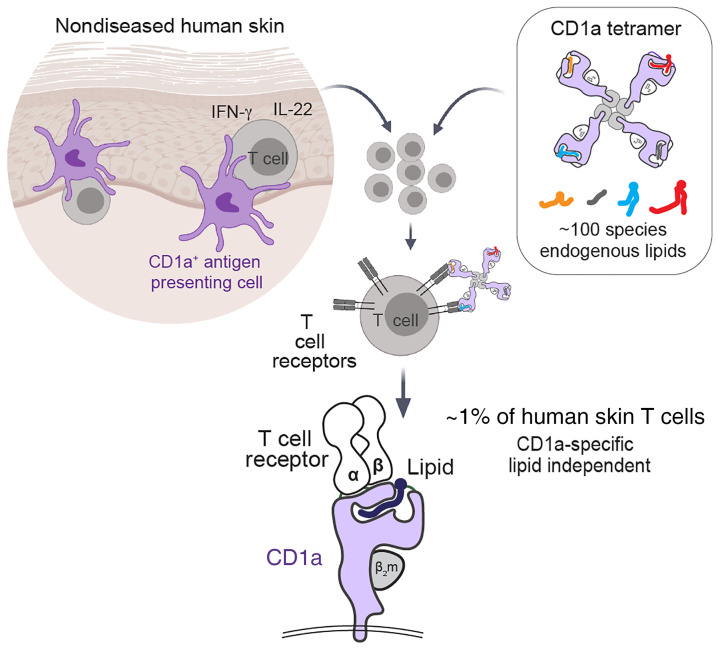
Abstract
CD1a-autoreactive T cells contribute to skin disease, but the identity of immunodominant self-lipid antigens and their mode of recognition are not yet solved. In most models, MHC and CD1 proteins serve as display platforms for smaller antigens. Here, we showed that CD1a tetramers without added antigen stained large T cell pools in every subject tested, accounting for approximately 1% of skin T cells. The mechanism of tetramer binding to T cells did not require any defined antigen. Binding occurred with approximately 100 lipid ligands carried by CD1a proteins, but could be tuned upward or downward with certain natural self-lipids. TCR recognition mapped to the outer A′ roof of CD1a at sites remote from the antigen exit portal, explaining how TCRs can bind CD1a rather than carried lipids. Thus, a major antigenic target of CD1a T cell autoreactivity in vivo is CD1a itself. Based on their high frequency and prevalence among donors, we conclude that CD1a-specific, lipid-independent T cells are a normal component of the human skin T cell repertoire. Bypassing the need to select antigens and effector molecules, CD1a tetramers represent a simple method to track such CD1a-specific T cells from tissues and in any clinical disease.
Keywords: Dermatology, Immunology
Keywords: Antigen presentation, Skin, T cells
Introduction
CD1a is an MHC class I–like protein that binds lipids in its hydrophobic cleft and stimulates T cells via T cell receptors (TCRs) (1–5). CD1-restricted autoreactive T cell clones were first derived from blood (6). More recently, pools of CD1a-autoreactive cells were found among circulating T cells expressing skin-homing receptors like cutaneous lymphocyte antigen (CLA), CCR4, CCR6, and CCR10 (7, 8), suggesting that they might be tropic for the skin. High expression of CD1a proteins on epidermal Langerhans cells suggests that CD1a-reactive T cells might play a role in cutaneous immune responses (1). Further, functional studies of anti-CD1a blockable cytokine responses from skin-derived T cells show that Th1 and Th2 T cell types enter the skin, and in many cases show higher response in patients with allergic dermatitis or psoriasis (9–13). In prior studies of CD1a-induced T cell activation, the identity of the lipid antigen presented by CD1a was not known, although lipid-dependent reactivity was suggested by the fact that phospholipases and lysophospholipids were required to trigger responses (9, 10). A role for CD1a-mediated cutaneous immune responses was further supported by findings that transgenic mice expressing human CD1a show increased skin inflammation in response to certain small molecules or lipids, including the topical immune modulator imiquimod and the poison ivy antigen urushiol (13). Recently, lipidic and nonlipidic contact allergens present in skin creams, such as balsam of Peru and farnesol, have been shown to activate CD1a-dependent T cell clones (14). However, any broadly acting, immunodominant antigen for CD1a, comparable to α-galactosylceramide for CD1d and NKT cells (15), remains unknown.
A fundamental difference between the MHC system for peptide presentation and the CD1 system for lipid presentation is that the former is the most polymorphic locus in the human genome, whereas CD1 genes are nearly monomorphic (16). The expression of almost identical CD1 proteins by all humans offers practical advantages for the basic study and potential therapeutic manipulation of such donor-unrestricted T cells. Human cohorts do not need to be MHC typed. Experiments can be accomplished with allogeneic or “off-the-shelf” antigen-presenting cells (APCs). One form of CD1 tetramer can be used for T cells from any donor (17).
Despite these advantages, there are currently no cell surface markers that reliably distinguish CD1a-autoreactive T cells from conventional MHC-restricted T cells, making it difficult to isolate and selectively study these cells. Current approaches to studying CD1a responses among polyclonal skin T cells focus on T cell activation–based assays, but this approach requires a priori selection of the antigens and the cytokines to be measured. T cell clones recognize squalene and related skin oils as CD1a-dependent self-antigens (18), but data do not currently address whether these antigens, balsam of Peru, or farnesol (14) might be broadly acting such that they can query the human CD1a-autoreactive T cell repertoire generally. The cytokine profiles produced by CD1a-autoreactive T cells are diverse (9–13, 19), and any single activation marker or cytokine that could broadly detect CD1a-autoreactive T cells remains to be identified. ELISPOT-based activation assays used to detect CD1a-dependent responses also destroy the cells under study, and T cells recovered from tissue are typically limited in number for capture and further study.
Therefore, we employed cell surface cytokine capture and CD1a tetramers to select T cells expressing CD1a-reactive or CD1a-binding TCRs in human skin. CD1a tetramers were previously validated using a foreign bacterial lipopeptide (20), so our studies represent, to our knowledge, the first use of CD1a tetramers with self-antigens or in the study of T cells from human tissues. Whereas a prior link of CD1a autoreactivity to IL-22 was based mainly on altered cytokine RNA increases, which are blocked by anti-CD1a (7), here we found that untreated CD1a tetramers carrying diverse endogenous lipids directly identified large numbers of CD4+ IL-22–producing CD1a-reactive T cells on a single-cell basis in healthy human skin. Lipidomic analysis of CD1a ligands and mutational analyses of CD1a showed that CD1a-restricted TCRs recognized CD1a itself in the presence of many types of self-lipids. Nearly all prior studies of T cell responses emphasize specific antigen recognition as the trigger for T cell activation. We demonstrated that the CD1a-specific TCRs had an intrinsic affinity for CD1a; the lipids loaded into the antigen cleft of CD1a either permitted or partially interfered with this interaction. This system allowed a nonpolymorphic antigen-presenting molecule, CD1a, to provide on-site and nuanced sensing by T cells of the skin environment.
Results
CD1a-autoreactive IL-22 responses in normal human skin.
The isolation of T cells in large numbers and at high purity from human skin was required to identify a putative CD1a-autoreactive T cell population of unknown frequency and antigen specificity. Conventional methods rely on tissue cutting, physical disaggregation, collagenases, and chelating agents to release T cells from tissues. These treatments damage cells and yield relatively low numbers, limiting downstream analyses (21, 22). An alternative outgrowth method takes advantage of the migratory tendency of skin T cells and enables the recovery of pure skin T cells (23, 24). Skin explants were cultured on collagen-seeded, 3D growth matrices (3D method) for 21–28 days, allowing T cells to emigrate in response to chemoattractants from skin fibroblasts and supplemental IL-2 and IL-15. Certain aspects of prior validation (23, 24) were relevant for the current study. IL-2 and IL-15 were chosen because they promote survival but are not strong T cell lineage–biasing cytokines. Conventional in vitro culture in the absence of collagen matrix results in phenotypic changes, including rapid downregulation of CLA-related addressins, but the combination of collagen matrix and fibroblasts preserves CLA expression on T cells. As compared with T cells directly isolated from skin, 3D culture–derived skin T cells have similar rates of CCR4, CCR6, and CCR8 expression. Another goal was to identify the targets of antigen-specific TCRs. Although 1 round of cytokine-mediated expansion might have caused loss of relevant TCRs, prior TCR spectratyping indicates a lack of detectable narrowing of the TCR repertoire (23, 24).
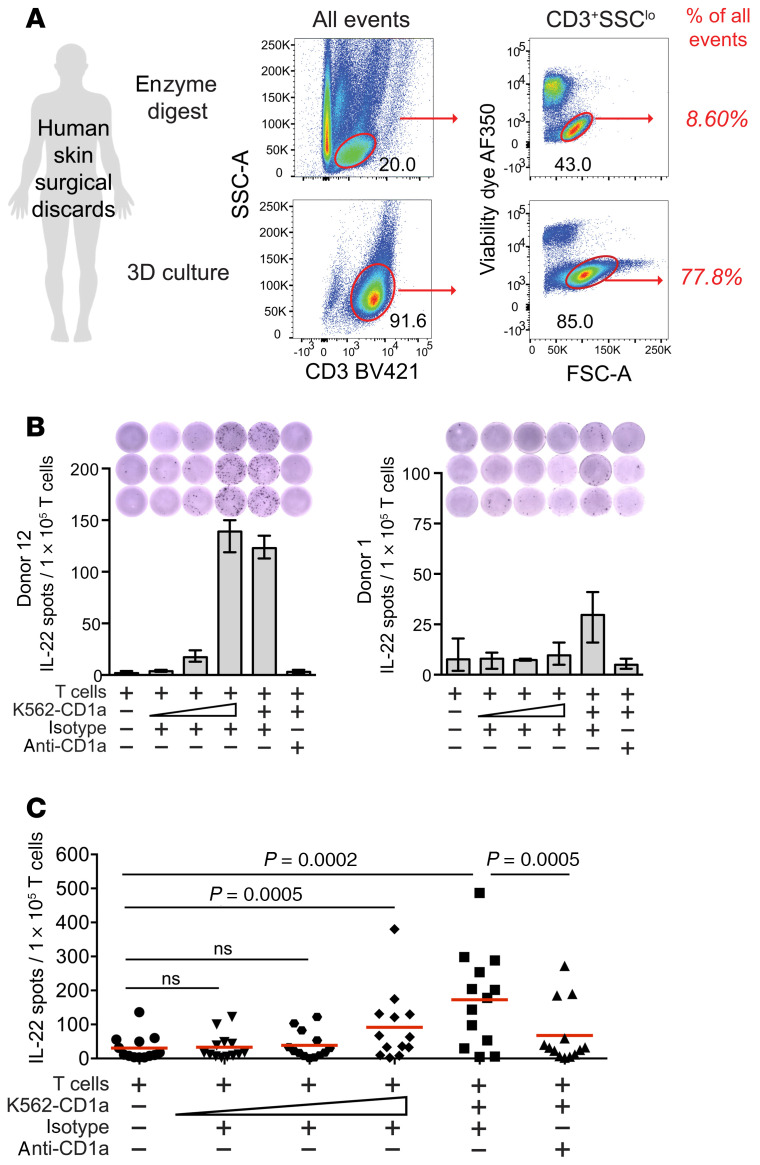
Studies have shown that this method increases T cell yield and quality compared with physical disruption methods (23, 24), and we routinely captured 107 or more pure T cells from skin explants. We compared T cell purity in a skin sample split between collagenase digestion and 3D culture, finding that the 3D method gave less subcellular debris and a 9-fold higher lymphocyte purity based on events entering live-dead stain exclusion, CD3+, and lymphocyte size and granularity gates (Figure 1A). We employed the 3D method to recover T cells from surgical discarded skin from 25 unrelated donors for use in CD1a-dependent response assays, CD1 tetramer staining, or both (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI140706DS1).
