Abstract
Background
Immune-related genes (IRGs) are closely related to the incidence and progression of tumors, potentially indicating that IRGs play an important role in laryngeal squamous cell carcinoma (LSCC).
Material/Methods
An RNA sequencing dataset containing 123 samples was collected from The Cancer Genome Atlas. Based on immune-related differentially expressed genes (IRDEGs), a potential molecular mechanism of LSCC was explored through analysis of information in the Gene Ontology (GO) resource and the Kyoto Encyclopedia of Genes and Genomes (KEGG), and protein-protein interactions (PPIs). A regulatory network of transcriptional regulators and IRDEGs was constructed to explore the underlying molecular mechanism of LSCC at the upstream level. Candidates from IRDEGs for signature were screened via univariate Cox analysis and using the least absolute shrinkage and selection operator (LASSO) technique. The IRDEG signature of LSCC was constructed by using a multivariate Cox proportional hazards model.
Results
GO and KEGG analysis showed that IRDEGs may participate in the progression of LSCC through immune-related reactions. PPI analysis demonstrated that, among the IRDEGs in LSCC, the Kininogen 1; C-X-X motif chemokine ligand 10; elastase, neutrophil expressed; and LYZ genes are hub genes in the development of LSCC. At the upstream level, SPI1, SP140, signal transducer and activator of transcription 4, zinc finger E-box binding homeobox, and Ikaros family zinc finger 2 are the hub transcriptional regulators of IRDEGs. The risk score based on the IRDEG signature was able to distinguish prognosis in patients with LSCC and represents an independent prognostic risk factor for LSCC.
Conclusions
From the perspective of IRGs, we first constructed an IRDEG signature related to the prognosis of LSCC, which can be used as a novel marker to predict prognosis in patients with LSCC.
MeSH Keywords: Carcinoma, Squamous Cell; Databases, Nucleic Acid; Immunity; Prognosis; Transcriptome
Background
Laryngeal squamous cell carcinoma (LSCC) is currently the most common pathological classification of laryngeal carcinoma (LC), accounting for more than 90% of cases [1–6]. Globally, more than 177 000 cases of LC are diagnosed every year, and more than 94 000 deaths from the disease [7]. Currently, treatments such as surgery, radiation therapy, and chemotherapy are used for early-stage LC [8,9], with favorable effects on survival [10–12]. However, more than two-thirds of patients are diagnosed with LC when the disease is already at an advanced stage [13–16]. This delay facilitates the growth and spread of LC cells [17], posing challenges for successful treatment of the disease.
Several pathogenic factors and mechanisms have been proposed for LC. Previous research has found that the incidence and development of it may be related to multiple risk factors, such as drinking [18–20], smoking [13], asbestos exposure [21], and human papillomavirus infection [22]. The mechanism may involve multiple molecules (such as MYC Target 1 [23], p16 [24], and NLK [25]), and multiple pathways (such as Wnt/β-catenin [26], ATR/p53/caspase-3 [27], and phosphatidylinositol-3-kinase/Akt/mTOR [28]); clearly, the onset and development of LC are complex. In summary, it is necessary to further explore the molecular mechanism of the pathogenesis and development of LC and identify novel and effective prognostic markers to assist in clinical screening and treatment of LC.
The immune system has been considered to affect the development, growth, and metastasis of tumors. Immune-related genes (IRGs) are considered to be relevant to tumor prognosis. For example, high expression of FGF19 may promote breast cancer progression by activating Akt signaling in cells, resulting in a discouraging prognosis for individuals with the disease [29]. Similarly, in esophageal squamous cell carcinoma, patients with TUBB3-negative carcinomas had higher rates of disease-free survival (DFS) and overall survival (OS) rates than those with TUBB3-positive disease [30]. In glioblastomas, another IRG, RBP1, is considered to be associated with poor prognosis [31]. Studies of LSCC and IRGs have found that programmed death-ligand 1 (PD-L1) expression is associated with favorable prognosis [32], and STC2 [33] overexpression is associated with poor prognosis. Clearly, IRGs are likely to contribute to the incidence, growth, and spread of LSCC. Therefore, exploring the relationship between IRGs and LSCC prognosis is crucial to understanding the pathogenesis of the disease and facilitating clinical screening and treatment.
High-throughput sequencing technologies are widely used in cancer multi-omics. In this study, we combined high-throughput data from public databases and published studies to explore the potential molecular mechanism of development of LSCC and to identify a prognostic signature based on IRGs to assist in clinical screening and treatment of LSCC.
Material and Methods
Collection of sample data and IRGs
The RNA sequencing (RNA-Seq) dataset from The Cancer Genome Atlas (TCGA) was obtained through the Genomic Data Commons Data Portal database (https://portal.gdc.cancer.gov/repository). The dataset contained 111 LSCC samples and 12 non-LSCC control samples. Clinical information from the samples in the RNA-Seq dataset was obtained from the University of California Santa Cruz Xena database (http://xena.ucsc.edu/). In addition, a list of 1811 IRGs was obtained from the ImmPort database (https://www.immport.org/home).
Differential expression analysis
RNA-Seq count data were input into R (v3.6.1), and through the edgeR package [34], differentially expressed genes (DEGs) that had a log2-fold change of 2 or more and an adjusted P<0.05 were selected for further study. Genes were identified as upregulated (up-DEGs) if they showed a log2-fold change of 2 or more, while genes with a log2 fold change less than −2 were considered downregulated (down-DEGs). The same methods were used to screen immune-related differentially expressed genes (IRDEGs), immune-related upregulated DEGs (up-IRDEGs), and immune-related downregulated DEGs (down-IRDEGs).
Analysis of the potential molecular mechanism of IRDEGs
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of up-IRDEGs and down-IRDEGs were performed using the clusterProfiler package [35] in R (v3.6.1). GO terms and KEGG signaling pathways were then filtered by identifying those that simultaneously satisfied the following 3 conditions: (1) adjusted P<0.05; (2) q<0.05; and (3) number of enriched genes ≥5. The STRING database (https://string-db.org/) was used for analysis of protein-protein interactions (PPIs) of up-IRDEGs and down-IRDEGs. The minimum required interaction score for a PPI was set at 0.7 in the STRING database, which is considered by the database as high confidence. The CytoHub plug-in in Cytoscape software (v3.7.2) was applied to screen for hub genes, and the hub genes of up-IRDEGs and down-IRDEGs were identified according to the degree algorithm.
Transcriptional regulator (TR) prediction for IRDEGs and construction of TR-mediated regulatory network
To further understand the potential molecular mechanisms of up-IRDEGs and down-IRDEGs in LSCC, we attempted to explore the upstream regulation mechanism of these genes through the correlation between the genes and transcriptional regulators (TRs). TRs for genes were predicted using Epigenetic Landscape In Silico (Lisa, http://lisa.cistrome.org/), a bioinformatics analysis tool [36] that contains a large amount of H3K27ac ChIP-seq data from Homo sapiens. We predicted TRs related to up-IRDEGs and down-IRDEGs with the Lisa tool and selected TRs with P<0.01. Pearson coefficients of expression levels of TRs and IRDEGs were calculated in R, while the TR-IRDEG regulatory network was constructed using Cytoscape.
Development and assessment of IRDEG signature
Univariate Cox analysis and least absolute shrinkage and selection operator (LASSO) were used to select the candidates for signature from IRDEGs, and a multivariate Cox proportional hazards model was applied to construct the IRDEG signature of LSCC based on candidate IRDEGs (Figure 1). Using univariate Cox analysis, IRDEGs related to the survival of patients with LSCC (P<0.05) were identified and named as preliminary candidate IRDEGs. Then, based on the LASSO regression analysis, which can quickly and effectively extract important variables from many variables, secondary candidate IRDEGs were obtained from the preliminary candidate IRDEGs. Eventually, through the multivariate Cox proportional hazard model, some of the secondary candidate IRDEGs were used to construct the IRDEG signature of LSCC.
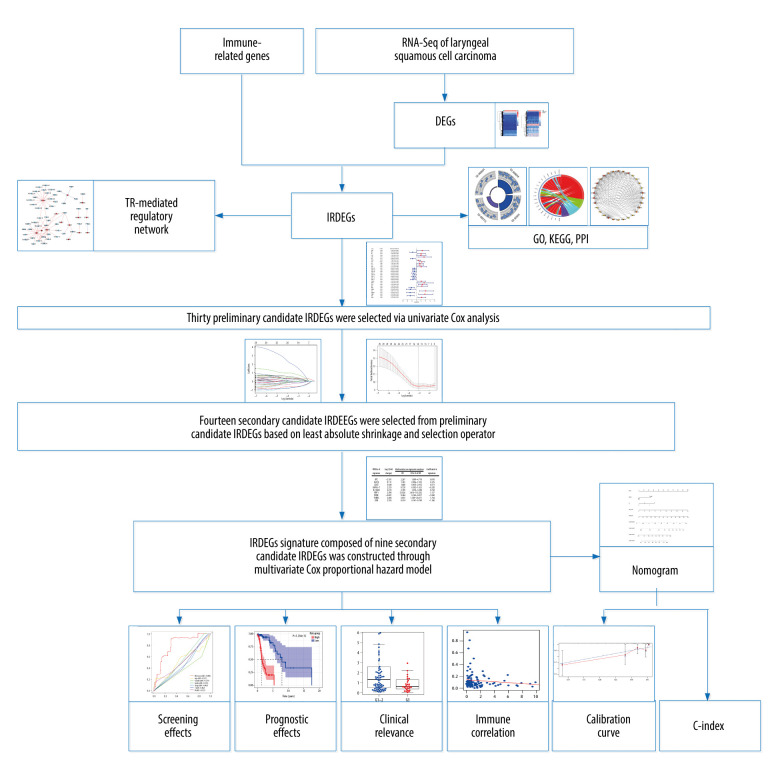
Figure 1.
The main processes of this study. DEGs – differentially expressed genes; IRDEGs – immune-related differentially expressed genes; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; PPI – protein–protein interaction; TR – transcriptional regulator; C-index – concordance index.
Curves for receiver operating characteristics (ROCs) and survival analysis (Kaplan-Meier) were used to evaluate the relationship between the IRDEG signature and the effects on screening and prognosis, respectively. Univariate and multivariate Cox regression analyses were used to identify factors related to independent prognoses in the IRDEG signature and clinical parameters. Development and assessment of the IRDEG signature were completed in R with the help of the glmnet [37], survival, and survivalROC package [38].
Relationship between risk score of IRDEG signature and immune infiltration level
The Tumor IMmune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) is an online tool for comprehensive analysis of tumor infiltration of immune cells. The table matrix of TCGA estimation in TIMER was used to analyze the correlation between the IRDEG signature infiltration level risk score and 6 types of immune cells: B cells, CD4 T cells, CD8 T cells, macrophages, neutrophils, and dendritic cells.
Application of IRDEG signature
Combining the IRDEG signature and the clinical parameters related to the independent prognosis of LSCC, a nomogram was constructed to quantify the risk of LSCC in individuals in a clinical environment. A calibration curve was used to detect the difference between the predicted and actual survival rates in the nomogram. The concordance index (C-index; range 0–1) was used to objectively evaluate the predictive power of the signature; the larger the C-index, the better the predictive power of the signature. Construction and verification of the nomogram were performed in R.
