Introduction
The transition of vascular smooth muscle cells (SMCs) from a quiescent, contractile phenotype to a motile, synthetic phenotype encompases dramatic changes in cell morphology and gene expression. It also is accompanied by equally dramatic changes in cell metabolism. Indeed, ultrastructural evidence of metabolic reprogramming was one of the first features recognized for the synthetic SMC phenotype.1 While our understanding of the transcriptional basis of SMC phenotypic switching has advanced significantly in recent decades,2 a similar understanding for how metabolic pathways are linked to these transcriptional changes lags behind.
Poldip2 and Mitochondrial Metabolism in SMCs
Polymerase delta interacting protein 2 (Poldip2) is a nuclear-encoded mitochondrial protein that controls the lipoylation and activation of two important mitochondrial enzymes; pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase.3 Poldip2 null mice are perinatal lethal exhibiting general fetal growth restriction.4 Poldip2 deficiency reduces the activity of the Krebs cycle and inhibits rates of oxidative metabolism while increasing rates of glycolytic activity in many different cell types, including vascular SMCs. While Poldip2 heterozygotes have no obvious phenotype, these mice are protected against the development of aortic aneurysm and the formation of a neointima after femoral artery wire injury.5
One explanation for the vascular wall protective effects could be that Poldip2 is somehow required for SMCs to undergo phenotypic switching. Paredes et al addressed this possibility by first asking what effect does Poldip2 down regulation have on expression levels of transcription factors (TFs) that control SMC phenotype.6 Knockdown of Poldip2 in human aortic SMCs (HASMCs) increased expression levels of serum response factor (SRF), myocardin (MYOCD), and myocardin-related transcription factor-A (MRTF-A), together with pronounced down regulation of krüppel-like factor-4 (KLF4).6 Poldip2 deficiency also inhibited PDGF-BB-induced down regulation of contractile proteins and blocked cholesterol-stimulated expression of macrophage markers CD68, LGALS3, and Arginase-1 in HASMCs. In a femoral artery wire injury model, medial SMCs from Poldip2+/− mice exhibited highly differentiated phenotypes at time points when SMCs from Poldip2+/+ mice exhibit strong down regulation of SMC marker proteins.6 Therefore, a lack of mitochondrial Poldip2 activity leads to maintenance of the SMC differentiated phenotype via effects on expression levels of nuclear transcription factors (TFs).
Poldip2 and the Ubiquitin-Proteasome Pathway in SMCs
In cultured HASMCs, Poldip2 knockdown led to accumulation of SRF even in the presence of cycloheximide, suggesting increased protein stability.6 Careful analysis of Poldip2-deficient HASMCs uncovered a general decrease in capacity of the ubiquitin-proteasome system (UPS) to degrade intracellular proteins, particularly in the nuclear compartment. Restoration of SRF turnover was observed upon forced expression of the nuclear UPS activator PSMC1/Rpt2 in HASMCs. These findings suggest that Poldip2 deficiency leads to reduced activity of a nuclear-localized UPS pathway for protein turnover.
Additional experiments showed that Poldip2 deficiency reduced mitochondrial respiration, increased glycolytic activity, and stimulated the activity of O-GlcNAc transferase (OGT).6 OGT-catalyzes the covalent modification of Ser/Thr residues with N-acetylglucosamine.7 Glycosylation of critical components of a nuclear UPS complex inhibited the turnover of nuclear TFs, including SRF, leading to their accumulation. Down regulation of OGT activity in Poldip2-deficient HASMCs restored nuclear UPS activity and prevented the accumulation of SRF, MYOCD, and MRTF-A, as well as reversed the down-regulation of KLF4.
Mechanistic Links between SMC Metabolism and Transcription
While reports of specific examples for metabolism-transcription coupling in SMCs are relatively few at the present time, it is worth recalling the often repeated quote by Max Delbrück (1949) that “any living cell today carries with it the experiences of a billion years of experimentation by its ancestors”.8 This experimentation has produced in mammalian cells a complex interconnecting web of thousands of coupled reactions. The many mechanisms linking metabolism to transcription progress from the simple direct binding of metabolites to nuclear receptors (lac repressor, retinoic acid receptors), to metabolic enzymes that can shuttle to the nucleus and themselves act as TFs (see, moonlightingproteins.org), and then to progressively more complex mechanisms including hypoxia sensing by prolylhydroxylases and hypoxia inducible factors,9 and the Poldip2 example reported by Paredes et al.6 In addition to directly acting on TFs, we also know many examples of metabolites acting at the epigenetic level of transcriptional regulation including fatty acid oxidation control of histone acetylation via AcetylCoA,10 S-adenosylmethionine regulation of methylation of histones and DNA,11 and alpha-ketoglutarate and nicotinamide regulation of DNA methylation and protein deacetylase activities respectively.12 Thus rather than distinct categories of metabolism and transcription, we are better served by conceptually merging these categories into one common and highly interwoven network for regulation of the mammalian genome.
