Abstract
Manganese (Mn) is an essential micronutrient required for the proper function of several enzymes. Accumulating evidence demonstrates a selective decrease of bioavailable Mn in vulnerable cell types of Huntington’s Disease (HD), an inherited progressive neurodegenerative disorder with no cure. Amelioration of underlying pathophysiology, such as alterations in Mn-dependent biology, may be therapeutic. We therefore sought to investigate global Mn-dependent and Mn-responsive biology following various Mn exposures in a mouse model of HD. YAC128 and wildtype (WT) littermate control mice received one of three different Mn exposure paradigms by subcutaneous injection of 50 mg/kg MnCl2•4(H2O) across two distinct HD disease stages. “Pre-manifest” (12-week old mice) mice received either a single (1 injection) or week-long (3 injections) exposure of Mn or vehicle (H2O) and were sacrificed at the pre-manifest stage. “Manifest” (32-week old) mice were sacrificed following either a week-long Mn or vehicle exposure during the manifest stage, or a 20-week-long chronic (2x weekly injections) exposure that began in the pre-manifest stage. Tissue Mn, mRNA, protein, and metabolites were measured in the striatum, the brain region most sensitive to neurodegeneration in HD. Across all Mn exposure paradigms, pre-manifest YAC128 mice exhibited a suppressed response to transcriptional and protein changes and manifest YAC128 mice showed a suppressed metabolic response, despite equivalent elevations in whole striatal Mn. We conclude that YAC128 mice respond differentially to Mn compared to WT as measured by global transcriptional, translational, and metabolomic changes, suggesting an impairment in Mn homeostasis across two different disease stages in YAC128 mice.
Graphical Abstract

Global manganese-responsive processes are suppressed in the YAC128 mouse model of Huntington’s Disease (HD) compared to wild type (WT).
Introduction
Manganese (Mn) is an essential metal required for several biological processes regulating human health. Mn functions as a co-factor for several enzymes, including glutamine synthetase (GS), superoxide dismutase 2 (MnSOD), pyruvate carboxylase (PC), and arginase 2 (ARG2)1. In addition, there are several canonical signaling pathways which are Mn-responsive, such as ATM2, p532, PI3K, insulin and IGF-13,4. Attention has been paid to the toxicological properties of Mn as it is a potent neurotoxicant with overexposure resulting in Manganism, which manifests with parkinsonian features5. Despite potential toxicity, it is clear that maintaining adequate levels of Mn is crucial for proper neurological function1 and therapeutic Mn exposures may ameliorate select neurodegenerative disorders in which Mn-dependent or Mn-responsive processes are suppressed or insufficient6. Mn deficiency is rare as the average human diet provides sufficient amounts of Mn1. There is, however, a consistent link between divalent metal homeostasis and neurodegenerative disorders7 – suggesting that neuronal metal biology may be impaired under these conditions.
HD is an autosomal dominant neurodegenerative disorder which develops as a result of a trinucleotide repeat expansion in the Huntingtin gene (HTT). The mutation results in selective degeneration of the medium spiny neurons within the striatum (caudate and putamen), as well as select other neurons, and often presents clinically with chorea, mood disorders, psychiatric symptoms and cognitive impairment. Symptom onset is largely dictated by length of CAG-repeat with longer expansions associated with earlier onset8,9; however, there is a great amount of variability10 which has been attributed to other genetic and environmental modifiers. There is clear evidence for a link between Mn and HD. Williams et al found reduced Mn-dependent neurotoxicity in HD mouse striatal cells compared to control striatal cells11. This neuroprotective effect of the HD genotype against Mn neurotoxicity has been replicated, with evidence for this phenomena occurring through an ATM-p53 mediated molecular pathway in the same mouse HD striatal cell model as well as human neuroprogenitors2.
We have also investigated the impact of Mn exposure on a well-established HD mouse model that replicates key features of HD pathobiology: the YAC128. The YAC128 line contains a mutant copy of the human Huntingtin gene and is described in greater detail below. YAC128 mice mirror the selective striatal and cortical degeneration observed in humans12 and furthermore, recapitulate several cognitive and motor symptoms observed in humans13. Specifically, YAC128 mice exhibit impairments in aspects of executive function (motor learning) as early as 2 months of age that progressively worsen13. At this early age, there are no consistently observed motor phenotypes – which is also commonly observed in prodromal HD patients. Starting around 4–6 months of age, YAC128 mice demonstrate more overt deficits in executive function (memory) and motor coordination and these also worsen with age13. This particular model is well suited for evaluating behavioral or pharmacokinetic interventions as disease progression is modest: 2-month old YAC128 mice resemble pre-manifest disease and 12-month old mice exhibit more severe manifest motor symptoms13. We have previously demonstrated that several HD phenotypes are attenuated following Mn exposure in YAC128s. For example, elevations of striatal urea cycle metabolites in a pre-manifest HD mouse model can be corrected with a 1-week Mn exposure14. Additionally, acute Mn treatment attenuates the “cargo-recognition failure” in autophagy and partially rescues the reduced glucose uptake phenotypes observed in HD cell lines15,16. It is worth noting that not all HD phenotypes examined are corrected by Mn exposure. For example, altered striatal sterols in HD model mice are not changed by acute Mn exposure17. Given the evidence for reduced bioavailable Mn in HD and that specific HD phenotypes can be ameliorated by Mn exposure, we further investigated the global striatal response to Mn in pre-manifest and manifest HD mice and WT controls using various lengths of Mn exposure.
The molecular mechanisms by which HD impairs Mn-biology are uncertain; we demonstrate here that the gene-environment interaction between HD and Mn are consistent across several stages of disease and diverse biological processes. We demonstrate that single, 1-week-long, and 20-week-long Mn exposures in pre-manifest and manifest HD model mice suppress multiple Mn-dependent and Mn-responsive biological processes at the gene expression (mRNA), protein and metabolic levels in the mouse brain. Further examination on the impact of these Mn exposures on HD phenotypes, including the behavioral manifestations observed in this HD mouse model are warranted in future studies.
Materials and Methods
Materials
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Optima grade LC-MS solvents for the mass spectrometry analysis were obtained from Thermo Fisher Scientific. (Fair Lawn, NJ). All sterol standards, natural and isotopically labeled, used in this study are available from Kerafast, Inc. (Boston, MA).
Animals
All animal use was approved by the Vanderbilt University Medical Center Institutional Animal Care and Use Committee and performed in compliance with all relevant laws and institutional guidelines. Animal husbandry and genotyping information has been previously described by Bichell et al14. Briefly, male and female FVB-Tg(YAC128)53Hay/J mice (Jackson Laboratory, Bar Harbor, ME; stock number: 027432) were weaned at 21 days and co-housed in groups of 2–5. These mice, which will be referred to as YAC128, contain two WT copies of the mouse Huntingtin (Htt) gene and are hemizygous mutant with one full-length copy of the human HTT gene12. Thus, our mutant YAC128 mice contain 3 copies of the Huntingtin gene compared to our wild-type (WT) littermates with 2 copies of mouse Htt. An additional mouse line of C57-YAC128 was generated by crossing the FVB-YAC128 mouse with the C57BL/6J mouse and backcrossing for at least 10 generations. These mice will be referred to as C57-YAC128. At weaning, mice were genotyped using Transnetyx qPCR-based genotyping as previously described14. Hemizygous mutant animals and WT littermates were used for experiments. Pre-manifest male C57-YAC128 and C57-WT mice are exclusively used for the single injection exposure experiment. Male and female YAC128 and WT mice at 12-weeks and 32-weeks of age were included in the 1-week and 20-week long Mn exposure studies. Data presented are the result of several independent experiments. Specifically, data presented in pre-manifest mice with 1-week Mn exposure are from three independent experiments (experiment 1: striatal Mn, Arginase 2 protein; experiment 2: BDNF, SLC30A10 mRNA, GLT-1 protein; experiment 3: RNAseq, qRT-PCR). Data presented in manifest mice receiving either 1-week or 20-week Mn exposure are from a fourth experiment. See Table 1.
