Abstract
Therapeutic strategies designed to target TP53-deficient cancer cells remain elusive. Here, we showed that TP53 loss initiated a pharmacologically actionable secretory process that drove lung adenocarcinoma (LUAD) progression. Molecular, biochemical, and cell biological studies showed that TP53 loss increased the expression of Golgi reassembly and stacking protein 55 kDa (G55), a Golgi stacking protein that maintains Golgi organelle integrity and is part of a GOLGIN45 (G45)–myosin IIA–containing protein complex that activates secretory vesicle biogenesis in the Golgi. TP53 loss activated G55-dependent secretion by relieving G55 and myosin IIA from miR-34a–dependent silencing. G55-dependent secreted proteins enhanced the proliferative and invasive activities of TP53-deficient LUAD cells and promoted angiogenesis and CD8+ T cell exhaustion in the tumor microenvironment. A small molecule that blocks G55-G45 interactions impaired secretion and reduced TP53-deficient LUAD growth and metastasis. These results identified a targetable secretory vulnerability in TP53-deficient LUAD cells.
Keywords: Cell Biology, Oncology
Keywords: Lung cancer, P53, Protein traffic
Introduction
The presence of a cytokine-rich, fibrotic stroma is correlated with advanced disease and poor clinical outcome in multiple cancer types (1). In one working hypothesis, prometastatic features of the tumor microenvironment are orchestrated by cancer cells that secrete cytokines, proteases, and growth factors that inhibit antitumor immunity, promote angiogenesis, and facilitate cancer cell dissemination (2–4). Transcriptomes activated by somatically acquired oncogenic mutations are enriched in secreted proteins that promote tumor growth and metastasis (5). Thus, a better understanding of the way in which oncogenic mutations activate secretion may identify therapeutic vulnerabilities in cancer.
Secreted proteins are transported as vesicular cargos from the endoplasmic reticulum to the cell surface through defined intracellular pathways. Proteins that lack N-terminal signal peptides are transported via unconventional protein secretion (UPS), a stress-activated, autophagy-dependent vesicular transport pathway that bypasses the Golgi (6). In autophagosomes, cargo loading, autophagosome-lysosome fusion, and autophagosome maturation are facilitated by Golgi reassembly and stacking protein 55 kDa (G55), a Golgi stacking protein that translocates to autophagosomes to facilitate UPS (7, 8). Proteins that contain signal peptides, however, are transported to the plasma membrane via the Golgi apparatus, a process called conventional protein secretion (CPS) (6). In the Golgi, vesicle budding, fission, and release are initiated by Golgi phosphoprotein 3 (GOLPH3), which bridges Golgi membranes to F-actin fibers that convey a tensile force required for secretory vesicle biogenesis (9, 10). On the basis of this conceptual framework, we evaluated the molecular underpinnings of secretion in cancer and identified a protumorigenic, G55-dependent secretory process that was activated by TP53 deficiency.
Results
TP53 deficiency activates a G55-dependent secretory program.
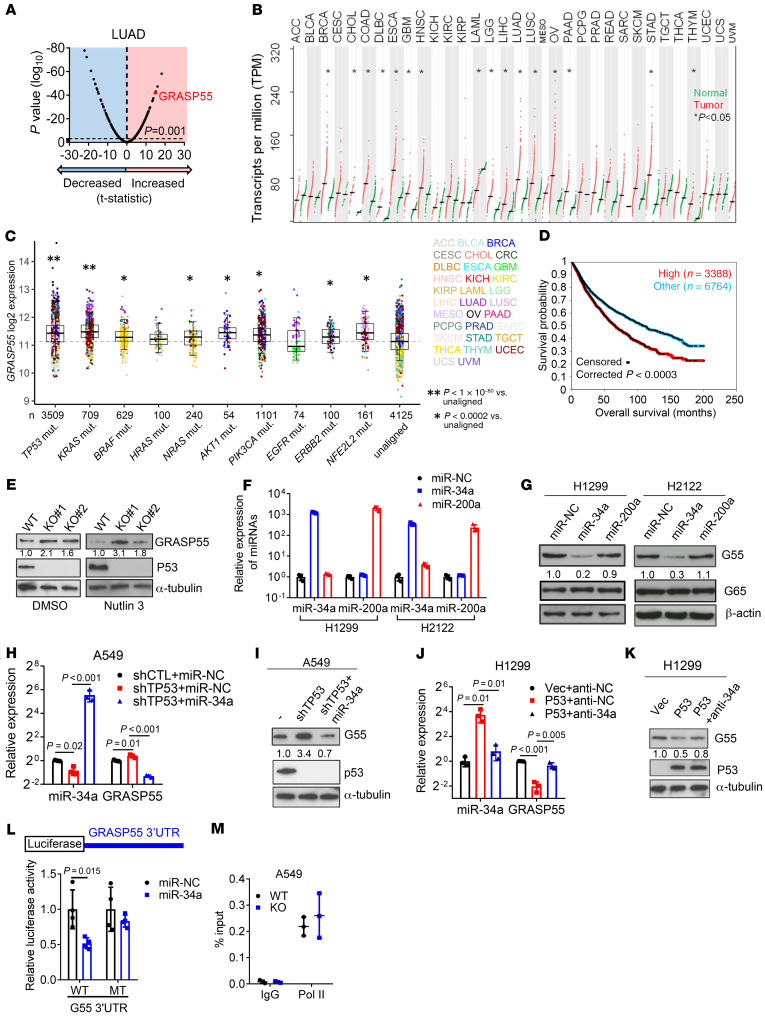
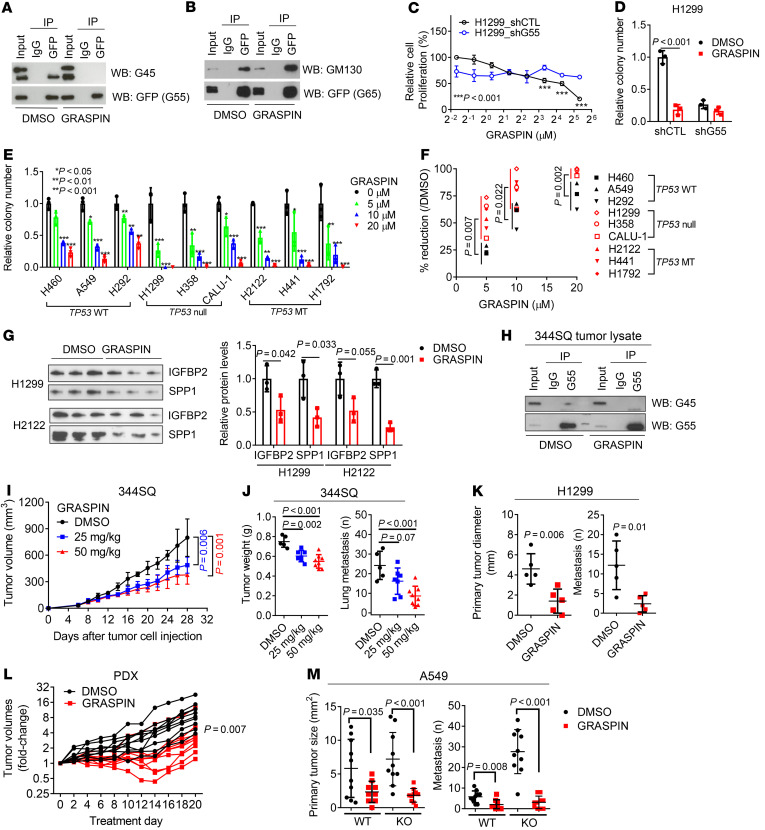
We queried the Gene Ontology term “Golgi apparatus” (n = 453 genes) in The Cancer Genome Atlas (TCGA) pan-cancer cohort and found that, compared with matched normal tissues, lung adenocarcinoma (LUAD) and other cancer types have higher expression of 201 genes (P < 0.001, fold change > 1.4), including G55, a Golgi stacking protein that promotes secretion (refs. 11, 12, and Figure 1, A and B). High G55 expression was correlated with multiple oncogenic driver mutations (TP53, KRAS, BRAF, NRAS, AKT1, PIK3CA, ERBB2, and NFE2L2) (Figure 1C) and shorter survival durations (Figure 1D). On the basis of these findings, we delved into the role of G55 as an effector of TP53-mutant LUAD progression.
Figure 1. p53 Deficiency relieves GRASP55 (G55) from miR-34a–dependent silencing.
(A) Volcano plot of mRNAs (dots). Differences between LUAD (n = 515) and normal lung (n = 51) in TCGA expressed as a P value (y axis) and a t-statistic (x axis) to identify genes upregulated (t > 0) or downregulated (t < 0) in cancer. G55 indicated. (B) Quantification of G55 mRNA in tumors (red dots) and normal tissues (green dots). Tumor types significantly different from normal (asterisks). Mean values (horizontal bars). (C) G55 mRNA levels in the TCGA pan-cancer cohort (dots) annotated on the basis of mutations. “Unaligned” indicates no correlation with mutations. Sample sizes indicated (n). Tumor types color coded. (D) Kaplan-Meier analysis of TCGA pan-cancer cohort (n = 9,502) scored based on G55 mRNA expression. High: top third. Other: middle and bottom thirds. (E) Western blot (WB) analysis of G55 in parental (WT) and TP53-KO A549 clones. Cells treated with vehicle (DMSO) or nutlin-3 to enhance p53 levels. α-Tubulin loading control. (F) Quantitative real-time PCR (qPCR) analysis of miR-transfected cells. Negative control (miR-NC). (G) WB analysis of miR-transfected cells. β-Actin loading control. (H and I) qPCR (H) and WB (I) analysis of A549 cells cotransfected with shRNAs and miRs. Control vectors indicated by (–). Values relative to control transfectants. (J and K) qPCR (J) and WB (K) analysis of H1299 cells cotransfected with p53 expression vector and antagomir-34a (anti-34a). Vec, empty vector; anti-NC, control antagomir. (L) Reporter assays on cells cotransfected with miRs and reporters containing G55 3′-UTRs with intact (WT) or mutant (MT) miR-34a binding sites. Values relative to miR-NC. n = 4. (M) RNA polymerase II ChIP assays on GORASP2 gene promoter. Values are percentage of input (total chromatin). All WB densitometric values are relative to control. Results represent mean ± SD. n = 3, unless otherwise indicated. P values: ANOVA (H and J), univariate Cox (D), 2-sided t test (all others).
