Abstract
A dairy-originated probiotic bacterium, Propionibacterium freudenreichii subsp. freudenreichii B3523 (PF) was found to be effective in reducing multidrug-resistant Salmonella Heidelberg (MDR SH) colonization in turkey poults (2-week-old) and growing (7-week-old) and finishing (12-week-old) turkeys. In this study, we explored the potential for microbiome modulation in the cecum of turkeys of different age groups due to PF supplementation in conjunction with MDR SH challenge. One-day-old commercial turkey poults were allocated to 3 treatment groups: negative control (N; turkeys without PF supplementation or SH challenge), SH control (S; turkeys challenged with SH without PF supplementation), and test group (P; turkeys supplemented with PF and challenged with SH). Turkeys were supplemented with 1010 CFU PF in 5-gallon (18.9 L) water until 7 or 12 week of age. At the 6th or 11th wk, turkeys were challenged with SH at 106 and 108 CFU/bird by crop gavage, respectively. After 2 and 7 d of challenge (2-d postinoculation [PI] and 7-d PI, respectively), cecal samples were collected and microbiome analysis was conducted using Illumina MiSeq. The experiments were repeated twice with 8 and 10 turkeys/group for 7- and 12-wk studies, respectively. Results indicated that the species richness and abundance (Shannon diversity index) was similar among the treatment groups. However, treatments caused apparent clustering of the samples among each other (P < 0.05). Firmicutes was the predominant phylum in the growing and finishing turkey cecum which was evenly distributed among the treatments except on wk 12 where the relative abundance of Firmicutes was significantly higher in P than in N (P = 0.02). The MDR SH challenge resulted in modulation of microflora such as Streptococcus, Gordonibacter, and Turicibacter (P < 0.05) in the S groups compared with the P and N groups, known to be associated with inflammatory responses in birds and mammals. The supplementation of PF increased the relative abundance of carbohydrate-fermenting and short-chain fatty acid–producing genera in the P group compared with the S group (P < 0.05). Moreover, the results revealed that PF supplementation potentially modulated the beneficial microbiota in the P group, which could mitigate SH carriage in turkeys.
Key words: Propionibacterium, Salmonella Heidelberg, microbiome, alternatives, dairy probiotic
Introduction
The poultry digestive tract harbors a diverse group of microorganisms. The complex interactions of the enteric microorganisms with the host and the environment maintain bird health by influencing the host immune system, modulating biochemical functions associated with break down of nutrients, strengthening intestinal morphology and physiology, and handling toxins and pathogens (Pan and Yu, 2014). The digestive tract of poultry is short compared with other livestock, and therefore, the full transit of digesta in poultry is less than 3.5 h (Hughes, 2008) favoring the survival of microorganisms with high adhesion capacity on the gut mucosa, especially in the cecum (Pan and Yu, 2014). The ceca are located between the small and large intestines and away from the main flow of the GIT. In addition, the discharge from the cecum is infrequent, happening twice daily in turkeys (Duke et al., 1969; Pan and Yu, 2014; Svihus, 2014). These factors make the cecum an ideal environment for survival of diverse microorganisms of nutritional and health importance to the birds, as high as 1010 to1011/g (Svihus, 2014).
The healthy cecal microflora reduces the likelihood of foodborne pathogens colonizing there (Salanitro et al., 1974; Beery et al., 1988; Hinton et al., 1990; Vasudevan et al., 2005). However, heavy loads of pathogenic microbes or infections result in alterations in intestinal immunity, potentially altering the microbial community. The production of antimicrobial substances and inflammatory response molecules such as nitric oxide radicals, reactive oxygen species, bacterial siderophores, chelators, and proteases act not only against invading pathogens but also against host-commensal microflora and affects intestinal integrity consequently resulting in an unbalanced microbiome, termed dysbiosis (Stecher et al., 2007). In addition, the external environment influences the poultry gut microbiome; most data are applied in this regard and geared toward better feed efficiency, especially with the use of feed additives (Wei et al., 2013).
Probiotic supplementation may help to maintain the normal microflora in the poultry gut and may enhance immunity (Callaway et al., 2008). Different challenge experiments in poultry have revealed the efficacy of therapeutic supplementation of probiotics against enteric pathogens, especially Salmonella in poultry (Pascual et al., 1999; La Ragione and Woodward, 2003; Tsai et al., 2005; Menconi et al., 2011; Yang et al., 2018). However, most of these challenge studies did not explore the effect of probiotic supplementation on poultry cecal microbiome populations, and their potential modulation in the presence of a pathogen challenge, especially in turkeys.
Previous studies conducted in our laboratory revealed that a dairy-originated probiotic bacterium, Propionibacterium freudenreichii subsp. freudenreichii B3523 (PF) was effective against multidrug-resistant Salmonella Heidelberg (MDR SH) in poults (2-week-old), growing (7-week-old), and finishing (12-week-old) turkeys (Nair et al., 2018a, 2019; Nair and Kollanoor Johny, 2018; Nair et al., 2020). The PF supplementation resulted in 1.6-2.2 log10 CFU/g reduction of MDR SH in the cecum of 2-week-old poults (Nair and Kollanoor Johny, 2018). Similarly, PF supplementation through water yielded 1 to 1.3 and 1.7 to 2.2 log10 CFU/g reduction of MDR SH in growing and finishing turkeys, respectively (Nair et al., 2018a, 2019, 2020). Even though the use of non–host-specific (allochthonous) and yet host gut-adaptable probiotic strains such as PF are encouraged over host-specific probiotics by the industry, the effect of such strains on the turkey microbiome and gut health has not been explored in relation to MDR SH carriage in turkeys.
The hypothesis of this study was that PF supplementation would modulate the beneficial microbiota in the cecum of turkeys to reduce MDR SH colonization. The objective was to determine the population shifts in the cecal microbiome of poults, growing and finishing turkeys (2-, 7-, and 12-wk-long studies in turkeys, respectively) in response to MDR SH challenge and PF supplementation.
Materials and methods
Bacterial Strains and Culture Conditions
Multidrug-Resistant Salmonella Heidelberg
After 3 subsequent subcultures of MDR SH in Trypticase soy broth (catalog no.C7141, CRITERION, Hardy Diagnostics, Santa Maria, CA), 16 h broth cultures grown to 109 CFU/mL MDR SH were washed in phosphate-buffered saline (PBS; pH=7.2) after centrifugation at 3,600 × g for 15 min at 4°C. The bacterial pellets were diluted in appropriate amount of PBS to obtain 105, 106, and 107 CFU/mL of SH. For turkey poults studies, 2 mL of 106 CFU/mL was administered to the turkey poults by passing a gavage tube through the mouth leading to the crop (crop gavage). Similarly, 10 mL of 105 and 107 CFU/mL MDR SH was administered to growing and finishing turkeys to obtain 106 CFU/growing turkey and 108 CFU/finishing turkey, respectively, by crop gavage (Nair and Kollanoor Johny, 2018; Nair et al., 2018a, Nair et al., 2018b; 2019).
Propionibacterium freudenreichii
Propionibacterium freudenreichii subsp. freudenreichii (B3523 USDA ARS NRRL Culture Collection, Peoria, IL) was used in the study. After 3 successive subcultures of PF in de Man, Rogosa and Sharpe (catalog no. C5932, CRITERION, Hardy Diagnostics, Santa Maria, CA) agar at 37°C, 24 h broth cultures of PF (approximately 1010 CFU/mL viable cells) in 1 L of de Man, Rogosa and Sharpe agar were washed in PBS centrifuging at 15,000 rpm for 15 min at 4°C. For the poults study, 1010 CFU/mL PF was supplemented per gallon of drinking water continuously for 14 d. For feeding growing and finishing turkeys, 1012 CFU PF was supplemented to the one-day-old turkeys through drinking water (5 gallons or 18.9 L) for 7 and 12 wk, respectively.
Experimental Design and Sample Collection
Bird Housing and Management
Turkey experiments were conducted with the approval from the IACUC at the University of Minnesota (Protocol#1803-35686A). For turkey poults study, day-of-hatch poults (Hybrid Converter), male and female in equal, were allocated to 3 different isolators in the Research Animal Resources biocontainment (isolation) units at the University of Minnesota for 14 d. For growing and finishing turkey studies, day-old female poults (Hybrid Converter) were housed in 3 different pens in the Poultry Teaching and Research Facility at the University of Minnesota until the end of 6 and 11 wk of age, respectively. On wk 7 and 12, these birds were transferred to isolation units for conducting the challenge experiments. The birds were transferred in sterile boxes in a sanitized truck to isolation units (0.5 miles away from the growth facility) and ensured that they were experiencing minimal stress during transfer and handling by trained assistants. In addition, similar conditions were provided in the climate-controlled isolation units specifically constructed for controlled challenge studies. In all experiments, the birds were provided with age-appropriate light, heat, and floor space (2 ft2/bird for poult studies, and 3 ft2/bird until 12 wk of age). The birds were supplied with Salmonella-free feed and water ad libitum specified by NRC recommendations.
