Abstract
Although often considered a single-entity, chronic kidney disease (CKD) comprises many pathophysiologically distinct disorders that result in persistently abnormal kidney structure and/or function, and encompass both monogenic and polygenic aetiologies. Rare inherited forms of CKD frequently span diverse phenotypes, reflecting genetic phenomena including pleiotropy, incomplete penetrance and variable expressivity. Use of chromosomal microarray and massively parallel sequencing technologies has revealed that genomic disorders and monogenic aetiologies contribute meaningfully to seemingly complex forms of CKD across different clinically defined subgroups and are characterized by high genetic and phenotypic heterogeneity. Investigations of prevalent genomic disorders in CKD have integrated genetic, bioinformatic and functional studies to pinpoint the genetic drivers underlying their renal and extra-renal manifestations, revealing both monogenic and polygenic mechanisms. Similarly, massively parallel sequencing-based analyses have identified gene- and allele-level variation that contribute to the clinically diverse phenotypes observed for many monogenic forms of nephropathy. Genome-wide sequencing studies suggest that dual genetic diagnoses are found in at least 5% of patients in whom a genetic cause of disease is identified, highlighting the fact that complex phenotypes can also arise from multilocus variation. A multifaceted approach that incorporates genetic and phenotypic data from large, diverse cohorts will help to elucidate the complex relationships between genotype and phenotype for different forms of CKD, supporting personalized medicine for individuals with kidney disease.
Chronic kidney disease (CKD) is a broad term that encompasses many complex disorders that collectively affect over 1 in 10 individuals worldwide, resulting in substantial morbidity and mortality and high health-care costs1. Despite the heavy economic and societal burden of CKD, its pathogenesis remains incompletely understood, impeding specific diagnosis and targeted therapy1. In addition to known Mendelian nephropathies2, multiple lines of evidence support a broader genetic contribution to the pathogenesis of CKD. Between 10 and 29% of adults with chronic kidney failure note a positive family history for nephropathy across different ethnicities and aetiologies3-5. Furthermore, glomerular filtration rate has a heritability of approximately 30–60% in the general population6-8, and other indices of kidney function, such as albuminuria and electrolyte excretion, show similarly significant heritability9-11.
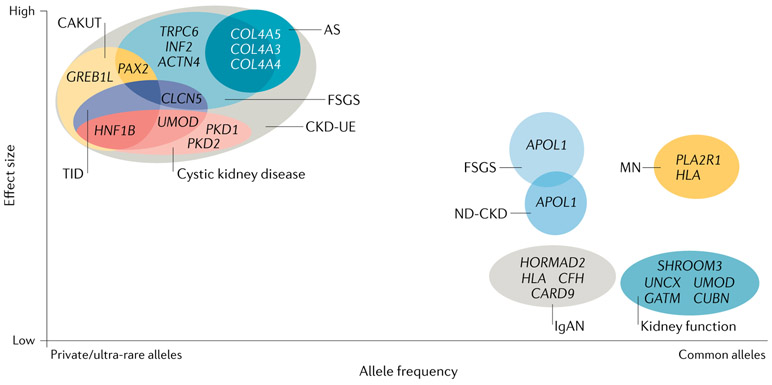
Prior research has shown that most common diseases result from a combination of genetic and environmental factors and have more complex genetic architectures than originally assumed. For example, investigations of epilepsy12,13, autism14,15 and coronary artery disease16,17 have demonstrated that these complex conditions encompass many distinct disorders, including both monogenic and polygenic subtypes. In addition, these studies have shown that, contrary to earlier assumptions, genetic variants spanning a wide spectrum of allele frequencies mediate an individual’s risk of developing complex disorders18,19. Typically, the effect of a variant on disease risk is inversely related to its frequency in the general population (FIG. 1). Thus, monogenic forms of disease are usually caused by rare or even private variants in a single gene that have a large effect size, whereas modifier alleles exhibit intermediate allele frequencies and more modest effect sizes. Polygenic subtypes of kidney disease usually reflect the collective contribution of multiple common variants, each of which has a small effect on disease risk; however, investigations of polygenic risk scores have shown that individuals who have many such common variants can have a similar disease risk to individuals with a monogenic form of the condition16 (BOX 1). Moreover, in some cases, relatively common variants have been shown to each have a large impact on disease risk, as illustrated by the association of APOL1 risk variants with various forms of non-diabetic CKD20,21 and the association of risk alleles at the HLA and PLA2R1 loci with idiopathic membranous nephropathy22,23.
Fig. 1 ∣. Chronic kidney disease as a complex disease.
Chronic kidney disease (CKD) is frequently considered to be a single disease entity, defined by persistent abnormalities of kidney structure and/or function26. However, these phenomena can result from many genetically distinct aetiologies, with disease risk shaped by the contributions of variants of varying frequencies and effect sizes. Causal variants for CKD include rare or even private variants with large effect sizes, which are generally associated with monogenic CKD subtypes, and more common alleles, which have been associated both with clinically distinct CKD subtypes, such as focal segmental glomerulosclerosis (FSGS), IgA nephropathy (IgAN), and membranous nephropathy (MN), and with kidney function traits, such as estimated glomerular filtration rate, serum creatinine level and albuminuria213. Although common alleles typically have small individual effect sizes, certain common alleles have a greater impact on disease risk, including APOL1 variants in FSGS and non-diabetic CKD (ND-CKD)20,21 and HLA and PLA2R1 variants in MN22. Moreover, variants within a single gene can have a role in both monogenic and polygenic forms of nephropathy. For example, rare variants in UMOD result in the monogenic nephropathy of autosomal-dominant tubulointerstitial kidney disease, whereas more common non-coding UMOD variants are associated with kidney function traits, such as estimated glomerular filtration rate and serum creatinine level46. In addition, the high genetic heterogeneity of CKD means that different single-gene mutations can yield the same clinical disease subtype as defined by clinical symptomatology and investigations of kidney function, imaging and/or histopathology. Conversely, the high phenotypic heterogeneity of CKD can also lead to mutations in the same gene, producing clinically distinct CKD subtypes. For example, the clinical disease entity of FSGS can result from mutations in any one of multiple different genes, including TRPC6, INF2, PAX2 and CLCN5. In many cases, mutations in these genes can also yield other clinical subtypes of CKD; for example, PAX2 mutations can cause congenital anomalies of the kidney and urinary tract (CAKUT) and CLCN5 mutations can cause tubulointerstitial disease (TID). Conversely, mutations in a single gene can produce a variety of CKD phenotypes. For instance, individuals with variants in HNF1B can present with CAKUT, TID or cystic kidney disease. As a result of such high phenotypic heterogeneity, many monogenic nephropathies can present as CKD of unknown aetiology (CKD-UE). Together, this genetic and phenotypic variability supports the notion that, as posited for other common diseases19, CKD might instead represent a wide array of rarer disorders, each of which has its own distinct genetic architecture.
Box 1 ∣. Opportunities for precision nephrology through polygenic risk scores and phenotypic risk scores.
The clinically non-specific and heterogeneous nature of many forms of chronic kidney disease (CKD) can make early diagnosis challenging. Moreover, such phenotypic heterogeneity can make it difficult to link genotype to phenotype, thereby impeding accurate prognosis and targeted management. Several findings point to the potential of genome-wide polygenic risk scores (GPS) and phenotypic risk scores (PheRS) to address these challenges and improve understanding of genotype–phenotype relationships in nephrology, supporting precision medicine for patients with kidney disease.
GPS represent a weighted sum of the risk alleles harboured by an individual for a given condition, with the weight assigned to a given allele corresponding to estimates of its effect on disease risk from prior genome-wide association studies (GWAS)214. These scores are then refined by assessing their performance in predicting the relative risk of the disease of interest in a second (validation) cohort. Typically, such common risk alleles each account for a small fraction of total disease risk, limiting their predictive value in clinical practice. However, when aggregated into a GPS, they can detect individuals at risk of a variety of complex disorders, including coronary artery disease, type 2 diabetes mellitus and breast cancer16. Importantly, disease risk for individuals at the uppermost GPS percentiles (first and second percentiles) was found to be comparable with that of individuals harbouring causal mutations for rare monogenic forms of the condition. Moreover, as GPS are derived from common variants, they have the potential to identify a greater proportion of individuals at risk of CKD than does screening for monogenic forms alone and could also help to explain the variable penetrance and expressivity observed for many monogenic nephropathies. However, owing to the great aetiological heterogeneity of CKD, the risk alleles thus far identified by GWAS explain a miniscule fraction of its heritability and are also often unique to ethnic subpopulations, both of which make it difficult to create a GPS that is sufficiently robust and broadly generalizable for use in clinical practice215. By focusing GWAS on specific (and thus more aetiologically homogeneous) forms of nephropathy in studies that include individuals of diverse ancestries, this challenge may be surmounted. In addition to being sensitive to ancestry, which limits their applicability across different ethnicities216, the performance of GPS might be impacted by other factors, including age, gender and socioeconomic status217. Thus, further research and integration of additional determinants of disease susceptibility, including demographic and lifestyle factors, into GPS may further augment their value, enabling prediction of absolute versus relative risk, and thereby supporting early disease detection and robust risk stratification in the greater CKD patient population.
As many monogenic nephropathies can manifest with clinically non-specific or ambiguous presentations, identifying affected individuals can be difficult. A 2018 study212 identified PheRS as a promising means of surmounting this diagnostic challenge. In that study, researchers mapped diagnostic codes from individuals’ electronic health records (EHRs) to the phenotypic manifestations of various monogenic disorders. Using these data, they constructed PheRS for these disorders, which was defined as a weighted summary of various traits associated with a given condition, with each trait weighted according to its specificity for that condition (that is, its prevalence among affected versus unaffected individuals). Individuals with high PheRS were found to harbour rare, putatively pathogenic variants in genes for the associated monogenic disorders, which included multiple monogenic causes of CKD, such as DGKE-associated nephrotic syndrome and AGXT-associated primary hyperoxaluria. Importantly, review of these individuals’ EHRs demonstrated that, despite their consistent clinical features, the majority of individuals were not clinically recognized to have the associated monogenic disorder. Together, these findings illustrate the potential to empower diagnosis of rare conditions through the leverage of genomic sequence and EHR data, supporting early recognition and targeted management for individuals who may otherwise have gone undiagnosed.
The spectrum of CKD consists of many clinically distinct disorders, including congenital anomalies of the kidney and urinary tract (CAKUT), diabetic kidney disease, hypertensive nephropathy and glomerulopathies, such as IgA nephropathy and focal segmental glomerulosclerosis (FSGS). Many of these disorders can arise from genetic aetiologies, but they can also occur from secondary factors, including metabolic disease, immune dysfunction, infection and/or toxic injury. Moreover, the mode of inheritance and underlying molecular mechanisms vary considerably within many CKD subtypes2,24,25. In epidemiological studies, however, CKD is often considered to be a single disease entity, defined as persistent abnormalities of kidney structure and/or function26. Using this definition, genome-wide association studies (GWAS) in large population cohorts have detected numerous loci associated with CKD and/or variation in kidney function; however, these loci explain a minority of the overall heritability of these traits8,27,28.
In this Review, we describe rare genetic causes of CKD and the insights that they offer into the genetic and phenotypic complexity of this group of disorders. We examine the phenotypic variability of recurrent hereditary nephropathies, including genomic disorders and monogenic aetiologies, and their underlying genetic mechanisms. To gain further understanding of the complexity of these diseases, we explore the phenotypes that result from pathogenic variants at multiple loci. Finally, we discuss novel approaches to help to address the challenges posed by the complexity of CKD, such as polygenic risk scores and phenotypic risk scores, which support a precision medicine approach for individuals with kidney disease.
CKD as a complex disease
Complex disorders are defined as conditions that have a strong heritable component, yet show considerable phenotypic heterogeneity owing to genetic and environmental modifiers19,29. Thus, although CKD and features of renal function show substantial heritability, the phenotypes associated with rarer genetic forms of disease can be highly variable and may overlap, which can complicate diagnosis based on clinical symptomatology alone. For example, patients with nephronophthisis-related ciliopathies and those with monogenic renal tubulopathies can both present with enhanced renal echogenicity30,31, and those with DGKE-associated atypical haemolytic uraemic syndrome, type IV collagen-associated nephropathy and Dent disease 2 caused by OCRL mutations can all present with steroid-resistant nephrotic syndrome (SRNS)32. Coupling genome-wide analysis with reassessment of clinical features consistent with the candidate genetic diagnosis (that is, ‘reverse phenotyping’) can help to surmount this diagnostic challenge and pinpoint individuals with phenocopies of the condition of interest. For instance, reverse phenotyping following exome sequencing among patients clinically diagnosed with SRNS identified variants diagnostic of other monogenic nephropathies that could phenocopy SRNS in 28% of cases33. Similarly, exome sequencing and reverse phenotyping of patients with presumed CAKUT found mutations in phenocopy genes in 4 of the 9 patients (44%) with diagnostic mutations34.