To investigate a causal relationship between p53 loss and increased G55 expression, we subjected A549 LUAD cells, which are TP53-WT, to CRISPR/Cas9-mediated TP53 KO and found that G55 expression was higher in TP53-KO cells (Figure 1E). Using a microRNA (miR) binding site prediction algorithm (www.targetscan.org), we identified a binding site for miR-34a, which is a p53 transcriptional target (13), in the G55 3′-untranslated region (3′-UTR), which led us to postulate that p53 loss upregulates G55 expression by silencing miR-34a. To test this hypothesis, we subjected TP53-mutant H1299 cells and H2122 cells to ectopic miR-34a expression and found that G55 levels were suppressed (Figure 1, F and G). Ectopic miR-34a expression abrogated G55 upregulation induced by shRNA-mediated p53 deficiency (Figure 1, H and I), and miR-34a antagomirs reversed ectopic p53-induced G55 suppression (Figure 1, J and K), indicating that G55 upregulation by TP53 deficiency is miR-34a dependent. To determine whether G55 is a miR-34a target, we performed G55 3′-UTR luciferase reporter assays and found that ectopic miR-34a expression inhibited the activity of a WT G55 3′-UTR luciferase reporter, whereas the activity of a mutant G55 3′-UTR reporter that lacked the miR-34a binding site was unaffected (Figure 1L). Demonstrating the specificity of this regulatory axis, ectopic miR-34a expression did not downregulate the G55 homolog GRASP65, which lacks miR-34a binding sites, and ectopic expression of miR-200a, which is not predicted to bind G55, did not inhibit G55 expression (Figure 1G). ChIP assays to quantify RNA polymerase II recruitment to the G55-encoding genomic locus (GORASP2) demonstrated no significant difference between parental and TP53-KO cells (Figure 1M), suggesting that p53 depletion did not increase GORASP2 gene transcription. These findings indicate that p53 deficiency relieved G55 from miR-34a–dependent silencing.
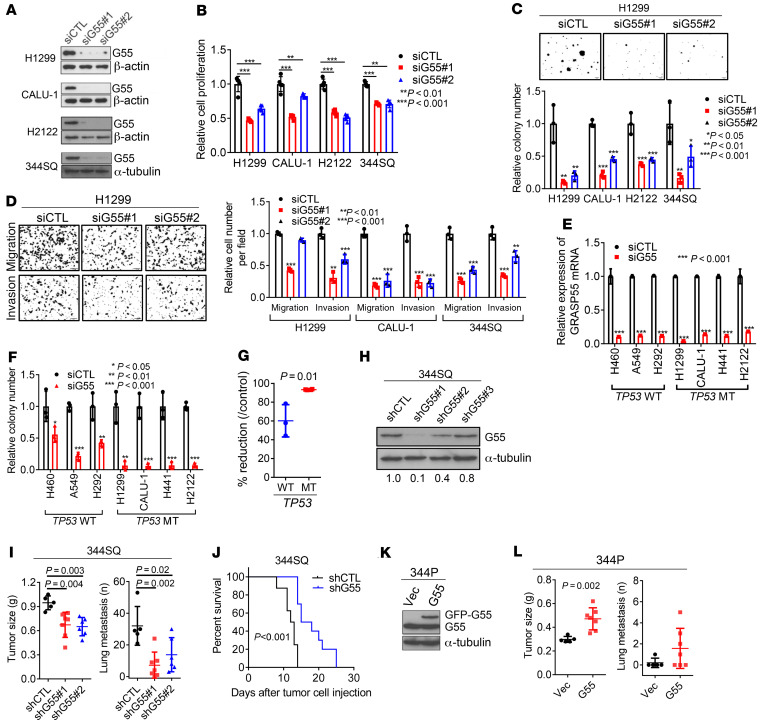
We postulated that high G55 expression is protumorigenic in TP53-mutant LUAD and tested this hypothesis in TP53-null LUAD cells (H1299, CALU-1) and LUAD cells that express mutant p53 proteins that do (H441, 344SQ) or do not (H2122) have gain-of-function properties (14, 15). Regardless of the type of TP53 mutation, siRNA-mediated G55 depletion impaired the proliferative, colony-forming, migratory, and invasive activities of these cells (Figure 2, A–D) without inducing apoptosis (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI137186DS1). In colony formation assays, G55 siRNAs demonstrated greater inhibitory activity against TP53-mutant than TP53-WT LUAD cell lines (Figure 2, E–G, and Supplemental Figure 1B), suggesting a selective vulnerability of TP53-mutant LUAD cells to G55 depletion. Conversely, ectopic G55 expression increased the migratory, invasive, and colony-forming activities of H1299 cells (Supplemental Figure 1, C–E) and reconstituted these activities in HeLa cells that had been subjected to CRISPR/Cas9-mediated GORASP2 KO (Supplemental Figure 1, F–H). Relative to G55-replete controls, orthotopic LUADs generated by G55-deficient 344SQ cells were smaller and less metastatic, and they caused delayed mortality in syngeneic, immunocompetent mice (Figure 2, H–J). Conversely, ectopic G55 expression accelerated the growth and enhanced the metastatic activity of subcutaneous tumors generated by 344P cells (Figure 2, K and L, and Supplemental Figure 2I). Thus, high G55 levels are required for, and may be sufficient to drive, LUAD progression. Moreover, G55 siRNAs reduced the proliferative and invasive activities of TP53-mutant breast and ovarian cancer cell lines (Supplemental Figure 1, J–N), suggesting a generalizable role for G55 in TP53-mutant cancer cells.
Figure 2. G55 is a driver of TP53-mutant LUAD progression.
(A) WB analysis of TP53-mutant LUAD cell lines transfected with G55 (siG55#1, #2) or control (siCTL) siRNA. (B) Cell proliferation in monolayer culture quantified at 4 days by WST-1 assays. Results relative to siCTL controls. n = 5. (C) Anchorage-independent colony formation quantified after 10 days in soft agarose. Results relative to siCTL controls. (D) Boyden chamber assays. FBS (10%) added to bottom chamber as chemoattractant. Cells that migrated or invaded through porous filter (Matrigel-coated for invasion assays) photographed 8 hours after seeding in upper chamber and counted manually. Results relative to siCTL controls. (E) qPCR analysis of TP53-WT and TP53-mutant (MT) LUAD cell lines. Results relative to siCTL. (F and G) Anchorage-dependent colony formation. Cells seeded at low density on plastic. Results expressed as fractions (F) or percentage reductions (G) relative to siCTL in TP53-WT and TP53-mutant (MT) cell lines (G). (H) WB analysis of 344SQ cells transfected with G55 (shG55#1, #2, #3) or control (shCTL) shRNAs. G55 densitometric values relative to shCTL. (I) Primary subcutaneous tumor weight (left plot) and lung metastasis numbers (right plot) per mouse (dots). Necropsies performed 5 weeks after subcutaneous injection of 344SQ cells into syngeneic, immunocompetent mice. (J) Kaplan-Meier analysis of mouse cohorts bearing G55-deficient (shG55) or G55-replete (shCTL) 344SQ orthotopic lung tumors (n = 10 mice per cohort). (K) WB analysis of 344P LUAD cells transfected with GFP-tagged G55 or empty (Vec) expression vectors. (L) Primary tumor weights (left plot) and lung metastasis numbers (right plot) determined at necropsy 5 weeks after subcutaneous injection of tumor cells. Results represent mean ± SD. n = 3, unless otherwise indicated. P values: ANOVA (B–D and I), log-rank test (J), 2-sided t test (all others).
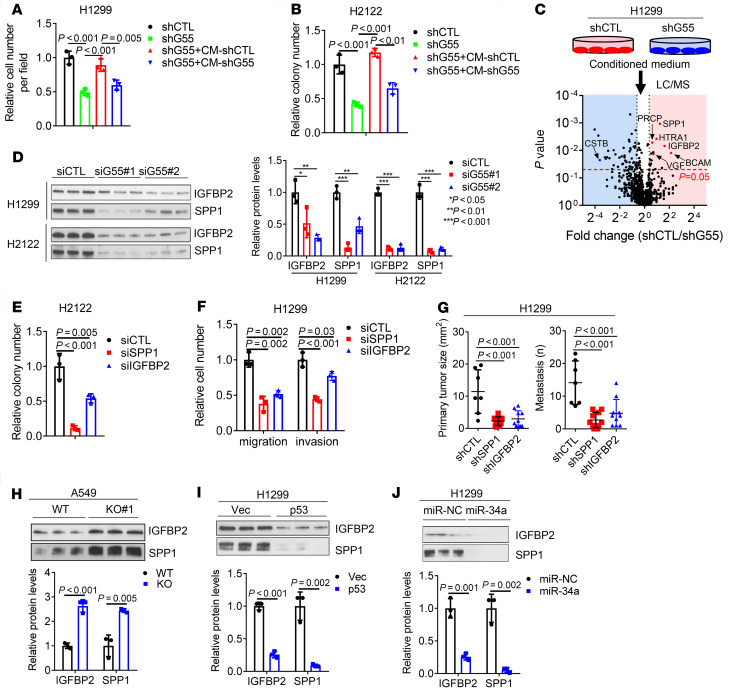
Given that G55 is a key secretory pathway component (8), we postulated that G55 drives tumor progression by activating the secretion of protumorigenic effector proteins. To test this hypothesis, we performed conditioned medium (CM) transfer experiments and found that the invasive and colony-forming activities of G55-deficient cancer cells were rescued by treatment with CM samples from G55-replete but not G55-deficient cancer cells (Figure 3, A and B, and Supplemental Figure 2, A–C). To identify G55-dependent secreted proteins, we performed liquid chromatography–mass spectrometry (LC-MS) analysis on CM samples from G55-deficient and G55-replete H1299 cells and identified 925 proteins (≥2 peptides per protein, 1% FDR), 18 of which were present at significantly lower concentrations (fold change > 1.4, P < 0.05) in G55-deficient cells (Figure 3C and Supplemental Table 1), including the matricellular protein osteopontin (SPP1), the IGF binding protein IGFBP2, the lysosomal prolylcarboxypeptidase PCRP, the serine peptidase HTRA1, the laminin receptor BCAM, and the secreted neurosecretory protein VGF, which are protumorigenic in cancer models (16–20). Western blot analysis confirmed SPP1 and IGFBP2 downregulation in CM samples but not lysates from G55-deficient H1299, H2122, and 344SQ cells (Figure 3D and Supplemental Figure 2, D–F). After G55 reconstitution in GORASP2-KO HeLa cells, IGFBP2 and SPP1 secretion were restored (Supplemental Figure 2, F and G), indicating that IGFBP2 and SPP1 are G55-dependent secreted proteins. We assessed the functional roles of IGFBP2 and SPP1 by performing siRNA- and neutralizing antibody–mediated depletion studies and found that they were required to varying degrees for H1299 cells and H2122 cells to form colonies, migrate, and invade (Figure 3, E and F, and Supplemental Figure 3, A–E) and for H1299 orthotopic LUADs to grow and metastasize (Figure 3G and Supplemental Figure 3F). Invasive activities of IGFBP2- and SPP1-deficient H1299 cells were rescued by treatment with IGFBP2 and SPP1 recombinant proteins (Supplemental Figure 3, G–I), demonstrating that colony-forming activity decreased as a result of on-target effects of the siRNAs.