Turkey Poults (2-Week-Long) Study
Day-old poults were purchased from a commercial hatchery in Minnesota, and the study was conducted for 14 d. The day-of-hatch poults (N = 36) were randomly distributed in 3 isolator pens with 12 birds each. The experiment was repeated twice. The treatment groups were negative control (N; poults without PF supplementation or SH challenge), SH control (S; poults challenged with SH and without PF supplementation), and test group (P; poults supplemented with PF and challenged with SH). On day 1, the poults in the P group were inoculated with 1010 CFU/mL PF using crop gavage method. On subsequent days, 1010 CFU/mL PF was supplemented per gallon of drinking water continuously for 14 d. On day 7, the S and P groups were challenged with SH at 106 CFU/mL through crop gavage. Two and 7 d post-SH inoculation (2-d postinoculation [PI] and 7-d PI, respectively), cecal contents were collected after euthanizing the poults (Nair and Kollanoor Johny, 2018). Cecal contents were collected from 3 and 4 poults from each group on 2-d PI and 7-d PI, respectively, for microbiome analysis.
Growing Turkey (7-Week-Long) Study
Day-old poults were purchased from a commercial hatchery in Minnesota, and the study was conducted for 7 wk. Day-old poults (N = 24) were randomly distributed into 3 pens with 8 birds each. The experiment was repeated twice. The 3 treatment groups were N, P, and S, similar to the poults study. The turkeys in the P groups were supplemented with 1012 CFU/mL of PF on alternate day through drinking water (5 gallons or 18.9 L) for 6 wk. The turkeys in the N and S groups did not receive any PF supplementation. At 6 wk of age, after moving turkeys to isolation facility, the S and P groups were challenged with SH (106 CFU/turkey) by crop gavage method. After the challenge, PF was supplemented to the P group daily for the next 7 d. Two d and 7 d after challenge, 4 turkeys from each group were euthanized and cecal samples were collected for microbiome analysis. Two independent studies were conducted. A total of 8 samples per group were collected for both sampling days for microbiome analysis (Nair et al., 2019).
Finishing Turkey (12-Week-Long) Study
Day-old poults were purchased from a commercial hatchery in Minnesota, and the study was conducted for 12 wk. Day-old poults (N = 30) were randomly distributed into 3 pens with 10 birds each. The experiment was repeated twice. The 3 treatment groups were N, P, and S. The turkeys in the P groups were provided with 1012 CFU/mL of PF on alternate days through drinking water (5 gallons or 18.9 L) for 11 wk in the Poultry Teaching and Research Facility. The turkeys in the N and S groups did not receive any PF supplementation. At 11 wk of age, the S and P groups were challenged with SH (108 CFU/bird) by crop gavage method at the isolation facility. After the challenge, PF was supplemented to the P group daily for the next 7 d. Two d and 7 d after challenge, 5 turkeys from each group were euthanized and cecum samples were collected for microbiome analysis. A total of 10 samples per group were collected for both sampling day in 2 independent experiments (Nair et al., 2018a).
Microbiome Analysis
After euthanizing the birds, necropsy was conducted under aseptic conditions in the Veterinary Diagnostic Laboratory. Cecal contents were aseptically collected in sterile 50-mL tubes and stored at −80°C until further analysis. The V4 hypervariable region of the 16S rRNA gene was amplified after extracting DNA from the cecal samples, individually, using PowerSoil DNA extraction kits (Qigong, Valencia, CA), following previously referenced protocols (Gohl et al., 2016; Johnson et al., 2018). Sequencing was performed at the University of Minnesota Genomic Center using Illumina MiSeq paired-end 2 × 250 bp technology.
Statistical Analysis
Separate experiments were conducted in poults, growing turkeys, and finishing turkeys to determine cecal microbiome modulations associated with MDR SH challenge and PF supplementation in turkeys. A completely randomized experimental design was followed to conduct the study and the experiments were repeated twice for each age group. The 2-wk long poult study was used as a preliminary study with 3 and 4 samples obtained from 2-d PI and 7-d PI, respectively. There were 48 and 60 cecal samples included for 7- and 12-wk studies, respectively.
For the microbiome analysis, sequencing was performed using Illumina MiSeq paired-end 2 × 250 bp technology. Sequences were filtered and clustered using DADA2 workflow (Callahan et al., 2016) and the downstream analysis of samples was performed using R version 3.3. Alpha diversity was calculated using Shannon indices to measure the species richness and evenness. The effect of treatment on α diversity was then analyzed using one-way ANOVA. Tukey's test was used to perform posthoc comparisons in growing and finishing turkey studies. For poults study, Kruskal-Wallis rank-sum test was used to analyze α diversity. Beta diversity was estimated as the difference in bacterial composition among different samples by coupling Bray-Curtis dissimilarity with principal coordinate analysis. Furthermore, the effect of treatments on the bacterial community composition was analyzed using permutational multivariate analysis (PERMANOVA, Adonis function, 99 permutations). Finally, the differentially abundant taxa driving the microbial shift between treatments were determined by characterizing species indicator values or Indval (Reis et al., 2019; Stubbington et al., 2019). From the probability table (Supplementary Tables 3A, 3B, 3C and 3D), a genus with indicator value ≥ 0.5 was selected, and Tukey's test was used to identify changes in the relative abundance of these indicator taxa among treatments. The significance was detected at P < 0.05.
Results
The quality details of the sequences are included in the Supplementary Table 1. The rarefaction curves tended to attain the saturation plateau, indicating that there were enough reads for each sample to represent most of microbiome community (Supplementary Figures 1–3).
Turkey Poults (2-Week-Long) Study
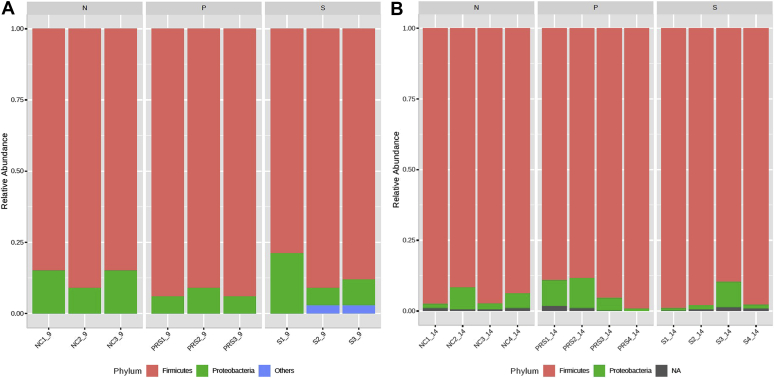
All treatment groups taken together, the most abundant phyla in poults at 2-d PI were Firmicutes and Proteobacteria, which occupied 88.6 and 10.8%, respectively (Figure 1A). Clostridia (95.8%) and Bacilli (4.2%) were the abundant classes among the Firmicutes. Gammaproteobacteria was most abundant among the Proteobacteria. Seven days after the challenge, Firmicutes and Proteobacteria continued as abundant phyla, where 94.6% bacterial population was Firmicutes, and 4.5% Proteobacteria, irrespective of treatments (Figure 1B). Among the Firmicutes, ∼99% abundance was observed for Clostridia.
Figure 1.
The relative abundance of major phyla in 2-d (A) and 7-d (B) postinoculation samples among different treatment groups in 2-week-long poult study. From the day of hatch, the poults were allocated into N, P, and S groups. Abbreviations: N, Negative control (poults without SH challenge or PF supplementation); P, test group (poults supplemented with PF for 14 d and challenged with SH on day 7); PF, Propionibacterium freudenreichii; S, SH control (poults challenged with SH on day 7 and without PF supplementation); SH, Salmonella Heidelberg.
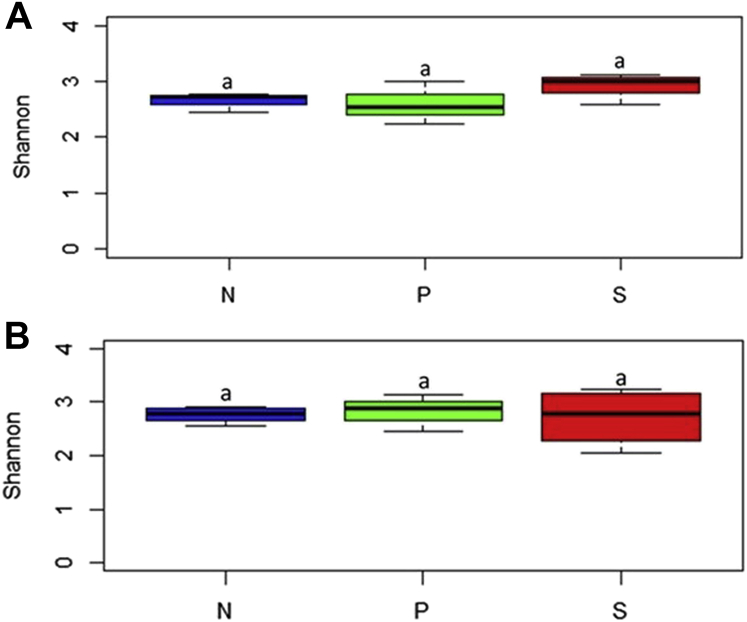
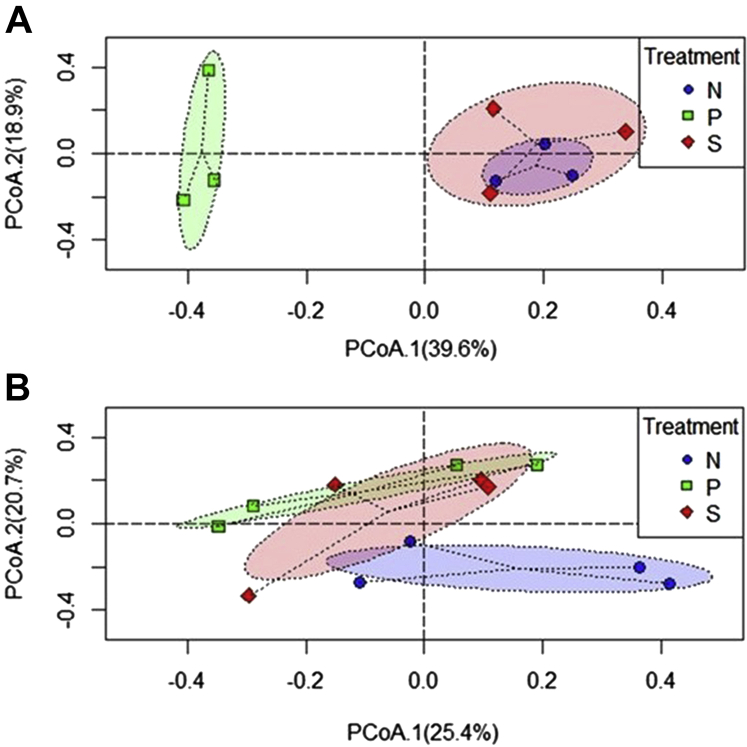
There was no significant difference in α diversity among the 3 treatment groups in the study (N, S, and P) at 2-d PI and 7-d PI (P > 0.05) (Figures 2A and 2B; Supplementary Table 2). The PERMANOVA analysis on the beta diversity indices at 2 d after SH challenge indicated that the treatments contributed to 46.8% variability among the samples resulting in apparent clustering with overlaps observed between the N and S groups which were different from the P group (P = 0.011) (Figure 3A). At 7-d PI also, the clustering was evident, and the treatment effects contributed to the variability of 27.3% to the samples (P = 0.034) (Figure 3B).