Statistical analysis
Except for differential expression analysis, for which original counts should be used, the RNA-Seq data used in the rest of the analysis were log2 (counts+1) converted data. All statistical analyses were performed in R.
The main processes in the present study are shown in Figure 1. Unless otherwise specified, P<0.05 for a difference was considered statistically significant.
Results
Identification of DEGs and IRDEGs in LSCC
After filtering, 4274 DEGs were included: 2990 up-DEGs and 1284 down-DEGs. Among the 1811 IRGs, there were 302 IRDEGs, including 224 up-IRDEGs and 78 down-IRDEGs. Supplementary Figure 1 shows the expression of DEGs and IRDEGs in LSCC.
GO, KEGG, and PPI analyses of IRDEGs
To analyze the potential molecular mechanism of LSCC, we conducted GO, KEGG, and PPI analyses of IRDEGs.
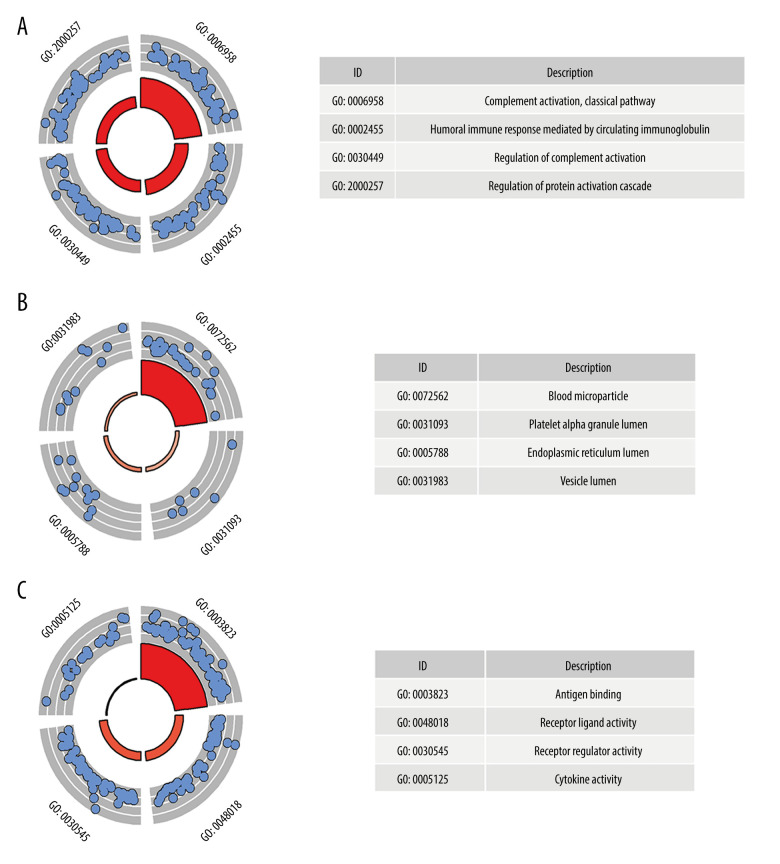
GO analysis was performed on 224 up-IRDEGs and 78 down-IRDEGs. Among up-IRDEGs, the most common biological terms were “complement activation, classical pathway” (“biological process”), “blood microparticle” (“cell component”), and “antigen binding” (“molecular function”) (Figure 2A–2C). For down-IRDEGs, the most common biological process, cell component, and molecular function terms were “antimicrobial humoral response,” “azurophil granule lumen,” and “receptor ligand activity,” respectively (Supplementary Figure 2A–2C). Table 1 shows the 4 most enriched terms among the 3 types of terms.
Figure 2.
Gene Ontology (GO) analysis for immune-related upregulated differentially expressed genes. (A) Biological process. (B) Cellular component. (C) Molecular function. The blue nodes in the concentric circles represent genes clustered in specific GO terms. The larger size and darker color of the internal departments represent more significant enrichment of GO terms.
Table 1.
Gene Ontology analysis (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis for immune-related up-regulated differentially genes and immune-related down-regulated differentially genes.
| Category | ID | Description | P. adjust | Q value | Count |
|---|---|---|---|---|---|
| GO-BP | GO: 0006958 | Complement activation, classical pathway | 3.57E-65 | 2.95E-65 | 46 |
| GO-BP | GO: 0002455 | Humoral immune response mediated by circulating immunoglobulin | 6.27E-62 | 5.17E-62 | 46 |
| GO-BP | GO: 0030449 | Regulation of complement activation | 2.89E-61 | 2.38E-61 | 47 |
| GO-BP | GO: 2000257 | Regulation of protein activation cascade | 3.59E-61 | 2.96E-61 | 47 |
| GO-CC | GO: 0072562 | Blood microparticle | 3.93E-29 | 3.70E-29 | 30 |
| GO-CC | GO: 0031093 | Platelet alpha granule lumen | 0.002598 | 0.002448 | 6 |
| GO-CC | GO: 0005788 | Endoplasmic reticulum lumen | 0.002598 | 0.002448 | 12 |
| GO-CC | GO: 0031983 | Vesicle lumen | 0.036346 | 0.034244 | 10 |
| GO-MF | GO: 0003823 | Antigen binding | 2.12E-88 | 1.81E-88 | 64 |
| GO-MF | GO: 0048018 | Receptor ligand activity | 2.65E-32 | 2.27E-32 | 45 |
| GO-MF | GO: 0030545 | Receptor regulator activity | 3.66E-31 | 3.13E-31 | 45 |
| GO-MF | GO: 0005125 | Cytokine activity | 5.77E-15 | 4.93E-15 | 20 |
| KEGG | hsa04060 | Cytokine-cytokine receptor interaction | 2.32E-22 | 2.09E-22 | 33 |
| KEGG | hsa04061 | Viral protein interaction with cytokine and cytokine receptor | 2.47E-11 | 2.23E-11 | 15 |
| KEGG | hsa04062 | Chemokine signaling pathway | 1.47E-06 | 1.33E-06 | 14 |
| KEGG | hsa04080 | Neuroactive ligand-receptor interaction | 1.88E-06 | 1.69E-06 | 18 |
| GO-BP | GO: 0019730 | Antimicrobial humoral response | 4.50E-08 | 3.61E-08 | 10 |
| GO-BP | GO: 0006959 | Humoral immune response | 2.16E-05 | 1.74E-05 | 11 |
| GO-BP | GO: 0009620 | Response to fungus | 3.92E-05 | 3.14E-05 | 6 |
| GO-BP | GO: 0031640 | Killing of cells of other organism | 6.00E-05 | 4.82E-05 | 6 |
| GO-CC | GO: 0035578 | Azurophil granule lumen | 1.25E-05 | 1.06E-05 | 7 |
| GO-CC | GO: 0005775 | Vacuolar lumen | 3.27E-05 | 2.76E-05 | 8 |
| GO-CC | GO: 0031983 | Vesicle lumen | 3.27E-05 | 2.76E-05 | 10 |
| GO-CC | GO: 0031012 | Extracellular matrix | 0.000352 | 0.000297 | 10 |
| GO-MF | GO: 0048018 | Receptor ligand activity | 1.81E-24 | 1.40E-24 | 28 |
| GO-MF | GO: 0030545 | Receptor regulator activity | 6.03E-24 | 4.65E-24 | 28 |
| GO-MF | GO: 0005125 | Cytokine activity | 1.04E-14 | 8.01E-15 | 15 |
| GO-MF | GO: 0008201 | Heparin binding | 1.07E-05 | 8.23E-06 | 8 |
| KEGG | hsa04060 | Cytokine-cytokine receptor interaction | 2.88E-12 | 2.72E-12 | 19 |
| KEGG | hsa04659 | Th17 cell differentiation | 0.02884 | 0.02727 | 5 |
| KEGG | hsa04630 | JAK-STAT signaling pathway | 0.02884 | 0.02727 | 6 |
| KEGG | hsa05321 | Inflammatory bowel disease (IBD) | 0.02884 | 0.02727 | 4 |
Count – count of genes enriched in the category; BP – biological process; CC – cellular component; MF – molecular function. The deep blue area was based on the analysis of immune-related up-regulated differentially genes while the light blue area was based on the analysis of immune-related down-regulated differentially genes.
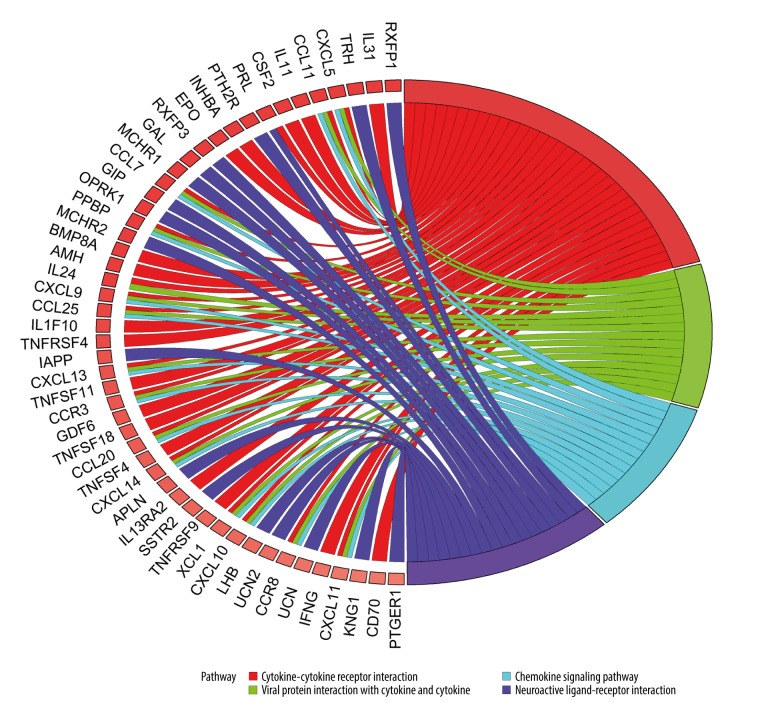
KEGG analysis results showed that the most enriched KEGG pathway for both up-IRDEGs (Figure 3) and down-IRDEGs (Supplementary Figure 3) was the “cytokine-cytokine receptor interaction.” The 4 most enriched pathways for up-IRDEGs and down-IRDEGs are shown in Figure 3, Supplementary Figure 3, and Table 1. The GO and KEGG analyses show that the biological terms enriched in IRDEGs and the KEGG signaling pathway are closely related to immunity. In fact, these results are not surprising, given that the IRDEGs used for KEGG analysis were originally genes closely related to immunity.
Figure 3.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of immune-related upregulated differentially expressed genes. Different color bands correspond to different KEGG enrichment pathways.