Figure 3. G55 activates the secretion of protumorigenic effector proteins.
(A) Invasion assays of H1299 cells in Boyden chambers performed after 24 hours of treatment with conditioned medium (CM) from G55-deficient or G55-replete cells. (B) Anchorage-dependent colony formation assays performed on G55-deficient (shG55) or G55-replete (shCTL) H2122 cells after treatment for 10 days with or without (–) CM from G55-deficient or G55-replete cells. (C) Volcano plot of proteins identified by LC-MS analysis of CM collected from H1299 cells (shG55 or shCTL). Proteins (dots) plotted by P value (y axis) and fold change (x axis). Fold change = 1.4, P = 0.05 indicated (vertical and horizontal dotted lines, respectively). Proteins of interest are labeled. (D) WB analysis of CM samples. Mean densitometric values of triplicate CM samples relative to control (siCTL) (bar graph). (E and F) Anchorage-independent colony formation (E) and cell migration and invasion (F) assays. Results expressed relative to control (siCTL). (G) Orthotopic lung tumor size and contralateral lung metastasis numbers at necropsy 5 weeks after injecting 1 × 106 tumor cells. (H–J) WB analysis of triplicate CM samples from parental and TP53-KO A549 cells (H) or H1299 cells subjected to ectopic expression of p53 (I) or miR-34a (J). Densitometric values relative to controls (bar graphs). Values are mean ± SD. n = 3, unless otherwise indicated. P values: ANOVA (A, B, and D–G), 2-sided t test (all others).
Proteins secreted by neuroendocrine and other secretory cells are stored in dense core vesicles that degranulate in response to external cues (21). However, by immunofluorescence staining, IGFBP2 and SPP1 did not localize in granules, which led us to speculate that secretion draws from newly synthesized proteins, a conclusion supported by evidence that cycloheximide treatment decreased IGFBP2 levels in cell lysates and CM medium samples and abrogated ectopic G55-induced IGFBP2 secretion (Supplemental Figure 4A). To determine whether TP53 loss coordinately increases the expression and secretion of IGFBP2 and SPP1, we quantified SPP1 and IGFBP2 levels in a panel of LUAD cell lines that were TP53-mutant or TP53-WT and found that SPP1 and IGFBP2 protein levels were higher in CM samples but not lysates from TP53-mutant cells (Supplemental Figure 4, B and C). In gain- and loss-of-function studies, p53 and its downstream mediator miR-34a suppressed the secretion but not the intracellular levels of IGFBP2 and SPP1 (Figure 3, H–J, and Supplemental Figure 4, D–H). We conclude that p53-dependent secretion drew on intracellular pools that were synthesized in a p53-independent manner.
Cancer cells secrete cytokines that drive inflammation, fibrosis, and angiogenesis in the tumor microenvironment (4). To determine whether G55-dependent secretion governs these processes, we performed flow cytometric analysis on flank tumors generated by G55-deficient or G55-replete 344SQ cells in syngeneic, immunocompetent mice and found that G55-deficient tumors had reduced numbers of endothelial cells, increased numbers of effector/memory CD8+ T cells, and reduced numbers of naive and exhausted CD8+ T cells (Supplemental Figure 5, A–I). To identify G55-dependent secreted cytokines that might mediate these activities, we performed multiplexed antibody-based assays on CM samples from G55-deficient and G55-replete 344SQ cells and found that concentrations of the proangiogenic cytokines IL-6, CXCL5/LIX, and VEGF-A were reduced by approximately 50% in G55-deficient cells (Supplemental Figure 5J). Consistent with these findings, G55 depletion in H1299 cells reduced IL-6 secretion (Supplemental Figure 5K) and inhibited the migratory and tube-forming activities of HUVECs in coculture assays (Supplemental Figure 5, L and M). Thus, G55-dependent secreted proteins drove protumorigenic processes in the tumor microenvironment.
A G55 antagonist inhibits secretion.
Cartier-Michaud et al. used a virtual screening strategy that combined high-throughput docking and pharmacophore filtering of chemical compounds to identify a small molecule (GRASPIN) that competes with binding proteins for insertion into the N-terminal PSD-95/discs large/ZO-1 (PDZ) domain of G55, which contains a surface groove and a hydrophobic cavity that serve as docking sites for binding partners (22, 23). GRASPIN inhibits G55-dependent spermatogenesis in mice and is under development as a male contraceptive (23). We tested the selectivity of this compound in H1299 cells and found that 10 μM GRASPIN treatment inhibited G55’s interactions with GOLGIN45 (G45) (Figure 4A), a known G55 binding partner (22), whereas binding of the G55 homolog GRASP65 to its binding partner GM130 was preserved (Figure 4B). GRASPIN exerted antiproliferative effects that were abrogated by shRNA-mediated G55 depletion in H1299 cells and H2122 cells (Figure 4, C and D, and Supplemental Figure 6, A–D). GRASPIN inhibited the colony-forming activities of TP53-mutant and TP53-WT LUAD cells, with IC50 values of approximately 5 and 10 μM, respectively (Figure 4, E and F, and Supplemental Figure 6, E and F), demonstrating enhanced sensitivity to GRASPIN in TP53-mutant LUAD cells. GRASPIN decreased IGFBP2 and SPP1 secretion (Figure 4G), and GRASPIN-dependent reductions in colony-forming activity were partially rescued by treatment with soluble IGFBP2 and SPP1 proteins (Supplemental Figure 6G), which had greater effects in combination than they did alone, suggesting that GRASPIN suppressed colony formation by inhibiting the secretion of IGFBP2, SPP1, and other proteins. Using a dose reported to inhibit spermatogenesis in vivo (23), we treated syngeneic, immunocompetent mice bearing subcutaneous 344SQ tumors for 3 weeks and found that GRASPIN treatment decreased G55/G45 interactions in tumor lysates (Figure 4H) and reduced 344SQ tumor size and metastatic activity (Figure 4, I and J). Antitumor activity was also observed in nu/nu mice bearing H1299 orthotopic lung tumors (Figure 4K) or TP53-mutant patient-derived LUAD xenografts (Figure 4L). GRASPIN sharply reduced the enhanced metastatic tumor burden driven by TP53-KO in A549 orthotopic lung tumors (Figure 4M). GRASPIN caused no weight loss (Supplemental Figure 6, H and I) or other signs of toxicity within the time frame of this study.
Figure 4. GRASPIN inhibits secretion and tumor growth.
(A and B) Co-IP of G55 and GOLGIN45 (G45) (A) or GRASP65 (G65) and GM130 (B). IP/WB analysis of H1299 cells transfected with GFP-tagged G55 (A) or G65 (B) and treated for 2 days with 10 μM GRASPIN or vehicle (DMSO). Whole cell lysates (input). (C) Cell proliferation after 4 days of GRASPIN treatment. Results relative to lowest GRASPIN dose in H1299_shCTL cells. n = 5. (D) Anchorage-independent colony formation. Colonies quantified after 14 days of GRASPIN or DMSO treatment. Results relative to controls (shCTL/DMSO). (E) Anchorage-dependent colony formation after 10 days of GRASPIN treatment. Results expressed relative to vehicle (0 μM). (F) The percentage reductions in E analyzed based on TP53 mutational status. TP53-WT, TP53-deleted (null), or TP53-missense-mutant (MT). (G) WB analysis of CM samples from indicated cells after 2 days of 10 μM GRASPIN or vehicle treatment. Densitometric values relative to DMSO (bar graph). (H) Co-IP of G55 and G45 in lysates of subcutaneous 344SQ tumors. Mice treated with GRASPIN or vehicle (DMSO) for 4 weeks beginning 7 days after tumor cell injection. (I and J) Tumor measurements on mice in H during treatment (I) and at necropsy (J). (K) Orthotopic lung tumor size and contralateral lung metastasis numbers in nu/nu mice treated for 3 weeks with 50 mg/kg GRASPIN or vehicle beginning 2 weeks after H1299 cell injection. (L) PDX tumor volumes in mice treated with 50 mg/kg GRASPIN or vehicle expressed as fold change relative to day 0. n = 10. (M) Orthotopic lung tumor size and contralateral lung metastasis numbers in nu/nu mice treated for 3 weeks with 50 mg/kg GRASPIN or vehicle beginning 2 weeks after A549 cell injection. n = 10. Results represent mean ± SD. n = 3, unless otherwise indicated. P values: ANOVA (E, I, and J), 2-sided t test (all others).
G55 activates CPS through a GOLPH3-independent mechanism.
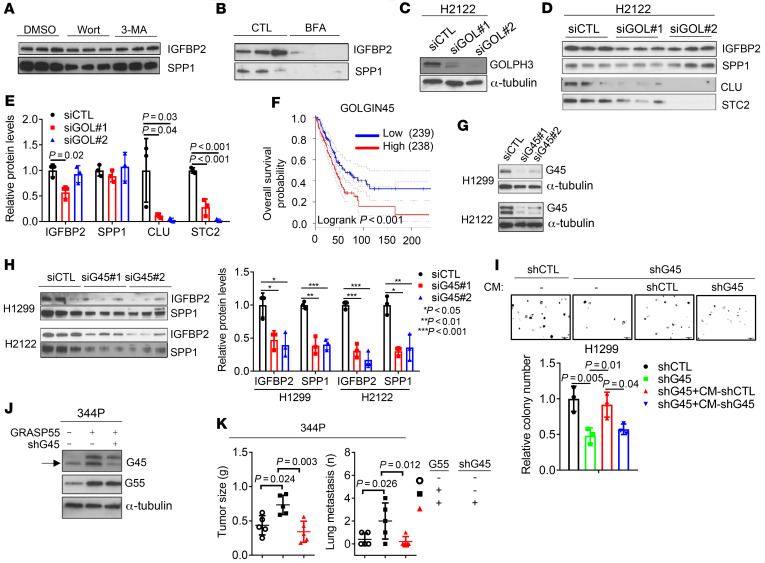
We sought to elucidate the molecular underpinnings of G55-dependent secretion. G55 is a key component of autophagy-dependent UPS, a mechanism by which proteins that lack signal peptides are transported to the cell surface (7, 8, 24). However, analysis of IGFBP2 and SPP1 protein sequences using prediction algorithms (https://www.uniprot.org, https://www.nextprot.org, https://elm.eu.org) identified classical N-terminal signal peptides in both proteins. Furthermore, the autophagy inhibitors wortmannin and 3′-methyladenine, which block UPS (25), had no detectable effect on IGFBP2 or SPP1 secretion in H1299 cells (Figure 5A), whereas brefeldin A, an ARF-1 inhibitor that blocks CPS (26), suppressed IGFBP2 and SPP1 secretion (Figure 5B), suggesting involvement of a CPS pathway.
Figure 5. G55 activates secretion via the Golgi.