Table 1.
Summary of Mn exposures and age of mice for each outcome measure.
| Age | Mn Exposure | Outcome measures | Figure |
|---|---|---|---|
| 13-weeks | 1-week-long | striatal Mn, ARG2 protein, RNAseq | 1A–E |
| 13-weeks | single exposure | Bdnf and Slc30a10 mRNA, ARG2 and GLT1 protein | 2A–C |
| 32-weeks | 1-week-long | striatal Mn, ARG2 protein, metabolomics | 3A–E |
| 32-weeks | 20-week-long | striatal Mn, ARG2 protein, metabolomics | 4A–E |
Manganese exposure paradigms
This study utilized three Mn exposure paradigms: 1) a single subcutaneous exposure of either a 1% solution of MnCl2•4(H2O) in filtered MilliQ water at 50 mg/kg body weight or vehicle (filtered water), 2) a 1-week, and 3) 20-week chronic exposure. The 1-week exposure is a previously published week-long exposure protocol (intermittent injections given on days 0, 3 and 6, with sacrifice on day 7) known to elevate brain Mn levels18. The 20-week exposure is a twice weekly injection (Mondays and Thursdays) based on the estimated half-life of 3.5 days for Mn administered via this route and pilot work suggesting the absence of observable toxicity with this exposure paradigm (including no weight loss or physical signs of distress).
Tissue Collection
For the single injection exposure, mice were sacrificed either 0, 1, 4, 12 or 24 hours after the injection to examine acute changes to Mn. For 1-week and 20-week Mn exposures, mice were sacrificed 24 hours after the final injection. For all studies, mice were sacrificed by cervical dislocation without anesthesia. Mice were decapitated with sharp scissors and the brain removed. The striatum was dissected out under a dissecting microscope, flash-frozen in liquid nitrogen and stored at −80°C until later analysis.
Striatal Mn levels
Striatal Mn concentrations were measured with graphite furnace atomic absorption spectrometry (GFAAS, Varian AA240, Varian, Inc., Palo Alto, CA). Protein lysates were prepared as described below (Western Blotting). These striatal protein lysates were further digested in ultrapure nitric acid (1:10 wt/vol dilution) for 48–72 h in a sand bath (60 °C); 100 μL of digested tissue was brought to 1 mL of total volume with 2% nitric acid and analyzed for Mn. This dilution ensured that all samples were analyzed in the middle of the standard curve of 0 μg Mn/L, 2.25 μg Mn/L, 4.5 μg Mn/L, and 9 μg Mn/L. The detection limit on the GFAAS for Mn is 5 ng/L. A bovine liver (NBS Standard Reference Material, USDC, Washington, DC) (10 μg Mn/g) was digested in ultra-pure nitric acid and used as an internal standard for analysis (final concentration 5 μg Mn/L).
RNA processing for RNA sequencing and qRT-PCR
RNA was extracted from frozen striatal tissue using Trizol (Invitrogen, cat # 15596026) following the standard protocol. Tissues were homogenized with plastic tissue grinders. RNA was quantified with a NanoDrop spectrophotometer, then converted to cDNA with the SuperScript III Reverse Transcriptase kit (Invitrogen 18080044) following manufacturer’s protocol.
Library Preparation
Library preparation was performed as previously described in Parmalee et al. (2015). Briefly, an Illumina RNASeq compatible library was prepared by ligating TruSeq adaptors to the cDNA and treating with UNG prior to amplification. RNA sequencing was performed at the Albert Einstein College of Medicine Center for Epigenomics / Computational Genomics Core. It was performed on an Illumina Hiseq2500 machine with 100bp single-end reads. All sample libraries were multiplexed with TruSeq indexed adaptors and ran on one lane. The output of the Illumina machine was raw FASTQ files. Quality control of raw FASTQ data files from the Illumina sequencing machine was done with FASTQC (Version 0.11.5; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trimming of low-quality base calls was done with cutadapt (Version 1.16), setting the quality cutoff to 20 and minimum read length to 20 (Martin, 2011). STAR (Version 2.5.3a) was used for alignment of 100bp reads to the mm10 reference genome, FVB/NJ genome (ftp://ftp.ensembl.org/pub/), and human HTT gene, all downloaded from ENSEMBL (Dobin & Gingeras, 2015). The corresponding gtf annotation files from ENSEMBL were used. The human HTT gene was manually changed to have 128 CAG repeats in exon 1, and the start and stop positions in its annotation file were modified according to the added bases. Alignment files were in BAM format sorted by coordinate. FeatureCounts from the subRead package (Version 1.6.1) was used to assign reads to genes, generating count tables19. Count tables were uploaded to R for differential expression analysis with DESeq220. The mouse ENSEMBL IDs (ENSMUSG) returned by DESeq2 were matched to their respective common gene names using the R package biomaRt.
Western Blotting
Striatal tissue lysates were prepared by homogenizing frozen tissue in 180μL Pierce RIPA lysis buffer (Thermo Scientific, Cat# 89900) with protein and phosphatase inhibitors (Sigma Aldrich, cat #P0044, #P2850, #P5726, and #P2714). Protein concentration was measured using a standard quantification protocol (Pierce BCA Protein Assay Kit, Thermo Scientific). Samples were prepared with denaturing sample buffer (Bio-Rad Cat# 0610747) and either 10 μg of protein (GLT1) or 40 μg of protein (ARG2) were loaded onto Mini-Protean TGX Pre-cast gels (Bio-Rad Cat# 4561094) and gel transfer was done using the iBlot™ system. Membranes were blocked with Odyssey blocking buffer (Odyssey Cat # 927–40000) for 1 hour prior to incubation with primary antibodies using Arginase 2 (ARG2) at 1:1,000 (Santa Cruz, sc-20151) and Glutamate Transporter 1 (GLT1) at 1:4,000 (Millipore Sigma, AB1783). Secondary antibodies were anti-rabbit or anti-guinea pig (Licor IRDye 700 or 800CW) diluted at 1:10,000 in Odyssey blocking buffer. Protein was detected using the Odyssey infrared system and analyzed with ImageStudio (www.licor.com) and normalized to total protein quantified from Coomassie stained gel signal.
Metabolomics
Previously frozen striatal tissue was lysed in 400μL ice-cold lysing buffer (1:1:2, ACN:MeOH:Ammonium Bicarbonate (0.1M, pH 8.0) (LC-MS grade). Individual samples were sonicated using a probe tip sonicator, 10 pulses, at 30% power and cooled down on ice between samples. A BCA protein assay was used to determine the protein concentration for each individual sample, and adjusted to a total amount of protein of 200μg in 200 μL of lysis buffer. Isotopically labeled standards, Creatinine-D3 and Lysine-D4, were added to each sample to assess sample processing steps (metabolite extraction and reconstitution). Following lysis and standard addition, protein precipitation was performed by adding 800μL of ice-cold methanol (4x by volume). Samples were incubated at −80°C overnight. Following incubation, samples were centrifuged at 10,000 rpm for 10 min to eliminate proteins. The supernatants containing metabolites were dried via speed-vacuum.
Dried metabolite extracts were stored frozen at −80°C until ready to use. Prior to mass spectrometry analysis, extracts were reconstituted in 100 μl of acetonitrile/ water (80:20, v/v) and centrifuged for 5 min at 15K rpm to remove insoluble material. Quality control samples were prepared by pooling equal volumes of each sample. Isotopically labeled standards, Valine-D8 and Inosine-4N15, were added to each sample to determine MS instrument reproducibility.
