Abstract
Metabolic reprogramming is a common hallmark of cancer, but a large variability in tumor bioenergetics exists between patients. Using high-resolution respirometry on fresh biopsies of human lung adenocarcinoma, we identified 2 subgroups reflected in the histologically normal, paired, cancer-adjacent tissue: high (OX+) mitochondrial respiration and low (OX–) mitochondrial respiration. The OX+ tumors poorly incorporated [18F]fluorodeoxy-glucose and showed increased expression of the mitochondrial trifunctional fatty acid oxidation enzyme (MTP; HADHA) compared with the paired adjacent tissue. Genetic inhibition of MTP altered OX+ tumor growth in vivo. Trimetazidine, an approved drug inhibitor of MTP used in cardiology, also reduced tumor growth and induced disruption of the physical interaction between the MTP and respiratory chain complex I, leading to a cellular redox and energy crisis. MTP expression in tumors was assessed using histology scoring methods and varied in negative correlation with [18F]fluorodeoxy-glucose incorporation. These findings provide proof-of-concept data for preclinical, precision, bioenergetic medicine in oxidative lung carcinomas.
Keywords: Metabolism, Oncology
Keywords: Bioenergetics
Introduction
Lung cancer is the leading cause of cancer death worldwide. Causative gene alterations have been identified in various signaling pathways including the MAPK, PI3K, oxidative stress response, RNA splicing, and processing and nucleosome remodeling pathways. Still, the majority of these genomic alterations remain distinct to specific tumor subsets, and genetically defined precision medicine is limited to small groups of non–small cell lung cancer (NSCLC) patients. Surgery remains the gold standard of nonadvanced NSCLC care, and the search for predictive biomarkers for conventional cytotoxic chemotherapy has proven to be challenging. However, immunotherapy recently emerged as a standard of care for the treatment of advanced NSCLC (1), and promising new KRASG12C inhibitors have demonstrated antitumor activity in advanced NSCLC patients (2). Currently, stage IV NSCLC has a 5-year survival rate of approximately 1%, and this dramatic situation fosters the need to identify early biomarkers, targets, and approaches to manage lung cancer.
Among the different emerging strategies for cancer therapy, targeting tumor bioenergetics may be promising since rewiring of the biochemical pathways that are involved in energy metabolism contributes to tumor initiation, survival, and resistance to therapy. Two main pathways transduce energy in cancer cells: the anaerobic pathway and the oxidative pathway. The former includes glycolysis that is linked to lactate production (Warburg effect). The oxidative pathway can include glycolysis but requires mitochondrial oxygen consumption and generates a higher yield of ATP using a variety of fuels. Metabolomic investigations performed on 9 patients with various types of lung cancer revealed a metabolic heterogeneity between tumors and demonstrated that oxidative lung tumors rely on a carbon source other than glucose to fuel the TCA cycle, given that only 8% of the acetyl-CoA originates from [13C]-glucose carbons (3). In particular, fatty acid oxidation (FAO) that is linked to oxidative phosphorylation could maximize ATP synthesis in oxidative lung tumors since this catabolic pathway is preferred in a large variety of tumor types, such as breast cancer and leukemia (4, 5). Here, we demonstrated the validity of inhibiting the mitochondrial trifunctional enzyme complex in oxidative lung tumors.
Results
Mitochondrial respiration defines 2 LUAD bioenergetic subsets.