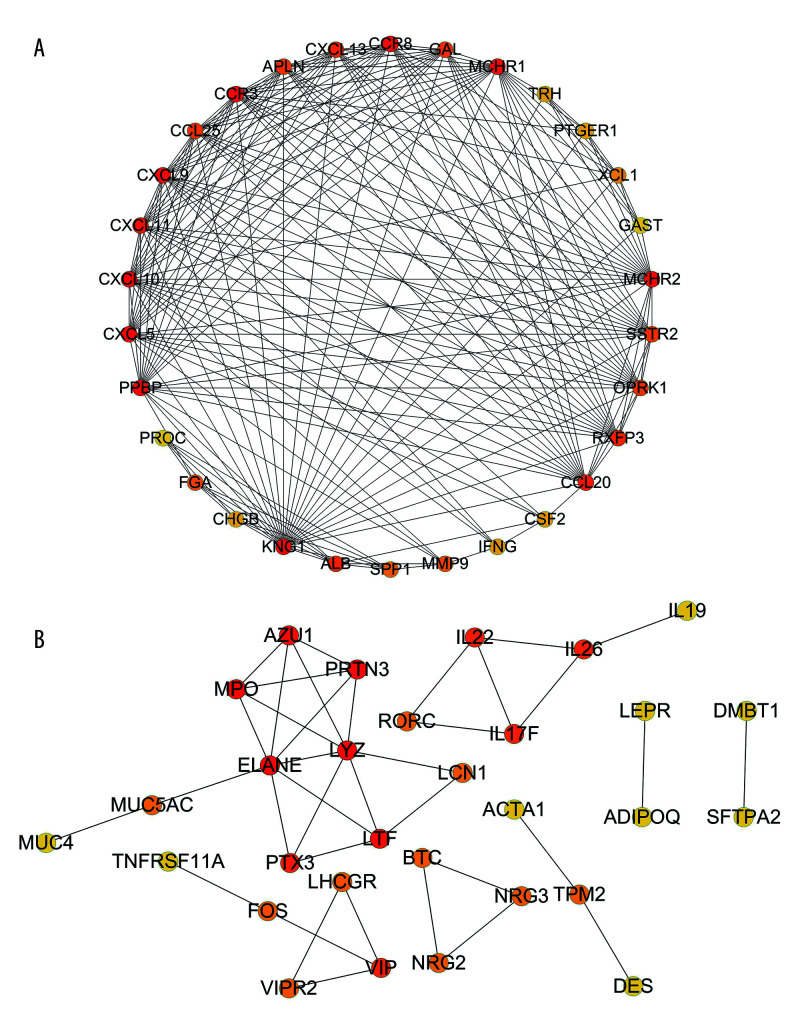
The results of the PPI analysis and the degree algorithm showed that in up-IRDEGs, Kininogen 1 (KNG1) and C-X-C motif chemokine ligand 10 (CXCL10) were hub genes in the development of LSCC, while in down-IRDEGs, the hub genes were elastase, neutrophil expressed (ELANE) and lysozyme (LYZ). Figure 4A and 4B, respectively, show the interaction between the first 30 up-IRDEGs and down-IRDEGs obtained with the Degree algorithm.
Figure 4.
Protein-protein interaction analysis. (A) Immune-related upregulated differentially expressed genes. (B) Immune-related downregulated differentially expressed genes.
Construction of a TR-mediated regulatory network
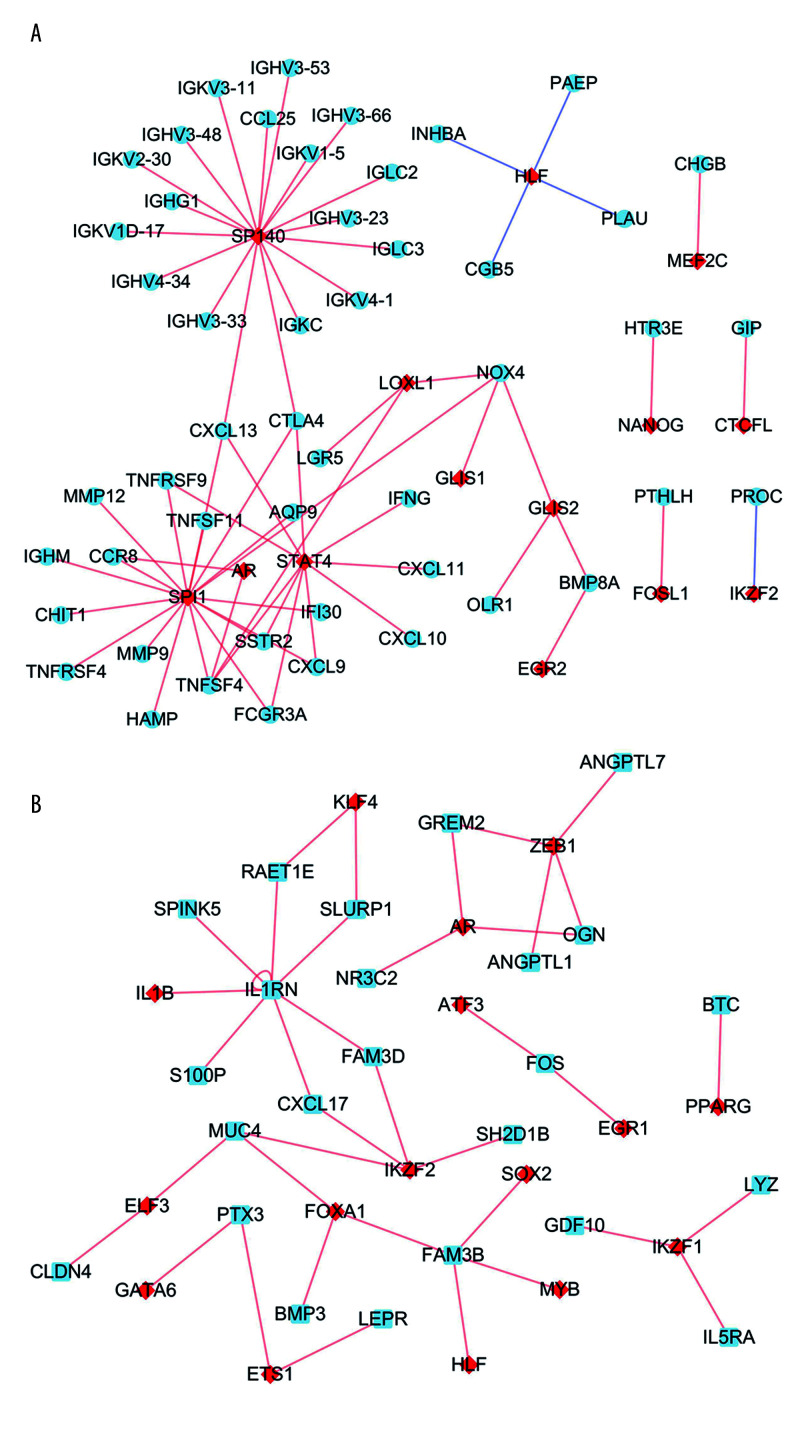
Based on the prediction results from the Lisa tool, we screened 103 up-TRs related to up-IRDEGs with a threshold of P<0.01. To reduce false positives in TR prediction results, we conducted a correlation analysis on the 96 up-TRs included in up-IRDEGs and RNA-Seq and finally selected 15 real up-TRs and 50 up-IRDEGs with Pearson correlation coefficients >0.5 and P<0.05 for constructing up-TR-IRDEGs. The results showed that SPI1, SP140, and signal transducer and activator of transcription 4 (STAT4) were likely to be the hub TRs regulating up-IRDEGs (Figure 5A).
Figure 5.
Transcriptional regulator (TR)-mediated regulatory networks. (A) Immune-related upregulated differentially expressed genes. (B) Immune-related downregulated differentially expressed genes. The red and blue nodes represent TRs and immune-related differentially expressed genes, respectively. The red and blue edges represent positive and negative correlation, respectively.
In the same way, we screened 159 down-TRs, 147 of which were included in the RNA-Seq dataset used in this study. Finally, we selected 17 real down-TRs and 24 down-IRDEGs with correlation coefficients >0.5 to construct down-TRs-IRDEGs. The results indicated that zinc finger E-box binding homeobox (ZEB1) and Ikaros family zinc finger 2 (IKZF2) may be important TRs in regulating down-IRDEGs (Figure 5B).
Construction of IRDEGs signature
Through univariate Cox analysis, 30 preliminary candidate IRDEGs related to the survival of patients with LSCC were obtained from 302 IRDEGs, as shown in Supplementary Figure 4. Based on LASSO regression analysis and selecting the largest lambda with an average error within 1 standard deviation, 14 secondary candidate IRDEGs for construction of an IRDEG signature were selected from as many as 30 preliminary IRDEGs (Supplementary Figure 5). In total, 14 secondary candidate IRDEGs – beta cellulin (BTC), EPO, fibroblast growth factor (FGF)19, FGF5, gastrin (GAST), immunoglobulin heavy chain variable (IGHV)3–7, IGHV6–1, interleukin 13 receptor subunit alpha 2 (IL13RA2, retinol binding protein 1 (RBP1), RAR related orphan receptor C (RORC), semaphoring 6C (SEMA6C), TNR receptor superfamily member 4 (TNFRSF4), TUBB3, and urocortin (UCN) – were used to construct the IRDEG signature. Finally, we used the multivariate Cox proportional hazards model to construct the IRDEG signature of LSCC. The final IRDEG signature contained 9 secondary candidate IRDEGs for LSCC. As shown in Table 2, of the 9 IRDEGs, 7 (FGF19, GAST, IGHV3–7, IL13RA2, RBP1, TUBB3, UCN) were highly expressed in LSCC, while IL13RA2, RBP1, and TUBB3 are prognostic risk factors for patients with LSCC (hazard ratio [HR] >1 and 95% confidence interval [CI] do not contain 1). Of the 9 IRDEGs, BTC and RORC are expressed at low levels in LSCC and BTC is a risk factor for patients with LSCC, while RORC is a prognostic protective factor (HR <1 and 95% CI do not contain 1). The 9 genes included in the IRDEG signature as well as the coefficients of each gene are listed in Table 2. The product of the expression value of each gene and the gene’s corresponding coefficient is the contribution of the gene to the risk score, and the sum of the contribution of all genes is the risk score, and we performed a risk score calculation for 111 LSCC samples.
Table 2.
Immune-related differentially expressed genes (IRDEGs) included in the signature.
| IRDEGs of signature | Log2 (fold change) | Multivariate Cox regression analyses | Coefficient in signature | |
|---|---|---|---|---|
| HR | 95% CI of HR | |||
| BTC | −2.181 | 2.267 | 1.089–4.718 | 0.818 |
| FGF19 | 8.111 | 1.383 | 0.996–1.922 | 0.325 |
| GAST | 5.928 | 1.668 | 0.955–2.912 | 0.511 |
| IGHV3–7 | 2.533 | 0.720 | 0.502–1.033 | −0.328 |
| IL13RA2 | 2.413 | 2.100 | 1.230–3.584 | 0.742 |
| RBP1 | 2.596 | 23.026 | 3.416–155.202 | 3.137 |
| RORC | −2.691 | 0.344 | 0.145–0.817 | −1.068 |
| TUBB3 | 2.400 | 5.835 | 1.380–24.671 | 1.764 |
| UCN | 2.183 | 0.314 | 0.141–0.700 | −1.160 |
HR – hazard ratio; CI – confidence interval.
Validation of IRDEG signature
To evaluate the screening and prognostic effects of the signature in LSCC prognosis, patients with LSCC were divided into high- and low-risk groups according to scores higher or lower than the median risk score (0.859).