Figure 1. CD1a-dependent IL-22 responses among human skin T cells.
(A) Human skin T cell collection recovered by a collagenase and DNAse digestion method at day 1 is shown in comparison to T cells collected at day 21 of 3D culture from the same donor. (B) After coculture with allogeneic in vitro–derived Langerhans-like cells plus HLA-blocking antibodies (W632, L243), polyclonal skin T cells (n = 13, 1 × 105/well) were tested in IL-22 ELISPOT assays for response to increasing numbers of CD1a-expressing K562 cells (400, 2,000, 10,000, or 50,000) and anti-CD1a (OKT6, 10 μg/mL) or isotype control (P3, 10 μg/mL) overnight. Two donors representing an intermediate and a low response pattern are shown. Error bars indicate mean with range of technical triplicates. (C) Summary data are from 13 donors, where each point represents the mean of technical triplicates from a single donor. P values were calculated using the Wilcoxon matched-pairs signed-rank test.
In general, IL-22–producing T cells are enriched among circulating CD4+ T cells that express skin-homing receptors (25, 26), and Th22 cells are more frequent in human skin compared with other tissue sites (27, 28). Although these CD4+ IL-22–secreting cells might recognize MHC II, formally demonstrating MHC restriction at the polyclonal level among genetically unrelated donors has been difficult. One study implicated CD1a as a target, based on high rates of CD1a-dependent IL-22 mRNA expression by blood-derived T cells that expressed skin-homing receptors, and the dominance of IL-22 signal when compared with other T cell lineage–defining cytokines (7). However, the actual homing of CD1a-autoreactive IL-22–producing T cells from blood to skin, and their contribution to the total skin T cell pool, are unknown. Therefore, we tested polyclonal skin T cells from 13 healthy donors by IL-22 ELISPOT using CD1a-expressing K562 (K562-CD1a) APCs in the presence of CD1a-blocking antibody (OKT6) or an isotype-matched control (P3). As shown in 2 representative donors, IL-22 responses were higher in response to K562-CD1a cells compared with baseline T cells alone, and stronger responses were seen with increasing numbers of K562-CD1a cells. As a second criterion for restriction, IL-22 secretion was blocked by anti-CD1a (Figure 1B).
The high absolute number of skin T cells obtained by 3D culture enabled triplicate measurements on a per donor basis (Figure 1C, n = 13), showing highly significant responses to K562-CD1a (P = 0.0002) and blockade of responses with anti-CD1a (P = 0.0005). We calculated precursor frequency as the difference in IL-22 spots appearing after addition of K562-CD1a cells versus T cells alone and as the decrement in spots blocked by anti-CD1a versus isotype control. Both measurements gave similar estimates across the cohort of 13 patients, placing the frequency of CD1a-autoreactive T cells at 0.10%–0.14% of skin T cells. This frequency is high for a single molecularly defined T cell epitope when measured without antigen-driven expansion. Previous studies have placed the frequency at 10–100 cells per million (29–31), similar to that of human type I NKT cells in the blood (32, 33). Thus, CD1a-autoreactive IL-22–producing T cells colonize human skin, fulfilling prior predictions based on skin-homing receptors present on circulating CD1a-restricted T cells (18).
Capture of polyclonal IL-22+ CD1a-autoreactive T cells from skin.
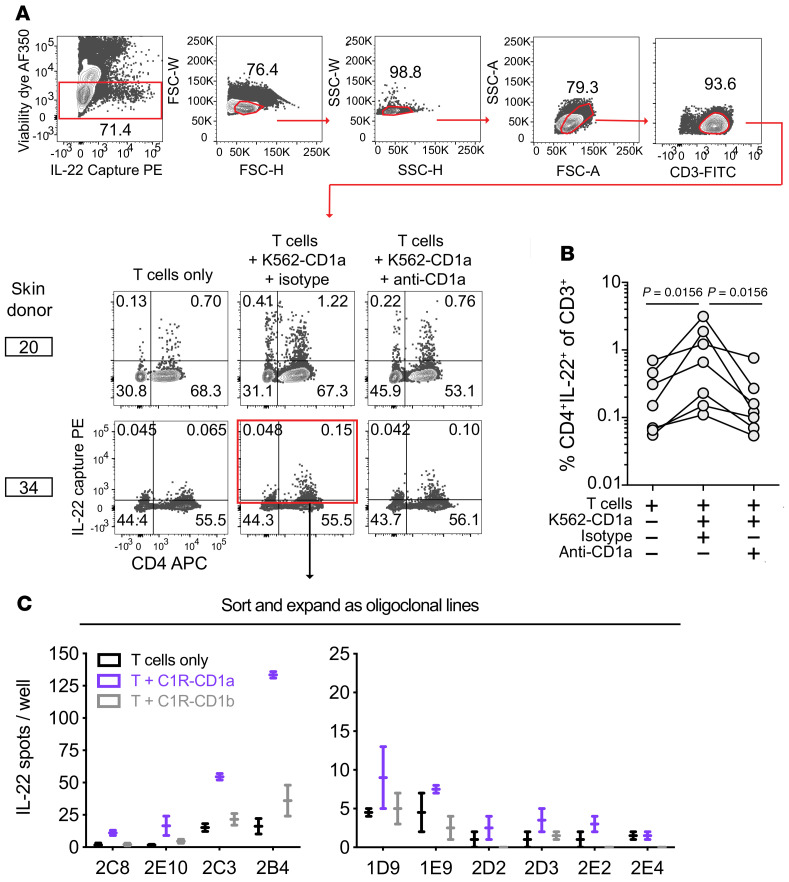
Because the destructive nature of ELISPOT studies did not allow capture and study of viable cells, we next employed IL-22 cell surface cytokine capture to detect and sort live CD1a-autoreactive cells. Using a bifunctional antibody specific for CD45 and IL-22 to preferentially coat cells that have secreted IL-22, we measured CD1a-dependent skin T cell responses from an additional 7 donors. In all donors tested, we detected elevated frequencies of IL-22–secreting T cells in response to K562-CD1a cells versus T cells alone or versus K562-CD1a cells pretreated with anti-CD1a (Figure 2, A and B). When calculated using the T cell–only condition as background, the median donor frequency was 0.35% of CD3+ T cells. When calculated using the CD1a-blocked condition as background, the median frequency was 0.46% (Figure 2B). Thus, the absolute frequencies were similar to but somewhat higher than those detected by IL-22 ELISPOT (Figure 1). Further, although we did observe IL-22–producing cells that were CD4–, the majority of IL-22–producing cells detected in all donors were CD4+ (Figure 2, A and B). Taken together, 20 individuals tested by IL-22 ELISPOT or surface capture demonstrated the clear presence of IL-22–producing CD1a-autoreactive T cells in human skin.
Figure 2. IL-22 surface capture selects CD1a-autoreactive skin T cells.
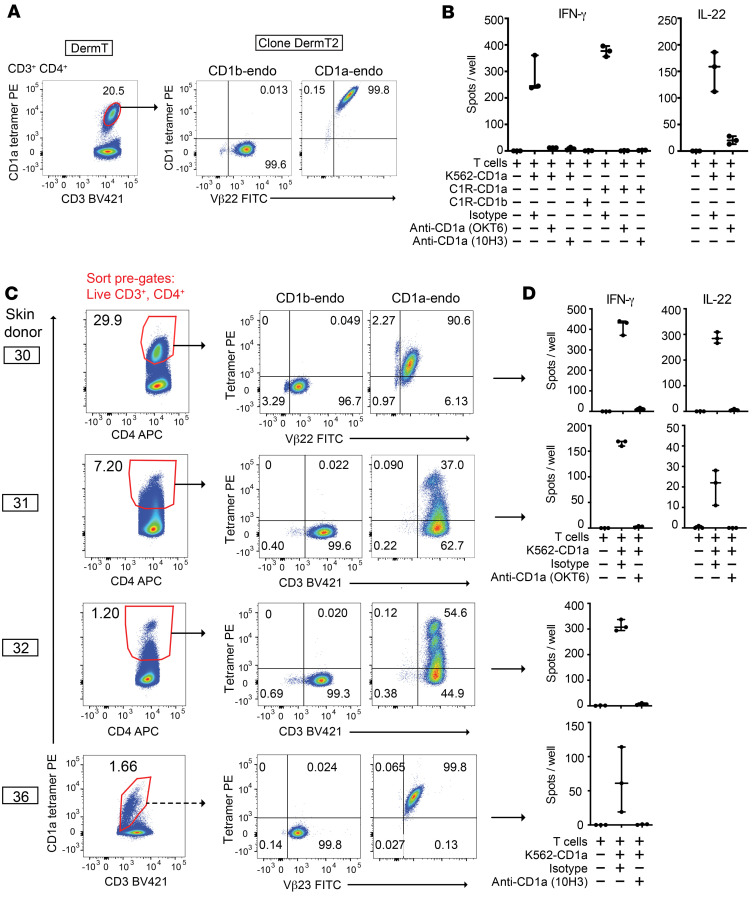
(A) Polyclonal skin T cells cultured with K562 cells overnight were labeled with the IL-22 capture reagent and pregated as shown for donor 20. (B) Frequencies of CD4+ IL-22+ cells among CD3+ cells (top right quadrant gate as in A; n = 7 donors). (C) After oligoclonal expansion of donor 34–sorted cells (red box indicates sort gate including CD4neg IL-22lo cells), lines were screened by IL-22 ELISPOT on C1R cells. Data from 10 lines shown as mean with range of technical duplicates.