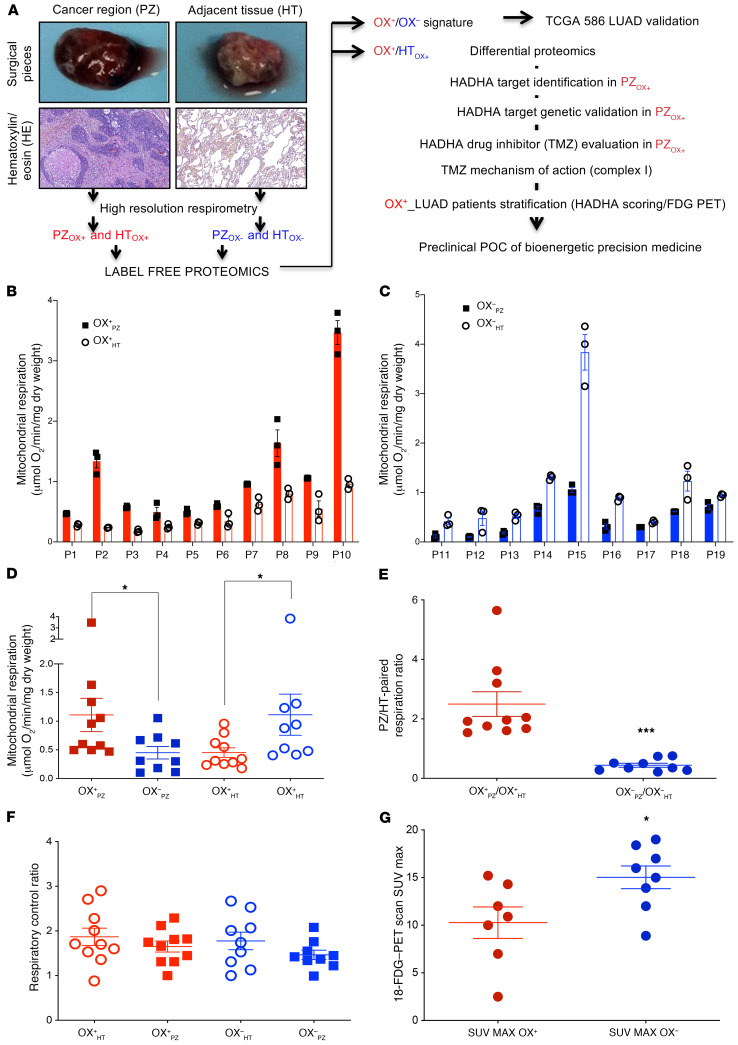
We analyzed mitochondrial respiration in intact surgical pieces obtained from 19 patients with NSCLC (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI133081DS1) after surgery and dissection by the pathologist in 2 zones: the Ki67-positive cancer region (proliferating zone, PZ) and the paired, cancer-adjacent, healthy tissue (HT) (Figure 1A). The high-resolution respirometry investigation revealed that for each patient, HT respiration systematically opposed that of the PZ (Figure 1, B and C), suggesting the existence of a “bioenergetic mispairing.” The group with a higher respiratory rate in the PZ compared with the HT was denoted OX+, and the group with tumor respiration lower than that of the HT was denoted OX–. Comparison of the respiratory rate between PZOX+ and PZOX– tumors showed a significant increase in the PZOX+ group, whereas the respiration of the HTOX+ was reduced compared with that of the HTOX– group (Figure 1D). Moreover, calculation of the ratio between tumor and paired-adjacent tissue respiration (PZ/HT) enabled a stronger (P = 0.0002) segregation of the surgical pieces in 2 bioenergetic groups (Figure 1E). This finding suggests that the intrinsic tumor bioenergetic profile is reflected in the histologically normal, cancer-adjacent tissue. Therefore, the strategy used to stratify OX+ from OX– LUAD tumors relied on 2 findings: (i) the systematic observation of a bioenergetic mispairing between OX+ and OX– tumors and their respective paired, adjacent tissue (HT) and (ii) a highly significant difference of the normalized respiration values (tumor/adjacent) between the 2 groups. No difference in the coupling degree of oxidative phosphorylation was found between the 2 groups (Figure 1F). Lastly, a lower (P = 0.0339) standardized uptake value (SUVmax) of [18F]fluorodeoxy-glucose (18-FDG) was measured in the OX+ group (Figure 1G).
Figure 1. Oxidative human lung tumor (OX+ LUAD) identification by high-resolution respirometry.
