Abstract
Noncoding RNAs, such as long noncoding RNAs (lncRNAs) and microRNAs (miRNAs), regulate gene expression in many physiological and pathological processes, including drug metabolism. Drug metabolizing enzymes (DMEs) are critical components in drug-induced liver toxicity. In this study, we used human hepatic HepaRG cells treated with 5 or 10 mM acetaminophen (APAP) as a model system and identified LINC00844 as a toxicity-responsive lncRNA. We analyzed the expression profiles of LINC00844 in different human tissues. In addition, we examined the correlations between the levels of LINC00844 and those of key DMEs and nuclear receptors (NRs) for APAP metabolism in humans. Our results showed that lncRNA LINC00844 is enriched in the liver and its expression correlates positively with mRNA levels of CYP3A4, CYP2E1, SULT2A1, pregnane X receptor (PXR), and hepatocyte nuclear factor (HNF) 4α. We demonstrated that LINC00844 regulates the expression of these five genes in HepaRG cells using gain- and loss-of-function assays. Further, we discovered that LINC00844 is localized predominantly in the cytoplasm and acts as an hsa-miR-486-5p sponge, via direct binding, to protect SULT2A1 from miRNA-mediated gene silencing. Our data also demonstrated a functional interaction between LINC00844 and hsa-miR-486-5p in regulating DME and NR expression in HepaRG cells and primary human hepatocytes. We depicted a LINC00844-mediated regulatory network that involves miRNA and NRs and influences DME expression in response to APAP toxicity.
Keywords: Long noncoding RNA, LINC00844, Liver toxicity, microRNA, Acetaminophen, Drug metabolism
Introduction
Acetaminophen (APAP) is a widely-used over-the-counter analgesic medication that may cause hepatic injury due to its intentional or inadvertent overdose (Yoon et al. 2016). APAP is primarily metabolized in the liver by sulfotransferases (SULTs) and UDP-glucuronosyltransferases (UGTs) and can be activated by cytochrome P450 (CYP) enzymes to generate the toxic metabolite, N-acetyl-p-benzoquinone imine (McGill and Jaeschke 2013). Key drug metabolizing enzymes (DMEs) involved in APAP metabolism are drastically downregulated upon APAP overdose as a consequence of cellular adaptation in response to stress, and multiple microRNAs (miRNAs) have been implicated in this “shut-down” response (Yu et al. 2018).
DMEs catalyze the metabolic biotransformation of xenobiotics, including drugs, thus influencing drug efficacy and toxicity (Sheweita 2000). These enzymes can be transcriptionally activated by nuclear receptors (NRs) under physiological conditions or in response to xenobiotic stimuli (Urquhart et al. 2007). Interindividual variability has been observed in the expression of DMEs due to genetic factors, such as variations in copy numbers and single nucleotide polymorphisms, or to epigenetic variations in DNA methylation, histone modifications, and noncoding RNA (ncRNA) expression (Ekstrom and Rane 2015; Nahar et al. 2014; Yang et al. 2013; Zanger and Schwab 2013).
Noncoding genes account for 98% of the human genome and produce a large number of various types of ncRNA transcripts, including miRNAs and long noncoding RNAs (lncRNAs) (Djebali et al. 2012). Typically, lncRNAs are > 200 nt in length and lack protein-coding potential; many are believed to fine-tune gene expression associated with normal biological or pathological processes, such as development and cancer (Marchese et al. 2017; Wang and Chang 2011). Increasing evidence in both human and animal models shows that lncRNAs play an important role in critical cellular events in response to stressful stimuli and toxicological conditions (Dempsey and Cui 2017; Valadkhan and Valencia-Hipolito 2016). Deregulation of lncRNAs has been implicated in a variety of human diseases, such as cardiovascular diseases, cancers, and Alzheimer’s disease (Dempsey and Cui 2017).
lncRNAs exhibit tissue-specific expression and function (Marchese et al. 2017) and regulate gene expression on various levels by diverse mechanisms (Wang and Chang 2011). Their functions are also largely dependent on their subcellular localization (Carlevaro-Fita and Johnson 2019). For example, nuclear lncRNAs participate in chromatin organization, transcription, and RNA splicing, while cytoplasmic lncRNAs may interfere with translation or act as competitive endogenous RNAs (ceRNAs) to sequester miRNAs from binding to their cognate mRNA targets (Salmena et al. 2011; Tay et al. 2014). Based on their genomic loci with respect to protein-coding genes, lncRNAs are categorized as long intergenic noncoding RNAs (lincRNAs), intronic lncRNAs, and sense or anti-sense lncRNAs (Ma et al. 2013; Ransohoff et al. 2018). Compared to other lncRNAs, lincRNAs have distinct features and functions due to their physical distance from protein-coding genes in the genome (Ransohoff et al. 2018).
Both miRNAs and lncRNAs can affect drug metabolism and toxicity by regulating the expression of DMEs (Li et al. 2019b). Many studies have shown that miRNAs target the mRNAs of DMEs for degradation or translational inhibition to suppress the expression of DMEs (Nakano and Nakajima 2018). For example, CYP3A4 mRNA is targeted at different loci within its 3′-UTR by at least a dozen miRNAs, including miR-1, miR-27a, and miR-122–5p (Li et al. 2019b). Our recent study reported that SULT2A1 expression is repressed by hsa-miR-495-3p and hsa-miR-486-5p, with the latter showing lower repression efficiency (Li et al. 2019a). Compared to miRNAs, lncRNAs have been reported to mediate DME expression in only a handful of studies (Li et al. 2019b). For instance, Li et al. (2015) discovered that lncLSTR, a liver-specific lncRNA, regulates the expression of Cyp8b1 in a systemic lipid metabolic pathway in mice. Another study (Chen et al. 2018) reported two anti-sense lncRNAs, HNF1α-AS1 and HNF4α-AS1, modulate the mRNA expression of several NRs and CYPs regulated by HNF1α and HNF4α, including PXR, constitutive androstane receptor (CAR), aryl hydrocarbon receptor (AhR), CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2E1, and CYP3A4. Moreover, knockdown of HNF1α-AS1 and HNF4α-AS1 affects susceptibility to APAP-induced toxicity in HepaRG cells (Chen et al. 2020). Our most recent report showed that LINC00574 and hsa-miR129–5p coordinately regulate UGT2B15 expression in HepaRG cells (Yu et al. 2020a).
Despite these findings, the vast majority of lncRNAs have not been characterized functionally. Their tissue-specific functions and responses to external stimuli make them potentially critical regulatory modules in drug-induced liver toxicity. In this study, we identified an intergenic lncRNA, LINC00844, that is responsive to hepatotoxicity induced by APAP in HepaRG cells. We demonstrated that LINC00844 modulates the expression of three DMEs, CYP3A4, CYP2E1, and SULT2A1, and two NRs, PXR and HNF4α. We also found that LINC00844 is a direct target of hsa-miR-486-5p and their physical and functional interaction influences the expression of the five genes regulated by LINC00844 in human liver cells.
Materials and methods
Cell culture
Human 293T embryonic kidney cells and HepG2 liver cancer cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). 293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, ATCC) supplemented with 10% fetal bovine serum (FBS), 1 × Non-Essential Amino Acids Solution, and 1 × glutamine (ThermoFisher, Waltham, MA). HepG2 cells were cultured in DMEM containing 10% FBS and 1 × Antibiotic–Antimycotic (ThermoFisher). 293T and HepG2 cells within passages 2–10 were used in designated experiments.
Terminally differentiated, cryopreserved HepaRG cells were obtained from ThermoFisher and cultured according to the manufacturer’s protocols with minor modifications. Briefly, HepaRG cells were thawed and resuspended in Williams’ E medium supplemented with 1 × GlutaMAX and the Thaw, Plate, and General Purpose Medium Supplement (ThermoFisher). Resuspended HepaRG cells were seeded in 24-well plates (4.8 × 105/well) and cultured for 24 h. Cells were then cultured in Williams’ E medium supplemented with 1 × GlutaMAX and the Maintenance/Metabolism Medium Supplement (ThermoFisher) for 7 days prior to designated experiments. Culture medium was refreshed every 2 days.
Cryopreserved primary human hepatocytes (10 donor pool) were obtained from In Vitro ADMET Laboratories (Columbia, MD). 24-well plates were pre-coated with rat tail Collagen I (ThermoFisher) following the manufacturer’s protocol. Cells were thawed and seeded at a density of 7 × 105 cells/well in 500 μl in the wells of 24-well plates following the supplier’s protocol. Cells were maintained in Universal Primary Cell Plating Medium (UPCM™) provided by the supplier. 16 h after seeding, cells were subjected to transfection.
All cells were maintained in a humidified incubator containing 5% CO2 at 37 °C.
lncRNA profiling
Total RNAs from HepaRG cells treated with 0.1% DMSO (the vehicle for APAP) or 5 or 10 mM APAP were subjected to RNA sequencing (RNA-seq) using an Illumina HiSeq1500 system (Illumina, San Diego, CA). The “long-noncoding RNA gene annotation” from the GENCODE human genome reference (v19) was used for mapping of paired-end sequencing reads to identify lncRNAs. Next, the term “LINCxxxxx” (“x” represents a single digit) was used to select lincRNAs among all lncRNAs. lncRNAs and lincRNAs with log2(fold change) > 1 or < − 1 and p value < 0.05 were considered significantly deregulated by APAP treatment compared to DMSO. Finally, lincRNAs with log2(fold change) > 5 or < − 5 and p value < 0.001 were considered substantially deregulated by APAP treatment.
