Abstract
Cell culture processes are used to produce the vast majority of protein therapeutics, valued at over US$180 billion per annum worldwide. For more than a decade now, these processes have become highly productive. To further enhance capital efficiency, there has been an increase in the adoption of disposable apparatus and continuous processing, as well as a greater exploration of in-line sensing, various -omic tools, and cell engineering to enhance process controllability and product quality consistency. These feats in cell culture processing for protein biologics will help accelerate the bioprocess advancements for virus and cell therapy applications.
1. Introduction
Mammalian cells have tremendous biosynthetic potential in producing complex proteins requiring difficult post-translational modifications that cannot be performed by microbial cells. For three decades, we have been extremely successful in harnessing this synthetic potential and have fostered a very large biomanufacturing sector. In this review, we highlight a number of innovations and renovations in cell culture process technology that facilitated the continued success of mammalian cell-based protein therapeutics, from process technology and product quality management, to improved cell lines and the employment of -omic tools and systems approaches (Figure 1). As the next generation of products for cell and gene therapy emerge, these new analytical assays and systems approaches will help shape the manufacturing process for these products to become robust in productivity and product quality.
Figure 1.
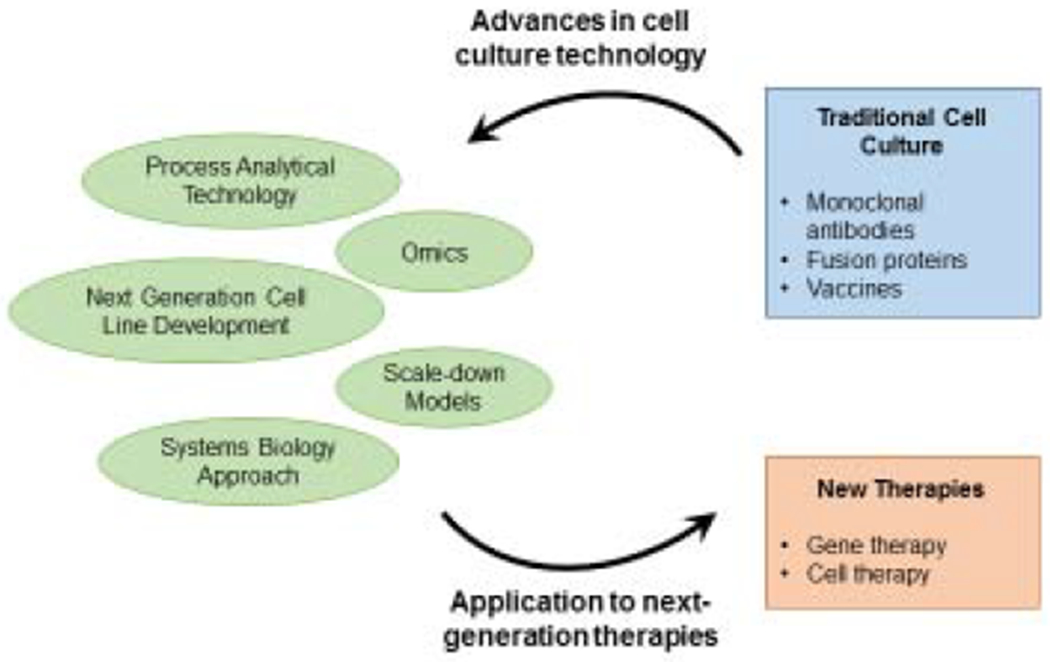
Major areas of advancement for cell culture bioprocessing.
2. Cell Culture Process Technology
A typical cell culture process starts with growing and expanding cells in suspension through a series of reactors of increasing size (also known as the seed train), until a sufficient number of cells have been produced to seed the production reactor. The process is very similar to microbial fermentation except for the longer time scale and the more complex medium needed to meet a mammalian cell’s nutritional requirements; a production reactor run may last more than ten days rather than a day or two in a typical microbial fermentation. During the production process, key process parameters such as dissolved oxygen (DO), pH, and temperature, are monitored in-line, while the concentrations of key metabolites, such as glucose, lactate, and CO2 are measured off-line.
For nearly three decades, most cell culture processes were practiced in fed-batch mode, in which concentrated nutrient feed is added during cell cultivation to extend the production period, allowing cell concentration to reach a high level and for the product to accumulate. In the past two decades, enhanced process technologies have allowed for higher cell densities and product concentrations. For products like IgGs, the titers can be in the 10 g/L range [1], a level unimaginable when antibody products began to take off in mid 1990s. Continuous cell culture, almost invariably coupled with cell retention using an internal or external cell settling device to increase cell concentration and productivity, was for many years relegated to products that are labile to degradation or are produced at very low levels. In the past ten years, there has been increased interest in smaller, disposable reactors and continuous culture, instead of using large stainless-steel based tanks for fed-batch culture.