(A) Tissue sampling and study workflow. The cancer region (PZ; proliferating zone) and the paired, cancer-adjacent tissue (HT; healthy tissue) were dissected by the pathologist and used for subsequent bioenergetic, proteomic, and immunohistology analyses. (B) Tissue respiration (PZ: plain bars and HT: empty bars) identified consistent bioenergetic mispairing between PZ and HT in the 2 groups of tumors: OX+ (PZOX+ > HTOX+; red) and (C) OX– (PZOX– < HTOX–; blue). (D) Comparison of the respiration of the OX+ LUAD group (red circles) versus the OX– LUAD (blue squares), for PZ (plain symbols) and HT (empty symbols), respectively. (E) PZ respiration normalized to HT respiration stratified the OX+ LUAD (red circles) and the OX– LUAD (blue squares) subgroups. (F) Respiratory control ratio (routine/oligomycin treated–respiration). (G) Glucose incorporation in the 2 subgroups of lung tumors as measured by the [18F]-FDG–PET scan. Differences between the OX+ and the OX– LUAD subgroups were compared using a 2-sided Student’s t test. Data are expressed as mean ± SEM. *P < 0.05, ***P < 0.001.
Proteome remodeling in OX+ and OX– LUADs.
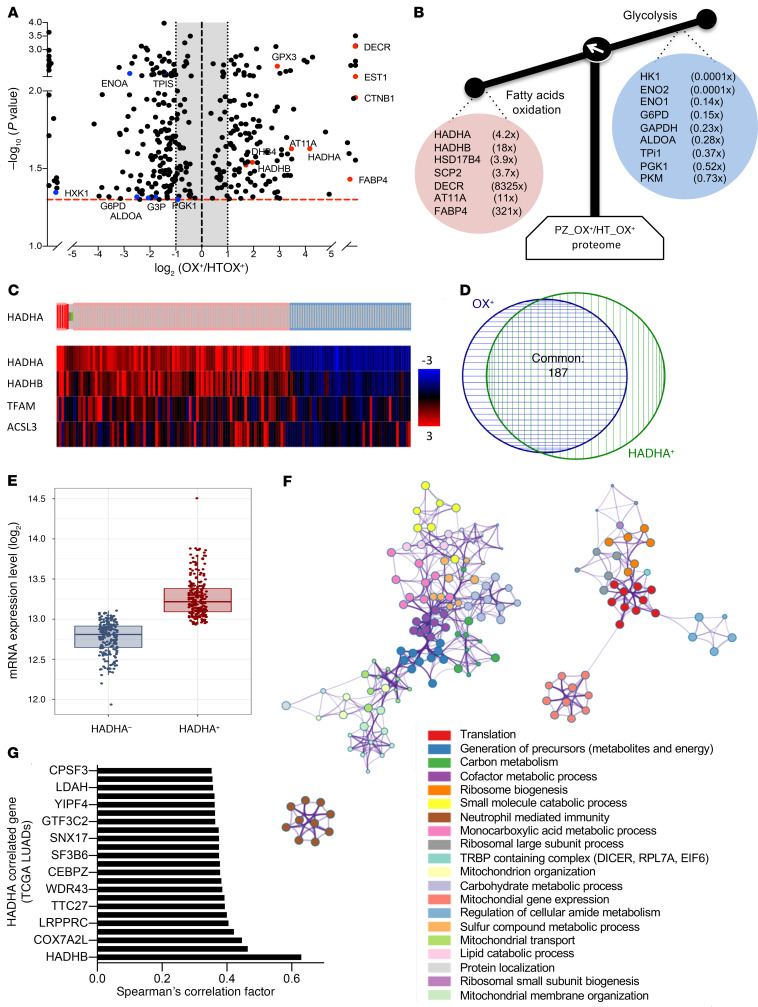
To investigate the molecular basis of the bioenergetic differences between PZOX+ and PZOX–, we performed a comparative molecular study using label-free proteomics. This analysis identified 333 proteins that differed in a significant manner (P < 0.05; 1-way ANOVA) (Supplemental Figure 1A and Supplemental Table 2, sheet 1). The cellular functions mostly affected included phagosome maturation, clathrin- and caveolar-mediated endocytosis signaling, and antioxidant response (Supplemental Figure 1B; Supplemental Table 2, sheet 2; Supplemental Table 2, sheet 3). Specifically, a strong reduction (≥65% inhibition) in 4 glucose-metabolizing enzymes was observed in PZOX+ compared with PZOX– (Supplemental Figure 1C), in full agreement with the lower 18-FDG–PET scan SUVmax values determined in OX+ tumors (Figure 1G). The differential proteomic signature of the PZOX+ versus PZOX– tumors (Supplemental Table 2, sheet 4) was used to stratify a larger panel of human lung adenocarcinomas (LUADs) from the National Cancer Institute Genomic Data Commons–The Cancer Genome Atlas (GDC-TCGA) portal (Supplemental Figure 2A). Hierarchical clustering revealed that the TCGA-OX+-LUAD tumors (58% of the total TCGA LUAD population; Supplemental Table 2, sheet 5) were associated with a reduced life expectancy (Supplemental Table 2, sheet 6). Genetic studies further revealed a higher level of DNA copy number alterations and of point mutations in the OX+ group, without differences in the EGFR, ELM-ALK, or KRAS mutational status (Supplemental Table 2, sheet 7). Gene Ontology (GO) terms and KEGG pathway analyses of the genes differentially expressed at the mRNA level (RNA-Seq) between OX+ LUAD and OX– LUAD tumors (adjusted P value DESeq2 < 0.05 and fold change > 1.5) indicated that the OX+ group was enriched for mRNAs involved in protein translation and oxidative phosphorylation (Supplemental Figure 2, B and C, and Supplemental Table 2, sheet 8). In contrast, OX– LUAD tumors showed enrichment in cell-adhesion processes and plasma membrane receptors (Supplemental Figure 2, D and E, and Supplemental Table 2, sheet 9).