A Revised View of SMC Phenotypic Diversity
The intricacy of SMC metabolic links to transcriptional activity suggests that the malfunction of a single protein within this web can have surprising and unpredictable phenotypes depending on connections it makes with other elements in the network. For most proteins, these connections are often transient with rapidly changing binding partners and characterized by fluxes of substrates and products through reactions that are highly compartmentalized within the cell. These principles of metabolic regulation can possibly explain the evolution of self-reinforcing feed-forward transcriptional networks that commit SMCs to long-term changes in cell fate and differentiation in, for example, vascular development. At the same time, activation of such feed-forward switches in the wrong place or at the wrong time could promote disease. Putting these concepts into motion suggests a revised model of SMC phenotypes in vivo. This revised model views SMCs in a common vessel wall as existing in transient heterogeneous subsets of metabolic-transcriptional states in oscillation between real or potential phenotypes. Any external change in environment (injury, inflammation, matrix disruption) therefore acts not on one type of SMC but a highly diverse population of cells with different potentials to respond to the change in environment.13 Many cells may either be unable to respond or fail to cross a “threshold” to respond based on their complement of links between metabolism and transcription. Under such hypothetical conditions, one can begin to see how only subsets of SMCs, or possibly even single SMCs, can participate in the response to vascular injury or perturbation.13–15 While there is much to learn about metabolism-transcription coupling in SMCs, the rapid advances in genome-wide and single cell methods to assess gene expression and epigenetic states suggests a new phase of discovery is upon the SMC field.
Summary
The findings reported by Paredes et al in this issue of Circulation Research link SMC metabolism with SMC phenotype via metabolic control of a nuclear ubiquitin-proteasome system that degrades TFs required to establish and maintain the differentiated SMC phenotype.6 They provide a glimpse of the intricate web of interconnected pathways that have evolved to ensure that changes in SMC phenotype are tightly coupled to coordinate changes in SMC metabolism. Given the rapid advances in methods available to assess whole genome transcriptomes, we can anticipate identification of many more examples of how SMC metabolic pathways are linked with SMC proliferative disorders in the near future. They also raise new questions about how diet and mitochondrial function interact with diverse human genomes carrying genetic variants in the proteins and cis-regulatory sequences that constitute the SMC metabolome.16
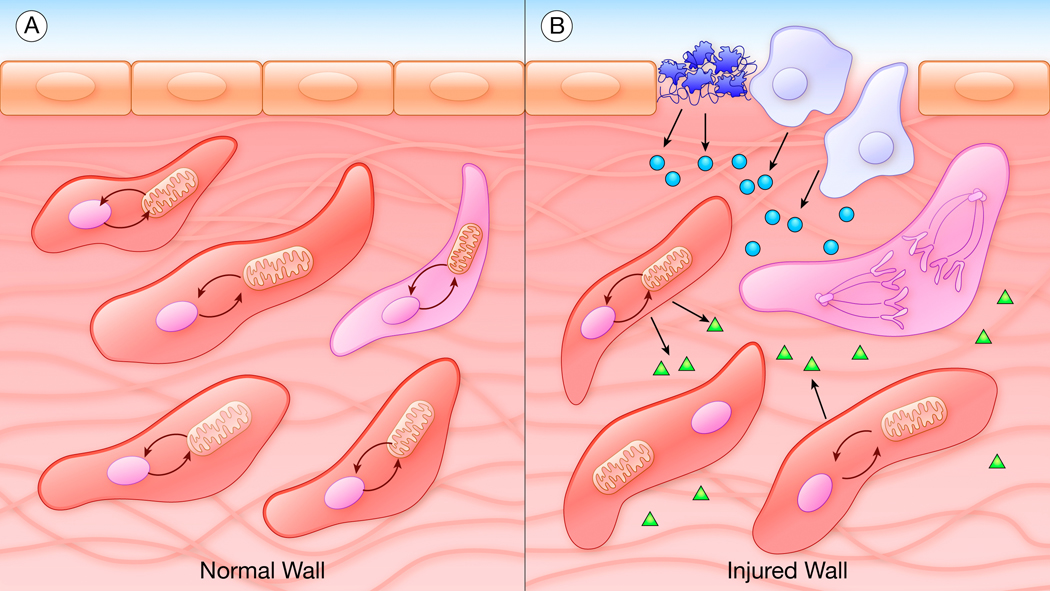
Figure.
Metabolic-transcriptional coupling in smooth muscle cells (SMCs). A, In the normal vessel wall, SMCs appear homogeneous but actually exist in many different real or potential phenotypes based on local variations in metabolic reactions and their links to transcriptional coupling. B, In the injured vessel wall, preexisting states of metabolic-transcriptional coupling mean that only a small number of “primed” SMCs are able to cross a critical threshold and respond to the change in environment. Platelets (dark blue), leukocytes (light blue), endothelial cells (tan), secreted factors (circles, triangles), SMCs (red), primed and dividing SMC (pink).