Disposable bioreactors, in the form of shaking bag or other types of containers, were used primarily for seed train preparation or small-scale production. As the protein product and its process technology became mature, the desire to reduce cost of goods also increased. A production plant employing disposable reactors requires less capital outlay and takes less time to construct than a traditional stainless-steel reactor based facility. It also allows for increased flexibility in production operations. However, disposable stirred tank reactors constructed with plastics are limited in their scalability [2]. To enhance the productivity of a disposable bioreactor, the solution is to operate it in a continuous mode. Adoption of continuous operation was also facilitated by reduced media protein content and reduced membrane fouling, together with the success of cell retention hollowfiber devices with alternating tangential flow (ATF) [3]. Hollow fiber systems used for cell retention are largely operated in laminar flow regions where a “tubular pinch” effect was predicted by fluid flow modeling, leading to larger, rigid spheres being lifted from the walls of the channel while submicron ones move to the membrane surface [4]. Whether tubular pinch occurs in the ATF device is not known as the direction of the axial flow is periodically reversed in the ATF system.
Studies have been performed to model the dynamics of fluid flow through ATF hollow fiber systems. The effect of cellular residence time in both the ATF device and the fluid transfer line on possible oxygen starvation and hydrodynamic damage to cells (via estimated energy dissipation) were evaluated [5]. A computational fluid dynamics (CFD) model was used to evaluate the effects of operating conditions on flux through an ATF hollow fiber and showed the presence of reverse flow across the membrane, a phenomenon known as Starling flow which is thought to reduce fouling [6].
A continuous culture can be operated at a steady state, thus keeping cell’s physiological state constant over time, and possibly delivering more consistent product quality. Most continuous cell culture processes are operated at a relatively slow growth rate compared to the maximal growth rate in a batch culture. At a low growth rate, a smaller cell purge rate can be used to maintain steady operation. Furthermore, a slower growth rate facilitates a switch to a low glycolytic flux state (for review see [7]). A strategy shown to improve perfusion cultures was to slow cell growth by reducing temperature or adding growth inhibitory valeric acid, reducing specific lactate production, increasing specific ammonia production, and increasing monoclonal antibody (mAb) productivity [8].
To maintain a constant cell concentration in a perfusion culture, some have adopted the classical turbidostat strategy, keeping the cell density constant by adjusting the dilution rate, feed nutrient concentration, or cell purge rate. An in-line capacitance probe that measures the viable cell concentration is often used for control [5]. Nonetheless, it is not unusual to see cell concentration and the various growth and metabolic indicators of specific rates fluctuate over a wide range. The complexity of allosteric regulations in cell metabolism allow for multiple metabolic states to exist for a given set of nutrient feed and dilution rate operating conditions. In such complex systems, the steady state that a system reaches is affected by the initial conditions and the culture trajectory. Manipulating the startup culture conditions may lead to different steady states [9]. However, systematic discussion of perfusion culture dynamics and control strategies has not emerged.
3. Product Quality and Process Analytical Technology
For nearly thirty years, the in-line sensors employed in cell culture bioreactors were limited to the traditional ones for pH, DO, and temperature. Chemical analysis was limited to off-line measurement or at-line measurement using an automatic sampling device which is used only in more sophisticated research reactors. Recently, Raman spectroscopy has become a commonly employed in-line sensor for monitoring various nutrient levels since its early application in cell culture processing [10]. The spectra acquired by an in-line Raman spectroscopic sensor are typically subjected to multivariate analysis or chemometrics to determine the concentration of the variable of interest using partial least squares. In the past few years, it has been adopted to monitor the concentrations of glucose, glutamate, glutamine, and ammonia. The concentration range of quantitative measurement is mostly in the ~1 mM range or higher. It has been shown that even with a relatively small number of training datasets, an online glucose measurement model could be established, and a Process Analytical Technology (PAT) controller was developed to keep glucose at a low level (2g/L), simultaneously reducing the level of glycation (i.e. the covalent binding of a glucose molecule to an amino group in a protein) in the product protein [11].
In addition to glycation, the abundance level of other types of structural variants, including glycosylation, disulfide bond scrambling, and various proteolytic cleavages of the product protein, must also be controlled. Like glycation, disulfide bond cleavage and reorganization occur after the product protein molecules are secreted to the medium. This happens in the product recovery stage when the dissolved oxygen level is low, or in the presence of high levels of reductive agents. It was reported that cell lysis from shear damage in depth filtration caused the release of enzymes and NADPH, resulting in disulfide bond cleavage [12]. This was mitigated by keeping the dissolved oxygen level high in the storage bag, thus decreasing the reducing environment in the clarified harvest [12]. Proteolytic cleavage by host cell enzymes may lead to loss of productivity and possible contamination of degradation fragments in the final product as seen in cell lines producing an IgG4-Fc fusion protein. Comparison of transcriptomes of cell lines with varying degree of proteolysis identified furin as the responsible protease, and subsequent use of a furin inhibitor in culture reduced the degradation of the fusion protein [13].
