Abstract
Abnormal expression of microRNAs (miRNAs), which are highlyconserved noncoding RNAs that regulate the expression of various genes post transcriptionally to control cellular functions, has been associated with the development of many diseases. In some cases, disease-promoting miRNAs are upregulated, while in other instances disease-suppressive miRNAs are downregulated. To alleviate this imbalanced miRNA expression, either antagomiRs or miRNA mimics can be delivered to cells to inhibit or promote miRNA expression, respectively. Unfortunately, the clinical translation of bare antagomiRs and miRNA mimics has been challenging because nucleic acids are susceptible to nuclease degradation, display unfavorable pharmacokinetics, and cannot passively enter cells. This review emphasizes the challenges associated with miRNA mimic delivery and then discusses the design and implementation of polymer nanocarriers to overcome these challenges. Preclinical efforts are summarized, and a forward-looking perspective on the future clinical translation of polymer nanomaterials as miRNA delivery vehicles is provided.
INTRODUCTION
MicroRNAs (miRNAs) are evolutionarily conserved, noncoding RNAs that regulate the expression of various genes by binding to messenger RNA (mRNA) molecules in the cell cytoplasm either with perfect complementarity, resulting in mRNA degradation, or with imperfect complementarity at the 3′ UTR (untranslated region), resulting in translational repression.1,2 Since the first miRNA, lin-4, was discovered in 1993,3 over 1917 human miRNAs have been identified, and these collectively regulate the expression of at least one-third of protein-coding genes.4–6 miRNAs play a pivotal role in controlling diverse cellular functions and metabolic pathways, and dysregulated miRNA expression has been implicated in diseases such as cancer, diabetes, heart disease, and more.1,2,5 Consequently, delivering antagomiRs that suppress the expression of overabundant disease-promoting miRNAs, or delivering miRNA mimics to restore the expression of downregulated disease-suppressive miRNAs, have become attractive therapeutic strategies to eradicate disease. Unfortunately, unmodified antagomiRs and miRNA mimics are not suitable for clinical use because they have poor stability in biological fluids, unfavorable pharmacokinetic profiles, and limited ability to penetrate across cellular membranes. Novel nanocarriers must be developed to protect these nucleic acids from degradation, improve their pharmacokinetics, and facilitate their intracellular delivery. Polymer-based delivery vehicles have received substantial attention owing to their favorable characteristics and promising preclinical results. In this review, we summarize the use of polymer nanocarriers specifically for delivery of miRNA mimics to diseased cells. We describe the biological function of miRNAs, discuss challenges associated with miRNA delivery, and emphasize the advantages of polymer-based miRNA mimic delivery vehicles. Additionally, we summarize current preclinical efforts and shed light on remaining challenges to be addressed in order for polymer-based miRNA nanocarriers to successfully reach the clinic.
MIRNA BIOGENESIS AND MECHANISM OF GENE REGULATION
miRNA biogenesis includes both canonical and noncanonical pathways. Here we describe the canonical pathway as it is used to process the majority of miRNAs. In canonical miRNA biogenesis (Figure 1), miRNAs are initially transcribed in the nucleus by RNA polymerase II, producing primary miRNAs (pri-miRNAs) that have an internal hairpin within their larger structure.7 These pri-miRNAs are cleaved by Drosha to remove each end, creating ~70 base pair (bp) long preliminary miRNAs (pre-miRNAs) with an imperfect stem-loop structure that are then exported from the nucleus into the cytoplasm by an exportin 5/RanGTP complex.8,9 Once pre-miRNAs reach the cytoplasm, the RNase III endonuclease Dicer removes their terminal loop, resulting in mature double-stranded miRNA duplexes that are ~22 bp long.8,10
Figure 1.
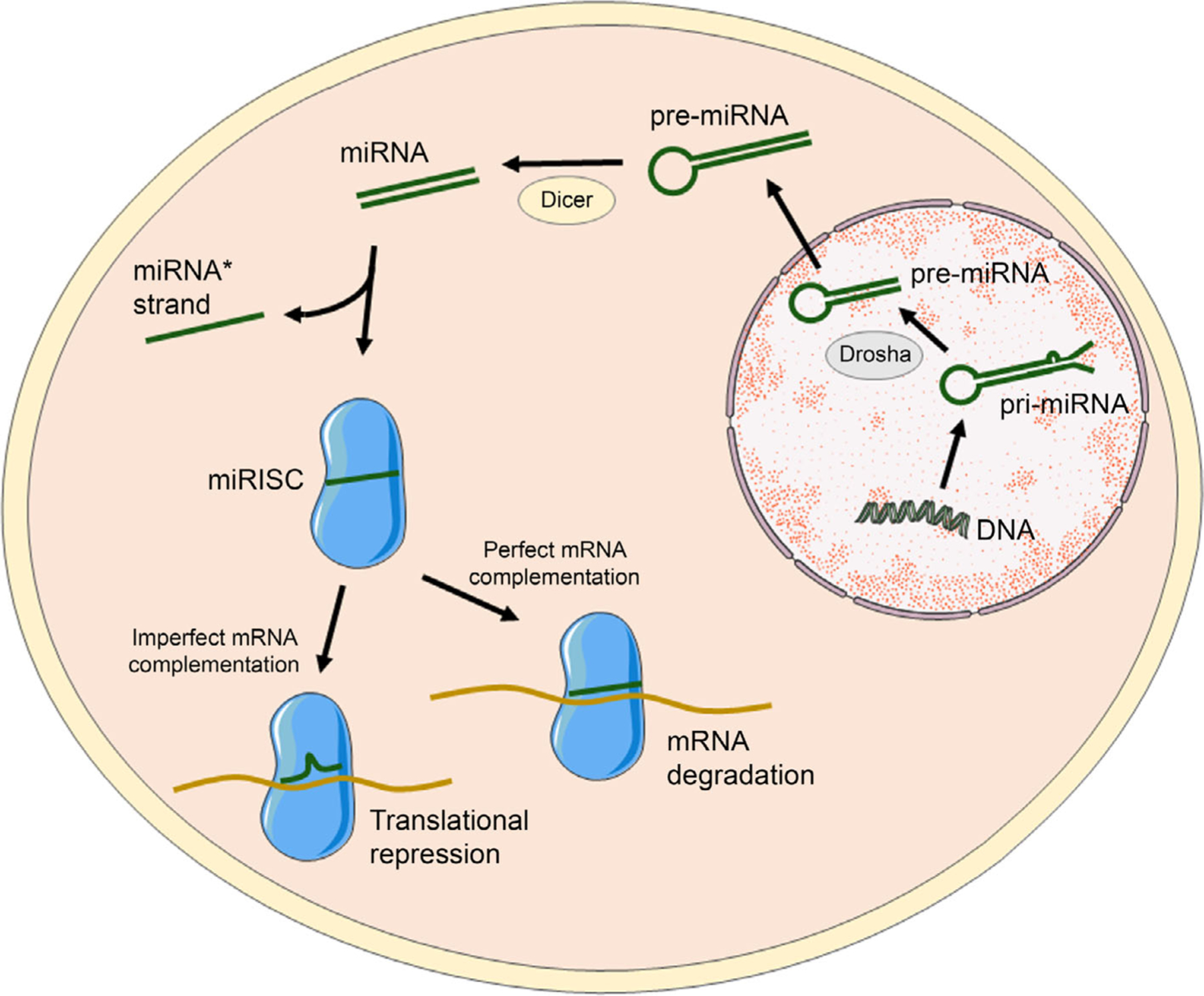
Scheme of miRNA biogenesis and mechanism of gene regulation. miRNAs are transcribed as pri-miRNAs in the nucleus that are cleaved by Drosha to form pre-miRNAs that are then transported to the cell cytoplasm by exportin proteins. Cytoplasmic pre-miRNAs with unstable stem-loop structures are further cleaved by dicer into small duplex RNA structures (miRNA:miRNA*) that contain both mature miRNA and its complementary strand (miRNA*). The miRNA* strand is typically degraded, and the mature miRNA strand assembles into RISC and guides it to specific messenger RNAs (mRNAs) to induce gene silencing.
After generation, one strand of the mature miRNA duplex, known as the guide strand or the miRNA strand, is incorporated into the miRNA-induced silencing complex (miRISC), which contains Argonaute proteins and is also known as the miRNA-containing ribonucleoprotein complex. The other strand, known as the passenger strand or miRNA* strand, is typically degraded, although there are instances where the miRNA* strand is loaded into miRISC to facilitate gene silencing.11 Once the mature miRNA is loaded into miRISC, it guides the complex to its cognate mRNA either with perfect complementarity or with imperfect base pairing. This results in either degradation or translation repression of the targeted mRNA, which thereby inhibits production of the encoded protein. Because miRNAs can regulate the expression of multiple genes, they play a fundamental and critical role in controlling cellular functions. Consequently, abnormal miRNA expression has been linked with the development of a broad spectrum of diseases, including neurodegenerative disorders, cardiovascular disorders, and cancers.1,2 Restoring the normal expression of disease-associated miRNAs is thus a promising therapeutic strategy, as outlined in the following section.
RESTORING NORMAL MIRNA EXPRESSION AND ACTIVITY THROUGH MIRNA MIMIC OR ANTAGOMIR DELIVERY—A THERAPEUTIC STRATEGY
In the past quarter century, the field of miRNA biology has expanded significantly. Due to global efforts in miRNA cloning and characterization and advances in technologies such as gene chips and real-time polymerase chain reaction, miRNA expression profiles have been rapidly established. As mentioned before, over 1917 human miRNAs have been identified to date and the abnormal over- or underexpression of specific miRNAs has been linked to the occurrence of various diseases. For example, several tumor-suppressive miRNAs, such as members of the miR-34,12,13 miR-200,14,15 let-7, and miR-15516–18 families (among others), have been identified as downregulated in cancers of the breast, lung, pancreas, bladder, ovary, and more. The loss of expression of these tumor-suppressive miRNAs allows cancer cells to proliferate uncontrollably, resist treatment, and invade foreign tissues, resulting in increased rates of metastasis and poorer prognoses.1,7 Moreover, multiple oncogenic miRNAs such as miR-21,19,20 miR-210,21,22 and miR-22123,24 have been identified as overexpressed in cancers including lymphoma, hepatocellular carcinoma, and non-small cell lung cancer, contributing to their progression. Apart from cancer, abnormal miRNA expression has been linked with the initiation and progression of cardiovascular disease,19,25–27 atherosclerosis,28–30 diabetes,31–33 hepatitis C,34–37 scleroderma,38 and more.1 For example, miR-122 upregulation increases the expression of hepatitis C virus RNA, and miR-21 upregulation in myocytes is linked with cardiac hypertrophy and fibrosis.1 Given that abnormal miRNA expression is linked with a number of devastating diseases, the development of technologies that can restore miRNA expression and activity to normal levels is a promising therapeutic strategy.
The two strategies that exist for restoring miRNA function to normal levels include (1) inhibiting miRNA activity through antagomiR delivery and (2) elevating miRNA expression through miRNA mimic delivery.39,40 In the first approach, antagomiRs (also known as anti-miRs), which are chemically modified synthetic antisense oligonucleotides that are complementary to their target miRNA, are delivered into cells and they sequester the mature miRNA to prevent mRNA binding, which thereby restores expression of the mRNAs that were previously silenced by the miRNA.41 The design and use of antagomiRs have been reviewed in detail elsewhere41; so this review focuses on discussion of miRNA mimics. miRNA mimics are synthetic double-stranded RNAs that are designed to replenish the activity of endogenous miRNAs whose expression is lost in disease states. These miRNA mimics are designed to have the same nucleotide sequence as the endogenous miRNA, and upon cellular delivery, they should load into miRISC and silence the same mRNAs that are targeted by the endogenous miRNA. While single-stranded RNAs that match the guide miRNA sequence could, in theory, achieve the same function as the endogenous miRNA, studies have shown that double-stranded miRNA mimics that contain both the guide and passenger strands are 100–1000 times more potent than single-stranded RNAs because they are better able to load into RISC.10,13 In the following sections, we discuss the challenges associated with enabling miRNA mimic delivery into diseased cells and tissues and introduce the design parameters that must be considered when developing miRNA mimic delivery vehicles.