Sources of phenotypic complexity in rare genetic nephropathies include pleiotropy, incomplete penetrance and/or variable expressivity. For example, mutations in PAX2 have pleiotropic effects as they can cause papillorenal syndrome, whose manifestations include ophthalmological anomalies, high frequency hearing loss and renal disease, reflecting the fact that PAX2 encodes a transcription factor that is involved in the development of the eye, ear and urinary tract35,36. However, owing to incomplete penetrance, not all individuals with a disease-causing PAX2 mutation will be symptomatic36. Variable expressivity can result in further phenotypic complexity, with the spectrum of disease manifestations and severity varying between affected individuals with mutations in the same gene. Hence, patients with PAX2 mutations can manifest with a wide spectrum of kidney diseases: although some present with the classic PAX2-associated phenotype of CAKUT35, some develop nephrotic syndrome37,38, and others display no signs of abnormal renal structure and/or function39. Such variability has even been reported among family members harbouring the same PAX2 mutation40.
The exact mechanisms underlying pleiotropy, incomplete penetrance and/or variable expressivity of rare genetic kidney and urological disorders are incompletely understood. In addition to being shaped by the type of genetic mutation and environmental factors, these phenomena can be mediated by the actions of modifier genes41. In the case of some purportedly monogenic conditions, the observed manifestations can vary substantially depending on the genotype at other loci, suggestive of oligogenic inheritance. For example, in sickle cell disease, which results from mutations in the gene that encodes the β-haemoglobin subunit, HBB, common variants at two other loci — BCL11A and HBS1L-MYB — lead to greater expression of (normal) fetal haemoglobin and thereby confer a milder phenotype, with fewer pain crises42. Similarly, in patients with NPHP1-associated nephronophthisis, the prevalence of retinal degeneration is over sevenfold higher among individuals who are heterozygous for a common missense variant in AH1 than among those without this missense variant43. Digenic inheritance has also been reported among patients with the renal tubulopathy Bartter syndrome type IV, with biallelic mutations in both CLCNKA and CLCNKB segregating with disease44,45. Common variants can also yield a genetic architecture similar to that of digenic inheritance resulting from two rare mutations: for example, individuals who harbour homozygous variants at the HLA and the PLA2R1 loci have a nearly 80-fold higher risk of developing idiopathic membranous nephropathy than those who do not have these risk variants22,23. Additional research will provide further insights into these and other complex models of inheritance and the role of modifier genes in mediating the clinical manifestations of nephropathies.
Insights from large-scale genetic studies
Hereditary kidney and urological disorders can arise from genomic disorders, which arise from structural variants, or from monogenic causes, which result from shorter variants (that is, single nucleotide variants or smaller insertions or deletions) in a single gene. Microarray and massively parallel sequencing (MPS)-based studies performed over the past 5 years (Supplementary Table 1) show that both genomic disorders and monogenic aetiologies contribute meaningfully across different clinical subtypes of CKD. Although common variants typically have small effects on disease risk, as noted above, exceptions do exist, including the APOL1 variants in FSGS and other forms of non-diabetic CKD20,21 and the HLA and PLA2R1 variants in membranous nephropathy22,23 (FIG. 1). Moreover, a single gene can contribute to both monogenic and polygenic forms of nephropathy. For instance, rare variants in UMOD are associated with a rare disease — autosomal-dominant tubulointerstitial kidney disease (ADTKD) — whereas more common UMOD variants confer increased risk of all-cause CKD46 (FIG. 1).
Genomic disorders
Several studies that have assessed the prevalence of genomic disorders in large CKD paediatric cohorts47-52 suggest that copy number variants (CNVs) in one or more syndrome-associated loci account for a meaningful proportion of disease cases. These investigations47-52 demonstrate a particularly high burden of genomic disorders among individuals with CAKUT, with pathogenic CNVs detected in approximately 4–10% of patients. Importantly, this enrichment was noted both among individuals reported to have isolated renal anomalies and among those with syndromic disease, reflecting the sensitivity of nephrogenesis to alterations in gene dosage53,54.
Review of these investigations also demonstrates a high level of heterogeneity with respect to both the spectrum of genomic disorders and the associated phenotypes observed. A microarray study of 178 individuals with all-cause CAKUT50, for instance, identified seven previously described genomic disorders, each of which was unique to a single patient. Similarly, assessment of nearly 3,000 patients with CAKUT49 detected 99 distinct genomic disorders among 159 patients, comprising 45 different known genomic disorders in 112 individuals and an additional 47 individuals harbouring 54 novel, large, rare, genic copy number variants. Of these 99 genomic disorders, 80 (81%) were singleton genetic diagnoses. Moreover, many of the recurrent genetic diagnoses spanned a diverse array of CAKUT phenotypes. For example, 16p11.2 microdeletions were identified in individuals with upper urinary tract anomalies, such as renal agenesis and obstructive uropathy, and in those with lower urinary tract anomalies, such as vesicoureteral valves and duplicated collecting systems.
Importantly, other studies have also noted enrichment of genomic disorders among individuals with other (non-CAKUT) kidney phenotypes, supporting a broader contribution of genomic imbalances to CKD. For example, a study of 419 paediatric patients with CKD of any cause identified pathogenic CNVs that were diagnostic for a known genomic disorder in 5% of non-CAKUT cases, and large, rare, gene-disrupting CNVs that were likely pathogenic in an additional 2.5% of patients48. These cases spanned a wide range of clinical disease subtypes, including glomerulopathy and thrombotic microangiopathy, as well as congenital renal disease. Furthermore, a 2019 assessment of nearly 400,000 adults from the UK Biobank, who were recruited for population sequencing analyses and unselected for CKD, found that multiple genomic disorders that had been previously noted to be enriched among patients with CAKUT, such as alterations at the 17q12 and 16p11.2 loci, were also significantly associated with all-cause kidney failure55.
In addition, owing to ascertainment bias in genetic testing practices, the above rates may in fact underestimate the true contribution of genomic disorders to CKD. Many of the genomic disorders noted above also involve extra-renal manifestations — such as neuro-developmental delay and congenital heart disease — that have been first-line indications for microarray testing. In these cases, patients’ renal function may have gone untested, thereby impeding accurate assessment of the prevalence of nephropathy. Accordingly, retrospective analyses of patients with genomic disorders noted to be enriched in CKD have revealed higher rates of kidney anomalies than previously thought. For example, in a medical record review of 42 patients with the 16p11.2 microdeletion syndrome, 9 of the 15 (60.0%) patients who had undergone nephrological work-up had CAKUT — a substantially higher prevalence than the 5.4% of cases with the 16p11.2 microdeletion syndrome noted in DECIPHER, a database that contains phenotypic data submitted for individuals with various genomic disorders49. Similarly, retrospective review of the medical records of 1,267 patients with 22q11.2 deletion syndrome demonstrated genitourinary anomalies in 162 (15.1%) of the 1,073 patients for whom renal imaging data were available56.
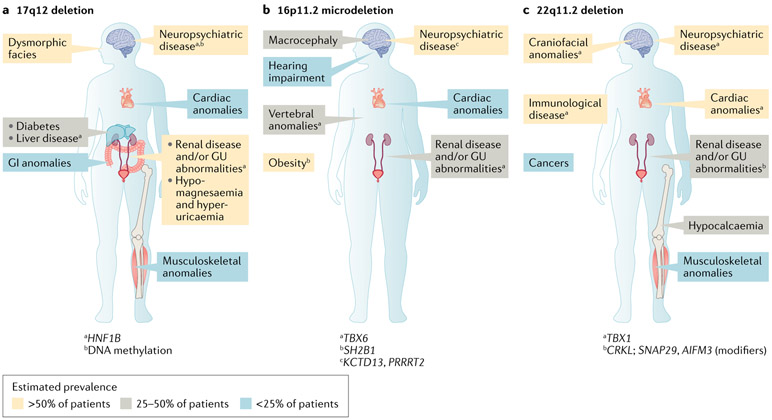
Of the genomic disorders identified in individuals with CKD, CNVs at the 17q12, 16p11.2 and 22q11.2 loci are among the most common, estimated to collectively occur in 2.9% of patients with CAKUT53. As these loci contain multiple genes that have pleiotropic effects, each of these disorders has diverse, multiorgan manifestations; however, they share the feature of being associated with highly variable neurodevelopmental, cardiac and renal anomalies53 (Fig. 2). This observed phenotypic complexity hinders full understanding of the molecular pathogenesis underlying these disorders and thereby impedes accurate clinical prognosis and targeted management. In some instances, phenotypic heterogeneity might in part result from the differing effects of a loss (for example, a deletion) versus a gain (for example, a duplication) at the locus. For example, deletions at the 16p11.2 locus lead to macrocephaly and obesity, whereas duplications at this same locus are associated with microcephaly and being underweight57. However, the highly variable penetrance and expressivity47-49 of these syndromes suggests that the phenotypic heterogeneity is a consequence of more complex genetic mechanisms than differences in gene dosage alone. Studies published over the past 4 years have utilized a multidisciplinary approach, integrating additional genetic studies, bioinformatics and functional modelling, to investigate the molecular pathogenesis of deletions at these loci and pinpoint genetic drivers of their renal and extra-renal manifestations.
Fig. 2 ∣. Genetic drivers of the complex phenotypes of common genomic disorders in chronic kidney disease.
Copy number variants (CNVs) at the 17q12, 16p11.2 and 22q11.2 loci are common genomic disorders detected among individuals with chronic kidney disease, altogether occurring in approximately 2.9% of individuals with congenital anomalies of the kidney and urinary tract53. These disorders are characterized by highly heterogeneous neurodevelopmental, cardiac and renal involvement, reflecting genetic pleiotropy and variable penetrance and expressivity. Multidisciplinary investigations integrating genetic, bioinformatic and functional studies have pinpointed key genetic mechanisms underlying the renal and extra-renal manifestations of these disorders. a ∣ In patients with deletions in 17q12, HNF1B haploinsufficiency mediates renal and endocrine anomalies and DNA methylation contributes to variable neuropsychiatric dysfunction. b ∣ In patients with 16p11.2 microdeletion, the degree of TBX6 gene inactivation mediates congenital anomalies of the kidney and urinary tract phenotypes. c ∣ In patients with deletion at the 22q11.2 locus, epistatic interactions between CRKL and other genes in the 22q11.2 locus (SNAP2 and AIFM3) are thought to drive the renal anomalies. GI, gastrointestinal; GU, genitourinary.
17q12 deletions.
Deletions at the 17q12 locus were first detected among individuals with renal cysts and/or CAKUT and maturity onset diabetes of the young58,59. They were subsequently found to be enriched among patients with neurological disorders, such as autism, schizophrenia and epilepsy; other manifestations can include dysmorphic facial features, liver disease, and cardiac, musculoskeletal and gastrointestinal anomalies60-62 (Fig. 2). A reciprocal duplication of this locus is less common, and more commonly presents with neuropsychiatric disease, with CAKUT reported in less than 25% of cases63.
The classic 17q12 deletion syndrome interval contains 15 genes, including HNF1B. In addition to the known importance of HNF1B in kidney and pancreas development64,65, findings of heterozygous intragenic HNF1B loss-of-function mutations among individuals with CAKUT and maturity onset diabetes of the young66,67 supported the notion that HNF1B haploinsufficiency drives the observed kidney and endocrine anomalies58,59. Although HNF1B participates in hindbrain development in mouse and zebrafish models68,69, the higher rates of neurodevelopmental and neuropsychiatric disease among patients with 17q12 deletions than among individuals with intragenic loss-of-function HNF1B mutations70 suggested that the neurological phenotype does not result from HNF1B haploinsufficiency alone. Two other genes in the 17q12 locus, LHX1 and ACACA, are thought to contribute to the neurological anomalies observed, based on the known role of LHX1 in brain development71,72 and the identification of smaller CNVs involving ACACA in patients with autism73. However, the dearth of individuals with neurological disease associated with mutations in either of these candidate genes and the variable severity of neurological disease among 17q12 deletion carriers74 support a more complex disease model.
An investigation published in 2018 suggested that this variability in neurological disease severity might reflect the role of epigenetic factors75. Analysis of methylation patterns revealed that these differed between patients with 17q12 deletions and those with HNF1B intragenic mutations. Within the 17q12 locus, top signals included sites upstream of LHX1 and ACACA; in addition, differential methylation was observed in another region, upstream of SLC1A3, a gene for which duplications have been associated with autism and attention deficit hyperactivity disorder76. Although further study is needed to fully understand the genetic basis of 17q12 deletion-associated neurological disease, these findings support a model wherein HNF1B haploinsufficiency is further mediated by altered methylation of other genes, both within and outside of the locus.
16p11.2 microdeletions.
16p11.2 microdeletions were first recognized as a recurrent genomic disorder among individuals with autism spectrum disorder77,78 and were subsequently found to have extra-neurological phenotypes including obesity, congenital heart defects, vertebral anomalies, macrocephaly and hearing impairment79-82 (FIG. 2). Although a 2012 investigation of 285 individuals with the 16p11.2 microdeletion reported CAKUT to be relatively infrequent, noting it in just 3.4% of the cases assessed80, 16p11.2 microdeletions have been detected in multiple patients with CAKUT in microarray studies of paediatric CKD cohorts48,52, pointing to a potential association.