Advances in analytical technology are now enabling the characterization of N-glycans on individual glycosylated peptides of protease digested erythropoietin-Fc fusion protein. The study revealed different glycan profiles, notably the abundance level of sialic acid and fucose, in different glycosylation sites [14]. Furthermore, the spatial distributions changed somewhat in different media and over different days of culture. The increased structural resolution of glycan heterogeneity will extend our understanding of their biological and clinical implications and enhance our capability to control them within an acceptable range. Spatial glycan distributions also attest to the complexity of manipulating glycosylation profiles, through either cell engineering or control of culture conditions. In this same study, o-glycan composition also changed over time and in different media [14]. The small number of o-glycans found on EPO-Fc was consistent with the limited o-glycosylation network predicted in Chinese Hamster Ovary (CHO) cells [15].
The level of host cell proteins (HCP) in the product must also be controlled to an acceptable regulatory level. Some enzymatic activity from residual HCPs may alter product characteristics over long-term storage. Proteomics were employed to identify contaminating HCPs, facilitating their chromatographic removal from the product [16] or knockout of the responsible gene to alleviate the problem [17]. Besides HCPs, endogenous retroviruses (ERVs) which are present in the genome of commonly used host cell lines are of safety and regulatory concern in biomanufacturing. ERVs have cryptic potential to generate infectious viral particles. They may also translocate, activate, or inactivate host cell genes and alter the host cell. Through the identification of integration loci of ERVs in Vero cells, it was shown that the likelihood of retro-translocation after the cell line is established is very low [18].
4. Cell lines
A number of mammalian host cell lines are commonly used for genetic modification to produce a product, including CHO cells, Human embryonic kidney cells (HEK-293), mouse myeloma (NS0), and baby hamster kidney (BHK) cells. However, the vast majority of therapeutic proteins are produced in CHO cells. Cell lines for therapeutic protein production were traditionally constructed by random integration of linearized plasmid DNA containing a gene of interest (GOI) into the host cell genome, followed by selection and amplification of GOI copy number to ensure a high transcript level. Using this approach generates concatemers of the plasmid DNA, either in its entirety or fragments of it, and not all copies of the GOI are active. More recent approaches preserve the integrity of the GOI sequence when inserting it into the host cell genome at random sites or at a chosen locus. Random integration of the GOI in its entirety can be done using transposase systems. Leap-in Transposase has been used to create higher producing pools with lower copy number than random plasmid integration [19], while piggyBac, Tol2, and Sleeping Beauty transposons were used to generate pools and clones with much higher volumetric productivity of TNFR-Fc than those made using plasmid only transfection [20].
Targeted integration approaches direct a GOI to a structurally stable, transcriptionally active location of the genome, with the aim of increasing the throughput and consistency of cell line development. A lentiviral vector was employed to integrate destabilized GFP (dGFP) and IgG along with sequence tags for recombinase mediated cassette exchange (RMCE) into the genome of CHO cells. After identifying a high producing clone with a single copy of the GOI through flowcytometric sorting and establishing the cell line, the RMCE site can be used to swap in a new target gene [21]. Another study utilized random integration to isolate a high producing, stable antibody producing clone, and then used RMCE to remove the antibody gene, creating a host cell line for targeted insertion of a new GOI [22]. CRISPR/Cas9 was later used to generate a landing pad for RMCE in this same site, creating a stable, high producing cell line [23]. In another case, multiple copies of GOI were inserted into a targeted integration host cell line to increase productivity [24].
A production cell line must give consistent productivity, growth characteristics, and product quality over a product’s life cycle. Lacking structural and physiological understanding of these complex traits, the stability of a production cell line has been evaluated empirically, focusing on easily observable phenomena such as productivity and structural chromosomal stability. CHO cells are aneuploid, having a wide range of chromosome number, ranging from fewer than 20 to over 60. A small number of those chromosomes appear normal microscopically, and the rest are aberrant or consist of fused fragments. The karyotypes of CHO cells are inherently variable among different cell lines, and change over time as cells undergo replication [25,26]. This appears to be different from a green monkey kidney cell line, Vero, which is frequently used in viral vaccine production [27]. Whether such karyotypic instability contributes to instability in productivity is not known. Another study showed that the distribution of chromosome number was quickly reestablished after repeated single cell cloning, and one copy of the integrated GOI was repeatedly lost, thus suggesting structural instability in certain genomic regions [28].
