Abstract
Compactness, among several others, is one unique and very attractive feature of a scanning fiber-optic two-photon endomicroscope. To increase the scanning area and the total number of resolvable pixels (i.e., the imaging throughput), it typically requires a longer cantilever which, however, leads to a much undesired, reduced scanning speed (and thus imaging frame rate). Herein we introduce a new design strategy for a fiber-optic scanning endomicroscope, where the overall numerical aperture (NA) or beam focusing power is distributed over two stages: 1) a mode-field focuser engineered at the tip of a double-clad fiber (DCF) cantilever to pre-amplify the single-mode core NA, and 2) a micro objective of a lower magnification (i.e., ~2× in this design) to achieve final tight beam focusing. This new design enables either an ~9-fold increase in imaging area (throughput) or an ~3-fold improvement in imaging frame rate when compared to traditional fiber-optic endomicroscope designs. The performance of an as-designed endomicroscope of an enhanced throughput-speed product was demonstrated by two representative applications: (1) high-resolution imaging of an internal organ (i.e., mouse kidney) in vivo over a large field of view without using any fluorescent contrast agents, and (2) real-time neural imaging by visualizing dendritic calcium dynamics in vivo with sub-second temporal resolution in GCaMP6m-expressing mouse brain. This cascaded NA amplification strategy is universal and can be readily adapted to other types of fiber-optic scanners in compact linear or nonlinear endomicroscopes.
Keywords: Biomedical optical imaging, Biophotonics, Endomicroscope, In vivo imaging, Nonlinear optics, Optical microscopy, Two-photon microscopy
I. Introduction
TWO-PHOTON microscopy (2PM), with its intrinsic depth-sectioning capability, deeper penetration and reduced photodamage, has become the three-dimensional microscopy method of choice for in vivo deep imaging of biological tissues [1], [2]. Among the numerous biomedical applications of 2PM, two particularly appealing categories are: 1) structural and functional imaging of unstained biological tissue using native (auto)fluorescence (especially from NADH and FAD) or second-harmonic generation signal from collagen fibers for translational clinical applications [3], [4], and 2) optical recording of neural dynamics from multiple neurons and other excitable glial cells (such as astrocytes) of mouse models with genetically encoded calcium indicators (GECIs), especially the family of GCaMP [5], [6]. The bulky footprint of conventional bench-top 2PM embodiments, however, limits its clinical usage to superficial tissues (such as skin or oral cavity), and necessitates constraining the head of mouse models beneath the objective for neural dynamics acquisition [7].
To further broaden the application of 2PM, a plethora of fiber-optic flexible two-photon endo- (or mini-) microscope had been prototyped during the past two decades [8]-[22]. The three central components shared among these prototypes are: 1) optical fibers to deliver both excitation beam and signal photons, 2) a high-NA miniature objective for tight focusing and efficient signal collection, and 3) a compact beam scanner for image forming. One common choice of the scanner is a two-axis microelectromechanical systems (MEMS) mirror, which is typically paired with two separate pieces of optical fibers: one single-mode fiber for delivering the excitation beam and one multimode fiber for effective epi-collection of signal photons [9], [15], [21], [22]. Another scanning mechanism is to drive a free-hanging double-clad fiber (DCF) cantilever at its mechanical resonant frequency using a tubular piezoelectric actuator, with the excitation beam delivered through the single-mode core and emission photons mainly collected/transmitted by the large-diameter inner clad of the DCF [8], [12], [13], [16]-[20], [23]. Compared with the MEMS mirror of a centimeter-scale footprint and beamfolding geometry, the imaging probe based on a piezoelectrically actuated single-DCF scanner can be made much more compact (down to ~2 mm in diameter) [19], [23]. It is not only well-suited for label-free endoscopic imaging of internal organs in translational applications like disease diagnosis and biopsy guidance, but also readily applicable as a flexible, light-weight head-mounted two-photon fiberscope to enable neural imaging of freely-behaving GCaMP mouse models and significantly expand the scope of behavioral paradigms that can be studied [7].
For both clinical and neuroscience applications, a large FOV and high imaging speed are always preferable (in addition to imaging signal-to-noise ratio). For a resonant fiber-optic scanner, however, there exists an intricate interplay between FOV and speed. To take a closer into this, note first that while the deflection of the PZT tube itself is limited (typically ±4 μm for the 1.3-mm-diameter PZT tube we used [24]), the tip displacement of a centimeter-long fiber cantilever gets magnified significantly via mechanical resonance to, for example, ±250 μm. Quantitatively, the resonant frequency f scales reciprocally with the square of the cantilever length L, while the displacement amplitude of a resonant fiber cantilever, denoted by Acant, scales proportionally with the driving voltage V (as the voltage determines bending angle of the PZT tube) and the cantilever length L, i.e.
| (1) |
The single-mode beam (with an ~0.12 numerical aperture) exiting the cantilever tip is focused by a miniature objective lens with a typical magnification M > 3× to ensure tight focusing and efficient two-photon excitation.
Similar to any imaging platform, there is an inherent tradeoff between field of view (FOV) and resolution for an endomicroscope. Denoting the displacement amplitude of the fiber cantilever tip by Acant and the numerical aperture (NA) of the excitation beam exiting the fiber tip by NAcant, the resultant focusing NA on the sample is NAsamp ≈ M × NAcant and the radius of the FOV is RFOV ≈ Acant / M. Evidently, increasing magnification M brings tighter beam focusing but shrinks FOV, while a smaller magnification M results in the opposite (i.e., an enlarged FOV with a lower focusing NA and thus a compromised lateral resolution). The invariant product of FOV and NA of imaging optics is well known as the Lagrange invariant or geometrical extent [25]-[27]. The total number of resolvable pixels per 2D image (i.e., the imaging throughput) for a given endomicroscope is thus an invariant, i.e.:
| (2) |
where δPSF denotes the full-width-at-half-maximum (FWHM) width of the diffraction-limited point spread function (PsF). For a typical endomicroscope prototype which has an ~0.7-0.8 μm lateral resolution (focusing NAsamp ~0.55) and an ~100-μm-diameter FOV [17]-[19], [23], the number of resolvable pixel number (throughput) is on the order of ~104.