RNA-seq data extraction and correlation analysis
The expression profiles of LINC00844 in non-tumor samples of 17 different human tissue types were extracted from The Cancer Genome Atlas (TCGA) database, which is provided by the TCGA Research Network https://www.cancer.gov/tcga. The tissue types, corresponding TCGA datasets, and sample numbers are listed as follows: liver (LIHC, liver hepatocellular carcinoma, n = 50; brain (GBM, glioblastoma multiforme), n = 5; prostate (PRAD, Prostate adenocarcinoma), n = 52; kidney (KICH, kidney chromophobe), n = 24; adrenal gland (PCPG, pheochromocytoma and paraganglioma), n = 3; rectum (READ, rectum adenocarcinoma), n = 1; breast (BRCA, breast invasive carcinoma), n = 113; cervix (CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma), n = 3; lung (LUSC, lung squamous cell carcinoma), n = 49; uterus (UCEC, uterine corpus endometrial carcinoma), n = 23; bladder (BLCA, bladder urothelial carcinoma), n = 19; thyroid (THCA, Thyroid carcinoma), n = 58; esophagus (ESCA, esophageal carcinoma), n = 11; stomach (STAD, stomach adenocarcinoma), n = 32; colon (COAD, colon adenocarcinoma), n = 41; pancreas (PAAD, pancreatic adenocarcinoma), n = 4; and skin (SKCM, skin cutaneous melanoma), n = 1. Publications by the TCGA Research Network on individual datasets are listed at https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/studied-cancers.
For correlation analysis, the levels of LINC00844 and mRNAs for nine key DMEs for APAP metabolism and three key NRs in 424 human liver samples (including tumor and non-tumor) were extracted from the TCGA LIHC dataset. GraphPad Prism 5 was used to perform Pearson’s correlation analysis. All RNA levels were presented as fragments per kilobase per million mapped reads (FPKM).
Lentiviral transduction of LINC00844
The lentiviral vector pLV-EF1a-MCS-IRES-Bsd (Biosettia, San Diego, CA) containing the gene sequence of LINC00844 (NR_108046) was prepared by GenScript (Piscataway, NJ) and is referred to as pLV-LINC00844 hereafter. The construct map and sequencing confirmation are shown in Supplemental Figs. 1 and 2. Lentivirus production was performed according to Biosettia’s protocols. Briefly, the empty vector (EV) or pLV-LINC00844 was transfected into 293T cells together with the lentiviral packaging mix (Biosettia) using Lipofectamine 2000 transfection reagent (ThermoFisher). Forty-eight hours after transfection, culture media containing viral particles were harvested and frozen at − 80°C for 1 day, followed by addition to HepG2 and HepaRG cells. To achieve efficient transduction, HepG2 or HepaRG cell cultures containing lentiviruses and 1 × polybrene (Biosettia) were centrifuged at 1000×g for 1 h at room temperature. HepG2 cells with stable overexpression of LINC00844 (HepG2-LINC00844) were then selected using blasticidin (ThermoFisher). Transduced HepaRG cells were cultured in fresh medium for 48 h before subsequent experiments. Three independent experiments were performed.
RNA fluorescence in situ hybridization (FISH)
HepG2-LINC00844 cells were seeded on collagen-coated Nunc™ Lab-Tek™ II 8-chamber slides (ThermoFisher) and cultured to 100% confluency. RNA FISH was performed using a ViewRNA™ Cell Plus Assay kit (ThermoFisher) according to the manufacturer’s instructions. Cells were first fixed and permeabilized with the ViewRNA™ Cell Plus Fixation/Permeabilization Solution for 30 min and then fixed with Fixation Solution for 1 h at room temperature. For target probe hybridization, fixed cells were incubated in Probe Set Solution containing the LINC00844 ViewRNA cell plus probe set conjugated with Alexa Fluor 488 (ThermoFisher) for 2 h at 40 ± 1 °C. Signal amplification was then conducted by subjecting cells to sequential incubations in pre-warmed (40 ± 1 °C) PreAmplifier Solution, Working Amplifier Solution, and Working Label Probe Mix Solution for 1 h each at 40 ± 1 °C. All incubations were performed in a humidified staining tray. Chambers were removed upon completion of the hybridization protocol, and slides were mounted with VECTASHIELD® ProLong™ Gold Antifade Mountant with DAPI (Vector Laboratories, Burlingame, CA) and covered with coverslips. Fluorescent images for LINC00844 detection and DAPI (a nucleus marker) staining were taken within the following 24 h using an Olympus FV1000 confocal microscope (Olympus, Center Valley, PA).
In silico analyses
RNAfold version 2.4.13 (https://rna.tbi.univie.ac.at/cgibin/RNAWebSuite/RNAfold.cgi) was used to predict the secondary RNA structure and free energy of thermodynamic ensemble of LINC00844. The coding potential of LINC00844 was assessed using CPC2 (Kang et al. 2017) at https://cpc2.cbi.pku.edu.cn/.
DIANA LncBase Predicted v.2 (https://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted) (Paraskevopoulou et al. 2016) was used to predict miRNA candidates targeting LINC00844. miRNAs targeting SULT2A1 were reported previously (Li et al. 2019a). The miRNA targeting network for SULT2A1 and LINC00844 was constructed using Cytoscape (Version 3.7.2.). RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) (Kruger and Rehmsmeier 2006) was used to calculate the MFE (minimum free energy) required for hsa-miR-486-5p and LINC00844 hybridization.
miRNA mimic or siRNA transfection of HepaRG cells and primary human hepatocytes
HepaRG cells and primary human hepatocytes were transfected with designated siRNAs and miRNA mimics at indicated concentrations using Lipofectamine RNAiMAX Reagent (ThermoFisher) according to the manufacturer’s instructions. Culture medium containing transfection reagent mixtures was replaced with fresh culture medium 16 h after transfection for HepaRG cells and 6 h for human primary hepatocytes. 48 h after transfection (i.e. the addition of transfection reagent mixtures), cells were harvested for RNA and protein extraction. Three independent experiments were performed.
Reverse transcription and real-time qPCR
Total RNAs, including mRNA, lncRNA, and miRNAs, were extracted from HepaRG cells, HepG2 cells, and primary human hepatocytes using miRNeasy Mini kits (Qiagen, Valencia, CA). To detect LINC00844 and mRNA expression, iScript™ cDNA Synthesis kits and SsoAdvanced Universal SYBR Green Supermix (BioRad, Hercules, CA) were used for reverse transcription and real-time qPCR, respectively. miRNA expression was measured using TaqMan MicroRNA Reverse Transcription kits and TaqMan Universal Master Mix II, no UNG (ThermoFisher) as described previously (Li et al. 2019a). A BioRad CFX96 Touch Real-Time PCR Detection System (BioRad) was used for qPCR assays. The expression of miRNAs or LINC00844 and mRNAs was normalized against that of RNU6 or GAPDH and analyzed using the 2–ΔΔCt method. Primers for LINC00844 and mRNA detection were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). Primer sequences are shown in Table 1.
Table 1.
Sequences of primers and probes
| Oligonucleotide | Sequence | Application |
|---|---|---|
| CYP2E1-F | ACGGTATCACCGTGACTGTGG | RT-qPCR |
| CYP2E1-R | GCATCTCTTGCCTATCCTTGA | RT-qPCR |
| CYP3A4-F | TTCAGCAAGAAGAACAAGGACAA | RT-qPCR |
| CYP3A4-R | GGTTGAAGAAGTCCTCCTAAGC | RT-qPCR |
| GAPDH-F | GAAATCCCATCACCATCTTCCAGG | RT-qPCR |
| GAPDH-R | GAGCCCCAGCCTTCTCCATG | RT-qPCR |
| HNF4α -F | GTACTCCTGCAGATTTAGCC | RT-qPCR |
| HNF4α -R | CTGTCCTCATAGCTTGACCT | RT-qPCR |
| PXR-F | TGCGAGATCACCCGGAAGAC | RT-qPCR |
| PXR-R | ATGGGAGAAGGTAGTGTCAAAGG | RT-qPCR |
| SULT2A1-F | TGAGTTCGTGATAAGGGATGAA | RT-qPCR |
| SULT2A1-R | CAGATGGGCACAGATTGGAT | RT-qPCR |
| hsa-miR-486-5p | Cy5.5-UCCUGUACUGAGCUGCCCCGAG | FREMSA |
| miR-486-MRE | IRDye800-GCUUGAUGCAGUCUGAUAGGAG | FREMSA |
| Cold hsa-miR-486-5p | UCCUGUACUGAGCUGCCCCGAG | FREMSA |
| Cold-NC | UCACAACCUCCUAGAAAGAGUAGA | FREMSA |
Western blotting
Protein lysates were extracted from HepaRG cells using RIPA Lysis and Extraction Buffer (ThermoFisher) and subjected to SDS–polyacrylamide gel electrophoresis (PAGE) and Western blotting. Antibodies against CYP3A4, CYP2E1, SULT2A1, PXR, HNF4α, and GAPDH (Abcam, Cambridge, MA) were used to immunoblot for the respective proteins. Blots were visualized and quantified using an Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) and Image Studio software. GAPDH was used as a loading control for normalization.
Fluorescence-based RNA electrophoretic mobility shift assays (FREMSAs)
Oligonucleotide probes for hsa-miR-486-5p were labeled with cy5.5™ dye at the 5′-end. Oligonucleotide probes for miR-486-MRE, an hsa-miR-486-5p recognition element on LINC00844, were 2′-O-methyl-modified and conjugated with IRDye800 at the 5′-end. The probes were synthesized by IDT; the sequences are shown in Table 1. FREMSAs were conducted using the EMSA binding buffer from a Light-Shift Chemiluminescent RNA EMSA kit (ThermoFisher) as described previously (Yu et al. 2020b) with minor modifications. Briefly, probes for hsa-miR-486-5p and miR-486-MRE of LINC00844 were incubated individually or together in the presence of 50 × cold hsa-miR-486-5p or cold negative control (cold-NC) at room temperature for 20 min. Reaction mixtures were then subjected to RNA electrophoresis on 12% native polyacrylamide gels. An Odyssey CLx Infrared Imaging System (LI-COR Biosciences) was used to detect the mobility shifts.
Statistical analyses
GraphPad Prism 5 was used for statistical analyses. Student t tests (two-tailed) were used to analyze two groups of data. Experiments containing more than two groups of data were analyzed using one-way ANOVA with Bonferroni’s Multiple Comparison Test. Data are displayed as mean with standard deviation (SD) from three independent experiments. p value < 0.05 was considered statistically significant.