With the advances in genome engineering, a variety of newer tools have been applied to engineer host cells. CRISPR interference (CRISPRi) utilizing dead Cas9 has been used to knockdown several genes associated with apoptosis (Bak, Bax, & Casp3) in CHO cells, decreasing caspase activity and increasing viable cell density [29]. CRISPR/Cas9 was used to knockout PKM1 (Pyruvate Kinase Muscle isoform 1), a minor isoform of a glycolytic enzyme in CHO cells, resulting in reduced lactogenic behavior in late stage culture [30]. Overexpression of enzymes in phenylalanine-tyrosine catabolism or knockout of BCAT1 in the branched chain amino acid catabolic pathway reduced the accumulation of inhibitory byproducts in fed-batch culture [31]. Genes predicted to code for endogenous retroviruses in the CHO genome were knocked out, causing a reduction in retroviral RNA present in cell culture supernatant [32]. Host cell engineering will likely become more prevalent, especially with multi-gene manipulation to engineer pathways or traits to create a cell line with ideal metabolic behavior, production capabilities, and product quality characteristics.
Some proteins are difficult to express at high levels for various reasons: toxicity to the cells, complex post-translational modifications or quaternary structure, or instability after secretion, to name a few. Cell engineering and protein engineering are tools that can facilitate the production of such proteins. For example, the membrane proteins which are a potential immunogen for vaccination against respiratory syncytial virus have been engineered and converted to soluble, secreted molecules for production. A highly glycosylated trimer of HIV envelope protein has been produced in CHO cells and is being explored as a candidate HIV vaccine [33]. BMP-4 (bone morphogenetic protein-4), a signaling molecule with potential to treat bone fractures, has low productivity in CHO cells due to its active re-internalization. Competitive inhibition of endocytosis by dextran sulfate increased its productivity [34]. Some difficult-to-express proteins, especially ones that are toxic to cells upon over-expression, such as ion channels, can be produced in a cell free system using CHO cell lysate, resulting in functional, properly folded proteins [35]. For such applications, CHO cells engineered to eliminate competing reactions and enhance the efficiency of expression will be in high demand.
5. Omics and Systems Approaches
In the past few years, genomic, transcriptomic, and some epigenomic assays have become widely applied to investigate different aspects of CHO cells in culture (reviewed in [36]). Sequence analysis of the mitochondrial genome of 22 CHO cell lines showed a large number of sequence variants, but most of them were cell line specific, including many in protein coding genes [37]. Not surprisingly, most variants are heteroplasmic with variant(s) existing in varying proportions within a cell line. Hence, even loss of function mutations are carried in some mitochondria within a cell line. These variants thus may or may not contribute to variability in cell lines or processes. Through transcriptome analysis of multiple high producing clones of two different products, a set of candidate genes associated with high productivity were identified [38]. Subsequent overexpression of various combinations of Erp27 and Erp57, both involved in protein folding and disulfide bond formation, and Foxa1, a transcription factor, resulted in increased productivity [38].
Besides CHO cell-based production of protein therapeutics, other cell culture processes, including virus production for vaccine and gene therapy applications, may benefit from -omic tools. Many cell lines used for vaccine production are of human origin, for which a well annotated genome is available. Two non-human cell lines frequently used in vaccine production, Vero cells and MDCK cells, were derived from African green monkey and dog respectively. The dog genome for many different breeds has long been available, and the genome of Vero cells has also been sequenced [27]. A 9 Mbp region of chromosome 12 was homozygously deleted in Vero cells, as compared to the reference genome of the African green monkey (from which Vero was originally derived). Among the genes lost was the type I interferon gene cluster, making the cells more susceptible to virus infections and more effective for virus production.
Increasingly, we shall be seeing multi-omic studies in cell bioprocessing. Using transcriptomic and metabolomic assays, the reduced supply of UDP-galactose was identified as the possible root cause of a changing glycosylation pattern in late stage culture, and this bottleneck was potentially due to a metabolic shift to a low glycolytic flux state. Supplementation of galactose in the late stage of culture increased the overall galactosylation level [39].
While the analysis of genomic and transcriptomic data requires bioinformatic tools, a model of the metabolic network is necessary to gain insight from metabolomic data. Most metabolic network models are based on stoichiometric balances. Since the system is invariably underdetermined, the calculated fluxes are dependent on the objective function selected and the solution algorithm used [40]. Carbon isotope labeling is used to determine the split of carbon flux between key metabolic branch reactions and to constrain flux analysis solutions [41]. Major strides have been made in this area, and a potentially previously neglected reaction has been identified [42]. Additionally, a genome scale metabolic model has been developed for several CHO host cell lines, which can be used to better understand the effects of different bioprocess treatments and cell line engineering efforts [43]. It is worth noting that the metabolism of mammalian cells is compartmentalized. Without considering compartmentalization, the redox balance and even the carbon flow is skewed. And yet, the compartmentalization for some key reactions in amino acid metabolism and anaplerosis is still not fully understood. These models thus need to be updated as new knowledge emerges. Some have taken a kinetic perspective to model metabolism using a mechanistic kinetic model. A mechanistic kinetic model of glycolysis, the Krebs cycle, and the pentose phosphate pathway was used to identify combinations of gene expression alterations that can rewire glucose metabolism to a low flux state while meeting the constraints stipulated as requirements for growth [44].