(A and B) WB analysis of triplicate CM samples from H1299 cells treated for 16 hours with 100 nM wortmannin (Wort) or 5 mM 3-methyladenine (3-MA) (A) or 1 μg/mL brefeldin A (BFA) (B). (C) WB analysis to confirm GOLPH3 depletion in H2122 cells transfected with GOLPH3 siRNAs (siGOL#1, #2). (D and E) WB analysis of triplicate CM samples from H2122 cells transfected with siRNAs against GOLPH3. Densitometric values relative to siCTL (E). (F) Kaplan-Meier analysis of the TCGA LUAD cohort (n = 477 patients) scored on the basis of G45 mRNA levels above (high) or below (low) the median value. (G) WB analysis of G45 protein levels in cells transfected with siRNAs against G45 (siG45) or control siRNA (siCTL). (H) WB analysis of triplicate CM samples from cells in G. Densitometric values relative to siCTL. (I) Anchorage-independent colony formation assays in soft agar. G45-deficient (shG45) or G45-replete (shCTL) H1299 cells cultured with or without (–) CM from shG45- or shCTL-transfected H1299 cells. (J) WB analysis to confirm G45 depletion or ectopic G55 expression. 344P cells transfected with G55 or shG45 expression vectors. Empty vectors indicated by (–). Arrow indicates the G45-specific band. (K) Primary tumor weights and lung metastasis numbers in syngeneic, immunocompetent mice determined at necropsy 5 weeks after subcutaneous injection of 1 × 106 cells in J. Results represent mean ± SD. n = 3, unless otherwise indicated. P values: log-rank test (F), ANOVA (all others).
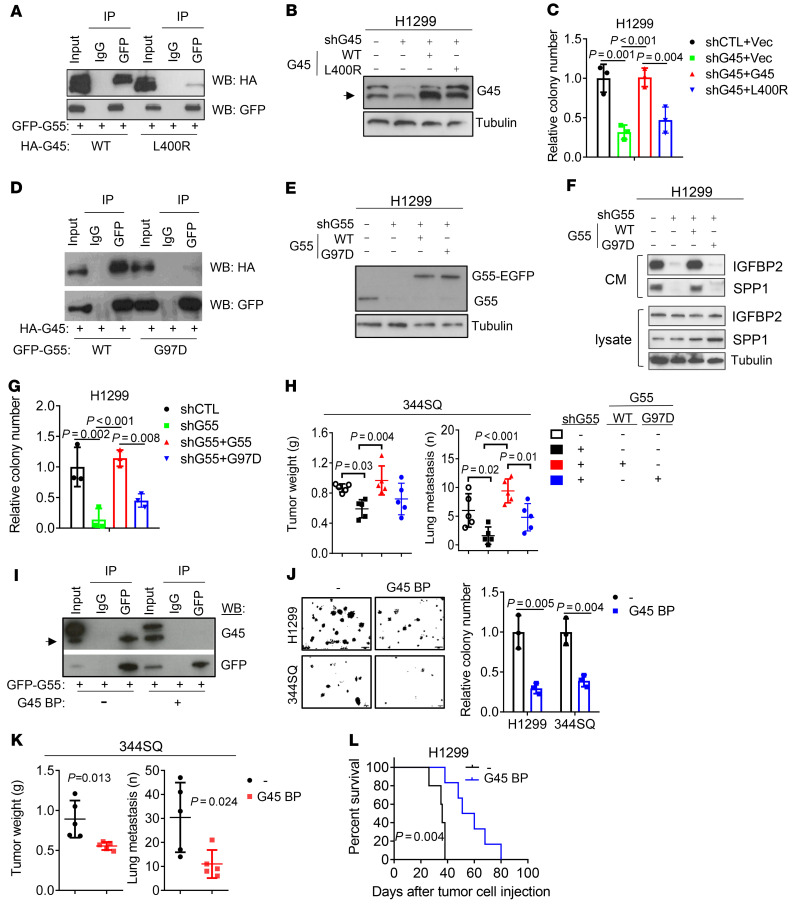
To elucidate how G55 activates CPS, we assessed whether G55-dependent secretion requires a GOLPH3–myosin-18A–RAB1B–containing protein complex that executes vesicular budding, fission, and release in the Golgi (9). Transfection of GOLPH3 or myosin-18A siRNAs in H2122 cells substantially reduced the secretion of clusterin (CLU) and stanniocalcin 2 (STC2), which are transported via CPS (25), but not IGFBP2 or SPP1 (Figure 5, C–E, and Supplemental Figure 7, A and B), suggesting that G55-driven CPS activation is GOLPH3 independent. To identify mediators of G55, we subjected H1299 cells to pulldown assays using GFP-tagged G55 as bait. Based on spectral counts of peptides identified by LC-MS analysis (Supplemental Figure 7C and Supplemental Table 2), G45 was the top hit among 88 proteins identified, an indication that G45 was a physiological binding partner of G55 in our model. G45 is a coiled-coil domain protein that facilitates SNARE-mediated vesicle fusion with Golgi membranes (12, 27). High G45 levels were correlated with shorter survival in the TCGA LUAD cohort (Figure 5F), and G45 is a known driver of vesicle biogenesis in the Golgi (12), but the mechanism by which G45 drives vesicle biogenesis and the biological significance of G45-dependent secretion in cancer remain unclear. We hypothesized that G45 mediates G55-dependent secretion and tested this hypothesis by performing siRNA-mediated G45 depletion studies in LUAD cell lines. G45 depletion in TP53-mutant LUAD cells impaired IGFBP2 and SPP1 secretion (Figure 5, G and H, and Supplemental Figure 7, D and E); reduced proliferative, migratory, invasive, and colony-forming activities (Supplemental Figure 7, F–H); and decreased HUVEC chemoattraction (Supplemental Figure 7I). The colony-forming activity of G45-deficient H1299 cells was rescued by treatment with CM from G45-replete but not G45-deficient H1299 cells (Figure 5I). G45 was required for ectopic G55 expression–driven 344P orthotopic lung tumor growth and metastasis (Figure 5, J and K). To determine whether G45 promotes secretion through its interactions with G55, we reconstituted G45-deficient H1299 cells with WT or mutant (L400R) G45 that cannot interact with G55 (ref. 22 and Figure 6, A and B) and found that only WT G45 reconstituted proliferative and colony-forming activities (Figure 6C and Supplemental Figure 8, A and B). Conversely, the secretory, colony-forming, invasive, and metastatic activities of G55-deficient cells could not be reconstituted with a G55 mutant (G97D) that cannot interact with G45 (ref. 22; Figure 6, D–H, and Supplemental Figure 8, C–F). Finally, ectopic expression of a 30–amino acid peptide that blocks G55-G45 interactions inhibited colony formation, reduced cell migration and invasion, blocked HUVEC recruitment, reduced tumor growth and metastasis, and prolonged the survival of tumor-bearing mice (Figure 6, I–L, and Supplemental Figure 8, G and H). Thus, G45 is a key component of the G55-dependent secretory pathway.
Figure 6. G55-dependent secretion and LUAD progression require G55/G45 interactions.
(A) Co-IP of G55 and G45. IP/WB assays on H1299 cells cotransfected with GFP-tagged G55 (GFP-G55) and HA-tagged WT or mutant (L400R) G45 (HA-G45). (B) WB analysis of H1299 cells cotransfected with G45 shRNA and WT or mutant (L400R) G45 (G45). Control vectors indicated by (–). Arrow indicates endogenous G45. (C) Anchorage-independent colony formation using cells in B. (D) Co-IP of G55 and G45. IP/WB analysis of H1299 cells cotransfected with HA-tagged G45 and GFP-tagged WT or mutant (G97D) G55. (E) WB analysis of H1299 cells cotransfected with G55 shRNA (shG55) and WT or mutant (G97D) G55. Endogenous (G55) and ectopic (G55-EGFP) G55. (F) WB analysis of CM samples from cells in E. (G) Anchorage-independent colony formation assays on cells in E. (H) Primary tumor weights and lung metastasis numbers at necropsy 5 weeks after subcutaneously injecting 1 × 106 344SQ cells into syngeneic, immunocompetent mice. Transfectants generated using the same vectors described in E. (I) Co-IP of G55 and G45. IP/WB assays on H1299 cells cotransfected with GFP-tagged G55 and a G45 peptide that blocks G55/G45 interactions (G45 BP). Empty vector indicated by (–). Arrow indicates endogenous G45. (J) Anchorage-independent colony formation using H1299 cells or 344SQ cells transfected with G45 BP or empty vector (–). (K) Primary tumor weights and lung metastasis numbers at necropsy 5 weeks after subcutaneous injection of 344SQ cells in J. (L) Kaplan-Meier analysis of nu/nu mice bearing orthotopic lung tumors generated by H1299 cells in J. Results represent mean ± SD. n = 3, unless otherwise indicated. P values: ANOVA (C, G, and H), log-rank test (L), 2-sided t test (all others).
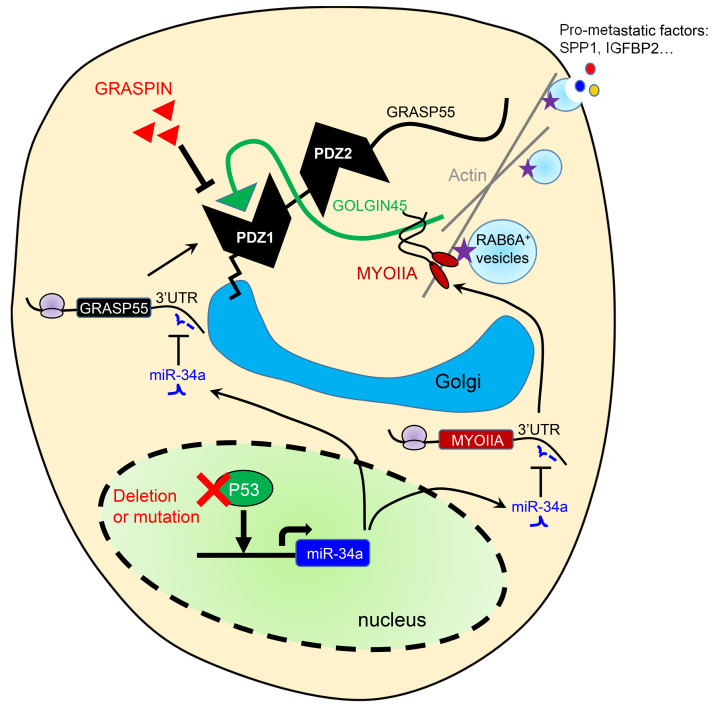
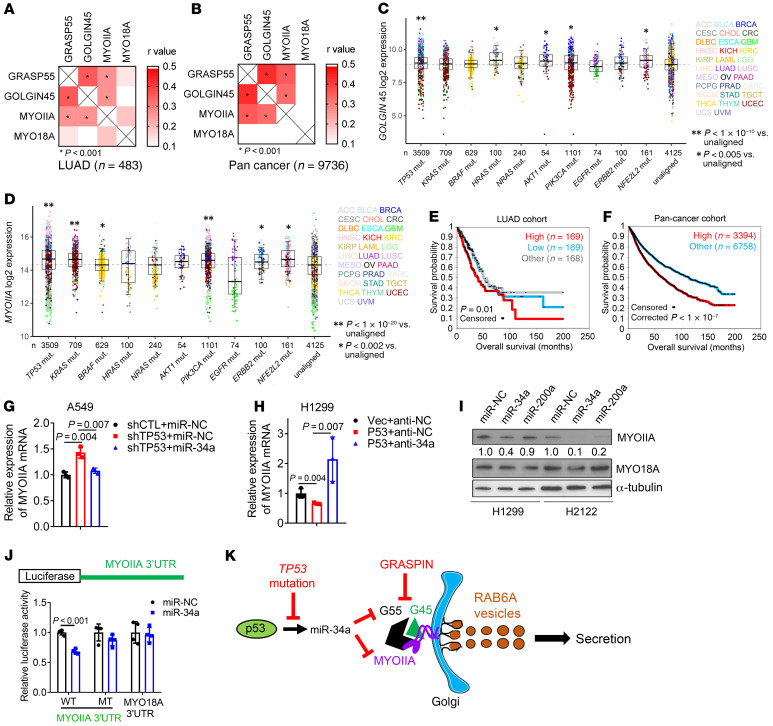
Figure 8. G55, G45, and MYOIIA are coordinately regulated in cancer.