CHALLENGES ASSOCIATED WITH ADMINISTRATION OF MIRNA MIMICS
To facilitate successful gene regulation, systemically administered miRNA mimics (and their carriers) must overcome many physiological barriers (Figure 2). Namely, they must be stable in circulation, cross vessel walls, penetrate through the extracellular matrix, be internalized by target cells, reach the cytosol, and be incorporated into RISC to induce downstream silencing of target mRNAs (Figure 2). Unfortunately, like small interfering RNAs and antisense oligonucleotides,14 naked miRNA mimics (i.e., miRNAs that are administered without any delivery vehicle) that are injected into the bloodstream are rapidly degraded by serum nucleases and cleared from the body. If any miRNA mimics reach the target site, they are at risk of instantaneous degradation by exonucleases and endonucleases present in the disease environment. Moreover, the hydrophilicity, negative charge, and large size (~14–15 kDa) of naked miRNA mimics limit their ability to cross negatively charged cell membranes via passive diffusion. Finally, naked miRNA mimics can engage toll-like receptors to activate the innate immune system, causing undesirable immunological side effects.42 Overcoming these barriers is a key for successful implementation of miRNA replacement therapy.
Figure 2.
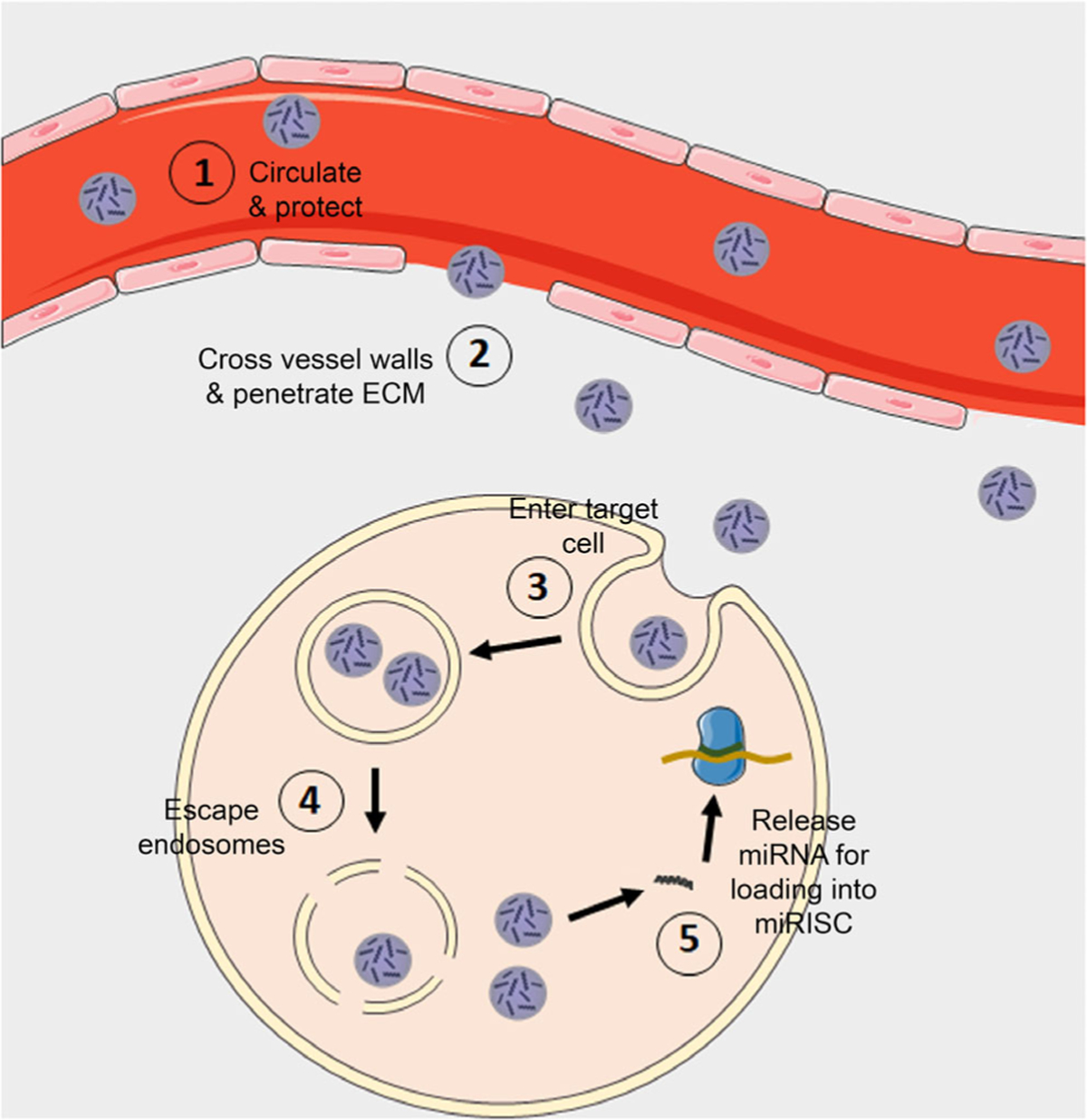
Scheme depicting the biological barriers that miRNA nanocarriers must overcome. miRNA nanocarriers must overcome various biological barriers to facilitate gene regulation. Specifically, they must (1) prevent miRNA degradation from serum nucleases and provide long circulation in the blood, (2) cross vessel walls and penetrate through the diseased tissue, (3) be efficiently internalized by target cells, (4) protect miRNA cargo in the harsh endo/lysosomal environment and escape endosomes, and (5) release miRNA into the cytoplasm so it can load into RISC to elicit gene knock-down
Historically, chemical modifications have been used to increase the stability and limit the immunogenicity of RNA therapeutics.43 Phosphorothioate modification of the RNA backbone, which substitutes a sulfur atom for an oxygen atom, reduces the ability of nucleases to degrade this bond.43 Likewise, replacing the 2′-OH with 2′-O-methyl, 2′-O-methoxyethyl, or 2′-fluoro groups can increase binding affinity to target mRNA,17 enhance nuclease stability,44 and extend serum stability.45 However, chemical modifications can also reduce the specificity and functionality of RNA therapeutics, and such alterations typically do not improve penetration across biological barriers.44 Therefore, there is a need to develop innovative delivery vehicles that can protect miRNA mimics from degradation and deliver them to target cells to induce robust gene regulation. Below, we introduce some of the key design parameters for miRNA mimic delivery vehicles.
DESIGN PARAMETERS FOR MIRNA MIMIC DELIVERY VEHICLES
As summarized above and illustrated in Figure 2, miRNA delivery vehicles must overcome many biological barriers to effectively deliver their cargo and regulate gene expression in target cells. Many characteristics influence the biodistribution, pharmacokinetics, and cellular uptake of materials, including size, charge, and surface functionality (Figure 3). With respect to size, nanoscale materials are ideally suited for RNA delivery because they can protect RNA from degradation, extend circulation half-life, facilitate cellular entry, and increase therapeutic index.46 Indeed, various nanoparticle (NP) delivery systems have shown considerable promise for the delivery of plasmid DNAs, antisense oligonucleotides, small interfering RNAs (siRNAs), and miRNAs to diseased cells in vitro and in vivo.47–61 When considering size as a design parameter for miRNA mimic delivery vehicles, it is important to note that NPs with diameters less than 5 nm rapidly undergo renal clearance upon intravenous administration and those with diameters greater than ~200 nm exhibit splenic filtration due to the 200–500 nm size range of interendothelial cell slits.62 Generally, NPs with diameters ranging from 10 to 100 nm are the longest circulating, with those less than 100 nm achieving the greatest tissue/cell uptake and transfection efficiency.62,63
Figure 3.
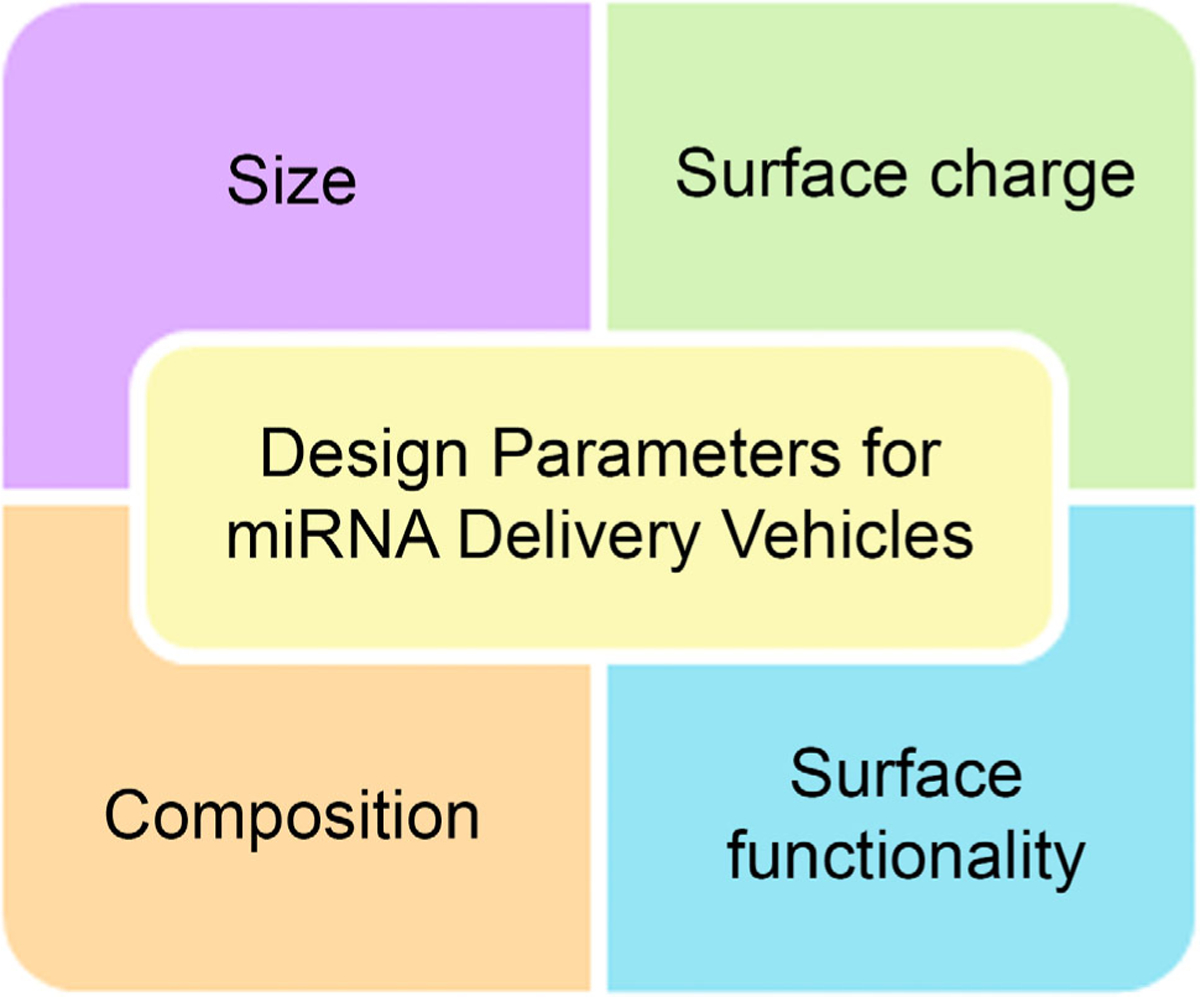
Design parameters for miRNA mimic delivery vehicles. The size, surface charge, surface functionality, and composition of miRNA nanocarriers influence their biodistribution, pharmacokinetics, and cellular trafficking, which ultimately define both safety and efficacy.