Figure 2.
Alpha diversity of the cecal microbiome of poults at 2 d (A) and 7 d (B) after inoculation represented as Shannon diversity index. From the day of hatch, the poults were allocated into N, P, and S groups for 14 d; a indicates P > 0.05. Abbreviations: N, Negative control (poults without SH challenge or PF supplementation); P, test group (poults supplemented with PF for 14 d and challenged with SH on d 7); PF, Propionibacterium freudenreichii; S, SH control (poults challenged with SH on d 7 and without PF supplementation); S, SH control (poults challenged with SH on d 7 and without PF supplementation)SH, Salmonella Heidelberg.
Figure 3.
Principal coordinates analysis showing microbial compositional difference among the cecal samples of poults at 2 d (A) and 7 d (B) after inoculation as determined by the beta diversity using the Bray-Curtis dissimilarity index. From the day of hatch, the poults were allocated into N, P, and S groups for 14 d. Abbreviations: N, Negative control (poults without SH challenge or PF supplementation); S, SH control (poults challenged with SH on d 7 and without PF supplementation); P, test group (poults supplemented with PF for 14 d and challenged with SH on d 7); PF, Propionibacterium freudenreichii; SH, Salmonella Heidelberg.
Growing Turkey (7-Week-Long) Study
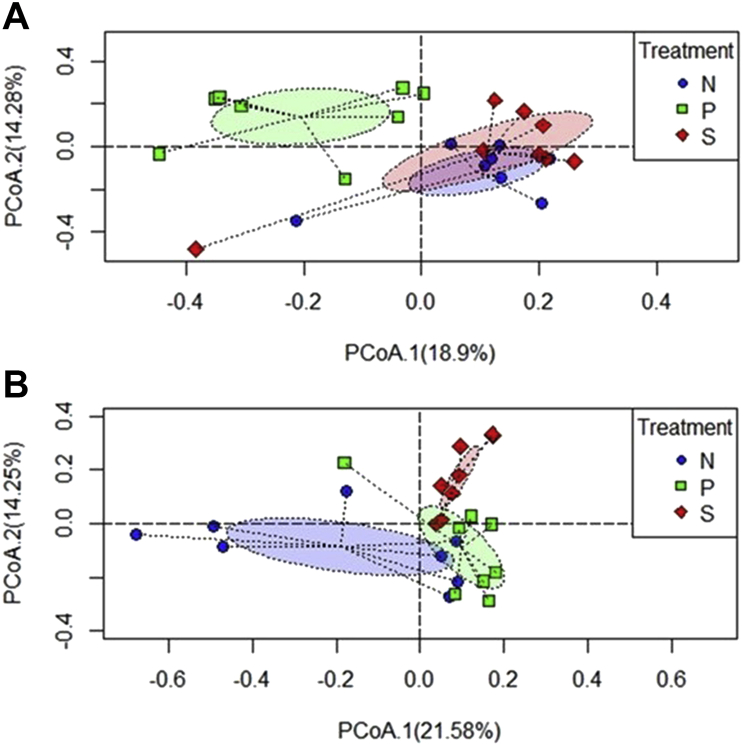
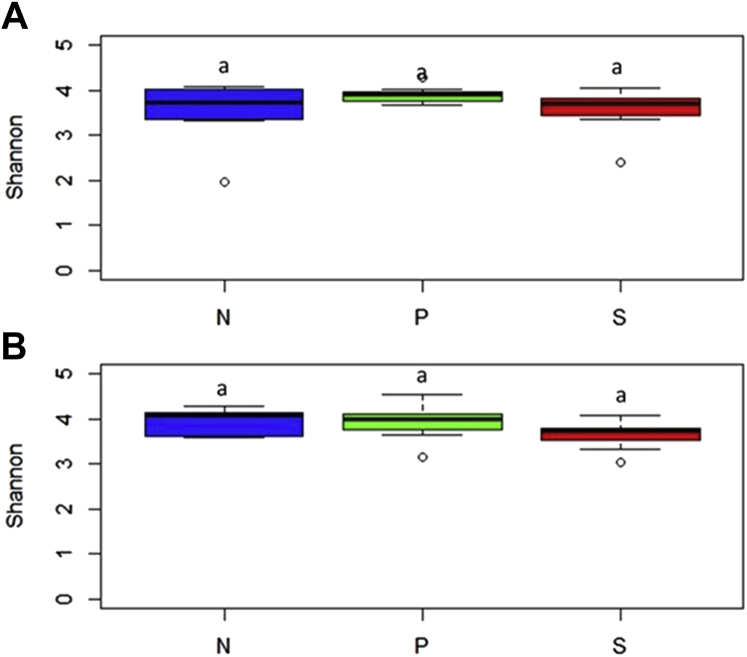
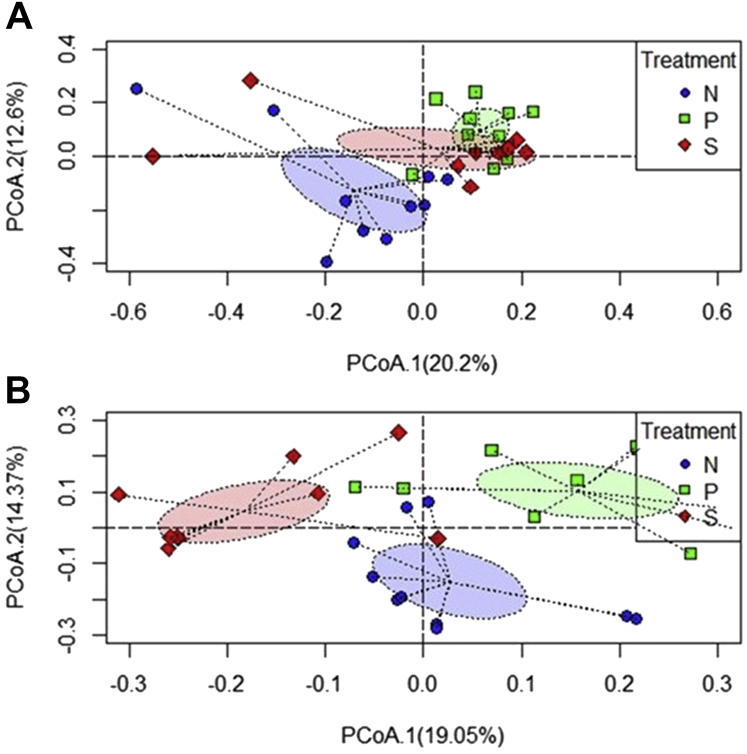
Shannon diversity index indicated that there was no significant difference in α diversity among the 3 treatment groups in the study after 2 d of SH challenge (P > 0.05) (Figure 4A; Supplementary Table 2). However, after 7 d of SH inoculation, an increase in α diversity was observed in the S group compared with the N group (P < 0.05) (Figure 4B; Supplementary Table 2). The beta diversity indices at 2 d after SH challenge indicated that treatments contributed 17.7% variability among the samples resulting in apparent clustering with overlaps between the N and S groups (P = 0.001) (Figure 5A). At 7 d after challenge, the clustering was more evident, and the treatment effects contributed to variability of 24.1% to the samples (P = 0.001) (Figure 5B).
Figure 4.
Alpha diversity of cecal microbiome of growing turkeys at 2 d (A) and 7 d (B) after inoculation represented as Shannon diversity index. From the day of hatch, the poults were allocated into N, P, and S groups for 7 wk; a,b indicate P < 0.05. Abbreviations: N, Negative control (turkeys without SH challenge or PF supplementation); P, test group (turkeys supplemented with PF for 7 wk and challenged with SH at wk 6); PF, Propionibacterium freudenreichii; S, SH control (turkeys challenged with SH at wk 6 and without PF supplementation); SH, Salmonella Heidelberg.