Cytokine capture to generate T cell lines.
We used IL-22 cytokine capture assays to isolate CD1a-responsive T cells from skin and generated T cell lines with the goal of making durable reagents to validate CD1a specificity and for molecular analyses of antigen responses and TCR expression. From donor 34, we set a broad sort gate to include all IL-22–producing cells (Figure 2A, red). We plated 60 sorted cells per well for expansion to oligoclonal lines. We considered the possibility that multiple rounds of activation on the K562 cell line, which expresses trace MHC class I, might lead to expansion and activation of alloreactive CD1a-nonspecific T cells or expansion of NK cells (34). Since cells had already been selected based on response to CD1a expressed on K562, we used a second CD1 cellular expression system on C1R cells for secondary testing. Among 64 lines tested for response to C1R-CD1a and C1R-CD1b cells, we selected 10 short-term T cell lines that had detectable IL-22 responses to C1R-CD1a greater than those seen with C1R-CD1b (Figure 2C). The remainder of cultures showed low absolute response to C1R cells regardless of CD1 expression, suggesting that weak alloreactivity or residual NK cell response were not confounding the results.
CD1 tetramer assays for autoreactive T cells from tissue.
Assays based on T cell activation require a priori selection of cytokines that may not reflect a dominant phenotype of responding T cells, so we next sought to optimize CD1a tetramers as a capture reagent based on CD1a-TCR binding. Human CD1a tetramer assays were initially developed in the context of loading CD1a with a foreign mycobacterial lipopeptide (20). However, human CD1a tetramers have not been applied to detection of autoreactive T cells or T cells derived from tissue. Therefore, we addressed key issues relating to possible false-positive or false-negative detection. First, tissue-derived T cells are often contaminated with debris and dead cells, which are highly autofluorescent and nonspecifically adhere to tetramer reagents, causing false-positive detection (35). Indeed, for T cells obtained by physical digestion of skin, we observed many suspected false-positive events located outside of size parameters for lymphocytes (Supplemental Figure 1). These events were less frequent in 3D cultures, which largely lack subcellular debris. Further nonspecific staining patterns could be reduced by removing autofluorescent events in the FITC channel and by live-dead cell exclusion reagents (Supplemental Figure 1). Combined, both the 3D method and the gating strategy greatly reduce noise and false-positive staining on skin T cells. Overall, we achieved similar staining results using CD1a proteins made at the NIH tetramer Facility and Monash University (Supplemental Figure 2).
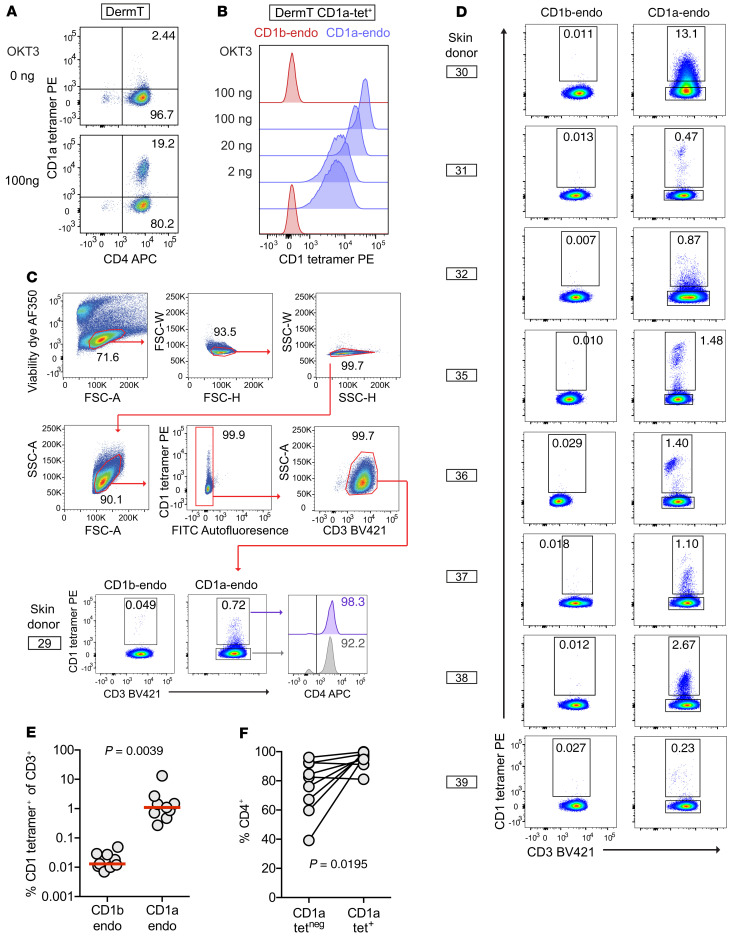
A known cause of false-negative detection by tetramers is the gap between the relatively high affinities typically needed for detectable TCR binding to tetramers versus a lower affinity threshold needed for T cell activation by APCs. This affinity gap is broadly documented for MHC (36, 37) and CD1 tetramers (38, 39). To mitigate false-negative results, unlabeled bivalent anti-CD3 monoclonal antibody (OKT3) was added during tetramer staining to cluster and retain surface TCR complexes, potentially increasing the stoichiometry of TCR interaction with tetramers. This approach produced striking improvements in sensitivity, revealing a previously obscured tetramer+ population in the oligoclonal CD1a-autoreactive skin T cell line DermT (7) (Figure 3A). Using a sorted CD1a-tetramer+ population, we observed a steep dose response and a 10-fold higher CD1a tetramer staining intensity using OKT3, with no observed effect on background CD1b tetramer staining (Figure 3B).
Figure 3. CD1a-endo tetramer staining of polyclonal skin T cells.
(A) Unlabeled OKT3 was added during CD1a-endo tetramer staining of the oligoclonal CD1a-autoreactive T cell line DermT, which was pregated to capture cells with low live-dead dye, low forward and side scatter typical of lymphyocytes, and CD3+ singlets. (B) Dose response of CD1 tetramer staining intensity with added OKT3 on a sorted DermT CD1a tetramer+ subpopulation. (C and D) Polyclonal skin T cells obtained from 3D culture were stained with CD1b-endo or CD1a-endo tetramers (n = 9) and representative pregating is shown for donor 29. (E) Summary data show the absolute tetramer detection rates and (F) rates of CD4 positivity among CD1a-endo tetramer+ and tetramer– gates.
CD1a-endo tetramers detect high levels of skin T cells.
Both MHC (40) and CD1 (20, 40–43) tetramers are typically treated with one known antigen to promote homogenous loading of multiple arms of the tetramer, leading to multivalent interactions with antigen-specific TCRs. Untreated or mock-treated tetramers are used as negative controls. Therefore, a basic unanswered feasibility question for CD1a tetramer detection relates to the choice of self-antigen to load. Although skin and plant oils activate T cell clones (13, 14, 18), no immunodominant lipid that broadly activates CD1a-dependent polyclonal T cells is known. However, there is some evidence that individual T cell lines can recognize CD1a or CD1c without adding exogenous lipids (18, 44). For example, the only available ternary CD1a-lipid-TCR structure shows that the BK6 TCR contacts CD1a itself, rather than its carried lysophosphatidylcholine (LPC) ligand (45). We reasoned that if CD1a-specific but lipid antigen–independent (hereafter known as CD1a-specific) TCRs like BK6 are common in vivo, untreated or mock-treated CD1a tetramers carrying many types of endogenous lipids from the expression system (CD1a-endo) might be sufficient to generate multivalent CD1a-TCR interactions and thereby detect polyclonal T cells (Figure 3C). CD1b-autoreactive cytokine responses in blood are rare (7), and our pilot studies found infrequent staining of polyclonal skin T cells with CD1b-endo tetramers. Both results indicate that CD1b-specific T cells are less common than CD1a-specific T cells and support the use of CD1b-endo tetramers as a negative control for staining (Figure 3, C and D).
Among polyclonal skin T cells from 9 donors tested (Supplemental Table 1), cells binding CD1a-endo tetramers were detected at rates above control CD1b-endo tetramer staining in all cases. The median rate of CD1a-endo tetramer staining as a percentage of T cells was high in absolute terms (1.1%), and approximately 100-fold higher than rates of CD1b-endo tetramer staining (median = 0.013%, P = 0.0039) (Figure 3E). Donor 30 was a notable outlier on the high end, where CD1a-endo tetramer+ cells represented 13% of CD3+ cells. This very high rate of staining was unlikely to be false positive, given the low rate of CD1b-endo tetramer staining in donor 30 and the low rate of CD1a-endo tetramer staining in the other subject tested in the same experiment (donor 29) (Figure 3C).
Coreceptor expression.
Prior analysis of blood-derived T cells showed increased CD1a-autoreactive response after selection on CD4, and 13 of 14 T cell lines expressed CD4 (7, 8). These data suggested that CD4 is linked to IL-22 secretion, which, if confirmed, would be consistent with a Th22 phenotype (25). CD3+ CD1a-endo tetramer+ cells were enriched for CD4 positivity (mean = 92%) versus the CD3+ CD1a-endo tetramer– fraction (Figure 3F, P = 0.0195). Thus, CD4 predominates on blood- and skin-derived autoreactive T cells, which is consistent with a Th22 phenotype.
Tetramer+ skin T cells recognize and respond to CD1a without added ligand.
Overall, the pattern in which approximately 1% of total skin T cells were stained with CD1a-endo tetramers represents a high rate of staining for any single MHC or CD1 epitope in polyclonal human T cells and is comparable to frequencies of MR1-reactive MAIT cells (~1%) (46) or CD1d-reactive NKT cells (0.1%) (47) in human blood. The higher measured frequency of CD1a autoreactivity using tetramers might have been seen because the cytokine methods used measure only IL-22 response, but tetramers measure TCR frequencies regardless of the activation outcome. The absence of high staining by CD1b tetramers ruled out many forms of nonspecific staining by CD1a tetramers, but CD1 proteins can bind non-TCR surface ligands on T cells (48). Therefore, to evaluate possible CD1a-TCR binding, we first bulk-sorted the oligoclonal CD1a-autoreactive line DermT, which was 20.5% CD1a tetramer+, to yield a clonal population DermT2, which was more than 99% tetramer+. TCR sequencing yielded 1 TCR-α and 1 TCR-β chain, where TRBV2*01 in the ImMunoGeneTics (IMGT) international database nomenclature encodes a variable region specifically recognized by the monoclonal antibody to TCR Vβ22 (49) (Figure 4A). Dual staining with tetramer and anti-TCR identified only 1 diagonally oriented population; the intensity of anti-TCR correlated with the intensity of tetramer staining, as expected if tetramers were binding the TCR complex. Also, the resulting clone was functionally CD1a-autoreactive by 2 different measures (Figure 4B). Thus, 1 clonotypic TCR was likely responsible for a CD1a-mediated functional response and binding to CD1a tetramers.