Global, untargeted mass spectrometry analyses was performed on a high resolution Q-Exactive HF hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a Vanquish UHPLC binary system (Thermo Fisher Scientific, Bremen, Germany) using an optimized parallel reaction monitoring (PRM) method. This method permits monitoring and detection of all metabolites as well as specific molecules (precursor ions) added to an inclusion list. In the same run (injection), associated fragment ions (MS2 scan) of individual ions are also detected. Particular to this study, parameters consisted of an MS1 (precursor ion) scan at 60,000 resolution with an automatic gain control (AGC) value of 1e6, max injection time (IT) of 100 ms, and scan range from m/z 70–1050 recorded as profile data. Following the MS1 scan, the method allows for up to six targeted MS2 scans at a resolution of 15,000 and with an AGC value of 1e5, a max injection time of 50 ms, a 1.3 m/z isolation window, with stepped collision energy of 20 and 40, and was performed and recorded as profile data. Lastly, the method was set with the following PRM parameters: a timed inclusion list containing the target precursor m/z value, charge, and a 2 min retention time window. These values were determined from prior analyses of synthetic standards.
After reconstitution, metabolite extracts (5uL injection volume) were separated on a SeQuant ZIC-HILIC 3.5-μm, 2.1 mm × 100 mm column (Millipore Corporation, Darmstadt, Germany) held at 40°C. Liquid chromatography was performed at a 200 μl min−1 using solvent A (5mM Ammonium formate in 90% water, 10% acetonitrile) and solvent B (5mM Ammonium formate in 90% acetonitrile, 10% water) with the following gradient: 90% B for 2 min, 90–40% B over 16 min, 40% B held 2 min, and 40–90% B over 10 min, 90% B held 10 min (gradient length 40 min).
Data Processing
Ultra-performance liquid chromatography - tandem mass spectrometry (UPLC-MS/MS) raw data were imported, processed, normalized and reviewed using Progenesis QI v.2.1 (Non-linear Dynamics, Newcastle, UK). Sample runs were aligned against a quality control pool reference run, and peak picking was performed on individual aligned runs to create an aggregate data set. Unique ions (retention time and m/z pairs) were grouped (a sum of the abundancies of unique ions) using both adduct and isotope deconvolutions to generate unique compounds (retention time and m/z pairs) representative of unannotated metabolites. Data were normalized to all compounds using Progenesis QI and normalized data was utilized for relative quantitation. Significance was assessed using p-values and fold changes calculated from normalized compound abundance data. Tentative and putative annotations were determined by using accurate mass measurements (< 5 ppm error), isotope distribution similarity, and fragmentation spectrum matching (when applicable) by searching the Human Metabolome Database (HMDB), METLIN, NIST, and an internal curated library21–23.
Metaboanalyst 4.0 (www.metaboanalyst.ca/) was used to perform pathway and metabolite enrichment analyses from annotated compounds with statistical significance (p<0.05) after Mn exposure in each genotype. Briefly, MetaboAnalyst assigns a pathway impact factor which reflects the location of an enriched compound within that particular biologic pathway and its impact on the downstream targets. Negative log p-values were calculated based upon the number of enriched compounds detected by UPLC-MS/MS within a particular metabolic pathway compared to the total number of compounds known to be present in that pathway. All metabolic pathways found to be significantly enriched after Mn exposure are annotated and circle color (yellow -> red) and circle size (small -> large) reflect increasing pathway impact and -log(p-value), respectfully. Each pathway network (Figure 3E and 4E) is scaled to a particular pathway impact and -log(p-value) in response to Mn; thus, pathway figures are not meant for direct visual comparisons between WT and YAC128.
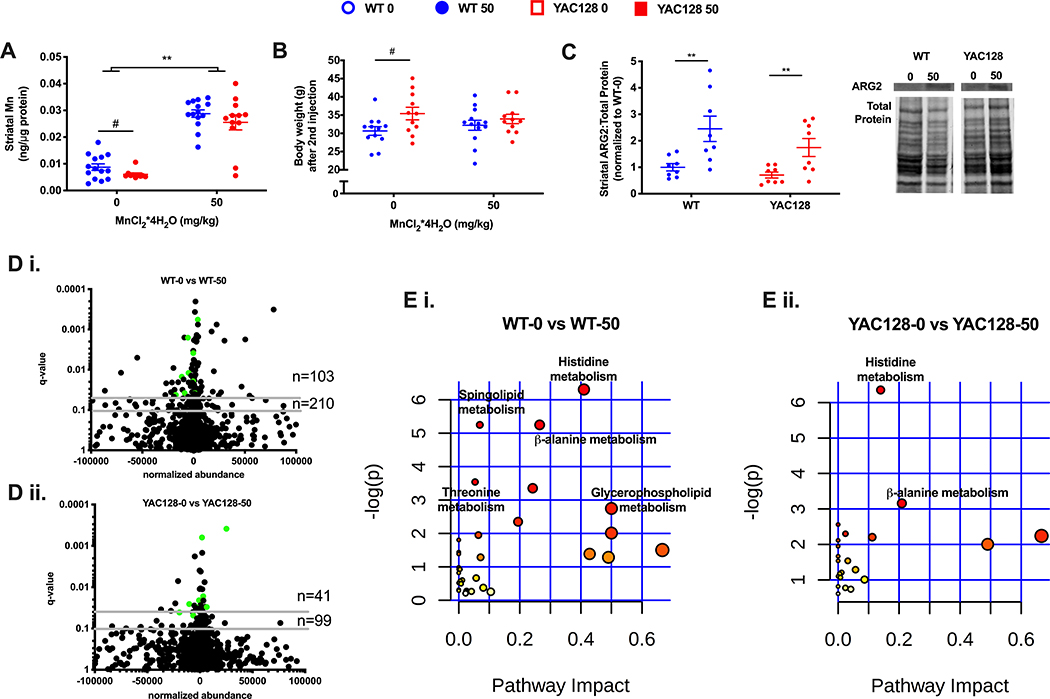
figure 3.
the effect of 1-week long mn exposure in aged mice on striatal mn (a), body weight (b), arginase 2 protein levels (c), striatal metabolome (d), and metabolic pathways (e) enriched in the striatum. data represented in a, b, and c as the mean ± sem and n=10–14 and an asterisk (*) indicates a mn effect and a hashtag (#) indicates a genotype effect at significance level of p<0.05. changes in the striatal metabolome (d) depicted in volcano plots indicate metabolites altered at q-value<0.05 and q-value<0.10 with horizontal grey lines. n=103 and n=210 indicate the number of significantly altered metabolites in wt (d i.) at q-value <0.05 and q-value 0.10, respectively. n=41 and n=99 indicate the number of significantly altered metabolites in hd (d ii.) at q-value <0.05 and q-value 0.10, respectively. the number of significantly altered metabolites is greater in wt than hd, χ2=14.0, df=1, p<0.001. green dots indicate those significantly altered metabolites in both wt and hd (15 metabolites). pathway analysis of striatal metabolites (e) indicates similar metabolic pathways. colored dots ranging from yellow to red indicate increasing significance by -log(p-value); biological pathways significantly enriched (p<0.05) are annotated. pathway impact relates to the ability of enriched metabolites to impact additional compounds within that pathway. circle color (yellow -> red) and circle size (small -> large) reflect an increasing pathway impact and −log (p-value), respectfully. metabolomic data represent feature changes in n=10-14 for each genotype and exposure.