(A and B) Pearson’s correlation of G55, G45, MYOIIA, and MYO18A in LUAD (A) and pan-cancer (B) TCGA cohorts. MYO18A included as negative control. (C and D) G45 (C) and MYOIIA (D) mRNA levels in tumors (dots) annotated on the basis of somatic DNA mutations in the TCGA pan-cancer cohort. Sample sizes indicated (n). Tumor types color-coded. “Unaligned” samples did not correlate with any of the indicated genetic mutations. (E and F) Kaplan-Meier analysis of TCGA cohorts on the basis of a 3-gene expression signature (G55, G45, MYOIIA). For the LUAD cohort (E), tumors in the top third (high), middle third (other), and bottom third (low) were compared. For the pan-cancer cohort (F), tumors in the top third (high) were compared with tumors in the middle and bottom third (other). (G) qPCR analysis of MYOIIA mRNA levels in A549 cells cotransfected with shp53 or shCTL and miR-34 or miR-NC. Results expressed relative to controls (shCTL/miR-NC). (H) qPCR analysis of MYOIIA mRNA levels in H1299 cells cotransfected with p53 or empty (Vec) expression vector and antagomir-34a (anti-34a) or control (anti-NC). Results expressed relative to controls (Vec/anti-NC). (I) WB analysis of H1299 cells and H2122 cells transfected with miR-34a, miR-200a, or control (miR-NC). Densitometric values relative to miR-NC. (J) Reporter assays on H1299 cells cotransfected with miR-34a or miR-NC and reporters containing MYOIIA 3′-UTRs with intact (WT) or mutant (MT) miR-34a binding sites. A MYO18A 3′-UTR reporter that lacks miR-34a binding sites included as a control. Values expressed relative to miR-NC. (K) Schematic illustration of our working model. Results represent mean ± SD. n = 3, unless otherwise indicated. P values: Pearson’s correlation (A and B), ANOVA (C, D, G, and H), univariate Cox (E and F), 2-way t test (J).
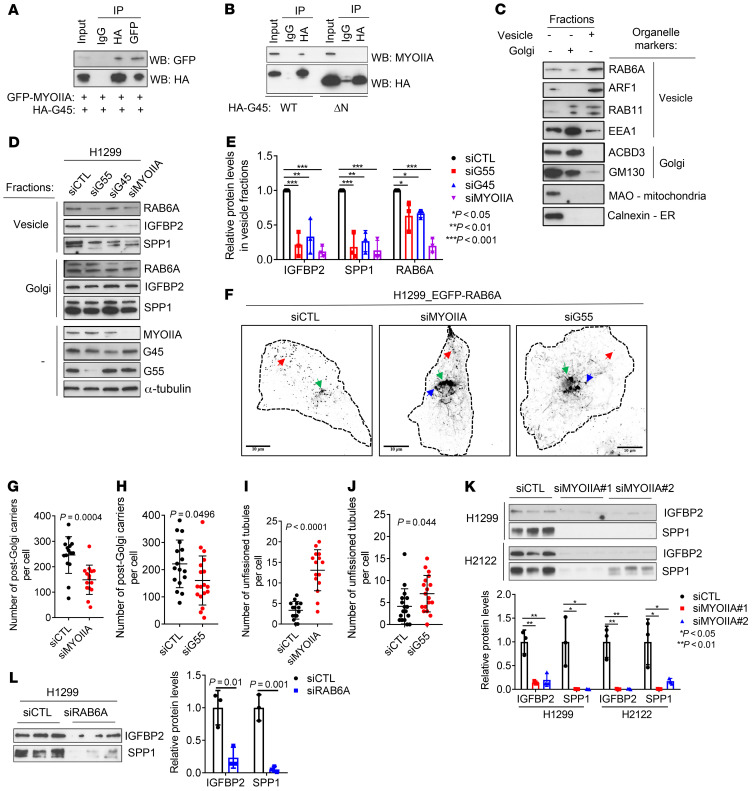
G55 and G45 lack contractile and GTPase activities required for vesicle biogenesis. To identify proteins that execute these functions, we performed pulldown assays on H1299 cell lysates using full-length HA-tagged G45 as bait. LC-MS of the immunoprecipitates identified 81 proteins (Supplemental Figure 8A and Supplemental Table 3), including G55 and myosin IIA (MYOIIA), a nonmuscle myosin that executes RAB6A vesicle fission in the Golgi (28). G45 coimmunoprecipitated with MYOIIA in cells transfected with tagged forms of MYOIIA and full-length but not N-terminally truncated G45 (Figure 7, A and B). Depletion of G55, G45, or MYOIIA in H1299 cells reduced RAB6A, IGFBP2, and SPP1 in vesicle-enriched but not Golgi-enriched cellular fractions (Figure 7, C–E). Depletion of G55 or MYOIIA decreased post-Golgi RAB6A vesicles and increased unfissioned Golgi-associated tubules (Figure 7, F–J, and Supplemental Video 1), an indication of impaired vesicle fission. In line with these findings, depletion of MYOIIA or RAB6A in LUAD cells decreased IGFBP2 and SPP1 secretion, colony formation, cell migration and invasion, and HUVEC recruitment (Figure 7, K and L, and Supplemental Figure 9, B–K), whereas siRNA-mediated depletion of other Golgi-associated RABs (RAB8, RAB10, RAB13) (29) did not inhibit IGFBP2 or SPP1 secretion (Supplemental Figure 9, L and M). Ectopic expression of full-length but not N-terminally truncated G45 enhanced H1299 cell colony formation, which was abrogated by siRNA-mediated MYOIIA depletion (Supplemental Figure 9N). Thus, G45 and MYOIIA are key mediators of G55.
Figure 7. G55 activates RAB6A vesicle biogenesis in the Golgi.
(A and B) Co-IP of G45 and myosin IIA (MYOIIA). IP/WB assays on H1299 cells cotransfected with GFP-tagged MYOIIA and HA-tagged G45 (A). H1299 cells transfected with WT or mutant (ΔN) HA-tagged G45 lacking N-terminal peptides (B). (C) WB analysis of total (–) and fractionated (+) H1299 cell lysates to confirm enrichment in “vesicle” and “Golgi” fractions on the basis of organelle markers (right). (D) WB analysis of RAB6A, IGFBP2, and SPP1 in Golgi- or vesicle-enriched fractions or total lysates (–) isolated from H1299 cells transfected with siRNA against G55, G45, or MYOIIA. (E) Densitometric analysis of results in D expressed relative to siCTL controls. (F) Contrast-adjusted confocal micrographs. RAB6A vesicles (red arrows) and unfissioned RAB6A tubules (blue arrows) emerge from the Golgi (green arrows). Cell boundary (dotted lines). Size bars: 10 μm. (G–J) Quantification of vesicles (G and H) and unfissioned tubules (I and J) per cell (dots). (K and L) WB analysis of CM samples isolated from H1299 cells and H2122 cells transfected with siRNAs against MYOIIA (K) or RAB6A (L). Densitometric values expressed relative to siCTL (bar graphs). Results represent mean ± SD. n = 3, unless otherwise indicated. P values: ANOVA (E and K), 2-way t test (all others).
p53 Governs a CPS regulatory axis.
To determine whether G55, G45, and MYOIIA are coordinately regulated in cancer, we queried the TCGA LUAD and pan-cancer cohorts and identified positive correlations between G55, G45, and MYOIIA (Figure 8, A and B). Like G55, G45 and MYOIIA expression was correlated with multiple oncogenic mutations (TP53, KRAS, BRAF, PIK3CA, ERBB2, and NFE2L2) (Figure 8, C and D). A 3-gene signature composed of G55, G45, and MYOIIA was negatively correlated with survival duration (Figure 8, E and F).
G45 protein expression was lower in G55-deficient than G55-replete LUAD cells (Supplemental Figure 10A), but G45 mRNA expression was unchanged (Supplemental Figure 10B), which led us to ask whether G55 regulates G45 protein stability. In the presence of cycloheximide, G45 protein stability was reduced by siRNA-mediated G55 depletion (Supplemental Figure 10C) or by a G45 mutation (L400R) that disrupts interactions with G55 (Supplemental Figure 10D). G45 ubiquitination was increased by siRNA-mediated G55 depletion (Supplemental Figure 10E) or the presence of L400R (Supplemental Figure 10F), and MG132 proteasome inhibitor treatment restored G45 protein expression in GORASP2-KO HeLa cells (Supplemental Figure 10G). Thus, G45 was degraded through the ubiquitin-mediated proteasomal pathway and was protected from proteasomal degradation by interactions with G55.
Based on the discovery of a predicted miR-34a binding site in the MYOIIA 3′-UTR (www.targetscan.org), we postulated that p53 deficiency relieves MYOIIA from miR-34a–dependent silencing. In A549 cells, shRNA-mediated p53 depletion increased MYOIIA expression, which was abrogated by ectopic miR-34a expression (Figure 8G). Conversely, ectopic p53 expression in H1299 cells downregulated MYOIIA expression, which was reversed by cotransfecting miR-34a antagomirs (Figure 8H). Demonstrating the specificity of this regulatory axis, MYOIIA expression was downregulated by ectopic expression of miR-34a but not miR-200a (Figure 8I), which is not predicted to bind the MYOIIA 3′-UTR. The functionality of the miR-34a binding site in the MYOIIA 3′-UTR was validated in H1299 cells cotransfected with a miR-34a expression vector and luciferase reporters containing WT or mutant MYOIIA 3′-UTRs or a MYO18A 3′-UTR that lacked predicted miR-34a binding sites (Figure 8J). Given that p53-dependent miR-34a silencing coordinately upregulated G55 and MYOIIA, we conclude that miR-34a is a nodal factor in the p53-dependent CPS regulatory axis.