Follow-up case-control genetic investigations and functional modelling have demonstrated that CAKUT is a major feature of the 16p11.2 microdeletion syndrome and identified TBX6 as a key driver of the renal phenotypes observed49. In that study49, a comparison of nearly 3,000 individuals with all-cause CAKUT with over 21,000 population controls found that 16p11.2 deletions were significantly enriched among patients with CAKUT. A retrospective review of clinical data from a separate cohort of 42 individuals with the 16p11.2 microdeletion syndrome further supported this association, revealing genitourinary anomalies in 9 of the 15 (40%) individuals with genitourinary tract imaging available49. Analysis of the 9 patients with CAKUT who were found to harbour 16p11.2 microdeletions demonstrated a minimal region of overlap containing 19 genes, including TBX6 (REF.49). In addition to being highly haploinsufficient, TBX6 was identified as a top candidate gene for 16p11.2-associated renal anomalies for two reasons. First, TBX6 has a known role in the development of the primitive streak83,84, which is the primary source of the embryonic mesoderm — a precursor tissue in the context of genitourinary development — and is expressed in the mesonephric duct and metanephric blastema, which give rise to the trigone of the urinary bladder and renal tubules, respectively85. Second, intragenic loss-of-function mutations in TBX6 had previously been detected in individuals with vertebral anomalies86, which are clinically associated with CAKUT87. Subsequent animal studies further supported TBX6 as a genetic driver of the renal manifestations in this syndrome49,88. In mice, inactivation of TBX6 resulted in CAKUT, with the degree of TBX6 gene inactivation mediating the penetrance and severity of the renal anomalies observed49,88.
Reports of CAKUT among individuals harbouring heterozygous loss-of-function TBX6 mutations88,89 further support a dosage effect of TBX6 on 16p11.2 microdeletion-associated renal manifestations. A 2019 case report89 of a Chinese patient with a duplex kidney and collecting system and mild vertebral anomalies detected two variants: an intronic variant that was experimentally demonstrated to be a loss-of-function mutation, resulting in altered splicing and premature protein truncation; and a known hypomorphic allele comprising three common polymorphisms previously found in Chinese individuals with congenital scoliosis86. Her unaffected father was homozygous for the hypomorphic allele and her unaffected mother was heterozygous for the intronic variant, with the other allele being wild type. A subsequent investigation detected loss-of-function TBX6 mutations in 7 individuals88; upon reverse phenotyping, 6 (86%) had phenotypes consistent with or suggestive of CAKUT. Although additional investigations will provide further insight, findings to date support gene dosage as a key mediator of TBX6-associated kidney disease in patients with 16p11.2 microdeletion, with the degree of TBX6 inactivation shaping its observed penetrance and expressivity.
22q11.2 deletions.
22q11.2 deletions are associated with DiGeorge syndrome, velocardiofacial syndrome and conotruncal anomaly face syndrome, the manifestations of which include congenital heart defects, immunological disease, hypocalcaemia, craniofacial and musculoskeletal anomalies, malignancies and neuropsychiatric disease; CAKUT has been reported in approximately 30% of patients90 (FIG. 2). The 22q11.2 locus is a 3-Mb region that includes four sets of low copy number repeats (LCRs), designated LCR22A, LCR22B, LCR22C and LCR22D, in which deletions can occur via meiotic non-allelic homologous recombination91,92. Heterozygous 2.5-Mb deletions spanning regions A–D (LCR22A–D) are detected in over 90% of affected individuals; the remainder harbour smaller deletions91. Prior studies identified TBX1, located in the LCR22A–B interval, as a candidate gene for DiGeorge syndrome-associated cardiac93,94 and potentially neuropsychiatric disease95. However, the fact that the syndrome can result from smaller 22q11.2 deletions that do not include TBX1 supports a role for other genes in the pathogenesis of the DiGeorge phenotype, including its renal manifestations.
Two studies in 2017 integrated case–control genetic analyses and animal models to identify candidate genetic drivers of 22q11.2 deletion-associated kidney anomalies47,96. Comparison of individuals with CAKUT with unselected population controls demonstrated a significant case-level enrichment for 22q11.2 deletions47; retrospective analyses showing high rates of genitourinary anomalies among individuals with a molecular diagnosis of DiGeorge syndrome (that is, harbouring 22q11.2 deletions)47,96 further supported this association. Comparison of the 22q11.2 deletions observed among patients with CAKUT revealed a minimal region of overlap in the LCRC–D interval, which includes CRKL. CRKL is a highly haploinsufficient gene that had been previously shown to confer DiGeorge-like features, including neurocristopathies97 and congenital heart defects98, in knockout mice; however, kidney phenotypes were not assessed in these previous mouse model studies. MPS-based sequencing demonstrated an excess of rare, putatively damaging intragenic CRKL variants among patients with CAKUT relative to those observed in exome data from population-based controls47, further supporting CRKL as a candidate gene. Subsequent studies in mice and zebrafish47,96 confirmed that CRKL is a genetic driver of CAKUT in DiGeorge syndrome and also suggested that the CAKUT phenotype is mediated by epistatic interactions between CRKL and AIFM3 and SNAP29 (REF.47) — two other genes within the LCRC–D interval that are shared among 22q11.2 deletion-carrying patients with CAKUT.
Monogenic aetiologies
MPS-based studies demonstrate that diagnostic yield differs considerably between various CKD subtypes (Supplementary Table 1), highlighting corresponding variability in genetic architecture. For example, detection rates of 60% or higher have been reported for renal cystic ciliopathies, compared with 5–20% for CAKUT and 10–30% for glomerulopathies, supporting the notion that the contribution of known monogenic causes differs for different forms of nephropathy. Importantly, high genetic heterogeneity has been noted across clinical disease categories, with sequencing studies of individuals sharing a single clinical diagnosis, such as SRNS or renal cystic ciliopathies, revealing the existence of many distinct single-gene aetiologies and a high rate of singleton genetic diagnoses. A review of large-scale MPS-based studies of patients with CKD of ‘unknown aetiology’ published over the past 2 years (Supplementary Table 1) further illustrates this heterogeneity99. Of the 443 individuals collectively included in the six studies assessed, monogenic forms of nephropathy were identified in 93 (21%), encompassing 47 distinct genes. Across these investigations, the majority of patients in whom a monogenic form of nephropathy was identified were singletons, with each study reporting that 61% or more of the monogenic nephropathies identified were unique to one individual. Together, the high detection rate and heterogeneity of genetic findings among patients with CKD of unknown aetiology reinforce the concept that CKD is a complex disease, with many monogenic aetiologies noted to result in clinically ambiguous presentations that cannot be resolved using traditional diagnostics alone.
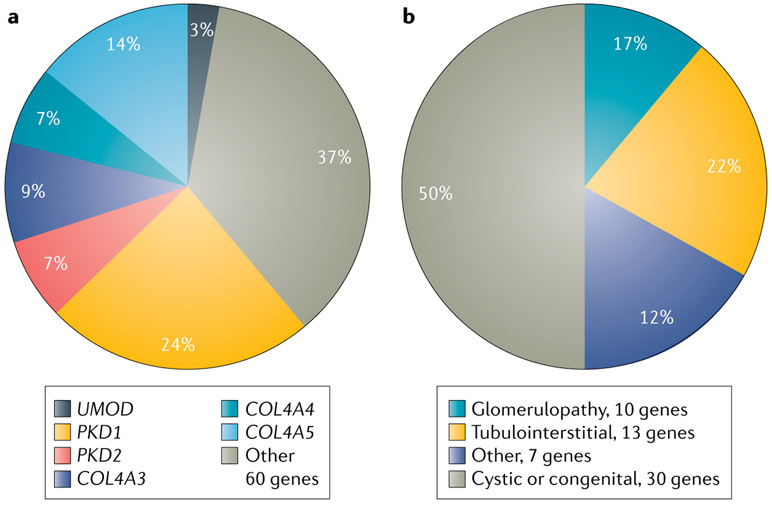
Investigations of individuals clinically diagnosed with CKD of various causes further support this concept. These studies report diagnostic findings for monogenic forms of kidney disease in 10–43% of patients100-105, consistent with a contribution of monogenic aetiologies to CKD across clinical disease subtypes (Supplementary Table 1). In addition to demonstrating a high level of genetic heterogeneity, with 55–77% of all diagnostic genetic findings occurring as singletons, these analyses also highlight the phenotypic complexity of CKD, with up to 78% of genetic findings recurring in patients within different clinical diagnostic categories. For example, diagnostic mutations in COL4A3, COL4A4 and COL4A5 are found not only in patients with clinically diagnosed Alport syndrome, thin basement membrane disease and hereditary FSGS106-110 but also among patients clinically diagnosed with other forms of CKD, such as CAKUT, cystic renal disease and hypertension-attributed nephropathy, and among patients with CKD of unknown aetiology100,101,103,104,111. This apparent phenotypic broadening may in part reflect that many monogenic nephropathies, including those associated with mutations in the type IV collagen genes101,103,107,108,112, can present with non-specific manifestations, which might impede accurate clinical diagnosis using traditional modalities alone. Wider usage of genetic testing could therefore contribute to such observations of a more diverse spectrum of phenotypes for type IV collagen-associated renal disease100,101,103,104,111. In addition to encompassing clinically distinct CKD subtypes, variation within a single gene may have different effects on disease risk. For example, although rare variants in COL4A3 are associated with an increased risk of type IV collagen-associated nephropathy, a 2019 study identified a common missense variant in COL4A3 that had a protective association with diabetic kidney disease, wherein mutation carriers showed lower rates of albuminuria and kidney failure than non-carriers113. Moreover, although certain monogenic nephropathies are recurrently found by exome sequencing of individuals with all-cause CKD, genetic findings among these diagnostic cases encompass a wide range of clinical CKD subtypes, and for each CKD subtype, many distinct single-gene aetiologies are observed (FIG. 3).
Fig. 3 ∣. Genetic and phenotypic heterogeneity observed on exome sequence analysis of patients with all-cause chronic kidney disease.
a ∣ Genetic spectrum of positive cases from exome sequence analysis of 3,315 patients with all-cause chronic kidney disease100. Of the 66 distinct monogenic disorders detected, findings in six genes accounted for 63% of all genetic diagnoses, with autosomal-dominant polycystic kidney disease resulting from mutations in PKD1 (75 cases; 24%) or PKD2 (22 cases; 7%); type IV collagen-associated nephropathy secondary to mutations in COL4A3 (27 cases; 9%), COL4A4 (21 cases; 7%) or COL4A5 (44 cases; 14%); and autosomal-dominant tubulointerstitial kidney disease caused by mutations in UMOD (9 cases; 3%). The remaining 37% of genetic diagnoses encompassed 60 different monogenic disorders. b ∣ The 60 other monogenic disorders spanned a diverse range of clinical chronic kidney disease subtypes, each of which included many distinct single-gene aetiologies, including cystic or congenital renal disease (30 genes; 50%); glomerulopathy (10 genes; 17%); tubulointerstitial disease (13 genes; 22%); and other causes of nephropathy (7 genes; 12%). Percentages do not total 100 because of rounding.
The variable detection rates of monogenic causes of kidney disease observed across studies of all-cause CKD probably reflect the corresponding variability in the clinical characteristics of the patients studied. For example, higher yields have been reported by studies of individuals preselected for features consistent with hereditary forms of nephropathy, such as familial and/or early-onset disease and/or a clinical diagnosis of a form of CKD thought to arise primarily from monogenic causes (for example, cystic or congenital renal disease). In a broad study of a largely adult cohort of over 3,300 individuals with all-cause renal disease100, diagnostic yield was nearly 10% — a rate similar to that observed for other complex diseases for which genetic testing is frequently used, such as all-cause cancer114,115. Moreover, diagnostic yield was higher among patients with familial CKD and/or those clinically diagnosed with cystic or congenital renal disease, consistent with the view that monogenic aetiologies contribute to nephropathy to a greater extent among patients with these features.
The majority of genetic studies to date have assessed individuals clinically diagnosed with CKD subtypes for which monogenic causes are thought to contribute substantially, such as SRNS or cystic renal disease. The knowledge gained from these investigations supports the use of precision diagnostics for patients clinically diagnosed with these forms of CKD. However, the majority of individuals with CKD are sporadic adult patients clinically diagnosed with CKD thought to be secondary to acquired causes, such as hypertension- or diabetes-associated renal disease, and scant data exist on the contribution of monogenic causes of disease in this population. Currently, a diagnosis of diabetic kidney disease or hypertensive kidney disease is often made presumptively on the basis of clinical history, without a confirmatory renal biopsy. Accordingly, review of biopsy samples from individuals clinically diagnosed with diabetic or hypertensive renal disease demonstrate that a substantial fraction of these individuals have biopsy findings consistent with other forms of CKD116-120. These findings support the hypothesis that a proportion of patients with presumed diabetic kidney disease or hypertensive kidney disease represent cases of undiagnosed genomic disorders or monogenic nephropathy. The lower yield of genetic findings observed for patients diagnosed with diabetic kidney disease or hypertensive kidney disease in the large-scale diagnostic sequencing studies of patients with all-cause CKD100, and the fact that relatively few causal loci with large effect size have been identified by genetic association studies in large cohorts121-124, both suggest that in many cases, these forms of CKD may have a complex, polygenic basis. Additional studies are needed to better understand the genetic architecture of the variety of clinically defined subtypes of CKD, including the extent to which monogenic causes contribute to the pathogenesis of each.
