10.1074/jbc.M114.603761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A., Brivanlou A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki N., Shiraki Y., Tsuyama T., Obata F., Miura M., Nagae G., Aburatani H., Kume K., Endo F., Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Shparberg R.A., Glover H.J., Morris M.B. Modeling mammalian commitment to the neural lineage using embryos and embryonic stem cells. Front. Physiol. 2019;10:705. doi: 10.3389/fphys.2019.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J., Wu C., Wu Y.T., Li Z.Y., Shao S.D., Zhao W.H., Tang X., Yang H., Shen L.J., Zuo X.H. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J., Zhang K., Zhang M.J., Yao A.Z., Shao S.D., Du F.X., Yang C.Y., Chen W.H., Wu C., Yang W.F. GATA family members as inducers for cellular reprogramming to pluripotency. Cell Res. 2015;25:169–180. doi: 10.1038/cr.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F., Jaenisch R. Stem cells, genome editing, and the path to translational medicine. Cell. 2018;175:615–632. doi: 10.1016/j.cell.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tam P.P., Loebel D.A. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A., Moulton H.M., Dejesus P., Che J., Mulder L.C. Meta- and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyas D.A., Simpson T.I., Carr C.B., Kleinjan D.A., Van Heyningen V., Mason J.O., Price D.J. Functional conservation of Pax6 regulatory elements in humans and mice demonstrated with a novel transgenic reporter mouse. BMC Dev. Biol. 2006;6:21. doi: 10.1186/1471-213X-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Alexander M., Pedersen R.A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Varum S., Rodrigues A.S., Moura M.B., Momcilovic O., Easley C.A., Ramalho-Santos J., Van Houten B., Schatten G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chen Y.G. Signaling control of differentiation of embryonic stem cells toward mesendoderm. J. Mol. Biol. 2016;428:1409–1422. doi: 10.1016/j.jmb.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Wang Z., Oron E., Nelson B., Razis S., Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Wu J., Ocampo A., Belmonte J.C.I. Cellular metabolism and induced pluripotency. Cell. 2016;166:1371–1385. doi: 10.1016/j.cell.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Xie W., Schultz M.D., Lister R., Hou Z., Rajagopal N., Ray P., Whitaker J.W., Tian S., Hawkins R.D., Leung D. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.H., Peck R.M., Li D.S., Feng X., Ludwig T., Thomson J.A. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat. Methods. 2005;2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- Xu R.H., Sampsell-Barron T.L., Gu F., Root S., Peck R.M., Pan G., Yu J., Antosiewicz-Bourget J., Tian S., Stewart R., Thomson J.A. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A., Peretz M., Aharony A., Sagi I., Benvenisty N. Defining essential genes for human pluripotent stem cells by CRISPR-Cas9 screening in haploid cells. Nat. Cell Biol. 2018;20:610–619. doi: 10.1038/s41556-018-0088-1. [DOI] [PubMed] [Google Scholar]
- Zhang J., Khvorostov I., Hong J.S., Oktay Y., Vergnes L., Nuebel E., Wahjudi P.N., Setoguchi K., Wang G., Do A. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells (vol 30, pg 4860, 2011) Embo J. 2016;35:899. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Li J., Tan Z., Wang C., Liu T., Chen L., Yong J., Jiang W., Sun X., Du L. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 2008;111:1933–1941. doi: 10.1182/blood-2007-02-074120. [DOI] [PubMed] [Google Scholar]
- Zhang S.C., Wernig M., Duncan I.D., Brustle O., Thomson J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Zhang X., Huang C.T., Chen J., Pankratz M.T., Xi J., Li J., Yang Y., Lavaute T.M., Li X.J., Ayala M. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li B., Li W., Ma L., Zheng D., Li L., Yang W., Chu M., Chen W., Mailman R.B. Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports. 2014;3:460–474. doi: 10.1016/j.stemcr.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Q., Zhang S.C. Differentiation of neural precursors and dopaminergic neurons from human embryonic stem cells. Methods Mol. Biol. 2010;584:355–366. doi: 10.1007/978-1-60761-369-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the correspondence authors upon reasonable request. The accession number for the CRISPR/Cas9 screening data reported in this paper is GEO: GSE132309.