Results
Identification of LINC00844 as a hepatotoxicity-associated lncRNA candidate
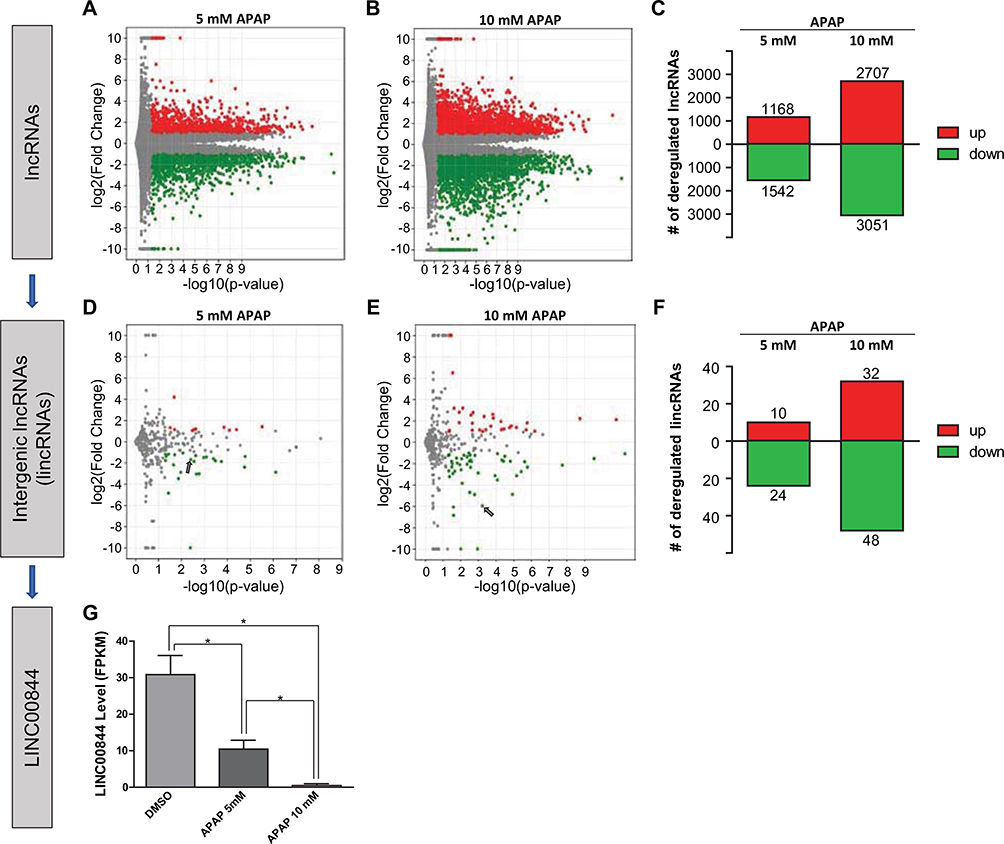
To identify lncRNA candidates associated with drug-induced hepatotoxicity, we used APAP-treated HepaRG cells as our model system. We treated HepaRG cells with the vehicle (0.1% DMSO) or 5 or 10 mM APAP, which are concentrations demonstrated to induce moderate or severe toxicity in hepatic cells (Yu et al. 2018). With total RNAs extracted from treated cells, we performed RNA-seq and screened differentially expressed lncRNAs. RNA-seq reads were mapped and quantified using the “long-noncoding RNA gene annotation” from the GENCODE human genome reference (v19), which contained the most comprehensive lncRNA datasets (Harrow et al. 2012). LncRNAs with log2(fold change) > 1 or < − 1 and p < 0.05 were considered significantly deregulated by APAP treatment. As shown in the volcano plots (Fig. 1a, b, each dot representing one lncRNA), we detected a total of 27,660 annotated lncRNAs. Among these, 1168 and 2707 were significantly upregulated (red dots), whereas 1542 and 3051 were significantly downregulated (green dots) in HepaRG cells treated with 5 and 10 mM APAP (Fig. 1a–c).
Fig. 1.
Identification of LINC00844 as a hepatotoxicity-associated lncRNA candidate. In HepaRG cells treated with 5 and 10 mM APAP, 27,660 lncRNAs were identified, among which 1168 and 2707 were significantly upregulated whereas 1542 and 3051 were significantly downregulated (a–c, log2(fold change) > 1 or < − 1 and p < 0.05). Among all lncRNAs, 559 lincRNAs were identified using the term “LINCxxxxx” (“x” represents a single digit). 10 and 32 lincRNAs were significantly upregulated, whereas 24 and 48 were significantly downregulated by 5 and 10 mM APAP treatment (d–f, log2(fold change) > 1 or < − 1 and p < 0.05). Among significantly deregulated lincRNAs, substantially deregulated lincRNAs were selected using more stringent cut-offs, log2(fold change) > 5 or < − 5 and p < 0.001. LINC00844 was the only lincRNA that met these criteria. In HepaRG cells, LINC00844 was significantly downregulated by APAP treatment in a concentration-dependent manner (d, e, arrow; and g). Results shown are mean ± SD from three independent experiments. *p < 0.05
Compared to other lncRNAs, lincRNA (intergenic lncRNA) genes do not overlap protein-coding genes in the genome and produce transcripts with fewer mRNA characteristics (Ransohoff et al. 2018). Therefore, we used the term “LINCxxxxx” (“x” represents a single digit) to select lincRNAs among all lncRNAs, and lincRNAs with log2(fold change) > 1 or < − 1 and p < 0.05 were considered significantly deregulated by APAP treatment. As shown in Fig. 1d, e (each dot representing one lincRNA), we identified 559 lincRNAs. Among these, 10 and 32 were significantly upregulated (red dots), whereas 24 and 48 were significantly downregulated (green dots) in HepaRG cells treated with 5 and 10 mM APAP (Fig. 1d–f).
Among the significantly deregulated lincRNAs, we selected substantially deregulated lincRNAs using more stringent cut-offs, log2(fold change) > 5 or < 5 and p < 0.001. LINC00844 was the only candidate that met these selection criteria (Fig. 1e, arrow). In HepaRG cells, LINC00844 was significantly downregulated by 5 and 10 mM APAP in a concentration-dependent manner (by 66% and 98%, respectively, Fig. 1g). The substantial downregulation of LINC00844 in response to APAP exposure suggests that LINC00844 might be involved in the metabolic “shut-down” response induced by APAP toxicity.
RNA feature characterization of LINC00844
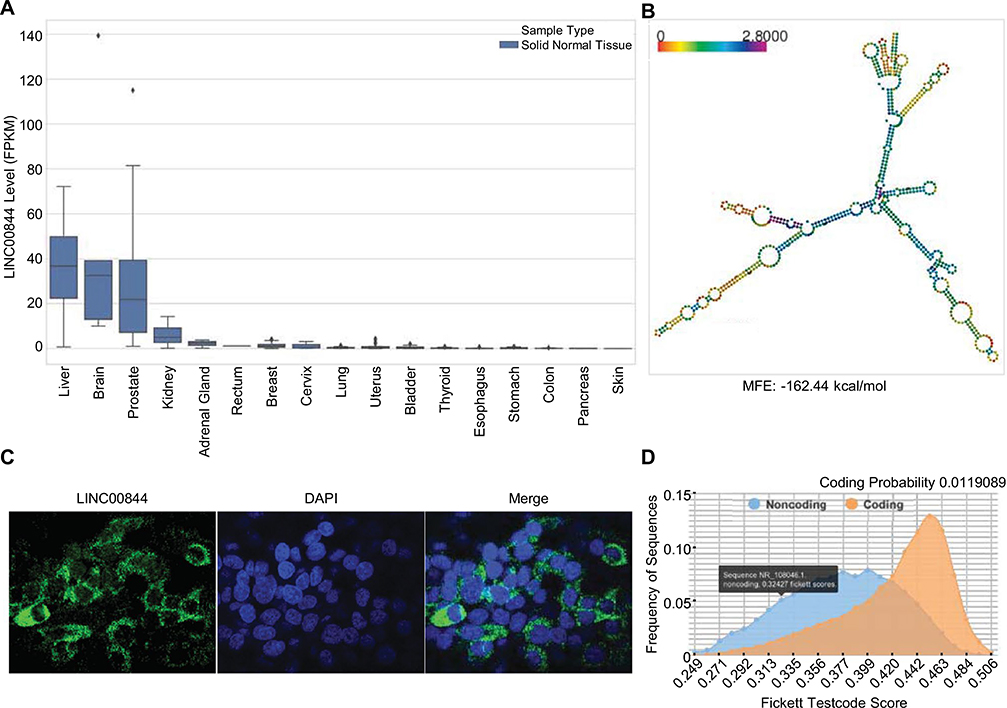
To understand the function of LINC00844, we first characterized basic RNA features of LINC00844. We analyzed the expression profiles of LINC00844 across 17 different human tissues by extracting RNA-seq data of solid non-tumor samples from the TCGA database. Figure 2a shows that LINC00844 was most enriched in the liver, brain, and prostate. The liver is a major target organ of drugs and other toxic xenobiotics and the center for xenobiotic metabolism with abundant DME and NR expression (Ballet 1997; Guo et al. 2011). Hepatic enrichment of LINC00844 indicates that LINC00844 may be involved in regulating DMEs and NRs in the liver.
Fig. 2.
RNA feature characterization of LINC00844. a LINC00844 is a liver-enriched lncRNA. Expression profiles of LINC00844 in non-tumor samples of 17 different human tissues (sample numbers described in the “Materials and methods”) were extracted from the TCGA database and show that LINC00844 is most enriched in the liver, brain, and prostate. b LINC00844 adopts a stable secondary structure. Secondary structure of LINC00844 was predicted by RNAfold with an MFE of − 162.44 kcal/mol for its thermodynamic ensemble. Color indicates positional entropy. c LINC00844 is predominantly localized in the cytoplasm of hepatic cells. HepG2 cells with stable LINC00844 overexpression and an Alexa Fluor 488 tagged probe set for LINC00844 were used to perform RNA FISH. DAPI was used to stain the nuclei. Confocal fluorescent images were taken at 40 × magnification. d LINC00844 is classified as a ncRNA species. Coding potential assessment by CPC2 showed that LINC00844 had a low coding probability of 0.0119089 and Fickett Testcode score of 0.32427 (≤ 0.74 indicates noncoding)
To evaluate whether LINC00844 possessed a stable structure, which is essential to functional molecules, we analyzed its secondary structure and folding energy. Our nucleotide sequence analysis, using RNAfold (2.4.13), revealed that LINC00844 (NR_108046) is 659 nucleotides long and that the free energy for its thermodynamic ensemble is − 162.44 kcal/mol (Fig. 2b), which is much lower than the threshold − 80 kcal/mol for stable RNAs (Mohammadin et al. 2015). This indicates that LINC00844 potentially adopts a stable secondary structure.