Metabolic vulnerability of OX+ LUADs.
To identify a potential therapeutic target that is specific to OX+ LUADs, we performed a comparative proteomic analysis of the tumor region (PZOX+) with the corresponding cancer-adjacent tissue (HTOX+) (Figure 2A). A similar comparison of the tumor and adjacent tissue respiration was used for the bioenergetic stratification of the OX+ and OX– groups (Figure 1). The proteomic study (OX+PZ/HT) revealed a specific enrichment in 7 proteins involved in fatty acid degradation and a reduction in 9 glycolytic enzymes (Figure 2B; Supplemental Table 3, sheet 1; Supplemental Table 3, sheet 2). We selected the mitochondrial trifunctional protein (MTP) enzyme complex as the target of choice because (i) this enzyme is the terminal step of FAO in the mitochondrion, (ii) the 2 subunits (HADHA and HADHB) are overrepresented in PZOX+ compared with the adjacent tissue HTOX+ (Figure 2, A and B), (iii) HADHA is overexpressed in cancer cells resistant to cisplatin and 5-fluorouracil (6) (see also the HADHA interactome in Supplemental Table 3, sheet 3), and (iv) an approved drug inhibitor of MTP (trimetazidine; TMZ) is available for eventual repurposing in oxidative lung cancer therapy. In silico analysis of MTP α-subunit (HADHA) expression in the TCGA cohort of human LUAD (Figure 2C and Supplemental Table 3, sheet 4), showed that 28% had a positive z score higher than 1.2 (HADHA+ LUADs). A strong overlap was observed between the OX+ LUAD specimens (Supplemental Table 2, sheet 4) and the HADHA+ LUADs (Figure 2D). Determination of HADHA tumor absolute expression using the RNA-Seq log2 FPKM (fragments per kb per million mapped reads) values also showed higher levels in the HADHA+ subgroup (Figure 2E). Further comparison of the transcriptome of HADHA+ LUADs and HADHA– LUADs allowed identification of all statistically enriched GO terms (Supplemental Table 3, sheet 5), hierarchical clustering, and visualization using Cytoscape (Figure 2F). The results showed that HADHA+ LUADs were enriched in mitochondrial transcripts and in metabolic and signaling pathways similar to those found in OX+ LUADs (see Supplemental Figure 2, B and C), namely protein translation, ribosomes, mitochondrial gene expression, and oxidative phosphorylation (Figure 2F and Supplemental Table 2, sheet 8). The coexpression analysis performed on the entire TCGA LUAD population revealed that HADHA tumor expression was correlated to that of respiratory chain proteins, such as COX15, NDUFS1, or ATP5F1B (Figure 2G and Supplemental Table 3, sheet 6). This correlation analysis also identified LRPPRC, a protein involved in the regulation of mitochondrial gene expression; COX7A2L, a protein necessary for the assembly of mitochondrial respiratory supercomplex; and IMMT (MICOS60), a component of the mitochondrial cristae organization system (Figure 2G).