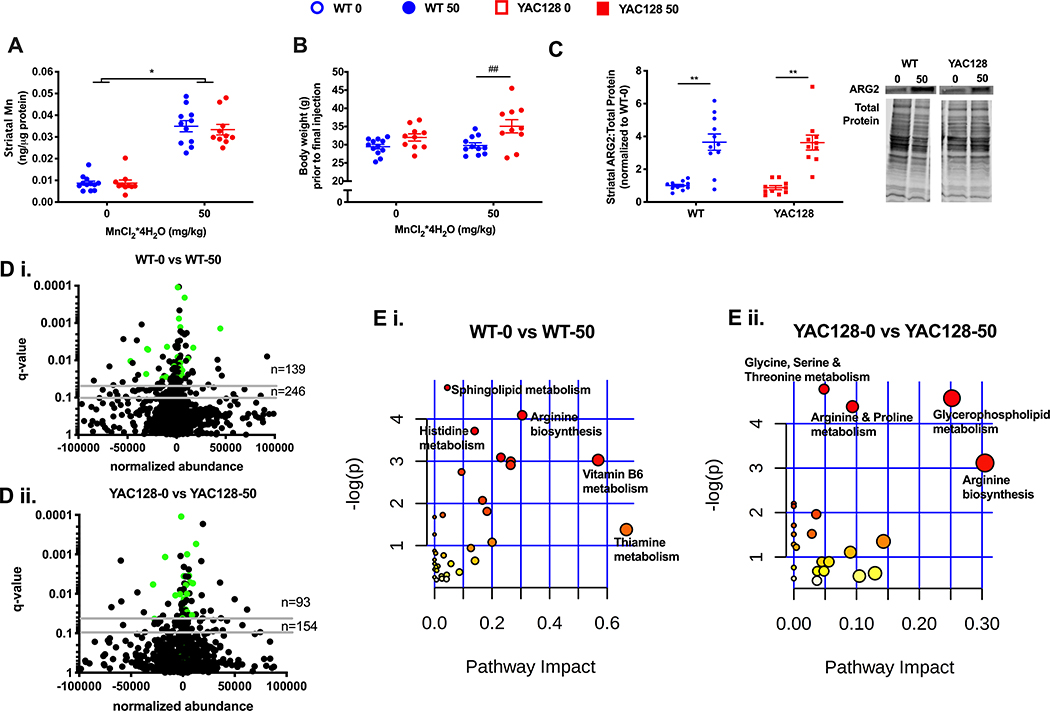
figure 4.
the effect of chronic (20-week long) mn exposure in aged wt and hd mice on striatal mn (a), body weights (b), striatal arginase 2 protein levels (c), striatal metabalome (d), and metabolic pathways (e). data for a, b, and c are represented as mean ± sem and n=10–12; an asterisk (*) indicates a significant mn effect and a hashtag (#) indicates a significant genotype effect at a significance level of p<0.05 (*, #) or p<0.01 (**, ##). volcano plots (d) indicate metabolites altered at q-value<0.05 and q-value<0.10 with horizontal grey lines. n=139 and n=246 indicate the number of significantly altered metabolites in wt (d i.) at q-value <0.05 and q-value 0.10, respectively. n=93 and n=154 indicate the number of significantly altered metabolites in hd (d ii.) at q-value <0.05 and q-value 0.10, respectively. more metabolites were altered by mn in wt than in hd, χ2=4.6, df=1, p<0.05. green dots indicate metabolites significantly altered with mn in both wt and hd (30 metabolites) at p<0.05. biological pathways (e) significantly enriched (p<0.05) are annotated in wt (e i.) and yac128 (e ii.), note the difference in x-axis scale (max pathway impact in yac128 is 0.3 compared to 0.6 in wt). pathway impact relates to the ability of enriched metabolites to impact additional compounds within that pathway. circle color (yellow -> red) and circle size (small -> large) reflect an increasing pathway impact and −log (p-value), respectfully. metabolomic data represent feature changes in n=10–12 for each genotype and exposure.
The metabolomics data is available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) Web site, the Metabolomics Workbench, https://www.metabolomicsworkbench.org where it has been assigned Project ID (PR001333).
Statistics
The statistical analyses for striatal Mn, body weights, mRNAs, ARG2 and GLT1 protein were conducted using a 2-way ANOVA with post hoc comparisons after determining either a main effect of either genotype or Mn exposure or a statistically significant (p<0.05) interaction between genotype and Mn exposure (GraphPad Prism 6.0). ARG2 and GLT1 protein levels were quantified by normalizing all samples to the mean of all WT-0 (WT-vehicle) samples within the same protein membrane. Data are represented as the mean ± standard error of the mean (sem).
Results
Pre-manifest YAC128 mice exhibit reduced striatal Mn uptake and transcriptional responses despite similar fold-changes in ARG2 protein levels after 1-week Mn exposure
Our initial experiments in pre-manifest mice utilized a 1-week Mn exposure first published by Dodd et al. to elevate levels of brain Mn18. This nearly 5-fold elevation in striatal Mn levels has been previously confirmed in our lab11. For reference, environmental exposures in young children living near oil refineries found approximately 50% higher levels of blood Mn compared to children living further away24. We found no difference in basal striatal Mn levels between WT and YAC128 mice at the pre-manifest age; however, after exposure, YAC128 animals had significantly less striatal Mn than compared to exposed WT animals (Figure 1A). A 1-week Mn exposure did not show any overt signs of toxicity or sickness indicated by the absence of weight loss in Mn-exposed animals in both genotypes (Figure 1B). As expected, YAC128 mice were heavier compared to WT25, with no significant effect of Mn on exposure. Additionally, the obligate Mn-dependent enzyme, arginase 2 (ARG2), was elevated nearly 3-fold after Mn-exposure in both WT and YAC128 in weeklong exposed pre-manifest mice (Figure 1C) with no basal difference between genotypes. Similar fold changes in ARG2 in response to Mn in YAC128 have been previously reported14.
figure 1.
striatal mn (a), body weights (b), striatal arginase 2 protein (c), volcano plots representing striatal rnaseq (d i. wt, d ii. yac128) and pathway analysis of differentially expressed (de) genes in pre-manifest wt (e i.) and yac128 (e ii.) male and female mice after 1-week subcutaneous mn exposure. weights collected after second subcutaneous injections of 0 or 50mg/kg mncl2*4h2o and demonstrate a main effect of genotype (p=0.04). data presented in a, b, and c are shown as mean ± sem and **** indicates significance level at p<0.001; n=6–8. data presented in d and e are from a second independent experiment with the same mn exposure and animal age. volcano plots (d) indicate fold change in mrna expression, black dots indicate a single mrna and horizontal grey lines mark q-values of 0.10 and 0.05. n=191 and n=239 indicate the number of significantly altered genes at q-value <0.05 and q-value 0.10, respectively. the number of de genes by mn exposure is greater in wt than yac128 by chi-square, χ2=66.3, df=1, p<0.0001. green dots represent significantly altered genes in both wt and yac128 (n=20). top biological networks enriched after mn exposure in wt (e i.; left) and yac128 (e ii.; right) mice. genes in green (upregulated) or red (downregulated) backgrounds indicate those that were significantly de after mn exposure while genes in grey were not de but function as genetic hubs through which de genes may signal or impact. the intensity of the color indicate the degree of fold change. solid grey lines indicate a direct relationship and dotted lines indicate an indirect relationship. arrows indicate the direction of the relationship. data represents gene changes in n=3 in each genotype and exposure.
We next sought to determine the extent to which Mn-dependent effects were suppressed by the HD genotype across several biological processes. Therefore, we next examined the effects of a 1-week Mn exposure in pre-manifest mice on the global transcriptome to capture differences in steady-state levels of mRNAs. RNA sequencing (RNAseq) of striatal tissue collected from YAC128 mice demonstrated a global reduction in the sensitivity to Mn-induced transcriptional changes compared to WT mice. Using two statistical thresholds of q<0.05 and q<0.10, the number of genes differentially expressed after Mn were significantly greater in WT at both cut-offs (with 191 and 239 differentially expressed genes, respectively) compared to YAC128 (n= 31, n=40). The volcano plots in Figure 1D demonstrate that although the range in mRNA fold changes after Mn exposure are similar for WT (Figure 1D i.) and YAC128 (Figure 1D ii.), a chi-square test confirms that a greater proportion of genes are altered in WT after exposure compared to YAC128 (p<0.0001). Pathway analysis of differentially expressed mRNAs identified from RNAseq indicates that the top biological pathway enriched in WT mice after Mn exposure was centered around brain derived neurotrophic factor (Bdnf) and related to “Neurological Disease, Organismal Injury and Abnormalities” (Figure 1E; Qiagen, Ingenuity Pathway Analysis). Interestingly, in YAC128 mice exposed to Mn, the top biological network altered after Mn exposure was focused upon the Htt gene and related to “Molecular Transport, Cellular Development, Cellular Growth and Proliferation”. Not surprisingly, there were a greater number of differentially expressed genes included in the WT network (Figure 1E i.) compared to the YAC128 top network (Figure 1E ii.). We sought to validate 9 genes of interest (10 comparisons) differentially expressed in our RNAseq analysis (Supplemental Table 1). These genes were selected due to their role in Mn-biology, HD pathology or degree of fold-change predicted by RNAseq. Only BDNF and IRS2 of the 10 comparisons identified from RNAseq were validated (q<0.05) by qRT-PCR in an independent set of samples. Although unexpected, this is likely the result of a small sample size (n=3 per group) and the difference in sensitivity between RNAseq and qRT-PCR methodology. However, both insulin receptor substrate 2 (Irs2) and Bdnf showed significant differences in expression by Mn in WT, but not in YAC128 mice (Supplemental Figure 1), despite RNAseq analysis identifying decreased Bdnf in YAC128 after Mn exposure (Supplemental Table 1).