Complexities of genetic analysis in CKD
Analytical challenges
Although MPS-based analysis of exonic (coding) regions frequently provides a genetic diagnosis, it has important technical limitations for identifying disease-causal variation. Thus, if a monogenic disease is strongly suspected but no diagnostic variants are detected by exome sequencing, these limitations should be considered and addressed as needed. First, some regions, such as the CAKUT-causing gene GREB1L, are not targeted by all exome sequencing kits125-127; thus, a negative test result should prompt assessment of whether all nephropathy-associated coding regions were adequately captured by the sequencing approach. In addition, as exome sequencing generally has more variable coverage and lower analytical sensitivity for insertion and deletion variants than does genome sequencing, it can therefore miss coding variants and moderate sized deletions and duplications that can be detected by genome sequencing128. Moreover, exome sequencing can miss certain classes of functional variants that can cause monogenic disease, such as intronic variants that affect splicing, mobile genetic elements that result in retrotransposition and mosaic variants129-133. Usage of genome sequencing and long-read technologies with specialized bioinformatics approaches can support identification of these types of variants.
In addition, integration of other omics technologies can further empower these analyses, helping to achieve a genetic diagnosis among patients for whom exome sequencing alone is insufficient130,131,134-136. For instance, although non-coding variants can be detected via genome sequencing, interpretation of their pathogenicity remains challenging. Integration of transcriptome sequencing with genome sequencing data can help to identify splice-altering intronic variants129,137 and has been shown to identify diagnostic variants in 7.5% and candidate causal variants in an additional 16.7% of patients with other, non-renal rare diseases who had negative findings by exome sequencing alone138. In one study, for example, among 4 patients with suspected type VI collagen-related muscular dystrophy, for whom targeted sequencing of the disease-associated genes (COL6A1, COL6A2 and COL6A3) had failed to detect a causative variant, use of exome sequencing, genome sequencing and RNA sequencing identified a splice-altering intronic variant in COL6A1 that resulted in insertion of 24 amino acids into the highly conserved N-terminal triple-helical collagenous domain of the encoded protein129. In this study, subsequent investigation of another 637 individuals with genetically unresolved type IV collagen-related muscular dystrophy detected an additional 27 patients who harboured this variant. Importantly, the mutant transcript was found to be present at appreciable levels in muscle, but at much lower levels in cultured skin fibroblasts, highlighting the importance of tissue-specific transcriptome analysis.
Phenotypes from multilocus variation
Several large-scale genome-wide sequencing studies of individuals with a variety of complex phenotypes have demonstrated that at least 5% of patients in whom diagnostic genetic findings are identified harbour dual genetic diagnoses, with pathogenic variants in two different disease-associated genes139-141. These findings challenge the principle of diagnostic parsimony that is frequently taught in clinical medicine. In support of these findings, sequencing investigations of individuals with CKD have also identified patients with diagnostic variants spanning multiple monogenic nephropathies25,48,102,103. Similarly, a large microarray study of patients with all-cause CAKUT49 found that 6.9% of individuals harboured multiple CNVs diagnostic for different genomic disorders. Importantly, these individuals manifested more severe forms of CAKUT and higher rates of extra-renal anomalies than those with CNV findings diagnostic for a single genomic disorder, suggesting that the aggregate burden of genomic disorders mediated the penetrance and expressivity of the disease.
Although the scale of the sequencing studies to date does not enable examination of genetic interactions between monogenic disorders, these observations collectively suggest that biological interactions might underlie the joint occurrence of these disorders. Phenotypes resulting from multilocus variation may present as a more severe form of a single, clinically defined disease, overlapping symptoms that are potentially attributable to one of a number of genetic conditions detected, or as an entirely novel condition that involves features unique to each of these genetic conditions139. These complicated presentations might delay diagnosis. Hence, dual diagnoses should be suspected and genome-wide approaches should be considered when a hereditary disease is suspected but the patient’s presentation cannot be explained by a single diagnosis.
Phenotypic heterogeneity
The genetic mechanisms of phenotypic heterogeneity in CKD can be explored by considering three disorders that are observed frequently in MPS-based studies of all-cause nephropathy100-104: autosomal-dominant polycystic kidney disease (ADPKD), type IV collagen-associated nephropathy and ADTKD. Although these monogenic nephropathies are traditionally considered to be clinically homogeneous conditions, investigations over the past 10 years have demonstrated substantial genetic and phenotypic complexity.
Autosomal-dominant polycystic kidney disease.
ADPKD classically presents with bilaterally enlarged, multicystic kidneys. Extra-renal manifestations include liver cysts, intracranial aneurysms and, less commonly, cysts in other organs and cardiac disease142. However, the severity of the renal and extra-renal manifestations vary substantially, from very early onset ADPKD, which presents in utero with oligohydramnios and hyperechogenic enlarged kidneys, to isolated bilateral renal cysts with normal kidney function143. Although previously thought to arise from mutations in only two genes — PKD1 and PKD2 — the disease has been found to be genetically as well as phenotypically heterogeneous, with identification of novel causal genes, including GANAB144, DNAJB11 (REF.145) and ALG9 (REF.146) in the past 5 years. ADPKD is traditionally diagnosed by imaging; however, the genetic and phenotypic heterogeneity of ADPKD, as well as findings of a strong genotype–phenotype relationship, have led some clinicians and researchers to call for a genetics-based framework for the clinical classification of ADPKD143. Importantly, however, genetic diagnosis of ADPKD remains challenging, as PKD1 — the most commonly mutated gene in ADPKD — is large, and has a high guanine–cytosine (GC) content with a high degree of pseudohomology. Use of long-range PCR or genome sequencing can increase the technical sensitivity of genetic testing for PKD1 (REF.147). Moreover, achieving a genetic diagnosis may require assessment of variants other than protein-altering mutations in the coding regions, as synonymous variants148 and intronic variants149 that lead to altered splicing of PKD1 can also result in ADPKD; integration of RNA-based functional assays can help to identify such variants and ascertain their effect on the encoded polycystin 1 (PC1) protein.
Investigations of familial cases of ADPKD150 and longitudinal studies of ADPKD case cohorts151,152 highlight the importance of gene- and allele-level variation in mediating disease severity, as well as the importance of identifying other genetic modifiers. ADPKD is thought to arise from insufficiency of the PC1 or polycystin 2 (PC2) proteins, which normally inhibit cyst formation142. Accordingly, disease most frequently results from loss-of-function mutations in the genes that encode these proteins — PKD1 and PKD2, respectively — which together account for at least 90% of ADPKD cases153. The finding that cysts only develop in a proportion of renal tubular epithelial cells has led to the proposal that cystogenesis in ADPKD occurs through a two-hit model154,155, whereby in addition to a heterozygous germline mutation, cyst formation requires a second somatic mutation in the normal allele, resulting in full insufficiency of PC1 or PC2.
In addition to mutations in PKD1 and PKD2, mutations in genes involved in the maturation and trafficking of PC1 (REFS142,156), including GANAB144, DNAJB11 (REF.145) and ALG9 (REF.146), can lead to ADPKD. However, despite the fact that mutations in these genes disrupt a common biological pathway, the phenotypes associated with mutations in these genes vary greatly. For example, mutations in PKD1 generally confer a severe form of the disease with enlarged, multicystic kidneys, hepatic cysts and a high risk of progression to kidney failure by 60 years of age, whereas mutations in GANAB144 seem to cause mild cystic renal disease with variable liver involvement that rarely progresses to kidney failure143,144. In addition to differing according to the gene involved, ADPKD phenotypes also vary by mutation type. For instance, protein-truncating mutations in PKD1 are associated with more severe disease, characterized by larger height-adjusted total kidney volume, lower estimated glomerular filtration rate and earlier age of onset of kidney failure151,157, than are non-truncating variants, reflecting that protein-truncating mutations inactivate PC1 to a greater extent. Such strong genotype–phenotype correlations have enabled the creation of prognostic algorithms such as the predicting renal outcomes in ADPKD (PROPKD) score158, which integrates knowledge of the causal gene (for instance, PKD1 versus PKD2) and mutation type (for instance, truncating versus non-truncating mutations) with other clinical features, such as gender and age at onset of hypertension, to predict renal outcomes in affected individuals.
ADPKD also shows substantial intrafamilial variability. For example, a 2019 study of over 600 families reported that 12% of families had members with mild disease and members with severe disease150. This heterogeneity might arise in part through complex genetic mechanisms. For example, individuals can present with ADPKD without a clinically apparent family history of disease as a result of parental mosaicism159. As mosaic variants are variably distributed and can be present in only a small fraction of an individual’s total cells, resulting in a low signal-to-noise (that is, the mutant to normal allele) ratio, they can be missed by traditional Sanger sequencing160. Screening of multiple cell types using MPS with very high sequencing coverage and read depths can help to surmount these challenges, supporting the detection of mosaic variants147,160. Digenic inheritance can also lead to intrafamilial variability. Thus, co-inheritance of PKD1 and PKD2 variants can yield severe, early-onset ADPKD, whereas family members who harbour only one of the two variants typically present with milder disease161,162. Similarly, in families with PKD1- and PKD2-associated ADPKD, individuals who have mutations in genes associated with other hereditary nephropathies, such as HNF1B163 and COL4A1 (REF.164), display more severe cystic disease and earlier onset of kidney failure than those who have only the PKD1 or PKD2 mutation.
Type IV collagen-associated nephropathy.
As described earlier, mutations in COL4A3, COL4A4 and COL4A5 are associated with type IV collagen-associated nephropathies, which include Alport syndrome, thin basement membrane disease and FSGS106-110. These genes encode the collagen IV α3, α4 and α5 proteins, which together form the collagen IV α3α4α5 trimer — a key constituent of basement membranes in the glomerulus, eye and inner ear165. Accordingly, pathogenic mutations in any one of the three genes can result in disease, which classically features haematuric nephropathy with progression to renal failure, ocular anomalies and sensorineural hearing loss107,108. MPS-based sequencing of the type IV collagen genes has been recommended as a time- and cost-effective means of diagnostic screening108. However, this approach has limited technical sensitivity for certain types of causal mutation, such as deep intronic variants that alter splicing and small genic deletions, and can also miss mosaic variants owing to insufficient sequencing depth of coverage108. Thus, more comprehensive high-coverage sequencing of non-coding regions, transcriptome analysis and functional splicing assays can help to resolve diagnosis in individuals who remain undiagnosed using standard MPS-based analysis of coding regions of type IV collagen genes166,167.
The phenotypic spectrum resulting from mutations in type IV collagen is diverse, with presentations ranging from isolated microhaematuria with normal renal function to early-onset kidney failure with ocular and audiological involvement107,108. This phenotypic heterogeneity is partially attributable to effects at the genotype level. Among individuals with kidney disease resulting from mutations in COL4A3 or COL4A4, those with (two) biallelic variants show more severe disease, with earlier-onset kidney failure and more frequent extra-renal involvement, than those who are heterozygous for a single variant168,169. Similarly, among individuals with kidney disease due to mutations in COL4A5, (hemizygous) males are more severely affected than (heterozygous) females170,171. Although variable, case reports172 and studies in mouse models173 suggest that phenotypes among heterozygous females are shaped by the pattern of X chromosome inactivation, with greater inactivation of the X chromosome harbouring the wild-type allele associated with more severe disease than with lesser inactivation of this chromosome172. Phenotypic variability can also occur among males with COL4A5-associated disease caused by somatic mosaicism, with these males presenting with atypically mild forms of kidney disease174. Mutation type also seems to have an important role in determining phenotype. Among individuals with type IV collagen variants in the same gene, those who harbour protein-truncating mutations or glycine substitutions in the collagenous domain generally manifest more severe disease than those who harbour mutations that result in premature protein truncation or a collagenous domain glycine substitution175,176. These more severe outcomes reflect the fact that these types of mutation either lead to absence of the encoded type IV collagen protein as a consequence of nonsense-mediated decay, or to structural instability resulting from interruption of the highly conserved Gly-Xaa-Yaa repeats, thereby impeding normal trimer assembly107,108,165.
To date, however, our understanding of the molecular basis of the phenotypic heterogeneity found among individuals heterozygous for COL4A3 and COL4A4 mutations is limited. Although heterozygotes generally show milder disease than those with autosomal-recessive or X-linked disease168,177,178, some individuals with heterozygous mutations show phenotypes of similar severity to those of patients with biallelic or hemizygous mutations179-181. Interestingly, a 2019 population-based genetic study suggested that type IV collagen mutations have highly variable penetrance as well as expressivity, reporting a heterozygous protein-truncating variant in COL4A3 among population controls without any apparent association with kidney impairment182. As heterozygous type IV collagen mutations encompass a diverse phenotypic spectrum and have a variable impact on kidney disease risk, interpretation of their pathophysiological relevance is challenging. Additional investigations, such as analyses that integrate genome-wide sequence data with longitudinal phenotypic data from patients with CKD and unselected population controls, as well as saturation editing183,184 studies to assess the functional consequences of a given mutation, will help to clarify the role of these variants in different forms of CKD, facilitating genetic diagnosis and supporting development of targeted therapies.