For a given endomicroscope, one straightforward approach to increase imaging throughput is to increase Acant, by either increasing the drive voltage V of the PZT actuator or the fiber cantilever length L. However, there are several limitations associated with an increased Acant, including the increased risk of vignetting within the micro objective, elevated off-axis optical aberrations (and associated deterioration in image quality), increased electrical hazard during in vivo imaging (if an increased drive voltage is used), and more importantly, the decrease of the beam scanning speed (when a longer cantilever is used, as shown in Eq. (1)).
To improve the throughput-speed product, in this paper we propose a new strategy by introducing the concept of cascaded NA amplification. Instead of amplifying the fiber core NA by ~5× entirely via a micro objective, our design distributes the amplification between two steps. First, a GRIN lens-based mode-field focuser is customized and concatenated to the DCF, yielding a composite cantilever with an output NAcant of ~0.35; then an ~2× micro objective further amplifies the beam NA for achieving tight beam focusing. In this way, the total number of resolvable pixels can be increased without the need for increasing (1) the drive voltage or the cantilever scan magnitude, and (2) the cantilever length which would otherwise compromise the imaging speed (see Eq. (1)).
II. Design Principles
In our two-photon endomicroscope, a double-clad fiber (DCF) is employed, which has a single-mode (sM) core with a mode-field diameter of about 3.4 μm and an NA of ~0.12. Although the NA of the DCF can be theoretically increased through customization, the mode-field diameter (MFD) will have to be decreased proportionally to maintain single-mode operation, but the difficulty in coupling light into the smaller-MFD sM core and more importantly the nonlinear effects on the femtosecond excitation light propagating in the sM core, will both elevate [28]. Thus, we decide to augment the NA of the output beam via fiber-tip engineering.
A. DCF-GRIN Composite Cantilever
To magnify the output NA of the fiber-optic cantilever, a tiny GRIN lens was attached to the end of the DCF to pre-focus the diverging single-mode beam from the DCF core and convert it to a beam of a higher diverging angle. Given the DCF of an ~250 μm diameter with an ~180 μm inner clad, we selected a commercial 350-μm-diameter, ~790-μm-long, 0.5-NA GRIN rod lens (GT-IFRL-035-005-50-570-NC, GRINTECH GmbH). With the length slightly longer than a quarter pitch, this GRIN lens features an effective focal length of ~0.30 mm. A silica rod was adopted as a spacer to connect the DCF and the GRIN lens (Fig. 1A), and the NA magnification factor (NAcant/NAcore in Fig. 1B) could be tuned by adjusting the spacer length. Evidently, a longer spacer leads to a tighter focus for the beam exiting the GRIN lens and a higher NAcant. The NA-enhanced (i.e., more diverging) beam from the cantilever, with its waist located close to the end face of the GRIN lens, was further focused by a 1.92× micro objective (GT-MO-080-0415-810, GRINTECH GmbH) onto the sample, resulting in a final beam NA of ~0.6 (water emersion) for efficient two-photon excitation.
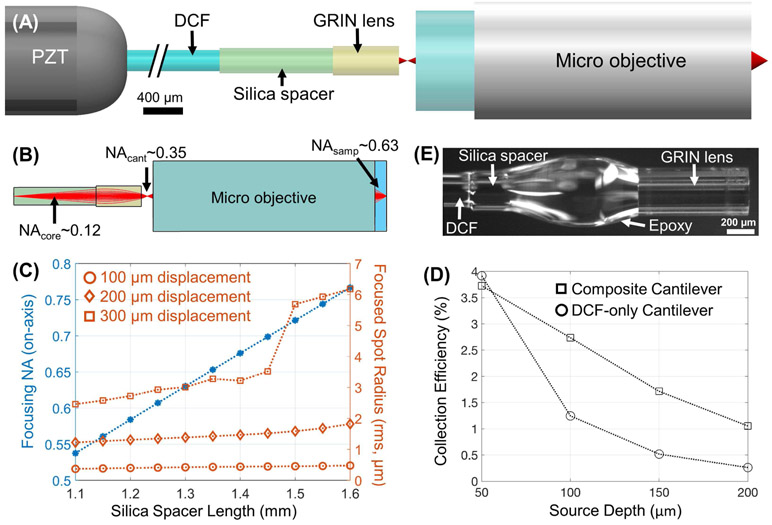
Fig. 1.
Design of the composite cantilever. (A) Schematic of the composite cantilever paired with a micro objective. (B) Illustration of the evolving beam NA from the DCF tip (and within the spacer), to the new beam focus near the exit surface of the GRIN rod lens, and the final beam focus in the sample (water immersion). (C) The dependence of the on-axis focusing NA and off-axis focused spot radii (root-mean-square, at three progressive tip displacements) on the silica spacer length extracted from ray-tracing simulations. For simplicity, we did not consider the angular deflection of the beam exiting the cantilever in our ZEMAX simulations since the deflection angle was very small (e.g., no larger than 1.7 degrees for both the long and short cantilevers) in comparison to the ~0.35 beam NA exiting the composite cantilever. (D) Collection efficiency comparison between two endomicroscope designs: 1) a traditional DCF only cantilever and 2) the proposed composite cantilever. Each data point shown here is the average value over 5 different scattering anisotropy factors. (E) Photograph of a finished composite cantilever tip.
The dependence of the final NA on the silica spacer length was simulated via ray tracing using ZEMAX (Optics Studio 16.5, Zemax LLC; see Supplementary Materials for details). While the final on-axis NA increases monotonically with the spacer length (blue asterisk curve, Fig. 1C), the off-axis aberration should also be taken into account. Herein the root-mean-square (rms) radius of the final focused spot diagram was adopted as a quick gauge of the overall aberration. With the cantilever tip displaced up to 200 μm off-axis, the rms radius of the focused spot diagram stays roughly stable for a spacer length ranging from 1.1 to 1.6 mm (see the circle- and diamond-marker curves in Fig. 1C); with the cantilever tip displaced by 300 μm off-axis, an abrupt increase in rms radius occurs when the spacer length surpasses 1.40 mm (see the square-marker curve in Fig. 1C). Since the off-axis aberration generally grows with the spacer length, a spacer ~1.3 mm in length was chosen in our design for a good trade-off between the final NA and the overall aberration across the entire FOV. In this configuration, the DCF core NA of ~0.12 is transformed to an ~0.35 beam NA out of the composite cantilever (DCF + GRIN rod lens), and finally to an ~0.63 focusing beam NA on the sample (Fig. 1B).