A consequence of employing -omic assays is the generally increased parameter space for any problem related to cells. Multiplexing of cell culture experimentation to explore a wider region of parameter space is now routinely practiced in industrial bioprocess development. This type of equipment, for example the ambr system, can be automated for feeding and sampling, and is capable of pH control. These multiplex culture devices have been used to simulate perfusion culture by periodic gravity cell settling and medium exchange [45]. Multiplexing instrumentation is frequently used in scale-down studies that aim to simulate conditions in manufacturing reactors [46]. To harness the power of high throughput experimentation, a systems approach should be taken to integrate bioreactor operation, cell physiology, and growth. When the scale changes, many physical parameters related to the bioreactor, including aeration rate, mechanical stress, and mass transfer rate, also change in different proportions. Changes to physical properties lead to changes in the chemical environment, which in turn alter the cell’s physiology. These physiological changes further modify the chemical environment, forming a feedback loop. A systems model can integrate the physical effects, the chemical environment, and the physiological state of the cells to simulate cell growth, metabolic state, and the reactor environment. It can be a powerful tool for assessing process performance in different bioreactors scales and can assist in the design of experiments that capture critical parameters in scale translation.
6. Looking forward – conclusion
Protein therapeutics currently constitute the largest proportion of cell culture products. Nevertheless, it is worth noting that cell culture processing was rooted in viral vaccines. Adherent cells were adapted to suspension growth seven decades ago for the production of foot and mouth disease virus. The emergence of the SARS-CoV-2 virus has renewed the focus on vaccine technology. In addition to vaccines, the development of viruses as a gene delivery vehicle, and of immune and stem cells for therapeutic use, is accelerating. The current process for production of autologous cell therapies is individualized, as the treatment of a patient requires their own cells. The culture volume required for producing one dose of product is on the order of a few mL for vaccines, and about a liter for therapeutic cells and viral vectors. For personalized autologous cell products such as CAR-T cells, the production scale is thus rather small. Conversely, some vaccines and viral vectors are produced at a similar scale to protein biologics.
For personalized applications, automation to reduce manual steps has drawn development efforts [47]. For cell therapy, vaccine, and viral vector applications, the cell culture process is largely similar to that for the manufacturing of biologics, with emphasis on deploying disposable apparatus, achieving high cell concentration, and for cell therapy applications, keeping the product cells at a high viability and potency state. For these processes, the technologies developed for traditional biologics production are readily adoptable, including devices for high density continuous perfusion and the in-line capacitance and Raman spectroscopic sensors.
In the emerging cell and gene therapy areas, scant public information is available on the process, such as the kinetics of cell growth and product cell quality. For personalized medicine applications, there is a great need to understand the inherent differences in cells from different individuals which may cause their drastically different expansion and clinical potential. For gene vector production processes, one needs better control of the virus production process to minimize the proportion of viral particles which are defective or are void of the recombinant virus genome. Increasing process research efforts to fill these knowledge gaps will expedite the transition from clinical research and production to full-fledged manufacturing.
In closing, the success of protein biologics continues to drive the advancement of process technology, and these process advances in turn fuel the success of the industry. We see opportunities for process innovation in this mutually facilitating cycle. We also see the potential of various -omic technologies and systems designs enabling innovation. Many production processes have accumulated a trove of data, information, and knowledge over the life of the product. These processes are ripe for machine learning for further exploitation of process potential. It goes without saying that systems analysis and design relies on a system model. At the foundation of a system model is the heart of the process, i.e. cell metabolism, synthesis, and growth. Linking biological kinetics to reactor and process dynamics will allow for further process enhancement and provide a framework for developing advanced manufacturing processes for emerging cell technologies.