Pre-manifest YAC128 mice exhibit a blunted transcriptional and protein response after a single Mn exposure
The 1-week exposure paradigm is likely to identify gene expression changes that are both directly tied to changes in brain Mn levels, as well as those that are indirectly responsive to direct effects of Mn on brain gene expression. Therefore, we sought to determine under more acute conditions, in which Mn-dependent changes are more likely to be directly related to changes in cell Mn levels, if HD suppression of Mn-dependent effects could be observed. Transcriptional and post-transcriptional consequences of a single subcutaneous Mn exposure were thus investigated to better understand how quickly Mn administration can impact Mn-dependent and Mn-responsive biology in the striatum. We report that as early as 1 hour after a single Mn injection, striatal mRNA levels of known Mn-responsive genes Solute Carrier 30 Family member 10 (Slc30a10) and Bdnf are significantly elevated in WT animals while there is no response at any time point in HD mice (Figure 2A and 2B). Consistent with previous literature on BDNF in HD26–28, we have also found that BDNF mRNA levels are significantly lower in the YAC128 striatum compared to WT (Figure 2A). The same single-injection paradigm had no impact on ARG2 protein expression up to 24 hours after Mn exposure (Figure 2C) with only notable changes in protein level after the 1-week exposure (data not included). Similarly, Glutamate Transporter 1 (GLT1) levels were not significantly altered after a single Mn exposure in YAC128 mice; however, WT levels show a small predicted reduction compared to YAC128 animals at 24 hours after Mn exposure in GLT1 (Figure 2D).
figure 2.
striatal mrna and protein changes in pre-manifest mice after a single subcutaneous mn exposure. an 'h' represents hour post-injection. data presented as mean ± sem and * indicates significant mn effect at p<0.05 and ** p<0.01 compared to control, # indicates significant genotype effect at p<0.05 and ## p<0.01; n=3–4. (a) brain-derived neurotrophic factor (bdnf) mrna, (b) solute carrier family 30 member 10 (slc30a10) mrna, (c) arginase 2 (arg2) protein, (d) glutamate transporter 1 (glt1) protein.
Manifest YAC128 mice exhibit reduced basal levels of striatal Mn and fewer changes in striatal metabolites following a 1-week Mn exposure
Previous studies of HD-Mn interaction have focused on this 12- to 13-week old YAC128 pre-manifest HD model. To determine if HD-Mn interaction effects persisted as disease processes progress, we investigated specific phenotypes at later disease manifest stages. To address this question, we exposed 32-week old manifest YAC128 mice to the same 1-week Mn exposure and examined the striatal metabolic and post-transcriptional consequences. In manifest mice, basal striatal Mn levels were lower in YAC128 mice compared to WT providing the first evidence in this HD model for a Mn deficiency phenotype without exogenous Mn exposure. These reduced HD striatal Mn levels were mitigated following a 1-week Mn exposure (Figure 3A). This novel finding may implicate that the impairment in Mn homeostasis in HD changes with disease state; namely that as the HD animals age, the uptake impairment observed in younger animals (Figure 1A) leads to a basal deficiency later in life. Mn administration in these manifest mice does not impact body weight – suggesting no general indication of toxicity (Figure 3B) similar to pre-manifest mice (Figure 1B). Mn-dependent ARG2 protein levels were elevated approximately 3-fold in WT and 2-fold in YAC128 (Figure 3C) with no differences in basal ARG2 levels.
To determine if the HD genotype suppression of global biological processes extends beyond the transcriptome, we performed a metabolomic analysis of HD and WT striatum at manifest stage (32-week old animals). This global analysis of the striatal metabolomic profile is highly similar to the HD-suppressed Mn-induced transcriptional changes in pre-manifest animals; far fewer metabolites are altered in YAC128 (Figure 3D ii.) with Mn exposure compared to WT mice (Figure 3D i.). Volcano plots indicate that the fold-change in metabolite abundance after Mn exposure were similar for WT and YAC128 mice although in WT mice, 103 and 210 metabolites were significantly dysregulated after Mn exposure at q<0.05 and q<0.10, respectively. In contrast, YAC128 mice exposed to 1-week Mn had only 41 and 99 metabolites significantly different detected at q<0.05 and q<0.10, respectively. A chi-square test further confirms that the proportion of significantly different striatal metabolites detected is indeed greater in WT mice (Figure 3E; χ= 14.00, z-statistic= 3.74) and that there are 15 metabolites altered by Mn in both WT (Figure 3D i.) and YAC128 mice (Figure 3D ii.). Pathway analysis of striatal metabolites significantly (q<0.05) altered with Mn indicates that histidine metabolism and β-alanine metabolism are enriched in both WT and YAC128 mice (Figure 3E) although a larger number of metabolites related to these pathways were observed in WT (4 and 3 metabolites, respectively) animals compared to YAC128 (3 and 2 metabolites, respectfully) in these pathways.
Manifest YAC128 mice exhibit similar increases in striatal Mn yet fewer changes in striatal metabolites following 20-week Mn exposure
Finally, we sought to determine if chronic elevations in systemic Mn would reveal HD-Mn interaction effects. Thus, another cohort of manifest mice were examined following chronic exposure to the same 50 mg/kg Mn dose for approximately 20-weeks (beginning at 12 weeks and sacrificed at 32 weeks of age). In this particular cohort of mice, striatal Mn levels were similarly elevated approximately 3-fold in both WT and YAC128 mice (Figure 4A). Since there is no difference in basal striatal Mn levels between WT and YAC128 manifest mice chronically exposed to vehicle, it was unexpected that chronic vehicle exposure would alter striatal Mn relative to the acute Mn exposure (Fig 3A vs 4A). As seen with acute Mn exposures, chronic Mn exposure did not result in a change in body weight for either WT or YAC128 mice (Figure 4B). In fact, Mn exposure slightly accentuated the genotype difference in body weight between WT and YAC128 animals. Striatal ARG2 levels were similarly elevated nearly 4-fold after Mn exposure in both WT and YAC128 mice versus vehicle exposure (Figure 4C). A second interrogation of the global striatal metabolomic profile was performed for mice after a 20-week Mn exposure. Similar to the 1-week exposure data in animals of the same age (Figure 3D–E), we observed that chronic Mn exposure also resulted in a greater number of differentially dysregulated metabolites in WT (n=139, n=246) compared to YAC128 (n=93, n=154) at two different significant levels, q<0.05 and q<0.10 (Figure 4D i. and D ii., respectively). Among the significantly changing metabolites in response to Mn in either genotype, the changes are greater in magnitude in WT compared to YAC128 mice for almost every metabolite (Supplemental Table 2). Among the metabolites altered by Mn at q<0.05, there were 30 significantly altered in both WT and YAC128 (Figure 4D). A chi-square test confirms these findings, in particular the proportion of differentially abundant striatal metabolites is greater in WT mice (χ= 4.60, z-statistic= 2.15). Pathway analysis of these animals indicates that both WT and YAC128 have similar metabolite enrichment profiles as arginine metabolism appears in both analyses (Figure 4E i. and E ii., respectively).