The spacer diameter should be larger than the beam diameter within the spacer to prevent the beam from reaching the spacer wall and thus to maintain the single-mode propagation of the beam within the spacer. At the end face of the 1.3-mm-long silica spacer, the diameter of the diverging beam grows to ~ 2 × 1.3 mm× NAcore/nsilica = 215 μm (taking the refractive index nsilica = 1.4533 at 800 nm), which prescribes the minimal outer diameter of the silica spacer. Given the DCF diameter of 250 μm and the GRIN lens diameter of 350 μm, we chose a silica rod of a 300 μm diameter as the spacer. Such progressive increase in physical diameter also helped ensure the mechanical strength and long-term stability of the composite cantilever.
To fabricate the composite cantilever, the 250-μm-diameter DCF was first spliced with a 6-inch-long silica rod as a spacer using a fusion splicer (FSU 925 PM, Ericsson). Excessive portion of the spacer was precisely cut off using a fiber cleaver (Vytran LDC-400, Thorlabs) to ensure the remaining spacer (fused to the DCF) of a proper length (i.e., ~1.30 mm according to the design). The tiny GRIN rod lens, held by a home-built kinematic micro-clamp, was then glued onto the spacer with UV-curable optical adhesive (NOA 68, Norland). Concentricity was maintained using a home-built three-dimensional manipulation platform along with visual inspection by a CCD camera from multiple angles. After the epoxy was cured, an endomicroscope was assembled with the composite cantilever, following the similar procedure as previously described [8], [23], [29].
B. Collection Efficiency
The affordable collection aperture of a miniature endomicroscope is inherently confined, and therefore signal collection efficiency is an essential design consideration [23]. Basically, fluorescence photons epi-collected by the micro objective and the scanning GRIN lens need to be captured by the spacer first, and then propagate through the inner clad of the DCF to the proximal end for detection. The governing factors here include the area and the acceptance NA of both the spacer and the DCF inner clad, and the chromatic focal shift of both the micro objective and the GRIN lens. Since the silica spacer works essentially as an air clad multimode fiber with an acceptance NA of > 1.0, the two leading limiting factors of collection loss inside the endomicroscope are: 1) chromatic aberration-induced back focal shift of the micro objective and the GRIN lens [12], [23]; 2) the size mismatch between the DCF inner clad (180 μm in diameter) and the silica spacer.
Numerical simulation was carried out to investigate the collection efficiency of the above scanning endomicroscope with a composite cantilever. The simulation assumed an isotropic point photon emitter situated at various depths (50, 100, 150, or 200 μm) beneath the surface of a semi-infinite tissue-mimicking phantom (as limited by the working distance of the micro objective). Monte Carlo was used to model photon propagation within the phantom until the photons emerged from the phantom surface [30] (see Supplementary Materials for details). The reduced scattering coefficient of the phantom was set to 40 cm−1, while the anisotropy factor g varied from 0.5 to 0.9 with a step size of 0.1, to mimic the scattering properties of various biological tissues [31]. Totally 106 photons were launched isotropically from an ideal focus point. Each photon was traced until it exited the phantom top surface, or was terminated by absorption, or stroke the boundary of the simulated cylindrical tissue volume (10 mm in diameter × 10 mm thick).
Photons emerging from the phantom top surface, tagged with their respective lateral position and emission angle information, were traced through the micro objective and the GRIN rod lens on cantilever tip by ZEMAX (see Supplementary Materials for details). Surviving photons were traced through the silica spacer using a home-developed MATLAB script which took care of total internal reflection (TIR) of skew rays on the cylindrical silica-air interface [32], and calculated for each input photon the exiting location and direction from the other end face of the spacer. A photon with exiting position falling inside the DCF inner clad (acceptance NA = 0.35) was further traced with the same MATLAB script until they bounced against the boundary of the DCF inner clad for the first time. If the TIR criteria were satisfied at the first bounce, this photon was considered successfully accepted. Collection efficiency was calculated as the number of photons collected by the DCF (most by its inner clad plus some by its core) divided by the total number of photons launched initially from the point photon emitter.
For comparison, the collection efficiency of a control design that employs a DCF-only cantilever (i.e. the same DCF without the GRIN lens-based mode-field focuser) and a commercial ~4.8× micro objective (GT-MO-080-018-810, GRINTECH GmbH) [17], [18], [33] was also simulated over the same configurations of the source depths and tissue scattering properties.
The collection efficiencies of both designs against various source depths are compared in Fig. 1D. Clearly, the composite cantilever-based design significantly outperforms (by almost 100%) the conventional DCF-only cantilever-based design for deeper sources (100-200 μm), while being comparable with the latter for shallower source depths (~50 μm).
III. Materials and Methods
A. System Setup
The new endomicroscopes described in this manuscript were integrated with our recently developed fiber-optic nonlinear endomicroscopy system for characterization and imaging performance test [23], [34]. Briefly, transform-limited near-infrared (NIR) laser pulses from a Ti:sapphire laser (Chameleon Vision II, Coherent Inc.), ~150 fs FWHM in pulsewidth and 80 MHz repetition rate, were launched into a piece of ~30-cm-long polarization-maintaining single-mode fiber (PM-SMF; PM780-HP, Thorlabs), in which the pulse spectrum broadened due to the nonlinear self-phase modulation (SPM) effect [28], [34]. Pulses out of the PM-SMF were then collimated, and went through (double pass) a pair of volume phase holographic transmission gratings (600 lpmm; Wasatch Photonics) to gain sufficient negative chirping, and finally were coupled into the core of the DCF (~1.2-meter long) and guided to the distal end of the endomicroscope. The pulse spectrum was recompressed in the DCF core due to the interaction between negative chirping and the SPM effect to achieve a short temporal pulsewidth when exiting the endomicroscope.