In addition to size, surface charge and surface chemistry also play a significant role in nanomaterial circulation time and cellular entry and should thus be considered when designing miRNA carriers. Since cells have negatively charged membranes, they can internalize positively charged nanomaterials more readily than negatively charged or neutral materials due to electrostatic interactions. However, when injected in the bloodstream, positively charged particles are cleared more rapidly than negative or neutral NPs and can induce hemolysis and platelet aggregation.64 Within the bloodstream, all NPs, regardless of charge, are masked by a biological corona (mainly consisting of opsonin proteins) that leads to their sequestration by the mononuclear phagocytic systems (MPS), reducing their distribution half-life.64,65 The rapid clearance of positively charged NPs is likely a consequence of the fact that the initial surface charge of an NP will dictate the specific makeup of the protein corona that forms on its surface, giving it a new “biological identity” that ultimately dictates pharmacokinetics and biodistribution.65
One strategy to minimize protein corona formation is to passivate the surface with poly(ethylene glycol) (PEG) polymer chains containing a methoxy end group. This provides a “stealth effect” in which a water hydration layer surrounds the NP to prevent opsonization and sequestration by the MPS. For example, adding PEG to liposomal doxorubicin increased its half-life from minutes to hours.66 While there are numerous examples of PEGylation being used to enhance NP circulation and target delivery, there are also some limitations to PEGylation.67,68 First, PEGylation can lead to an immune response, as up to 70% of people have anti-PEG antibodies existing in their body.69 Additionally, PEGylation can limit the cellular uptake of NPs. This can be overcome by simultaneously decorating NPs with ligands designed to bind specific receptors that are overexpressed on the surface of the target cells in order to facilitate receptor-mediated endocytosis.70,71 We recently reviewed the different types of targeting strategies that are utilized in nanomedicine72 and note that there is currently substantial debate regarding the benefits of targeting agents. For targeting agents to effectively mediate cell-specific binding and internalization, the NPs must first reach the diseased tissue, penetrate through the tissue to interact with the desired cells, and then engage the targeted receptors. This requires that the protein corona formed around the NP does not limit the effectiveness of the targeting agent, which is difficult to achieve. Recent meta-analyses and experimental studies have emphasized this point by showing that targeting agents only modestly improve the percentage of NPs that reach their target cells.73,74 These findings, and the challenges associated with the PEG immune response, have led researchers to begin exploring alternative strategies to enhance NP delivery to target cells, such as designing NPs to mimic cells within the body to hide them from the immune system and increase target delivery.72,75–77
Once an miRNA nanocarrier has overcome the aforementioned barriers to reach its target cell, it must then be internalized. Cells can take up NPs through either receptor-mediated or nonreceptor-mediated endocytosis, and the specific mechanism is influenced by both NP size and surface functionality. The mechanism of uptake is critical since it dictates the microenvironment the NPs will ultimately face after internalization. If NPs are internalized by clathrin-mediated endocytosis, the classic method of uptake, then they will be routed to endosomes and then to lysosomes, where the harsh acidic and enzyme-rich environment will degrade nucleic acid cargo.46,62,63 Alternatively, if NPs are internalized by caveolin-mediated endocytosis, they will be trafficked to neutral caveosomes and bypass the acidic lysosomal environment.46,62,63 The ultimate fate of nanomaterials in caveosomes is still being explored, but some evidence suggests caveosomes route their cargo to the endoplasmic reticulum and the cytoplasm.78 Regardless of the mechanism of uptake, RNA nanocarriers must ultimately reach the cytosol to deliver their miRNA cargo to miRISC.
Since clathrin-mediated uptake is the primary method by which NPs enter cells, an extraordinary amount of research has explored ways to enable endosomal escape of NPs prior to lysosomal fusion.79–81 Studies have shown that materials with a high buffering capacity (such as polyethylenimine [PEI]) can be incorporated into NPs to induce swelling and rupture of endosomes. This is often attributed to the “proton sponge effect,” wherein proton absorption by the material leads to an influx of water and rupture of the endosomal compartment, but recent studies have called into question the accuracy of this proposed phenomenon.82,83 An alternative strategy is to coat RNA carriers with fusogenic peptides or lipids or membrane-destabilizing peptides.62 Finally, cationic polymers can be incorporated into NPs to mediate endosome escape by causing the positive surface of the NP to interact with the negative outer surface of the endosome, leading to membrane flipping and cargo release via the “flip-flop” mechanism.84,85
The final, but perhaps most important, design parameter that one might consider in creating an miRNA mimic delivery vehicle is the composition of the carrier. Composition will dictate whether the miRNA is loaded on the surface of the carrier or encapsulated within the carrier, and it will also dictate the degradation profile (if any), as well as the rate of cargo release. Metal-based NPs,56–59 liposomes,50,86,87 polymer nanocarriers88–95 (including polymer NPs, polyplexes, and dendrimers), lipid NPs,96,97 hydrogels,98–100 and more have been explored for the delivery of siRNA and miRNA. The benefits and limitations of lipid NPs and liposomes and of metal NPs as miRNA delivery vehicles have been reviewed elsewhere.39,63,90,101 In the following sections, we review polymer nanocarriers that have been explored for delivering miRNA therapeutics, highlighting their preclinical success and challenges remaining in the transition to the clinic.
POLYMER NANOCARRIERS FOR MIRNA DELIVERY
Polymer materials have been widely explored as tools to carry therapeutic cargo (e.g., small molecules, peptides, proteins, DNAs, siRNAs, and miRNAs) to specific tissues/cells to intervene in disease.47,52–55,102,103 The types of polymers that have been incorporated into delivery vehicles include poly(lactic-co-glycolic acid) (PLGA), PEG, poly-L-lysine (PLL), poly-L-arginine (PLA), PEI, and more. The advantages of polymer carriers over other material choices include ease of synthesis, biodegradability, tailorability, and the ability to overcome certain biological barriers (Figure 2) to extend circulation and facilitate cell uptake and endosomal escape.104,105 With respect to miRNA mimic delivery, the most widely studied delivery vehicles include polyplexes, PLGA NPs, and dendrimers, whose structures are depicted in Figure 4. The following sections highlight the accomplishments made with each of these systems.
Figure 4.
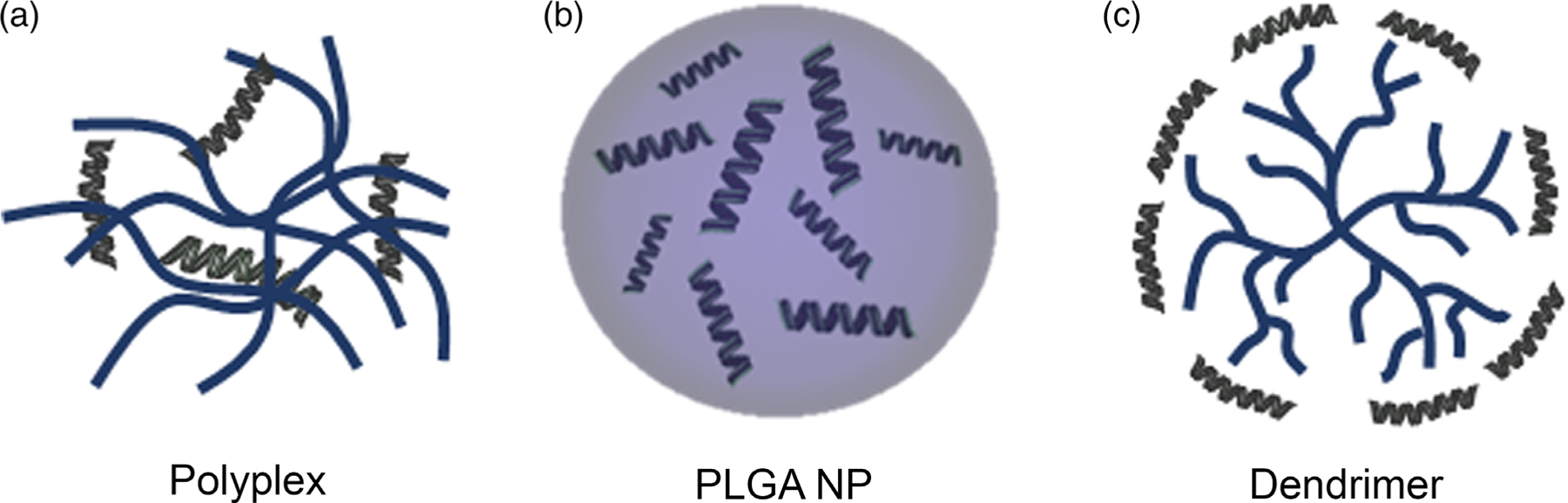
Artistic rendering of the structure of the three main types of polymer miRNA nanocarriers. (a) Polyplexes consist of nucleic acids (green) that are electrostatically complexed with cationic polymers (blue). (b) PLGA NPs can encapsulate miRNA molecules in their interior to protect them from premature degradation during transport. (c) Dendrimers are hyperbranched polymers with tree-like structures that can be complexed with miRNAs electrostatically or that can be chemically linked to miRNAs through their end-terminal functional groups.
Polyplexes
Polyplexes are nanocarriers that consist of negatively charged nucleic acids that are complexed with positively charged polymers (polycations) through electrostatic interactions [Figure 4(a)]. Various cationic polymers such as PEI, chitosan, and N-(2-hydroxypropyl) methacrylamide (HPMA) have been explored for intracellular delivery of different miRNAs.106–110 Compared with viral vectors, polyplexes have several advantages, including ease of manufacturing and low immunogenicity. Accordingly, they have been widely explored for miRNA delivery, as summarized below.
PEI is one of the most commonly used polycations for miRNA delivery. It consists of repeating units of CH2CH2NH and can have either a linear or a branched structure. Ibrahim et al. used branched PEI to deliver miR-33a or miR-145 (both are known for their tumor-suppressive role) in a murine colorectal carcinoma model.111 Intraperitoneal injections of PEI/miR-145 or PEI/miR-33a in nude mice with subcutaneous LS174T colorectal tumors significantly attenuated tumor growth and reduced the expression of the target genes c-Myc and ERK5, as compared with mice that received control PEI/miRNA polyplexes.111 Others have also prepared PEI polyplexes to deliver miRNA mimics to treat triple-negative breast cancer (TNBC).112,113 One drawback associated with PEI is that it can bind to negatively charged cytosolic proteins and cause aggregation resulting in cytotoxicity.110 To overcome this limitation, researchers have utilized PEI modified with either disulfide groups113 or PLL,112 which can maintain or enhance transfection efficiency while reducing toxicity. Hwang et al. explored a similar approach to enable miRNA delivery to the brain, wherein disulfide-modified PEI (SSPEI) was conjugated to rabies virus glycoprotein (RVG) to form RVG-SSPEI that could target acetylcholine receptors on neural cells.114 Compared with unmodified PEI/miRNA polyplexes, the RVG-SSPEI polyplexes were less toxic in acetylcholine receptor-positive Neuro2a cells and they also greatly enhanced the delivery of miR-124a to the brain parenchyma following intravenous injection into Balb/c mice.114
The above studies demonstrate that the toxicity of PEI can be lessened by cleverly engineering the polymer design. However, additional studies need to be performed to further optimize the therapeutic ratio of PEI polyplexes. In general, as the molecular weight of PEI increases, so does its toxicity.109,115 Additionally, the structure of PEI affects its toxicity, as linear versus branched PEIs interact differently with cells owing to differences in the types of amine groups they contain (linear PEIs contain strictly primary and secondary amino groups, while branched PEIs also contain tertiary amino groups).116,117 Understanding structure/function relationships is critical to maximize the potential for PEI/miRNA polyplexes to have success in the clinic.