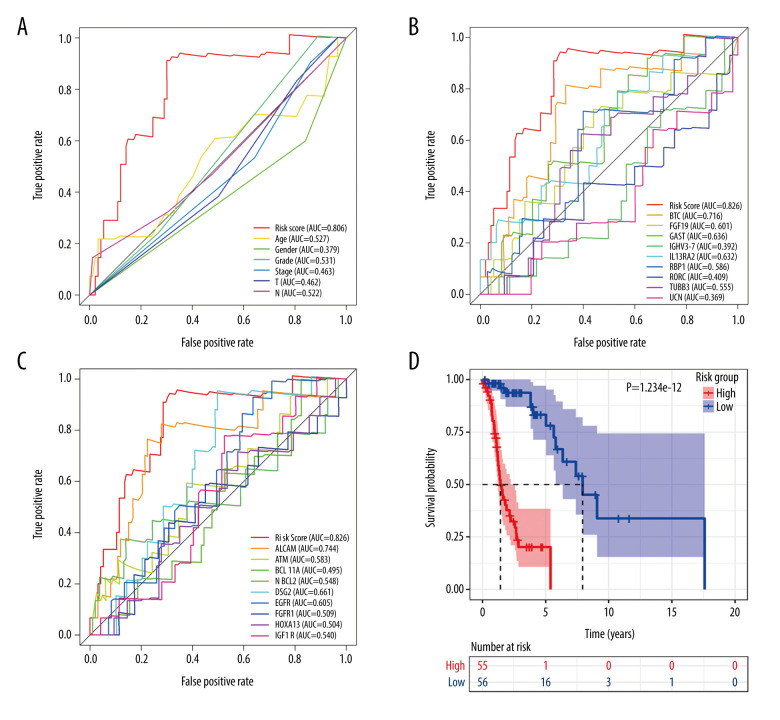
In terms of screening, the ROC curve showed that the risk score calculated according to the signature was the best measure for determining the survival of patients with LSCC, with a higher area under the curve than any clinical parameter (age, sex, neoplasm histologic grade, stage, tumor stage, node stage; Figure 6A) and any single gene included in the IRDEG signature (Figure 6B). Moreover, we also compared the performance of the signature to potential markers related to the prognosis of LSCC identified in research recently added to PubMed (published between January 1, 2020 and May 15, 2020), while 9 of the potential markers – activated leukocyte cell adhesion molecular (ALCAM) [39], ATM [40], BAF chromatin remodeling complex subunit (BCL11A) [41], B-cell lymphoma 2 (BCL-2) [42], desmoglein 2 (DSG2) [43], epidermal growth factor receptor (EGFR) [44], FGFR1 [45], homeobox A13 (HOXA13) [46], insulin like growth factor 1 receptor (IGF-1R) [47] – were included in the RNA-Seq data used in this study. Compared with any gene from these 9 potential prognostic markers, the risk score based on the IRDEG signature showed the best ability to screen for prognosis in patients with LSCC (Figure 6C).
Figure 6.
Signature of immune-related differentially expressed genes (IRDEGs). Receiver operating characteristic curves. (A) Risk score and clinical parameters. (B) IRDEGs. (C) Nine prognostic markers. (D) Kaplan-Meier curve of high- and low-risk groups, which were classified based on median risk score of IRDEGs signature.
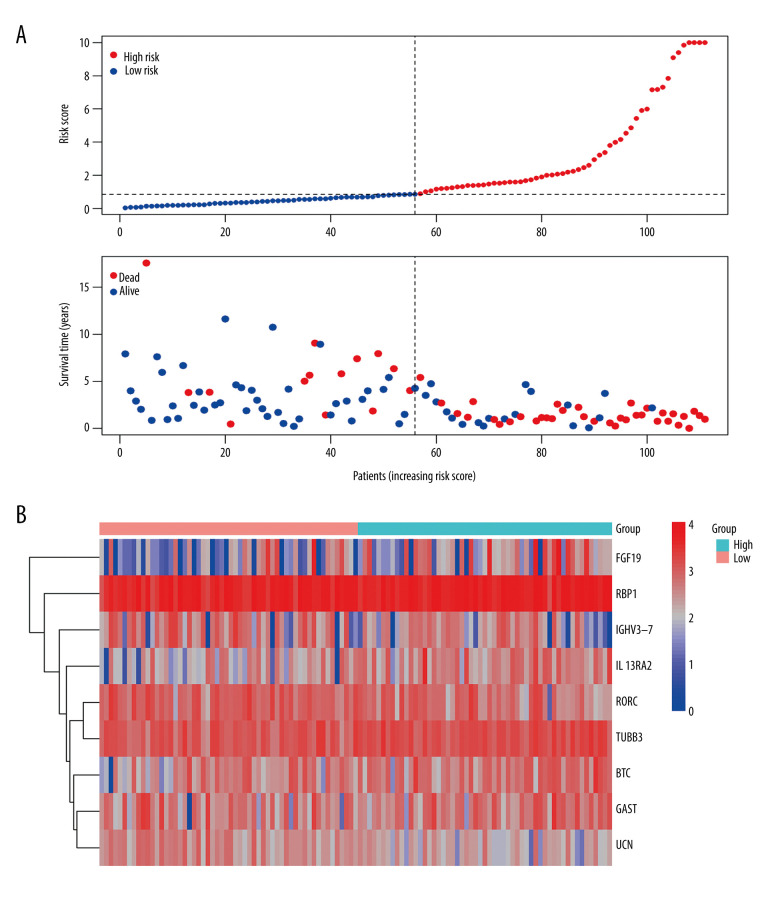
In terms of prognosis, the OS rate for the high-risk group was significantly lower than that for the low-risk group (Figure 6D, P=1.234e-12). Moreover, the risk curve showed that the majority of patients with LSCC who died were those with a higher risk score, whereas there were significantly fewer deaths in the group with low-risk scores (Figure 7A). Figure 7B shows the expression of the 9 IRDEGs in the signature in the high- and low-risk groups.
Figure 7.
(A) Risk plots for high- and low-risk groups. Each blue dot represents a living patient, whereas each red dot represents a dead patient. (B) Heatmap of expression levels of the 9 genes in response to IRDEGs signature in patients.
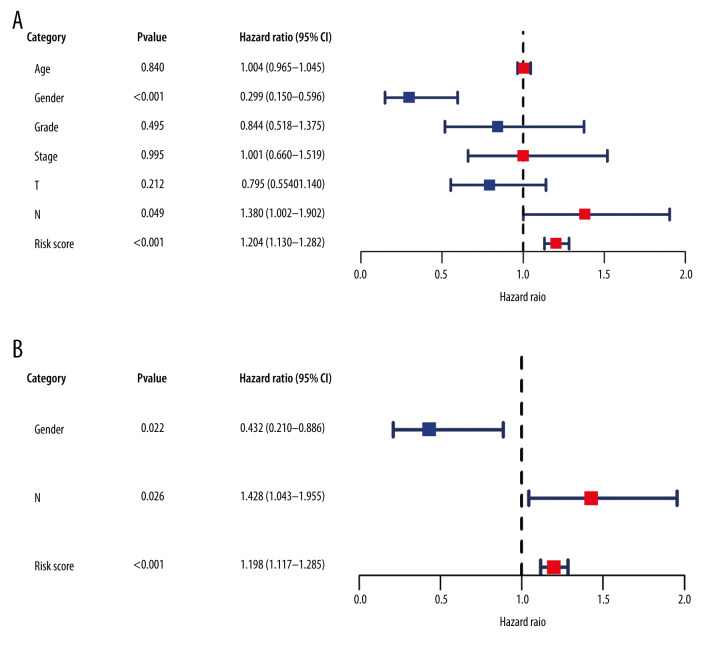
In addition, we used Cox regression analysis to explore prognostic factors in LSCC. The results of univariate and multivariate Cox regression analysis demonstrated that the risk score based on the IRDEG signature can be considered a risk factor for the independent prognosis of LSCC (Figure 8A, 8B). In terms of risk of death, the risk for men with LSCC was 0.432 times that for women, whereas the node stage of the tumor was a risk factor related to the prognosis of LSCC (Figure 8B).
Figure 8.
Forest plots reflecting factors related to independent prognosis of laryngeal squamous cell carcinoma. (A) Plot based on univariate Cox regression analysis. (B) Plot based on multivariate Cox regression analysis.
Relationship between IRDEG signature and clinical parameters in patients with LSCC
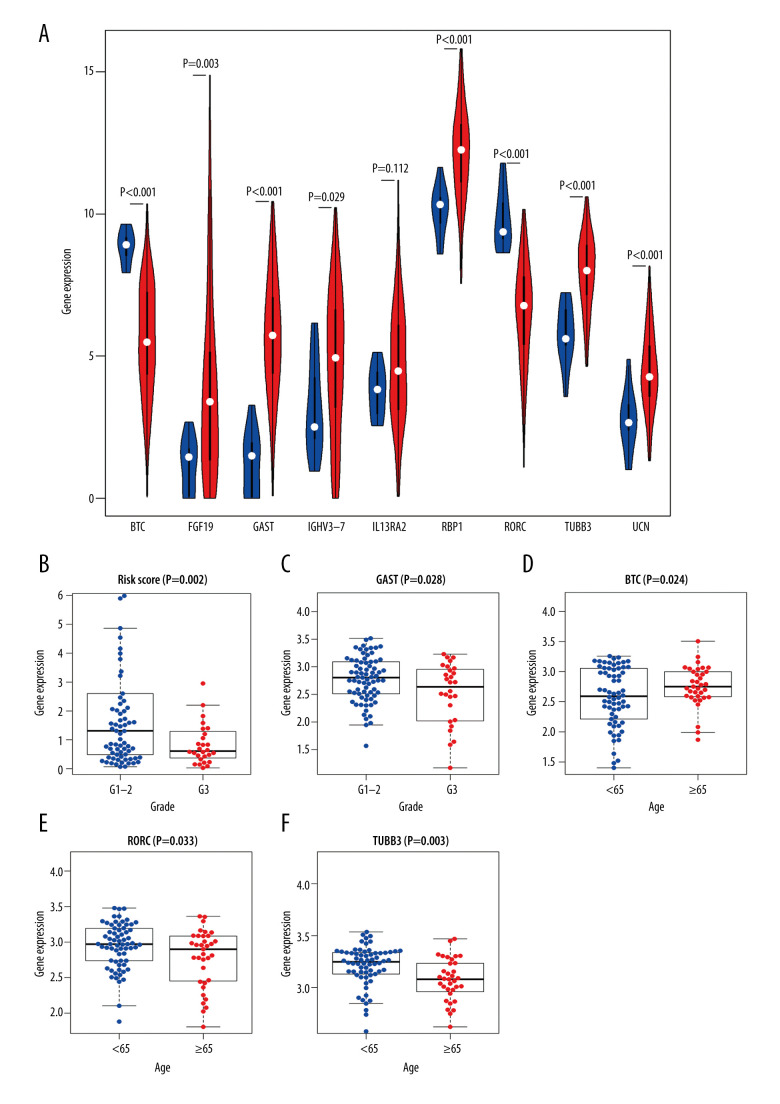
To explore the clinical significance of IRDEGs in the signature of LSCC, we studied the expression of each IRDEG in the signature and its relationship with clinical parameters. Among the 9 IRDEGs included in the signature, BTC and RORC had low levels of expression in LSCC and the remaining 7 genes were highly expressed in LSCC. A Wilcoxon test showed that there were statistically significant differences in expression of 6 of the remaining 7 genes in the LSCC and non-LSCC samples. The exception was IL13RA2 (Figure 9A). Subsequently, based on data from 102 patients with LSCC for whom we had complete clinical information (Supplementary Table 1), we explored the relationship between risk score and expression level of the 9 genes and the clinical parameters in the patients (age, sex, neoplasm histologic grade, stage, and tumor and node stage), and found that the risk score and GAST were related to the neoplasm histologic grade of LSCC (Figure 9B, 9C), while the levels of expression of BTC, RORC, and TUBB3 were related to the age of patients with LSCC (Figure 9D–9F).