Fluorescence photons were epi-collected into the DCF, predominately into the inner clad, and guided back to the proximal end of the endomicroscope, and then separated from the excitation beam via a dichroic filter (FF665-Di02-25 × 36, Semrock). A short-pass optical filter (FF01-680/SP-25, Semrock) was installed in front of the photomultiplier (PMT, H10771P-40, Hamamatsu) to eliminate residual excitation photons. Extra optical filters adopted to further define fluorescence detection bands included: 1) a 496-nm longpass optical filter (FF01-496/LP-25, Semrock) to detect the total fluorescence of Alexa Fluor 488 and MitoTracker Red CMXRos of the bovine pulmonary artery endothelial (BPAE) slide (F36924, Thermo Fisher Scientific) with 750-nm excitation; 2) a 417-477-nm band-pass optical filter (FF02-447/60-25, Semrock) for NADH imaging of mouse kidney with 730-nm excitation; 3) a 490–580-nm band-pass filter (FF01-538/84-25, Semrock) for GCaMP-based dendritic Ca2+ imaging of mouse brain with 920-nm excitation.
B. Resolution Measurement
The point spread function (PSF) of the enlarged-FoV endomicroscope was measured using 0.5-μm-diameter Fluoresbrite yellow green microspheres (17152-10, Polysciences, Inc.). The fluorescent beads were dispersed sparsely across the 300-μm imaging field, and each image was acquired with 1024 spiral traces to ensure an adequate sampling density. Richardson-Lucy deconvolution was carried out in MATLAB using an open-source library [35], where the ideal fluorescence intensity profile of a 500-μm fluorescent bead [36] was deconvoluted from the measured bead intensity profiles. The extracted intensity profile, closer to the true PSF, was then fitted to a 2D Gaussian profile for FWHM estimation. Multiple beads were selected from several different FOVs, and the average PSF FWHM and standard deviation values at different off-axis regions were reported.
C. Animal Preparation
For in vivo kidney imaging, a BALB/c mouse 10-12 weeks in age (Taconic Biosciences) was anesthetized using isoflurane (Baxter International) flowed in O2 (~4% at ~1.0 liter/min for induction and ~1.5% at ~0.5 liter/min for maintenance, targeting at ~1 breath per second), with its body temperature maintained at ~37.5 ° using a feed-back controller (TC-1000, CWE Inc.). The mouse was positioned on the heat pad, lying on its right side, and then, after shaving the left flank and disinfecting the skin using 70% ethanol, an ~8-mm-long incision, parallel and ventral to the spine and midway between the last rib and the iliac crest, was made with fine scissors. The left kidney was then exteriorized through the incision using cotton swabs and curved forceps. After carefully separating the kidney from surrounding fatty tissues using cotton swabs, a home-built kidney rack was inserted beneath the left kidney to hold it and maximally isolate breathing-induced motion artefacts. The endomicroscope was placed gently against the kidney surface to access the renal cortex. Warm sterile saline was used to keep the kidney moist and to maintain water immersion for the micro objective lens.
For in vivo Ca2+ imaging, a 4 week-old male Camk2-Cre mouse on the 129S6SvTac strain (JAX#005359, The Jackson Laboratory) was used. For preparation, the mouse was anesthetized following the same aforementioned protocol and secured onto the stereotaxic equipment. A 4 mm-wide craniotomy was then drilled over the motor cortex using a high-performance surgical drill. The center of the craniotomy was ~1.6 mm lateral to bregma [37]. Using a 1.0-mm-diameter glass microneedle with a 10-20 μm diameter tip and a microinject pump (Nanoject II, Drummond), about 300 nL of AAV/DJ-flex-GCaMP6m virus was injected into the forelimb area of the right motor cortex (1.5 mm lateral and 0.3 mm anterior to bregma, and 300 μm in depth [38]). The surgical site was then enclosed with a #0 round coverglass (80-120 μm thick) using vet glue. A custom titanium head-restraining bar was then attached to the skull with “cold cure” denture material. Sterile surgical techniques were practiced throughout the procedure using a hot bead sterilizer (Fine Science Tools), and once exposing subcutis, the surgical field was constantly irrigated and submerged in sterile 0.9% NaCl (Baxter Healthcare). The transgene expression was checked three weeks after the surgery with a benchtop two-photon microscope through the cranial window. For imaging, the mouse with good expression was positioned on a custom head-restraining apparatus. The endomicroscope featuring the faster composite fiber-optic cantilever/scanner and a water-immersion micro-objective was then placed above the cranial window to record spontaneous dendritic dynamics in cortical layer I while the mouse stayed awake.
All animal housing and experimentation procedures were performed in accordance with the standards of humane animal care described in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Protocols were approved by the institutional animal care and use committees of the Johns Hopkins University and of George Washington University.
D. Spatiotemporal Demixing of Dendritic Calcium Data
The dendritic calcium imaging data were analyzed using a well-established, sparse, noise-constrained non-negative matrix factorization (CNMF) algorithm which expresses the observed spatiotemporal fluorescence data Y into the product of a spatial footprint matrix A (with each column encoding the spatial footprint of a dendritic component) and a temporal fluorescence trace matrix C (with each row modeling the temporal calcium fluorescence profile of a dendritic component) [39]. Technically, the CNMF algorithm decomposes the spatiotemporal observation data YN×T into YN×T = AN×K × CK×T + bN×1 ×f1×T + EN×T, where N denotes the number of pixels in each frame (i.e., the dimension of linear space spanned by 2D images), T the number of frames acquired, K the number of dendritic components to retrieve, b the spatial structure and f the temporal profile of a time-varying rank-1 background component, and E the residual error (mainly noise). Note that by writing A = [a1, a2, …, aK] and C = [c1, c2, …, cK]T, we have , reflecting that each extracted dendritic component corresponds to an ensemble of dendritic segments (with spatial footprint defined by vectors ai) that exhibit highly synchronous calcium dynamics over time (described by vectors ci). Both vectors ai and vectors ci are sparse in their respective domains[39, 40]. Dendritic components can overlap with each other in space, while still be demixed based on their distinct temporal fluorescence activities.
Custom processing code was implemented in MATLAB based on the open-source CaImAn toolbox [40]. The entire data set consisted of T = 1000 fluorescence frames (totally ~160 sec in duration), each containing N = 512 × 512 pixels. To better infer the sparse spatiotemporal structures of underlying dendritic components, the FOV was split into 25 spatially overlapping square patches, each of 128 × 128 pixels with the overlapping margin between adjacent patches measuring 32 pixels in width. For each patch, a sparse NMF method was employed for initialization of the dendritic components, and then ~10 local dendritic components were extracted per patch. Finally, spatially overlapped local dendritic components from adjacent patches with highly correlated temporal activities were identified and merged into an extended, global component. This eventually yielded ~150 global components, whose spatial footprints and temporal profiles were further refined via alternate optimization as final results.