Molecular functions of lncRNAs are primarily determined by their subcellular localization (Carlevaro-Fita and Johnson 2019). Therefore, we performed RNA FISH to examine the subcellular localization of LINC00844 in hepatic cells. To maximize the fluorescence signal from RNA in situ hybridization, we generated a HepG2 cell line with stable overexpression of LINC00844 using a lentiviral vector containing LINC00844 nucleotide sequence (pLV-LINC00844). An RNA hybridization probe set conjugated with Alexa Fluor 488 was used to specifically detect LINC00844. As shown in Fig. 2c, LINC00844 probe hybridization produced green fluorescence in puncta that were observed within, but more intensely surrounding the nuclei (marked by DAPI), suggesting that LINC00844 is localized predominantly in the cytoplasm of hepatic cells.
The conventional function of RNAs that are translocated from the nucleus to the cytoplasm is the translation for protein synthesis; thus, there is a possibility that at least some cytoplasmic lncRNAs have protein-coding capacity. To test whether the cytoplasmic pool of LINC00844 is for translation, we analyzed the coding potential of LINC00844 based on its nucleotide sequence using CPC2 at https://cpc2.cbi.pku.edu.cn/. As shown in Fig. 2d, LINC00844 was classified as a ncRNA species with a low coding probability (0.0119089) and Fickett Testcode score (0.32427), which is a linguistic feature that distinguishes protein-coding genes and ncRNAs (≤ 0.74 indicates noncoding) based on a calculation of codon usage bias and nucleotide composition (Fickett 1982).
In summary, our data showed that LINC00844 is a liver-enriched lncRNA species distributed predominantly in the cytoplasm with a stable secondary structure. These results suggest that LINC00844 may regulate hepatic expression of DMEs and NRs and influence miRNA-mediated gene silencing, mRNA stability, or translation in the cytoplasm.
The level of LINC00844 is positively correlated with mRNA levels of CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α in the human liver
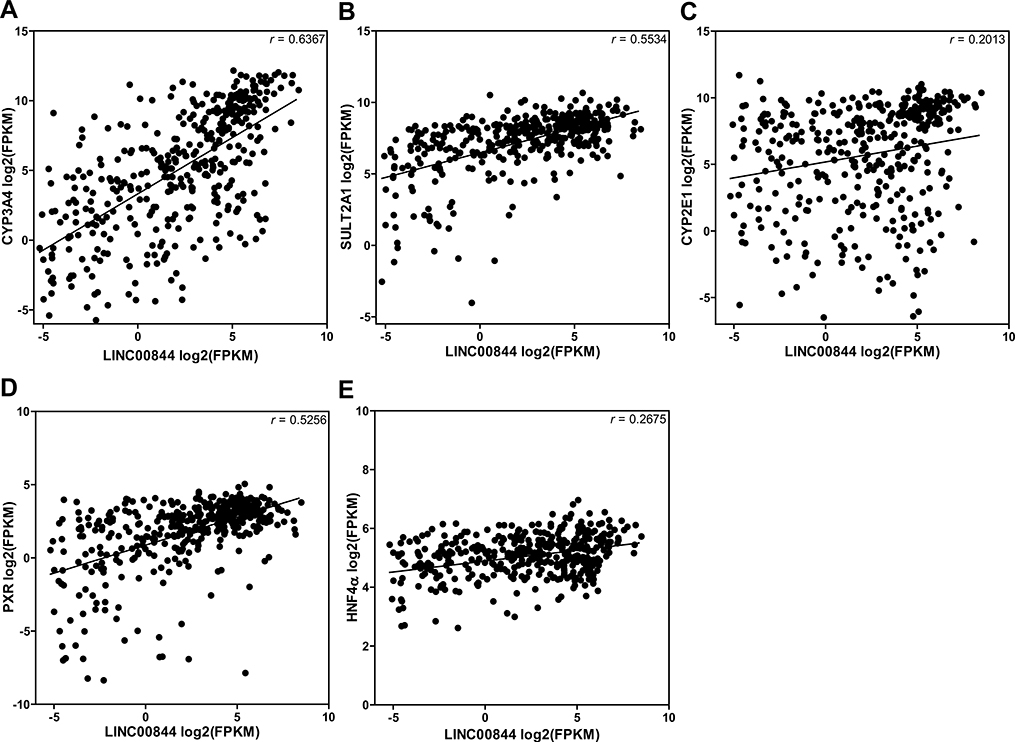
To explore the potential function of LINC00844 in regulating drug metabolism and toxicity, we tested whether the level of LINC00844 was correlated with those of nine key DMEs (CYP2A6, CYP2E1, CYP3A4, GSTT1, GSTM1, SULT1A1, SULT2A1, UGT1A1, and UGT2B15) for APAP metabolism and three key NRs (HNF1α, HNF4α, and PXR). We extracted the expression profiles of these genes in both tumor and non-tumor liver samples (n = 424) from the TCGA-LIHC dataset and performed Pearson’s correlation analysis. The results showed that the level of LINC00844 was positively correlated with those of three DMEs (CYP3A4, r = 0.6367; SULT2A1, r = 0.5534; and CYP2E1, r = 0.2013) and two NRs (PXR, r = 0.5256; and HNF4α, r = 0.2675), respectively, in the liver (p < 0.05, Fig. 3), which indicates that LINC00844 may play a regulatory role in the expression of these genes.
Fig. 3.
The level of LINC00844 is positively correlated with those of CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α in the human liver. Gene expression profiles were extracted from the TCGA-LIHC dataset including both tumor and non-tumor human liver samples (n = 424). Scatter plots from Pearson’s correlation analyses showed that the level of LINC00844 was positively correlated with mRNA levels of CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α, respectively, with varying r values. All correlations were statistically significant (p < 0.05)
Overexpression of LINC00844 upregulates CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α in HepaRG cells
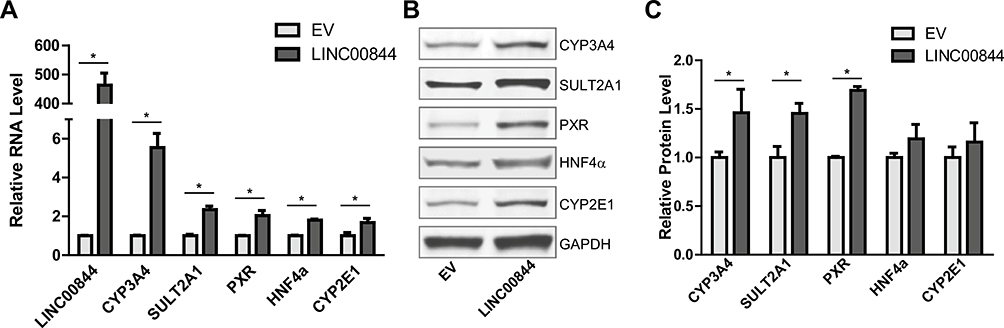
To evaluate whether LINC00844 regulates the expression of CYP3A4, SULT2A1, CYP2E1, PXR, or HNF4α, we performed gain-of-function assays by transducing HepaRG cells with a lentiviral vector carrying the sequence of LINC00844 (pLV-LINC00844) or the empty vector (EV) as control. The level of LINC00844 was significantly increased in HepaRG cells transduced with pLV-LINC00844, compared to those with EV. The overexpression of LINC00844 significantly increased the mRNA levels of CYP3A4 (by 5.5-fold), SULT2A1 (by 2.3-fold), PXR (by 2.0-fold), HNF4α (by 1.8-fold), and CYP2E1 (by 1.7-fold), compared to control (Fig. 4a). We also analyzed the effect of LINC00844 overexpression on the protein levels of these genes. Our data showed that LINC00844 overexpression increased the protein levels of CYP3A4 (by 1.5-fold), SULT2A1 (by 1.5-fold), PXR (by 1.7-fold), HNF4α (by 1.2-fold), and CYP2E1 (by 1.2-fold), and the increases of CYP3A4, SULT2A1, and PXR were statistically significant (Fig. 4b, c). These data suggest that LINC00844 positively regulates CYP3A4, CYP2E1, SULT2A1, PXR, and HNF4α to different extent.
Fig. 4.
LINC00844 overexpression upregulates CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α in HepaRG cells. HepaRG cells were transduced with a lentiviral vector pLV-LINC00844 to overexpress LINC00844. LINC00844 overexpression increased CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α at both the mRNA (a) and protein (b, c) level to different extents. Results shown are mean ± SD from three independent experiments. *p < 0.05
Knockdown of LINC00844 downregulates CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α in HepaRG cells
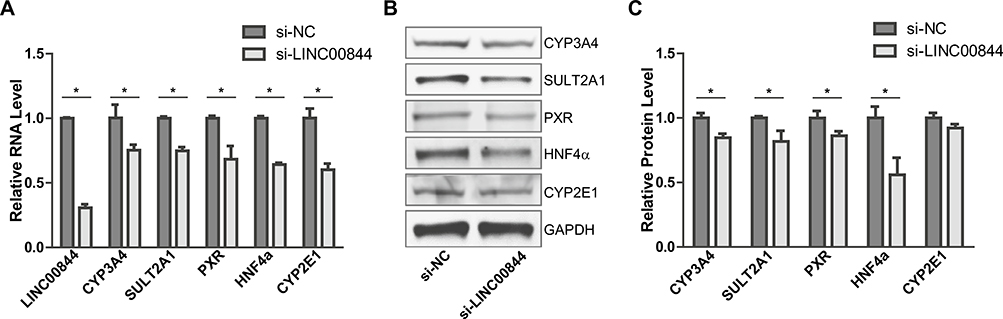
We then examined the effect of LINC00844 knockdown on the expression of CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α in HepaRG cells via RNAi-mediated gene silencing. lncRNA expression is commonly suppressed using two types of RNAi agents: anti-sense oligonucleotides (ASOs) and small interfering RNAs (siRNAs); the former is more effective for nuclear lncRNAs, whereas the latter is more effective for cytoplasmic lncRNAs (Lennox and Behlke 2016). As LINC00844 is distributed predominantly in the cytoplasm (Fig. 2c), we transfected HepaRG cells with an siRNA specifically against LINC00844 (si-LINC00844) to knock down LINC00844. An si-NC was used as a negative control. As shown in Fig. 5a, the level of LINC00844 was significantly decreased by 69% in HepaRG cells transfected with 25 nM si-LINC00844 compared to si-NC, which in turn significantly reduced the mRNA levels of CYP3A4 by 25%, SULT2A1 by 25%, PXR by 32%, HNF4α by 36%, and CYP2E1 by 40% (Fig. 5a). Under the same conditions, LINC00844 knockdown also reduced the protein levels of CYP3A4 (by 15%), SULT2A1 (by 18%), PXR (by 14%), HNF4α (by 44%), and CYP2E1 (8%) (Fig. 5b, c), and the decreases in CYP3A4, SULT2A1, PXR, and HNF4α protein were statistically significant.