Figure 9.
Clinical significance of immune-related differentially expressed genes (IRDEGs) signature in laryngeal squamous cell carcinoma (LSCC). (A) Violin plot showing the difference in expression levels of 9 IRDEGs between LSCC samples (red violin) and control samples (blue violin), based on Wilcoxon test. (B–F) relationship between IRDEGs of signature and clinical parameters. “Grade” means neoplasm histologic grade. Age was in years.
In addition, we analyzed the relationship between the IRDEG signature and the infiltration levels of B and CD4 and CD8 T cells; macrophages; neutrophils; and dendritic cells and found that the risk score based on the IRDEG signature was negatively correlated with the level of B-cell immune infiltration (Supplementary Figure 6).
Construction and verification of nomogram based on IRDEG signature
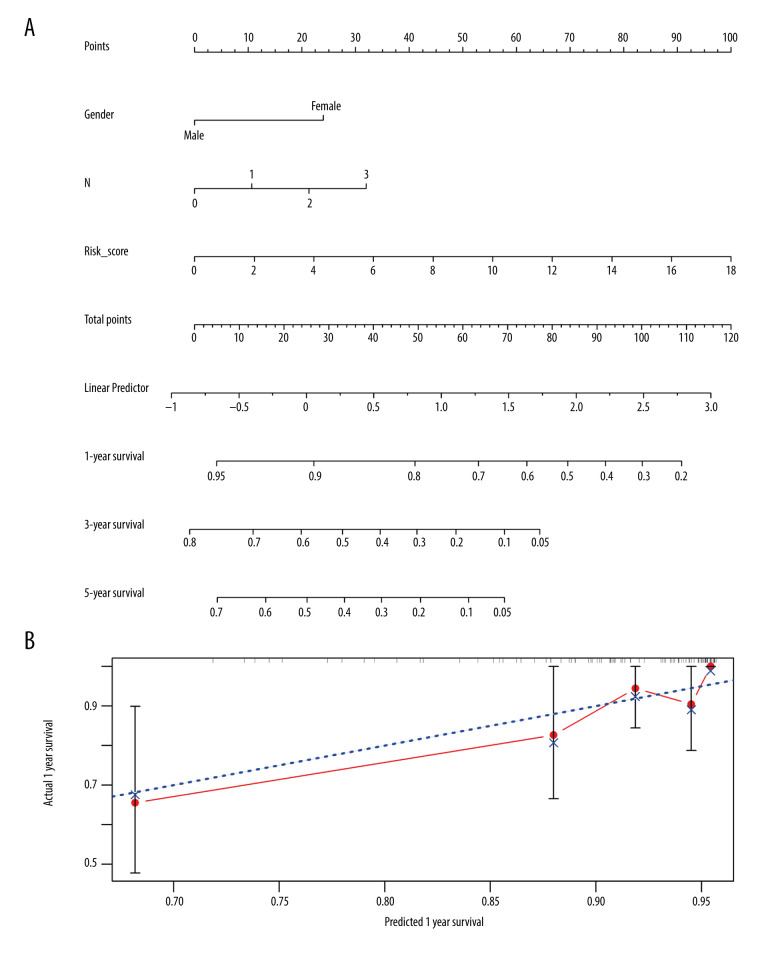
To investigate a preliminary application of the IRDEG signature, we constructed a nomogram of it and the clinical parameters related to independent prognosis of LSCC and predicted 1-, 3-, and 5-year survival rates in patients with LSCC (Figure 10A).
Figure 10.
Nomogram and corresponding verification. (A) Nomogram based on immune-related differentially expressed genes and clinical parameters. (B) Calibration curve with a horizontal axis that represents the predicted value of the nomogram and a vertical axis representing the actual survival rate of patients. N, nodes stage.
The calibration curve revealed that although the 3- and 5-year survival rate predictions by the nomogram were not in good agreement with actual patient survival (data not shown), the 1-year survival rate predictions were close to actual survival (Figure 10B). Moreover, the more objective C-index was 0.777 (standard deviation=0.031), indicating that the nomogram could predict the survival rate with moderate accuracy.
Discussion
According to current research, a number of IRGs are closely related to the incidence and progression of tumors, potentially indicating that IRGs play an important role in cancer [32]. Therefore, new markers for screening and treatment of tumors could be discovered through exploration of and research about IRGs.
Using the RNA-Seq dataset containing 123 samples, we first explored the potential molecular mechanism of LSCC from the perspective of IRGs, and for the first time, constructed an IRDEG signature related to the prognosis of LSCC based on IRGs. Through GO and KEGG analysis, we found, as expected, that IRDEGs may participate in the progression of LSCC through immune-related reactions. PPI analysis identified hub genes among IRDEGs in LSCC. We then constructed a regulatory network of TRs and IRDEGs to explore the underlying molecular mechanism of LSCC at the upstream level. In addition, the risk score based on the IRDEG signature that we constructed did well in distinguishing the prognosis of patients with LSCC. The risk score based on the IRDEG signature was an independent prognostic risk factor for LSCC, indicating that the IRDEG signature can be used as a novel marker to predict prognosis in patients with LSCC.
Initially, we explored the underlying molecular mechanism of LSCC from the perspective of IRGs. To this end, we conducted enrichment and PPI analyses on IRDEGs. In up-IRDEGs, the most common GO terms in biological processes, cellular components, and molecular functions were “complement activation, classical pathway,” “blood microparticle,” and “antigen binding,” respectively, while in down-IRDEGs, the terms were “antimicrobial humoral response,” “azurophil granule lumen,” and “receptor ligand activity,” respectively. The KEGG analysis revealed that the most enriched pathways for both up-IRDEGs and down-IRDEGs was the “cytokine-cytokine receptor interaction.” It is worth mentioning that quite a few GO terms and KEGG signaling pathways enriched in IRDEGs were closely related to tumor progression and immune response [48,49]; this, to a certain extent, indicates that IRDEGs may be involved in immune response, and thus, they participate in the occurrence and progression of LSCC. Through PPI analysis, we found that in up-IRDEGs, KNG1 and CXCL10 were likely the hub genes in the development of LSCC, and in down-IRDEGs, ELANE and LYZ were the hub genes. Interestingly, each of these 4 hub genes were considered a potential marker for screening or treating at least 1 cancer [50–53], suggesting that they are essential genes in LSCC. In addition, to explore the upstream regulatory mechanism of IRDEGs, we predicted TRs related to up-REDEGs and down-IRDEGs and plotted a TR-mediated regulatory network according to the co-expression relationship between TRs and IRDEGs. SPI1, SP140, and STAT4 were likely to be key TRs regulating up-IRDEGs; ZEB1 and IKZF2 may be the important TRs regulating down-IRDEGs. Although in previous studies, SPI1 [54] and STAT4 [55] have been identified as influencing tumors, insight into the regulatory mechanisms of these 5 TRs in LSCC is limited in the literature and more research is needed to verify their role.
To explore new markers that can be used for the screening and treatment of LSCC, we constructed a signature from 9 IRDEGs (BTC, FGF19, GAST, IGHV3-7, IL13RA2, RBP1, RORC, TUBB3, and UCN), and the screening and prognostic effects of it in LSCC were evaluated. In terms of screening, according to the ROC curves, the risk score calculated for the signature proved better at predicting the prognosis of patients with LSCC than any single parameter including age, sex, neoplasm histologic grade, stage, or tumor or node stage, as well as any single IRDEG used to create the signature. Moreover, the risk score also was better for screening and predicting prognosis than any of the recently reported prognostic markers for LSCC, namely, ALCAM [39], ATM [40], BCL11A [41], BCL-2 [42], DSG2 [43], EGFR [44], FGFR1 [45], HOXA13 [46], and IGF-1R [47]. In terms of prognosis, not only was the OS rate in the high-risk score group significantly lower than in low-risk score group, but the risk curve showed that there were significantly fewer deaths in patients with lower risk scores. The risk score also proved to be an independent prognostic factor for LSCC in multivariate Cox regression analysis. In summary, the IRDEG signature may be a novel marker for the screening and treatment of LSCC.
All the IRDEGs included in the signature other than IGHV3-7 have been identified as playing a role in tumors. Among them, BTC is considered to promote the development of head and neck squamous cell carcinoma (HNSCC) and has been shown to be a prognostic risk factor in patients with the disease [56]; this was confirmed by our research. We also observed a high level of expression of FGF19, which also has been identified in research about HNSCC [57] and which activates the ERK signaling cascade to stimulate the progression of nasopharyngeal carcinoma [58]. Another IRDEG signature gene, GAST, may contribute to the progression of gastric cancer at the hormone level [59] through participation in neuroendocrine function. In papillary thyroid carcinoma, with which IL13RA2 has been associated, the gene may be involved in cell migration by enhancing the epithelial-mesenchymal transition. RBP1 may be a factor associated with poor prognosis in glioblastoma [31]. RORC is thought to trigger proliferation of bladder cancer cells, and this process likely results from its ability to inhibit the PD-L1/integrin beta-6/signal transducer and activator of transcription 3 signal axis [60]. In patients with esophageal squamous cell carcinoma, those who were TUBB3-negative had considerably higher DFS and OS rates than those who were TUBB3-positive [30]. UCN has a dual effect on liver cancer cell metastasis due to its potential involvement in the regulation of cytosolic phospholipase A2 (cPLA2) and calcium-independent phospholipase A2 (iPLA2), that is, contributing to cell migration via promoting cPLA2 expression while preventing cell migration through reducing iPLA2 expression [61]. It is worth mentioning that, with the exception of TUBB3 [62] (which contributes to shorter progression-free survival and cancer-specific survival in patients), none of the remaining 8 IRDEGs have been reported in LSCC. This demonstrates the novelty of our work and reveals that further research into these 9 IRDEGs in LSCC is still needed.
To further explore the clinical significance of the IRDEG signature in LSCC, we analyzed the relationship between the signature, clinical parameters, and immune cell infiltration level in patients with LSCC. The results demonstrated that the risk score and the levels of expression of GAST were related to the neoplasm histologic grade of LSCC, while the levels of expression of BTC, RORC, and TUBB3 were related to the age of patients with LSCC; furthermore, neither the IRDEG signature nor the 9 IRDEGs were relevant to sex, stage, or tumor or nodes stage, but larger samples would be required to verify these observations. At the same time, the risk score based on the IRDEG signature was negatively correlated with the level of B-cell immune infiltration, suggesting that the signature may affect the occurrence and progression of LSCC by affecting the B-cell infiltration level. In short, we found that the clinical significance of the IRDEG signature in LSCC was mainly reflected in the neoplasm histologic grade and the B-cell immune infiltration level.