Another polycation that is frequently used for miRNA delivery is chitosan, which is a naturally derived linear disaccharide composed of N-acetyl-D-glucosamine and D-glucosamine.118 Chitosan/miRNA polyplexes have been investigated to treat a broad spectrum of disorders, including spinal cord injury and cancer, with impressive results. For example, delivery of miR-124 via chitosan polyplexes reduced neuroinflammation in a rat with spinal cord injury.107 Moreover, ex vivo transfection of chitosan/miR-124 polyplexes in rat microglia reduced the secretion of pro-inflammatory factors [e.g., TNF-α (tumor necrosis factor alpha)] and decreased expression of major histocompatibility complex-II. Furthermore, injured rats that received chitosan/miR-124 polyplexes had significantly reduced macrophage activation as compared with injured rats that received chitosan/miR-CTRL polyplexes. This drastic in vivo attenuation of neuroinflammation suggests that chitosan nanocarriers could prove useful as miRNA delivery vehicles for brain disorders.107
With respect to cancer, chitosan-based polyplexes that delivered miR-200a or miR-200b to tumor vasculature in a metastatic HeyA8 ovarian cancer model reduced metastatic burden by 92% compared with control miRNA.14 Likewise, miR-34a/chitosan polyplexes reduced bone metastasis of both breast and skin cancers in murine models,95 and they also attenuated prostate tumor growth in the bone in an intrafemoral model that represents established prostate cancer bone metastasis.94 Given that metastasis is the leading cause of cancer-related deaths, these results are extremely exciting and warrant further investigation into the use of chitosan-based miRNA delivery vehicles.
In recent years, some less common polymers have begun to be explored for miRNA delivery. For instance, Peng et al. conjugated a CXCR4 antagonist, AMD3465 to HPMA via a disulfide linker and prepared polyplexes to deliver miR-200c into U20S human osteosarcoma cells.106 U20S cells that were treated with P-SS-AMD/miR-200c polyplexes had significantly reduced migration and lower levels of ZEB1 (a known miR-200c target) compared with U20S cells that received negative control P-SS-AMD/miR-NC polyplexes. While these results are promising, the lack of in vivo studies with P-SS-AMD polyplexes limits the potential impact of these findings.106 A different imine backbone-based polymer, TPSP, was used by Luo et al. to synthesize polyplexes for treatment of diabetic peripheral neuropathy (DPN) through delivery of miR-146a-5p, an miRNA that has anti-inflammatory effects in DPN.108 TPSP consists of imine, spermidine, and 1,4-phthalaldehyde linkers with an external shell of PEG-PCL-maltotriose-COO−. Compared with DPN rats that received no treatment or empty TPSP polyplexes via intramuscular injection, those that received TPSP/miR-146a-5p polyplexes had significantly lower levels of the pro-inflammatory cytokines interleukin (IL)-6 and IL-10.108 Additionally, Western blotting on explanted sciatic nerve tissue samples showed that DPN rats that were treated with TPSP/miR-146a-5p polyplexes had significantly decreased expression of the proapoptotic factor TNF-α and cleaved caspase-3 compared with DPN rats that received no treatment or empty TPSP polyplexes.108 These results suggest that TPSP/miR-146a-5p polyplexes can attenuate DPN-associated inflammation and sciatic nerve damage.108 They also support further investigation into this polymer for delivery of other therapeutic miRNAs.
While the above studies demonstrate that polyplexes can offer good transfection efficiency in vitro and elicit gene regulation in vivo, there is always a dilemma when using polyplexes for RNA delivery because their in vivo efficacy and clinical development have been thwarted due to their high toxicity, which limits the range of practical doses that may be safely applied.109,110 Accordingly, other polymer-based materials, such as those described in the following sections, may be more likely to reach the clinic in a timely manner.
PLGA Nanoparticles
PLGA NPs [Figure 4(b)] are attractive tools for drug and gene delivery owing to their biodegradability, biocompatibility, and ability to encapsulate and protect either hydrophobic or hydrophilic cargo. PLGA consists of lactic and glycolic acid monomers that are coupled by ester linkages, and the ratio of the monomers identifies the type of PLGA (e.g., 50:50 PLGA, the most commonly employed in nanomedicine, consists of 50% lactic acid and 50% glycolic acid). To produce PLGA NPs, either single- or double-emulsion solvent evaporation is used. The single emulsion, or oil-in-water emulsification method, is used to encapsulate water-insoluble drugs, while the double-emulsion water-in-oil-in-water method is used to encapsulate water-soluble cargo such as nucleic acids.119 In biological environments, PLGA degrades by hydrolysis of the ester linkages and the rate of degradation depends on the monomer ratio, with 50:50 PLGA displaying the fastest degradation. As hydrolysis of the ester linkages yields the original monomers that can be metabolized in the body by the Krebs cycle, PLGA has minimal toxicity.119 Given these desirable properties, PLGA has been approved by the Food and Drug Administration (FDA) for parenteral administration for many decades, with many products available on the market. Most products have focused on hydrophobic cargo delivery, with researchers only recently turning their attention to miRNA delivery.
To facilitate miRNA delivery, PLGA NP formulations are typically prepared with an additional positively charged molecule such as PEI120 or chitosan,92 which can increase loading by condensing the nucleic acids through electrostatic interactions. In some cases, this leads to RNA loading within the PLGA NP core,120 while in others the RNA is bound to the exterior of the PLGA NP.92 In an alternative approach, copolymers of PEI-PLGA,93 PLL-PLGA,121 or PLA-PLGA122 can be used to synthesize miRNA-loaded NPs. With these various formulations, encapsulation efficiencies ranging from 78.3%120 up to 95.1%92 have been reported. Further, all these different synthesis strategies can achieve sustained miRNA release, which is important for therapeutic utility. Arora et al. showed that PLGA NPs loaded with miR-150 mimics and PEI as a condensing agent could release ~60% of their cargo within 14 days at 37 °C in Tris-ethylene diamine tetraacetic acid (TE) buffer containing 10% serum,120 while Cai et al. showed that their NPs made with mPEG-PLGA-PLL-lactobionic acid polymers could release ~85% of their miR-99a cargo within 6 days at 37 °C in TE buffer.121 Besides enhancing RNA loading, another advantage of using PEI, PLL, PLA, or similar polycations to prepare PLGA NPs is that these molecules can enhance endosome escape, as described in preceding sections. Achieving cytosolic delivery is imperative for miRNA mimics to load into RISC and elicit therapeutic gene silencing.
As with other miRNA nanocarriers, PLGA NPs have been most widely studied for their use as tools to fight cancer. Those that incorporate PEI have been tested only in vitro, with Arora et al. showing they could deliver miR-150 mimics to pancreatic cancer cells to reduce cell motility and invasion120 and Wang et al. demonstrating they could deliver miR-542–3p to TNBC cells to promote apoptosis by activating p53 and inhibiting survivin expression.93 Notably, Wang et al. co-encapsulated doxorubicin in their NPs, which were coated with hyaluronic acid to target CD44 receptors on TNBC cells, and showed that they could increase drug uptake and cytotoxicity in MDA-MB-231 TNBC cells as compared with MCF7 cells that have low CD44 expression.93 Excitingly, several other PLGA NP miRNA nanocarriers have achieved in vivo success. For example, PLGA NPs have been used to treat multiple myeloma via miR-34a delivery,92 hepatic carcinoma via miR-99a delivery,121 and colon cancer via miR-204 delivery122 in murine models. These studies demonstrate the substantial potential of PLGA NPs as miRNA delivery vehicles.
PLGA-based miRNA nanocarriers have also been used for applications beyond cancer. Intimal hyperplasia is the thickening of a blood vessel, typically in response to injury, and miR-145 is downregulated in hyperplastic vascular smooth muscle cells. Nishio et al. showed that they could use PLGA nanocarriers to increase miR-145 expression by 18.5-fold in vascular smooth muscle cells in vitro, and this translated to attenuated intimal hyperplasia in a rabbit model.123 In another study, McKiernan et al. showed PLGA microparticles could deliver pre-miR-19b-3p to macrophages, which are cells that play a critical role in inflammatory diseases such as cystic fibrosis.124 Although this research is in its early stages, it is exciting given that macrophages are difficult to transfect.
Importantly, some of the above studies that included in vivo analyses have indicated PLGA is a safe material to use for miRNA delivery, as animals treated with PLGA NPs have shown no signs of tissue toxicity, changes in blood chemistry, or alterations in body weight.92,121 A limitation of these studies is that they were restricted to short time frames, so future studies should examine long-term safety and the body’s response to multiple injections of PLGA NPs. Additionally, future studies should directly compare the potency of PLGA NPs versus other carriers as miRNA delivery vehicles.
Dendrimers
Dendrimers are hyperbranched polymers with symmetric tree-like structures [Figure 4(c)]. All dendrimers have the same structural makeup, which is a central core surrounded by three or more branches of repeating monomer units.125 At the ends of the branches are terminal functional groups that provide ease of conjugation.126 The number of repeating branch units defines a dendrimer’s “generation” and dictates its overall globular structure. Dendrimers are attractive as miRNA delivery vehicles because they have a combination of properties that distinguish them from other polymer delivery systems. The main advantage of dendrimers is that they have precise and well-defined size, which is controlled by the choice of monomeric unit used to make them.127 In addition to controlling size and shape, the monomer unit building blocks also define the resulting dendrimer’s charge and solubility. A further advantage of dendrimers is that they can be conjugated to many desired end moieties due to the sheer amount of functional terminal groups they contain. Overall, dendrimers are customizable and can thus be designed with specific desired properties to enable various therapeutic applications. When complexed with biological molecules, dendrimers are called dendriplexes, and cationic dendrimers are a practical choice of carrier for miRNA because they can readily associate with negatively charged nucleic acids. While many dendrimer types have been explored for siRNA delivery128–133 and antagomiR delivery,134–138 little research has examined the use of dendrimers for miRNA mimic delivery. Below, we describe some of the findings in this field to date.
Polyamidoamine (PAMAM) dendrimers have been the most widely studied for delivery of siRNAs, antagomiRs, and miRNA mimics due to their low toxicity in comparison to other classes.126 To facilitate miRNA delivery to specific cells, PAMAM dendrimers are often coated with targeting ligands.139–141 For example, Duan and coworkers conjugated chondroitin sulfate (CS) to PAMAM dendrimers to form CS-PAMAM that could deliver miR-34a to CD44 overexpressing pancreatic cancer cells.139 MiaPaCa-2 human pancreatic carcinoma cells treated with CS-PAMAM/miR-34a dendriplexes showed moderately higher apoptosis (21.3%) and cell cycle arrest in G1 phase (75.4%) than cells treated with nontargeted dendriplexes, which exhibited 17.5% apoptosis and 70.7% G1 arrest. To put these numbers in perspective, untreated control cells displayed 6% apoptosis and 55.2% G1 arrest.139 In a similar strategy, Liu et al. modified PAMAM dendrimers with folic acid (FA),140 which can bind folate receptors that are overexpressed by many cancer cell types. Using these FA/PAMAM dendrimers, they showed they could deliver miR-7 to U251 glioma cells in vitro to suppress cell proliferation and invasion by decreasing the expression of the miR-7 target genes EGFR, PI3K, and AKT2.140 When U251 cells that were treated in vitro with FA/PAMAM/miR-7 dendriplexes were subsequently inoculated into the right caudate, tumor growth was significantly delayed as compared with tumors that were formed by untreated cells or cells treated with miR-7-loaded nontargeted liposomes.140 Further, the FA/PAMAM-miR-7 tumors had decreased expression of markers indicative of proliferation and invasion, including proliferating cell nuclear antigen, and matrix metalloproteinases 2 and 9.140Moving forward, these studies should be extended to more clinically relevant models where tumor treatment is performed by systemic or intratumoral injection of the dendriplexes. Additionally, targeted versus nontargeted dendriplexes should be compared, and miR-7 cargo should be compared against scrambled control miRNA.