IV. Results
A. High-Resolution, Label-Free Imaging of Internal Organs With a Large FOV
The first prototype scanner was built with the overall length of the composite cantilever set to ~13.0 mm. The resonant frequency of the cantilever was measured to be approximately 830 Hz, which is lower than a typical resonant frequency of a 13-mm-DCF-only cantilever (i.e., ~1.0 kHz [19], [20], [23]); this is expected since the spacer and GRIN lens are much thicker than the DCF, thus adding additional weight to the cantilever tip [41]. By applying sine and cosine waveforms of an ~48 Vpp in amplitude to the two pairs of orthogonal electrodes on the PZT actuator, respectively, the composite cantilever tip could be actuated into a circular scan of an ~600 μm diameter. By modulating the drive waveforms, a spiral scan pattern covering an ~300-μm-diameter FOV was achieved (Fig. 2A).
Fig. 2.
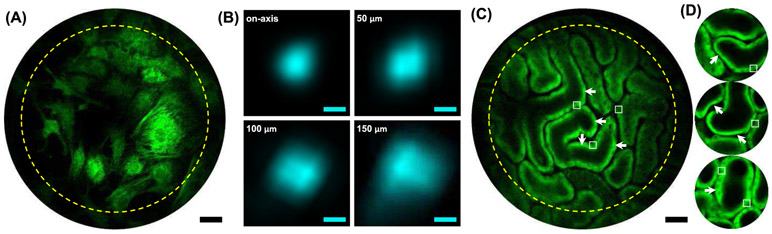
Imaging characterization of the high-resolution, large-FOV endomicroscope based on a composite cantilever. (A) Large FOV imaging of stained bovine pulmonary artery endothelial cells with the cytoskeletal filamentous actins labeled by Alexa Flour 488 Phalloidin and the intracellular mitochondrial network labeled with MitoTracker Red CMXRos. The excitation power was ~10 mW at 750 nm. (B) Representative images of 500-nm fluorescent beads at progressively increasing field heights (i.e., off-axis distances, annotated at the upper-left corner). The full-width-at-half-maximum (FWHM) of point-spread function (PSF) grows with off-axis distance. The mean and standard deviation of FWHM PSF size are (from on-axis to peripheral regions; 4 beads measured for each location): 0.66 ± 0.019 μm, 0.81 ± 0.029 μm, 0.88 ± 0.058 μm, and 1.12 ± 0.075 μm, respectively, before deconvolution, and 0.61 ± 0.024 μm, 0.77 ± 0.033 μm, 0.84 ± 0.057 μm and 1.10 ± 0.073 μm, respectively, after deconvolution. Scale bars: 0.5 μm. (C) Endomicroscopic label-free two-photon NADH imaging of mouse kidney in vivo. The dark round-to-elliptical signal void regions scattered along the renal tubule walls (square boxes) are nuclei of epithelial cells, while the predominant concentration of NADH fluorescence appeared along the basolateral side of renal tubules (arrows). Scale bars: 30 μm. For both (A) and (C), the dimmer signal outside the yellow dash circle results mainly from vignetting of the current micro objective. (D) Three label-free two-photon NADH images of mouse kidney in vivo acquired with the traditional DCF-only cantilever-based endomicroscope with the same scale bar as in (C). The increased FOV (shown in (C)) afforded by the new endomicroscope based on the composite cantilever is very pronounced (by almost 3 fold in length or 9 fold in area). Nuclei of epithelial cells and the predominant concentration of NADH along the basolateral side of renal tubules are also indicated by square boxes and arrows, respectively.
Lateral resolution at different off-axis locations within the FOV was measured by imaging 0.5-μm-diameter fluorescent beads (Fig. 2B). The FWHM lateral resolution was ~0.61 ± 0.02 μm near the FOV center, where the deviation from the theoretical two-photon resolution (~0.50 μm for a 0.6 focusing NA could result from 1) the actual DCF core NA is slightly less than the value 0.12 as used in simulation and 2) the actual spacer length is slightly shorter than 1.3 mm. The resolution deteriorated gradually towards the peripheral zone (i.e., up to ~1.10 ± 0.07 μm), which was primarily caused by the increasing off-axis aberrations (predominantly spherical aberration from the micro-objective), as consistent with ZEMAX simulations shown in Fig. 1C. The degrading off-axis resolution, as well as the discernible vignetting effects (Figs. 2A and 2C), can potentially be mitigated with new micro objective designs to support diffraction-limited performance over an extended FOV [42].
In vivo two-photon fluorescence imaging of an unstained mouse kidney was conducted with the composite-cantilever-based endomicroscope to investigate its label-free imaging capability (based on NADH fluorescence). With an ~30 mW average incident power at 730-nm excitation wavelength, subcellular structures of renal tubules in the kidney cortex were clearly delineated (Fig. 2C). Note that many renal tubule sections were captured within a single 300-μm FOV, thereby representing an ~9-fold enlargement in the total imaging area (of ~ 2 × 105 resolvable pixels) compared with typical FOVs afforded by our previous prototype [23] (Fig. 2D). Nuclei of epithelial cells lining the tubular wall were discernible as round dark spots across the image (indicated by square boxes in Figs. 2C and 2D). Also noticeable was the predominant concentration of NADH along the basolateral side of the tubule (indicated by arrows in Figs. 2C and 2D), matching the characteristic basolateral mitochondrial distribution in renal tubular cells [43]. In summary, the experimental results confirmed the capability of the endomicroscope based on a NA-augmented composite cantilever for label-free two-photon fluorescence imaging of biological tissues in vivo with an extended FOV.