Acknowledgements
SAO was supported in part by the NIGMS Biotechnology Training Program (T32GM008347-22). This work was supported in part by NIIMBL PC2.1-042.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Huang YM, Hu W, Rustandi E, Chang K, Yusuf-Makagiansar H, Ryll T: Maximizing productivity of CHO cell-based fed-batch culture using chemically defined media conditions and typical manufacturing equipment. Biotechnol Prog 2010, 26:1400–1410. [DOI] [PubMed] [Google Scholar]
- 2.Le TS, McCann M, Azarin SM, Hu W-S: An introduction to mammalian cell culture. 2016. [Google Scholar]
- 3.Bielser JM, Wolf M, Souquet J, Broly H, Morbidelli M: Perfusion mammalian cell culture for recombinant protein manufacturing - A critical review. Biotechnol Adv 2018, 36:1328–1340. [DOI] [PubMed] [Google Scholar]
- 4.Altena FW, Belfort G: Lateral migration of spherical particles in porous flow channels: application to membrane filtration. Chemical Engineering Science 1984, 39:343–355. [Google Scholar]
- 5.Walther J, McLarty J, Johnson T: The effects of alternating tangential flow (ATF) residence time, hydrodynamic stress, and filtration flux on high-density perfusion cell culture. Biotechnol Bioeng 2019, 116:320–332. [DOI] [PubMed] [Google Scholar]
- 6.Radoniqi F, Zhang H, Bardliving CL, Shamlou P, Coffman J: Computational fluid dynamic modeling of alternating tangential flow filtration for perfusion cell culture. Biotechnol Bioeng 2018, 115:2751–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien CM, Mulukutla BC, Mashek DG, Hu W-S: Regulation of Metabolic Homeostasis in Cell Culture Bioprocesses. Trends in Biotechnology 2020:S0167779920300354. [DOI] [PubMed] [Google Scholar]
- 8.Wolf MKF, Closet A, Bzowska M, Bielser JM, Souquet J, Broly H, Morbidelli M: Improved Performance in Mammalian Cell Perfusion Cultures by Growth Inhibition. Biotechnol J 2019, 14:e1700722. [DOI] [PubMed] [Google Scholar]
- 9.Yongky A, Lee J, Le T, Mulukutla BC, Daoutidis P, Hu WS: Mechanism for multiplicity of steady states with distinct cell concentration in continuous culture of mammalian cells. Biotechnol Bioeng 2015, 112:1437–1445. [DOI] [PubMed] [Google Scholar]
- 10.Abu-Absi NR, Kenty BM, Cuellar ME, Borys MC, Sakhamuri S, Strachan DJ, Hausladen MC, Li ZJ: Real time monitoring of multiple parameters in mammalian cell culture bioreactors using an in-line Raman spectroscopy probe. Biotechnol Bioeng 2011, 108:1215–1221. [DOI] [PubMed] [Google Scholar]
- 11.Berry BN, Dobrowsky TM, Timson RC, Kshirsagar R, Ryll T, Wiltberger K: Quick generation of Raman spectroscopy based in-process glucose control to influence biopharmaceutical protein product quality during mammalian cell culture. Biotechnol Prog 2016, 32:224–234. [DOI] [PubMed] [Google Scholar]
- 12.O’Mara B, Gao ZH, Kuruganti M, Mallett R, Nayar G, Smith L, Meyer JD, Therriault J, Miller C, Cisney J, et al. Impact of depth filtration on disulfide bond reduction during downstream processing of monoclonal antibodies from CHO cell cultures. Biotechnol Bioeng 2019, 116:1669–1683. [DOI] [PubMed] [Google Scholar]; Reported that the release of certain NADPH dependent enzymes from lysed cells after depth filtration led to disulfide bond cleavage in the antibody product. Increasing oxygen content in the gas phase of the storage bag for the clarified harvest reduced antibody reduction over several days.
- 13.Clarke C, Gallagher C, Kelly RM, Henry M, Meleady P, Frye CC, Osborne MD, Brady CP, Barron N, Clynes M: Transcriptomic analysis of IgG4 Fc-fusion protein degradation in a panel of clonally-derived CHO cell lines using RNASeq. Biotechnol Bioeng 2019, 116:1556–1562. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Yang G, Wang T, Yang W, Betenbaugh MJ, Zhang H: Characterization of intact glycopeptides reveals the impact of culture media on site-specific glycosylation of EPO-Fc fusion protein generated by CHO-GS cells. Biotechnol Bioeng 2019, 116:2303–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]; N-glycan profiles on four glycosylation sites in EPO-Fc were identified by intact glycopeptide analysis using LC-MS/MS. Both N- and O-glycan distributions over culture time and in different media were determined.