Figure 5.
Principal coordinates analysis showing microbial compositional difference among the cecal samples of growing turkeys at 2 d (A) and 7 d (B) after inoculation as determined by the beta diversity using Bray-Curtis dissimilarity index. From the day of hatch, the poults were allocated into N, P, and S groups for 7 wk. Abbreviations: N, Negative control (turkeys without SH challenge or PF supplementation); P, test group (turkeys supplemented with PF for 7 wk and challenged with SH at wk 6); PF, Propionibacterium freudenreichii; S, SH control (turkeys challenged with SH at wk 6 and without PF supplementation); SH, Salmonella Heidelberg.
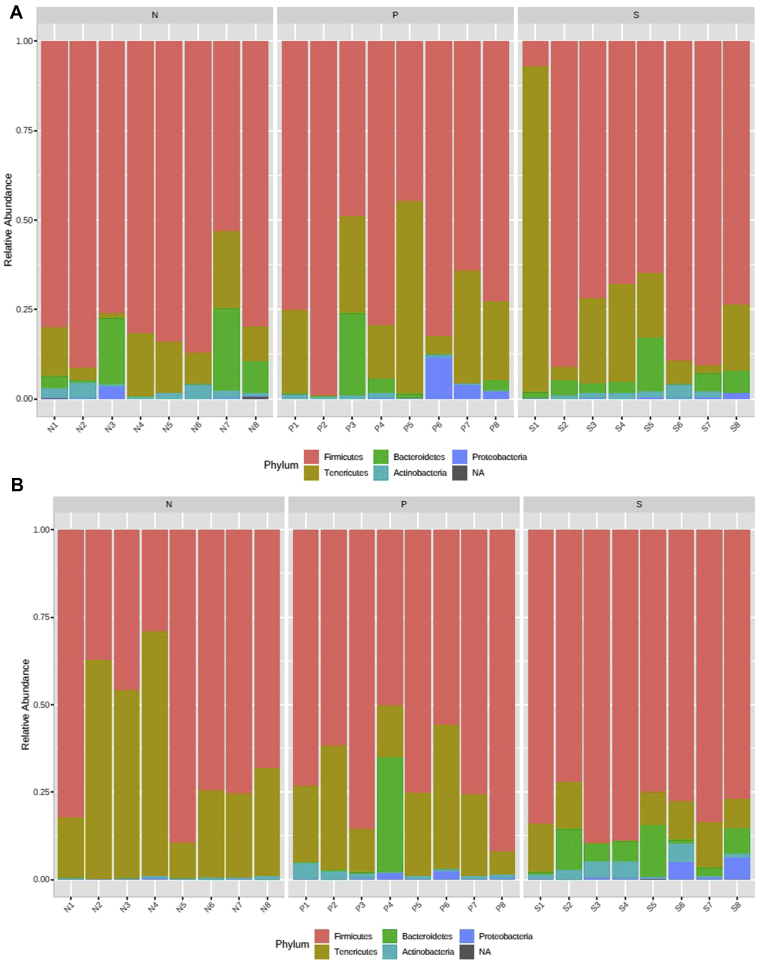
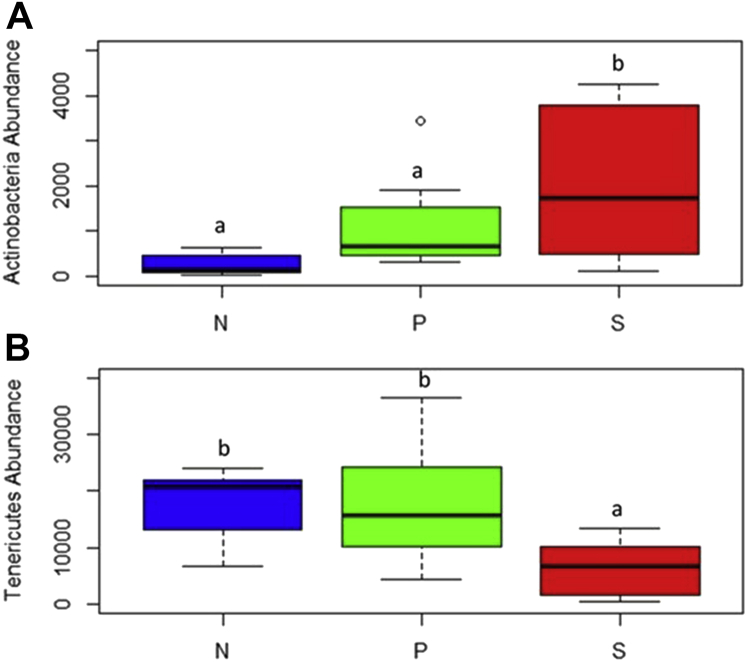
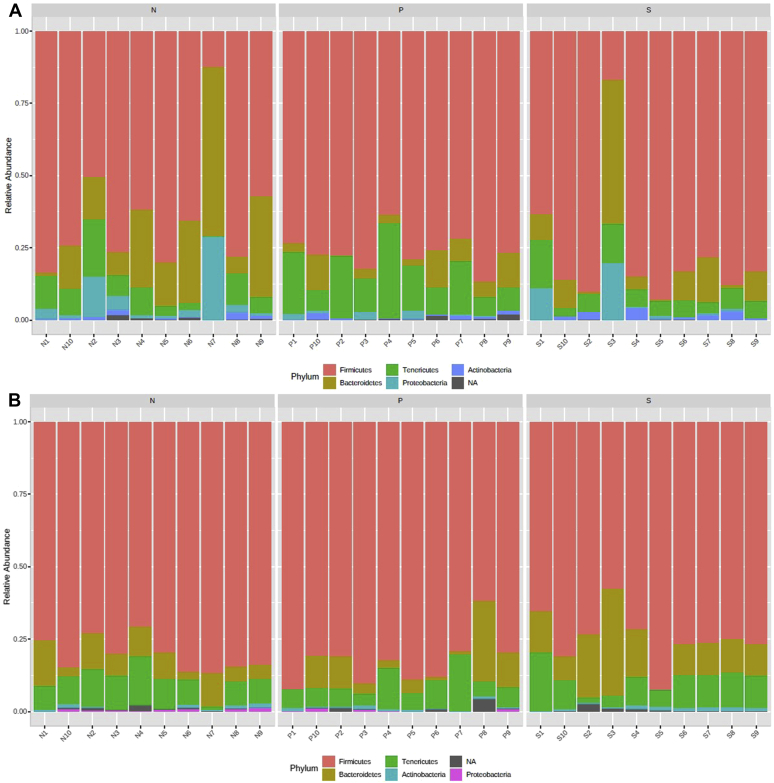
The top 5 abundant phyla at 2-d PI samples in growing turkeys were Firmicutes, Tenericutes, Bacteroidetes, Actinobacteria, and Proteobacteria with a relative abundance of 73, 19.2, 5.2, 1.5, and 1.1%, respectively (Figure 6A). In 7-d PI samples, the relative abundance of those major phyla was 71.6, 22.5, 3.5, 1.5, and 0.9%, respectively (Figure 6B). The treatments (SH challenge, or SH challenge with PF supplementation) did not affect the abundance of Firmicutes in the growing turkey cecum at 2 or 7 d after SH challenge (P > 0.05). The other phyla such as Tenericutes, Bacteroidetes, Actinobacteria, and Proteobacteria did not show any difference in abundance at 2 d after SH challenge when turkeys were inoculated with the pathogen on wk 6 (P > 0.05). However, in 7-d PI samples, the abundance of the phylum Actinobacteria was high in the S group compared with the N group (P = 0.015) (Figure 7A), whereas the distribution of Tenericutes was less in the S groups compared with the other 2 groups (P = 0.02) (Figure 7B).
Figure 6.
The relative abundance of major phyla in 2 d (A) and 7 d (B) after inoculation samples among different treatment groups in growing turkeys. From the day of hatch, the poults were allocated into N, P, and S groups for 7 wk. Abbreviations: N, Negative control (turkeys without SH challenge or PF supplementation); P, test group (turkeys supplemented with PF for 7 wk and challenged with SH at wk 6); PF, Propionibacterium freudenreichii; S, SH control (turkeys challenged with SH at wk 6 and without PF supplementation); SH, Salmonella Heidelberg.
Figure 7.
The distribution of Actinobacteria (A) and Tenericutes (B) in 7 d after inoculation samples among different treatment groups in growing turkeys. From the day of hatch, the poults were allocated into N, P, and S groups for 7 wk; a,b indicate P < 0.05. Abbreviations: N, Negative control (turkeys without SH challenge or PF supplementation); S, SH control (turkeys challenged with SH at wk 6 and without PF supplementation); P, test group (turkeys supplemented with PF for 7 wk and challenged with SH at wk 6); PF, Propionibacterium freudenreichii; S, SH control (turkeys challenged with SH at wk 6 and without PF supplementation); SH, Salmonella Heidelberg.
Furthermore, a potential indicator genus among the 2-d PI samples was explored. The relative abundance of Subdoligranulum was higher in the P group than in the N (P = 0.02) and S (P = 0.004) groups. Similarly, the relative abundance of genus Faecalibacterium was higher in the P group than in the N (P = 0.03) and S (P = 0.004) groups. However, the relative abundance of Streptococcus was lower in the P group than in the N (P = 0.001) and S (P = 0.037) groups. A higher relative abundance of Turicibacter was observed in the S group than in the N (P = 0.02) and P (P = 0.03) groups in growing turkeys at 2-d PI (Table 1).