B. High-Speed Recording of Calcium Dynamics in Vivo
Besides enhancing the imaging FOV, the above NA-augmented composite cantilever design also offers an avenue for increasing the endomicroscope scanning speed without sacrificing the size of imaging FOV. In the traditional DCF-only cantilever design, an increase in scanning speed requires shortening of the cantilever length, and according to Eq. (1), this will lead to a reduced imaging FOV for a given PZT actuator drive voltage [44]. Considering that the NA-augmented composite cantilever allows the use of a micro objective of a lower magnification (i.e., ~2× instead of ~4.8× in the traditional design), a sizeable FOV can still be achieved even though the tip sweeping range is reduced for an overall shorter composite cantilever. This design principle is also implied in Eq. (2), where, with the total number of resolvable pixels kept constant (i.e., for a fixed FOV and resolution), an enlarged NAcant corresponds to an increased scan frequency f. Based on this principle, we prototyped a second endomicroscope for which the composite cantilever length was shortened to ~7.0 mm, resulting in an increased mechanical resonant frequency of approximately 3.1 kHz. With the same PZT actuator drive waveforms (of ~48 Vpp in amplitude), this shortened cantilever could sweep over ~320 μm in diameter, thus yielding an imaging FOV of ~160 μm in diameter. Although the FOV is relatively smaller than the one described above, this new composite cantilever increased the scanning speed by about 3-fold, and therefore the frame rate could readily reach ~6 fps without sacrificing the spatial sampling density.
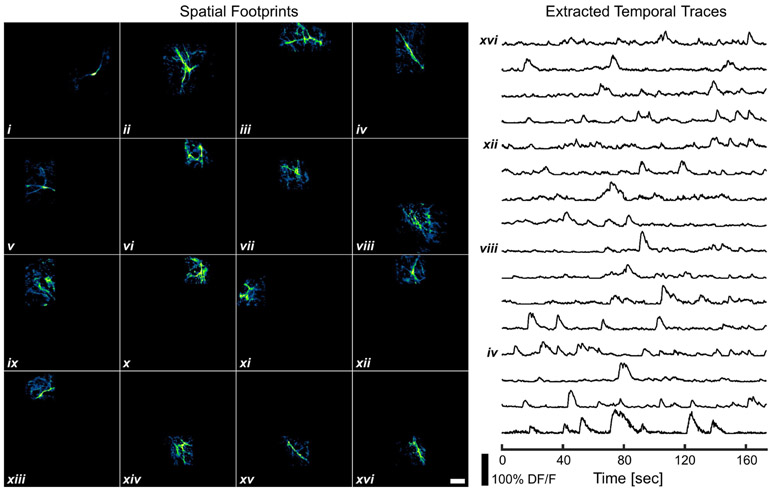
The performance of this faster endomicroscope was tested via in vivo two-photon dendritic calcium imaging of a head-fixed GCaMP6m-expressing transgenic mouse model. Using an ~45 mW incident power at 920-nm excitation wavelength, spontaneous calcium dynamics of dendrites in the primary motor cortex layer I (~50 μm beneath the cortical surface) were recorded at ~6 fps continuously for ~160 sec. Spatial overlapping and crossing among densely packed dendrite branches are commonly seen in the imaging FOV (Supplementary Video I). To carry out connectivity inference and functional analysis of the underlying dendritic structures and neural circuits, it is necessary to demix the interleaved spatiotemporal calcium dynamics data into multiple compact source components, with each component corresponding to an ensemble of dendritic segments that exhibit highly synchronous temporal profiles. This is accomplished via a sparse, noise-constrained non-negative matrix factorization algorithm, which essentially decomposes the observation data into the product of a spatial matrix that delineates the spatial footprint of each dendritic component and a temporal matrix that encodes the corresponding fluorescence fluctuation over time (see Methods) [39]. Shown in Fig. 3 are the spatial footprints and the corresponding temporal fluorescence traces (presented in DF/F units) of the selected, prominent dendritic components. Note that these extracted components are frequently overlapped in space, a scenario in which successful component demixing relies significantly on accurately resolving and distinguishing temporal activities of all dendrites, underscoring the critical importance of a sufficient temporal sampling density. Compared with traditional DCF-only cantilever designs with an ~1.3 kHz resonant frequency [19], [23], this new composite cantilever enabled an ~3-fold increase in imaging frame rate, thus affording markedly improved spatiotemporal resolution for better resolving calcium-based neural dynamics.
Fig. 3.
Dendritic source components extracted from two-photon calcium imaging data captured at ~6 fps with the high-speed prototype endomicroscope based on a shorter composite cantilever. Related data processing follows the details described in Section III-D. Spatial footprints and temporal traces of 16 representative dendritic components (i – xvi) extracted by using a sparse noise-constrained non-negative matrix factorization algorithm are presented. Scale bar: 20 μm.
V. Discussion
Compared with previous endomicroscopes based on a DCF-only cantilever, the NA-augmented composite cantilever enabled ~3-fold increase in FOV diameter (i.e., ~9-fold in FOV area) while maintaining a comparable beam scanning speed. The use of a composite cantilever also enabled an ~3-fold acceleration in beam scanning (and imaging frame rate) while maintaining a similar FOV. Both advancements were achieved without sacrificing imaging resolution and two-photon excitation/collection efficiency. Given the typical response kinetics of prevalent fast genetically-encoded calcium indicators to single action potential (half rise/decay time =~26/~140 ms for GCaMP6f, and ~26 /~265 ms for jGCaMP7f, respectively, commonly used in neuroscience) [5], [45], a frame rate of 6 fps is essential for resolving calcium dynamics with common fast and slow GCaMP variants. By tailoring design parameters such as the output NA and total length of the composite cantilever, and the magnification of the micro objective, one can come up with different prototypes to offer even more resolvable pixels (i.e., throughput) and/or faster beam scanning speed according to specific imaging applications. Examples include enlarging the FOV for high-throughput diagnostic imaging of subepithelial tissue histology, or further increasing the frame rate for spatiotemporally resolving neuronal activities with the aid of ever-improving genetically encoded calcium or other indicators.
While this manuscript focuses on boosting the throughput-speed product via cascaded NA amplification, we would like to point out that there indeed exist other approaches towards the same goal. For example, besides increasing the drive voltage as aforementioned following Eq. (2), one could also use a longer and thicker PZT tube to increase the cantilever angular deflection range per drive voltage and subsequently the achievable FOV size and imaging throughput. These approaches, albeit straightforward, could be ill-suited for miniaturization. Our strategy, instead, breaks the long-standing natural equivalence of the cantilever NA (i.e., NAcant in Eq. (2)) to the NA of DCF core. it is universally applicable, and can readily be combined with other approaches to boosting the imaging throughput and speed. Moreover, the basic design concept of the composite cantilever can be easily extended to other fiber types such as the photonics crystal fiber and fiber bundles, and is compatible with other beam scanning strategies such as raster [17] or Lissajous scans [29], [46].