- 15.Le T, O’Brien C, Gupta U, Sousa G, Daoutidis P, Hu WS: An integrated platform for mucin-type O-glycosylation network generation and visualization. Biotechnol Bioeng 2019, 116:1341–1354. [DOI] [PubMed] [Google Scholar]
- 16.Lavoie RA, di Fazio A, Williams TI, Carbonell R, Menegatti S: Targeted capture of Chinese hamster ovary host cell proteins: Peptide ligand binding by proteomic analysis. Biotechnol Bioeng 2020, 117:438–452. [DOI] [PubMed] [Google Scholar]
- 17.Chiu J, Valente KN, Levy NE, Min L, Lenhoff AM, Lee KH: Knockout of a difficult-to-remove CHO host cell protein, lipoprotein lipase, for improved polysorbate stability in monoclonal antibody formulations. Biotechnol Bioeng 2017, 114:1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakuma C, Sekizuka T, Kuroda M, Kasai F, Saito K, Ikeda M, Yamaji T, Osada N, Hanada K: Novel endogenous simian retroviral integrations in Vero cells: implications for quality control of a human vaccine cell substrate. Sci Rep 2018, 8:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balasubramanian S, Peery RB, Minshull J, Lee M, White R, Kelly RM, Barnard GC: Generation of High Expressing Chinese Hamster Ovary Cell Pools Using the Leap-In Transposon System. Biotechnol J 2018, 13:e1700748. [DOI] [PubMed] [Google Scholar]
- 20.Balasubramanian S, Rajendra Y, Baldi L, Hacker DL, Wurm FM: Comparison of three transposons for the generation of highly productive recombinant CHO cell pools and cell lines. Biotechnol Bioeng 2016, 113:1234–1243. [DOI] [PubMed] [Google Scholar]
- 21.O’Brien SA, Lee K, Fu HY, Lee Z, Le TS, Stach CS, McCann MG, Zhang AQ, Smanski MJ, Somia NV, et al. Single Copy Transgene Integration in a Transcriptionally Active Site for Recombinant Protein Synthesis. Biotechnol J 2018, 13:e1800226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Inniss MC, Han S, Moffat M, Jones H, Zhang B, Cox WL, Rance JR, Young RJ: Recombinase-mediated cassette exchange (RMCE) for monoclonal antibody expression in the commercially relevant CHOK1SV cell line. Biotechnol Prog 2015, 31:1645–1656. [DOI] [PubMed] [Google Scholar]
- 23.Inniss MC, Bandara K, Jusiak B, Lu TK, Weiss R, Wroblewska L, Zhang L: A novel Bxb1 integrase RMCE system for high fidelity site-specific integration of mAb expression cassette in CHO Cells. Biotechnol Bioeng 2017, 114:1837–1846. [DOI] [PubMed] [Google Scholar]; Used CRISPR/Cas9 to insert a landing pad flanked by recombination sites into a genomic locus previously found to be stable. Integrated monoclonal antibody genes using cassette exchange, generating clones with high specific productivity.
- 24.Carver J, Ng D, Zhou M, Ko P, Zhan D, Yim M, Shaw D, Snedecor B, Laird MW, Lang S, et al. Maximizing antibody production in a targeted integration host by optimization of subunit gene dosage and position. Biotechnol Prog 2020:e2967. [DOI] [PubMed] [Google Scholar]; Utilized recombinase mediated cassette exchange to integrate multiple copies of IgG into a targeted integration host cell with a defined landing pad. Increased copy number at a single locus improved specific productivity and may help overcome some limitations with single copy targeted integration.
- 25.Vcelar S, Jadhav V, Melcher M, Auer N, Hrdina A, Sagmeister R, Heffner K, Puklowski A, Betenbaugh M, Wenger T, et al. Karyotype variation of CHO host cell lines over time in culture characterized by chromosome counting and chromosome painting. Biotechnol Bioeng 2018, 115:165–173. [DOI] [PubMed] [Google Scholar]
- 26.Vcelar S, Melcher M, Auer N, Hrdina A, Puklowski A, Leisch F, Jadhav V, Wenger T, Baumann M, Borth N: Changes in Chromosome Counts and Patterns in CHO Cell Lines upon Generation of Recombinant Cell Lines and Subcloning. Biotechnol J 2018, 13:e1700495. [DOI] [PubMed] [Google Scholar]
- 27.Osada N, Kohara A, Yamaji T, Hirayama N, Kasai F, Sekizuka T, Kuroda M, Hanada K: The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res 2014, 21:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandyopadhyay AA, O’Brien SA, Zhao L, Fu HY, Vishwanathan N, Hu WS: Recurring genomic structural variation leads to clonal instability and loss of productivity. Biotechnol Bioeng 2019, 116:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong K, Marquart KF, la Cour Karottki KJ, Li S, Shamie I, Lee JS, Gerling S, Yeo NC, Chavez A, Lee GM, et al. Reduced apoptosis in Chinese hamster ovary cells via optimized CRISPR interference. Biotechnol Bioeng 2019, 116:1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang D, Subramanian J, Haley B, Baker J, Luo L, Hsu W, Liu P, Sandoval W, Laird MW, Snedecor B, et al. Pyruvate Kinase Muscle-1 Expression Appears to Drive Lactogenic Behavior in CHO Cell Lines, Triggering Lower Viability and Productivity: A Case Study. Biotechnol J 2019, 14:e1800332. [DOI] [PubMed] [Google Scholar]
- 31.Mulukutla BC, Mitchell J, Geoffroy P, Harrington C, Krishnan M, Kalomeris T, Morris C, Zhang L, Pegman P, Hiller GW: Metabolic engineering of Chinese hamster ovary cells towards reduced biosynthesis and accumulation of novel growth inhibitors in fed-batch cultures. Metab Eng 2019, 54:54–68. [DOI] [PubMed] [Google Scholar]
- 32.Duroy PO, Bosshard S, Schmid-Siegert E, Neuenschwander S, Arib G, Lemercier P, Masternak J, Roesch L, Buron F, Girod PA, et al. Characterization and mutagenesis of Chinese hamster ovary cells endogenous retroviruses to inactivate viral particle release. Biotechnol Bioeng 2020, 117:466–485. [DOI] [PMC free article] [PubMed] [Google Scholar]; Utilized genome sequencing and RNAseq to identify expressed endogenous retrovirus (ERV) sequences in CHO that are secreted in retroviral-like particles. CRISPR/Cas9 was then used to knockout an essential gene for retroviral budding, reducing RNA-containing ERV particles in the cell culture supernatant.