Figure 4. CD1a tetramers select for CD1a-autoreactive T cell cytokine response.
(A) A CD1a tetramer+ subpopulation from the CD1a-autoreactive DermT line was sorted (pregated live, lymphocyte, and singlet forward and side scatter gates, CD3+, autofluoresence [FITC]neg, and CD4+) and bulk expanded. The sorted line, DermT2, was stained with the TCR Vβ22 antibody and tetramers (pregated: live, lymphocytes, singlets, CD3+, and CD4+). (B) IL-22 and IFN-γ ELISPOTs of DermT2 cells. (C) Bulk expanded cells (presorted on forward and side scatter, CD3, viability dye, and CD4) were subjected to tetramer and Vβ antibody staining by flow cytometry and (D) CD1a-dependent activation in IL-22 and IFN-γ ELISPOT assays. Dashed-line arrow from donor 36 indicates that a second round of sorting and expansion was done prior to shown tetramer staining versus Vβ23. Solid-line arrows for all other lines reflect a single sort and expansion. Individual data points are shown with range indicated. Lines were separately generated and tested at different times using cytokine ELISPOTs (IFN-γ and IL-22) chosen based on cytokine profiles observed during outgrowth.
To more broadly test whether tetramer-stained T cells expressed TCRs with functional recognition of CD1a, we sorted CD1a tetramer+ cells from polyclonal skin T cells from 4 donors (Figure 4C). When tested after the expansion of sorted T cells, the resulting T cell lines did not bind CD1b-endo tetramers but were strongly enriched for CD1a-endo tetramer binding in all cases (Figure 4C). We plotted CD1a-endo tetramer staining versus anti-CD3 (donors 31, 32) or anti-TCR antibodies that recognized TCR variable regions as predicted by sequencing of sorted cells (Supplemental Table 3), which were TCR Vβ22 (TRBV2) in donor 30 and TCR Vβ23 (TRBV13) in donor 36. This approach revealed multiple distinct populations (Figure 4C). First, populations separated by CD1a tetramer intensity were consistent with tetramer binding to clonally distributed ligands. Second, the diagonal staining pattern within each dual-staining population indicated that tetramer staining correlated with anti-CD3 or particular anti-TCR Vβ intensity. Third, the limited, nearly clonal narrowing of TCR repertoire after sorting with CD1a-endo tetramers in donor 30 and line 36 suggests selection based on TCR binding. Last, TCRs typically determined the antigen specificity of T cells. In all cases, we observed strong cytokine release and a high stimulation index, and the response was largely or completely blocked with anti-CD1a or use of cells not expressing CD1a (Figure 4, B and D). Therefore, measured on a multidonor basis and at polyclonal, oligoclonal, and clonal T cell levels, CD1a tetramers selected skin T cells with anti-CD3 and anti-TCR binding properties expected for TCRs binding CD1a, as well as functional reactivity toward CD1a.
Thus, CD1a-autoreactive T cells are abundant in human skin and can be identified and enumerated using CD1a tetramers without any knowledge of lipid ligands bound to CD1a. Further, on a molecular basis, these unexpected findings suggest that CD1a tetramers carrying diverse self-ligands bind to TCRs and that TCR binding mainly to CD1a itself is a common mechanism of CD1a autoreactivity. Because CD1-reactive T cells were generally thought to require a specific lipid antigen, we investigated this new and controversial hypothesis in detail.
Molecular analysis of CD1a-endo tetramers.
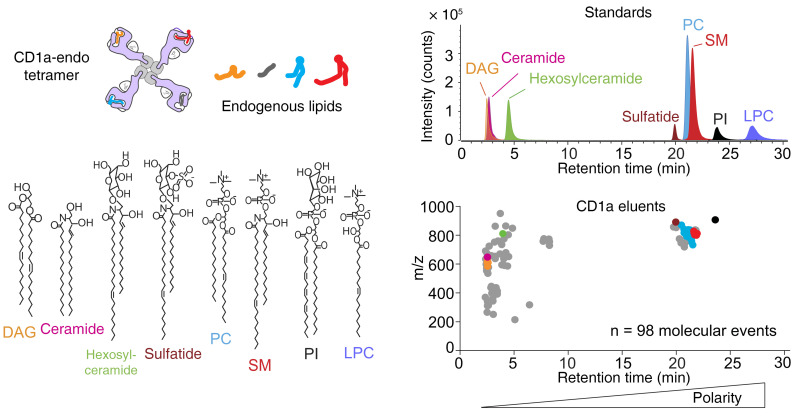
Next, we sought to determine whether the spectrum of endogenous cellular lipids bound to CD1a monomers during their production in human HEK293 cells were homogenous or heterogeneous, such that individual arms of a tetramer might carry the same or different lipids (Figure 5). We measured the lipids bound in CD1a-endo monomers from the NIH Tetramer Facility using a normal-phase lipidomics method based on HPLC-TOF mass spectrometry (MS). We detected approximately 600 ion chromatograms in eluents of CD1a treated with chloroform and methanol, but this was likely an overestimate of ligand heterogeneity. We censored ions corresponding to redundant detection of isotopes, recognizable high-mass lipid dimers, and alternate adducts of any molecule M ([M + H]+, [M + Na]+, [M + NH4]+). We further censored ions present in solvents, ions also eluting from an MHC II control protein, or ions matching patterns for polyethylene glycol–based detergents. This process identified 98 high-quality mass chromatograms with unique m/z and retention time values, as a conservative estimate of the number of distinct lipids bound to CD1a (Figure 5).
Figure 5. CD1a endogenous lipid tetramers bind autoreactive T cells despite lipid heterogeneity.
Tetramers formed on streptavidin have 4 CD1 arms, which can capture lipids. Positive-mode, normal-phase HPLC-QToF-MS lipidomics of lipids eluted from CD1a monomer (bottom) and standards for each lipid class (top). Gray points indicate unique ion chromatograms, which were not solved as named lipids. Colored dots were solved as named compounds based on m/z values that matched literature values within 10 parts per million and showed coelution with named standards. Structures of solved lipids are shown where the overall length and saturation status of alkyl chains can be reliably deduced, but the position and absolute stereochemistry at the saturation as well as the length of individual alkyl chains are not known.
Normal-phase chromatography is known to provide low-resolution separation, such that small changes in retention time correlate with differing lipid classes, and retention time correlates directly with polarity (50). The 98 unnamed compounds (Figure 5) spanned nearly the complete range of polarity of natural lipid classes present in HEK293 cells, and retention times were consistent with neutral lipids, glycolipids, phospholipids, and charged lipids. These singly charged ions differed in mass from approximately 200 to 1000 amu, which approximates the molecular size of most known CD1a antigens (9, 10, 14, 18, 45, 51, 52). Assuming random tetramerization, these data predict on a statistical basis that differentially loaded monomers exist among the 4 arms of nearly all tetramers. Such diversely liganded CD1a-endo tetramers (Figure 5) brightly stained DermT (Figure 3, A and B) and identified subpopulations among polyclonal skin T cells from all 9 donors tested (Figure 3, C and D). As hinted by the binding of 1 TCR to CD1a (45) and the isolation of CD1d- and CD1c-restricted T cell lines that do not absolutely require antigen (44, 53, 54), these results could be explained if CD1a-autoreactive TCRs showed CD1a-specific binding and largely ignore the carried lipid.
To more directly investigate molecular mechanisms, we first identified a subset of the total CD1a-lipid interactome by matching the detected m/z values with masses of known lipids in databases (Supplemental Table 2). These tentative compound identifications were further confirmed by measuring retention time in comparison with lipid standards (Figure 5). From 98 unknown lipids, we identified and annotated 30 molecular species across 7 families: 4 diacylglycerols (DAGs), 1 ceramide, 1 hexosylceramide, 1 sulfatide, 16 phosphatidylcholines (PCs), 6 sphingomyelins (SM), and 1 phosphatidylinositol (PI).
Although absolute quantification of lipids from microscale elution from proteins is challenging, we recently validated a method for estimating molar ratios of bound lipids (55). Intensity values were calculated from the area under chromatograms, which were converted to absolute mass based on external standard curves to estimate the absolute molar ratio of eluted lipids in each class (Supplemental Figure 3). This method can rank the named ligands bound to CD1a from highest to lowest abundance: PCs, DAG, sphingomyelins, PI, ceramide, hexosylceramide, sulfatide. Whereas compound identification initially focused on the most intense ions in eluates, we also specifically scanned for and identified LPC bound to CD1a. This additional targeted lipid search was conducted because the structure of CD1a-LPC complexes is known (45). Although LPC was not identified by automated peak picking methods because of low signal, manual analysis identified a low-intensity ion chromatogram that was detected with mass and retention time matching the LPC standard (m/z 522.36).
These studies confirmed that CD1a carries diverse families of ligands, including neutral lipids, glycolipids, phospholipids, lysophospholipids, sulfolipids, and sphingolipids. These lipids are composed of distinct polar head groups and nearly all liganded CD1 structures show that such head groups are exposed on the surface of CD1. Therefore, this result implied TCR binding to CD1a when differing head groups are exposed, raising questions about whether and how such exposed carbohydrate, phospho-, and sulfo-structures could be equivalently recognized or ignored. The identification of named lipids in CD1a complexes allowed us to test this hypothesis in several ways, including measuring CD1a tetramer binding to T cells after CD1a-endo complexes were treated with known CD1a ligands.