Figure 2. The MTP subunit HADHA is a priority target in OX+ LUAD.
(A) Volcano plot of the differential label-free proteomic analysis performed between PZOX+ and HTOX+ LUADs. (B) Proteins involved in glycolysis were repressed in PZOX+, whereas proteins involved in FAO were upregulated. The mean fold change ratio between OX+ PZ and OX+ HT is indicated. (C) Analysis of HADHA expression in 586 human LUAD samples (https://www.cbioportal.org/). Tumors with a positive HADHA z score higher than 1.2 (HADHA+) are indicated in red; tumors with HADHA z score less than 0.8 (HADHA–) are shown in blue. The heatmap gives the z score for the genes listed on the left. (D) Venn diagram showing the overlap between the TCGA LUAD OX+ tumors and the TCGA LUAD HADHA+ tumors identified in panel C. (E) HADHA absolute mRNA expression in HADHA+ and HADHA– LUAD tumors. (F) RNA-Seq data from the HADHA+ and the HADHA– LUADs were analyzed using DESeq2 to generate the list of genes that differed between the 2 groups (adjusted P < 0.005). This list was further analyzed using Metascape (metascape.org) to identify all statistically enriched GO/KEGG terms. The significant terms were then hierarchically clustered into a tree and converted into a network layout. Each term is represented by a circle node, where its size is proportional to the number of genes, and its color represents its cluster. Terms with a similarity score higher than 0.3 are linked by an edge (the thickness represents the similarity score) and visualized with Cytoscape (v3.1.2). One term from each cluster was selected as label. (G) Genes coexpressed with HADHA in TCGA lung tumors. The top genes with a Pearson coefficient higher than 0.35 are shown in the bar graph. Data are expressed as mean ± SEM.
HADHA histology scoring for OX+ LUAD stratification.
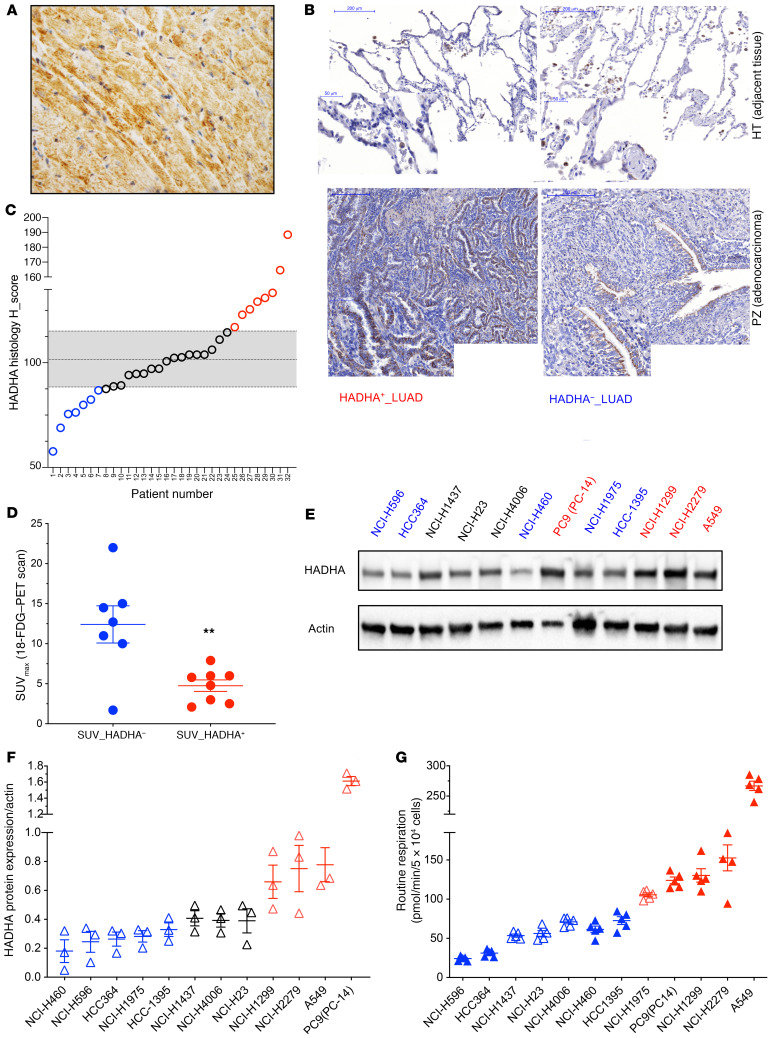
To verify the existence of HADHA+ and HADHA– LUADs in tumor samples obtained from the University Hospital of Bordeaux, departments of thoracic surgery and oncology and pathology, we developed a HADHA histology scoring method using a monoclonal antibody. First, we used mouse hearts as a positive control for HADHA staining since the MTP is a central enzyme complex for cardiomyocyte bioenergetics (Figure 3A). Then, we applied this method to study HADHA expression in human LUAD paraffin-embedded sections. Typical staining showed cytosolic HADHA expression in the bronchiolar and the alveolar epithelia (Figure 3B). This method was used to semi-quantify HADHA expression on a series of 32 