Table 1.
Differentially expressed indicator genus in the P and S groups.
| Study | Differentially expressed genus in the P group compared with the S and N groups (P < 0.05) | Differentially expressed genus in the S group compared with the P and N groups (P < 0.05) |
|---|---|---|
| Growing turkey; 2-d PI |
|
|
| Growing turkey; 7-d PI | No differentially expressed indicator genus were identified. |
|
| Finishing turkey; 2-d PI |
|
|
| Finishing turkey; 7-d PI |
|
|
N, Negative control (turkeys without SH challenge or PF supplementation); S, SH control (turkeys challenged with SH without PF supplementation); P, (turkeys supplemented with PF and challenged with SH). 2-d PI and 7-d PI indicate 2- and 7-d postinoculation samples, respectively.
Abbreviations: PF, Propionibacterium freudenreichii; SH, Salmonella Heidelberg.
At 7-d PI, the relative abundance of Ruminococcaceae_UCG.014 was significantly higher in the N group than in the S group (P = 0.027) without a difference observed between the N and P groups (P > 0.05). The relative abundance of Erysipelatoclostridium was higher in the P group than in the N (P = 0.028) group. Similarly, a higher relative abundance of Romboutsia was also observed in the P group than in the S group (P = 0.01) at 7-d PI in growing turkeys.
Similarly, in 7-d PI samples, the relative abundance of Flavonifractor, Streptococcus, and Gordonibacter were significantly higher in the S group than the N and P groups (P < 0.05). Similar to the 2-d PI samples, a higher relative abundance of Turicibacter was observed in the S group than in the N and P groups (P < 0.05) (Table 1).
Finishing Turkey (12-Week-Long) Study
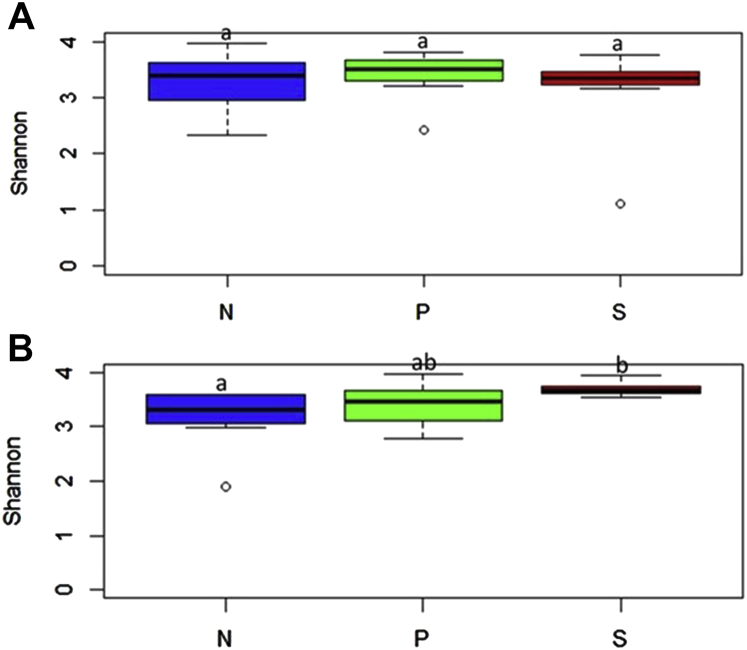
There was no difference in α diversity among the treatment groups 2- and 7-d after SH challenge (P < 0.05) (Figures 8A and 8B; Supplementary Table 2). However, at 7-d PI, the S group showed a trend of lower α diversity than the P (P = 0.08) and N (P = 0.07) groups (Figure 8B). The beta diversity determination 2 d after SH challenge revealed apparent clustering of samples based on the treatments with some overlap between the P and S groups (Figure 9A). The treatments contributed to a variability of 15.4% among the samples (P = 0.001). In 7 d after SH challenge, the clustering was more evident, and the treatments contributed to variability of 23.7% to the samples (P = 0.001) (Figure 9B).
Figure 8.
Alpha diversity of cecal microbiome of finishing turkeys at 2 d (A) and 7 d (B) after inoculation represented as Shannon diversity index. From the day of hatch, the poults were allocated into N, P, and S groups for 12 wk; a indicates P > 0.05. Abbreviations: N, Negative control (turkeys without SH challenge or PF supplementation); S, SH control (turkeys challenged with SH at wk 11 and without PF supplementation); P, test group (turkeys supplemented with PF for 12 wk and challenged with SH at wk 11); PF, Propionibacterium freudenreichii; S, SH control (turkeys challenged with SH at wk 6 and without PF supplementation); SH, Salmonella Heidelberg.
Figure 9.
Principal coordinates analysis showing microbial compositional difference among the cecal samples of finishing turkeys at 2 d (A) and 7 d (B) after inoculation as determined by the beta diversity using Bray-Curtis dissimilarity index. From the day of hatch, the poults were allocated into N, P, and S groups for 12 wk. Abbreviations: N, Negative control (turkeys without SH challenge or PF supplementation); P, test group (turkeys supplemented with PF for 12 wk and challenged with SH at wk 11); PF, Propionibacterium freudenreichii; S, SH control (turkeys challenged with SH at wk 11 and without PF supplementation); SH, Salmonella Heidelberg.
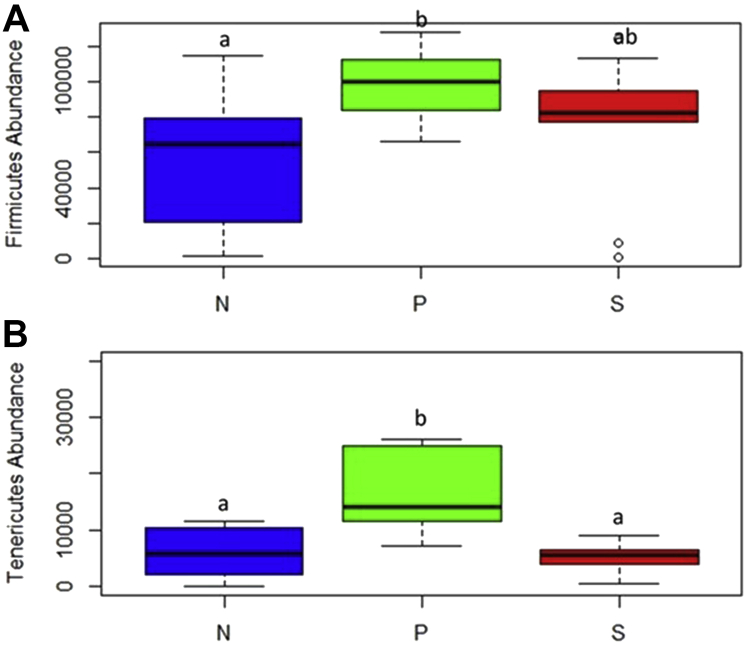
The top 5 abundant phyla at 2-d PI samples were Firmicutes, Bacteroidetes, Tenericutes, Proteobacteria, and Actinobacteria with a relative abundance of 72.2, 12.7, 10.3, 3.5, and 1.0%, respectively (Figure 10A). In 7-d PI samples, the relative abundance of Firmicutes, Bacteroidetes, Tenericutes, Actinobacteria, and Proteobacteria was 79.1, 10.0, 9.2, 0.75, and 0.31%, respectively (Figure 10B). The abundance of Firmicutes was significantly higher in the P group than in the N group (P = 0.02) at 2 d after SH challenge (Figure 11A). In addition, the distribution of the phylum Tenericutes was found to be elevated in the P group than in the N (P = 0.003) and S (P = 0.002) groups (Figure 11B).
Figure 10.
The relative abundance of major phyla at 2 d (A) and 7 d (B) after inoculation samples among different treatment groups in finishing turkeys. From the day of hatch, the poults were allocated into N, P, and S groups for 12 wk. Abbreviations: N, Negative control (turkeys without SH challenge or PF supplementation); P, test group (turkeys supplemented with PF for 12 wk and challenged with SH at wk 11); PF, Propionibacterium freudenreichii; S, SH control (turkeys challenged with SH at wk 11 and without PF supplementation); SH, Salmonella Heidelberg.
Figure 11.
The distribution of Firmicutes (A) and Tenericutes (B) in 7-d postinoculation samples among different treatment groups in finishing turkeys. From the day of hatch, the poults were allocated into N, P, and S groups for 12 wk; a,b indicate P < 0.05. Abbreviations: N, Negative control (turkeys without SH challenge or PF supplementation); P, test group (turkeys supplemented with PF for 12 wk and challenged with SH at wk 11); PF, Propionibacterium freudenreichii; S, SH control (turkeys challenged with SH at wk 11 and without PF supplementation); SH, Salmonella Heidelberg.
The genera in Firmicutes such as Ruminococcaceae_NK4A214_group, Ruminococcaceae_UCG.010, and Lactobacillus showed the higher distribution in the P group than in the N and S groups (P < 0.05) at 2 d after SH challenge in finishing turkeys. In addition, similar to growing turkeys, the genus Turicibacter showed higher relative abundance in the S group than in the N and P groups (P < 0.05) (Table 1).