Autosomal-dominant tubulointerstitial kidney disease.
ADTKD describes a set of rare nephropathies characterized by the non-specific findings of progressive CKD with tubulointerstitial fibrosis on renal biopsy185,186. To date, mutations in any one of five genes — UMOD, MUC1, REN, HNF1B and SEC61A1 — have been implicated in ADTKD185,186; an ADTKD-like phenotype has also been noted among families with segregating mitochondrial gene mutations187. Although defined by a common histopathology, ADTKD can vary substantially by molecular subtype, corresponding to biological functions specific to the given gene. For example, individuals with disease caused by mutations in the gene that encodes renin, REN (ADTKD-REN), display the additional features of early-onset anaemia as well as mild hypotension and hyperkalaemia, reflecting the involvement of the renin-angiotensin-aldosterone system in erythropoietin production, blood-pressure regulation and potassium homeostasis185,188. Similarly, the phenotype resulting from mutations in SEC61A1 can include primary immunodeficiency189,190, consistent with the role of the encoded protein in plasma cell differentiation and immunoglobulin synthesis190.
Given its non-specific symptomatology and histopathology, ADTKD can be difficult to detect using traditional clinical diagnostics alone, supporting the use of genetic testing185. Moreover, the fact that some of the major known causal genes have mutational hot spots can facilitate diagnostic interpretation of variants. For example, the majority of disease-causal variants in ADTKD-UMOD are missense mutations that cluster in exons 3–4 (REF.46). However, in certain cases, causal variants cannot be detected using standard MPS-based testing. For example, ADTKD-MUC1 results from a cytosine insertion mutation in a long, GC-rich variable-number tandem repeat sequence of the MUC1 gene191. The high GC content and repetitive nature of the region impede detection using MPS-based sequencing and, instead, molecular diagnosis requires other assays, including long-range PCR191 and immunostaining for the encoded (mucin 1) protein192. Broader adoption of these alternative methodologies will facilitate identification of disease cases, supporting further longitudinal study of ADTKD and providing greater insight into genotype–phenotype correlations for the disorder.
Towards targeted therapies
Despite CKD being a collectively common disease, there is a paucity of targeted therapies. In part, this paucity reflects the challenge posed by the high aetiological and phenotypic heterogeneity of the disease, which makes it difficult to identify, understand and treat disease subtypes on the basis of their molecular pathogenesis193,194. Genomics and other omics technologies have the potential to surmount this challenge by supporting discernment of different CKD subtypes at the molecular level, thereby enabling focused studies of affected individuals and the development of targeted therapies.
The use of pharmacological chaperone therapy, whereby small-molecule ligands known as pharmacological chaperones are used to stabilize and correct misfolded proteins, in Fabry disease195-197 illustrates the potential of this type of targeted approach. Fabry disease results from mutations in GLA, which encodes the α-galactosidase enzyme. Fabry disease encompasses a diverse clinical spectrum: patients can present with the more severe, classic phenotype, a systemic disorder whose manifestations include progressive kidney failure, cardiac disease, cerebrovascular disease, peripheral neuropathy, skin lesions and ocular anomalies, or with the milder, non-classic phenotype, characterized by isolated cardiac disease and/or kidney disease. To date, 268 known disease-causal mutations that cause the enzyme to be misfolded but enable it to retain residual catalytic activity, and thus remain amenable to rescue by the pharmacological chaperone migalastat, have been identified198. Importantly, these migalastat-responsive GLA mutations are found across the phenotypic spectrum of Fabry disease, among both patients with the classic phenotype and those with the non-classic phenotype, making it difficult to discern who may be a suitable candidate for pharmacological chaperone therapy on the basis of clinical presentation alone. Accordingly, a cellular assay198 screen for patients with migalastat-responsive mutations has been used in clinical trials to identify which patients could benefit from this therapy195-197.
In addition to targeting the encoded protein, precision therapies can also directly act at the genomic or transcriptomic level. For example, in sickle cell disease, which arises from the haemoglobin S (p.Glu6Val) variant in HBB, gene therapy via lentiviral transfer of a modified copy of HBB that encodes an anti-sickling allele has been shown to result in full remission199. Among patients with hereditary transthyretin amyloidosis, use of oligonucleotide agents that target the mutant TTR RNA transcript has been shown to halt or reverse the progression of multiple disease manifestations, by inhibiting the synthesis and thereby systemic accumulation of misfolded transthyretin200,201. In primary hyperoxaluria type 1, in which impaired glyoxylate metabolism leads to systemic oxalosis, CRISPR–Cas9-mediated gene editing202 and mRNA silencing using small interfering RNAs203 are being investigated as potential approaches with which to inhibit synthesis of glycolate oxidase, the enzyme involved in glyoxylate production. Growing usage of genomic sequencing and other omics tools among patients with CKD will support the further development of precision therapeutics in nephrology.
Conclusions
Genomics medicine aims to provide personalized care based on an individual’s genetic information. However, understanding the relationship between genotype and phenotype for many forms of CKD is complicated by the complexity of rare genetic causes of CKD, impeding accurate assessment of an individual’s risk of disease and the identification of approaches for targeted management. Many monogenic nephropathies demonstrate variable and multisystem manifestations, reflecting phenomena including genetic pleiotropy, dosage sensitivity, incomplete penetrance and/or variable expressivity. Such phenotypic heterogeneity can hinder our ability to pinpoint a causal aetiology. Moreover, as patients can present atypically, individuals with monogenic forms of CKD can go clinically unrecognized. The high genetic heterogeneity observed within a single disease subtype also complicates the choice of optimal genetic testing modality. Thus, in many cases, genome-wide approaches such as exome sequencing or genome sequencing may provide superior diagnostic sensitivity over targeted panels100, as the patient may have a subtype of CKD other than that clinically suspected or have one that has a multitude of potential causal genes. In either of these situations, use of a targeted gene panel may lead to a diagnostic genetic variant being missed, as the gene causal for the patient’s disease may not be included on the panel. However, the expanded scope of analysis offered by exome or genome sequencing also confers a greater burden of variant interpretation, which is augmented by the phenotypic complexity of CKD. The abundance of rare, putatively deleterious variations within any human genome results in a high potential for false-positive results204,205. Moreover, both the variable penetrance and expressivity of many rare genetic causes of CKD and the lack of detailed, longitudinal clinical data available for control cohorts can make it difficult to discern whether findings of such variants among self-declared healthy individuals indicate that they are in fact benign, or instead result from cases of undiagnosed monogenic nephropathy206. Beyond impeding diagnosis, such phenotypic heterogeneity can make it difficult to provide a clear prognosis upon detecting a disease-causing variant. As many investigations of rare genetic causes of CKD have been performed in highly preselected populations, comprising individuals with clinical features consistent with the disorder under study, knowledge of the true prevalence, penetrance and frequency of the manifestations of these variants — which often encompass multiple organ systems — remains incomplete. This limited knowledge prevents accurate assessment of disease severity and rate of progression and thereby hinders targeted work-up, surveillance and management.
Addressing these challenges will require a multifaceted approach. Use of unbiased, genome-wide sequencing approaches and reverse phenotyping will increase diagnostic sensitivity and help to distinguish hereditary nephropathies that overlap in clinical presentation. Inclusion of multiple types of omics data in the diagnostic work-up, as well as in the research-level study of individuals with suspected monogenic nephropathies, will not only support the identification of disease-causing variants but also offer deeper insight into the molecular pathogenesis of these disorders129,207. Refinement of existing guidelines for clinical sequence interpretation by working groups, such as those led by ClinGen208, will also facilitate the discernment of pathogenic versus benign variation. Similarly, as further large-scale genetic studies are performed in nephrology, regular review of the literature by such expert panels will help the field to maintain an up-to-date understanding of the contribution of the genetic and phenotypic heterogeneity of CKD (Supplementary Table 1) and support the creation of evidence-based guidelines for the use of genetic testing in nephrology. Moreover, periodic reanalysis of sequence data may enable the reclassification of variants of unknown significance or the detection of pathogenic variants in newly discovered genes, as illustrated by studies from the past few years, where reanalysis of exome data identified disease-causal variants in patients who had been previously sequenced, with non-diagnostic results209-211. In addition, novel techniques that integrate genetic and phenotypic data from large, diverse cohorts, including genome-wide polygenic risk scores16 and phenotypic risk scores212 (BOX 1), will deepen our understanding of genotype–phenotype relationships, enabling accurate assessment of disease risk across different subtypes of CKD. Incorporation of these emerging methodologies with traditional tools such as clinical history and renal histopathology will help to unravel the complex phenotypes encompassed by CKD, empowering both genetic diagnosis and novel gene discovery for patients with nephropathy.
Supplementary Material
Key points.
Chronic kidney disease (CKD) is a complex disorder comprising many rarer, pathophysiologically distinct conditions that encompass both monogenic and polygenic forms, which share the common feature of leading to persistent anomalies in renal structure and/or function.
Rare hereditary causes of CKD often show high phenotypic heterogeneity, which can result from pleiotropy, incomplete penetrance or variable expressivity.
Microarray and massively parallel sequencing studies have shown that both genomic disorders and monogenic diseases account for a meaningful proportion of cases across different clinical subtypes of CKD.
Copy number variants at the 17q12, 22q11.2 and 16p11.2 loci are recurrent genomic disorders among patients with CKD and display diverse and highly variable multiorgan manifestations, which can reflect gene dosage sensitivity and epistatic and epigenetic effects.
Although classically considered to be clinically homogeneous, common monogenic causes of CKD, such as autosomal-dominant polycystic kidney disease, type IV collagen-associated nephropathy and autosomal-dominant tubulointerstitial kidney disease, display variable penetrance and expressivity, in part due to gene-and allele-level variation.
Multilocus variation, involving variants for multiple genetic conditions, can confer complex phenotypes and has been detected in at least 5% of positive cases from genome-wide testing.
Novel techniques that integrate genetic sequencing, experimental assays and clinical data, such as reverse phenotyping, functional screening of potentially pathogenic variants, and genetic and phenotypic risk scores, will support greater understanding of the phenotypic complexity of different forms of CKD.
Acknowledgements
The work of the authors is supported by grants from the US National Institutes of Health (1F30DK116473 (E.E.G.) and 2R01DK080099, R01DK082753 and U54DK104309 (A.G.G.)).
Glossary
- Heritability
The proportion of interindividual variation in a trait that is caused by genetic factors.
- Private variants
Genetic variants unique to a single individual (that is, variants not observed in other individuals).
- Phenocopy
A phenotype that bears resemblance to the phenotype resulting from a particular genotype but occurs in an individual who does not harbour that genotype.
- Pleiotropy
Variation in a single gene influencing multiple phenotypic traits.
- Incomplete penetrance
Penetrance is the proportion of individuals with a particular genotype who display the associated phenotype. In the case of incomplete penetrance, not all individuals with the genotype manifest the associated phenotype.
- Variable expressivity
Expressivity refers to the extent to which individuals with a given genetic disease display the associated phenotype and thus reflects the range of phenotypes associated with the genotype. Variable expressivity describes a situation wherein affected individuals display the associated phenotype to differing extents.
- Papillorenal syndrome
Also known as renal coloboma syndrome. A disorder resulting from pathogenic variants in PAX2 characterized by kidney disease and ophthalmological anomalies; other manifestations may include sensorineural hearing loss, soft skin and ligamentous laxity.
- Oligogenic inheritance
A form of inheritance in which transmission of a phenotypic trait is mediated by multiple different genetic loci
- Digenic inheritance
A form of inheritance in which transmission of a phenotypic trait is mediated by two different genetic loci.
- Structural variants
Large (≥1-kb) DNA variants; these alterations can be balanced (for example, inversions or reciprocal translocations, with no overall change in the amount of DNA at the relevant locus) or imbalanced (for example, copy number variants), which lead to gain or loss of DNA at the relevant locus.
- Single nucleotide variants
Alterations of single bases (nucleotides) in a DNA sequence; single nucleotide variants can lead to a different amino acid sequence in the encoded protein (non-synonymous variants) or leave the sequence unchanged (synonymous variants).
- Insertions or deletions
Gains or losses in the number of bases in a DNA sequence versus the reference sequence at that site, producing a different amino acid sequence in the encoded protein.
- Copy number variants
(CNVs). Structural variants that lead to gain (duplications) or loss (deletions) of DNA at the relevant locus.
- Genic copy number variants
Copy number variants that contain protein-coding regions of the genome (that is, genes).
- Locus
A specific site in the genome (plural: loci).
- Haploinsufficiency
A condition resulting from inactivation of one copy of a gene for which two copies are needed for normal gene function; for such haploinsufficient genes, gene function is thus altered in heterozygotes, as the remaining (functional) copy does not produce sufficient gene product for normal function.
- Meiotic non-allelic homologous recombination
Exchange of genetic material (DNA) between homologous regions located on different loci during meiosis.
- Epistatic interactions
Interaction between multiple different loci, wherein the phenotypic impact of the genotype at one locus depends on the genotype at other loci.