human LUADs (Supplemental Table 1). Little HADHA staining was observed in the HT compared with the tumor region (Figure 3B). Moreover, significant differences in HADHA tumor expression were observed between the samples (Figure 3, B and C). The HADHA histology score was used to segregate tumors above (HADHA+ LUADs) or below (HADHA– LUADs) the mean absolute deviation (Figure 3C). This analysis showed that 18% of the tested LUADs were HADHA+. Representative images of HADHA+ LUAD and HADHA– LUAD tumors are shown in Figure 3B. Comparison of the 18-FDG–PET scan SUVmax values between the 2 subgroups of tumors also showed lower values (P < 0.05) in the HADHA+ LUADs (Figure 3D). These findings suggest that the clinical 18-FDG–PET scan analysis combined with HADHA tumor IHC scoring could be used to stratify HADHA+ LUADs.
Figure 3. HADHA+ LUAD tumor and cell line stratification.
(A) HADHA immunohistology staining in mouse heart (40× zoom). A strong HADHA cytosolic staining (brown) can be observed in the myofibers (nuclei were stained in blue). (B) This method was applied to study HADHA expression in paraffin-embedded sections of lung tumors stained with hematoxylin (blue), eosin (red), and with a monoclonal antibody recognizing HADHA (brown staining). Representative tumors with either high HADHA expression (HADHA+ LUAD; left panel) or low HADHA expression (HADHA– LUAD; right panel) are shown. (C) HADHA tissue expression in the tumor and the noncancer tissue was used to calculate the HADHA histology score in 32 tumor samples. Tumors with HADHA score greater than median absolute deviation (MAD) were denominated HADHA+ LUADs (shown in red) and tumors with HADHA score less than MAD were denoted HADHA– LUADs (shown in blue). (D) Distribution of the [18F]-FDG–PET scan SUVmax values. (E) HADHA expression was determined by Western blot on a panel of 12 human lung cancer cell lines. (F) HADHA expression normalized to actin levels (mean expression value at dashed line) was used to segregate the cell lines with high (HADHA+; red) or low HADHA expression (HADHA–; blue). Cells with a black symbol show no difference to the median value. (G) Mitochondrial respiration was measured in the 12 human lung cancer cell lines using the Seahorse extracellular flux analyzer. The mean (dashed line) was used to segregate the cell lines with high (red) or low (blue) respiration. Cells with a plain symbol correspond to the HADHA–/OX– group (plain blue) or to the HADHA+/OX+ group (plain red). Data are expressed as mean ± SEM. **P < 0.01.