In an intriguing study, Gray et al. presented a novel targeting approach wherein they constructed “bowtie” shaped PAMAM dendrimers in which one side was polyplexed with miR-126 and the other side was conjugated to either polyarginine (polyR), a cell-penetrating peptide, or to RGD peptides, which can also facilitate cell uptake.141 Since miR-126 regulates vascular integrity, endothelial cell proliferation, and neovascularization, the bowtie dendrimers were examined for their interactions with human umbilical vein endothelial cells (HUVECs).141 When HUVECs were treated with either the polyR-bowties or the RGD-bowties, they experienced significantly enhanced tube formation as compared with nontreated cells or cells treated with undecorated bowties. qPCR validated that the bowtie formulations reduced the expression of SPRED1 mRNA, a known target of miR-126.141
Besides being coupled to targeting agents, dendrimers have also been coupled to other entities to enable multifaceted therapy and imaging. For example, Yang et al. modified gadolinium-functionalized nanographene oxide (Gd-NGO) with PAMAM and showed that these materials could be complexed with both the anticancer drug epirubicin (EPI) and Let-7g miRNA for regulation of the Ras pathway.142 Accordingly, these materials could act as dual drug/miRNA delivery vehicles and also be imaged by magnetic resonance imaging. When U87 human glioma cells were treated with Gd-NGO alone, the expression levels of Pan-Ras proteins were unaltered, but treatment with Gd-NGO/Let-7g significantly reduced Pan-Ras expression. Additionally, incubation of U87 cells with naked Let-7g miRNA in the presence of transfection reagent or with Gd-NGO/scrambled miRNA had no effect on Pan-Ras expression.142 The impact of the different materials on U87 cell viability was assessed through XTT (2,3-bis-[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide) assay, which quantifies cellular proliferation based on a colored product that forms when mitochondrial enzymes reduce extracellular XTT. This showed Gd-NGO and pure Let-7g were not cytotoxic toward U87 cells. This assay also revealed that the concentration of EPI required for 50% inhibition of cellular growth (IC50) was higher for free EPI than for Gd-NGO/EPI. Furthermore, the IC50 of Gd-NGO/Let-7g/EPI was significantly lower than that of Gd-NGO/EPI or Gd-NGO/Let-7g.142 These findings led the authors to conclude that inhibition of oncogenic Ras signaling by Let-7g can sensitize U87 cells to EPI chemotherapy. These results remain to be validated in in vivo models, but overall, this approach to enhance drug potency through miRNA delivery has notable potential.
Beyond PAMAM, other dendrimers have also been explored for miRNA delivery. One, developed by Satchi-Fainaro and team, consists of polyglycerolamine (dPG-NH2).89,91 In an initial study, the team showed that human peripheral blood mononuclear cells that were treated with dPG-NH2 dendrimers carrying tumor-suppressive miR-34a exhibited minimal secretion of TNF-alpha and IL-6, demonstrating that dPG-NH2/miR-34a polyplexes have low immunogenicity.91 Further, intratumoral injection of dPG-NH2/miR-34a polyplexes in mice with glioblastoma significantly attenuated tumor growth and prolonged survival compared with mice treated with control dPG-NH2/NC-miR polyplexes or phosphate-buffered saline.91
In an exciting related study, the team demonstrated that these dendrimers could deliver miR-34a, miR-93, or miR-200c to osteosarcoma to regulate dormancy and progression.89 Polyplexes composed of dPG-NH2 dendrimers and each respective miRNA mimic reduced the levels of the target genes cMET, hypoxia-inducible factor 1α (HIF1α), and moesin in two different aggressive osteosarcoma cell lines, Saos-2 and MG-63.89 The authors further demonstrated that delivery of miR-93 and miR-200c via these dendrimers reduced the level of vascular endothelial growth factor (VEGF) and thus attenuated the angiogenic capabilities of fast-growing osteosarcoma in vitro and in vivo.89 Furthermore, intratumoral administration of the dendriplexes in Saos-2-E tumor-bearing SCID mice yielded ~100 000-fold increased expression of miR-34a and miR-93 and ~1000-fold increased expression of miR-200c.89 This increased expression converted Saos-2-E tumors into a phenotype that resembles dormant tumors, which was maintained for ~40 days.89 The median survival of the mice was noticeably extended by 55 days following treatment with miR-93 and by 74 days following treatment with miR-34a or miR-200c.89
Together, the above studies demonstrate the exciting potential of dendrimers as miRNA delivery vehicles. However, like other polymer carriers, toxicity is a concern blocking the clinical translation of dendrimers. All classes of highly branched dendrimers are cationic at physiological pH and therefore present cytotoxic and hemolytic properties.143 The cationic properties and toxicity will vary depending on the makeup of the dendrimers and the end functional groups.144 The main reason for this toxicity is that the positive charge of the dendrimers, which enhances cellular uptake, also leads to destabilization of negatively charged cell membranes.144 When dendrimers are conjugated to targeting antibodies, drug molecules, or nucleic acids, this decreases the number of reactive groups, which subsequently reduces the charge and associated toxicity.145–148 One strategy to improve dendrimers’ safety is to modify their surface with PEG moieties, which reduces cytotoxicity and extends circulation.149–152 Other methods include shielding of the positive charges via acetylation153 and hydroxylation.151,154 Moving forward, minimizing toxicity will be imperative for dendrimer therapeutics to reach their full clinical potential.
FUTURE OUTLOOK: REMAINING CHALLENGES TO ADDRESS TO ENABLE CLINICAL TRANSLATION OF POLYMERIC MIRNA NANOCARRIERS
A substantial body of work indicates polymer-based miRNA nanocarriers have great promise to treat a variety of diseases. Table I summarizes some of the systems that have been developed and examined in preclinical studies. The beauty of miRNA replacement therapy is that it offers the ability to target virtually any gene known to contribute to disease progression. If this vision can become a reality, it will transform medical practice. However, there are many substantial challenges that remain to be overcome in order for polymer-based miRNA therapeutics to reach their therapeutic potential (Figure 5).
Table I.
Polymer-Based miRNA Nanocarriers in Preclinical Animal Studies
| Delivery vehicle (polycation) | miRNA | Disease/Target | Delivery route | Animal model | Reference |
|---|---|---|---|---|---|
| Polyplexes | |||||
| PEI | miR-145, miR-33a | Colon carcinoma | Intraperitoneal injection | Subcutaneous inoculation of LS174T or HCT116 cells in athymic nude mice | 111 |
| RVG-SSPEI | miR-124a | Acetylcholine receptor positive euro2a cells | Tail vein injection | Male BALB/c mice | 114 |
| Chitosan | miR-124 | Macrophages, microglia | Spinal cord microinjection | Female, Sprague-Dawley rats | 107 |
| Dendrimers | |||||
| Polyglycerol (dPG-NH2) | miR-34a | Glioblastoma | Intratumoral injection | Subcutaneous inoculation of human U87 MG cells in male SCID mice | 91 |
| TPSP (imine backbone-based polymer) | miR-146a-5p | DPN | Intramuscular injection | Male Wistar rats | 108 |
| Polyglycerol (dPG-NH2) | miR-34a, miR-93, and miR-200c | Osteosarcoma | Intratumoral injection | Subcutaneous inoculation of Saos-2-E cells in SCID mice | 89 |
| FA-conjugated PAMAM | miR-7 | Glioma | U251 cells were treated with the dendrimers In vitro before inoculation in mice | Intracranial tumors formed from pretreated U251 cells | 140 |
| PLGA NPs | |||||
| Chitosan/PLGA NPs | miR-34a | Multiple myeloma (SKMM1 cells) | Intravenous injection | Interscapular injection of SKMM1 cells in NOD-SCID mice | 92 |
| mPEG-PLGA-PLL-LA/VEGFab NPs | miR-99a | Hepatic carcinoma | Intravenous injection | Subcutaneous inoculation of HepG2 cells in BALB/c mice | 121 |
| PLGA/PLA-PEG-FA NPs with spermidine | miR-204–5p | Colon cancer | Intravenous injection | Subcutaneous Luc-HT-29 tumors in female athymic BALB/c nu/nu mice | 122 |
| Chitosan/PLGA NPs | miR-145 | Venous intimal hyperplasia | Incubation of rabbit jugular vein grafts in various treatment groups | Implantation of jugular vein grafts in the ipsilateral carotid artery | 123 |
mPEG-PLGA-PLL-LA/VEGFab NPs, monomethoxy (polyethylene glycol)-poly(D,L-lactide-co-glycolide)-poly(L-lysine)-lactobionic acid-anti-vascular endothelial growth factor antibody NPs; PLGA/PLA-PEG-FA NPs, poly(D,L-lactide-co-glycolide)/poly(L-lactide)-block-poly(ethylene glycol)-folate polymer NPs.
Figure 5.

Remaining challenges to address to enable clinical translation of polymeric miRNA nanocarriers
One key challenge for miRNA replacement therapy is to identify a target miRNA whose loss of expression is specific to disease and whose replacement with miRNA mimics will halt disease progression without causing off-target effects. Bioinformatic prediction of miRNA targets is challenging because miRNAs can bind hundreds of mRNA molecules and miRNA expression can be heterogeneous across tissues. With the advent of human genome and Gene Chip technology, our understanding of various miRNAs, their targets, and their role in normal physiology and disease has significantly improved. By coupling knowledge derived from databases such as The Cancer Genome Atlas, miRTarBase, or StarBase with technologies such as miR-CHIP sequencing and biochemical assays, our ability to identify miRNA–mRNA associations will continuously improve. Using prediction tools such as TargetScan will help researchers more accurately predict mRNA targets of miRNAs and enable more efficient identification of promising miRNA mimics for disease intervention.
A second challenge associated with miRNA replacement therapy is toxicity related to nonspecific gene silencing and immune reaction. With regard to nonspecific gene silencing, this includes both miRNA-induced silencing of previously unknown mRNA targets, as well as the silencing of known mRNA targets inside healthy cells rather than diseased cells due to off-site delivery. It is also possible that overloading RISC with exogenous miRNA can hinder the ability of endogenous miRNAs to achieve their normal cellular function and/or hyperactivate cellular pathways to reduce the viability of normal cells.155 It is difficult to predict the magnitude of off-target toxicity for miRNA mimic delivery vehicles because few studies have evaluated the effect of miRNA therapeutics in vivo. Most studies performed in animals have concluded that miRNA mimics and their carriers are well tolerated.156–160 It was therefore extremely surprising when the phase I clinical trial of MRX34, a liposomal miR-34a formulation, was halted due to cases of cytokine release syndrome, an immune-related toxicity, occurring in patients.161 However, it is not known whether the adverse effects were due to off-target effects of the miRNA or due to the design of the liposomal delivery vehicles. Many scientists predict that therapeutic miRNA mimics will be better tolerated in humans than siRNA because the endogenous miRNAs that are being replaced in diseased cells are still expressed in healthy cells. Thus, any accumulation of miRNA mimics in normal cells will be an insignificant increase relative to what is already present.162 In contrast, since diseased cells have low to no expression of the targeted miRNA, accumulation of miRNA mimics in these cells should have substantial impacts. Nevertheless, it is of the utmost importance to design miRNA sequences with the goal of minimizing side effects.