In this study, although we identified potential molecules involved in LSCC from the perspective of IRGs and established the first IRDEG signature related to the prognosis of the disease, there were several limitations. For example, the sample size was relatively small (n=123), and a study with a larger sample would be needed to verify the results. Moreover, because of the limited sample data, we could not externally verify the IRDEG signature, nor could we identify the relationship between the IRDEG signature and specific treatment methods, such as chemotherapy. Further experimental research is needed to verify the clinical significance and molecular mechanisms of the IRDEG signature in LSCC.
Conclusions
The IRDEG signature constructed in this study had a strong effect on the screening and prognosis of patients with LSCC and it may be considered an independent prognostic factor for use as a new marker in early screening and treatment of LSCC.
Supplementary Data
Differentially expressed genes (DEGs) and immune-related differentially expressed genes (IRDEGs) in laryngeal squamous cell carcinoma (LSCC). Heatmaps demonstrating differentially expressed genes. (A) Comparison between LSCC and non-tumor tissues of DEGs. (B) Comparison between LSCC and non-tumor tissues of IRDEGs. The depth of color represents the level of gene expression.
Gene Ontology (GO) analysis for immune-related downregulated differentially expressed genes. (A) Biological process. (B) Cellular component. (C) Molecular function. The blue nodes in the concentric circles represent genes clustered in specific GO terms. The larger size and darker color of the internal departments represent more significant enrichment of GO term.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of immune-related downregulated differentially expressed genes. Different color bands correspond to different KEGG enrichment pathways.
Forest plot of screening of preliminary candidate immune-related differentially expressed genes (IRDEGs) from 302 IRDEGs.
Screening of secondary candidate immune-related differentially expressed genes (IRDEGs) for a signature. (A) With the continuous increase in lambda, the absolute value of the first 30 preliminary candidate IRDEGs was compressed accordingly. In this process, the absolute value of some relatively unimportant preliminary candidate IRDEGs was compressed to 0. (B) Plot of partial likelihood deviance. Each red point corresponds to a lambda value on the lower horizontal axis and a partial likelihood deviance on the vertical axis. The first vertical dashed line on the left corresponds to the highest lambda value, with an average error within 1 standard deviation; from the upper horizontal axis in the figure, this lambda value is based on 14 IRDEGs, which were chosen as secondary candidate IRDEGs.
(A–F) Relationship between the IRDEG signature and the infiltration level of 6 kinds of immune cells. Cor, correlation coefficient.
Supplementary Table 1.
Clinical information for 102 patients with laryngeal squamous cell carcinoma.
| Sample ID of patients | Age | Gender | Grade | Stage | Tumor stage | Nodes stage | Risk score |
|---|---|---|---|---|---|---|---|
| TCGA-BA-4076-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 1.994791 |
| TCGA-BA-4078-01 | ≥65 | Male | G1–2 | Stage III–IV | T1–2 | N2–3 | 7.175697 |
| TCGA-BA-5555-01 | <65 | Male | G1–2 | Stage III–IV | T1–2 | N2–3 | 0.613095 |
| TCGA-BA-6868-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 9.846364 |
| TCGA-BA-6869-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.212306 |
| TCGA-BA-6870-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 2.463018 |
| TCGA-BA-A6DA-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 0.180343 |
| TCGA-BA-A6DI-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 4.540384 |
| TCGA-BB-4217-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.479486 |
| TCGA-CN-4722-01 | <65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 0.391675 |
| TCGA-CN-4723-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.676743 |
| TCGA-CN-4727-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.858521 |
| TCGA-CN-4735-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 1.057629 |
| TCGA-CN-4738-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.380504 |
| TCGA-CN-4739-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.209937 |
| TCGA-CN-5355-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.015923 |
| TCGA-CN-5356-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.22631 |
| TCGA-CN-5360-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.150048 |
| TCGA-CN-5363-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 1.581695 |
| TCGA-CN-6010-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.485192 |
| TCGA-CN-6012-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.698411 |
| TCGA-CN-6021-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 7.842248 |
| TCGA-CN-6022-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 2.948932 |
| TCGA-CN-6988-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.143416 |
| TCGA-CN-6989-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 4.86375 |
| TCGA-CN-6992-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.676387 |
| TCGA-CN-6997-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 1.203079 |
| TCGA-CN-A497-01 | <65 | Male | G1–2 | Stage III–IV | T1–2 | N2–3 | 0.070669 |
| TCGA-CN-A49B-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 2.197291 |
| TCGA-CN-A63T-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 1.399677 |
| TCGA-CN-A63U-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.641362 |
| TCGA-CN-A63W-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 2.018491 |
| TCGA-CN-A641-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 1.561376 |
| TCGA-CN-A6V3-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.086898 |
| TCGA-CR-6474-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 9.085744 |
| TCGA-CR-7364-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.727247 |
| TCGA-CR-7370-01 | ≥65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 2.24474 |
| TCGA-CR-7371-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 3.987309 |
| TCGA-CR-7374-01 | ≥65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 2.598793 |
| TCGA-CR-7388-01 | ≥65 | Female | G1–2 | Stage III–IV | T1–2 | N2–3 | 2.350348 |
| TCGA-CR-7389-01 | <65 | Male | G1–2 | Stage III–IV | T1–2 | N0–1 | 1.464127 |
| TCGA-CR-7398-01 | <65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 1.308124 |
| TCGA-CR-7399-01 | <65 | Female | G3 | Stage III–IV | T3–4 | N2–3 | 0.832214 |
| TCGA-CR-7402-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.304261 |
| TCGA-CV-5430-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.323973 |
| TCGA-CV-5431-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.58173 |
| TCGA-CV-5432-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.455714 |
| TCGA-CV-5434-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.572978 |
| TCGA-CV-5435-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.822588 |
| TCGA-CV-5440-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.576644 |
| TCGA-CV-5441-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.028728 |
| TCGA-CV-5443-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.148582 |
| TCGA-CV-5444-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.209379 |
| TCGA-CV-5978-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 3.792788 |
| TCGA-CV-6935-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 1.824833 |
| TCGA-CV-6962-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 9.405468 |
| TCGA-CV-7089-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 0.886302 |
| TCGA-CV-7101-01 | ≥65 | Male | G1–2 | Stage I–II | T1–2 | N0–1 | 1.524558 |
| TCGA-CV-7177-01 | ≥65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 14.47409 |
| TCGA-CV-7242-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.393045 |
| TCGA-CV-7245-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 7.160395 |
| TCGA-CV-7247-01 | <65 | Male | G1–2 | Stage I–II | T1–2 | N0–1 | 1.302915 |
| TCGA-CV-7248-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 5.899374 |
| TCGA-CV-7250-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.765147 |
| TCGA-CV-7261-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.778688 |
| TCGA-CV-7415-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 2.108499 |
| TCGA-CV-7418-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 5.986007 |
| TCGA-CV-7421-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 12.39132 |
| TCGA-CV-7422-01 | <65 | Female | G3 | Stage III–IV | T3–4 | N0–1 | 1.383207 |
| TCGA-CV-7424-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 1.60996 |
| TCGA-CV-7430-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 16.21789 |
| TCGA-CV-7433-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 7.315585 |
| TCGA-CV-A45W-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.269141 |
| TCGA-CV-A45Y-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.689461 |
| TCGA-CV-A45Z-01 | ≥65 | Male | G3 | Stage I–II | T1–2 | N0–1 | 0.854492 |
| TCGA-CV-A460-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.546034 |
| TCGA-CV-A461-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.546608 |
| TCGA-CV-A6K1-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.360293 |
| TCGA-D6-6517-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.68767 |
| TCGA-D6-6824-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.495199 |
| TCGA-D6-6826-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 1.514392 |
| TCGA-D6-8568-01 | <65 | Male | G1–2 | Stage I–II | T1–2 | N0–1 | 0.417747 |
| TCGA-D6-A6EK-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.189092 |
| TCGA-D6-A6EQ-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.545436 |
| TCGA-D6-A6ES-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.191914 |
| TCGA-D6-A74Q-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.22864 |
| TCGA-DQ-5629-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 2.062312 |
| TCGA-F7-7848-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.690225 |
| TCGA-F7-8298-01 | <65 | Male | G1–2 | Stage I–II | T1–2 | N0–1 | 0.30976 |
| TCGA-KU-A66S-01 | ≥65 | Female | G1–2 | Stage III–IV | T1–2 | N0–1 | 4.15964 |
| TCGA-QK-A8Z8-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.330232 |
| TCGA-QK-A8ZB-01 | v65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.588431 |
| TCGA-QK-AA3J-01 | ≥65 | Male | G3 | Stage I–II | T1–2 | N0–1 | 0.419213 |
| TCGA-T3-A92M-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 3.215592 |
| TCGA-UF-A718-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.79612 |
| TCGA-UF-A71D-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.063101 |
| TCGA-UF-A7J9-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 3.368041 |
| TCGA-UF-A7JF-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 0.350744 |
| TCGA-UF-A7JH-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.220921 |
| TCGA-UF-A7JJ-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.834466 |
| TCGA-UF-A7JK-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.902253 |
Grade – neoplasm histologic grade.
Acknowledgment
The results of the study are based, in part, upon data generated by The Cancer Genome Atlas Program Research Network: https://www.cancer.gov/tcga.
Footnotes
Conflicts of interest
None.