Fig. 5.
LINC00844 knockdown downregulates CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α in HepaRG cells. HepaRG cells were transfected with an si-RNA against LINC00844 to specifically knock down LINC00844. LINC00844 expression was significantly reduced by 69% in cells transfected with si-LINC00844 compared to those with si-NC (a). CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α were downregulated to different extents due to LINC00844 knockdown at both the mRNA (a) and protein (b, c) levels. Results shown are mean ± SD from three independent experiments. *p < 0.05
These results further confirmed that LINC00844 positively regulates the expression of CYP3A4, SULT2A1, CYP2E1, PXR, and HNF4α.
LINC00844 acts as an hsa-miR-486-5p sponge and protects SULT2A1 from hsa-miR-486-5p-mediated downregulation in HepaRG cells
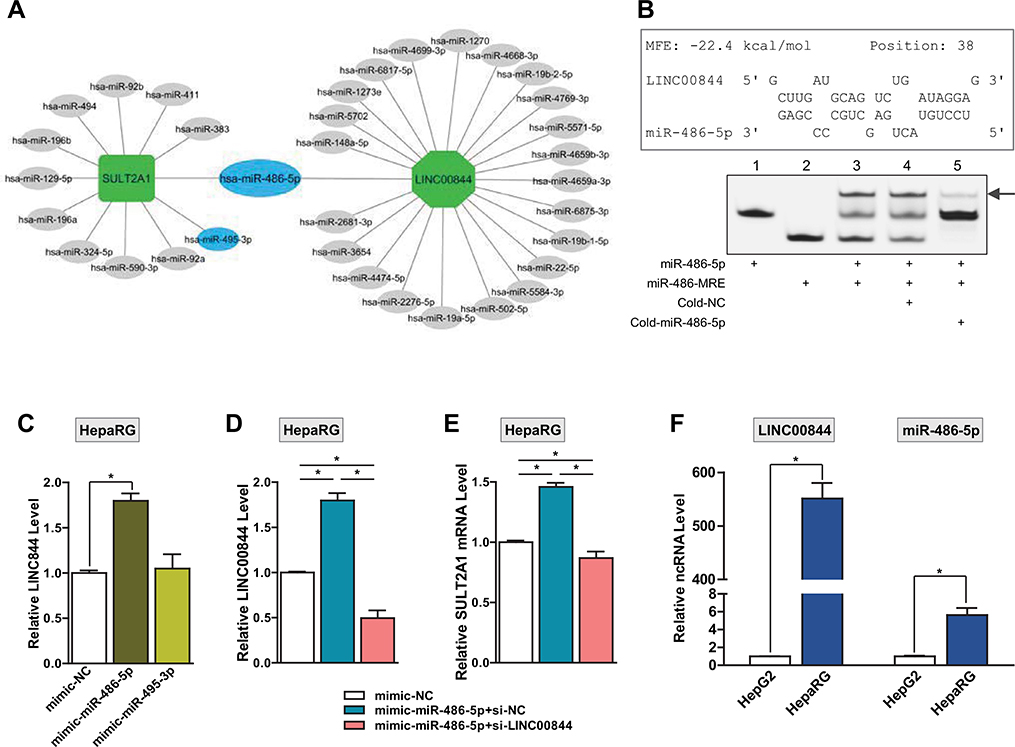
Using the miRanda prediction algorithm, we recently identified 12 miRNA candidates that target SULT2A1 mRNA and demonstrated in HepG2 cells that hsa-miR-486-5p and hsa-miR-495-3p downregulate SULT2A1 expression via direct binding to SULT2A1 mRNA to promote mRNA degradation (Li et al. 2019a). In addition, we found that although hsa-miR-486-5p is expressed at a higher level than hsa-miR-495-3p in human liver tissues, hsa-miR-486-5p appears to have less suppression efficiency than hsa-miR-495-3p in regulating SULT2A1 expression. We speculated that the less potent effect of hsa-miR-486-5p on silencing SULT2A1 expression might be due to the involvement of other regulators. To test whether LINC00844 interferes with hsa-miR-486-5p-mediated regulation of SULT2A1 expression by acting as a miRNA sponge, we evaluated potential overlap of miRNA candidates that target SULT2A1 and LINC00844. We first used LncBase (Predicted v.2) and identified 23 human miRNA candidates that target LINC00844. Using Cytoscape (Version 3.7.2.), we constructed a miRNA targeting network (Fig. 6a) consisting of the miRNAs (oval) targeting LINC00844 (octagon) and SULT2A1 (rectangle) mRNA. As shown in the network, hsa-miR-486-5p was the only miRNA that targets both LINC00844 and SULT2A1, suggesting that LINC00844 may competitively bind to hsa-miR-486-5p and attenuate the intracellular availability of hsa-miR-486-5p that promotes SULT2A1 mRNA degradation.
Fig. 6.
LINC00844 serves as an hsa-miR-486-5p sponge to protect SULT2A1 from hsa-miR-486-5p-mediated suppression in HepaRG cells. a A miRNA targeting network for SULT2A1 and LINC00844. LncBase Predicted v.2 predicted that LINC00844 (octagon) was potentially targeted by 23 miRNAs (oval), including hsa-miR-486-5p, which was among the 12 miRNA candidates for SULT2A1 (rectangle) predicted by miRanda. hsa-miR-486-5p and hsa-miR-495-3p are two experimentally-verified miRNAs (blue oval) that repress SULT2A1 in HepG2 cells. b LINC00844 directly interacts with hsa-miR-486-5p. Top, RNAhybrid analysis predicted that hsa-miR-486-5p may hybridize with LINC00844 beginning at position 38 (transcriptional start site as position 1) with an MFE of −22.4 kcal/mol. Bottom, FREMSAs verified the direct binding between miR-486-MRE on LINC00844 and hsa-miR-486-5p. Individually incubated probes for hsa-miR-486-5p (Lane 1) and miR-486-MRE (Lane 2) exhibited single bands. Co-incubation of hsa-miR-486-5p and miR-486-MRE probes resulted in a gel shift (Lane 3, arrow) and reduction of the band for either hsa-miR-486-5p and miR-486-MRE. The intensity of the shifted band was reduced by the addition of cold-miR-486–5p (Lane 5) at a 50 × concentration, but not by cold-NC (Lane 4). c LINC00844 is a specific target of hsa-miR-486-5p. LINC00844 was significantly upregulated in HepaRG cells transfected with hsa-miR-486-5p mimics, but not hsa-miR-495-3p, compared to control. d Confirmation of LINC00844 knockdown in HepaRG cells overexpressing hsa-miR-486-5p. Transfection of si-LINC00844 (pink bar), but not si-NC (blue bar), significantly reduced the level of LINC00844 in HepaRG cells overexpressing hsa-miR-486-5p. e LINC00844 protects SULT2A1 from hsa-miR-486-5p-mediated gene suppression. The level of SULT2A1 mRNA significantly increased in HepaRG cells overexpressing hsa-miR-486-5p (blue bar), compared to mimic-NC (white bar). However, when LINC00844 was knocked down, SULT2A1 mRNA expression was suppressed by hsa-miR-486-5p overexpression (pink bar). f Differential expression of hsa-miR-486-5p and LINC00844 between HepaRG and HepG2 cells. LINC00844 expression was > 500-fold higher in HepaRG cells than HepG2 cells, whereas hsa-miR-486-5p expression only exhibited a 5.6-fold elevation in HepaRG cells compared to HepG2 cells. Results shown are mean ± SD from three independent experiments. *p < 0.05 (color figure online)
To evaluate whether LINC00844 binds directly to hsa-miR-486-5p, we calculated the MFE for hsa-miR-486-5p/LINC00844 hybridization using RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid). As shown in Fig. 6b (top), our analysis revealed that a miRNA recognition element (MRE) for hsa-miR-486-5p within the RNA sequence of LINC00844 beginning at position 38 (transcription start site as position 1, referred to as miR-486-MRE hereafter). The MFE for the hsa-miR-486-5p/miR-486-MRE hybridization was − 22.4 kcal/mol, which is lower than the minimum ΔG of − 20 kcal/mL for RNA duplex formation (Yu et al. 2015), indicating that LINC00844 may bind directly to hsa-miR-486-5p. To verify this interaction, we performed FREMSAs using RNA probes for hsa-miR-486-5p and miR-486-MRE labeled with cy5.5 and IRDye800 fluorophores, respectively. The probe sequences are shown in Fig. 6b (top) and Table 1. As shown in Fig. 6b (bottom), hsa-miR-486-5p (Lane 1) or miR-486-MRE (Lane 2) exhibited single bands when incubated alone. Co-incubation of the two probes resulted in a gel shift (Lane 3, arrow), indicating duplex formation between hsa-miR-486-5p and miR-486-MRE of LINC00844. Addition of a cold hsa-miR-486-5p probe (without Cy5.5 label) at a 50 × concentration reduced the intensity of the shifted band (Lane 5) that represented duplex formation between two fluorophore-tagged probes, compared to that in Lane 3. The cold-NC probe at the same competing concentration had little effect on the hybridization of hsa-miR-486-5p and miR-486-MRE (Lane 4). These results indicate that LINC00844 specifically and directly interacts with hsa-miR-486-5p.