- Mobile genetic elements
DNA sequences that can move from their original site and integrate into another, different region of the genome
- Retrotransposition
Insertion of a DNA sequence at a new site in the genome using an RNA intermediate: the DNA sequence is first transcribed into RNA, and the RNA is then reverse transcribed into DNA, which is then inserted at the new site.
- Mosaic variants
Genetic variants that are present in a mosaic form — that is, the genotype at that locus is present within a certain proportion of an individual’s cells, such that they have multiple genetically different cell populations, with different genotypes at that locus.
- Pseudohomology
The state in which the DNA sequence of a (protein-coding) gene is highly similar to the DNA sequence of a pseudogene (that is, a copy of the gene that is not transcribed or translated, and thus does not yield a protein), because they both originate from the same ancestral gene.
- Synonymous variants
DNA sequence changes in the protein-coding (exonic) regions of the genome that do not alter the amino acid in the associated encoded proteins.
- Sanger sequencing
A method of DNA sequencing that uses labelled chain-terminating dideoxynucleotides to detect the nucleotides in the DNA strand being sequenced. This method yields a sequence chromatogram, which can subsequently be analysed to identify genetic variants.
- Sequencing coverage and read depths
In this Review, sequencing coverage refers to the percentage of bases in the DNA region targeted by sequencing that is sequenced a given number of times. Sequencing depth denotes the average number of times that a given nucleotide is read in a set of DNA sequence reads. Higher coverage and depth means that more of the targeted genomic region has been sampled a greater number of times, increasing the technical accuracy of the resulting data.
- Hemizygous
The condition in which an individual has a single copy of a pair of chromosomes or a segment of a chromosome pair, rather than two copies. This occurs for genes located on the X chromosome among human males, as their sex chromosomes comprise one X chromosome and one Y chromosome
- Saturation editing
A technique that replaces a DNA sequence in a gene of interest with variant DNA sequences encoding alternative amino acids in order to generate all possible amino acid substitutions at that site in the protein. The impact of each of the mutations on wild-type (non-mutated/normal) protein function can then be evaluated through various high-throughput screening assays (for example, for a gene encoding a kinase enzyme, assessing its ability to phosphorylate its substrate).
- Transthyretin amyloidosis
A disorder resulting from systemic amyloid deposition in organs including the heart, liver, nervous system and kidney owing to mutations in TTR.
Footnotes
Competing interests
A.G.G. has served as a consultant for the AstraZeneca Center for Genomics Research and for Goldfinch Bio. The other authors declare no competing interests.
Peer review information
Nature Reviews Nephrology thanks R. Gbadegesin, A. Mallett and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41581-020-0325-2.
References
- 1.Webster AC, Nagler EV, Morton RL & Masson P Chronic kidney disease. Lancet 389, 1238–1252 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Devuyst O et al. Rare inherited kidney diseases: challenges, opportunities, and perspectives. Lancet 383, 1844–1859 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skrunes R, Svarstad E, Reisaeter AV & Vikse BE Familial clustering of ESRD in the Norwegian population. Clin. J. Am. Soc. Nephrol 9, 1692–1700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connaughton DM et al. The Irish kidney gene project-prevalence of family history in patients with kidney disease in ireland. Nephron 130, 293–301 (2015). [DOI] [PubMed] [Google Scholar]
- 5.McClellan WM et al. Individuals with a family history of ESRD are a high-risk population for CKD: implications for targeted surveillance and intervention activities. Am. J. Kidney Dis 53, S100–S106 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Fox CS et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framingham Heart Study. J. Am. Soc. Nephrol 15, 2457–2461 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Arpegard J et al. Comparison of heritability of Cystatin C- and creatinine-based estimates of kidney function and their relation to heritability of cardiovascular disease. J. Am. Heart Assoc 4, e001467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorski M et al. 1000 genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci. Rep 7, 45040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moulin F et al. A population-based approach to assess the heritability and distribution of renal handling of electrolytes. Kidney Int. 92, 1536–1543 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Lieske JC, Turner ST, Edeh SN, Smith JA & Kardia SL Heritability of urinary traits that contribute to nephrolithiasis. Clin. J. Am. Soc. Nephrol 9, 943–950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanetti D et al. Identification of 22 novel loci associated with urinary biomarkers of albumin, sodium, and potassium excretion. Kidney Int. 95, 1197–1208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leu C et al. Polygenic burden in focal and generalized epilepsies. Brain 142, 3473–3481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speed D et al. Describing the genetic architecture of epilepsy through heritability analysis. Brain 137, 2680–2689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Fishawy P & State MW The genetics of autism: key issues, recent findings, and clinical implications. Psychiatr. Clin. North. Am 33, 83–105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorstman JAS et al. Autism genetics: opportunities and challenges for clinical translation. Nat. Rev. Genet 18, 362–376 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Khera AV et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 50, 1219–1224 (2018).This study created and validated genome-wide polygenic risk scores for multiple common conditions, including coronary artery disease, type 2 diabetes and breast cancer, and demonstrated that individuals at the uppermost risk percentiles had disease risk equivalent to those with pathogenic mutations for rare monogenic forms of these conditions.
- 17.Khera AV et al. Whole-genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation 139, 1593–1602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katsanis N The continuum of causality in human genetic disorders. Genome Biol. 17, 233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupski JR, Belmont JW, Boerwinkle E & Gibbs RA Clan genomics and the complex architecture of human disease. Cell 147, 32–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genovese G et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzur S et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum. Genet 128, 345–350 (2010).Together with Genovese et al., this investigation demonstrated that two common missense variants in the apoliproprotein L1 gene significantly increased the risk of a variety of forms of non-diabetic CKD among individuals of sub-Saharan African descent, and point to a potential mechanism for their being positively selected for over the course of human evolution.
- 22.Stanescu HC et al. Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N. Engl. J. Med 364, 616–626 (2011).This genome-wide association study found that homozygotes for risk alleles at the HLA-DQA1 and PLA2R1 loci had nearly an 80-fold higher odds of developing idopathic membranous nephropathy than those without these alleles. These findings highlight that common variants can result in a genetic architecture similar to that of digenic inheritance arising from rare mutations at two different loci.
- 23.Xie J et al. The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis. Nat. Commun 11, 1600 (2020).This investigation identified two additional risk alleles for idiopathic membranous nephropathy and examines the ancestry-specific effects of variation at the HLA locus. Using the genome-wide significant risk loci detected, they create a genetic risk score that can correctly reclassify up to 37% of affected individuals with negative results on serological screening, emphasizing the potential of genetic risk score to empower non-invasive diagnosis of kidney disease.
- 24.Connaughton DM & Hildebrandt F Personalized medicine in chronic kidney disease by detection of monogenic mutations. Nephrol. Dial. Transplant 35, 390–397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groopman EE, Rasouly HM & Gharavi AG Genomic medicine for kidney disease. Nat. Rev. Nephrol 14, 83–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eknoyan G et al. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. Suppl 3, 19–62 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Graham SE et al. Sex-specific and pleiotropic effects underlying kidney function identified from GWAS meta-analysis. Nat. Commun 10, 1847 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wuttke M et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet 51,957–972 (2019).This genome-wide association study of a combined cohort of over one million individuals identified 166 novel kidney function-associated loci and utilized gene expression data to prioritize candidate genes for future study.
- 29.Mitchell KJ What is complex about complex disorders? Genome Biol. 13, 237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braun DA et al. Whole exome sequencing identifies causative mutations in the majority of consanguineous or familial cases with childhood-onset increased renal echogenicity. Kidney Int. 89, 468–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gee HY et al. Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int. 85, 880–887 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bierzynska A et al. Genomic and clinical profiling of a national nephrotic syndrome cohort advocates a precision medicine approach to disease management. Kidney Int. 91,937–947 (2017).This investigation integrated exome sequencing and detailed phenotyping to identify monogenic forms of early-onset steroid-resistant nephrotic syndrome and develop an evidence-based framework for the diagnosis and management of this clinically and aetiologically heterogeneous disorder.
- 33.Landini S et al. Reverse phenotyping after whole-exome sequencing in steroid-resistant nephrotic syndrome. Clin. J. Am. Soc. Nephrol 15, 89–100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vivante A et al. Exome sequencing discerns syndromes in patients from consanguineous families with congenital anomalies of the kidneys and urinary tract. J. Am. Soc. Nephrol 28, 69–75 (2017).This study applied exome sequencing and reverse phenotyping to differentiate individuals with monogenic forms of congenital anomalies of the kidney and urinary tract from those with mutations in phenocopy genes, pointing to the diagnostic value of genome-wide sequencing among this patient population.
- 35.Schimmenti LA Renal coloboma syndrome. Eur. J. Hum. Genet 19, 1207–1212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eccles MR & Schimmenti LA Renal-coloboma syndrome: a multi-system developmental disorder caused by PAX2 mutations. Clin. Genet 56, 1–9 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Vivante A et al. Dominant PAX2 mutations may cause steroid-resistant nephrotic syndrome and FSGS in children. Pediatr. Nephrol 34, 1607–1613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barua M et al. Mutations in PAX2 associate with adult-onset FSGS. J. Am. Soc. Nephrol 25, 1942–1953 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bower M et al. Update of PAX2 mutations in renal coloboma syndrome and establishment of a locus-specific database. Hum. Mutat 33, 457–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adam J et al. A wide spectrum of phenotypes in a family with renal coloboma syndrome caused by a mutation. Clin. Kidney J 6, 410–413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riordan JD & Nadeau JH From peas to disease: modifier genes, network resilience, and the genetics of health. Am. J. Hum. Genet 101, 177–191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lettre G et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl Acad. Sci. USA 105, 11869–11874 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Louie CM et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat. Genet 42, 175–180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nozu K et al. Molecular analysis of digenic inheritance in Bartter syndrome with sensorineural deafness. J. Med. Genet 45, 182–186 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Schlingmann KP et al. Salt wasting and deafness resulting from mutations in two chloride channels. N. Engl. J. Med 350, 1314–1319 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Devuyst O, Olinger E & Rampoldi L Uromodulin: from physiology to rare and complex kidney disorders. Nat. Rev. Nephrol 13, 525–544 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Rivera E et al. Genetic drivers of kidney defects in the DiGeorge syndrome. N. Engl. J. Med 376, 742–754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verbitsky M et al. Genomic imbalances in pediatric patients with chronic kidney disease. J. Clin. Invest 125, 2171–2178 (2015).This study demonstrates that genomic disorders can be found across paediatric patients clinically diagnosed with a variety of forms of CKD, supporting that genomic disorders contribute broadly to early-onset CKD.
- 49.Verbitsky M et al. The copy number variation landscape of congenital anomalies of the kidney and urinary tract. Nat. Genet 51, 117–127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caruana G et al. Copy-number variation associated with congenital anomalies of the kidney and urinary tract. Pediatr. Nephrol 30, 487–495 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Faure A et al. DNA copy number variants: a potentially useful predictor of early onset renal failure in boys with posterior urethral valves. J. Pediatr. Urol 12, 227.E1–227.E7 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Sanna-Cherchi S et al. Copy-number disorders are a common cause of congenital kidney malformations. Am. J. Hum. Genet 91,987–997 (2012).This analysis of chromosomal microarray data from nearly 500 patients with renal hypodysplasia found that 16.6% of them harboured diagnostic copy number variants. Importantly, the majority of known genomic disorders identified were associated with developmental delay or neuropsychiatric disease, pointing to the utility of chromosomal microarray among individuals with congenital anomalies of the kidney and urinary tract for early detection and targeted management in individuals with such syndromic forms of disease.