As noted throughout this review, the clinical translation of miRNA mimics relies heavily on the development of safe and efficient carriers. Several systems have shown promise in preclinical scenarios, but one challenge remaining to be addressed is scaling up the manufacturing of NP delivery vehicles. Currently, many NPs are formed via self-assembly and mixing of various components, and it is difficult to produce consistent size, quality, and stability as the batch size increases, which is necessary for large clinical trials. New microfluidic methods have been developed to provide better control over manufacturing of polymeric NPs comprised of three to four chemical components, resulting in less batch-to-batch variability.163–166 Implementing techniques such as this will be imperative for the field moving forward.
Beyond manufacturing, several challenges related to enabling nanocarriers to overcome biological barriers remain to be addressed. PEGylation is the primary method of enhancing nanomaterial stability and circulation, but recent studies showed the body can produce a substantial anti-PEG immune response.167,168 This may severely limit the use of PEGylated carriers in the clinic. Therefore, innovative design strategies are required to abrogate or minimize the immunogenicity associated with PEGylated carriers or to enhance circulation and stability without PEG coatings.169 Additionally, most nanocarriers are trafficked to the liver and spleen following intravenous delivery. Minimizing the percentage of MPS clearance and maximizing delivery to desired tissues will be critical for the success of miRNA nanocarriers. Finally, endosomal escape is a major bottleneck for the translation of miRNA therapeutics. According to Gilleron et al., only 1–2% of siRNAs conjugated to colloidal gold NPs achieve endosome escape,170 and it is likely that similar levels are achieved for polymer-based miRNA carriers. Improving this efficiently even slightly may dramatically increase the potency of miRNA therapeutics.
In summary, miRNA mimics are exciting tools to combat disease. Although some challenges remain to be addressed regarding miRNA selection and the synthesis and optimization of delivery carriers, polymer-based miRNA nanocarriers have a promising path forward toward clinical translation. There are multiple studies investigating the therapeutic potential of miRNA mimics in preclinical trials, and some commercial efforts are already under way.171–173 As we continue to learn more about basic miRNA biology and to develop new knowledge in the fields of biomaterials and nanomedicine, the potential of miRNA replacement therapy will become a reality.
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under award number R35GM119659. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIH.
Biographies
Chintan Kapadia is a postdoctoral researcher in Dr. Emily Day’s lab at the University of Delaware. He received his Ph.D. in Pharmaceutical Sciences from University of North Carolina at Chapel Hill. He has extensive experience in formulation development and biophysical characterization of various biotherapeutics and delivery carriers for small molecules, peptides, and oligonucleotides. His current work focuses on engineering polymer carriers to deliver microRNAs for the treatment of triple-negative breast cancer.

Benjamin Luo is a graduate research assistant at the University of Delaware, where he is currently developing nanocarriers for intracellular RNA delivery. He received his bachelor’s degree in bioengineering from the University of California, Berkeley.

Megan Dang is a graduate student in Dr. Emily Day’s lab in Biomedical Engineering at the University of Delaware. She received her undergraduate degree in Bioengineering from the University of Maryland, College Park. Her current work focuses on utilizing gold nanoshells as carriers to deliver antibodies and/or RNA molecules to triple-negative breast cancer to suppress tumor growth.

N’Dea Irvin-Choy is a doctoral student in the Biomedical Engineering Department at the University of Delaware. Her research aims to develop polymeric nanoparticles as therapeutic carriers to improve maternal and fetal health during pregnancy. Her honors and awards include the University of Delaware Biomedical Engineering Chair’s Fellowship and the University Graduate Scholars Award.

Danielle Valcourt is a graduate student in Dr. Emily Day’s lab in Biomedical Engineering at the University of Delaware. She received her undergraduate degree in Chemical and Biomolecular Engineering from the University of Notre Dame. Her current work focuses on treating triple-negative breast cancer using polymer-based nanomedicines.

Dr. Emily Day is an Assistant Professor of Biomedical Engineering at the University of Delaware. Her lab engineers nanoparticles with unique architectures to enable high precision therapy of cancer and other diseases. She has received numerous awards for her independent research spanning photothermal therapy, gene regulation, and biomimetic cargo delivery, including the 2018 Rita Schaffer Award from the Biomedical Engineering Society, an NSF CAREER Award, an NIH R01, and an NIH R35 MIRA Award.

REFERENCES
- 1.Rupaimoole R; Slack FJ Nat. Rev. Drug Discov 2017, 16, 203. [DOI] [PubMed] [Google Scholar]
- 2.He L; Hannon GJ Nat. Rev. Genet 2004, 5, 522. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC; Feinbaum RL; Ambros V Cell. 1993, 75, 843. [DOI] [PubMed] [Google Scholar]
- 4. http://www.mirbase.org/
- 5.Cha W; Fan R; Miao Y; Zhou Y; Qin C; Shan X; Wan X; Cui T MedChemComm. 2018, 9, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alles J; Fehlmann T; Fischer U; Backes C; Galata V; Minet M; Hart M; Abu-Halima M; Grässer FA; Lenhof H-P; Keller A; Meese E Nucleic Acids Res. 2019, 47, 3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macfarlane LA; Murphy PR Curr. Genomics 2010, 11, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim VN Nat. Rev. Mol. Cell Biol 2005, 6, 376. [DOI] [PubMed] [Google Scholar]
- 9.Nakielny S; Dreyfuss G Cell. 1999, 99, 677. [DOI] [PubMed] [Google Scholar]
- 10.Murchison EP; Hannon GJ Curr. Opin. Cell Biol 2004, 16, 223. [DOI] [PubMed] [Google Scholar]
- 11.Guo L; Lu Z PLOS ONE. 2010, 5, e11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misso G; Di Martino MT; De Rosa G; Farooqi AA; Lombardi A; Campani V; Zarone MR; Gulla A; Tagliaferri P; Tassone P; Caraglia M Mol. Ther. Nucleic Acids 2014, 3, e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slabakova E; Culig Z; Remsik J; Soucek K Cell Death Dis. 2017, 8, e3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pecot CV; Rupaimoole R; Yang D; Akbani R; Ivan C; Lu C; Wu S; Han H-D; Shah MY; Rodriguez-Aguayo C; Bottsford-Miller J; Liu Y; Kim SB; Unruh A; Gonzalez-Villasana V; Huang L; Zand B; Moreno-Smith M; Mangala LS; Taylor M; Dalton HJ; Sehgal V; Wen Y; Kang Y; Baggerly KA; Lee J-S; Ram PT; Ravoori MK; Kundra V; Zhang X; Ali-Fehmi R; Gonzalez-Angulo A-M; Massion PP; Calin GA; Lopez-Berestein G; Zhang W; Sood AK Nat. Commun 2013, 4, 2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortez MA; Valdecanas D; Zhang X; Zhan Y; Bhardwaj V; Calin GA; Komaki R; Giri DK; Quini CC; Wolfe T; Peltier HJ; Bader AG; Heymach JV; Meyn RE; Welsh JW Mol. Ther 2014, 22, 1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tili E; Croce CM; Michaille JJ Int. Rev. Immunol 2009, 28, 264. [DOI] [PubMed] [Google Scholar]
- 17.Faraoni I; Antonetti FR; Cardone J; Bonmassar E Biochim. Biophys. Acta 2009, 1792, 497. [DOI] [PubMed] [Google Scholar]
- 18.Gironella M; Seux M; Xie M-J; Cano C; Tomasini R; Gommeaux J; Garcia S; Nowak J; Yeung ML; Jeang K-T; Chaix A; Fazli L; Motoo Y; Wang Q; Rocchi P; Russo A; Gleave M; Dagorn J-C; Iovanna JL; Carrier A; Pébusque M-J; Dusetti NJ Proc. Natl. Acad. Sci 2007, 104, 16170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y; Zhang CJ Cardiovasc. Transl. Res 2010, 3, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan LX; Huang XF; Shao Q; Huang MY; Deng L; Wu QL; Zeng YX; Shao JY RNA. 2008, 14, 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W; Sun T; Cao J; Liu F; Tian Y; Zhu W Exp. Cell Res 2012, 318, 944. [DOI] [PubMed] [Google Scholar]
- 22.Puissegur MP; Mazure NM; Bertero T; Pradelli L; Grosso S; Robbe-Sermesant K; Maurin T; Lebrigand K; Cardinaud B; Hofman V; Fourre S; Magnone V; Ricci JE; Pouyssegur J; Gounon P; Hofman P; Barbry P; Mari B Cell Death Differ. 2011, 18, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pineau P; Volinia S; McJunkin K; Marchio A; Battiston C; Terris B; Mazzaferro V; Lowe SW; Croce CM; Dejean A Proc. Natl. Acad. Sci. U.S.A 2010, 107, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garofalo M; Di Leva G; Romano G; Nuovo G; Suh SS; Ngankeu A; Taccioli C; Pichiorri F; Alder H; Secchiero P; Gasparini P; Gonelli A; Costinean S; Acunzo M; Condorelli G; Croce CM Cancer Cell. 2009, 16, 498. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Thum T; Gross C; Fiedler J; Fischer T; Kissler S; Bussen M; Galuppo P; Just S; Rottbauer W; Frantz S; Castoldi M; Soutschek J; Koteliansky V; Rosenwald A; Basson MA; Licht JD; Pena JT; Rouhanifard SH; Muckenthaler MU; Tuschl T; Martin GR; Bauersachs J; Engelhardt S Nature. 2008, 456, 980. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda S; He A; Kong SW; Lu J; Bejar R; Bodyak N; Lee KH; Ma Q; Kang PM; Golub TR; Pu WT Mol. Cell. Biol 2009, 29, 2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan ZX; Lin QX; Fu YH; Deng CY; Zhou ZL; Zhu JN; Liu XY; Zhang YY; Li Y; Lin SG; Yu XY Biochem. Biophys. Res. Commun 2009, 381, 597. [DOI] [PubMed] [Google Scholar]