Acknowledgments
I thank my colleagues in the Center for Developmental Biology and Regenerative Medicine, Seattle Children’s Research Institute, and in the Department of Pathology, University of Washington, for their insights and helpful discussions.
Sources of Funding
This work was supported by National Institutes of Health grants 1RO1HL121877, 1RO1HL133723, and the Loie Power Robinson Stem Cell and Regenerative Medicine Fund, and the Seattle Children’s Research Institute.
Footnotes
Disclosures
None
References
- (1).Campbell JH, Campbell GR. Smooth muscle phenotypic modulation - a personal experience. Arterioscler Thromb Vasc Biol 2012;32:1784–1789. doi: 10.1161/ATVBAHA.111.243212. pmid: 22815344. [DOI] [PubMed] [Google Scholar]
- (2).Bennett MR, Sinha S, Owens GK. Vascular smooth muscle cells in atherosclerosis. Circ Res 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. pmid: 26892967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Paredes F, Sheldon K, Lassegue B, Williams HC, Faidley EA, Benavides GA, Torres G, Sanchueza-Olivares F, Yeligar SM, Griendling KK, et al. Poldip2 is an oxygen-sensitive protein that controls PDH and αKGDH lipoylation and activation to support metabolic adaptation in hypoxia and cancer. Proc Natl Acad Sci USA 2018;115:1789–1794. doi: 10.1073/pnas.1720693115. pmid: 29434038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Brown DI, Lasseque B, Lee M, Zafari R, Long JS, Saavedra HI, Griendling KK. Poldip2 knockout results in perinatal lethality, reduced cellular growth and increased autophagy of mouse embryonic fibroblasts. PLoS One 2014;9:e96657. doi: 10.1371/journal.pone.0096657. pmid: 24797518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Datla SR, Hilenski L, Seidel-Rogol B, Dikalova AE, Harousseau M, Punkova L, Joseph G, Taylor WR, Lassegue B, Griendling KK. Poldip2 knockdown inhibits vascular smooth muscle proliferation and neointima formation by regulating the expression of PCNA and p21. Lab Invest 2019; 99:387–398. doi: 10.1038/s41374-018-0103-y. pmid: 30237457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Paredes F, Williams HC, Quintana RA, San Martin A. Mitochondrial protein Poldip2 controls VSMC differentiated phenotype by O-linked GlcNAc transferase-dependent inhibition of a ubiquitin proteasome system. Circ Res 2019;vol:page-page. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Aquino-Gil M, Pierce A, Perez-Cervera Y, Zenteno E, Lefebvre T. OGT: a short overview of an enzyme standing out from usual glycosyltransferases. Biochem Soc Trans 2017;45:365–370. doi: 10.1042/BST20160404. pmid: 28408476. [DOI] [PubMed] [Google Scholar]
- (8).Strauss BS. A physicist’s quest in biology: Max Delbrück and “complementarity”. Genetics 2017;206:641–650. doi: 10.1534/genetics.117.201517. pmid: 28592501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. pmid: 22304911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Rohlenova K, Veys K, Miranda-Santos I, De Bock K, Carmeliet P. Endothelial cell metabolism in health and disease. Trends Cell Biol 2018;28:224–236. doi: 10.1016/j.tcb.2017.10.010. pmid: 29153487. [DOI] [PubMed] [Google Scholar]
- (11).Reid MA, Dai Z, Locasale JW. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol 2017;19:1298–1306. doi: 10.1038/ncb3629. pmid: 29058720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mottis A, Herzig S, Auwerx J. Mitocellular communication: shaping health and disease. Science 2019;366:827–832. doi: 10.1126/science.aax3768. pmid: 31727828. [DOI] [PubMed] [Google Scholar]
- (13).Dobnikar L, Taylor AL, Chappell J, Oldach P, Harman JL, Oerton E, Dzierzak E, Bennett MR, Spivakov M, Jorgensen HF. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat Commun 2018;9:4567. doi: 10.1038/s41467-018-06891-x. pmid: 30385745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Benditt EP, Benditt JM. Evidence for a monoclonal origin of human atherosclerotic plaques. Proc Natl Acad Sci USA 1973;70:1753–1756. doi: 10.1073/pnas.70.6.1753. pmid: 4515934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jacobsen K, Lund MB, Shim J, Gunnersen S, Füchtbauer EM, Kjolby M, Carramolino L, Bentzon JF. Diverse cellular architecture of atherosclerotic plaque derives from clonal expansion of a few medial SMCs. J Clin Invest-Insight 2017;2:19. doi: 10.1172/jci.insight.95890. pmid: 28978793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Harrington DM, Mao C, Parker SJ, Fu Z, Yu G, Chen L, Venkatraman V, Fu Y, Wang Y, Howard TD, et al. Proteomic architecture of human coronary and aortic atherosclerosis. Circulation 2018;137:2741–2756. doi: 10.1161/CIRCULATIONAHA.118.034365. pmid: 29915101. [DOI] [PMC free article] [PubMed] [Google Scholar]