- 53.Sanna-Cherchi S, Westland R, Ghiggeri GM & Gharavi AG Genetic basis of human congenital anomalies of the kidney and urinary tract. J. Clin. Invest 128, 4–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicolaou N, Renkema KY, Bongers EM, Giles RH & Knoers NV Genetic, environmental, and epigenetic factors involved in CAKUT. Nat. Rev. Nephrol 11, 720–731 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Crawford K et al. Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J. Med. Genet 56, 131–138 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Van Batavia JP et al. Anomalies of the genitourinary tract in children with 22q11.2 deletion syndrome. Am. J. Med. Genet. A 179, 381–385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacquemont S et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature 478, 97–102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bellanne-Chantelot C et al. Large genomic rearrangements in the hepatocyte nuclear factor-1beta (TCF2) gene are the most frequent cause of maturity-onset diabetes of the young type 5. Diabetes 54, 3126–3132 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Mefford HC et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am. J. Hum. Genet 81, 1057–1069 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moreno-De-Luca D et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am. J. Hum. Genet 87, 618–630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagamani SC et al. Clinical spectrum associated with recurrent genomic rearrangements in chromosome 17q12. Eur. J. Hum. Genet 18, 278–284 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitchel MW et al. 17q12 recurrent deletion syndrome. GeneReviews https://www.ncbi.nlm.nih.gov/books/NBK401562/ (8 December 2016). [Google Scholar]
- 63.Mefford H, Mitchell E & Hodge J 17q12 recurrent duplication. GeneReviews https://www.ncbi.nlm.nih.gov/books/NBK344340/ (25 February 2016). [Google Scholar]
- 64.Clissold RL, Hamilton AJ, Hattersley AT, Ellard S & Bingham C HNF1B-associated renal and extra-renal disease-an expanding clinical spectrum. Nat. Rev. Nephrol 11, 102–112 (2015). [DOI] [PubMed] [Google Scholar]
- 65.El-Khairi R & Vallier L The role of hepatocyte nuclear factor 1beta in disease and development. Diabetes Obes. Metab 18, 23–32 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Bingham C et al. Mutations in the hepatocyte nuclear factor-1beta gene are associated with familial hypoplastic glomerulocystic kidney disease. Am. J. Hum. Genet 68, 219–224 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horikawa Y et al. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat. Genet 17, 384–385 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Choe SK, Hirsch N, Zhang X & Sagerstrom CG hnf1b genes in zebrafish hindbrain development. Zebrafish 5, 179–187 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makki N & Capecchi MR Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev. Biol 357, 295–304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clissold RL et al. Chromosome 17q12 microdeletions but not intragenic HNF1B mutations link developmental kidney disease and psychiatric disorder. Kidney Int. 90, 203–211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bedont JL et al. Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep. 7, 609–622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao Y et al. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc. Natl Acad. Sci. USA 104, 13182–13186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Girirajan S et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am. J. Hum. Genet 92, 221–237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laffargue F et al. Towards a new point of view on the phenotype of patients with a 17q12 microdeletion syndrome. Arch. Dis. Child 100, 259–264 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Clissold RL et al. Genome-wide methylomic analysis in individuals with HNF1B intragenic mutation and 17q12 microdeletion. Clin. Epigenetics 10, 97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Amen-Hellebrekers CJ et al. Duplications of SLC1A3: associated with ADHD and autism. Eur. J. Med. Genet 59, 373–376 (2016). [DOI] [PubMed] [Google Scholar]
- 77.Weiss LA et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med 358, 667–675 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Kaminsky EB et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med 13, 777–784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bochukova EG et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature 463, 666–670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zufferey F et al. A 600kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet 49, 660–668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller DT et al. 16p11.2 recurrent microdeletion. GeneReviews https://www.ncbi.nlm.nih.gov/books/NBK11167 (updated 10 Dec 2015). [Google Scholar]
- 82.Steinman KJ et al. 16p11.2 deletion and duplication: characterizing neurologic phenotypes in a large clinically ascertained cohort. Am. J. Med. Genet. A 170, 2943–2955 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Chapman DL, Agulnik I, Hancock S, Silver LM & Papaioannou VE Tbx6, a mouse T-Box gene implicated in paraxial mesoderm formation at gastrulation. Dev. Biol 180, 534–542 (1996). [DOI] [PubMed] [Google Scholar]
- 84.Chapman DL & Papaioannou VE Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature 391,695–697 (1998). [DOI] [PubMed] [Google Scholar]
- 85.Concepcion D et al. Cell lineage of timed cohorts of Tbx6-expressing cells in wild-type and Tbx6 mutant embryos. Biol. Open 6, 1065–1073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu N et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med 372, 341–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.MacEwen GD, Winter RB & Hardy JH Evaluation of kidney anomalies in congenital scoliosis. J. Bone Jt. Surg. Am 54, 1451–1454 (1972). [PubMed] [Google Scholar]
- 88.Yang N et al. Human and mouse studies establish TBX6 in Mendelian CAKUT and as a potential driver of kidney defects associated with the 16p11.2 microdeletion syndrome. Kidney Int. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dong S et al. Noncoding rare variants of TBX6 in congenital anomalies of the kidney and urinary tract. Mol. Genet. Genomics 294, 493–500 (2019). [DOI] [PubMed] [Google Scholar]
- 90.McDonald-McGinn DM et al. 22q11.2 deletion syndrome. GeneReviews https://www.ncbi.nlm.nih.gov/books/NBK1523/ (updated 27 Feb 2020). [Google Scholar]
- 91.Morrow BE, McDonald-McGinn DM, Emanuel BS, Vermeesch JR & Scambler PJ Molecular genetics of 22q11.2 deletion syndrome. Am. J. Med. Genet. A 176, 2070–2081 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shaikh TH et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet 9, 489–501 (2000). [DOI] [PubMed] [Google Scholar]
- 93.Merscher S et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104, 619–629 (2001). [DOI] [PubMed] [Google Scholar]
- 94.Yagi H et al. Role of TBX1 in human del22q11.2 syndrome. Lancet 362, 1366–1373 (2003). [DOI] [PubMed] [Google Scholar]
- 95.Paylor R et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc. Natl Acad. Sci. USA 103, 7729–7734 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haller M, Mo Q, Imamoto A & Lamb DJ Murine model indicates 22q11.2 signaling adaptor CRKL is a dosage-sensitive regulator of genitourinary development. Proc. Natl Acad. Sci. USA 114, 4981–4986 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guris DL, Fantes J, Tara D, Druker BJ & Imamoto A Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat. Genet 27, 293–298 (2001). [DOI] [PubMed] [Google Scholar]
- 98.Racedo SE et al. Mouse and human CRKL is dosage sensitive for cardiac outflow tract formation. Am. J. Hum. Genet 96, 235–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hays T, Groopman EE & Gharavi AG Genetic testing for kidney disease of unknown etiology. Kidney Int. 10.1016/j.kint.2020.03.031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Groopman EE et al. Diagnostic utility of exome sequencing for kidney disease. N. Engl. J. Med 380, 142–151 (2019).This study found that exome sequencing in a combined cohort of 3,315 individuals with CKD of diverse causes identified a monogenic cause in nearly 1 in 10 cases, highlighting the diagnostic utility of genome-wide sequencing for patients with kidney disease.
- 101.Lata S et al. Whole-exome sequencing in adults with chronic kidney disease: a pilot study. Ann. Intern. Med 168, 100–109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mallett AJ et al. Massively parallel sequencing and targeted exomes in familial kidney disease can diagnose underlying genetic disorders. Kidney Int. 92, 1493–1506 (2017).This investigation applied targeted massively parallel sequencing and phenotype-driven analysis led by a multidisciplinary team to identify a monogenic cause in over 40% of individuals referred for evaluation of familial kidney disease. The high detection rate supports the efficacy of this approach for the work-up of patients suspected to have an inherited form of CKD.
- 103.Connaughton DM et al. Monogenic causes of chronic kidney disease in adults. Kidney Int. 95, 914–928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mansilla MA et al. Targeted broad-based genetic testing by next-generation sequencing informs diagnosis and facilitates management in patients with kidney diseases. Nephrol. Dial. Transplant 10.1093/ndt/gfz173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rao J et al. Genetic spectrum of renal disease for 1001 Chinese children based on a multicenter registration system. Clin. Genet 96, 402–410 (2019). [DOI] [PubMed] [Google Scholar]
- 106.Malone AF et al. Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int. 86, 1253–1259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kashtan CE et al. Alport syndrome: a unified classification of genetic disorders of collagen IV alpha345: a position paper of the Alport Syndrome Classification Working Group. Kidney Int. 93, 1045–1051 (2018). [DOI] [PubMed] [Google Scholar]
- 108.Savige J et al. Expert consensus guidelines for the genetic diagnosis of Alport syndrome. Pediatr. Nephrol 34, 1175–1189 (2019). [DOI] [PubMed] [Google Scholar]
- 109.Gast C et al. Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol. Dial. Transpl 31, 961–970 (2016). [DOI] [PubMed] [Google Scholar]
- 110.Yao T et al. Integration of genetic testing and pathology for the diagnosis of adults with FSGS. Clin. J. Am. Soc. Nephrol 14, 213–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gulati A et al. Collagen IV gene mutations in adults with bilateral renal cysts and CKD. Kidney Int. Rep 5, 103–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wuttke M et al. A COL4A5 mutation with glomerular disease and signs of chronic thrombotic microangiopathy. Clin. Kidney J 8, 690–694 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Salem RM et al. Genome-wide association study of diabetic kidney disease highlights biology involved in glomerular basement membrane collagen. J. Am. Soc. Nephrol 30, 2000–2016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang KL et al. Pathogenic germline variants in 10,389 adult cancers. Cell 173, 355–370 e314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang J et al. Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med 373, 2336–2346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fiorentino M et al. Renal biopsy in patients with diabetes: a pooled meta-analysis of 48 studies. Nephrol. Dial. Transpl 32, 97–110 (2017). [DOI] [PubMed] [Google Scholar]
- 117.Sharma SG et al. The modern spectrum of renal biopsy findings in patients with diabetes. Clin. J. Am. Soc. Nephrol 8, 1718–1724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fogo A et al. Accuracy of the diagnosis of hypertensive nephrosclerosis in African Americans: a report from the African American study of kidney disease (AASK) trial. AASK Pilot Study Investigators. Kidney Int. 51, 244–252 (1997). [DOI] [PubMed] [Google Scholar]
- 119.Freedman BI, Iskander SS, Buckalew VM Jr Burkart JM & Appel RG Renal biopsy findings in presumed hypertensive nephrosclerosis. Am. J. Nephrol 14, 90–94 (1994). [DOI] [PubMed] [Google Scholar]
- 120.Marcantoni C, Ma LJ, Federspiel C & Fogo AB Hypertensive nephrosclerosis in African Americans versus caucasians. Kidney Int. 62, 172–180 (2002). [DOI] [PubMed] [Google Scholar]
- 121.Liu C et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat. Genet 48, 1162–1170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Warren HR et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet 49, 403–415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sandholm N et al. The genetic landscape of renal complications in type 1 diabetes. J. Am. Soc. Nephrol 28, 557–574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van Zuydam NR et al. A genome-wide association study of diabetic kidney disease in subjects with type 2 diabetes. Diabetes 67, 1414–1427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.De Tomasi L et al. Mutations in GREB1L cause bilateral kidney agenesis in humans and mice. Am. J. Hum. Genet 101,803–814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sanna-Cherchi S et al. Exome-wide association study identifies GREB1L mutations in congenital kidney malformations. Am. J. Hum. Genet 101, 789–802 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kingsmore SF et al. A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am. J. Hum. Genet 105, 719–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Goodwin S, McPherson JD & McCombie WR Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet 17, 333–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cummings BB et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med 9, eaal5209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.King DA et al. Detection of structural mosaicism from targeted and whole-genome sequencing data. Genome Res. 27, 1704–1714 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wright CF et al. Clinically-relevant postzygotic mosaicism in parents and children with developmental disorders in trio exome sequencing data. Nat. Commun 10, 2985 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gardner EJ et al. Contribution of retrotransposition to developmental disorders. Nat. Commun 10, 4630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ju YS et al. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature 543, 714–718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lionel AC et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med 20, 435–443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Redin C et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet 49, 36–45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vabres P et al. Postzygotic inactivating mutations of RHOA cause a mosaic neuroectodermal syndrome. Nat. Genet 51, 1438–1441 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mohammadi P et al. Genetic regulatory variation in populations informs transcriptome analysis in rare disease. Science 366, 351–356 (2019).This study incorporated tissue-specific and population-level RNA sequencing data to identify causal genes among patients with genetically unresolved disease, demonstrating the utility of transcriptomics for the diagnosis of rare disease.
- 138.Fresard L et al. Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat. Med 25, 911–919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Posey JE et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med 376, 21–31 (2017).This retrospective analysis of exome sequence data from over 7,000 individuals found that among those with diagnostic genetic findings, nearly 5% had genetic diagnoses involving multiple disease-associated loci. These findings illustrate the importance of considering multilocus variation and using genome-wide sequencing approaches among patients whose presentation is consistent with a hereditary disease but cannot be explained by a single genetic cause.