Collection efficiency simulation also revealed that around two thirds of photons captured by the spacer were lost at the DCF-spacer interface. A composite cantilever adopting a DCF with an even larger inner clad to further promote the overall collection efficiency warrants future investigation. Another method to increase the collection efficiency is to employ an achromatized micro objective [23], [47]. Besides, the vignetting towards the periphery of the ~300-μm-diameter FOV results predominantly from the gradually broadening off-axis PSF (see Fig. 1C and Fig. 2B) and subsequently deteriorating two-photon excitation efficiency. Therefore, we anticipate that both off-axis aberration and imaging signal-to-noise ratio could potentially be further improved by employing a customized, achromatized micro objective that supports diffraction-limited performance over an extended FOV [42].
Supplementary Material
Acknowledgment
We thank Bernhard Messerschmidt and Herbert Stürmer from GRINTECH GmbH for technical assistance with ZEMAX simulations of the GRIN lenses.
This work was supported in part by the National Institutes of Health (NIH) under Grant R01 CA153023 and P50 CA103175, the National Science Foundation: Major Research Instrumentation (MRI) grant (CBET-1430040), and the Hartwell Foundation.
Contributor Information
Wenxuan Liang, Dept. of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, 21205 USA..
Hyeon-Cheol Park, Dept. of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, 21205 USA..
Kaiyan Li, Dept. of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, 21205 USA..
Ang Li, Dept. of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, 21205 USA..
Defu Chen, Dept. of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, 21205 USA..
Honghua Guan, Dept. of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, 21205 USA..
Yuanlei Yue, Department of Pharmacology and Physiology, George Washington University, Washington, DC, 20052 USA..
Yung-Tian A. Gau, Dept. of Neuroscience, Johns Hopkins University, Baltimore, MD 21205 USA.
Dwight E. Bergles, Dept. of Neuroscience, Johns Hopkins University, Baltimore, MD 21205 USA.
Ming-Jun Li, Science and Technology Division, Corning Incorporated, Corning, NY 14831..
Hui Lu, Department of Pharmacology and Physiology, George Washington University, Washington, DC, 20052 USA..
Xingde Li, Dept. of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, 21205 USA..
References
- [1].Zipfel WR, Williams RM, and Webb WW, "Nonlinear magic: multiphoton microscopy in the biosciences," Nat Biotechnol, vol. 21, no. 11, pp. 1369–77, 2003 [DOI] [PubMed] [Google Scholar]
- [2].Helmchen F and Denk W, "Deep tissue two-photon microscopy," Nat Methods, vol. 2, no. 12, pp. 932–40, 2005 [DOI] [PubMed] [Google Scholar]
- [3].Campagnola PJ and Loew LM, “Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms," Nat Biotechnol, vol. 21, no. 11, pp. 1356–60, 2003 [DOI] [PubMed] [Google Scholar]
- [4].Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, and Webb WW, "Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation," Proc Natl Acad Sci USA, vol. 100, no. 12, pp. 7075–80, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen TW et al. , "Ultrasensitive fluorescent proteins for imaging neuronal activity," Nature, vol. 499, no. 7458, pp. 295–300, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nimmerjahn A and Bergles DE, "Large-scale recording of astrocyte activity," Curr Opin Neurobiol, vol. 32, pp. 95–106, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Silva AJ, "Miniaturized two-photon microscope: seeing clearer and deeper into the brain," Light-Sci Appl, vol. 6, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Myaing MT, MacDonald DJ, and Li XD, "Fiber-optic scanning two-photon fluorescence endoscope," Opt. Lett, vol. 31, no. 8, pp. 1076–1078, 2006 [DOI] [PubMed] [Google Scholar]
- [9].Hoy CL et al. , "Miniaturized probe for femtosecond laser microsurgery and two-photon imaging," Opt Express, vol. 16, no. 13, pp. 9996–10005, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Le Harzic R, Weinigel M, Riemann I, Konig K, and Messerschmidt B, "Nonlinear optical endoscope based on a compact two axes piezo scanner and a miniature objective lens," Opt Express, vol. 16, no. 25, pp. 20588–96, 2008 [DOI] [PubMed] [Google Scholar]
- [11].Engelbrecht CJ, Johnston RS, Seibel EJ, and Helmchen F, "Ultracompact fiber-optic two-photon microscope for functional fluorescence imaging in vivo," Opt. Express, vol. 16, no. 8, pp. 5556–5564, 2008 [DOI] [PubMed] [Google Scholar]
- [12].Wu YC, Leng YX, Xi JF, and Li XD, "Scanning all-fiber-optic endomicroscopy system for 3D nonlinear optical imaging of biologicaltissues," Opt. Express, vol. 17, no. 10, pp. 7907–7915, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wu YC, Xi JF, Cobb MJ, and Li XD, "Scanning fiber-optic nonlinear endomicroscopy with miniature aspherical compound lens and multimode fiber collector," Opt. Lett, vol. 34, no. 7, pp. 953–955, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tang S et al. , "Design and implementation of fiber-based multiphoton endoscopy with microelectromechanical systems scanning," J. Biomed. Opt, vol. 14, no. 3, p. 034005, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Piyawattanametha W et al. , "In vivo brain imaging using a portable 2.9 g two-photon microscope based on a microelectromechanical systems scanning mirror," Opt. Lett, vol. 34, no. 15, pp. 2309–2311, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhao Y, Nakamura H, and Gordon RJ, "Development of a versatile two-photon endoscope for biological imaging," Biomed. Opt. Express, vol. 1, no. 4, pp. 1159–1172, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rivera DR et al. , "Compact and flexible raster scanning multiphoton endoscope capable of imaging unstained tissue," Proceedings of the National Academy of Sciences, vol. 108, no. 43, pp. 17598–17603, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brown CM et al. , "In vivo imaging of unstained tissues using a compact and flexible multiphoton microendoscope," J Biomed Opt, vol. 17, no. 4, p. 040505, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang Y et al. , "A compact fiber-optic SHG scanning endomicroscope and its application to visualize cervical remodeling during pregnancy," Proceedings of the National Academy of Sciences, vol. 109, no. 32, pp. 12878–12883, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ducourthial G et al. , "Development of a real-time flexible multiphoton microendoscope for label-free imaging in a live animal," Scientific Reports, vol. 5, p. 18303, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zong W et al. , "Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice," Nat Meth, vol. 14, no. 7, pp. 713–719, 2017 [DOI] [PubMed] [Google Scholar]