The genera such as Leuconostoc, Erysipelatoclostridium, and Lactococcus showed higher relative abundance in the P group than in the N and S groups (P < 0.05) at 7-d PI in finishing turkeys. In addition, Butyricicoccus showed higher relative abundance in the P group than in the S group (P = 0.012). As previously detected, the relative abundance of Streptococcus and Turicibacter was high in the S group compared with the N and P groups (P < 0.05) (Table 1).
Discussion
In light of the Federal regulations on the use of antibiotics in the United States, including turkeys, effective alternative approaches for pathogen control are warranted. Although many alternatives have been tested to control foodborne Salmonella in poultry, very rarely, they have discussed the microbiome shifts and potential restoration of imbalanced microbiota in response to the pathogen challenge. In addition, studies focusing beyond the growth phase in turkeys (7 wk) are rare (Wilkinson et al., 2017). In the present study, we have focused on a dairy-origin allochthonous probiotic, PF, on its ability to restore the microbial balance after MDR SH challenge.
The 2-week-long turkey poult study was intended to understand the general trend in the compositional microbiome matrix and clustering as a result of the treatments. In this study, the Shannon diversity index was used for deciphering α diversity among the treatment groups 2 and 7 d after SH challenge in poults. The α diversity indices represent species richness and abundance in an ecosystem (Morgan and Huttenhower, 2012). We found that the treatments did not significantly change α diversity after 2 d of inoculation, indicating that an apparent effect on the evenness and richness of the microbial population in cecum due to SH inoculation was not an immediate event in turkeys despite their age difference. However, after 7 d of inoculation in the growing and finishing turkeys, a potential shift due to SH challenge was evident. Because a significant increase in the α diversity in the 7-week-old turkeys was observed, it can be speculated that the SH colonization over 7-d period might have altered the commensal network existed in the turkey cecum to accommodate the imminent pathogen challenge which was later restored by the supplementation of PF. Argüello et al. (2018) have reported that pathogens could result in apparent changes in the microbial flora. For example, Salmonella Typhimurium infection resulted in decreased abundance of multiple healthy commensal bacterial genus, including Bifidobacterium, Lactobacillus, and Ruminococcus in ileal mucosa of pigs. We also observed that the PF supplementation resulted in 1.0-1.3 log10 CFU/g reduction of MDR SH in the P group (Nair et al., 2019), indicating a potential beneficial microbial modulation in the P group to reduce the colonization of MDR SH. However, in 12-week-old turkeys, a decreasing trend in α diversity was observed (P = 0.08) in the S group with SH inoculation, potentially associated with an increased inoculum level (108 CFU/bird) used in the 12-week-old turkeys compared with their 7-week-old counterparts (106 CFU/bird). Despite the higher inoculation of MDR SH in P, the PF supplementation resulted in 1.7-2.3 log10 CFU/g reduction of MDR SH colonization in the cecum (Nair et al., 2018a) which might have resulted in a balanced microbiome in the P group similar to that of the N group. However, potential changes in the gut microbiome as a result of differences in SH inoculum levels needs more focused investigations.
Apparent clustering was observed with both 2- and 7-d postchallenge phases in poults, growing, and finishing turkeys as determined by the principal coordinate analysis plots, indicating that the treatments resulted in compositional changes in microbial population among the samples. Generally, at the phylum level, Firmicutes, Tenericutes, Bacteroidetes, Actinobacteria, and Proteobacteria dominated within the microbiomes of growing and finishing turkeys. This is in line with previously published research, although we observed more abundance for Firmicutes than Bacteroidetes in the turkey ceca in this study (Yeoman et al., 2012; Oakley et al., 2014; Pan and Yu, 2014; Shaufi et al., 2015; Thibodeau et al., 2015; Borda-Molina et al., 2016; Wilkinson et al., 2017; Wei et al., 2018). The phylum Firmicutes includes genera that are beneficial to the hosts and economically important such as Clostridium, Ruminococcus, and Lactobacillus which are the major short-chain fatty acid (SCFA) producers in the cecum (Wei et al., 2018).
The difference in the relative abundance of major phyla was not observed among the treatment groups at 2 d PI in growing turkeys (Figure 8A). However, in 7-d PI samples, the relative abundance of the phylum Actinobacteria was high in the S group compared with the N and P groups (P = 0.015), whereas the distribution of Tenericutes was less in the S group than in the other 2 groups (P = 0.02).
While exploring at the genus level, a higher relative abundance of Subdoligranulum and Faecalibacterium was observed in the P group than in the S and N groups at 2 d PI in growing turkeys. These genera are part of Firmicutes, found in the healthy chicken cecum, and the members are known as butyrate producers (Bjerrum et al., 2006; Eeckhart et al., 2011). Ruminococcaceae_UCG.014, a member of the Firmicutes, was abundant in the P and N groups, whereas a decrease in abundance was noticed in the S group at 7-d PI in growing turkeys. Ruminococcaceae_UCG.014 is also a butyrate producer in Ruminococcaceae family and is considered a part of a healthy microbiome in animals (Huang et al., 2017; Gao et al., 2017). In addition, this bacteria has a role in reducing high-fat diet–induced obesity in rats (Zhao et al., 2017) and the relative abundance of these genera was found to be decreased during intestinal dysfunction and inflammation in rats (Huang et al., 2017). Moreover, the higher abundance of Romboutsia was observed in the PF-supplemented groups. Romboutsia belongs to the phylum Firmicutes (Lu et al., 2017a) and is associated with healthy intestinal tract of turkeys, humans, and rats (Ricaboni et al., 2016; Lu et al., 2017b). The increased relative abundance of these genera could potentially be associated with reduced MDR SH colonization and restoration of impaired microbiota in the P group compared with the S group.
Furthermore, a higher relative abundance of Streptococcus was found in the SH control (S group) in growing turkeys. Although Streptococcus are normal inhabitants in the turkey cecum (Schulz et al., 2015; Wilkinson et al., 2017), they are opportunistic pathogens (Collins et al., 2002; Pan and Yu, 2014). The high relative abundance of Streptococcus in the S group could be due to the change in the commensal network associated with SH-induced changes in the turkey cecum (Stecher et al., 2007; Argüello et al., 2018). In addition, a higher abundance of Turicibacter in the S group would indicate a potential impairment in the gut homeostasis. Turicibacter has been previously isolated from inflammatory conditions associated with human ulcerative colitis, human appendicitis, and swine infections with S. Typhimurium and Lawsonia intracellularis infections. In addition, these genera have been reported to produce anti-inflammatory effects in mice (Bosshard et al., 2002; Falk et al., 2007; Werner et al., 2011; Borewicz et al., 2015). Gordonibacter was also identified from inflammatory bowel disease conditions in humans (Würdemann et al., 2009). The higher relative abundance of these genera in the SH challenge group could potentially indicate a resident inflammatory response to SH challenge. However, further research is warranted to specifically delineate the function of these intestinal genera as a result of pathogen challenge in turkeys.
As observed with growing turkeys, some of the beneficial genera in Firmicutes were significantly higher in abundance in the P groups in finishing turkeys at 2 d PI. Among the beneficial genera, Ruminococcaceae are cellulose and starch degraders which are associated with SCFA production (Duncan et al., 2007). Ruminococcaceae is generally present in the turkey gut. More specifically, Ruminococcaceae UCG-010 and Ruminococcaceae_UCG.014 are mucosa-attaching carbohydrate-utilizing bacteria which are associated with elevated SCFA production (Song et al., 2017). Ruminococcaceae_NK4A214_group is also health-related bacteria that ferment carbohydrates in digesta (Song et al., 2017). Lactobacillus in the phylum Firmicutes include beneficial bacteria and are found to be effective against enteric pathogens, including Salmonella (Pascual et al., 1999; Higgins et al., 2008; Kizerwetter-Świda and Binek, 2009). A higher abundance of Turicibacter in the S group was also observed, indicating a potentially impaired cecal microbiome as a result of SH inoculation in the absence of PF supplementation.
In finishing turkeys, at 7 d PI, a significantly higher abundance of Firmicutes and Tenericutes populations were observed in the PF-supplemented group (P group). As indicated above, these 2 are associated with the healthy gut of animals and have a significant role in apparent crude fiber digestibility (Niu et al., 2015). Higher abundances of Leuconostoc, Lactococcus, and Butyricoccus were observed in the P group at 7 d PI in finishing turkeys. Leuconostoc is a genus that is normally isolated from poultry and is known for their inhibitory activity against enteric pathogens such as Salmonella (Sorrells and Speck, 1970). In addition, Leuconostoc ferments carbohydrates and modulate other beneficial microorganisms such as Bifidobacterium and Lactobacillus (Chung, 1995). In addition, these butyrate-producing bacteria have a role in improving GI epithelial barrier function in humans and have shown promising results in inflammatory bowel syndrome (Eeckhaut et al., 2013; Jin et al., 2014). The relative abundance of Butyricicoccus was previously positively correlated with anti-inflammatory cytokine, interleukin-10 in the poultry cecum, that controls the inflammatory response associated pathogen infections in mammals (Oakley and Kogut, 2016; Wu et al., 2016). In the present study, the relative abundance of Erysipelatoclostridium genus was found to be increased when dairy-originated PF supplemented in turkeys and the finding is corroborated with the previous study where consumption of dairy-based foods such as milk, cheese, and yogurt was positively correlated with increased abundance of Erysipelatoclostridium in children (Smith-Brown et al., 2016). As previously observed with growing turkeys and finishing turkeys, Streptococcus and Turicibacter were present in the S groups.