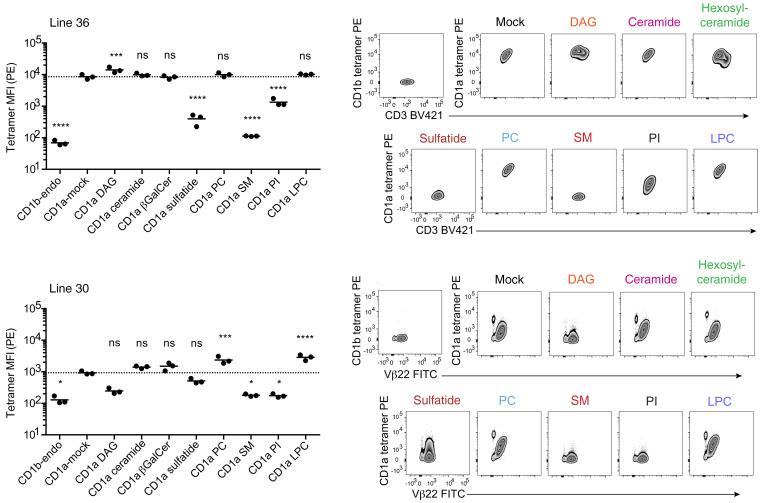
TCR recognition of CD1a when treated with diverse ligands.
Using line 30 and line 36, we next determined whether T cells showed stain equivalently with CD1a tetramers treated with synthetic ligands that matched defined endogenous ligands, including neutral lipids (DAG; ceramide), a glycolipid (β-galactosyl ceramide, β-GalCer), a sulfolipid (sulfatide), a sphingolipid (sphingomyelin), phospholipids (PC, PI), and a lysophospholipid (LPC) (Figure 6). These lipids are capable of efficient CD1a binding as endogenous cellular ligands based on the data in Figure 5. Also, in 6 cases, the exogenously applied lipids altered T cell staining by CD1a tetramers, implying that lipids normally loaded in cells can also be loaded exogenously (Figure 6). However, for ceramide and β-GalCer, altered T cell staining was not seen, so preserved staining might have occurred through nonloading of lipids onto CD1a. As a further control, we treated CD1a with these 2 ligands and measured lipid eluents by mass spectrometry (Supplemental Figure 4). We detected ceramide and β-GalCer in eluents, as well as evidence for loss of most endogenously bound lipids. Therefore, lipid-dependent changes in T cell staining by lipid-treated CD1a tetramers (Figure 6) can be interpreted as a correlate of lipid compatibility with CD1a-TCR binding.
Figure 6. T cell staining by CD1a tetramers treated with lipid ligands.
CD1a tetramers treated with pure synthetic ligands corresponding to endogenously bound lipid families shown in Figure 5 were for used staining of line 36 and line 30, with replicates corresponding to 3 independent experiments. Significant difference from CD1a mock was tested by 1-way ANOVA and Holm-Sidak multiple-comparison test. Multiplicity-adjusted P values indicated (NS P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Line 36 revealed equivalent, high-intensity staining with untreated CD1a tetramers and tetramers treated with DAG, ceramide, β-GalCer, PC, and LPC. Staining was reduced with PI and sulfatide treatment, with nearly complete blockade of staining with sphingomyelins. For line 30, baseline staining was much lower in intensity and detected 2 cell populations. Alternately treated tetramers showed a pattern for both populations that was similar to line 36 except that DAG showed some staining suppression and increases in PC and LPC. Overall, a diversely loaded tetramer stained many types of T cells brightly. Also, many individual lipids with differing head groups did not alter the CD1-TCR interaction, consistent with the interpretation that lipids are ignored. However, certain lipids, especially sulfatide and sphingomyelin, partially blocked CD1a tetramer binding, likely through effects on CD1a structure or steric hindrance of the TCR approach (18, 45).
Role of the A′ roof of CD1a.
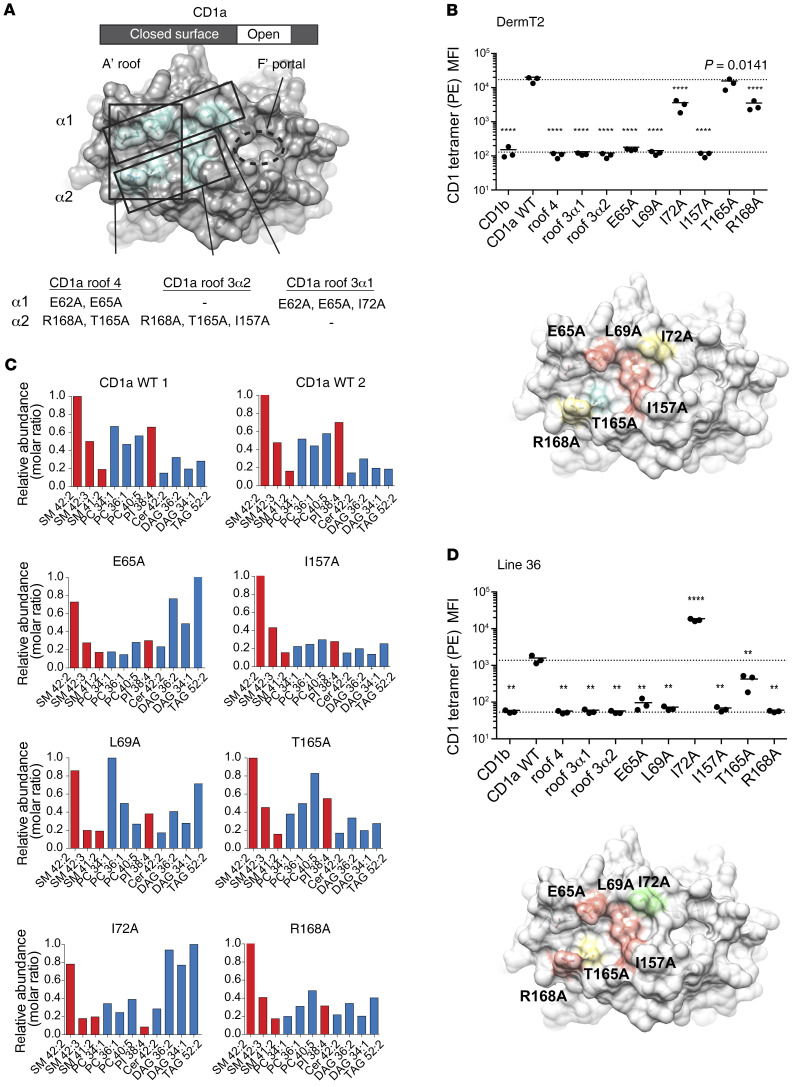
A testable hypothesis for explaining how TCR binding could absolutely require CD1a, yet ignore carried ligands, was suggested by the crystal structure of the BK6 TCR bound to a CD1a-LPC complex (45). In this structure, the LPC head group minimally protrudes to the CD1a surface through a portal on the right side of the platform. The TCR is shifted to the left, where it lacks direct contact with LPC, but extensively contacts the outer surface of CD1a, where tethering residues link the α1 and α2 helices to create a flat surface known as the A′ roof (Figure 7A). The A′ roof is considerably larger than the typical 600–900 Å2 area typical of TCR footprints, so it could provide a TCR contact platform (56). This CD1a-centric mode of recognition is known for only 1 TCR (45), and is clearly distinct from the many examples of dual TCR contact with CD1 and lipid (57–59) or MHC and peptide (60) and MAIT TCR-MR1 (61, 62).
Figure 7. CD1a mutant tetramers point to direct CD1a-TCR contacts for recognition.
(A) Summary of CD1a A′ roof multimutants, which contained 4 mutations on both helices (roof 4), 3 mutations on the α1 helix (roof 3α1), or 3 mutations in α2 (roof 3α2) shown in aqua, overlaid on PDB: 4X6E. (B) CD1b, CD1a-WT, and CD1a mutant tetramer staining on T cell line DermT2, shown as tetramer MFI. Each point represents an individual experiment, and solid lines indicate the mean of replicates. Dashed line is drawn from the mean of CD1a-WT staining. Significant difference from CD1a-WT was tested by 1-way ANOVA and Holm-Sidak multiple-comparison test. Multiplicity-adjusted P values indicated (**P < 0.01, ****P < 0.0001). Relative contributions of each mutant to tetramer staining intensity are summarized as overlaid on PDB 4X6E (green = increased staining, red = low or absent staining, yellow = intermediate/reduced staining, aqua = similar to WT). (C) Positive- mode, reversed-phase HPLC-QToF-MS analysis of lipids eluted from CD1a monomers. Lipids are named according to the class X:Y, where X and Y are the total number of methylene units and unsaturations in the alkyl chains. Data shown as relative abundance (molar ratio) relative to the most abundant species. (D) CD1 mutant tetramer staining of line 36, as in B.
To determine the role of the A′ roof, we generated alanine mutations in CD1a, including triple mutants that cluster on the α1 (E62A, E65A, I72A) and α2 (R168A, T165A, I157A) helices, as well as a tetramutant at the extreme left margin of both helices (E62A, E65A, R168A, T165A) (Figure 7A). In all cases, these multimutants failed to stain T cells (Figure 7, B–D), consistent with TCR binding to the A′ roof. However, simultaneous changes in 3 or 4 residues may alter domains distant from the site of mutation, so the single-site mutants dispersed throughout the CD1a roof represented a more rigorous test of binding (E65A, L69A, I72A, R168A, T165A, I157A). Because the main goal was to distinguish TCR contact with the roof versus the roles of bound lipids located inside CD1a, we further screened these single-site mutants with HPLC-TOF-MS to determine the relative abundance of a selected panel of 11 bound ligands determined from the elution experiments (Figure 5). All 6 mutants detectably bound all 11 lipids eluted from CD1a, although some differences in the relative abundance profiles were seen (Figure 7C). For example, I72A, which is located nearer the antigen portal, showed moderately increased binding of DAG and triacylglycerol (TAG) species (Figure 7C).
Testing of single-site mutant tetramers on DermT2 (Figure 7B) and line 36 (Figure 7D) showed a complete loss of staining for mutations located near the center of the A′ roof: E65A, L69A, and I157A. Two other mutants (T165A, R168A) located on the α2 helix and at the left-central portion of the A′ roof showed partial or complete binding loss, which varied according to the line tested. Unexpectedly, mutation at I72A showed increased tetramer binding compared with WT CD1a. Whereas the other residues were located nearer the center of the CD1a roof, I72 was a special case because it was located at the rim of the antigen portal (Figure 7D), where I72 might have been responsible for the increased the binding of DAG and decreased binding of PI as observed in Figure 7C. DAG itself caused a slight increase in binding of CD1a tetramers to line 30, whereas PI was inhibitory (Figure 6). Thus, the periportal location of I72 might have affected lipids in ways that indirectly affect the TCR. However, all other mutants located near the center of the roof had no effect or inhibitory effect on tetramers, consistent with TCR binding to the A′ roof. The role of the A′ roof in TCR contact (Figure 7) can be distinguished from TCR binding that covers the CD1d antigen exit portal, as seen for invariant NKT cells (57) or a newly identified mode of TCR binding on the underside of MR1 proteins (63).