The existence of HADHA+ and HADHA– LUADs was further investigated on a panel of 12 human NSCLC cell lines with variable mutational status regarding P53, KRAS, EGFR, or BRAF (Supplemental Table 1, sheet 2). Measurement of HADHA protein expression revealed 2 subgroups of lung cancer cell lines that also differed for mitochondrial respiration (Figure 3, E–G; see complete unedited blots in the supplemental material). The cell lines were defined as HADHA+ or OX+ when the expression of HADHA and the respiratory rate were above the median value. Cells with both HADHA+ and OX+ characteristics were defined as HADHA+/OX+. Four cell lines belonged to the HADHA+/OX+ group (H1299, H2279, A549, and PC9) and 4 to the HADHA–/OX– (H460, H596, HCC364, and HCC1395; Supplemental Table 1, sheet 2). As for the tumors, no difference in the mutational status of p53, KRAS, EGFR, or BRAF could be observed between these 2 subgroups (Supplemental Table 1, sheet 2). Likewise, no difference in glucose dependency was observed (Supplemental Figure 3A). Last, exploration of the data obtained on 80 NSCLC cell lines by Chen et al. (7) revealed no difference in cell malignancy between the OX+ and the OX– groups identified in our study (Supplemental Table 1, sheet 3).
The stimulation of HADHA expression was previously explained by the activation of β-catenin signaling in human hepatocytes, osteoblasts, dendritic cells, hepatocellular carcinomas, and breast carcinomas (8–12). In agreement with this mechanism, the proteomic analysis of human OX+ LUAD surgical pieces showed an increase of β-catenin (CTNNB1) protein levels in OX+ LUADs (Figure 2A). Moreover, β-catenin activation through GSK3β inhibition using lithium chloride (LiCl) or using siRNAs stimulated the expression of HADHA in A549 cells (Supplemental Figure 3B). Likewise, the ectopic expression of a constitutively active form of β-catenin (activating T41A mutant) in A549 cells triggered the upregulation of HADHA transcript (Supplemental Figure 3B). Conversely, inhibition of β-catenin transcriptional activity using methyl 3-{[(4-methylphenyl)sulfonyl]amino}benzoate (MSAB) or siRNA reduced the HADHA expression level in A549 cells (Supplemental Figure 3C) and altered cell viability, without an effect in H460 cells (Supplemental Figure 3C). Lastly, the ectopic expression of the β-catenin T41A mutant in OX– (H460) cells increased their sensitivity to TMZ treatment (Supplemental Figure 3D). The β-catenin activation status determined by reverse-phase protein arrays on 80 NSCLC cell lines (7) did not reveal a higher level of CTNNB1 in the OX+ NSCLC cell lines compared with the OX– cells (Supplemental Table 1, sheet 3), suggesting that canonical mutational activation of CTNNB1 is not the sole mechanism for HADHA stimulation in OX+ cells.