Source of support: The study was supported by funding from the National Natural Science Foundation of China (81460479)
References
- 1.Gamez ME, Blakaj A, Zoller W, et al. Emerging concepts and novel strategies in radiation therapy for laryngeal cancer management. Cancers (Basel) 2020;12(6):1651. doi: 10.3390/cancers12061651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elicin O, Giger R. Comparison of current surgical and non-surgical treatment strategies for early and locally advanced stage glottic laryngeal cancer and their outcome. Cancers (Basel) 2020;12(3):732. doi: 10.3390/cancers12030732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Qian J, Xia X, et al. Long non-coding RNA OIP5-AS1 serves as an oncogene in laryngeal squamous cell carcinoma by regulating miR-204-5p/ZEB1 axis. Naunyn Schmiedebergs Arch Pharmacol. 2020 doi: 10.1007/s00210-020-01811-7. [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Yang T, Li S, Liu J, et al. Long non-coding RNA KRT16P2/miR-1294/EGFR axis regulates laryngeal squamous cell carcinoma cell aggressiveness. Am J Transl Res. 2020;12(6):2939–55. [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Y, Huang Q, Zheng J, et al. Diagnostic role of dysregulated circular RNA hsa_circ_0036722 in laryngeal squamous cell carcinoma. Onco Targets Ther. 2020;13:5709–19. doi: 10.2147/OTT.S231076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang X-Z, Zhou X-G, Zhang X-G, et al. The clinical significance of Interleukin 24 and its potential molecular mechanism in laryngeal squamous cell carcinoma. Cancer Biomark. 2020;29(1):111–24. doi: 10.3233/CBM-201441. [DOI] [PubMed] [Google Scholar]
- 7.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 8.Dietz A, Wichmann G, Kuhnt T, et al. Induction chemotherapy (IC) followed by radiotherapy (RT) versus cetuximab plus IC and RT in advanced laryngeal/hypopharyngeal cancer resectable only by total laryngectomy-final results of the larynx organ preservation trial DeLOS-II. Ann Oncol. 2018;29(10):2105–14. doi: 10.1093/annonc/mdy332. [DOI] [PubMed] [Google Scholar]
- 9.Steuer CE, El-Deiry M, Parks JR, et al. An update on larynx cancer. Cancer J Clin. 2017;67(1):31–50. doi: 10.3322/caac.21386. [DOI] [PubMed] [Google Scholar]
- 10.Pedregal-Mallo D, Sánchez Canteli M, López F, et al. Oncological and functional outcomes of transoral laser surgery for laryngeal carcinoma. Eur Arch Otorhinolaryngol. 2018;275(8):2071–77. doi: 10.1007/s00405-018-5027-z. [DOI] [PubMed] [Google Scholar]
- 11.Giotakis AI, Pototschnig C. Use of erbium laser in the treatment of persistent post-radiotherapy laryngeal edema: A case report and review of the literature. World J Surg Oncol. 2018;16(1):176. doi: 10.1186/s12957-018-1480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietz A, Wiegand S, Kuhnt T, et al. Laryngeal preservation approaches: Considerations for new selection criteria based on the DeLOS-II Trial. Front Oncol. 2019;9:625. doi: 10.3389/fonc.2019.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menach OP, Patel A, Oburra HO. Demography and histologic pattern of laryngeal squamous cell carcinoma in kenya. Int J Otolaryngol. 2014;2014 doi: 10.1155/2014/507189. 507189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bree R. The current indications for non-surgical treatment of hypopharyngeal cancer. Adv Otorhinolaryngol. 2019;83:76–89. doi: 10.1159/000492314. [DOI] [PubMed] [Google Scholar]
- 15.Fulcher CD, Haigentz M, Ow TJ. AHNS Series: Do you know your guidelines? Principles of treatment for locally advanced or unresectable head and neck squamous cell carcinoma. Head Neck. 2018;40(4):676–86. doi: 10.1002/hed.25025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mannelli G, Lazio MS, Luparello P, et al. Conservative treatment for advanced T3–T4 laryngeal cancer: Meta-analysis of key oncological outcomes. Eur Arch Otorhinolaryngol. 2018;275(1):27–38. doi: 10.1007/s00405-017-4799-x. [DOI] [PubMed] [Google Scholar]
- 17.Nocini R, Molteni G, Mattiuzzi C, et al. Updates on larynx cancer epidemiology. Chin J Cancer Res. 2020;32(1):18–25. doi: 10.21147/j.issn.1000-9604.2020.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Praud D, Rota M, Rehm J, et al. Cancer incidence and mortality attributable to alcohol consumption. Int J Cancer. 2016;138(6):1380–87. doi: 10.1002/ijc.29890. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad Kiadaliri A, Jarl J, Gavriilidis G, et al. Alcohol drinking cessation and the risk of laryngeal and pharyngeal cancers: A systematic review and meta-analysis. PLoS One. 2013;8(3):e58158. doi: 10.1371/journal.pone.0058158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakita D, Matsuo K. Alcohol and head and neck cancer. Cancer Metastasis Rev. 2017;36(3):425–34. doi: 10.1007/s10555-017-9690-0. [DOI] [PubMed] [Google Scholar]
- 21.Kang D-M, Kim J-E, Kim Y-K, et al. Occupational burden of asbestos-related diseases in Korea, 1998–2013: Asbestosis, mesothelioma, lung cancer, laryngeal cancer, and ovarian cancer. J Korean Med Sci. 2018;33(35):e226. doi: 10.3346/jkms.2018.33.e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang D, Shi Y, Tang Y, et al. Effect of HPV Infection on the occurrence and development of laryngeal cancer: A review. J Cancer. 2019;10(19):4455–62. doi: 10.7150/jca.34016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H-T, Tong X, Zhang Z-X, et al. MYCT1 represses apoptosis of laryngeal cancerous cells through the MAX/miR-181a/NPM1 pathway. FEBS J. 2019;286(19):3892–908. doi: 10.1111/febs.14942. [DOI] [PubMed] [Google Scholar]
- 24.Dogantemur S, Ozdemir S, Uguz A, et al. Assessment of HPV 16, HPV 18, p16 expression in advanced stage laryngeal cancer patients and prognostic significance. Braz J Otorhinolaryngol. 2020;86(3):351–57. doi: 10.1016/j.bjorl.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen N, Duan XH, Wang XL, et al. Effect of NLK on the proliferation and invasion of laryngeal carcinoma cells by regulating CDCP1. Eur Rev Med Pharmacol Sci. 2019;23(14):6226–33. doi: 10.26355/eurrev_201907_18441. [DOI] [PubMed] [Google Scholar]
- 26.Tang X, Sun Y, Wan G, et al. Knockdown of YAP inhibits growth in Hep-2 laryngeal cancer cells via epithelial-mesenchymal transition and the Wnt/β-catenin pathway. BMC Cancer. 2019;19(1):654. doi: 10.1186/s12885-019-5832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Zhou L, Zhang Y, et al. DIAPH1 is upregulated and inhibits cell apoptosis through ATR/p53/Caspase-3 signaling pathway in laryngeal squamous cell carcinoma. Dis Markers. 2019;2019 doi: 10.1155/2019/6716472. 6716472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu X, Zhu R. Curcumin suppresses the progression of laryngeal squamous cell carcinoma through the upregulation of miR-145 and inhibition of the PI3K/Akt/mTOR pathway. Onco Targets Ther. 2018;11:3521–31. doi: 10.2147/OTT.S159236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X, Xu F, Dominguez NP, et al. FGFR4 provides the conduit to facilitate FGF19 signaling in breast cancer progression. Mol Carcinog. 2018;57(11):1616–25. doi: 10.1002/mc.22884. [DOI] [PubMed] [Google Scholar]
- 30.Gong L, Mao W, Chen Q, et al. Analysis of SPARC and TUBB3 as predictors for prognosis in esophageal squamous cell carcinoma receiving nab-paclitaxel plus cisplatin neoadjuvant chemotherapy: A prospective study. Cancer Chemother Pharmacol. 2019;83(4):639–47. doi: 10.1007/s00280-019-03769-7. [DOI] [PubMed] [Google Scholar]
- 31.Erhart F, Hackl M, Hahne H, et al. Combined proteomics/miRNomics of dendritic cell immunotherapy-treated glioblastoma patients as a screening for survival-associated factors. NPJ Vaccines. 2020;5:5. doi: 10.1038/s41541-019-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birtalan E, Danos K, Gurbi B, et al. Expression of PD-L1 on Immune cells shows better prognosis in laryngeal, oropharygeal, and hypopharyngeal cancer. Appl Immunohistochem Mol Morphol. 2018;26(7):e79–85. doi: 10.1097/PAI.0000000000000590. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Guan Z, Peng J. [The correlation between stanniocalcin 2 expression and prognosis in laryngeal squamous cell cancer]. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2015;29(2):102–7. [in Chinese] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu G, Wang L-G, Han Y, et al. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–87. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin Q, Fan J, Zheng R, et al. Lisa: Inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol. 2020;21(1):32. doi: 10.1186/s13059-020-1934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engebretsen S, Bohlin J. Statistical predictions with glmnet. Clin Epigenetics. 2019;11(1):123. doi: 10.1186/s13148-019-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61(1):92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 39.Nicolau-Neto P, de Souza-Santos PT, Severo Ramundo M, et al. Transcriptome analysis identifies ALCAM overexpression as a prognosis biomarker in laryngeal squamous cell carcinoma. Cancers (Basel) 2020;12(2):470. doi: 10.3390/cancers12020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruine de Bruin L, Schuuring E, de Bock GH, et al. High pATM is associated with poor local control in supraglottic cancer treated with radiotherapy. Laryngoscope. 2020;130(8):1954–60. doi: 10.1002/lary.28641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou J, Zhou L, Zhang D, et al. BCL11A promotes the progression of laryngeal squamous cell carcinoma. Front Oncol. 2020;10:375. doi: 10.3389/fonc.2020.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lovato A, Franz L, Carraro V, et al. Maspin expression and anti-apoptotic pathway regulation by bcl2 in laryngeal cancer. Ann Diagn Pathol. 2020;45:151471. doi: 10.1016/j.anndiagpath.2020.151471. [DOI] [PubMed] [Google Scholar]
- 43.Cury SS, Lapa RML, de Mello JBH, et al. Increased DSG2 plasmatic levels identified by transcriptomic-based secretome analysis is a potential prognostic biomarker in laryngeal carcinoma. Oral Oncol. 2020;103:104592. doi: 10.1016/j.oraloncology.2020.104592. [DOI] [PubMed] [Google Scholar]
- 44.Leesutipornchai T, Ratchataswan T, Vivatvakin S, et al. EGFR cut-off point for prognostic impact in laryngeal squamous cell carcinoma. Acta Otolaryngol. 2020;140(7):610–14. doi: 10.1080/00016489.2020.1738548. [DOI] [PubMed] [Google Scholar]
- 45.Klobučar M, Visentin S, Jakovčević A, et al. Expression of polysialic acid in primary laryngeal squamous cell carcinoma. Life Sci. 2017;173:73–79. doi: 10.1016/j.lfs.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Ye M, Zhou C. Expression profile and prognostic values of family members in laryngeal squamous cell cancer. Front Oncol. 2020;10:368. doi: 10.3389/fonc.2020.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ben Elhadj M, Goucha A, Fourati A, et al. The prognostic significance of IGF-1R and the predictive risk value of circulating IGF-1 in Tunisian patients with laryngeal carcinoma. Cancer Invest. 2020;38(5):289–99. doi: 10.1080/07357907.2020.1758711. [DOI] [PubMed] [Google Scholar]
- 48.Szczerba BM, Castro-Giner F, Vetter M, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566(7745):553–57. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 49.Macor P, Capolla S, Tedesco F. Complement as a biological tool to control tumor growth. Front Immunol. 2018;9:2203. doi: 10.3389/fimmu.2018.02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu J, Fang J, Cheng Z, et al. Overexpression of the Kininogen-1 inhibits proliferation and induces apoptosis of glioma cells. J Exp Clin Cancer Res. 2018;37(1):180. doi: 10.1186/s13046-018-0833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karin N. Chemokines and cancer: New immune checkpoints for cancer therapy. Curr Opin Immunol. 2018;51:140–45. doi: 10.1016/j.coi.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Sarvestani SK, Signs SA, Lefebvre V, et al. Cancer-predicting transcriptomic and epigenetic signatures revealed for ulcerative colitis in patient-derived epithelial organoids. Oncotarget. 2018;9(47):28717–30. doi: 10.18632/oncotarget.25617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei Z, Wu B, Wang L, et al. A large-scale transcriptome analysis identified ELANE and PRTN3 as novel methylation prognostic signatures for clear cell renal cell carcinoma. J Cell Physiol. 2020;235(3):2582–89. doi: 10.1002/jcp.29162. [DOI] [PubMed] [Google Scholar]
- 54.Luo D, Ge W. MeCP2 promotes colorectal cancer metastasis by modulating ZEB1 transcription. Cancers (Basel) 2020;12(3):758. doi: 10.3390/cancers12030758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Zhong J-H, Gong F-S, et al. MiR-141-3p suppresses gastric cancer induced transition of normal fibroblast and BMSC to cancer-associated fibroblasts via targeting STAT4. Exp Mol Pathol. 2019;107:85–94. doi: 10.1016/j.yexmp.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 56.Gao J, Ulekleiv CH, Halstensen TS. Epidermal growth factor (EGF) receptor-ligand based molecular staging predicts prognosis in head and neck squamous cell carcinoma partly due to deregulated EGF-induced amphiregulin expression. J Exp Clin Cancer Res. 2016;35(1):151. doi: 10.1186/s13046-016-0422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao L, Lang L, Zhao X, et al. FGF19 amplification reveals an oncogenic dependency upon autocrine FGF19/FGFR4 signaling in head and neck squamous cell carcinoma. Oncogene. 2019;38(13):2394–404. doi: 10.1038/s41388-018-0591-7. [DOI] [PubMed] [Google Scholar]
- 58.Shi S, Zhang Q, Xia Y, et al. Mesenchymal stem cell-derived exosomes facilitate nasopharyngeal carcinoma progression. Am J Cancer Res. 2016;6(2):459–72. [PMC free article] [PubMed] [Google Scholar]
- 59.Speck O, Tang W, Morgan DR, et al. Three molecular subtypes of gastric adenocarcinoma have distinct histochemical features reflecting Epstein-Barr virus infection status and neuroendocrine differentiation. Appl Immunohistochem Mol Morphol. 2015;23(9):633–45. doi: 10.1097/PAI.0000000000000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao D, Qi Z, Pang Y, et al. Retinoic acid-related orphan receptor c regulates proliferation, glycolysis, and chemoresistance via the PD-L1/ITGB6/STAT3 signaling axis in bladder cancer. Cancer Res. 2019;79(10):2604–18. doi: 10.1158/0008-5472.CAN-18-3842. [DOI] [PubMed] [Google Scholar]
- 61.Zhu C, Sun Z, Li C, et al. Urocortin affects migration of hepatic cancer cell lines via differential regulation of cPLA2 and iPLA2. Cell Signal. 2014;26(5):1125–34. doi: 10.1016/j.cellsig.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Koh Y, Kim TM, Jeon YK, et al. Class III beta-tubulin, but not ERCC1, is a strong predictive and prognostic marker in locally advanced head and neck squamous cell carcinoma. Ann Oncol. 2009;20(8):1414–19. doi: 10.1093/annonc/mdp002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes (DEGs) and immune-related differentially expressed genes (IRDEGs) in laryngeal squamous cell carcinoma (LSCC). Heatmaps demonstrating differentially expressed genes. (A) Comparison between LSCC and non-tumor tissues of DEGs. (B) Comparison between LSCC and non-tumor tissues of IRDEGs. The depth of color represents the level of gene expression.