- [22].Li A et al. , ”A biopsy-needle compatible varifocal multiphoton rigid probe for depth-resolved optical biopsy," Journal of Biophotonics, vol. 12 no. 1, p. e201800229, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liang W, Hall G, Messerschmidt B, Li M-J, and Li XD, "Nonlinear optical endomicroscopy for label-free functional histology in vivo,” Light: Science & Applications, vol. 6, p. e17082, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen CJ, "Electromechanical deflections of piezoelectric tubes with quartered electrodes," Applied Physics Letters, vol. 60, no. 1, pp. 132–134, 1992 [Google Scholar]
- [25].Steel WH, "Luminosity, Throughput, or Etendue?," Appl. Opt, vol. 13 no. 4, pp. 704–705, 1974 [DOI] [PubMed] [Google Scholar]
- [26].Mertz J, Introduction to Optical Microscopy. Freeman WH, 2009. [Google Scholar]
- [27].Brady DJ and Hagen N, "Multiscale lens design," Opt. Express, vol. 17, no. 13, pp. 10659–10674, 2009 [DOI] [PubMed] [Google Scholar]
- [28].Agrawal GP, Nonlinear Fiber Optics, 3rd ed. San Diego: Academic Press, 2001. [Google Scholar]
- [29].Liang W, Murari K, Zhang Y, Chen Y, Li MJ, and Li XD, "Increased illumination uniformity and reduced photodamage offered by the Lissajous scanning in fiber-optic two-photon endomicroscopy," J Biomed Opt, vol. 17, no. 2, p. 021108, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang L, Jacques SL, and Zheng L, "MCML—Monte Carlo modeling of light transport in multi-layered tissues," Computer Methods and Programs in Biomedicine, vol. 47, no. 2, pp. 131–146, 1995 [DOI] [PubMed] [Google Scholar]
- [31].Jacques SL, "Optical properties of biological tissues: a review," Phys Med Biol, vol. 58, no. 11, pp. R37–61, 2013 [DOI] [PubMed] [Google Scholar]
- [32].Ang AS, Sugon QM, and McNamara DJ, "Skew ray tracing in a step-index optical fiber using geometric algebra," Appl. Opt, vol. 54, no. 12, pp. 3764–3773, 2015 [Google Scholar]
- [33].Wang Y, Li Z, Liang X, and Fu L, "Four-plate piezoelectric actuator driving a large-diameter special optical fiber for nonlinear optical microendoscopy," Opt. Express, vol. 24, no. 17, pp. 19949–60, 2016 [DOI] [PubMed] [Google Scholar]
- [34].Liang W, Hall G, and Li XD, "Spectro-temporal dispersion management of femtosecond pulses for fiber-optic two-photon endomicroscopy," Opt. Express, vol. 26, no. 18, pp. 22877–22893, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sage D et al. , "DeconvolutionLab2: An open-source software for deconvolution microscopy," Methods, vol. 115, pp. 28–41, 2017 [DOI] [PubMed] [Google Scholar]
- [36].Yoo H, Song I, and Gweon D-G, "Measurement and restoration of the point spread function of fluorescence confocal microscopy," J Microscopy, vol. 221, no. 3, pp. 172–176, 2006 [DOI] [PubMed] [Google Scholar]
- [37].Ash RT, Fahey P,G, Park J, Zoghbi HY, and Smirnakis SM, "Increased Axonal Bouton Stability during Learning in the Mouse Model of MECP2 Duplication Syndrome," eNeuro, vol. 5, no. 3, pp. e0056–17, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Masamizu Y et al. , "Two distinct layer-specific dynamics of cortical ensembles during learning of a motor task," Nat Neurosci, vol. 17, no. 7, pp. 987–94, 2014 [DOI] [PubMed] [Google Scholar]
- [39].Pnevmatikakis EA et al. , "Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data," Neuron, vol. 89, no. 2, pp. 285–99, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Giovannucci etal A., "CaImAn an open source tool for scalable calcium imaging data analysis," Elife, vol. 8, p. e38173, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Huo L, Xi J, Wu Y, and Li XD, "Forward-viewing resonant fiber-optic scanning endoscope of appropriate scanning speed for 3D OCT imaging," Opt. Express, vol. 18, no. 14, pp. 14375–14384, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Matz G, Messerschmidt B, and Gross H, "Design and evaluation of new color-corrected rigid endomicroscopic high NA GRIN-objectives with a sub-micron resolution and large field of view," Opt. Express, vol. 24, no. 10, pp. 10987–11001, 2016 [DOI] [PubMed] [Google Scholar]
- [43].Hall AM, Rhodes GJ, Sandoval RM, Corridon PR, and Molitoris BA, "In vivo multiphoton imaging of mitochondrial structure and function during acute kidney injury," Kidney Int, vol. 83, no. 1, pp. 72–83, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shabana AA, “Vibration of Continuous Systems,” in Vibration of Discrete and Continuous Systems, Shabana AA Ed. New York, NY: Springer New York, 1997, pp. 188–267. [Google Scholar]
- [45].Dana H et al. , "High-performance GFP-based calcium indicators for imaging activity in neuronal populations and microcompartments," bioRxiv, p. 434589, 2018 [DOI] [PubMed] [Google Scholar]
- [46].Park HC, Seo YH, and Jeong KH, "Lissajous fiber scanning for forward viewing optical endomicroscopy using asymmetric stiffness modulation," Opt Express, vol. 22, no. 5, pp. 5818–25, 2014 [DOI] [PubMed] [Google Scholar]
- [47].Wu T-Y, Gmitro AF, and Rouse AR, "An Achromatized Miniature Objective for Microendoscopy," in Classical Optics 2014, OSA Technical Digest (online) (Optical Society of America, 2014), paper IW4A.2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.