Discussion
The skin of an adult human contains an estimated 20 billion resident T cells (23, 24). The antigen specificity and immune repertoires of T cells in human skin have not been comprehensively studied. Prior studies support a role for CD1a-reactive T cells in skin immune responses, including the high CD1a expression in the epidermis (1), the existence of CD1a-autoreactive cells with skin-homing receptors in blood (7, 8), and the reduction of some skin T cell responses after CD1a blockade (9–13). We used CD1a tetramers to physically capture autoreactive cells for study and determined the molecular basis of their response. Taken together, the patterns of staining with diversely loaded CD1a tetramers, with tetramers treated with alternate ligands, and with CD1a mutant tetramers collectively indicated that CD1a-specific TCRs bound the A′ roof in a way that was tolerated by many types of lipids carried in the cleft. As compared with a prior study of CD1a-autoreactive blood T cells detected using cytokine RNA (7), the precursor frequency observed here in skin was about 10-fold higher. Both studies demonstrated CD1a autoreactivity in all or nearly all subjects studied, and both studies provided evidence for CD4 predominance and a linkage of CD1a autoreactivity to IL-22. Whereas the prior study found skin addressins on blood T cells, this study identified T cells in the skin.
The concept of a CD1a-specific TCR repertoire in human skin contrasts with prior models that have emphasized the role of CD1 proteins in lipid antigen display. Further, the consistent detection of approximately 100-fold increased staining with CD1a-endo compared with CD1b-endo strongly suggests that CD1a specificity is greater than CD1b specificity, at least in skin T cells. This new perspective regarding a CD1a-centric response is plausible because CD1 proteins, unlike MHC I and II, lack an open groove spanning the length of the platform. Instead, CD1a residues form tethers between the α1 and α2 helices, which create a roof over the A′ pocket (51, 52). This closed roof blocks direct TCR contact to antigens located underneath the roof, simultaneously forming an outer, antigen-free surface for TCRs to bind. Lipids are sequestered inside CD1 proteins and emerge to the surface shifted toward the right edge of the CD1 display platform (64, 65). Currently, direct contact of lipid-free surfaces on CD1 is known for 2 TCRs (44, 45). Also, 1 TCR has been shown to recognize CD1d in a manner that minimally requires antigen (54). However, many counterexamples exist in which the approaching TCR does indeed contact protruding antigen (57, 58, 66, 67). Accordingly, models of MHC peptide response (61, 62) and CD1 lipid response (57, 58) strongly emphasize TCR corecognition of antigens and antigen-presenting molecules. Unlike these many previously known “CD1-restricted” T cell responses where CD1a serves as a bridge to recognize something else, “CD1-specific” T cells identified here directly recognize the outer surface of CD1a. Because this mode of CD1a-TCR binding was seen in clones, lines, and polyclonal T cells and among all subjects tested, we conclude that a repertoire of CD1a-specific T cells represents a normal part of the human immune system. CD1a-specific T cells are particularly abundant in skin, where they represent 1 in 100 T cells on average.
Unlike CD1b, CD1c, and CD1d, the cytoplasmic tail of CD1a lacks an adaptor protein binding domain, which sorts CD1 for trafficking to late endosomes, where exchange for exogenous antigens normally occurs. Thus, the unique trafficking pattern of CD1a is consistent with capture of endogenous self-lipids in preference to exogenous antigen presentation (3, 4, 65, 68). Furthermore, CD1a-autoreactive T cell responses were readily detected using nonprofessional APCs, like human erythroleukemia K562 cells or B cell–like C1R cells, suggesting that CD1a itself is necessary and sufficient to induce response without costimulation. Thus, CD1a-autoreactive cells might normally be subject to some kind of negative regulation, be chronically stimulated to serve some homeostatic immunoregulatory role, or both. CD1a-endo tetramer staining of polyclonal skin T cells across individuals directly rules in skin residence of CD1a-reactive T cells, as well as their IL-22 production and CD4+ predominance, suggesting that CD1a is a natural target of Th22 cells. Both CD1a itself and bone fide Th22 cells, which secrete IL-22 in the absence of IL-17 (26), are absent in mice but present in humans. This correlative evidence makes it tempting to speculate on the role of CD1a in promoting the Th22 cell program and skin residence.
Most models of TCR antigen recognition on MHC or CD1 emphasize the high specificity of lock-and-key type interactions that occur only in the setting of rare antigens. T cells are off until a rare event turns them on. Opposite to this model, patterns of CD1a specificity observed here predict that the default response among the repertoire of autoreactive TCRs is activation when contacting the surface of CD1a. Extending recent studies (18, 45), here we found that sulfatide and sphingomyelin were natural endogenous ligands bound to CD1a, and that they partially blocked polyclonal skin T cell responses to CD1a. Added lipids are not required for activation, but endogenous lipid blockers might dampen or tune this “on until off” recognition system. The picture that emerges here is of a nonpolymorphic cell surface protein that can nonetheless provide nuanced immunological signals. For T cells that bind the combination of CD1a and exogenous ligand, CD1a can act as an antigen-presenting molecule, facilitating specific detection of foreign lipids. For the many T cells in skin that mainly bind directly to CD1a and are mildly tuned by certain lipids, it likely serves as an innate-like sensor of tissue damage, where self-derived lipids tune response.
Finally, these various results validate CD1a tetramers in practical ways for new uses in clinical studies of T cell–mediated autoimmunity. Major causes of false-positive and false-negative detection by tetramers on tissue-derived T cells have been addressed here, and because of low polymorphism, the same CD1a tetramer can be used for any patient. Compared with the common use of cytokines as readouts, CD1a tetramers bypass the need to understand the expected effector function in advance. The common CD1-centric mode of activation seen here bypasses the need to know the identity of immunodominant lipids prior to planning clinical studies. Untreated CD1a-endo tetramers represent a straightforward and feasible approach to study CD1-specific T cell populations in the skin and potentially other tissues in any disease state.
Methods
Recovery of skin T cells by 3D culture or enzymatic digestion
Skin T cells were recovered after culture for 21–28 days on 3D cell foam growth matrices (Cytomatrix) seeded with collagen I (Thermo Fisher Scientific, 354236). Skin T cell culture media [IMDM, 10%–20% FCS, L-glutamine, penicillin-streptomycin, 2-mercaptoethanol] was supplemented with IL-2 (BWH or Peprotech) and recombinant human IL-15 (10 ng/mL, Peprotech, 200-15) as previously described (24). Fresh skin cells were isolated by enzymatic digestion using a mixture of 30 Kunitz units/mL DNase I from bovine pancreas (Sigma-Aldrich) and 0.2% collagenase type I in skin T cell culture media, incubated for up to 2 hours at 37°C with shaking at 300–500 rpm.
T cell activation assays
Prior to IL-22 ELISPOT or IL-22 capture assays, polyclonal skin T cells were cocultured 5:1 with irradiated allogeneic Langerhans-like cells and HLA-blocking antibodies for 7–10 days in complete T cell media with 2 nM IL-2. In vitro Langerhans cells were derived from adherent PBMCs over 6 days as previously described (69, 70), using recombinant human TGF-β1 (100-21), GM-CSF (300-3), and IL-4 (200-4) from Peprotech. Langerhans-like cells were irradiated to prevent proliferation of any remnant lymphocytes and treated with antibody clones W632 (10 μg/mL) and L243 (10 μg/mL) for 1 hour prior to coculture with allogeneic skin T cells.
Cell lines and in-house antibodies.
K562 (7) or C1R (6) cells transduced to express CD1a or CD1b were used as APCs. Antibodies to HLA-A, -B, and -C (W6/32); HLA-DR (L243); CD1a (OKT6, 10H3); and CD3 (OKT3) and a mouse IgG1 isotype control (P3) were made in-house and added at 10 μg/mL for 1 hour prior to adding T cells.
ELISPOT.
IL-22 and IFN-γ ELISPOT assays were done according to the manufacturer’s instructions (Mabtech) using anti–IL-22 (MT12A3), biotin anti–IL-22 (MT7B27), anti–IFN-γ (1-D1K), and biotin anti–IFN-γ (7-B6-1). Polyclonal T cells were tested in triplicate at 1 × 105 T cells per well. T cell lines and clones were tested in ELISPOT assays directly without prior coculture on Langerhans-like cells and at lower cell numbers, typically a ratio of 10,000 T cells to 20,000 K562 or C1R cells.
IL-22 capture assay.
The IL-22 secretion assay (MACS Miltenyi) was used with skin T cells restimulated by overnight coculture with K562-CD1a (2:1) in T cell media with 4% human AB serum. The next day, IL-22–secreting cells were labeled over a 45-minute secretion period at 37°C according to the manufacturer’s instructions with minor modifications, which included lower cell concentration (1 × 105 to 5 × 105 cells/mL) during the secretion period. Cells were also labeled with anti–CD3-BV421 and anti–CD4-APC during secondary labeling with anti–IL-22 biotin and labeled just prior to flow cytometry analysis with a viability dye.
CD1-endo tetramer staining and flow cytometry
Biotinylated human CD1a and CD1b monomers (NIH Tetramer Core Facility and Monash University) were produced in HEK293-derived cell lines in 4 versions, which were generally found to have equivalent results, but differed in the origin or design of the CD1a protein (Supplemental Figure 2). Version 1 CD1a monomer from NIH was previously described (20, 43, 71), and is produced in lentivirus-transduced HEK293T cells (used in Figure 3A and Supplemental Figure 2). Versions 2 and 3 of NIH CD1a monomers were expressed in HEK293T cells deficient in MGAT and TAP2 genes and used a lentiviral vector for separate expression of β2m and CD1a with a double hexahistidine tag. Only version 3 contained a cysteine to serine swap meant to reduce aggregation, and these were used interchangeably for polyclonal skin T cell staining and sorting. Version 4, produced at Monash University, was expressed in HEK293S GnTI cells as previously described (45) and treated with endoglycosidase H (New England Biolabs). Version 4 is shown in analytical polyclonal T cell staining in Supplemental Figure 2 and Figure 3 (donors 31, 35, 36, 37).