Overall, the results indicated that PF supplementation maintained the ecological balance and diversity in the growing and finishing turkeys after MDR SH challenge. Continuous supplementation of PF in turkeys could also enrich carbohydrate-fermenting and SCFA-producing genera in the turkey ceca. These beneficial organisms could be playing significant roles in reducing Streptococcus, Gordonibacter, and Turicibacter, which are associated with an inflammatory response in birds and mammals. The results suggest that PF, an allochthonous dairy-origin probiotic bacterium, could be used as an antibiotic alternative to control MDR SH infections in turkeys by potentially modulating beneficial microbiota.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.09.091.
Supplementary data
References
- Argüello H., Estellé J., Zaldívar-López S., Jiménez-Marín Á., Carvajal A., López-Bascón M.A., Crispie F., O’Sullivan O., Cotter P.D., Priego-Capote F., Morera L., Garrido J.J. Early Salmonella Typhimurium infection in pigs disrupts microbiome composition and functionality principally at the ileum mucosa. Sci. Rep. 2018;8:7788. doi: 10.1038/s41598-018-26083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beery J.T., Hugdahl M.B., Doyle M.P. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 1988;54:2365–2370. doi: 10.1128/aem.54.10.2365-2370.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerrum L., Engberg R.M., Leser T.D., Jensen B.B., Finster K., Pedersen K. Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture-based techniques. Poult. Sci. 2006;85:1151–1164. doi: 10.1093/ps/85.7.1151. [DOI] [PubMed] [Google Scholar]
- Borda-Molina D., Vital M., Sommerfeld V., Rodehutscord M., Camarinha-Silva A. Insights into broilers’ gut microbiota fed with phosphorus, calcium, and phytase supplemented diets. Front. Microbiol. 2016;7:2033. doi: 10.3389/fmicb.2016.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borewicz K.A., Kim H.B., Singer R.S., Gebhart C.J., Sreevatsan S., Johnson T., Isaacson R.E. Changes in the porcine intestinal microbiome in response to infection with Salmonella enterica and Lawsonia intracellularis. PLoS One. 2015;10:e0139106. doi: 10.1371/journal.pone.0139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosshard P.P., Altwegg M., Zbinden R. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 2002;52:1263–1266. doi: 10.1099/00207713-52-4-1263. [DOI] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway T.R., Edrington T.S., Anderson R.C., Harvey R.B., Genovese K.J., Kennedy C.N., Venn D.W., Nisbet D.J. Probiotics, prebiotics and competitive exclusion for prophylaxis against bacterial disease. Anim. Heal. Res. Rev. 2008;9:217–225. doi: 10.1017/S1466252308001540. [DOI] [PubMed] [Google Scholar]
- Chung C.-H. A Potential Nutraceutical from Leuconostoc mesenteroides B-742 (ATCC 13146); Production and Properties. 1995. http://digitalcommons.lsu.edu/gradschool_dissertations
- Collins M.D., Hutson R.A., Falsen E., Ingana S E., Bisgaard M. Streptococcus gallinaceus sp. nov., from chickens. Int. J. Syst. Evol. Microbiol. 2002;52:1161–1164. doi: 10.1099/00207713-52-4-1161. [DOI] [PubMed] [Google Scholar]
- Duke G.E., Dziuk H.E., Hawkins L. Gastrointestinal transit-times in normal and bluecomb diseased turkeys. Poult. Sci. 1969;48:835–842. doi: 10.3382/ps.0480835. [DOI] [PubMed] [Google Scholar]
- Duncan S.H., Louis P., Flint H.J. Cultivable bacterial diversity from the human colon. Lett. Appl. Microbiol. 2007;44:343–350. doi: 10.1111/j.1472-765X.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- Eeckhaut V., Machiels K., Perrier C., Romero C., Maes S., Flahou B., Steppe M., Haesebrouck F., Sas B., Ducatelle R., Vermeire S., Van Immerseel F. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut. 2013;62:1745–1752. doi: 10.1136/gutjnl-2012-303611. [DOI] [PubMed] [Google Scholar]
- Eeckhaut V., Van Immerseel F., Croubels S., De Baere S., Haesebrouck F., Ducatelle R., Vandamme P. Butyrate production in phylogenetically diverse Firmicutes isolated from the chicken caecum. Microb. Biotechnol. 2011;4:503–512. doi: 10.1111/j.1751-7915.2010.00244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A., Olsson C., Ahrné S., Molin G., Adawi D., Jeppsson B. Ileal pelvic pouch microbiota from two former ulcerative colitis patients, analysed by DNA-based methods, were unstable over time and showed the presence of Clostridium perfringens. Scand. J. Gastroenterol. 2007;42:973–985. doi: 10.1080/00365520701204238. [DOI] [PubMed] [Google Scholar]
- Gao R., Zhu C., Li H., Yin M., Pan C., Huang L., Kong C., Wang X., Zhang Y., Qu S., Qin H. Dysbiosis signatures of gut microbiota along the sequence from healthy, young patients to those with overweight and obesity. Obes. 2017;26:351–361. doi: 10.1002/oby.22088. [DOI] [PubMed] [Google Scholar]
- Gohl D.M., Vangay P., Garbe J., MacLean A., Hauge A., Becker A., Gould T., Clayton J.B., Johnson T.J., Hunter R., Knights D., Beckman K.B. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016;34:942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- Higgins S.E., Higgins J.P., Wolfenden A.D., Henderson S.N., Torres-Rodriguez A., Tellez G., Hargis B. Evaluation of a Lactobacillus-based probiotic culture for the reduction of Salmonella Enteritidis in neonatal broiler chicks. Poult. Sci. 2008;87:27–31. doi: 10.3382/ps.2007-00210. [DOI] [PubMed] [Google Scholar]
- Hinton A., Corrier D.E., Spates G.E., Norman J.O., Ziprin R.L., Beier R.C., DeLoach J.R., DeLoach J.R. Biological control of Salmonella Typhimurium in young chickens. Avian Dis. 1990;34:626. [PubMed] [Google Scholar]
- Huang C., Chen J., Wang J., Zhou H., Lu Y., Lou L., Zheng J., Tian L., Wang X., Cao Z., Zeng Y. Dysbiosis of intestinal microbiota and decreased antimicrobial peptide level in Paneth cells during hypertriglyceridemia-related acute necrotizing pancreatitis in rats. Front. Microbiol. 2017;8:776. doi: 10.3389/fmicb.2017.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R.J. Relationship between digesta transit time and apparent metabolisable energy value of wheat in chickens. Br. Poult. Sci. 2008;49:716–720. doi: 10.1080/00071660802449145. [DOI] [PubMed] [Google Scholar]
- Jin D., Zhang H., Sun J. Manipulation of microbiome, a promising therapy for inflammatory bowel diseases. J. Clin. Cell Immunol. 2014;5:234. [Google Scholar]
- Johnson T.J., Youmans B.P., Noll S., Cardona C., Evans N.P., Karnezos T.P., Ngunjiri J.M., Abundo M.C., Lee C. A consistent and predictable commercial broiler chicken bacterial microbiota in antibiotic-free production displays strong correlations with performance. Appl. Environ. Microbiol. 2018;84:e00362-18. doi: 10.1128/AEM.00362-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizerwetter-Świda M., Binek M. Protective effect of potentially probiotic Lactobacillus strain on infection with pathogenic bacteria in chickens. Pol. J. Vet. Sci. 2009;12:15–20. [PubMed] [Google Scholar]
- La Ragione R.M., Woodward M.J. Competitive exclusion by Bacillus subtilis spores of Salmonella enterica serotype Enteritidis and Clostridium perfringens in young chickens. Vet. Microbiol. 2003;94:245–256. doi: 10.1016/s0378-1135(03)00077-4. [DOI] [PubMed] [Google Scholar]
- Lu C., Sun T., Li Y., Zhang D., Zhou J., Su X. Microbial diversity and composition in different gut locations of hyperlipidemic mice receiving krill oil. Appl. Microbiol. Biotechnol. 2017;102:355–366. doi: 10.1007/s00253-017-8601-1. [DOI] [PubMed] [Google Scholar]
- Lu C., Sun T., Li Y., Zhang D., Zhou J., Su X. Modulation of the gut microbiota by krill oil in mice fed a high-sugar high-fat diet. Front. Microbiol. 2017;8:905. doi: 10.3389/fmicb.2017.00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menconi A., Wolfenden A.D., Shivaramaiah S., Terraes J.C., Urbano T., Kuttel J., Kremer C., Hargis B.M., Tellez G. Effect of lactic acid bacteria probiotic culture for the treatment of Salmonella enterica serovar Heidelberg in neonatal broiler chickens and Turkey poults. Poult. Sci. 2011;90:561–565. doi: 10.3382/ps.2010-01220. [DOI] [PubMed] [Google Scholar]
- Morgan X.C., Huttenhower C. Chapter 12: human microbiome analysis. Plos Comput. Biol. 2012;8:e1002808. doi: 10.1371/journal.pcbi.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D.V.T., Kollanoor Johny A. Characterizing the antimicrobial function of a dairy-originated probiotic, Propionibacterium freudenreichii, against multidrug-resistant Salmonella enterica serovar Heidelberg in Turkey poults. Front. Microbiol. 2018;9:1475. doi: 10.3389/fmicb.2018.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D.V.T., Vazhakkattu Thomas J., Dewi G., Langlie J., Kollanoor Johny A. Effect of a Dairy – Origin Probiotic Bacterium, Propionibacterium freudenreichii Subsp. Freudenreichii NRRL 3523 against Multidrug-Resistant Salmonella enterica Serovar Heidelberg in Turkeys. 2018. International Association for Food Protection annual meeting abstract. Accessed Jul. 2020. 2018. https://iafp.confex.com/iafp/2018/meetingapp.cgi/Paper/18780