To test whether hsa-miR-486-5p regulates the expression of LINC00844, we transfected HepaRG cells with mimics for hsa-miR-486-5p, hsa-miR-495-3p, or the negative control (NC) at 25 nM. The overexpression of hsa-miR-486-5p, but not hsa-miR-495-3p, significantly increased the level of LINC00844 (by 1.8-fold) compared to NC (Fig. 6c). This suggests that LINC00844 is a specific target of hsa-miR-486-5p and may regulate other target genes of hsa-miR-486-5p as a miRNA sponge.
To test the role of LINC00844 in hsa-miR-486-5p-dependent regulation of SULT2A1, we knocked down LINC00844 in HepaRG cells overexpressing hsa-miR-486-5p via co-transfection of si-LINC00844 with hsa-miR-486-5p mimics. si-NC and mimic-NC were used as negative controls for si-LINC00844 and hsa-miR-486-5p, respectively. The knockdown of LINC00844 was confirmed in Fig. 6d when transfection of si-LINC00844 (pink bar), but not si-NC (blue bar), significantly reduced the level of LINC00844 in HepaRG cells overexpressing hsa-miR-486-5p. We then examined the expression of SULT2A1 under the same transfection conditions. The level of SULT2A1 mRNA was significantly increased by overexpression of hsa-miR-486-5p in HepaRG cells (Fig. 6e, blue bar), in contrast to the SULT2A1 suppression by hsa-miR-486-5p that we observed in HepG2 cells (Li et al. 2019a). However, when LINC00844 was knocked down in HepaRG cells, hsa-miR-486-5p overexpression significantly decreased the level of SULT2A1 mRNA (Fig. 6e, pink bar), which was consistent with our results in HepG2 cells. To test whether the level of LINC00844 contributes to the disparity between HepG2 and HepaRG cells regarding hsa-miR-486-5p-directed SULT2A1 suppression, we compared the levels of hsa-miR-486-5p and LINC00844 between the two cell lines. The level of LINC00844 was 551-fold higher in HepaRG cells than HepG2 cells, whereas hsa-miR-486-5p only exhibited a 5.6-fold expression elevation in HepaRG cells compared to HepG2 cells (Fig. 6f). This further confirms that LINC00844 competitively occupies hsa-miR-486-5p in hepatic cells and shows that the net effect of hsa-miR-486-5p on SULT2A1 expression is determined by the relative abundance of LINC00844.
In summary, we demonstrate that LINC00844 directly interacts with hsa-miR-486-5p which positively regulates LINC00844 expression. Our data suggest that LINC00844 exerts a protective effect on SULT2A1 expression by acting as an hsa-miR-486-5p sponge and that this effect is more prominent with a higher abundance of LINC00844 in HepaRG cells compared to HepG2 cells.
hsa-miR-486-5p indirectly regulates the expression of CYP3A4, PXR, and HNF4α via LINC00844 in HepaRG cells
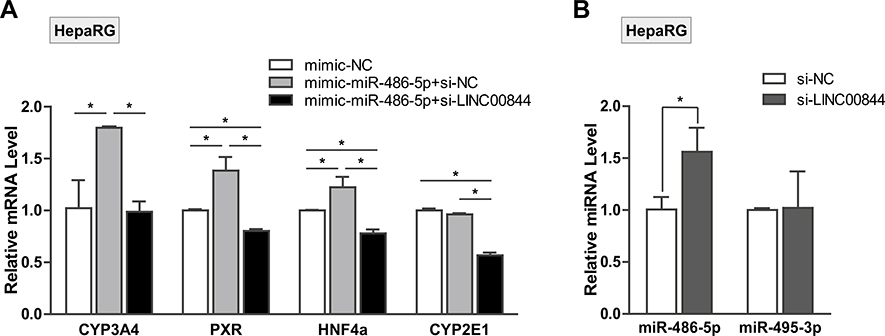
To test whether hsa-miR-486-5p modulates the expression of CYP3A4, CYP2E1, PXR, or HNF4α via LINC00844, we transfected HepaRG cells with mimics for NC or hsa-miR-486-5p in the presence of si-LINC00844 or si-NC. As Fig. 7a shows, the overexpression of hsa-miR-486-5p significantly upregulated the mRNA levels of CYP3A4, PXR, and HNF4α (gray bars) compared to NC (white bars). Introduction of si-LINC00844 abolished the upregulation of CYP3A4, PXR, and HNF4α caused by the hsa-miR-486-5p overexpression (black bars). Although the hsa-miR-486-5p overexpression had no significant effect on CYP2E1 mRNA level, si-LINC00844 transfection significantly reduced CYP2E1 expression, which is consistent with our results in Fig. 5a. Additionally, we found that si-LINC00844 transfection significantly increased the level of hsa-miR-486-5p but not hsa-miR-495-3p (Fig. 7b). Since hsa-miR-486-5p did not have targeting sites on the mRNA of CYP3A4, PXR, or HNF4α, according to miRanda prediction, these results indicate that hsa-miR-486-5p modulates the expression of CYP3A4, PXR, and HNF4α via LINC00844.
Fig. 7.
hsa-miR-486-5p indirectly regulates the expression of CYP3A4, PXR, and HNF4α via LINC00844 in HepaRG cells. a The mRNA levels of CYP3A4, PXR, and HNF4α were significantly upregulated in HepaRG cells transfected with hsa-miR-486-5p mimics (gray bars) compared to mimic-NC (white bars). Co-transfection of si-LINC00844 with hsa-miR-486-5p mimics abolished the upregulation of CYP3A4, PXR, and HNF4α caused by hsa-miR-486-5p overexpression (black bars). hsa-miR-486-5p overexpression had no significant effect on CYP2E1 mRNA level in the presence of si-NC. However, the addition of si-LINC00844 significantly reduced CYP2E1 expression, which was consistent with results in Fig. 5a. b LINC00844 knockdown specifically reduced the level of hsa-miR-486-5p but not hsa-miR-495-3p in HepaRG cells. Results shown are mean ± SD from three independent experiments. *p < 0.05
LINC00844 and hsa-miR-486-5p regulate DME expression in primary human hepatocytes
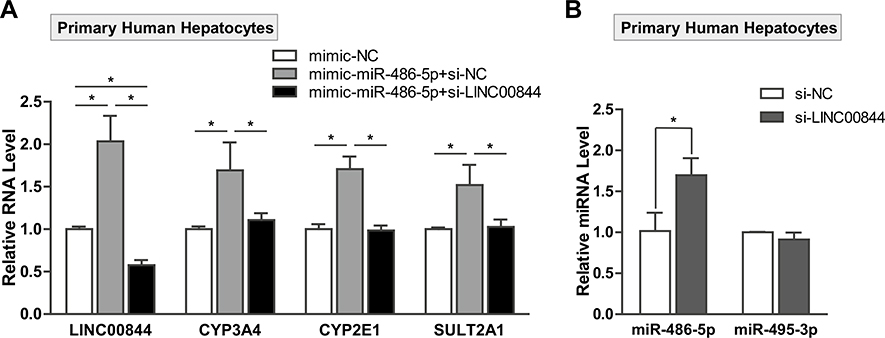
To further confirm the functional interaction of LINC00844 and hsa-miR-486-5p in regulating DME expression, we transfected 10-donor pooled primary human hepatocytes with 10 nM mimic-NC or mimic-hsa-miR-486-5p in the presence of 50 nM si-NC or si-LINC00844. In Fig. 8a, the overexpression of hsa-miR-486-5p significantly increased the levels of CYP3A4, CYP2E1, and SULT2A1 mRNAs (gray bars) compared to control (white bars). However, the hsa-miR-486-5p-induced upregulation for these DMEs was abolished by si-RNA knockdown of LINC00844 (black bars). Our results in primary human hepatocytes are consistent with those in HepaRG cells (Figs. 6, 7) except for CYP2E1. We speculate that interindividual variation may have led to the difference in CYP2E1 regulation by LINC00844 and hsa-miR-486-5p between HepaRG cells (single donor) and primary human hepatocytes (10 donors). In addition, the knockdown of LINC00844 specifically increased the level of hsa-miR-486-5p but not hsa-miR-495-3p (Fig. 8b), which is consistent with our results in HepaRG cells (Fig. 7b). Together, our data in both liver cell culture systems demonstrated a functional LINC00844/hsa-miR-486-5p interaction in regulating DME expression.
Fig. 8.
Functional interaction of LINC00844 and hsa-miR-486-5p in regulating DME expression in primary human hepatocytes. a The mRNA levels of CYP3A4, CYP2E1, and SULT2A1 were significantly upregulated in primary human hepatocytes transfected with hsa-miR-486-5p mimics (gray bars) compared to mimic-NC (white bars). Co-transfection of si-LINC00844 with hsa-miR-486-5p SULT2A1 induced by hsa-miR-486-5p overexpression (black bars). b LINC00844 knockdown specifically reduced the level of hsa-miR-486-5p but not hsa-miR-495-3p in primary human hepatocytes. Results shown are mean ± SD from three independent experiments. *p < 0.05
Discussion
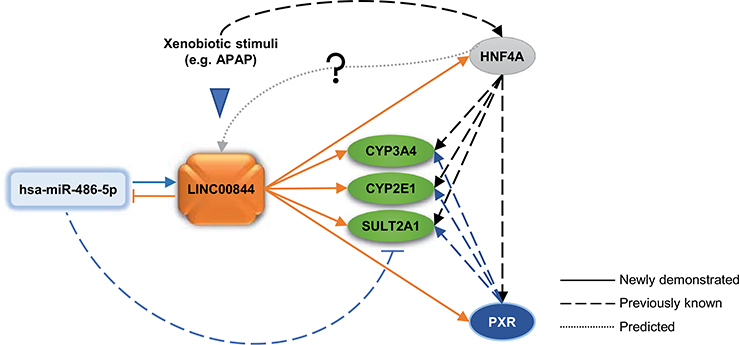
Among 167,150 annotated lncRNAs (Zhao et al. 2016), only a limited number of them have been functionally characterized, with a handful implicated in drug metabolism and toxicity (Li et al. 2019b). In this work, we identified LINC00844 that is substantially downregulated in response to APAP toxicity in human hepatic HepaRG cells (Fig. 9). We show that LINC00844 is enriched in the human liver and is predominantly localized in the cytoplasm. As summarized and indicated by solid lines in Fig. 9, our mechanistic study revealed that LINC00844 regulates the expression of three DMEs (CYP3A4, CYP2E1, and SULT2A1) and two NRs (PXR and HNF4α). PXR and HNF4α are well-known transactivators of CYP3A4, CYP2E1, SULT2A1, and many other DMEs (dashed lines, Fig. 9) (Chen et al. 2018; Echchgadda et al. 2007; Prakash et al. 2015; Yu et al. 2018). Moreover, LINC00844 antagonizes hsa-miR-486-5p through direct interaction to protect SULT2A1 from hsa-miR-486-5p-induced suppression. hsa-miR-486-5p also positively regulates the expression of CYP3A4, CYP2E1, PXR, and HNF4α via LINC00844. We propose that the LINC00844-mediated molecular network may contribute to a metabolic “shut-down” upon APAP toxicity and interfere with drug metabolism, drug-drug interactions, and adverse drug reactions by modulating the expression of CYP3A4, CYP2E1, SULT2A1, PXR, and HNF4α.