Discussion
As an essential co-factor for several enzymes regulating neuronal health, impairments in Mn handling and utilization may play a part in the pathophysiology of neurodegenerative diseases. Others have investigated the impact of Mn exposure on a wild-type transcriptome29,30 and metabolome in rats31 or cells. Differentially expressed genes following a single injection of a Mn dose that was half the one used here (25 vs 50 mg/kg MnCl2* 4(H2O)) have been reported. Many of the genes altered by Mn related to the dopaminergic system and oxidative stress pathways. In addition, the transcriptome is differentially altered by toxic compared to physiological Mn exposures30. Given the difference detected in striatal Mn uptake (and hypothesized differences in intracellular compartmentalization) between WT and YAC128 mice following our 1-week Mn exposure, this may explain why only a small subset of the same genes were differentially altered by Mn in both genotypes (Figure 1D). Metabolite changes by Mn detected in our studies are similar to others. Specifically, amino acid metabolism was altered by a single intravenous injection of Mn in rats31 and SH-SY5Y cells exposed to both physiological and toxic concentrations of Mn32. While others have determined transcriptomic and metabolomic changes by Mn exposure, the current study is unique in that we investigated how the HD genotype alters the global reponses to Mn exposure across different disease stages and Mn exposure paradigms. These experiments sought to further eluicdiate the mechanisms of the gene-environment interaction between HD and Mn. However, they also reveal the nature of striatal metabolomic and transcriptomic changes to Mn exposure under new and differing conditions.
Mn levels within the brain are most concentrated in the basal ganglia33, which include the specific region that degenerates in HD – the striatum. In HD mouse models, the tissue distribution of Mn is not consistent, so it appears to be dependent upon the different mouse model used and Mn exposure paradigm. It is noteworthy to mention that the effect size of Mn exposure on transcriptional, translational and metabolic markers is largely dependent on the mechanism of exposure. For instance, we utilize a subcutaneous Mn exposure, although dietary and genetic manipulations to alter brain Mn content have also been utilized34,35. Models utilizing dietary or subcutaneous administration exhibit 2–4 fold increases in brain Mn14,36,37 whereas mice with mutations SLC30A10 exhibit approximately 10-fold increases in brain Mn36. Despite the exposure mechanism, a compelling argument for the role of Mn in HD pathology comes from the observed reductions in the level of or activity of many manganoproteins in HD38,39. Our data presented here further support a link between Mn biology and disease pathology at the systemic level of the transcriptome and metabolome by examining the impact of Mn exposure at both pre-manifest and manifest stages of disease. In addition to different disease stages, we also investigated the impact of different exposure paradigms on the interaction between Mn and HD pathophysiology. Our data demonstrate suppression of global Mn-responsive biology in the YAC128 mouse model of HD. Specifically, we observed differences in Mn-responsiveness across different molecular processes governing transcription, protein translation and metabolic function.
In pre-manifest mice exposed to Mn for 1-week, we observed a reduction in the uptake of striatal Mn in YAC128 mice compared to WT animals (Figure 1A), corroborating previous findings by our lab11. There was no basal difference in striatal Mn between WT and pre-manifest age YAC128 mice suggesting that at this stage in disease and under this exposure paradigm, there is an impairment in Mn homeostasis only following Mn exposure, although the exact mechanism is unknown. Furthermore, at this younger age and under this sub-chronic exposure paradigm, Mn elevated the protein expression of the Mn-dependent enzyme Arginase 2 (ARG2) nearly 3-fold in both WT and YAC128 mice. We did not see a reduction in ARG2 levels at baseline in YAC128; however, this might be expected given that Mn predominantly regulates ARG2 activity, not expression14. Additionally, we see no genotype difference in ARG2 levels after exposure, despite a reduction in Mn uptake. This discrepancy may be due to a difference in cellular composition (i.e. increase in astrocytes in HD compared to control)40. Prior work found that the same Mn exposure paradigm rescued elevated levels of striatal arginine, citrulline and ornithine14 suggesting that there are potentially pathogenic metabolic alterations in pre-manifest HD which can be corrected with Mn. In our RNAseq transcriptome analysis, we found that fewer genes were altered by Mn exposure in the striatum of YAC128 (Figure 1D ii.) compared to WT (Figure 1D i.) mice at both q-values of <0.10 and <0.05. Furthermore, the transcriptional effect of Mn on YAC128 appears unique as only 20 mRNAs are differentially expressed in both YAC128 and WT mice (at q<0.05). It is noteworthy to acknowledge that our attempt to validate select transcripts of interest using qRT-PCR was minimally successful—with only 27% of genes replicating our RNAseq results. While this finding is uncommon, we suspect it is largely due to relatively high biological variability between animals, and the relatively small sample size (n=3 for each study arm). However, this study was designed to discover global effects rather than be powered to identify specific single gene changes. Furthermore, although the effect size as it relates to fold-change in our RNAseq data is relatively modest, a previous study examining the impact of Cadmium exposure in mice on mRNA profile reports similar fold-changes41. It is also important to note that even modest changes in these biological pathways can have robust functional consequences. Interestingly, one of the genes that we did validate was brain derived neurotrophic factor (Bdnf) which has been previously shown to be lower in HD42 and reduced after Mn administration43. Pathway analysis of genes differentially expressed after Mn exposure revealed that Bdnf was a central target for mRNAs in WT and related to “Neurological disorders” while Htt was the central target in the top pathway for YAC128 and was most related to “Cellular development and proliferation”. Our pathway analysis provides additional evidence of a global transcriptional response difference to Mn exposure in WT versus YAC128. Specifically, it suggests that Mn significantly impacts BDNF signaling in healthy animals and this, coupled with our qRT-PCR validation of Bdnf mRNA levels indicates that Mn may be a potential modifier of BDNF levels (an established pathological mechanism in HD44). We found that basal levels of Bdnf were lower in the YAC128 striatum compared to WT, as previously reported by us and others, and interestingly that Mn increased Bdnf in WT yet reduced it in YAC128 animals. The interaction between HD and Mn on Bdnf provides additional evidence for the role of Mn homeostasis in HD pathology and has clinically meaningful implications as BDNF protein is reduced in HD patient postmortem striatal tissue42. Given that BDNF is synthesized in the cerebral cortex and transported into the striatum, future studies need to examine the impact of Mn on BDNF in the cortex of these animals. BDNF overexpression has been shown to prevent striatal atrophy in the YAC128 mouse model45. The ability of a naturally occurring supplement, such as Mn, to increase BDNF expression and potentially ameliorate an HD molecular phenotype is worthy of additional investigation.
The 1-week Mn exposure paradigm has been previously shown to elevate brain Mn levels; however, it is unknown how quickly Mn can alter relevant biologic pathways within the striatum. Thus, we investigated the transcriptional impact after a single Mn exposure and observed differences in Mn-responsive genes in the striatum as early as 1-hour post subcutaneous injection in WT animals whereas there were no differences in mRNA levels of two well-known Mn-responsive genes: Bdnf and Solute Carrier Family 30 member 10 (Slc30a10) (Figure 2A). Consistent with previous literature, we confirmed that BDNF levels are significantly lower in the striatum of YAC128 animals compared to WT27. We found that BDNF levels were decreased in WT after Mn exposure up to 12 hours after exposure whereas there was no detectable change in YAC128 mice (Figure 2A). Our observed decrease in BDNF after Mn exposure in WT animals is consistent with previous reports as Guilarte et al demonstrate a reduction in BDNF levels after Mn exposure in primates27. Slc30a10 mRNA increased and remained elevated in WT mice for 24 hours post-exposure while YAC128 had no change within 24 hours (Figure 2B). Slc30a10 is a known systemic Mn efflux transporter34,36,46,47 and has been shown to be elevated after various Mn exposures. The change in expression of both Bdnf and Slc30a10 in WT animals only following Mn exposure further supports the notion that YAC128 mice exhibit a suppressed sensitivity to Mn compared to WT with clearly established protein markers of cellular Mn status. We also investigated the impact of a single Mn exposure on Mn-responsive protein levels in YAC128 mice compared to WT. At the time of these experiments, there was no direct assay to measure SLC30A10 protein in mice and BDNF is notoriously difficult to quantify with standard protein quantification techniques. As previously shown in Figure 1, ARG2 has a robust response to Mn in addition to Glutamate transporter 1 (GLT1), documented previously in cultured astrocytes48 and mouse striatum49. However, in this experiment, ARG2 remained unchanged 24 hours after a single exposure providing greater insight into the time course and Mn exposure required to achieve the protein changes observed after 3-injections. We observed a reduction in GLT1 levels in WT striatum whereas this trend was suppressed in YAC128 mice (Figure 2D). The molecular mechanism for this phenomenon remains unclear, but Aschner et al. suggest that GLT1 levels are mediated through an interaction with select forms of Protein Kinase C50. This reduction in GLT1 levels may be detrimental to neurons because of excitotoxicity in response to impaired synaptic glutamate clearance and as such, we may in future investigate GLT1 levels after lower Mn exposures to evaluate harmful or toxic effects; however, the primary focus of our current work is to demonstrate that Mn biology or at least Mn responsiveness is suppressed in HD.