- 33.Dey AK, Cupo A, Ozorowski G, Sharma VK, Behrens AJ, Go EP, Ketas TJ, Yasmeen A, Klasse PJ, Sayeed E, et al. cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate. Biotechnol Bioeng 2018, 115:885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim CL, Jung MY, Kim YS, Jang JW, Lee GM: Improving the production of recombinant human bone morphogenetic protein-4 in Chinese hamster ovary cell cultures by inhibition of undesirable endocytosis. Biotechnol Bioeng 2018, 115:2565–2575. [DOI] [PubMed] [Google Scholar]
- 35.Thoring L, Dondapati SK, Stech M, Wustenhagen DA, Kubick S: High-yield production of “difficult-to-express” proteins in a continuous exchange cell-free system based on CHO cell lysates. Sci Rep 2017, 7:11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stolfa G, Smonskey MT, Boniface R, Hachmann AB, Gulde P, Joshi AD, Pierce AP, Jacobia SJ, Campbell A: CHO-Omics Review: The Impact of Current and Emerging Technologies on Chinese Hamster Ovary Based Bioproduction. Biotechnol J 2018, 13:e1700227. [DOI] [PubMed] [Google Scholar]
- 37.Kelly PS, Clarke C, Costello A, Monger C, Meiller J, Dhiman H, Borth N, Betenbaugh MJ, Clynes M, Barron N: Ultra-deep next generation mitochondrial genome sequencing reveals widespread heteroplasmy in Chinese hamster ovary cells. Metab Eng 2017, 41:11–22. [DOI] [PubMed] [Google Scholar]
- 38.Berger A, Le Fourn V, Masternak J, Regamey A, Bodenmann I, Mermod N: Overexpression of transcription factor Foxa1 and target genes remediates therapeutic protein production bottlenecks in Chinese hamster ovary cells. Biotechnol Bioeng 2020, 20:20. [DOI] [PMC free article] [PubMed] [Google Scholar]; Used RNAseq to identify differentially expressed genes between high and low producing CHO cells, and overexpressed several candidate genes that were upregulated in high producing cells, resulting in increased VCD and titer.
- 39.Sumit M, Dolatshahi S, Chu AA, Cote K, Scarcelli JJ, Marshall JK, Cornell RJ, Weiss R, Lauffenburger DA, Mulukutla BC, et al. Dissecting N-Glycosylation Dynamics in Chinese Hamster Ovary Cells Fed-batch Cultures using Time Course Omics Analyses. iScience 2019, 12:102–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang Z, Lee DY, Yoon S: Quantitative intracellular flux modeling and applications in biotherapeutic development and production using CHO cell cultures. Biotechnol Bioeng 2017, 114:2717–2728. [DOI] [PubMed] [Google Scholar]
- 41.Antoniewicz MR: A guide to (13)C metabolic flux analysis for the cancer biologist. Exp Mol Med 2018, 50:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn WS, Crown SB, Antoniewicz MR: Evidence for transketolase-like TKTL1 flux in CHO cells based on parallel labeling experiments and (13)C-metabolic flux analysis. Metab Eng 2016, 37:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hefzi H, Ang KS, Hanscho M, Bordbar A, Ruckerbauer D, Lakshmanan M, Orellana CA, Baycin-Hizal D, Huang Y, Ley D, et al. A Consensus Genome-scale Reconstruction of Chinese Hamster Ovary Cell Metabolism. Cell Syst 2016, 3:434–443 e438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Brien C, Allman A, Daoutidis P, Hu WS: Kinetic model optimization and its application to mitigating the Warburg effect through multiple enzyme alterations. Metab Eng 2019, 56:154–164. [DOI] [PubMed] [Google Scholar]
- 45.Sewell DJ, Turner R, Field R, Holmes W, Pradhan R, Spencer C, Oliver SG, Slater NK, Dikicioglu D: Enhancing the functionality of a microscale bioreactor system as an industrial process development tool for mammalian perfusion culture. Biotechnol Bioeng 2019, 116:1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandner V, Pybus LP, McCreath G, Glassey J: Scale-Down Model Development in ambr systems: An Industrial Perspective. Biotechnol J 2019, 14:e1700766. [DOI] [PubMed] [Google Scholar]
- 47.Vormittag P, Gunn R, Ghorashian S, Veraitch FS: A guide to manufacturing CAR T cell therapies. Curr Opin Biotechnol 2018, 53:164–181. [DOI] [PubMed] [Google Scholar]