Fig. 9.
A proposed model of a LINC00844-mediated regulatory network that influences DME and NR gene expression. Our data showed that LINC00844 is a lncRNA responsive to APAP-induced hepatotoxicity and regulates the expression of CYP3A4, CYP2E1, SULT2A1, PXR, and HNF4α. PXR and HNF4α have been shown to transactivate CYP3A4, CYP2E1, and SULT2A1, while HNF4α can also activate PXR. LINC00844 interacts with hsa-miR-486-5p and functionally antagonizes hsa-miR-486-5p as a miRNA sponge to protect SULT2A1 from hsa-miR-486-5p-mediated gene suppression. Also, hsa-miR-486-5p indirectly regulates the expression of CYP3A4, CYP2E1, PXR, and HNF4α via LINC00844. The Genecards database shows the LINC00844 gene has response elements for 15 transcription factors including HNF4α (dotted line), which has been shown to respond to xenobiotic stimuli such as drugs and diet. Solid lines indicate regulation paths newly demonstrated in this study; dashed lines indicate regulation paths that are previously known. All lines are in the same colors as the corresponding regulatory modules/factors
lncRNAs constitute a large portion of differentially expressed genes in response to stressful stimuli (Valadkhan and Valencia-Hipolito 2016). The Genecards database (https://www.genecards.org) shows that the LINC00844 gene has response elements for 15 transcription factors, including HNF4α (Fig. 9, dotted line), HNF4G, retinoid X receptor (RXR)α, RXRβ, and peroxisome proliferator-activated receptor γ (PPARγ), which can be activated or inhibited under various external stimuli, such as drugs and diet (Hwang-Verslues and Sladek 2010; Ouamrane et al. 2003). A previous study reported that LINC00844 was a transcriptional target of androgen receptor (AR) and responsive to androgen stimulation in a time- and concentration-dependent manner in AR-dependent cells (Lingadahalli et al. 2018). Moreover, the study showed that LINC00844 modulated AR-regulated genes and prostate cancer progression. The functions of LINC00844 were otherwise largely unknown before our current study. Based on our data, we propose that LINC00844 responds to xenobiotic stress, and in turn, influences xenobiotic metabolism catalyzed by target enzymes of PXR and HNF4α. It is likely that the feedback loop between LINC00844 and NRs may mediate the ultimate levels of DMEs and affect drug resistance or drug-drug interactions.
lncRNAs have been found in both the cytoplasm and nucleus and regulate gene expression with localization-related mechanisms (Chen 2016); thus, functional characterization of LINC00844 relies heavily on accurate assessment of its subcellular distribution. Cytoplasmic/nuclear RNA fractionation followed by qPCR has been used for detecting subcellular enrichment; however, cross-contamination is often introduced during fractionation and easily detected by qPCR (Gagnon et al. 2014; Zaghlool et al. 2013). To avoid this issue, we conducted RNA FISH for LINC00844 in hepatic cells. LncRNAs generally exhibit lower expression than protein-coding genes (Derrien et al. 2012). The expression of LINC00844 in HepaRG cells, although higher than in HepG2 cells (Fig. 6f), was still low (Figs. 1g) and barely detectable in RNA FISH (data not shown). We overcame this limitation by overexpressing LINC00844 using lentivirus in HepG2 cells, which were much more transducible than HepaRG cells.
Many factors have been shown to regulate the expression of DMEs, including environmental stimuli, genetic variations, NRs, miRNAs, and lncRNAs; crosstalk between different regulatory pathways often exists (Koturbash et al. 2015; Li et al. 2019c; Ning et al. 2005, 2008, 2019; Yang et al. 2012). We demonstrated that LINC00844 regulates the expression of three DMEs—CYP3A4, CYP2E1, and SULT2A1. LINC00844 may exert a regulatory function toward these DMEs either directly or indirectly through regulating PXR and HNF4α. lncRNAs can regulate gene expression at the transcriptional and posttranscription level by interacting with other molecules in the cells, including DNA, other RNAs, and proteins (Wang and Chang 2011). Functionally, lncRNAs may serve as a signal, decoy, guide, or scaffold to modulate chromatin structure and transcription in the nucleus and mediate mRNA stability and translation in the cytoplasm (Wang and Chang 2011). The molecular mechanisms for LINC00844′s regulation of CYP3A4, CYP2E1, SULT2A1, PXR, and HNF4α requires further investigation. LINC00844 may also affect other target genes of PXR and HNF4α, including other NRs, such as HNF1α, or other DMEs, such as CYP2B6 (Chen et al. 2018; Hariparsad et al. 2009; Yu et al. 2018); however, LINC00844′s indirect gene regulation via PXR and HNF4α needs to be further validated.
We reported a physical and functional interaction between hsa-miR-486-5p and LINC00844 in modulating DME expression. The direct miRNA-lncRNA binding was demonstrated in the gel shift assay using 22-nt long probes for hsa-miR-486-5p and miR-486-MRE of LINC00844 (Fig. 6b). One caveat of this assay is that the miR-486-MRE probe lacks the secondary structure of the full-length LINC00844 that may interfere with the RNA binding in cells. It is worth noting, however, that RNA binding proteins are often present in cells and involved in altering RNA secondary structures for binding site accessibility and molecular interaction (van Kouwenhove et al. 2011). Previously, miRNA regulation of DMEs directly or indirectly through NRs have been well established (Li et al. 2019b; Nakano and Nakajima 2018). We demonstrated here that LINC00844 serves as a ceRNA in hepatic cells to antagonize hsa-miR-486-5p-mediated suppression of SULT2A1, while hsa-miR-486-5p also positively regulates CYP3A4, CYP2E1, PXR, and HNF4α through LINC00844. Although miRNAs are widely known for their role in gene silencing, miRNA-mediated gene upregulation has also been recognized (Vasudevan 2012). For example, miR-145 upregulates myocardin during muscle differentiation (Cordes et al. 2009). One of the major factors contributing to miRNA-mediated gene upregulation is the cell state, such as G0 or G0-like state, in which cells operate a different gene expression program than proliferating cells (Valinezhad Orang et al. 2014). Also, miRNA-mediated gene upregulation is specific to miRNA species, target transcripts, cell types, cell environment, and the presence of special factors (e.g. miRNA binding proteins) (Valinezhad Orang et al. 2014; Vasudevan 2012). This might explain our observation of gene upregulation by hsa-miR-486-5p in HepaRG cells, which are terminally differentiated and non-proliferating. However, the underlying mechanism requires additional investigation.
Levels of lncRNAs and miRNAs are responsive to drugs and are associated with drug resistance (Liu et al. 2019; Pan et al. 2015). They have also been explored as biomarkers for drug safety and toxicity assessment (Koturbash et al. 2015; Vrijens et al. 2015) or as drug targets (Matsui and Corey 2017). Understanding the mechanisms of ncRNAs in regulating DMEs may provide insight on interindividual variability in drug efficacy and adverse drug reactions.
Supplementary Material
Acknowledgements
This study was supported by the NCTR/FDA project E0753201 of the USA. Dongying Li and Dianke Yu were sponsored by the Oak Ridge Institute for Science and Education (ORISE) fellowships.
This article is not an official guidance or policy statement of the U.S. Food and Drug Administration (FDA). No official support or endorsement by the U.S. FDA is intended or should be inferred.
Abbreviations
- AhR
Aryl hydrocarbon receptor
- APAP
Acetaminophen
- CAR
Constitutive androstane receptor
- CYP
Cytochrome P450
- DAPI
4ʹ,6-Diamidino-2-phenylindole
- DME
Drug metabolizing enzyme
- DMEM
Dulbecco’s Modified Eagle’s Medium
- DMSO
Dimethyl sulfoxide
- FISH
Fluorescence in situ hybridization
- FPKM
Fragments per kilobase per million mapped reads
- FREMSA
Fluorescence-based RNA electrophoretic mobility shift assay
- GST
Glutathione S-transferase
- HNF
Hepatocyte nuclear factor
- LIHC
Liver hepatocellular carcinoma
- lincRNA
Long intergenic noncoding RNA
- lncRNA
Long noncoding RNA
- MFE
Minimum free energy
- miRNA
microRNA
- MRE
microRNA recognition element
- NC
Negative control
- PAGE
Polyacrylamide gel electrophoresis
- PBS
Phosphate-buffered saline
- PPAR
Peroxisome proliferator-activated receptor
- PXR
Pregnane X receptor
- qPCR
Quantitative polymerase chain reaction
- RXR
Retinoid X receptor
- SDS
Sodium dodecyl sulfate
- SULT
Sulfotransferase
- TCGA
The Cancer Genome Atlas
- UGT
UDP-glucuronosyltransferase
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare no competing financial interests.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00204-020-02706-5) contains supplementary material, which is available to authorized users.