Finally, we investigated whether the suppression of Mn biology was specific for pre-manifest HD by utilizing manifest mice which consistently exhibit impairments in motor function13,51,52. After a 1-week exposure in manifest mice, we observed a similar increase in striatal Mn after exposure in both WT and YAC128; however, we also detected a difference in basal Mn levels at this age (Figure 3A). Differences in basal levels of striatal Mn has never been documented in HD rodent models and might reflect a change in Mn handling over the course of HD pathology. It may also reflect the change in cellular composition within the striatum with a reduction in neurons and rise in astrogliosis with the progression of HD40. Although Mn is preferentially accumulated within astrocytes and one might anticipate a corresponding rise in Mn, there is also considerable cell death within the striatum which might prevent notable rises in cellular Mn. It should be noted that Mn administration did not appear overtly toxic to either WT or YAC128 mice as indicated by stable body weights (Figure 3B), however specific cell death or morphological changes were not examined in the present study. After Mn exposure, there remains a significant increase in ARG2 expression; however, the fold change appears diminished in the YAC128 mice (Figure 3C) compared to the Mn response in pre-manifest YAC128 mice (Figure 1C). The suppression of Mn-responsive biology in YAC128 animals is also apparent in the global metabolomic signature. For instance, Figure 3E shows that an untargeted metabolomic profile identified significantly more metabolites altered in WT mice compared to YAC128 after Mn exposure at both q-values of <0.10 and <0.05. It is worthwhile to note that the effect size of our Mn exposure on metabolite profile resembles that of mice in previous studies being exposed to Cadmium37 or Lead53 in drinking water. Interestingly, YAC128 mice exposed to a chronic Mn paradigm no longer exhibit uptake deficiencies (Figure 4A) or differences in ARG2 fold-change (Figure 4C). Despite the chronic exposure to Mn, mice maintained their body weight and interestingly, the genotype effect on body weight no longer exists in vehicle-treated manifest mice (Figure 4B). The untargeted metabolomic profile depicted in Figure 4E suggests that chronic exposure in YAC128 (E ii.) results in a suppressed response to Mn compared to WT mice (E i.). The metabolites altered by chronic Mn in YAC128 are largely different from those altered in WT – as there is only a small overlap (20% of WT metabolites; Figure 4E). In addition, of the 139 metabolites altered in WT (at q<0.05), all but 3 of them had a greater fold change in WT than HD (Supplemental Table 2). These findings are further indicative of both a unique and shared metabolic response to Mn in YAC128 mice is suppressed and provides additional evidence to support the role of Mn biology in HD pathology at multiple stages of disease.
Summary
Our work provides evidence of a global interaction between Mn biology and Huntington’s disease pathobiology, versus the pathway selective effects previously reported. We demonstrate a Mn by HD genotype interaction under multiple Mn exposure paradigms in both pre-manifest and manifest disease models. Although there are currently in vivo and in vitro data to support a Mn and HD interaction in the literature, this study provides additional evidence in support of global molecular change of Mn biology in HD. Specifically, subcutaneous administration of Mn impacts both transcription and translation of Mn-dependent and Mn-responsive biology as quickly as one hour post-exposure in WT mice. Additionally, Mn has a more substantial impact on the metabolomic profile in WT compared to HD animals. These immediate molecular consequences are not observed in HD mice suggesting a resistance to or a suppression of normal Mn-responsive or homeostatic mechanisms. Although the exact molecular mechanisms linking Mn biology to HD pathology remain unknown, this study provides evidence that targetting a Mn-BDNF interaction may provide valuable insight into the pathophysiology of HD.
Supplementary Material
Significance to Metallomics.
Manganese (Mn) is an essential metal required for diverse biological processes, and deficiency of this micronutrient is rarely observed. Our lab has previously idenitified a gene-environment interaction between Mn and the inherited neurodegenerative disease Huntington’s disease (HD). Our current work expands this evidence by demonstrating in a mouse model of HD that this mutant genotype suppresses alterations in Mn-dependent and Mn-responsive processes across several Mn exposure lengths and stage of disease. Further investigation of this gene-environment interaction has the potential to reveal underlying mechanisms of both HD pathophysiology and Mn homeostasis.
Acknowledgements
This work was supported by R01 ES016931 (ABB), R01 ES010563 (ABB and MA) and T32 ES007028 (TJVB, ACP, JMW).
Footnotes
Conflicts of Interest
There are no conflicts of interest to declare.