- 140.Retterer K et al. Clinical application of whole-exome sequencing across clinical indications. Genet. Med 18, 696–704 (2016). [DOI] [PubMed] [Google Scholar]
- 141.Farwell KD et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med 17, 578–586 (2015). [DOI] [PubMed] [Google Scholar]
- 142.Cornec-Le Gall E, Alam A & Perrone RD Autosomal dominant polycystic kidney disease. Lancet 393, 919–935 (2019). [DOI] [PubMed] [Google Scholar]
- 143.Cornec-Le Gall E, Torres VE & Harris P C. Genetic complexity of autosomal dominant polycystic kidney and liver diseases. J. Am. Soc. Nephrol 29, 13–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Porath B et al. Mutations in GANAB, encoding the glucosidase IIalpha subunit, cause autosomal-dominant polycystic kidney and liver disease. Am. J. Hum. Genet 98, 1193–1207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cornec-Le Gall E et al. Monoallelic mutations to DNAJB11 cause atypical autosomal-dominant polycystic kidney disease. Am. J. Hum. Genet 102, 832–844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Besse W et al. ALG9 mutation carriers develop kidney and liver cysts. J. Am. Soc. Nephrol 30, 2091–2102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lanktree MB, Iliuta IA, Haghighi A, Song X & Pei Y Evolving role of genetic testing for the clinical management of autosomal dominant polycystic kidney disease. Nephrol. Dial. Transpl 34, 1453–1460 (2019). [DOI] [PubMed] [Google Scholar]
- 148.Gonzalez-Paredes FJ, Ramos-Trujillo E & Claverie-Martin F Defective pre-mRNA splicing in PKD1 due to presumed missense and synonymous mutations causing autosomal dominant polycystic disease. Gene 546, 243–249 (2014). [DOI] [PubMed] [Google Scholar]
- 149.Xu P et al. A novel splicing mutation in the PKD1 gene causes autosomal dominant polycystic kidney disease in a Chinese family: a case report. BMC Med. Genet 19, 198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lanktree MB et al. Intrafamilial variability of ADPKD. Kidney Int. Rep 4, 995–1003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cornec-Le Gall E et al. Type of PKD1 mutation influences renal outcome in ADPKD. J. Am. Soc. Nephrol 24, 1006–1013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hwang YH et al. Refining genotype-phenotype correlation in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol 27, 1861–1868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Harris PC & Torres VE Polycystic kidney disease, autosomal dominant. GeneReviews https://www.ncbi.nlm.nih.gov/books/NBK1246/ (updated 19 Jul 2018). [Google Scholar]
- 154.Qian F, Watnick TJ, Onuchic LF & Germino GG The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell 87, 979–987 (1996). [DOI] [PubMed] [Google Scholar]
- 155.Reeders ST Multilocus polycystic disease. Nat. Genet 1, 235–237 (1992). [DOI] [PubMed] [Google Scholar]
- 156.Antignac C et al. The future of polycystic kidney disease research–as seen by the 12 Kaplan awardees. J. Am. Soc. Nephrol 26, 2081–2095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Heyer CM et al. Predicted mutation strength of nontruncating PKD1 mutations aids genotype-phenotype correlations in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol 27, 2872–2884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cornec-Le Gall E et al. The PROPKD score: a new algorithm to predict renal survival in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol 27, 942–951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Iliuta IA et al. Polycystic kidney disease without an apparent family history. J. Am. Soc. Nephrol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Song X, Haghighi A, Iliuta IA & Pei Y Molecular diagnosis of autosomal dominant polycystic kidney disease. Expert. Rev. Mol. Diagn 17, 885–895 (2017). [DOI] [PubMed] [Google Scholar]
- 161.Elisakova V et al. Bilineal inheritance of pathogenic PKD1 and PKD2 variants in a Czech family with autosomal dominant polycystic kidney disease — a case report. BMC Nephrol. 19, 163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pei Y et al. Bilineal disease and trans-heterozygotes in autosomal dominant polycystic kidney disease. Am. J. Hum. Genet 68, 355–363 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bergmann C et al. Mutations in multiple PKD genes may explain early and severe polycystic kidney disease. J. Am. Soc. Nephrol 22, 2047–2056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cornec-Le Gall E et al. The value of genetic testing in polycystic kidney diseases illustrated by a family with PKD2 and COL4A1 mutations. Am. J. Kidney Dis 72, 302–308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kashtan CE Alport syndrome. An inherited disorder of renal, ocular, and cochlear basement membranes. Medicine 78, 338–360 (1999). [DOI] [PubMed] [Google Scholar]
- 166.Chiereghin C et al. Alport syndrome cold cases: missing mutations identified by exome sequencing and functional analysis. PLoS One 12, e0178630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Daga S et al. Urine-derived podocytes-lineage cells: a promising tool for precision medicine in Alport syndrome. Hum. Mutat 39, 302–314 (2018). [DOI] [PubMed] [Google Scholar]
- 168.Kamiyoshi N et al. Genetic, clinical, and pathologic backgrounds of patients with autosomal dominant Alport syndrome. Clin. J. Am. Soc. Nephrol 11, 1441–1449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Storey H, Savige J, Sivakumar V, Abbs S & Flinter FA COL4A3/COL4A4 mutations and features in individuals with autosomal recessive Alport syndrome. J. Am. Soc. Nephrol 24, 1945–1954 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jais JP et al. X-linked Alport syndrome: natural history and genotype-phenotype correlations in girls and women belonging to 195 families: a “European community Alport syndrome concerted action” study. J. Am. Soc. Nephrol 14, 2603–2610 (2003). [DOI] [PubMed] [Google Scholar]
- 171.Savige J et al. Alport syndrome in women and girls. Clin. J. Am. Soc. Nephrol 11, 1713–1720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Guo C et al. Severe alport phenotype in a woman with two missense mutations in the same COL4A5 gene and preponderant inactivation of the X chromosome carrying the normal allele. J. Clin. Invest 95, 1832–1837 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rheault MN et al. X-inactivation modifies disease severity in female carriers of murine X-linked Alport syndrome. Nephrol. Dial. Transpl 25, 764–769 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Krol RP et al. Somatic mosaicism for a mutation of the COL4A5 gene is a cause of mild phenotype male Alport syndrome. Nephrol. Dial. Transpl 23, 2525–2530 (2008). [DOI] [PubMed] [Google Scholar]
- 175.Gross O, Netzer KO, Lambrecht R, Seibold S & Weber M Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: impact on clinical counselling. Nephrol. Dial. Transpl 17, 1218–1227 (2002). [DOI] [PubMed] [Google Scholar]
- 176.Jais JP et al. X-linked Alport syndrome: natural history in 195 families and genotype–phenotype correlations in males. J. Am. Soc. Nephrol 11, 649–657 (2000). [DOI] [PubMed] [Google Scholar]
- 177.Marcocci E et al. Autosomal dominant Alport syndrome: molecular analysis of the COL4A4 gene and clinical outcome. Nephrol. Dial. Transpl 24, 1464–1471 (2009). [DOI] [PubMed] [Google Scholar]
- 178.Longo I et al. COL4A3/COL4A4 mutations: from familial hematuria to autosomal-dominant or recessive Alport syndrome. Kidney Int. 61, 1947–1956 (2002). [DOI] [PubMed] [Google Scholar]
- 179.Fallerini C et al. Unbiased next generation sequencing analysis confirms the existence of autosomal dominant Alport syndrome in a relevant fraction of cases. Clin. Genet 86, 252–257 (2014). [DOI] [PubMed] [Google Scholar]
- 180.Rosado C, Bueno E, Felipe C, Valverde S & Gonzalez-Sarmiento R Study of the true clinical progression of autosomal dominant Alport syndrome in a European population. Kidney Blood Press. Res 40, 435–442 (2015). [DOI] [PubMed] [Google Scholar]
- 181.Temme J et al. Incidence of renal failure and nephroprotection by RAAS inhibition in heterozygous carriers of X-chromosomal and autosomal recessive Alport mutations. Kidney Int. 81,779–783 (2012). [DOI] [PubMed] [Google Scholar]
- 182.Wright CF et al. Assessing the pathogenicity, penetrance, and expressivity of putative disease-causing variants in a population setting. Am. J. Hum. Genet 104, 275–286 (2019).This study examines genetic and phenotypic data from nearly 400,000 individuals to evaluate the prevalence and penetrance of putatively pathogenic variants across a variety of monogenic diseases, including type IV collagen-associated nephropathy.
- 183.Findlay GM et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Starita LM et al. A multiplex homology-directed DNA repair assay reveals the impact of more than 1,000 BRCA1 missense substitution variants on protein function. Am. J. Hum. Genet 103, 498–508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Devuyst O et al. Autosomal dominant tubulointerstitial kidney disease. Nat. Rev. Dis. Prim 5, 60 (2019). [DOI] [PubMed] [Google Scholar]
- 186.Eckardt KU et al. Autosomal dominant tubulointerstitial kidney disease: diagnosis, classification, and management — a KDIGO consensus report. Kidney Int. 88, 676–683 (2015). [DOI] [PubMed] [Google Scholar]
- 187.Connor TM et al. Mutations in mitochondrial DNA causing tubulointerstitial kidney disease. PLoS Genet. 13, e1006620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gomez RA & Sequeira-Lopez MLS Renin cells in homeostasis, regeneration and immune defence mechanisms. Nat. Rev. Nephrol 14, 231–245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bolar NA et al. Heterozygous loss-of-function SEC61A1 mutations cause autosomal-dominant tubulo-interstitial and glomerulocystic kidney disease with anemia. Am. J. Hum. Genet 99, 174–187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schubert D et al. Plasma cell deficiency in human subjects with heterozygous mutations in Sec61 translocon alpha 1 subunit (SEC61A1). J. Allergy Clin. Immunol 141, 1427–1438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kirby A et al. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat. Genet 45, 299–303 (2013).This investigation applied specialized sequencing methods to identify causal variation for MUC1-associated autosomal-dominant tubulointerstitial kidney disease, illustrating the limitations of massively parallel sequencing-based analysis alone for the diagnosis of rare monogenic disorders.
- 192.Zivna M et al. Noninvasive immunohistochemical diagnosis and Novel MUC1 mutations causing autosomal dominant tubulointerstitial kidney disease. J. Am. Soc. Nephrol 29, 2418–2431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Baigent C et al. Challenges in conducting clinical trials in nephrology: conclusions from a kidney disease-improving global outcomes (KDIGO) controversies conference. Kidney Int. 92, 297–305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Breyer MD & Susztak K Developing treatments for chronic kidney disease in the 21st century. Semin. Nephrol 36, 436–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Germain DP et al. Treatment of Fabry’s disease with the pharmacologic chaperone migalastat. N. Engl. J. Med 375, 545–555 (2016).This clinical trial demonstrated that the pharmacological chaperone migalastat reduced the severity of renal, cardiac and gastrointestinal manifestations among patients with Fabry disease with responsive GLA mutations. These findings point to the importance of incorporating genotype-level knowledge when assessing the efficacy of novel therapeutic agents.
- 196.Germain DP et al. Efficacy of the pharmacologic chaperone migalastat in a subset of male patients with the classic phenotype of Fabry disease and migalastat-amenable variants: data from the phase 3 randomized, multicenter, double-blind clinical trial and extension study. Genet. Med 21, 1987–1997 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hughes DA et al. Oral pharmacological chaperone migalastat compared with enzyme replacement therapy in Fabry disease: 18-month results from the randomised phase III ATTRACT study. J. Med. Genet 54, 288–296 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Benjamin ER et al. The validation of pharmacogenetics for the identification of Fabry patients to be treated with migalastat. Genet. Med 19, 430–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ribeil JA et al. Gene therapy in a patient with sickle cell disease. N. Engl. J. Med 376, 848–855 (2017). [DOI] [PubMed] [Google Scholar]
- 200.Adams D et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med 379, 11–21 (2018). [DOI] [PubMed] [Google Scholar]
- 201.Benson MD et al. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N. Engl. J. Med 379, 22–31 (2018). [DOI] [PubMed] [Google Scholar]
- 202.Zabaleta N et al. CRISPR/Cas9-mediated glycolate oxidase disruption is an efficacious and safe treatment for primary hyperoxaluria type I. Nat. Commun 9, 5454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liebow A et al. An investigational RNAi therapeutic targeting glycolate oxidase reduces oxalate production in models of primary hyperoxaluria. J. Am. Soc. Nephrol 28, 494–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Goldstein DB et al. Sequencing studies in human genetics: design and interpretation. Nat. Rev. Genet 14, 460–470 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rasouly HM et al. The burden of candidate pathogenic variants for kidney and genitourinary disorders emerging from exome sequencing. Ann. Intern. Med 170, 11–21 (2019). [DOI] [PubMed] [Google Scholar]
- 207.Ramoni RB et al. The undiagnosed diseases network: accelerating discovery about health and disease. Am. J. Hum. Genet 100, 185–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rivera-Munoz EA et al. ClinGen variant curation expert panel experiences and standardized processes for disease and gene-level specification of the ACMG/AMP guidelines for sequence variant interpretation. Hum. Mutat 39, 1614–1622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ewans LJ et al. Whole-exome sequencing reanalysis at 12 months boosts diagnosis and is cost-effective when applied early in Mendelian disorders. Genet. Med 20, 1564–1574 (2018). [DOI] [PubMed] [Google Scholar]
- 210.Liu P et al. Reanalysis of clinical exome sequencing data. N. Engl. J. Med 380, 2478–2480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wenger AM, Guturu H, Bernstein JA & Bejerano G Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet. Med 19, 209–214 (2017). [DOI] [PubMed] [Google Scholar]
- 212.Bastarache L et al. Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science 359, 1233–1239 (2018).This study leveraged electronic health record data to develop phenotypic risk scores and used these scores to detect individuals with undiagnosed rare diseases, including multiple monogenic forms of kidney disease.
- 213.Wuttke M & Kottgen A Insights into kidney diseases from genome-wide association studies. Nat. Rev. Nephrol 12, 549–562 (2016). [DOI] [PubMed] [Google Scholar]
- 214.Torkamani A, Wineinger NE & Topol EJ The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet 19, 581–590 (2018). [DOI] [PubMed] [Google Scholar]
- 215.Liu L & Kiryluk K Genome-wide polygenic risk predictors for kidney disease. Nat. Rev. Nephrol 14, 723–724 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591 (2019).This incisive commentary discusses how the variable performance of genome-wide polygenic risk scores across different ethnic groups could exacerbate existing health disparities, highlighting the need to prioritize greater diversity in genetic research.
- 217.Mostafavi H et al. Variable prediction accuracy of polygenic scores within an ancestry group. eLife 9, e48376 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.