References
- Ballet F (1997) Hepatotoxicity in drug development: detection, significance and solutions. J Hepatol 26(Suppl 2):26–36. 10.1016/s0168-8278(97)80494-1 [DOI] [PubMed] [Google Scholar]
- Carlevaro-Fita J, Johnson R (2019) Global positioning system: understanding long noncoding RNAs through subcellular localization. Mol Cell 73(5):869–883. 10.1016/j.molcel.2019.02.008 [DOI] [PubMed] [Google Scholar]
- Chen L, Bao Y, Piekos SC, Zhu K, Zhang L, Zhong XB (2018) A transcriptional regulatory network containing nuclear receptors and long noncoding RNAs controls basal and drug-induced expression of cytochrome P450s in HepaRG cells. Mol Pharmacol 94(1):749–759. 10.1124/mol.118.112235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang P, Manautou JEE, Zhong XB (2020) Knockdown of LncRNAs HNF1alpha-AS1 and HNF4alpha-AS1 Alters susceptibility of acetaminophen-induced cytotoxicity in HepaRG cells. Mol Pharmacol. 10.1124/mol.119.118778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL (2016) Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci 41(9):761–772. 10.1016/j.tibs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP et al. (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460(7256):705–710. 10.1038/nature08195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey JL, Cui JY (2017) Long non-coding RNAs: a novel paradigm for toxicology. Toxicol Sci 155(1):3–21. 10.1093/toxsci/kfw203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22(9):1775–1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A et al. (2012) Landscape of transcription in human cells. Nature 489(7414):101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Oh T, Ahmed M, De La Cruz IJ, Chatterjee B (2007) The xenobiotic-sensing nuclear receptors pregnane X receptor, constitutive androstane receptor, and orphan nuclear receptor hepatocyte nuclear factor 4alpha in the regulation of human steroid-/bile acid-sulfotransferase. Mol Endocrinol 21(9):2099–2111. 10.1210/me.2007-0002 [DOI] [PubMed] [Google Scholar]
- Ekstrom L, Rane A (2015) Genetic variation, expression and ontogeny of sulfotransferase SULT2A1 in humans. Pharmacogenomics J 15(4):293–297. 10.1038/tpj.2015.18 [DOI] [PubMed] [Google Scholar]
- Fickett JW (1982) Recognition of protein coding regions in DNA sequences. Nucleic Acids Res 10(17):5303–5318. 10.1093/nar/10.17.5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Janowski BA, Corey DR (2014) Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat Protoc 9(9):2045–2060. 10.1038/nprot.2014.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Dial S, Shi L et al. (2011) Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab Dispos 39(3):528–538. 10.1124/dmd.110.035873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariparsad N, Chu X, Yabut J et al. (2009) Identification of pregnane-X receptor target genes and coactivator and corepressor binding to promoter elements in human hepatocytes. Nucleic Acids Res 37(4):1160–1173. 10.1093/nar/gkn1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22(9):1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang-Verslues WW, Sladek FM (2010) HNF4alpha–role in drug metabolism and potential drug target? Curr Opin Pharmacol 10(6):698–705. 10.1016/j.coph.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YJ, Yang DC, Kong L et al. (2017) CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res 45(W1):W12–W16. 10.1093/nar/gkx428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Tolleson WH, Guo L et al. (2015) microRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomark Med 9(11):1153–1176. 10.2217/bmm.15.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M (2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34(Web Server issue):W451–W454. 10.1093/nar/gkl243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KA, Behlke MA (2016) Cellular localization of long noncoding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 44(2):863–877. 10.1093/nar/gkv1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Knox B, Chen S et al. (2019a) MicroRNAs hsa-miR-495-3p and hsa-miR-486-5p suppress basal and rifampicin-induced expression of human sulfotransferase 2A1 (SULT2A1) by facilitating mRNA degradation. Biochem Pharmacol. 10.1016/j.bcp.2019.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Tolleson WH, Yu D et al. (2019b) Regulation of cytochrome P450 expression by microRNAs and long non coding RNAs: epigenetic mechanisms in environmental toxicology and carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 1:1–35. 10.1080/10590501.2019.1639481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Tolleson WH, Yu D et al. (2019c) Regulation of cytochrome P450 expression by microRNAs and long noncoding RNAs: Epigenetic mechanisms in environmental toxicology and carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 37(3):180–214. 10.1080/10590501.2019.1639481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ruan X, Yang L et al. (2015) A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab 21(3):455–467. 10.1016/j.cmet.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang S, Zhou S et al. (2019) Drug Resistance-Related Competing Interactions of lncRNA and mRNA across 19 Cancer Types. Mol Ther Nucleic Acids 16:442–451. 10.1016/j.omtn.2019.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Bajic VB, Zhang Z (2013) On the classification of long noncoding RNAs. RNA Biol 10(6):925–933. 10.4161/rna.24604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese FP, Raimondi I, Huarte M (2017) The multidimensional mechanisms of long noncoding RNA function. Genome Biol 18(1):206 10.1186/s13059-017-1348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Corey DR (2017) Non-coding RNAs as drug targets. Nat Rev Drug Discov 16(3):167–179. 10.1038/nrd.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MR, Jaeschke H (2013) Metabolism and disposition of acetaminophen: recent advances in relation to hepatotoxicity and diagnosis. Pharm Res 30(9):2174–2187. 10.1007/s11095-013-1007-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadin S, Edger PP, Pires JC, Schranz ME (2015) Positionally-conserved but sequence-diverged: identification of long non-coding RNAs in the Brassicaceae and Cleomaceae. BMC Plant Biol 15:217 10.1186/s12870-015-0603-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar MS, Kim JH, Sartor MA, Dolinoy DC (2014) Bisphenol A-associated alterations in the expression and epigenetic regulation of genes encoding xenobiotic metabolizing enzymes in human fetal liver. Environ Mol Mutagen 55(3):184–195. 10.1002/em.21823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Nakajima M (2018) Current knowledge of microRNA-mediated regulation of drug metabolism in humans. Expert Opin Drug Metab Toxicol. 10.1080/17425255.2018.1472237 [DOI] [PubMed] [Google Scholar]
- Ning B, Dial S, Sun Y, Wang J, Yang J, Guo L (2008) Systematic and simultaneous gene profiling of 84 drug-metabolizing genes in primary human hepatocytes. J Biomol Screen 13(3):194–201. 10.1177/1087057108315513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning B, Nowell S, Sweeney C et al. (2005) Common genetic polymorphisms in the 5’-flanking region of the SULT1A1 gene: haplotypes and their association with platelet enzymatic activity. Pharmacogenet Genomics 15(7):465–473 [DOI] [PubMed] [Google Scholar]
- Ning B, Yu D, Yu AM (2019) Advances and challenges in studying noncoding RNA regulation of drug metabolism and development of RNA therapeutics. Biochem Pharmacol 169:113638 10.1016/j.bcp.2019.113638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouamrane L, Larrieu G, Gauthier B, Pineau T (2003) RXR activators molecular signalling: involvement of a PPAR alpha-dependent pathway in the liver and kidney, evidence for an alternative pathway in the heart. Br J Pharmacol 138(5):845–854. 10.1038/sj.bjp.0705113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JJ, Xie XJ, Li X, Chen W (2015) Long non-coding RNAs and drug resistance. Asian Pac J Cancer Prev 16(18):8067–8073. 10.7314/apjcp.2015.16.18.8067 [DOI] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Vlachos IS, Karagkouni D et al. (2016) DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res 44(D1):D231–D238. 10.1093/nar/gkv1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash C, Zuniga B, Song CS et al. (2015) Nuclear Receptors in drug metabolism, drug response and drug interactions. Nucl Receptor Res. 10.11131/2015/101178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff JD, Wei Y, Khavari PA (2018) The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19(3):143–157. 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 146(3):353–358. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheweita SA (2000) Drug-metabolizing enzymes: mechanisms and functions. Curr Drug Metab 1(2):107–132 [DOI] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP (2014) The multilayered complexity of ceRNA crosstalk and competition. Nature 505(7483):344–352. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart BL, Tirona RG, Kim RB (2007) Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J Clin Pharmacol 47(5):566–578. 10.1177/0091270007299930 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Valencia-Hipolito A (2016) lncRNAs in stress response. Curr Top Microbiol Immunol 394:203–236. 10.1007/82_2015_489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M (2014) Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014:970607 10.1155/2014/970607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R (2011) MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11(9):644–656. 10.1038/nrc3107 [DOI] [PubMed] [Google Scholar]
- Vasudevan S (2012) Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA 3(3):311–330. 10.1002/wrna.121 [DOI] [PubMed] [Google Scholar]
- Vrijens K, Bollati V, Nawrot TS (2015) MicroRNAs as potential signatures of environmental exposure or effect: a systematic review. Environ Health Perspect 123(5):399–411. 10.1289/ehp.1408459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43(6):904–914. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li Y, Hong H et al. (2012) Sex differences in the expression of drug-metabolizing and transporter genes in human liver. J Drug Metab Toxicol 3(3):1000119 10.4172/2157-7609.1000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Price ET, Chang CW et al. (2013) Gene expression variability in human hepatic drug metabolizing enzymes and transporters. PLoS ONE 8(4):e60368 10.1371/journal.pone.0060368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon E, Babar A, Choudhary M, Kutner M, Pyrsopoulos N (2016) Acetaminophen-induced hepatotoxicity: a comprehensive update. J Clin Transl Hepatol 4(2):131–142. 10.14218/JCTH.2015.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen J, Chen S et al. (2020a) Coordinated regulation of UGT2B15 expression by long noncoding RNA LINC00574 and hsa-miR-129–5p in HepaRG cells. Drug Metab Dispos. 10.1124/dmd.119.090043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen S, Li D, Knox B, Guo L, Ning B (2020b) FREMSA: A method that provides direct evidence of the interaction between microRNA and mRNA. Methods Mol Biol 2102:557–566. 10.1007/978-1-0716-0223-2_30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Green B, Marrone A et al. (2015) Suppression of CYP2C9 by microRNA hsa-miR-128–3p in human liver cells and association with hepatocellular carcinoma. Sci Rep 5:8534 10.1038/srep08534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Wu L, Gill P et al. (2018) Multiple microRNAs function as self-protective modules in acetaminophen-induced hepatotoxicity in humans. Arch Toxicol 92(2):845–858. 10.1007/s00204-017-2090-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghlool A, Ameur A, Nyberg L et al. (2013) Efficient cellular fractionation improves RNA sequencing analysis of mature and nascent transcripts from human tissues. BMC Biotechnol 13:99 10.1186/1472-6750-13-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138(1):103–141. 10.1016/j.pharmthera.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li H, Fang S et al. (2016) NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 44(D1):D203–D208. 10.1093/nar/gkv1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.