- 28.Feinberg MW; Moore KJ Circ. Res 2016, 118, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rayner KJ; Esau CC; Hussain FN; McDaniel AL; Marshall SM; van Gils JM; Ray TD; Sheedy FJ; Goedeke L; Liu X; Khatsenko OG; Kaimal V; Lees CJ; Fernandez-Hernando C; Fisher EA; Temel RE; Moore KJ Nature. 2011, 478, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goedeke L; Salerno A; Ramírez CM; Guo L; Allen RM; Yin X; Langley SR; Esau C; Wanschel A; Fisher EA; Suárez Y; Baldán A; Mayr M; Fernández-Hernando C EMBO Mol. Med 2014, 6, 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long J; Wang Y; Wang W; Chang BH; Danesh FR J. Biol. Chem 2011, 286, 11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Putta S; Lanting L; Sun G; Lawson G; Kato M; Natarajan RJ Am. Soc. Nephrol 2012, 23, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trajkovski M; Hausser J; Soutschek J; Bhat B; Akin A; Zavolan M; Heim MH; Stoffel M Nature. 2011, 474, 649. [DOI] [PubMed] [Google Scholar]
- 34.Thibault PA; Huys A; Amador-Canizares Y; Gailius JE; Pinel DE; Wilson JA J. Virol 2015, 89, 6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jopling CL; Yi M; Lancaster AM; Lemon SM; Sarnow P Science. 2005, 309, 1577. [DOI] [PubMed] [Google Scholar]
- 36.Elmen J; Lindow M; Silahtaroglu A; Bak M; Christensen M; Lind-Thomsen A; Hedtjarn M; Hansen JB; Hansen HF; Straarup EM; McCullagh K; Kearney P; Kauppinen S Nucleic Acids Res. 2008, 36, 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elmen J; Lindow M; Schutz S; Lawrence M; Petri A; Obad S; Lindholm M; Hedtjarn M; Hansen HF; Berger U; Gullans S; Kearney P; Sarnow P; Straarup EM; Kauppinen S Nature. 2008, 452, 896. [DOI] [PubMed] [Google Scholar]
- 38.Maurer B; Stanczyk J; Jungel A; Akhmetshina A; Trenkmann M; Brock M; Kowal-Bielecka O; Gay RE; Michel BA; Distler JH; Gay S; Distler O Arthritis Rheum. 2010, 62, 1733. [DOI] [PubMed] [Google Scholar]
- 39.Lam JK; Chow MY; Zhang Y; Leung SW Mol. Ther. Nucleic Acids 2015, 4, e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakraborty C; Sharma AR; Sharma G; Doss CGP; Lee S-S Mol. Ther. Nucleic Acids 2017, 8, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenvang J; Petri A; Lindow M; Obad S; Kauppinen S Silence. 2012, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabbri M; Paone A; Calore F; Galli R; Croce CM RNA Biol. 2013, 10, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lennox KA; Behlke MA Gene Ther. 2011, 18, 1111. [DOI] [PubMed] [Google Scholar]
- 44.Watts JK; Deleavey GF; Damha MJ Drug Discov. Today 2008, 13, 842. [DOI] [PubMed] [Google Scholar]
- 45.Layzer JM; McCaffrey AP; Tanner AK; Huang Z; Kay MA; Sullenger BA RNA. 2004, 10, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petros RA; DeSimone JM Nat. Rev. Drug Discov 2010, 9, 615. [DOI] [PubMed] [Google Scholar]
- 47.Knipe JM; Strong LE; Peppas NA Biomacromolecules. 2016, 17, 788. [DOI] [PubMed] [Google Scholar]
- 48.Yin H; Kanasty RL; Eltoukhy AA; Vegas AJ; Dorkin JR; Anderson DG Nat. Rev. Genet 2014, 15, 541. [DOI] [PubMed] [Google Scholar]
- 49.Gao K; Huang L Mol. Pharm 2009, 6, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanasty R; Dorkin JR; Vegas A; Anderson D Nat. Mater 2013, 12, 967. [DOI] [PubMed] [Google Scholar]
- 51.Amalvy JI; Wanless EJ; Li Y; Michailidou V; Armes SP; Duccini Y Langmuir. 2004, 20, 8992. [DOI] [PubMed] [Google Scholar]
- 52.Liechty WB; Scheuerle RL; Vela Ramirez JE; Peppas NA Int. J. Pharm 2019, 562, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forbes DC; Peppas NA Macromol. Biosci 2014, 14, 1096. [DOI] [PubMed] [Google Scholar]
- 54.Forbes DC; Peppas NA ACS Nano. 2014, 8, 2908. [DOI] [PubMed] [Google Scholar]
- 55.Creixell M; Peppas NA Nano Today. 2012, 7, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riley RS; Dang MN; Billingsley MM; Abraham B; Gundlach L; Day ES Nano Lett. 2018, 18, 3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goyal R; Kapadia CH; Melamed JR; Riley RS; Day ES Cell. Mol. Bioeng 2018, 11, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kouri FM; Hurley LA; Daniel WL; Day ES; Hua Y; Hao L; Peng CY; Merkel TJ; Queisser MA; Ritner C; Zhang H; James CD; Sznajder JI; Chin L; Giljohann DA; Kessler JA; Peter ME; Mirkin CA; Stegh AH Genes Dev. 2015, 29, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jensen SA; Day ES; Ko CH; Hurley LA; Luciano JP; Kouri FM; Merkel TJ; Luthi AJ; Patel PC; Cutler JI; Daniel WL; Scott AW; Rotz MW; Meade TJ; Giljohann DA; Mirkin CA; Stegh AH Sci. Transl. Med 2013, 5, 209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melamed JR; Ioele SA; Hannum AJ; Ullman VM; Day ES Mol. Pharm 2018, 15, 5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melamed JR; Kreuzberger NL; Goyal R; Day ES Mol. Ther. Nucleic Acids 2018, 12, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blanco E; Shen H; Ferrari M Nat. Biotechnol 2015, 33, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahmadzada T; Reid G; McKenzie DR Biophys Rev. 2018, 10, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Albanese A; Tang PS; Chan WC Annu. Rev. Biomed. Eng 2012, 14, 1. [DOI] [PubMed] [Google Scholar]
- 65.Lazarovits J; Chen YY; Sykes EA; Chan WC Chem. Commun. (Camb.) 2015, 51, 2756. [DOI] [PubMed] [Google Scholar]
- 66.Hamilton A; Biganzoli L; Coleman R; Mauriac L; Hennebert P; Awada A; Nooij M; Beex L; Piccart M; Van Hoorebeeck I; Bruning P; de Valeriola D Ann. Oncol 2002, 13, 910. [DOI] [PubMed] [Google Scholar]
- 67.Verhoef JJ; Anchordoquy TJ Drug Deliv. Transl. Res 2013, 3, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsieh YC; Wang HE; Lin WW; Roffler SR; Cheng TC; Su YC; Li JJ; Chen CC; Huang CH; Chen BM; Wang JY; Cheng TL; Chen FM Theranostics. 2018, 8, 3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Q; Lai SK Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 2015, 7, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brannon-Peppas L; Blanchette JO Adv. Drug Deliv. Rev 2012, 64, 206. [DOI] [PubMed] [Google Scholar]
- 71.Wang M; Thanou M Pharmacol. Res 2010, 62, 90. [DOI] [PubMed] [Google Scholar]
- 72.Valcourt DM; Harris J; Riley RS; Dang M; Wang J; Day ES Nano Res. 2018, 11, 4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilhelm S; Tavares AJ; Dai Q; Ohta S; Audet J; Dvorak HF; Chan WC W. Nat. Rev. Mater 2016, 1, 16014. [Google Scholar]
- 74.Dai Q; Wilhelm S; Ding D; Syed AM; Sindhwani S; Zhang Y; Chen YY; MacMillan P; Chan WC W. ACS Nano 2018, 12, 8423. [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez PL; Harada T; Christian DA; Pantano DA; Tsai RK; Discher DE Science. 2013, 339, 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fang RH; Hu C-MJ; Luk BT; Gao W; Copp JA; Tai Y; O’Connor DE; Zhang L Nano Lett. 2014, 14, 2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao W; Zhang LJ Drug Target. 2015, 23, 619. [DOI] [PubMed] [Google Scholar]
- 78.El-Sayed A; Harashima H Mol. Ther 2013, 21, 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dominska M; Dykxhoorn DM J. Cell Sci 2010, 123, 1183. [DOI] [PubMed] [Google Scholar]
- 80.Lönn P; Kacsinta AD; Cui X-S; Hamil AS; Kaulich M; Gogoi K; Dowdy SF Sci. Rep 2016, 6, 32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma D Nanoscale. 2014, 6, 6415. [DOI] [PubMed] [Google Scholar]
- 82.Cho YW; Kim J-D; Park KJ Pharm. Pharmacol 2003, 55, 721. [DOI] [PubMed] [Google Scholar]
- 83.Benjaminsen RV; Mattebjerg MA; Henriksen JR; Moghimi SM; Andresen TL Mol. Ther 2013, 21, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Varkouhi AK; Scholte M; Storm G; Haisma HJ J. Control. Release 2011, 151, 220. [DOI] [PubMed] [Google Scholar]
- 85.Medina-Kauwe LK; Xie J; Hamm-Alvarez S Gene Ther. 2005, 12, 1734. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y; Huang L Mol. Ther 2010, 18, 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bochicchio S; Dalmoro A; Barba AA; Grassi G; Lamberti G Curr. Drug Metab 2014, 15, 882. [DOI] [PubMed] [Google Scholar]
- 88.Ban E; Kwon TH; Kim A Drug Deliv. Transl. Res 2019. 10.1007/s13346-019-00645-y [DOI] [PubMed] [Google Scholar]
- 89.Tiram G; Segal E; Krivitsky A; Shreberk-Hassidim R; Ferber S; Ofek P; Udagawa T; Edry L; Shomron N; Roniger M; Kerem B; Shaked Y; Aviel-Ronen S; Barshack I; Calderon M; Haag R; Satchi-Fainaro R ACS Nano. 2016, 10, 2028. [DOI] [PubMed] [Google Scholar]
- 90.Ben-Shushan D; Markovsky E; Gibori H; Tiram G; Scomparin A; Satchi-Fainaro R Drug Deliv. Transl. Res 2014, 4, 38. [DOI] [PubMed] [Google Scholar]
- 91.Ofek P; Calderon M; Mehrabadi FS; Krivitsky A; Ferber S; Tiram G; Yerushalmi N; Kredo-Russo S; Grossman R; Ram Z; Haag R; Satchi-Fainaro R Nanomedicine. 2016, 12, 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cosco D; Cilurzo F; Maiuolo J; Federico C; Di Martino MT; Cristiano MC; Tassone P; Fresta M; Paolino D Sci. Rep 2015, 5, 17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang S; Zhang J; Wang Y; Chen M Nanomedicine. 2016, 12, 411. [DOI] [PubMed] [Google Scholar]
- 94.Gaur S; Wen Y; Song JH; Parikh NU; Mangala LS; Blessing AM; Ivan C; Wu SY; Varkaris A; Shi Y; Lopez-Berestein G; Frigo DE; Sood AK; Gallick GE Oncotarget. 2015, 6, 29161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krzeszinski JY; Wei W; Huynh H; Jin Z; Wang X; Chang TC; Xie XJ; He L; Mangala LS; Lopez-Berestein G; Sood AK; Mendell JT; Wan Y Nature. 2014, 512, 431. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Tam YYC; Chen S; Cullis PR Pharmaceutics. 2013, 5, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Whitehead KA; Dorkin JR; Vegas AJ; Chang PH; Veiseh O; Matthews J; Fenton OS; Zhang Y; Olejnik KT; Yesilyurt V; Chen D; Barros S; Klebanov B; Novobrantseva T; Langer R; Anderson DG Nat. Commun 2014, 5, 4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dunn SS; Tian S; Blake S; Wang J; Galloway AL; Murphy A; Pohlhaus PD; Rolland JP; Napier ME; DeSimone JM J. Am. Chem. Soc 2012, 134, 7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang LL; Burdick JA Adv. Healthc. Mater 2017, 6 (1). 10.1002/adhm.201601041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Krebs MD; Jeon O; Alsberg EJ Am. Chem. Soc 2009, 131, 9204. [DOI] [PubMed] [Google Scholar]