CD1 monomers were adjusted to 0.2 μg/μL in 50 mM Tris-buffered saline pH 8.0 (Sigma-Aldrich) with or without 0.5% CHAPS (Sigma-Aldrich) prior to tetramer assembly 1:5 w/w with streptavidin-phycoerythrin (streptavidin-PE) (BD Biosciences) for a final concentration of 0.1 μg/μL with respect to CD1a monomer. Antibodies and reagents were Blue Live/Dead (Life Technologies), Near IR Live/Dead (Life Technologies), CD4-APC (clone RPA-T4, BD Biosciences), CD4-APC-Cy7 (clone OKT4), CD8α-BV711 (clone RPA-T8, Biolegend), CD3-BV421 (clone UCHT1, Biolegend), TCR Vβ22-FITC (clone IMMU546, Beckman Coulter), and TCR Vβ23-FITC (clone REA497, Miltenyi). Cells were washed twice in PBS and pulsed with Live/Dead stain in PBS for 15 minutes at room temperature, washed once with PBS 1% BSA ± 0.01% sodium azide (FACS buffer), and resuspended in FACS buffer at a maximum of 1 million cells/50 μL. Per 50 μL staining volume, 1 μL tetramer PE was added to cells and incubated for 25–30 mins at room temperature in the dark. Without washing, 0.1 μg unlabeled anti-CD3 (OKT3) was added and incubated for an additional 10 minutes at room temperature. Labeled surface stains were then added for 15 minutes on ice. Cells were washed once in FACS buffer and resuspended in PBS for immediate acquisition on an LSR Fortessa or a custom-modified FACSAria II flow cytometer (BD Biosciences) and analyzed with FlowJo version 10.5.3 (Tree Star).
Lipid sources
PC (850475), C18:1 LPC (845875), C24:1 sphingomyelin (860593), milk sphingomyelins (860063), C24:1 β-GalCer (860546), and PI (840042) were purchased from Avanti Polar Lipids. Sulfatides (1049) were from Matreya. Diacylglycerol (D0138) was from Sigma-Aldrich.
Loading CD1a monomers with defined lipids for tetramers
Lipids stored in chloroform and methanol were transferred to new borosilicate glass tubes, dried under nitrogen gas, and reconstituted to 400 uM in TBS pH 8.0, 0.5% CHAPS buffer by sonication in a 37°C water bath for approximately 1 hour. Lipid-buffer sonicates or a buffer-only control were transferred to 1.5 mL Eppendorf tubes on a 37°C heat block to which CD1a monomer was added to a final concentration of 0.2 μg/μL and incubated for 2 hours at 37°C, then overnight at room temperature. Loaded monomers were stored at 4°C and used for tetramer assembly and staining within 3 months.
Generation of mutant CD1a tetramers
WT and mutant CD1a proteins were recombinantly produced in human embryonic kidney cells (HEK293 GnTI–). Both β2-microglobulin and the heavy chain of CD1a were expressed as a single construct from a modified pHLsec vector, in which the original signal peptide was substituted by the Igκ leader sequence and the P2A self-cleavage site was introduced between both genes. Both chains carried leucine zippers Fos (β2m) and Jun (CD1a). Additional BirA- and 6xHis- tags were fused to the carboxy-terminus of CD1a. The CD1a/β2m heterodimer was purified by nickel affinity and size exclusion chromatography. Pure protein was biotinylated using BirA ligase followed by a gel filtration run.
Generation of T cell lines
The DermT parent line was previously derived by limiting dilution expansion from PBMCs (7, 18). Sorting from the DermT line to generate DermT2 was done with LPC-treated CD1a tetramers, which previous experiments showed to be similar to CD1a-endo. For 2 donors (29, 30), skin T cells were cocultured with allogeneic Langerhans-like cells as for activation assays prior to tetramer staining. Live CD3+ CD4+ CD1a-LPC tetramer+ cells were sorted and then expanded over 14–18 days by stimulation with 25 × 106 irradiated, allogeneic PBMCs; 5 × 106 irradiated Epstein-Barr virus–transformed B cells; and 30 ng/mL anti-CD3 (OTK3); and with 2 nM IL-2 added on day 2 of the culture.
T cell receptor PCR and sequencing
RNA was isolated from sorted T cell populations using the RNeasy kit (QIAGEN) and cDNA synthesized using the Quantitect Reverse Transcription Kit (QIAGEN). TCR V gene usage was determined by PCR using primer sets IPS000029 and IPS000030 as described at www.imgt.org/IMGTPrimerDB in combination with TCR-α constant region reverse primer 5′GTGGTAGCAGCTTTCACCTCCTTGG 3′ and TCR-β constant region reverse primer 5′ GGTGGCAGACAGGACCCCTTGC 3′.
HPLC-MS analysis of eluents from CD1a monomers
Version 2 CD1a protein and HLA-DRB1*03:01 protein (80 μg) were extracted in triplicate in 15 mL glass tubes using chloroform, methanol, and water (72). The organic phase was recovered and dried under nitrogen gas. Eluent residue was redissolved in chloroform-methanol, normalized to 20 μM based on input protein, and stored at –20°C. We injected 20 μL for Q-ToF HPLC-MS positive ion mode analysis using an Agilent 6530 Accurate-Mass Q-TOF mass spectrometer and 1260 series HPLC system with a normal-phase Inertsil Diol column (150 mm × 2 mm, GL Sciences), running at 0.15 mL/minute as described (50, 73, 74).
For the reversed-phase HPLC-MS, an Agilent Poroshell 120 A, EC-C18, 3 × 50 mm, 1.9 μm column coupled with an Agilent EC-C18, 3 × 5 mm, 2.7 μm guard column were used based on published methods (75): mobile phase (A) methanol /water (95/5; v/v) supplemented with 2 mM ammonium-formate and mobile phase (B) 1-propanol/cyclohexane/water (90/10/0.1; v/v/v) supplemented with 3 mM ammonium-formate. The solvent gradient changes in a 20-minute run: 0–4 minutes, 100% A; 4–10 minutes, from 100% A to 100% B; 10–15 minutes, 100% B; 15–16 minutes, from 100% B to 100% A; 16–20 minutes, 100% A. The columns were equilibrated by running 100% A for 5 minutes before the next run. The lipid quantification was determined by the individual ion chromatogram peak area compared with the external standard curves (50).
Statistics
Statistical analyses were done in GraphPad Prism 6 using the Wilcoxon matched-pairs signed-rank test unless otherwise stated. Tetramer staining on T cell lines and clones (Figure 5, Figure 6) was tested by ordinary 1-way ANOVA and the Holm-Sidak multiple-comparison test for multiplicity-adjusted P values. Significance was defined as P < 0.05.
Study approval
Discarded skin from cosmetic surgeries was obtained through the Human Skin Disease Resource Center at Harvard Medical School and Brigham and Women’s Hospital. Human PBMCs were obtained from leukoreduction collars provided by Brigham and Women’s Hospital. All studies were approved by the Partners Healthcare Institutional Review Board.
Author contributions
The indicated individuals carried out project oversight and direction (DBM, RAC, GO, JR), T cell and tetramer assays (RNC, TYC, MW), and lipid mass spectrometry (TYC); contributed reagents, cells and tissues, and original ideas (MW, JLN, JR, RAW, JDA, ADJ, DPO, SGT, RAC); and prepared the manuscript (RNC, TYC, DBM) with input from all authors.
Supplementary Material
Acknowledgments
We thank the NIH Tetramer Core Facility staff for biotinylated CD1 proteins and HLA-DRB1 monomer and matched HEK293T cells. J. Teague provided advice on human skin culture, and BWH Division of Plastic and Reconstructive Surgery clinical staff provided surgical discarded tissue. The work is supported by the NIH (R01 AR048632, R01 AI127654, P30 AR069625, T32 AR007530), the Wellcome Trust Collaborative Award in Science, the UK Medical Research Council, the NIHR Oxford Biomedical Research Centre, the National Health and Medical Research Council of Australia, and the Australian Research Council (ARC) (CE140100011). JLN is supported by an ARC Future Fellowship (FT160100074); JR is supported by an Australian ARC Laureate Fellowship. ADJ is supported by NIH K01 AR068475.
Version 1. 01/04/2021
Print issue publication
Funding Statement
Wellcome Trust Collaborative Award in Science
Footnotes
Conflict of interest: RNC declares income from MPM Capital. GO declares laboratory research support from UCB through Oxford University.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e140706.https://doi.org/10.1172/JCI140706.
Contributor Information
Rachel N. Cotton, Email: rcotton@g.harvard.edu.
Tan-Yun Cheng, Email: tcheng@rics.bwh.harvard.edu.
Marcin Wegrecki, Email: marcin.wegrecki@monash.edu.
Jérôme Le Nours, Email: jerome.lenours@monash.edu.
Dennis P. Orgill, Email: dorgill@bwh.harvard.edu.
Bohdan Pomahac, Email: BPOMAHAC@PARTNERS.ORG.
Simon G. Talbot, Email: SGTALBOT@bwh.harvard.edu.
Richard A. Willis, Email: richard.willis@emory.edu.
John D. Altman, Email: jaltman@emory.edu.
Annemieke de Jong, Email: ad2952@cumc.columbia.edu.
Graham Ogg, Email: graham.ogg@ndm.ox.ac.uk.
Ildiko Van Rhijn, Email: I.vanRhijn@uu.nl.
Jamie Rossjohn, Email: jamie.rossjohn@monash.edu.