References
- 1.Horning KJ, Caito SW, Tipps KG, Bowman AB and Aschner M, Annu. Rev. Nutr, 2015, 35, 71–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tidball AM, Bryan MR, Uhouse MA, Kumar KK, Aboud AA, Feist JE, Ess KC, Diana Neely M, Aschner M and Bowman AB, Hum. Mol. Genet, 2014, 24, 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan MR, Uhouse MA, Nordham KD, Joshi P, Rose DIR, O’Brien MT, Aschner M and Bowman AB, Neurotoxicology, 2018, 64, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryan MR and Bowman AB, Adv. Neurobiol, 2017, 18, 113–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cersosimo MG and Koller WC, Neurotoxicology, 2006, 27, 340–346. [DOI] [PubMed] [Google Scholar]
- 6.Pfalzer AC and Bowman AB, Curr. Environ. Heal. Reports, 2017, 4, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cicero CE, Mostile G, Vasta R, Rapisarda V, Signorelli SS, Ferrante M, Zappia M and Nicoletti A, Environ. Res, 2017, 159, 82–94. [DOI] [PubMed] [Google Scholar]
- 8.Lee JM, Ramos EM, Lee JH, Gillis T, Mysore JS, Hayden MR, Warby SC, Morrison P, Nance M, Ross CA, Margolis RL, Squitieri F, Orobello S, Di Donato S, Gomez-Tortosa E, Ayuso C, Suchowersky O, Trent RJA, McCusker E, Novelletto A, Frontali M, Jones R, Ashizawa T, Frank S, Saint-Hilaire MH, Hersch SM, Rosas HD, Lucente D, Harrison MB, Zanko A, Abramson RK, Marder K, Sequeiros J, Paulsen JS, Landwehrmeyer GB, Myers RH, MacDonald ME and Gusella JF, Neurology,, DOI: 10.1212/WNL.0b013e318249f683. [DOI] [Google Scholar]
- 9.Walker FO, Lancet, 2007, 369, 218–228. [DOI] [PubMed] [Google Scholar]
- 10.Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA, Graham RK and Hayden MR, Nat. Genet, 1993, 4, 398–403. [DOI] [PubMed] [Google Scholar]
- 11.Williams BB, Li D, Wegrzynowicz M, Vadodaria BK, Anderson JG, Kwakye GF, Aschner M, Erikson KM and Bowman AB, J. Neurochem, 2010, 112, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, Oh R, Bissada N, Hossain SM, Yang YZ, Li XJ, Simpson EM, Gutekunst CA, Leavitt BR and Hayden MR, Hum. Mol. Genet, 2003. [DOI] [PubMed] [Google Scholar]
- 13.Van Raamsdonk JM, Neurosci J.,, DOI: 10.1523/JNEUROSCI.0590-05.2005. [DOI] [Google Scholar]
- 14.Bichell TJV, Wegrzynowicz M, Tipps KG, Bradley EM, Uhouse MA, Bryan M, Horning K, Fisher N, Dudek K, Halbesma T, Umashanker P, Stubbs AD, Holt HK, Kwakye GF, Tidball AM, Colbran RJ, Aschner M, Neely MD, Di Di Pardo A, Maglione V, Osmand A and Bowman AB, Biochim. Biophys. Acta - Mol. Basis Dis, 2017, 1863, 1596–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryan MR, O’Brien MT, Nordham KD, Rose DIR, Foshage AM, Joshi P, Nitin R, Uhouse MA, Di Pardo A, Zhang Z, Maglione V, Aschner M and Bowman AB, Hum. Mol. Genet,, DOI: 10.1093/hmg/ddz209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan MR, Nordham KD, Rose DIR, O’Brien MT, Joshi P, Foshage AM, Gonçalves FM, Nitin R, Uhouse MA, Aschner M and Bowman AB, Mol. Neurobiol,, DOI: 10.1007/s12035-019-01824-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfalzer AC, Wages PA, Porter NA and Bowman AB, J. Huntingtons. Dis,, DOI: 10.3233/JHD-180321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodd CA, Ward DL and Klein BG, Int. J. Toxicol, 2005, 24, 389–397. [DOI] [PubMed] [Google Scholar]
- 19.Liao Y, Smyth GK and Shi W, Bioinformatics,, DOI: 10.1093/bioinformatics/btt656. [DOI] [Google Scholar]
- 20.Love MI, Huber W and Anders S, Genome Biol,, DOI: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R and Siuzdak G, in Therapeutic Drug Monitoring, 2005. [DOI] [PubMed] [Google Scholar]
- 22.Jablonski FSA LA, Powell CJ, Natl. Inst. Stand. Technol [Google Scholar]
- 23.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R and Scalbert A, Nucleic Acids Res,, DOI: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Röllin H, Mathee A, Levin J, Theodorou P and Wewers F, Environ. Res,, DOI: 10.1016/j.envres.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Carroll JB, Deik A, Fossale E, Weston RM, Guide JR, Arjomand J, Kwak S, Clish CB and Macdonald ME,, DOI: 10.1371/journal.pone.0134465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massaro AN, Wu YW, Bammler TK, Comstock B, Mathur A, McKinstry RC, Chang T, Mayock DE, Mulkey SB, Van Meurs K and Juul S, J. Pediatr,, DOI: 10.1016/j.jpeds.2017.10.060. [DOI] [PubMed] [Google Scholar]
- 27.Stansfield KH, Bichell TJ, Bowman AB and Guilarte TR, J. Neurochem,, DOI: 10.1111/jnc.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu C, Li CH, Chen S, Yoo H, Qin X and Park H, Sci. Rep,, DOI: 10.1038/s41598-018-34883-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian Y, Guo S, Chen C, Zhao L, Li Z and Yan Y, Environ. Toxicol. Pharmacol,, DOI: 10.1016/j.etap.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes J, Chandler JD, Lili LN, Uppal K, Hu X, Hao L, Go YM and Jones DP, Front. Genet, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neth K, Lucio M, Walker A, Zorn J, Schmitt-Kopplin P and Michalke B, PLoS One,, DOI: 10.1371/journal.pone.0138270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandes J, Uppal K, Liu KH, Hu X, Go Y-M and Jones D, Free Radic. Biol. Med,, DOI: 10.1016/j.freeradbiomed.2018.10.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen NA, Pakkenberg H, Damsgaard E and Heydorn K, J. Neurol. Sci,, DOI: 10.1016/0022-510X(79)90173-4. [DOI] [PubMed] [Google Scholar]
- 34.Hutchens S, Liu C, Jursa T, Shawlot W, Chaffee BK, Yin W, Gore AC, Aschner M, Smith DR and Mukhopadhyay S, J. Biol. Chem, 2017, 292, 9760–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aschner M, Liu C, Smith DR, Shawlot W, Taylor CA, Mukhopadhyay S, Hutchens S and Jursa T, J. Biol. Chem,, DOI: 10.1074/jbc.ra118.005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor CA, Hutchens S, Liu C, Jursa T, Shawlot W, Aschner M, Smith DR and Mukhopadhyay S, J. Biol. Chem,, DOI: 10.1074/jbc.RA118.005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutchens S, Liu C, Jursa T, Shawlot W, Chaffee BK, Yin W, Gore AC, Aschner M, Smith DR and Mukhopadhyay S, J. Biol. Chem, 2017, 292, 9760–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butterworth J, J. Neurochem,, DOI: 10.1111/j.1471-4159.1986.tb04539.x. [DOI] [Google Scholar]
- 39.Carter CJ, Lancet, 1981. [Google Scholar]
- 40.Gray M, Springer, Singapore, 2019, pp. 355–381. [Google Scholar]
- 41.Go YM, Sutliff RL, Chandler JD, Khalidur R, Kang BY, Anania FA, Orr M, Hao L, Fowler BA and Jones DP, Toxicol. Sci,, DOI: 10.1093/toxsci/kfv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gauthier LR, Charrin BC, Borrell-Pagès M, Dompierre JP, Rangone H, Cordelières FP, De Mey J, MacDonald ME, Leßmann V, Humbert S and Saudou F, Cell,, DOI: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 43.Liang G, Qin H, Zhang L, Ma S, Huang X, Lv Y, Qing L, Li Q, xiong Y, Huang Y, Chen K, Huang Y, Shen Y, Nong J, Yang X and Zou Y, Food Chem. Toxicol, 2015, 83, 261–267. [DOI] [PubMed] [Google Scholar]
- 44.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S and Cattaneo E, Science (80-. ).,, DOI: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 45.Xie Y, Hayden MR and Xu B, J. Neurosci,, DOI: 10.1523/JNEUROSCI.1637-10.2010. [DOI] [Google Scholar]
- 46.Chen P, Bowman AB, Mukhopadhyay S and Aschner M, Worm,, DOI: 10.1080/21624054.2015.1042648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mercadante CJ, Prajapati M, Conboy HL, Dash ME, Herrera C, Pettiglio MA, Cintron-Rivera L, Salesky MA, Rao DB and Bartnikas TB, J. Clin. Invest,, DOI: 10.1172/JCI129710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karki P, Webb A, Smith K, Johnson J, Lee K, Son D-S, Aschner M and Lee E, Mol. Cell. Biol,, DOI: 10.1128/mcb.01176-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pajarillo E, Johnson J, Kim J, Karki P, Son DS, Aschner M and Lee E, Neurotoxicology,, DOI: 10.1016/j.neuro.2017.11.008. [DOI] [Google Scholar]
- 50.Sidoryk-Wegrzynowicz M, Lee E and Aschner M, J. Neurochem,, DOI: 10.1111/j.1471-4159.2012.07835.x. [DOI] [Google Scholar]
- 51.Moreno CL, Ehrlich ME and V Mobbs C, Neurobiol. Dis, 2016, 85, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopes C, Ribeiro M, Duarte AI, Humbert S, Saudou F, Pereira De Almeida L, Hayden M and Rego AC, Mol. Neurobiol, 2014, 49, 1126–1142. [DOI] [PubMed] [Google Scholar]
- 53.Gao B, Chi L, Mahbub R, Bian X, Tu P, Ru H and Lu K, Chem. Res. Toxicol,, DOI: 10.1021/acs.chemrestox.6b00401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.