- 101.Campani V; De Rosa G; Misso G; Zarone MR; Grimaldi A Curr. Pharm. Biotechnol 2016, 17, 741. [DOI] [PubMed] [Google Scholar]
- 102.Panyam J; Labhasetwar V Adv. Drug Deliv. Rev 2003, 55, 329. [DOI] [PubMed] [Google Scholar]
- 103.Vasir JK; Labhasetwar V Expert Opin. Drug Deliv 2006, 3, 325. [DOI] [PubMed] [Google Scholar]
- 104.Neuse EW Met. Based Drugs 2008, 2008, 469531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liechty WB; Kryscio DR; Slaughter BV; Peppas NA Annu. Rev. Chem. Biomol. Eng 2010, 1, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peng Z-H; Xie Y; Wang Y; Li J; Oupický D Mol. Pharm 2017, 14, 1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Louw AM; Kolar MK; Novikova LN; Kingham PJ; Wiberg M; Kjems J; Novikov LN Nanomedicine. 2016, 12, 643. [DOI] [PubMed] [Google Scholar]
- 108.Luo Q; Feng Y; Xie Y; Shao Y; Wu M; Deng X; Yuan W-E; Chen Y; Shi X Nanomed-Nanotechnol. 2019, 17, 188. [DOI] [PubMed] [Google Scholar]
- 109.Hall A; Lachelt U; Bartek J; Wagner E; Moghimi SM Mol. Ther 2017, 25, 1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ulasov AV; Khramtsov YV; Trusov GA; Rosenkranz AA; Sverdlov ED; Sobolev AS Mol. Ther 2011, 19, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ibrahim AF; Weirauch U; Thomas M; Grunweller A; Hartmann RK; Aigner A Cancer Res. 2011, 71, 5214. [DOI] [PubMed] [Google Scholar]
- 112.Gao S; Tian H; Guo Y; Li Y; Guo Z; Zhu X; Chen X Acta Biomater. 2015, 25, 184. [DOI] [PubMed] [Google Scholar]
- 113.Dai Y; Zhang X Macromol. Biosci 2019, 19, e1800445. [DOI] [PubMed] [Google Scholar]
- 114.Hwang DW; Son S; Jang J; Youn H; Lee S; Lee D; Lee YS; Jeong JM; Kim WJ; Lee DS Biomaterials. 2011, 32, 4968. [DOI] [PubMed] [Google Scholar]
- 115.Grayson AC; Doody AM; Putnam D Pharm. Res 2006, 23, 1868. [DOI] [PubMed] [Google Scholar]
- 116.Wiseman JW; Goddard CA; McLelland D; Colledge WH Gene Ther. 2003, 10, 1654. [DOI] [PubMed] [Google Scholar]
- 117.Wightman L; Kircheis R; Rossler V; Carotta S; Ruzicka R; Kursa M; Wagner EJ Gene Med. 2001, 3, 362. [DOI] [PubMed] [Google Scholar]
- 118.Mao S; Sun W; Kissel T Adv. Drug Deliv. Rev 2010, 62, 12. [DOI] [PubMed] [Google Scholar]
- 119.Lu JM; Wang X; Marin-Muller C; Wang H; Lin PH; Yao Q; Chen C Expert Rev. Mol. Diagn 2009, 9, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arora S; Swaminathan SK; Kirtane A; Srivastava SK; Bhardwaj A; Singh S; Panyam J; Singh AP Int. J. Nanomed 2014, 9, 2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cai C; Xie Y; Wu L; Chen X; Liu H; Zhou Y; Zou H; Liu D; Zhao Y; Kong X; Liu P Sci. Rep 2017, 7, 46250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zheng B; Chen L; Pan CC; Wang JZ; Lu GR; Yang SX; Xue ZX; Wang FY; Xu CL Nanomedicine (Lond.) 2018, 13, 769. [DOI] [PubMed] [Google Scholar]
- 123.Nishio H; Masumoto H; Sakamoto K; Yamazaki K; Ikeda T; Minatoya KJ Thorac. Cardiovasc. Surg 2018, 45, 298. [DOI] [PubMed] [Google Scholar]
- 124.McKiernan PJ; Lynch P; Ramsey JM; Cryan SA; Greene CM Medicines. 2018, 5, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tomalia DA; Baker H; Dewald J; Hall M; Kallos G; Martin S; Roeck J; Ryder J; Smith P Polymer J. 1985, 17, 117. [Google Scholar]
- 126.Dzmitruk V; Apartsin E; Ihnatsyeu-Kachan A; Abashkin V; Shcharbin D; Bryszewska M Pharmaceutics. 2018, 10, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mintzer MA; Grinstaff MW Chem. Soc. Rev 2011, 40, 173. [DOI] [PubMed] [Google Scholar]
- 128.Patil ML; Zhang M; Betigeri S; Taratula O; He H; Minko T Bioconjug. Chem 2008, 19, 1396. [DOI] [PubMed] [Google Scholar]
- 129.Waite CL; Sparks SM; Uhrich KE; Roth CM BMC Biotechnol. 2009, 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou J; Wu J; Hafdi N; Behr JP; Erbacher P; Peng L Chem. Commun. (Camb.) 2006, 22, 2362. [DOI] [PubMed] [Google Scholar]
- 131.Taratula O; Garbuzenko OB; Kirkpatrick P; Pandya I; Savla R; Pozharov VP; He H; Minko TJ Control. Release 2009, 140, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Taratula O; Garbuzenko O; Savla R; Andrew Wang Y; He H; Minko T Curr. Drug Deliv 2011, 8, 59. [DOI] [PubMed] [Google Scholar]
- 133.Biswas S; Torchilin VP Pharmaceuticals (Basel). 2013, 6, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang F; Zhang B; Zhou L; Shi Y; Li Z; Xia Y; Tian J ACS Appl. Mater. Interfaces 2016, 8, 9014. [DOI] [PubMed] [Google Scholar]
- 135.Qian X; Ren Y; Shi Z; Long L; Pu P; Sheng J; Yuan X; Kang C Mol. Pharm 2012, 9, 2636. [DOI] [PubMed] [Google Scholar]
- 136.Qian X-M; Shi Z-D; Ren Y; Liu C-Y; Ji Y-R; Long L-X; Pu P; Sheng J; Yuan X-B; Kang C-SJ Appl. Polym. Sci 2013, 127, 570. [Google Scholar]
- 137.Mei M; Ren Y; Zhou X; Yuan X-B; Li F; Jiang L-H; Kang C-S; Yao ZJ Appl. Polym. Sci 2009, 114, 3760. [Google Scholar]
- 138.Ren Y; Kang CS; Yuan XB; Zhou X; Xu P; Han L; Wang GX; Jia Z; Zhong Y; Yu S; Sheng J; Pu PY J. Biomater. Sci. Polym. Ed 2010, 21, 303. [DOI] [PubMed] [Google Scholar]
- 139.Duan Y; Xing Z; Yang J; Wang Y; Chen J; Zhang Y; Shi W; Li Q RSC Adv. 2016, 6, 70870. [Google Scholar]
- 140.Liu X; Li G; Su Z; Jiang Z; Chen L; Wang J; Yu S; Liu Z Oncol. Rep 2013, 29, 1387. [DOI] [PubMed] [Google Scholar]
- 141.Gray WD; Wu RJ; Yin X; Zhou J; Davis ME; Luo Y Biomacromolecules. 2013, 14, 101. [DOI] [PubMed] [Google Scholar]
- 142.Yang H-W; Huang C-Y; Lin C-W; Liu H-L; Huang C-W; Liao S-S; Chen P-Y; Lu Y-J; Wei K-C; Ma C-CM Biomaterials. 2014, 35, 6534. [DOI] [PubMed] [Google Scholar]
- 143.Duncan R; Izzo L Adv. Drug Deliv. Rev 2005, 57, 2215. [DOI] [PubMed] [Google Scholar]
- 144.Madaan K; Kumar S; Poonia N; Lather V; Pandita DJ Pharm. Bioallied Sci 2014, 6, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Arima H; Yoshimatsu A; Ikeda H; Ohyama A; Motoyama K; Higashi T; Tsuchiya A; Niidome T; Katayama Y; Hattori K; Takeuchi T Mol. Pharm 2012, 9, 2591. [DOI] [PubMed] [Google Scholar]
- 146.Sunoqrot S; Bugno J; Lantvit D; Burdette JE; Hong SJ Control. Release 2014, 191, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jain K; Kesharwani P; Gupta U; Jain NK Int. J. Pharm 2010, 394, 122. [DOI] [PubMed] [Google Scholar]
- 148.Kulhari H; Pooja D; Shrivastava S; Kuncha M; Naidu VGM; Bansal V; Sistla R; Adams DJ Sci. Rep 2016, 6, 23179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Suk JS; Xu Q; Kim N; Hanes J; Ensign LM Adv. Drug Deliv. Rev 2016, 99, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Luo D; Haverstick K; Belcheva N; Han E; Saltzman WM Macromolecules. 2002, 35, 3456. [Google Scholar]
- 151.Palmerston Mendes L; Pan J; Torchilin VP Molecules. 2017, 22, 1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bugno J; Hsu HJ; Hong SJ Drug Target. 2015, 23, 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kolhatkar RB; Kitchens KM; Swaan PW; Ghandehari H Bioconjug. Chem 2007, 18, 2054. [DOI] [PubMed] [Google Scholar]
- 154.Mastorakos P; Kambhampati SP; Mishra MK; Wu T; Song E; Hanes J; Kannan RM Nanoscale. 2015, 7, 3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ishida M; Selaru FM Curr. Anesthesiol. Rep 2013, 1, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Esquela-Kerscher A; Trang P; Wiggins JF; Patrawala L; Cheng A; Ford L; Weidhaas JB; Brown D; Bader AG; Slack FJ Cell Cycle. 2008, 7, 759. [DOI] [PubMed] [Google Scholar]
- 157.Montgomery RL; Yu G; Latimer PA; Stack C; Robinson K; Dalby CM; Kaminski N; van Rooij E EMBO Mol. Med 2014, 6, 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wiggins JF; Ruffino L; Kelnar K; Omotola M; Patrawala L; Brown D; Bader AG Cancer Res. 2010, 70, 5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kota J; Chivukula RR; O’Donnell KA; Wentzel EA; Montgomery CL; Hwang HW; Chang TC; Vivekanandan P; Torbenson M; Clark KR; Mendell JR; Mendell JT Cell. 2009, 137, 1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Takeshita F; Patrawala L; Osaki M; Takahashi RU; Yamamoto Y; Kosaka N; Kawamata M; Kelnar K; Bader AG; Brown D; Ochiya T Mol. Ther 2010, 18, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. https://www.businesswire.com/news/home/20160920006814/en/Mirna-Therapeutics-Halts-Phase-1-Clinical-Study.
- 162.Bader AG; Brown D; Winkler M Cancer Res. 2010, 70, 7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xu J; Zhang S; Machado A; Lecommandoux S; Sandre O; Gu F; Colin A Sci. Rep 2017, 7, 4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xu Z; Lu C; Riordon J; Sinton D; Moffitt MG Langmuir. 2016, 32, 12781. [DOI] [PubMed] [Google Scholar]
- 165.Li X; Jiang X Adv. Drug Deliv. Rev 2018, 128, 101. [DOI] [PubMed] [Google Scholar]
- 166. https://www.dolomite-microfluidics.com/
- 167.Ishida T; Kiwada H Int. J. Pharm 2008, 354, 56. [DOI] [PubMed] [Google Scholar]
- 168.Ishida T; Atobe K; Wang X; Kiwada HJ Control. Release 2006, 115, 251. [DOI] [PubMed] [Google Scholar]
- 169.Abu Lila AS; Kiwada H; Ishida TJ Control. Release 2013, 172, 38. [DOI] [PubMed] [Google Scholar]
- 170.Gilleron J; Querbes W; Zeigerer A; Borodovsky A; Marsico G; Schubert U; Manygoats K; Seifert S; Andree C; Stöter M; Epstein-Barash H; Zhang L; Koteliansky V; Fitzgerald K; Fava E; Bickle M; Kalaidzidis Y; Akinc A; Maier M; Zerial M Nat. Biotechnol 2013, 31, 638. [DOI] [PubMed] [Google Scholar]
- 171. http://regulusrx.com/
- 172. http://www.miragen.com/
- 173. https://engeneic.com/