Gene Ontology (GO) analysis for immune-related downregulated differentially expressed genes. (A) Biological process. (B) Cellular component. (C) Molecular function. The blue nodes in the concentric circles represent genes clustered in specific GO terms. The larger size and darker color of the internal departments represent more significant enrichment of GO term.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of immune-related downregulated differentially expressed genes. Different color bands correspond to different KEGG enrichment pathways.
Forest plot of screening of preliminary candidate immune-related differentially expressed genes (IRDEGs) from 302 IRDEGs.
Screening of secondary candidate immune-related differentially expressed genes (IRDEGs) for a signature. (A) With the continuous increase in lambda, the absolute value of the first 30 preliminary candidate IRDEGs was compressed accordingly. In this process, the absolute value of some relatively unimportant preliminary candidate IRDEGs was compressed to 0. (B) Plot of partial likelihood deviance. Each red point corresponds to a lambda value on the lower horizontal axis and a partial likelihood deviance on the vertical axis. The first vertical dashed line on the left corresponds to the highest lambda value, with an average error within 1 standard deviation; from the upper horizontal axis in the figure, this lambda value is based on 14 IRDEGs, which were chosen as secondary candidate IRDEGs.
(A–F) Relationship between the IRDEG signature and the infiltration level of 6 kinds of immune cells. Cor, correlation coefficient.
Supplementary Table 1.
Clinical information for 102 patients with laryngeal squamous cell carcinoma.
| Sample ID of patients | Age | Gender | Grade | Stage | Tumor stage | Nodes stage | Risk score |
|---|---|---|---|---|---|---|---|
| TCGA-BA-4076-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 1.994791 |
| TCGA-BA-4078-01 | ≥65 | Male | G1–2 | Stage III–IV | T1–2 | N2–3 | 7.175697 |
| TCGA-BA-5555-01 | <65 | Male | G1–2 | Stage III–IV | T1–2 | N2–3 | 0.613095 |
| TCGA-BA-6868-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 9.846364 |
| TCGA-BA-6869-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.212306 |
| TCGA-BA-6870-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 2.463018 |
| TCGA-BA-A6DA-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 0.180343 |
| TCGA-BA-A6DI-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 4.540384 |
| TCGA-BB-4217-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.479486 |
| TCGA-CN-4722-01 | <65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 0.391675 |
| TCGA-CN-4723-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.676743 |
| TCGA-CN-4727-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.858521 |
| TCGA-CN-4735-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 1.057629 |
| TCGA-CN-4738-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.380504 |
| TCGA-CN-4739-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.209937 |
| TCGA-CN-5355-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.015923 |
| TCGA-CN-5356-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.22631 |
| TCGA-CN-5360-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.150048 |
| TCGA-CN-5363-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 1.581695 |
| TCGA-CN-6010-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.485192 |
| TCGA-CN-6012-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.698411 |
| TCGA-CN-6021-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 7.842248 |
| TCGA-CN-6022-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 2.948932 |
| TCGA-CN-6988-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.143416 |
| TCGA-CN-6989-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 4.86375 |
| TCGA-CN-6992-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.676387 |
| TCGA-CN-6997-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 1.203079 |
| TCGA-CN-A497-01 | <65 | Male | G1–2 | Stage III–IV | T1–2 | N2–3 | 0.070669 |
| TCGA-CN-A49B-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 2.197291 |
| TCGA-CN-A63T-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 1.399677 |
| TCGA-CN-A63U-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.641362 |
| TCGA-CN-A63W-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 2.018491 |
| TCGA-CN-A641-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 1.561376 |
| TCGA-CN-A6V3-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.086898 |
| TCGA-CR-6474-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 9.085744 |
| TCGA-CR-7364-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.727247 |
| TCGA-CR-7370-01 | ≥65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 2.24474 |
| TCGA-CR-7371-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 3.987309 |
| TCGA-CR-7374-01 | ≥65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 2.598793 |
| TCGA-CR-7388-01 | ≥65 | Female | G1–2 | Stage III–IV | T1–2 | N2–3 | 2.350348 |
| TCGA-CR-7389-01 | <65 | Male | G1–2 | Stage III–IV | T1–2 | N0–1 | 1.464127 |
| TCGA-CR-7398-01 | <65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 1.308124 |
| TCGA-CR-7399-01 | <65 | Female | G3 | Stage III–IV | T3–4 | N2–3 | 0.832214 |
| TCGA-CR-7402-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.304261 |
| TCGA-CV-5430-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.323973 |
| TCGA-CV-5431-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.58173 |
| TCGA-CV-5432-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.455714 |
| TCGA-CV-5434-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.572978 |
| TCGA-CV-5435-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.822588 |
| TCGA-CV-5440-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.576644 |
| TCGA-CV-5441-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.028728 |
| TCGA-CV-5443-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.148582 |
| TCGA-CV-5444-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.209379 |
| TCGA-CV-5978-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 3.792788 |
| TCGA-CV-6935-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 1.824833 |
| TCGA-CV-6962-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 9.405468 |
| TCGA-CV-7089-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 0.886302 |
| TCGA-CV-7101-01 | ≥65 | Male | G1–2 | Stage I–II | T1–2 | N0–1 | 1.524558 |
| TCGA-CV-7177-01 | ≥65 | Female | G1–2 | Stage I–II | T1–2 | N0–1 | 14.47409 |
| TCGA-CV-7242-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.393045 |
| TCGA-CV-7245-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 7.160395 |
| TCGA-CV-7247-01 | <65 | Male | G1–2 | Stage I–II | T1–2 | N0–1 | 1.302915 |
| TCGA-CV-7248-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 5.899374 |
| TCGA-CV-7250-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.765147 |
| TCGA-CV-7261-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.778688 |
| TCGA-CV-7415-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 2.108499 |
| TCGA-CV-7418-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 5.986007 |
| TCGA-CV-7421-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 12.39132 |
| TCGA-CV-7422-01 | <65 | Female | G3 | Stage III–IV | T3–4 | N0–1 | 1.383207 |
| TCGA-CV-7424-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 1.60996 |
| TCGA-CV-7430-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 16.21789 |
| TCGA-CV-7433-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 7.315585 |
| TCGA-CV-A45W-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.269141 |
| TCGA-CV-A45Y-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.689461 |
| TCGA-CV-A45Z-01 | ≥65 | Male | G3 | Stage I–II | T1–2 | N0–1 | 0.854492 |
| TCGA-CV-A460-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N2–3 | 0.546034 |
| TCGA-CV-A461-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.546608 |
| TCGA-CV-A6K1-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.360293 |
| TCGA-D6-6517-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.68767 |
| TCGA-D6-6824-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.495199 |
| TCGA-D6-6826-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N2–3 | 1.514392 |
| TCGA-D6-8568-01 | <65 | Male | G1–2 | Stage I–II | T1–2 | N0–1 | 0.417747 |
| TCGA-D6-A6EK-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.189092 |
| TCGA-D6-A6EQ-01 | <65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.545436 |
| TCGA-D6-A6ES-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.191914 |
| TCGA-D6-A74Q-01 | ≥65 | Male | G3 | Stage III–IV | T3–4 | N0–1 | 0.22864 |
| TCGA-DQ-5629-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 2.062312 |
| TCGA-F7-7848-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.690225 |
| TCGA-F7-8298-01 | <65 | Male | G1–2 | Stage I–II | T1–2 | N0–1 | 0.30976 |
| TCGA-KU-A66S-01 | ≥65 | Female | G1–2 | Stage III–IV | T1–2 | N0–1 | 4.15964 |
| TCGA-QK-A8Z8-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.330232 |
| TCGA-QK-A8ZB-01 | v65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.588431 |
| TCGA-QK-AA3J-01 | ≥65 | Male | G3 | Stage I–II | T1–2 | N0–1 | 0.419213 |
| TCGA-T3-A92M-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 3.215592 |
| TCGA-UF-A718-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.79612 |
| TCGA-UF-A71D-01 | <65 | Female | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.063101 |
| TCGA-UF-A7J9-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 3.368041 |
| TCGA-UF-A7JF-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N2–3 | 0.350744 |
| TCGA-UF-A7JH-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.220921 |
| TCGA-UF-A7JJ-01 | ≥65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 0.834466 |
| TCGA-UF-A7JK-01 | <65 | Male | G1–2 | Stage III–IV | T3–4 | N0–1 | 1.902253 |
Grade – neoplasm histologic grade.