Discussion
Here, we showed that p53 loss activated the secretion of protumorigenic effector proteins to create a secretory vulnerability that was actionable using a small molecule inhibitor of G55 (Figure 8K). Our findings suggest that therapeutic strategies designed to suppress G55-dependent secretion may be applicable to diverse oncogenic contexts in which G55 levels are upregulated. For example, activating BRAF mutations, which were correlated with high G55 levels, was reported to silence miR-34a (30) and may thereby activate G55-dependent secretion.
The findings presented here build on a growing body of evidence that p53 is a key secretory regulator in cancer. Loss of p53 enhances SDF-1 secretion by cancer-associated fibroblasts to promote tumor growth and metastasis (31). Deficiency of p53 in hepatic stellate cells releases factors that stimulate macrophage polarization into a tumor-promoting M2 state and enhances the proliferation of premalignant hepatic epithelial cells (32). Transcriptomes activated by TP53 gain-of-function mutations in cancer cells are enriched in secreted proteins, including heat shock protein 90α, which promotes metastasis (14, 33, 34). Thus, secretion underlies p53’s contextual cellular functions in cancer.
The N-terminal PDZ domain of G55 has been the focus of intense study. The PDZ domain facilitates the formation of G55 homodimers and transoligomers that maintain cisternal stacking and Golgi organelle integrity (35) and is the insertion site for other binding partners that carry out various cellular functions, including junctional adhesion molecule-C, which is essential for germ/Sertoli cell contacts and spermatogenesis (23), and the autophagy effector proteins lysosomal membrane protein 2 and microtubule-associated protein 1A/1B-light chain 3, which promote autophagosome-lysosome fusion and cargo transport via UPS (24). Here, we showed that G55-G45 interactions were required for G55-dependent RAB6A vesicle formation, which extends evidence that G55-G45 interactions generate a RAB2 effector complex that drives anterograde vesicle transport (12). Thus, G55 influences cellular functions through PDZ domain–binding proteins that play pleiotropic roles in coordinating vesicular transport through CPS and UPS pathways.
In the Golgi, proteins destined for secretion are sorted and loaded into vesicles through a complex process involving secretory carrier membrane proteins (e.g., CARTS, LAMP1-RUSH, RAB6, and ARF1) (36), transmembrane cargo receptors (e.g., Sortlin and Wntless), and cargo adaptors (e.g., Golgi-localized, γ-ear–containing, ADP-ribosylation factor binding proteins) (37). However, the way in which oncogenic mutations regulate secretion at the level of cargo sorting and loading remains unclear. Although our study does not directly address this question, several findings presented here support the existence of an oncogene-regulated cargo-sorting apparatus in the Golgi. For example, in addition to decreasing the secretion of protumorigenic effector proteins, G55 depletion increased the secretion of other proteins, including cystatin-B, a potent tumor suppressor (38). Furthermore, the G55-dependent secreted proteins identified here do not overlap with GOLPH3-dependent secreted proteins in the same LUAD cell lines (39). Notably, the cargo-sorting protein ARFGAP1 (40) was identified in G45 pulldown assays (Supplemental Table 3).
There are important limitations to our study. Our focus was on TP53 mutations, but the extent to which co-occurring mutations influence p53-regulated secretion remains unclear. We demonstrated enhanced vulnerability of TP53-mutant LUAD cells to GRASPIN, but determining whether TP53 mutations predict enhanced clinical benefit from GRASPIN treatment awaits clinical trials. Our biochemical, cellular, and in vivo studies with GRASPIN revealed no evidence of off-target effects, but GRASPIN’s selectivity, long-term toxicities, and mechanisms of de novo and acquired resistance were not addressed.
In conclusion, we have identified an actionable secretory vulnerability in p53-deficient malignancies. Basket clinical trials are warranted to test the efficacy of G55 antagonists in TP53-mutant malignancies and other conditions in which secretion plays a contributing role, including acquired resistance to targeted therapies in RTK-mutant cancers and fibrotic and inflammatory diseases (41–43).
Methods
Animal husbandry.
Mice received standard care and were euthanized according to the standards set forth by the IACUC at the University of Texas MD Anderson Cancer Center. Syngeneic, immunocompetent mice (n = 8–10 per group) were injected with 1 × 106 murine LUAD cells in the right flank and necropsied after 5 weeks. Primary tumors were weighed, and lung metastases on the pleural surfaces were counted. To generate orthotopic lung tumors, we subjected nu/nu mice (n = 10 mice per cohort) to intrathoracic injection with 1 × 106 human LUAD cells, necropsied them after 5 weeks, and measured primary tumor size and the number of metastases on the contralateral lung surface. For GRASPIN treatment experiments, we injected syngeneic, immunocompetent mice or nu/nu mice with 1 × 106 murine or human LUAD cells or lung cancer patient–derived xenografts subcutaneously or intrathoracically and initiated treatment with GRASPIN (25 mg/kg or 50 mg/kg) or vehicle (10% DMSO and 90% corn oil) 1 week after tumor cell injection. Drugs were administered intraperitoneally every 2 days. The mice were checked daily and necropsied at first sign of morbidity or after the last treatment to measure primary tumor size, count metastases to the contralateral lung, and obtain tumor tissues for analysis.
Cell lines.
Murine LUAD cells generated in mice that expressed K-rasG12D and p53R172H (344P, 344SQ, 531LN2, and 393P) (44), human LUAD cells (A549, H1299, H460, H596, H23, H1792, H441, H358, H157, H226, H2122, CALU-1, H292, and H520), and CAOV-4 human ovarian cancer cell lines were cultured in RPMI 1640 containing 10% FBS. MDA-MB-468 cell lines were cultured in DMEM containing 10% FBS. HUVECs were cultured in endothelial cell growth medium-2 BulletKit (CC-5035, Lonza). Cells were maintained at 37°C in an incubator with a humidified atmosphere containing 5% CO2. Cells were transfected with jetPRIME Versatile DNA/siRNA transfection reagent (Polyplus). Stable cell transfectants were selected by using puromycin (for pLVX or pLKO.1 vectors) or G418 (for pcDNA3.1 and pEGFP-C3 vectors). TP53-KO A549 cells (guide RNA sequence: 5′-GAGCGCTGCTCAGATAGCGA-3′) were generated in the Cell-Based Assay Screening Service Core Facility at Baylor College of Medicine. Parental and CRISPR/Cas9-mediated GORASP2-KO HeLa cells were a gift from Yanzhuang Wang (University of Michigan, Ann Arbor, Michigan, USA) (45).
Reagents.
We purchased SYBR Green, FBS, HEPES-buffered media, DMEM, RPMI 1640, Alexa Fluor–tagged secondary antibodies, and DAPI from Life Technologies; puromycin from InvivoGene; paraformaldehyde from Electron Microscopy Sciences; Transwell and Matrigel-coated Boyden chambers from BD Biosciences; G418 from Corning; GRASPIN (PubChem CID: 3113208) from Vitas-M Laboratory (STK700118); and wortmannin (W1628), 3-methyladenine (SAE0107), nutlin-3 (444151), and brefeldin A (B7651) from MilliporeSigma. We purchased the following from MilliporeSigma: shRNAs against murine G55 (TRCN0000278363,TRCN0000077518, and TRCN0000077520) and murine G45 (TRCN0000096503), shRNAs against human G55 (TRCN0000278363 and TRCN0000130898), human IGFBP2 (TRCN0000318660, TRCN0000318716, TRCN0000318659, and TRCN0000006576), human SPP1 (TRCN0000342616, TRCN0000342561, TRCN0000004878 and TRCN0000004875), human G45 (TRCN0000013243); siRNAs against human G55 (SASI_Hs01_00012371 and SASI_Hs02_00348040), human IGFBP2 (SASI_Hs02_00302878 and SASI_Hs01_00039595), human SPP1 (SASI_Hs01_00174866 and SASI_Hs01_00174867), human G45 (SASI_Hs01_00073080 and SASI_Hs01_00073083), human MYOIIA (SASI_Hs01_00197338 and SASI_Hs01_00197339), human MYO18A (SASI_Hs01_00224320 and SASI_Hs01_00224321), GOLPH3 (SASI_Hs02_00355527 and SASI_Hs01_00133692); esiRNAs against human RAB6A (EHU113451), human RAB8A (EHU054251), human RAB10 (EHU131761), human RAB13 (EHU107931), and EGFP (EHUEGFP). We purchased primary antibodies against G55 (NB100-74431) from Novus Biologicals; against GM130 (560066) from BD Transduction Laboratories; against G45 (OAAN01805) from Aviva Systems Biology; against GOLPH3 (ab98023) from Abcam; against IGFBP2 (11065-3-AP), SPP1 (22952-1-AP), MYO18A (14611-1-AP), MAOA (10539-1-AP), and RAB6A (10187-2-AP) from Proteintech; against α-tubulin (T9026), EGFP (G6539) from MilliporeSigma; against PARP-1 (9542), hemagglutinin (3724 and 2367), β-actin (4970), p53 (9282), GM130 (12480), EEA1 (3288), ubiquitin (43124), and MYOIIA (3403) from Cell Signaling Technology; against ACBD3 (sc-101277), GRASP65 (sc-374423), and p53 (sc-126) from Santa Cruz Biotechnology; against calnexin (ab22595) and RAB11 (ab95375) from Abcam; and against RNA polymerase II (05-623, clone CTD4H8) from MilliporeSigma. We purchased neutralizing antibodies against IGFBP2 (AF674) and SPP1 (AF1433) from R&D Systems. We purchased recombinant IGFBP2 (ab63223) and SPP1 (ab92964) proteins from Abcam. We purchased a human IL-6 ELISA kit (ab178013) from Abcam. rG55-pEGFP-N3 was a gift from Yanzhuang Wang at the University of Michigan Medical School. Myosin-IIA-GFP was a gift from Matthew Krummel (Addgene plasmid 38297). EGFP-RAB6A was a gift from Marci Scidmore (Addgene plasmid 49469).
Vector construction.
The human G55 and G45 coding sequences were isolated by performing PCR on cDNA prepared from H1299 cells, and then cloned into pLVX-Puro or pEGFP-C3 (Clontech) or pcDNA3.1 (Invitrogen). To generate a G45 BP expression vector for transfection studies in cells, the DNA sequence encoding the last 30 amino acids of the G45 C-terminus was amplified by PCR and cloned into the pEGFP-C3 vector. The G55, MYH9, and MYO18A 3′-UTR sequences were amplified from genomic DNA from H1299 cells by PCR and cloned into pCI-neo-hRL vector. Mutations were introduced by PCR method. PCR primers are listed in Supplemental Table 4.