- Nair D.V.T., Vazhakkattu Thomas J., Noll S., Porter R., Kollanoor Johny A. Effect of various inoculum levels of multidrug-resistant Salmonella enterica serovar Heidelberg (2011 ground Turkey outbreak isolate) on cecal colonization, dissemination to internal organs, and deposition in skeletal muscles of commercial turkeys after experimental oral challenge. Front. Microbiol. 2018;8:2680. doi: 10.3389/fmicb.2017.02680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D.V.T., Vazhakkattu Thomas J., Dewi G., Noll S., Brannon J., Kollanoor Johny A. Reduction of multidrug-resistant Salmonella enterica serovar Heidelberg using a dairy-originated probiotic bacterium, Propionibacterium freudenreichii freudenreichii B3523, in growing turkeys. J. Appl. Poult. Res. 2019;28:356–363. [Google Scholar]
- Nair D.V.T., Vazhakkattu Thomas J., Dewi G., Brannon J., Noll S.L., Johnson T.J., Cox R.B., Kollanoor Johny A. Propionibacterium freudenreichii freudenreichii B3523 reduces cecal colonization and internal organ dissemination of multidrug-resistant Salmonella Heidelberg in Finishing Turkeys. J. Appl. Poult. Res. 2020 (in press) [Google Scholar]
- Niu Q., Li P., Hao S., Zhang Y., Kim S.W., Li H., Ma X., Gao S., He L., Wu W., Huang X., Hua J., Zhou B., Huang R. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 2015;5:9938. doi: 10.1038/srep09938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B.B., Kogut M.H. Spatial and temporal changes in the broiler chicken cecal and fecal microbiomes and correlations of bacterial taxa with cytokine gene expression. Front. Vet. Sci. 2016;3:11. doi: 10.3389/fvets.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B.B., Buhr R.J., Ritz C.W., Kiepper B.H., Berrang M.E., Seal B.S., Cox N.A. Successional changes in the chicken cecal microbiome during 42 days of growth are independent of organic acid feed additives. BMC Vet. Res. 2014;10:282. doi: 10.1186/s12917-014-0282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Yu Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microb. 2014;5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M., Hugas M., Badiola J.I., Monfort J.M., Garriga M. Lactobacillus salivarius CTC2197 prevents Salmonella Enteritidis colonization in chickens. Appl. Environ. Microbiol. 1999;65:4981–4986. doi: 10.1128/aem.65.11.4981-4986.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis F., Soares-Castro P., Costa D., Tavares R.M., Baptista P., Santos P.M., Lino-Neto T. Climatic impacts on the bacterial community profiles of cork oak soils. Appl. Soil Ecol. 2019;143:89–97. [Google Scholar]
- Ricaboni D., Mailhe M., Khelaifia S., Raoult D., Million M. Romboutsia timonensis, a new species isolated from human gut. New Microb. New Infect. 2016;12:6–7. doi: 10.1016/j.nmni.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanitro J.P., Fairchilds I.G., Zgornicki Y.D. Isolation, culture characteristics, and identification of anaerobic bacteria from the chicken cecum. Appl. Microbiol. 1974;27:678–687. doi: 10.1128/am.27.4.678-687.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz J., Dumke J., Hinse D., Dreier J., Habig C., Kemper N. Organic Turkey flocks: a reservoir of Streptococcus gallolyticus subspecies gallolyticus. PLoS One. 2015;10:e0144412. doi: 10.1371/journal.pone.0144412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaufi M., Sieo C., Chong C., Gan H., Ho Y. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog. 2015;7:4. doi: 10.1186/s13099-015-0051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Brown P., Morrison M., Krause L., Davies P.S.W. Dairy and plant based food intakes are associated with altered faecal microbiota in 2 to 3 year old Australian children. Sci. Rep. 2016;6:32385. doi: 10.1038/srep32385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Malmuthuge N., Steele M.A., Guan L.L. Shift of hindgut microbiota and microbial short chain fatty acids profiles in dairy calves from birth to pre-weaning. FEMS Microbiol. Ecol. 2017;94:fix179. doi: 10.1093/femsec/fix179. [DOI] [PubMed] [Google Scholar]
- Sorrells K.M., Speck M.L. Inhibition of Salmonella gallinarum by culture filtrates of Leuconostoc citrovorum. J. Dairy Sci. 1970;53:239–241. doi: 10.3168/jds.S0022-0302(70)86186-0. [DOI] [PubMed] [Google Scholar]
- Stecher B., Robbiani R., Walker A.W., Westendorf A.M., Barthel M., Kremer M., Chaffron S., Macpherson A.J., Buer J., Parkhill J., Dougan G., von Mering C., Hardt W.-D. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. Plos Biol. 2007;5:e244. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbington R., Amael P., Judy E., Amélie B., Agnès B., Frédéric R., Sánchez-Montoya M.M., Westwood C.G., Datry T. A comparison of biotic groups as dry-phase indicators of ecological quality in intermittent rivers and ephemeral streams. Ecol. Indic. 2019;97:165–174. [Google Scholar]
- Svihus B. Function of the digestive system. J. Appl. Poult. Res. 2014;23:306–314. [Google Scholar]
- Thibodeau A., Fravalo P., Yergeau É., Arsenault J., Lahaye L., Letellier A. Chicken caecal microbiome modifications induced by Campylobacter jejuni colonization and by a non-antibiotic feed additive. PLoS One. 2015;10:e0131978. doi: 10.1371/journal.pone.0131978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-C., Hsih H.-Y., Chiu H.-H., Lai Y.-Y., Liu J.-H., Yu B., Tsen H.-Y. Antagonistic activity against Salmonella infection in vitro and in vivo for two Lactobacillus strains from swine and poultry. Int. J. Food Microbiol. 2005;102:185–194. doi: 10.1016/j.ijfoodmicro.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Vasudevan P., Marek P., Nair M.K.M., Annamalai T., Darre M., Khan M., Venkitanarayanan K. In vitro inactivation of Salmonella Enteritidis in autoclaved chicken cecal contents by caprylic acid. J. Appl. Poult. Res. 2005;14:122–125. [Google Scholar]
- Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2018;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- Werner T., Wagner S.J., Martínez I., Walter J., Chang J.-S., Clavel T., Kisling S., Schuemann K., Haller D. Depletion of luminal iron alters the gut microbiota and prevents Crohn’s disease-like ileitis. Gut. 2011;60:325–333. doi: 10.1136/gut.2010.216929. [DOI] [PubMed] [Google Scholar]
- Wilkinson T.J., Cowan A.A., Vallin H.E., Onime L.A., Oyama L.B., Cameron S.J., Gonot C., Moorby J.M., Waddams K., Theobald V.J., Leemans D., Bowra S., Nixey C., Huws S.A. Characterization of the microbiome along the gastrointestinal tract of growing turkeys. Front. Microbiol. 2017;8:1089. doi: 10.3389/fmicb.2017.01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Hu T., Rothwell L., Vervelde L., Kaiser P., Boulton K., Nolan M.J., Tomley F.M., Blake D.P., Hume D.A. Analysis of the function of IL-10 in chickens using specific neutralizing antibodies and a sensitive capture ELISA. Dev. Comp. Immunol. 2016;63:206–212. doi: 10.1016/j.dci.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würdemann D., Tindall B.J., Pukall R., Lünsdorf H., Strömpl C., Namuth T., Nahrstedt H., Wos-Oxley M., Ott S., Schreiber S., Timmis K.N., Oxley A.P.A. Gordonibacter pamelaeae gen. nov., sp. nov., a new member of the Coriobacteriaceae isolated from a patient with Crohn’s disease, and reclassification of Eggerthella hongkongensis Lau et al. 2006 as Paraeggerthella hongkongensis gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2009;59:1405–1415. doi: 10.1099/ijs.0.005900-0. [DOI] [PubMed] [Google Scholar]
- Yang Y., Latorre J.D., Khatri B., Kwon Y.M., Kong B.W., Teague K.D., Graham L.E., Wolfenden A.D., Mahaffey B.D., Baxter M., Hernandez-Velasco X., Merino-Guzman R., Hargis B.M., Tellez G. Characterization and evaluation of lactic acid bacteria candidates for intestinal epithelial permeability and Salmonella Typhimurium colonization in neonatal Turkey poults. Poult. Sci. 2018;97:515–521. doi: 10.3382/ps/pex311. [DOI] [PubMed] [Google Scholar]
- Yeoman C.J., Chia N., Jeraldo P., Sipos M., Goldenfeld N.D., White B.A. The microbiome of the chicken gastrointestinal tract. Anim. Heal. Res. Rev. 2012;13:89–99. doi: 10.1017/S1466252312000138. [DOI] [PubMed] [Google Scholar]
- Zhao L., Zhang Q., Ma W., Tian F., Shen H., Zhou M. A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 2017;8:4644–4656. doi: 10.1039/c7fo01383c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.