D. Branch Moody, Email: bmoody@bwh.harvard.edu.
References
- 1.Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells. Curr Top Microbiol Immunol. 2007;314:113–141. doi: 10.1007/978-3-540-69511-0_5. [DOI] [PubMed] [Google Scholar]
- 2.Furue M, Nindl M, Kawabe K, Nakamura K, Ishibashi Y, Sagawa K. Epitope mapping of CD1a, CD1b, and CD1c antigens in human skin: differential localization on Langerhans cells, keratinocytes, and basement membrane zone. J Invest Dermatol. 1992;99(5):23S–26S. doi: 10.1111/1523-1747.ep12668273. [DOI] [PubMed] [Google Scholar]
- 3.Hunger RE, et al. Langerhans cells utilize CD1a and langerin to efficiently present nonpeptide antigens to T cells. J Clin Invest. 2004;113(5):701–708. doi: 10.1172/JCI19655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peña-Cruz V, Ito S, Dascher CC, Brenner MB, Sugita M. Epidermal Langerhans cells efficiently mediate CD1a-dependent presentation of microbial lipid antigens to T cells. J Invest Dermatol. 2003;121(3):517–521. doi: 10.1046/j.1523-1747.2003.12429.x. [DOI] [PubMed] [Google Scholar]
- 5.Rosat JP, et al. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ alpha beta T cell pool. J Immunol. 1999;162(1):366–371. [PubMed] [Google Scholar]
- 6.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature. 1989;341(6241):447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 7.de Jong A, Peña-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat Immunol. 2010;11(12):1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lalla C, et al. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. Eur J Immunol. 2011;41(3):602–610. doi: 10.1002/eji.201041211. [DOI] [PubMed] [Google Scholar]
- 9.Bourgeois EA, et al. Bee venom processes human skin lipids for presentation by CD1a. J Exp Med. 2015;212(2):149–163. doi: 10.1084/jem.20141505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung KL, et al. Psoriatic T cells recognize neolipid antigens generated by mast cell phospholipase delivered by exosomes and presented by CD1a. J Exp Med. 2016;213(11):2399–2412. doi: 10.1084/jem.20160258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarrett R, et al. Filaggrin inhibits generation of CD1a neolipid antigens by house dust mite-derived phospholipase. Sci Transl Med. 2016;8(325):325ra18. doi: 10.1126/scitranslmed.aad6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramaniam S, et al. Elevated and cross-responsive CD1a-reactive T cells in bee and wasp venom allergic individuals. Eur J Immunol. 2016;46(1):242–252. doi: 10.1002/eji.201545869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JH, et al. CD1a on Langerhans cells controls inflammatory skin disease. Nat Immunol. 2016;17(10):1159–1166. doi: 10.1038/ni.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicolai S, et al. Human T cell response to CD1a and contact dermatitis allergens in botanical extracts and commercial skin care products. Sci Immunol. 2020;5(43):eaax5430. doi: 10.1126/sciimmunol.aax5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278(5343):1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 16.Joosten SA, et al. Harnessing donor unrestricted T-cells for new vaccines against tuberculosis. Vaccine. 2019;37(23):3022–3030. doi: 10.1016/j.vaccine.2019.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Rhijn I, Moody DB. Donor unrestricted T cells: a shared human T cell response. J Immunol. 2015;195(5):1927–1932. doi: 10.4049/jimmunol.1500943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong A, et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol. 2014;15(2):177–185. doi: 10.1038/ni.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagchi S, et al. CD1b-autoreactive T cells contribute to hyperlipidemia-induced skin inflammation in mice. J Clin Invest. 2017;127(6):2339–2352. doi: 10.1172/JCI92217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasmar AG, et al. Cutting Edge: CD1a tetramers and dextramers identify human lipopeptide-specific T cells ex vivo. J Immunol. 2013;191(9):4499–4503. doi: 10.4049/jimmunol.1301660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell JJ, et al. CCR7 expression and memory T cell diversity in humans. J Immunol. 2001;166(2):877–884. doi: 10.4049/jimmunol.166.2.877. [DOI] [PubMed] [Google Scholar]
- 22.Schaerli P, et al. A skin-selective homing mechanism for human immune surveillance T cells. J Exp Med. 2004;199(9):1265–1275. doi: 10.1084/jem.20032177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark RA, et al. The vast majority of CLA+ T cells are resident in normal skin. J Immunol. 2006;176(7):4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- 24.Clark RA, et al. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J Invest Dermatol. 2006;126(5):1059–1070. doi: 10.1038/sj.jid.5700199. [DOI] [PubMed] [Google Scholar]
- 25.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10(8):857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 26.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol. 2009;10(8):864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- 27.Eyerich S, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119(12):3573–3585. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong MT, et al. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity. 2016;45(2):442–456. doi: 10.1016/j.immuni.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Geiger R, Duhen T, Lanzavecchia A, Sallusto F. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. J Exp Med. 2009;206(7):1525–1534. doi: 10.1084/jem.20090504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkins MK, Chu HH, McLachlan JB, Moon JJ. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol. 2010;28:275–294. doi: 10.1146/annurev-immunol-030409-101253. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins MK, Moon JJ. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J Immunol. 2012;188(9):4135–4140. doi: 10.4049/jimmunol.1102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan AC, et al. Ex-vivo analysis of human natural killer T cells demonstrates heterogeneity between tissues and within established CD4(+) and CD4(-) subsets. Clin Exp Immunol. 2013;172(1):129–137. doi: 10.1111/cei.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melum E, et al. Control of CD1d-restricted antigen presentation and inflammation by sphingomyelin. Nat Immunol. 2019;20(12):1644–1655. doi: 10.1038/s41590-019-0504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein E, et al. Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int J Cancer. 1976;18(4):421–431. doi: 10.1002/ijc.2910180405. [DOI] [PubMed] [Google Scholar]
- 35.Moon JJ, et al. Tracking epitope-specific T cells. Nat Protoc. 2009;4(4):565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolton G, et al. Optimized peptide-MHC multimer protocols for detection and isolation of autoimmune T-cells. Front Immunol. 2018;9:1378. doi: 10.3389/fimmu.2018.01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rius C, et al. Peptide-MHC class I tetramers can fail to detect relevant functional T cell clonotypes and underestimate antigen-reactive T cell populations. J Immunol. 2018;200(7):2263–2279. doi: 10.4049/jimmunol.1700242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Rhijn I, et al. CD1d-restricted T cell activation by nonlipidic small molecules. Proc Natl Acad Sci U S A. 2004;101(37):13578–13583. doi: 10.1073/pnas.0402838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanderson JP, et al. CD1d protein structure determines species-selective antigenicity of isoglobotrihexosylceramide (iGb3) to invariant NKT cells. Eur J Immunol. 2013;43(3):815–825. doi: 10.1002/eji.201242952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 41.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192(5):741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191(11):1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasmar AG, et al. CD1b tetramers bind αβ T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. 2011;208(9):1741–1747. doi: 10.1084/jem.20110665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wun KS, et al. T cell autoreactivity directed toward CD1c itself rather than toward carried self lipids. Nat Immunol. 2018;19(4):397–406. doi: 10.1038/s41590-018-0065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birkinshaw RW, et al. αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol. 2015;16(3):258–266. doi: 10.1038/ni.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dusseaux M, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117(4):1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 47.Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, Godfrey DI. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur J Immunol. 2005;35(5):1399–1407. doi: 10.1002/eji.200425958. [DOI] [PubMed] [Google Scholar]
- 48.Li D, et al. Ig-like transcript 4 inhibits lipid antigen presentation through direct CD1d interaction. J Immunol. 2009;182(2):1033–1040. doi: 10.4049/jimmunol.182.2.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei S, Charmley P, Robinson MA, Concannon P. The extent of the human germline T-cell receptor V beta gene segment repertoire. Immunogenetics. 1994;40(1):27–36. doi: 10.1007/BF00163961. [DOI] [PubMed] [Google Scholar]
- 50.Layre E, et al. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem Biol. 2011;18(12):1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zajonc DM, et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22(2):209–219. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4(8):808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 53.Guo T, et al. A subset of human autoreactive CD1c-restricted T cells preferentially expresses TRBV4-1+ TCRs. J Immunol. 2018;200(2):500–511. doi: 10.4049/jimmunol.1700677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mallevaey T, et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 2011;34(3):315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van ‘t Klooster JS, Cheng TY, Sikkema HR, Jeucken A, Moody DB, Poolman B. Membrane lipid requirements of the lysine transporter Lyp1 from Saccharomyces cerevisiae. J Mol Biol. 2020;432(14):4023–4031. doi: 10.1016/j.jmb.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB. Lipid and small-molecule display by CD1 and MR1. Nat Rev Immunol. 2015;15(10):643–654. doi: 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448(7149):44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 58.Gras S, et al. T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nat Commun. 2016;7:13257. doi: 10.1038/ncomms13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy S, et al. Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by αβ T cells. Proc Natl Acad Sci U S A. 2014;111(43):E4648–E4657. doi: 10.1073/pnas.1408549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 61.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384(6605):134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 62.Garcia KC, et al. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274(5285):209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 63.Le Nours J, et al. A class of γδ T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science. 2019;366(6472):1522–1527. doi: 10.1126/science.aav3900. [DOI] [PubMed] [Google Scholar]
- 64.Cotton RN, Shahine A, Rossjohn J, Moody DB. Lipids hide or step aside for CD1-autoreactive T cell receptors. Curr Opin Immunol. 2018;52:93–99. doi: 10.1016/j.coi.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moody DB, Cotton RN. Four pathways of CD1 antigen presentation to T cells. Curr Opin Immunol. 2017;46:127–133. doi: 10.1016/j.coi.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shahine A, et al. A molecular basis of human T cell receptor autoreactivity toward self-phospholipids. Sci Immunol. 2017;2(16):eaao1384. doi: 10.1126/sciimmunol.aao1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shahine A, et al. A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids. Nat Commun. 2019;10(1):56. doi: 10.1038/s41467-018-07898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cernadas M, Cavallari M, Watts G, Mori L, De Libero G, Brenner MB. Early recycling compartment trafficking of CD1a is essential for its intersection and presentation of lipid antigens. J Immunol. 2010;184(3):1235–1241. doi: 10.4049/jimmunol.0804140. [DOI] [PubMed] [Google Scholar]
- 69.Geissmann F, Prost C, Monnet JP, Dy M, Brousse N, Hermine O. Transforming growth factor beta1, in the presence of granulocyte/macrophage colony-stimulating factor and interleukin 4, induces differentiation of human peripheral blood monocytes into dendritic Langerhans cells. J Exp Med. 1998;187(6):961–966. doi: 10.1084/jem.187.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cotton RN, McDonald-Fleming R, Boyd A, Spates K, Nutman TB, Tolouei Semnani R. Brugia malayi infective larvae fail to activate Langerhans cells and dermal dendritic cells in human skin. Parasite Immunol. 2015;37(2):79–91. doi: 10.1111/pim.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ly D, et al. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J Exp Med. 2013;210(4):729–741. doi: 10.1084/jem.20120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 73.Huang S, et al. Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc Natl Acad Sci U S A. 2011;108(48):19335–19340. doi: 10.1073/pnas.1112969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madigan CA, et al. Lipidomic discovery of deoxysiderophores reveals a revised mycobactin biosynthesis pathway in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2012;109(4):1257–1262. doi: 10.1073/pnas.1109958109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van ‘t Klooster JS, Cheng TY, Sikkema HR, Jeucken A, Moody B, Poolman B. Periprotein lipidomes of Saccharomyces cerevisiae provide a flexible environment for conformational changes of membrane proteins. Elife. 2020;9:e57003. doi: 10.7554/eLife.57003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.