Cell proliferation, colony formation, apoptosis, migration, and invasion assays.
Cell proliferation assays were performed using Cell Proliferation Reagent WST-1 (Roche) according to the manufacturer’s protocol. Colony formation at low density on plastic and in soft agarose were performed as described previously (46). Apoptosis was determined by flow cytometry using a Dead Cell Apoptosis Kit (Thermo Fisher Scientific, V13242) according to the manufacturer’s instructions. Migration and invasion assays were performed in Transwell and Matrigel-coated Boyden chambers, respectively, as we have described (47). For endothelial recruitment assays, 2 × 104 HUVECs seeded in the top chamber in Transwell plates, cocultured with H1299 cells for 12 hours, and migrated HUVECs were stained with 0.1% crystal violet, photographed, and quantified.
Tube formation assays.
As previously described (39), HUVECs (1 × 105/well) were seeded into the lower chambers of Transwell plates containing Matrigel-coated filters, and tumor cells (2 × 104/well) were seeded in the upper chambers. After 8 hours, HUVECs were imaged and tube-like structures were quantified.
ChIP assays.
A549 cell lysates were formaldehyde fixed and digested using SimpleChIP Enzymatic Chromatin IP Kit (9002, Cell Signaling Technology) following the manufacturer’s instructions. Resulting chromatin was immunoprecipitated with anti–RNA polymerase II or anti–rabbit IgG antibodies (Santa Cruz Biotechnology). DNA was eluted and purified with the MinElute Reaction Cleanup Kit (Qiagen) and subjected to quantitative PCR. PCR primers are listed in Supplemental Table 4.
Luciferase reporter assays.
3′-UTR luciferase reporters (10 ng), pGL3-enhancer control (100 ng), and miR mimics (20 nM) were cotransfected into H1299 cells. After 24 hours, luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega).
Western blot and IP assays.
For Western blot analysis, protein lysates were separated on a 4% to 20% Bis-Tris gel and transferred to PVDF membranes using Trans-Blot Turbo Transfer System (Bio-Rad). The membranes were blocked in 5% milk and probed with primary antibodies following a standard protocol. For IP followed by Western blot analysis, H1299 cells were transfected with indicated expression vectors, lysed in 1× radioimmunoprecipitation assay buffer (RIPA buffer, Cell Signaling Technology) 48 hours later, and incubated with antibodies or control IgG at 4°C overnight. The immune complex was captured with protein G agarose beads (GE Healthcare Life Sciences), washed with lysis buffer 4 times, and boiled in 1× SDS loading buffer for 5 minutes. The resulting samples were analyzed by Western blot using relevant antibodies.
Quantitative RT-PCR assays.
Total RNA was isolated from cells using an RNeasy Mini Kit (Qiagen, 74106) and subjected to reverse transcription using the qScript cDNA superMix (Quanta Biosciences). mRNA levels were determined using SYBR Green Real-Time PCR Master Mixes (Thermo Fisher Scientific) and normalized on the basis of ribosomal protein L32 (Rpl32) mRNA. PCR primers are listed in Supplemental Table 4.
CM assays.
As previously described (48), CM samples were isolated, filtered through a 0.45-μm filter, mixed with an equal volume of complete growth medium, and applied to cells. For Transwell experiments, cells that had been incubated in serum-free medium for 16 hours were seeded in the top chambers in serum-free medium, and CM samples were added to the bottom wells. For colony formation assays, cells were incubated in CM samples that were refreshed every 2 days.
IL-6 ELISA.
H1299 cells in a 6-well plate were transfected with siRNAs against G55 or control siRNA. After 48 hours, cells were incubated in free medium for 16 hours. CM samples were collected and subjected to ELISA assay to quantify IL-6 concentrations using a Human IL-6 ELISA Kit (ab46027, Abcam) according to the manufacturer’s instructions.
LC-MS.
To identify G55- and G45-interacting proteins, H1299 cells were transfected with GFP-G55 or HA-G45, lysed in 1× RIPA buffer, and subjected to IP with anti-GFP or anti-HA antibodies, respectively, linked to agarose G beads. Immune complexes were subjected to LC-MS. To identify G55-dependent secreted proteins, CM samples were collected, concentrated, and subjected to LC-MS as previously described (49). Samples were solubilized with 5% SDS, 50 mM TEAB, pH 7.55, final volume 25 μL. The sample was then centrifuged at 17,000g for 10 minutes to remove any debris. Proteins were reduced by making the solution 20 mM TCEP (Thermo Fisher Scientific, 77720) and incubated at 65°C for 30 minutes. The sample was cooled to room temperature and 1 μL of 0.5 M iodoacetamide acid added and allowed to react for 20 minutes in the dark. Next, 2.75 μL of 12% phosphoric acid was added to the protein solution, and 165 μL of binding buffer (90% methanol, 100 mM TEAB, final pH 7.1) was then added to the solution. The resulting solution was added to S-Trap spin column (protifi.com) and passed through the column using a benchtop centrifuge (30-second spin at 4,000g). The spin column was washed with 400 μL of binding buffer and centrifuged. This was repeated 2 more times. Trypsin was added to the protein mixture in a ratio of 1:25 in 50 mM TEAB, pH 8, and incubated at 37°C for 4 hours. Peptides were eluted with 80 μL of 50 mM TEAB, followed by 80 μL of 0.2% formic acid, and finally 80 μL of 50% acetonitrile, 0.2% formic acid. The combined peptide solution was then dried in a speed vac and resuspended in 2% acetonitrile, 0.1% formic acid, 97.9% water and placed in an autosampler vial.
Cytokine and chemokine analysis.
Cells were seeded (5 × 105 per well) in triplicate in a 6-well plate. CM samples were collected and analyzed using a multiplex magnetic bead–based assay (Luminex 200 System).
Isolation of Golgi and vesicle fractions.
A Minute Golgi Apparatus Enrichment Kit (GO-037, Invent Biotechnologies) was used to enrich cell lysates in Golgi and vesicle fractions following the manufacturer’s instructions. Briefly, 3 × 107 cells were suspended in 550 μL buffer A. The cell suspension was loaded into a filter cartridge, centrifuged at 16,000g for 30 seconds and then for 5 minutes at 5,000g at 4°C without removing the filter. After centrifugation, the filter was removed and the supernatants were transferred to a fresh 1.5 mL tube and centrifuged at 4°C for 30 minutes at 16,000g. After centrifugation, 400 μL supernatant was transferred to a fresh 1.5 mL tube. The pellet contained mainly mitochondria, endoplasmic reticulum, lysosomes, and plasma membranes. Next, 400 μL buffer B was added to the tube containing 400 μL supernatant. The tube was placed on ice for 10 minutes and centrifuged at 8,000g for 5 minutes at 4°C. Supernatants containing secretory vesicles associated with trans-Golgi membranes were transferred to a new tube. The remaining pellet was resuspended in 200 μL cold buffer A and centrifuged at 8,000g for 5 minutes. The supernatant was transferred to a fresh 1.5 mL tube and 100 μL cold buffer C was added to the tube and mixed by vortexing. The tube was incubated on ice for 20 minutes and centrifuged at 8,000g for 10 minutes to isolate the pellet, which constituted the “Golgi fraction.” To isolate vesicle fractions, 100 μL buffer D was added to supernatants containing secretory vesicles associated with trans-Golgi membranes, and the mixture was incubated on ice for 20 minutes and centrifuged at 16,000g for 5 minutes at 4°C. The supernatant was removed to isolate the pellet, which constituted the “vesicle fraction.” The pellets containing Golgi and vesicle fractions were resuspended in 50 μL SDS loading buffer, heated for 10 minutes at 98°C, and analyzed by Western blot.
RAB6A vesicle trafficking.
Live-cell imaging was performed on H1299 cells stably expressing RAB6A-EGFP. Cells were plated on 35-mm glass-bottom plates (MatTek), transfected with siRNAs, and imaged 48 to 72 hours later using a Nikon A1+ confocal microscope equipped with 488 nm laser line, GaAsP detectors, and ×100 1.45 NA objective. Live-cell imaging conditions (5% CO2, 37°C, and 90% humidity) were maintained on stage using an Okolab incubation chamber. For optimal spatial and temporal resolution, Nyquist sampling was applied in bidirectional image acquisition mode without averaging. Images were acquired at 1 fps for 2.5 to 3 minutes. After acquisition, images were bleach-corrected (Huygens Professional). RAB6A vesicles were identified and tracked using the spots module in Imaris (Bitplane). A spot diameter of 500 nm was applied to segment the vesicles before tracking them using an autoregressive motion algorithm. To avoid unnecessary splitting of tracks due to uneven illumination or out-of-plane movement, a gap-closing algorithm was used. As described previously (28), unfissioned tubular structures were defined as RAB6A-expressing tubular structures that were attached to the Golgi. For each cell, a mask that isolated the Golgi and connected tubular structures was generated, and unfissioned tubules were counted and measured.
Data and materials availability.
All data associated with this study are present in the paper or the Supplementary Materials.
Statistics.
For analysis of G55 mRNA levels in tumors annotated on the basis of somatic DNA mutations in the TCGA pan-cancer cohort (50), oncogenic mutations were defined by single nucleotide variant or indel falling within a “hotspot” residue as defined by Chang et al. (51). For pan-cancer TCGA cohort analyses, P values are by univariate Cox, stratified by cancer type. For LUAD cohort analyses, P values are by univariate Cox. For computing multiple-gene signature scores, we first normalized each gene to standard deviations from the median across the pan-cancer cohort (using log2-transformed expression values), and then took the average of the 3 normalized values. Unless stated otherwise, the results shown are representative of replicated experiments and are the mean ± SD from triplicate samples or randomly chosen cells within a field. Statistical evaluations were carried out with Prism 6 (GraphPad Software, Inc.). Unpaired 2-tailed Student’s t tests were used to compare the mean values of 2 groups. One-way ANOVA with Dunnett’s test was used for comparing multiple treatments to a control. P values less than 0.05 were considered statistically significant. Plots were generated using software (http://gepia.cancer-pku.cn/). Box plots: 5%, 25%, 50%, 75%, 95%. Tumor type abbreviations are described here (http://cancergenome.nih.gov).
Study approval.
All mouse studies were approved by the IACUC at the University of Texas MD Anderson Cancer Center.
Author contributions
XT conceived, designed, executed, and interpreted cell culture and in vivo experiments. PB conceived, designed, executed, and interpreted all of the experiments analyzing vesicular trafficking by confocal microscopy. LS assisted XT with IP experiments and performed the HUVEC tube formation assay. XL assisted XT with the in vivo experiments. JY bred mice for the in vivo studies. NBR performed cell culture assays. HFG assisted with G55 protein crystal structure analysis. BLR and NBR performed flow cytometric analysis of tumor tissues. DLG supervised the efforts of BLR. WKR directed and interpreted mass spectrometry experiments. CJC directed and interpreted bioinformatic analyses. JMK conceived and supervised the project and contributed to the design and interpretation of all experiments.
Supplementary Material
Acknowledgments
We thank Fengju Chen for technical assistance. This work was supported by the NIH through R01 CA181184 (to JMK), R01 CA2111125 (to JMK), P30 CA125123 (to CJC), K99 CA225633 (to HFG), and Lung Cancer Research Foundation FP 00005299 (to XT). JMK holds the Gloria Lupton Tennision Distinguished Endowed Professorship in Lung Cancer. DLG is an R. Lee Clark Fellow of the University of Texas MD Anderson Cancer Center, supported by the Jeane F. Shelby Scholarship Fund. The work was also supported by the generous philanthropic contributions to the University of Texas MD Anderson Lung Cancer Moon Shots Program.
Version 1. 09/15/2020
In-Press Preview
Version 2. 01/04/2021
Print issue publication
Footnotes
Conflict of interest: DLG serves on scientific advisory committees for AstraZeneca, GlaxoSmithKline, Sanofi, and Janssen; provides consultations for Ribon Therapeutics; and has research support from Janssen, Takeda, and AstraZeneca. JMK has received consulting fees from Halozyme.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e137186.https://doi.org/10.1172/JCI137186.
See the related Commentary at Targeting the mutant p53 secretome.
Contributor Information
Xiaochao Tan, Email: xiaochtan@163.com.
Lei Shi, Email: LShi2@mdanderson.org.
Priyam Banerjee, Email: PBanerjee@mdanderson.org.
Xin Liu, Email: XLiu9@mdanderson.org.
Hou-Fu Guo, Email: HGuo5@mdanderson.org.
Jiang Yu, Email: JYu1@mdanderson.org.
Neus Bota-Rabassedas, Email: MBota@mdanderson.org.
B. Leticia Rodriguez, Email: BLRodriguez@mdanderson.org.
Don L. Gibbons, Email: dlgibbon@mdanderson.org.
William K. Russell, Email: wirussel@utmb.edu.
Chad J. Creighton, Email: creighto@bcm.edu.
Jonathan M. Kurie, Email: jkurie@mdanderson.org.
References
- 1.Werb Z, Lu P. The role of stroma in tumor development. Cancer J. 2015;21(4):250–253. doi: 10.1097/PPO.0000000000000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prager BC, Xie Q, Bao S, Rich JN. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019;24(1):41–53. doi: 10.1016/j.stem.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Stanger BZ. The tumor as organizer model. Science. 2019;363(6431):1038–1039. doi: 10.1126/science.aau9861. [DOI] [PubMed] [Google Scholar]
- 4.Paltridge JL, Belle L, Khew-Goodall Y. The secretome in cancer progression. Biochim Biophys Acta. 2013;1834(11):2233–2241. doi: 10.1016/j.bbapap.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295(5564):2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Gee HY, Lee MG. Unconventional protein secretion — new insights into the pathogenesis and therapeutic targets of human diseases. J Cell Sci. 2018;131(12):jcs213686. doi: 10.1242/jcs.213686. [DOI] [PubMed] [Google Scholar]
- 7.Chiritoiu M, Brouwers N, Turacchio G, Pirozzi M, Malhotra V. GRASP55 and UPR control interleukin-1β aggregation and secretion. Dev Cell. 2019;49(1):145–155.e4. doi: 10.1016/j.devcel.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011;30(23):4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halberg N, Sengelaub CA, Navrazhina K, Molina H, Uryu K, Tavazoie SF. PITPNC1 recruits RAB1B to the Golgi network to drive malignant secretion. Cancer Cell. 2016;29(3):339–353. doi: 10.1016/j.ccell.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dippold HC, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139(2):337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, et al. Monomerization and ER relocalization of GRASP is a requisite for unconventional secretion of CFTR. Traffic. 2016;17(7):733–753. doi: 10.1111/tra.12403. [DOI] [PubMed] [Google Scholar]
- 12.Short B, Preisinger C, Körner R, Kopajtich R, Byron O, Barr FA. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001;155(6):877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeudall WA, et al. Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis. 2012;33(2):442–451. doi: 10.1093/carcin/bgr270. [DOI] [PubMed] [Google Scholar]
- 15.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119(6):861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Drewniok C, et al. Molecular interactions of B-CAM (basal-cell adhesion molecule) and laminin in epithelial skin cancer. Arch Dermatol Res. 2004;296(2):59–66. doi: 10.1007/s00403-004-0481-4. [DOI] [PubMed] [Google Scholar]
- 17.Duan L, Motchoulski N, Danzer B, Davidovich I, Shariat-Madar Z, Levenson VV. Prolylcarboxypeptidase regulates proliferation, autophagy, and resistance to 4-hydroxytamoxifen-induced cytotoxicity in estrogen receptor-positive breast cancer cells. J Biol Chem. 2011;286(4):2864–2876. doi: 10.1074/jbc.M110.143271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shevde LA, Samant RS. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klose R, et al. Inactivation of the serine protease HTRA1 inhibits tumor growth by deregulating angiogenesis. Oncogene. 2018;37(31):4260–4272. doi: 10.1038/s41388-018-0258-4. [DOI] [PubMed] [Google Scholar]
- 20.Yao X, Sun S, Zhou X, Guo W, Zhang L. IGF-binding protein 2 is a candidate target of therapeutic potential in cancer. Tumour Biol. 2016;37(2):1451–1459. doi: 10.1007/s13277-015-4561-1. [DOI] [PubMed] [Google Scholar]
- 21.Revelo NH, Ter Beest M, van den Bogaart G. Membrane trafficking as an active regulator of constitutively secreted cytokines. J Cell Sci. 2019;133(5):jcs234781. doi: 10.1242/jcs.234781. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, Li B, Huang X, Morelli X, Shi N. Structural basis for the interaction between Golgi reassembly-stacking protein GRASP55 and Golgin45. J Biol Chem. 2017;292(7):2956–2965. doi: 10.1074/jbc.M116.765990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cartier-Michaud A, et al. Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis. PLoS Genet. 2017;13(6):e1006803. doi: 10.1371/journal.pgen.1006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Wang L, Lak B, Li J, Jokitalo E, Wang Y. GRASP55 senses glucose deprivation through O-GlcNAcylation to promote autophagosome-lysosome fusion. Dev Cell. 2018;45(2):245–261.e6. doi: 10.1016/j.devcel.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noh SH, et al. Specific autophagy and ESCRT components participate in the unconventional secretion of CFTR. Autophagy. 2018;14(10):1761–1778. doi: 10.1080/15548627.2018.1489479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orci L, et al. Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell. 1991;64(6):1183–1195. doi: 10.1016/0092-8674(91)90273-2. [DOI] [PubMed] [Google Scholar]
- 27.Tiwari N, et al. Golgin45-Syntaxin5 interaction contributes to structural integrity of the Golgi stack. Sci Rep. 2019;9(1):12465. doi: 10.1038/s41598-019-48875-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol. 2010;12(7):645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- 29.Goud B, Liu S, Storrie B. Rab proteins as major determinants of the Golgi complex structure. Small GTPases. 2018;9(1–2):66–75. doi: 10.1080/21541248.2017.1384087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christoffersen NR, et al. p53-Independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17(2):236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 31.Addadi Y, et al. p53 Status in stromal fibroblasts modulates tumor growth in an SDF1-dependent manner. Cancer Res. 2010;70(23):9650–9658. doi: 10.1158/0008-5472.CAN-10-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lujambio A, et al. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153(2):449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solomon H, et al. Various p53 mutant proteins differently regulate the Ras circuit to induce a cancer-related gene signature. J Cell Sci. 2012;125(pt 13):3144–3152. doi: 10.1242/jcs.099663. [DOI] [PubMed] [Google Scholar]
- 34.Ubertini V, et al. Mutant p53 gains new function in promoting inflammatory signals by repression of the secreted interleukin-1 receptor antagonist. Oncogene. 2015;34(19):2493–2504. doi: 10.1038/onc.2014.191. [DOI] [PubMed] [Google Scholar]
- 35.Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188(2):237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stalder D, Gershlick DC. Direct trafficking pathways from the Golgi apparatus to the plasma membrane. Semin Cell Dev Biol. 2020;107:112–125. doi: 10.1016/j.semcdb.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Y, Sirkis DW, Schekman R. Protein sorting at the trans-Golgi network. Annu Rev Cell Dev Biol. 2014;30:169–206. doi: 10.1146/annurev-cellbio-100913-013012. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Shi Z, Huang J, Zou X. CSTB downregulation promotes cell proliferation and migration and suppresses apoptosis in gastric cancer SGC-7901 cell line. Oncol Res. 2016;24(6):487–494. doi: 10.3727/096504016X14685034103752. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Tan X, et al. PI4KIIIβ is a therapeutic target in chromosome 1q-amplified lung adenocarcinoma. Sci Transl Med. 2020;12(527):eaax3772. doi: 10.1126/scitranslmed.aax3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SY, Yang JS, Hong W, Premont RT, Hsu VW. ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J Cell Biol. 2005;168(2):281–290. doi: 10.1083/jcb.200404008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones VS, Huang RY, Chen LP, Chen ZS, Fu L, Huang RP. Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochim Biophys Acta. 2016;1865(2):255–265. doi: 10.1016/j.bbcan.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Gong B, et al. Secreted PD-L1 variants mediate resistance to PD-L1 blockade therapy in non-small cell lung cancer. J Exp Med. 2019;216(4):982–1000. doi: 10.1084/jem.20180870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson MB, et al. Stress hormones promote EGFR inhibitor resistance in NSCLC: implications for combinations with β-blockers. Sci Transl Med. 2017;9(415):eaao4307. doi: 10.1126/scitranslmed.aao4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23(18):2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bekier ME, et al. Knockout of the Golgi stacking proteins GRASP55 and GRASP65 impairs Golgi structure and function. Mol Biol Cell. 2017;28(21):2833–2842. doi: 10.1091/mbc.E17-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn YH, et al. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest. 2012;122(9):3170–3183. doi: 10.1172/JCI63608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan X, et al. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J Clin Invest. 2018;128(4):1267–1282. doi: 10.1172/JCI97225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brady JJ, et al. An Arntl2-driven secretome enables lung adenocarcinoma metastatic self-sufficiency. Cancer Cell. 2016;29(5):697–710. doi: 10.1016/j.ccell.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson BA, et al. Peptidoglycan-associated cyclic lipopeptide disrupts viral infectivity. J Virol. 2019;93(22):e01282-19. doi: 10.1128/JVI.01282-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen F, et al. Pan-cancer molecular classes transcending tumor lineage across 32 cancer types, multiple data platforms, and over 10,000 cases. Clin Cancer Res. 2018;24(9):2182–2193. doi: 10.1158/1078-0432.CCR-17-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang MT, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34(2):155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.