MTP inhibition alters OX+ cancer cell proliferation and bioenergetics.
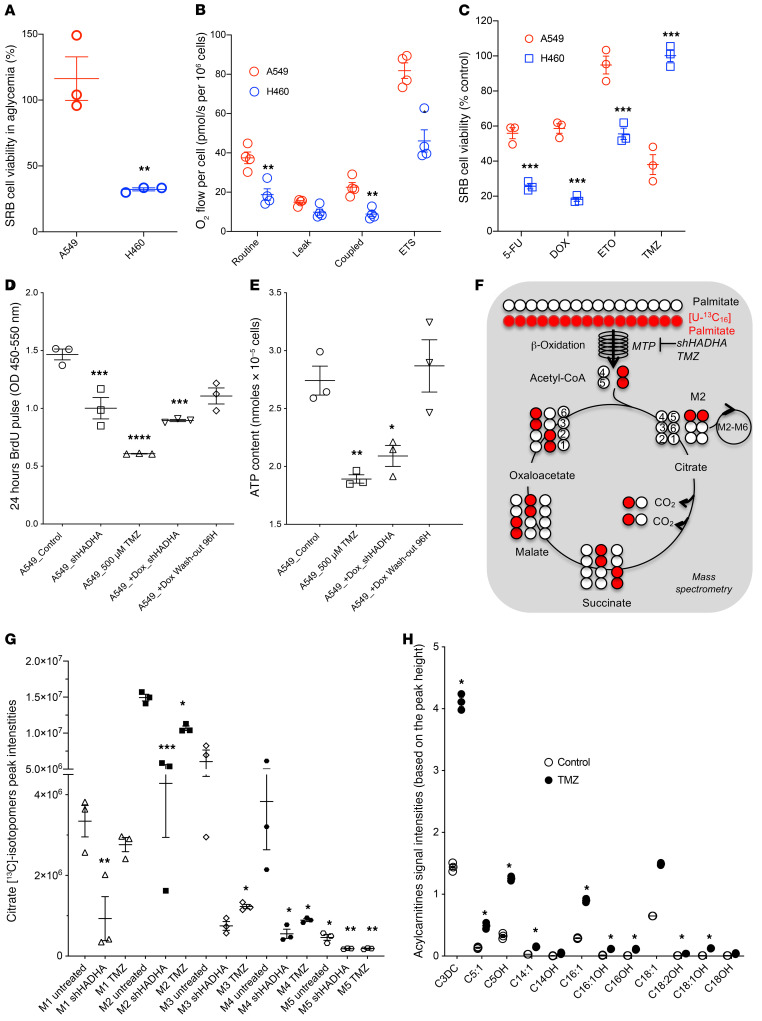
To perform mechanistic studies, we investigated the anticancer effects of HADHA inhibition in 2 human NSCLC cell lines, A549 (HADHA+/OX+) and H460 (HADHA–/OX–), with contrasting addiction to glucose, bioenergetics (13), and HADHA expression (see Figure 3, F and G). First, we observed that H460 cells (OX–) showed a strong dependency for growth on glucose availability in striking contrast with A549 cells (Figure 4A). Mitochondrial respiration was also higher in A549 cells (2-fold) compared with H460 cells (Figure 4B; see also Figure 3G). These 2 bioenergetic disparities mirrored the differences found between OX+ and OX– LUAD tumors (Figure 1, D and G). Blockade of FAO using the MTP inhibitor antianginal drug TMZ (14) altered cell viability specifically in A549 cells, without effect in H460 cells (Figure 4C). Likewise, a higher sensitivity to TMZ was observed in HADHA+/OX+ lung cancer cell lines compared with HADHA–/OX– cell lines (Supplemental Figure 4A). In striking contrast, no difference in the sensitivity to 20 different chemotherapy treatments was observed between these OX+ and OX– cells by Chen et al. (7) (Supplemental Table 1, sheet 3). TMZ could also potentiate the cancer-killing effect of cisplatin in 2 OX+ cell lines (Supplemental Figure 4B). Conversely, H460 cells showed a higher sensitivity to the chemotherapeutic drugs 5-fluorouracil, doxorubicin, and etoposide (Figure 4C). Investigation of the anticancer effect of MTP inhibition in OX+ cells using a HADHA-shRNA (Supplemental Figure 4, C and D; see complete unedited blots in the supplemental material) or TMZ showed a reduction of A549 cell proliferation, as assessed by BrdU incorporation (Figure 4D), and a significant decrease in the steady-state level of ATP (Figure 4E). To verify the specificity of these findings and their reversibility, we performed a genetic rescue experiment using a doxycycline-inducible HADHA knockdown system in A549 cells (Supplemental Figure 4C). The results revealed that doxycycline activation of the HADHA-targeting shRNA-miR altered cell proliferation and bioenergetics in A549 cells, while 96 hours doxycycline washout rescued these 2 parameters (Figure 4, D and E, and Supplemental Figure 4, C and D). Likewise, OXPHOS inhibition using the complex I inhibitor rotenone specifically altered cell viability in the OX+ cells (Supplemental Figure 4E), which also showed a higher capacity to oxidize palmitate in comparison with OX– cells (Supplemental Figure 4F).
Figure 4. Metabolic impact of HADHA inhibition in OX+ LUADs.
(A) Impact of glucose withdrawal on A549 (HADHA+/OX+; red) and H460 (HADHA–/OX–; blue) cell viability. (B) High-resolution respirometry comparative analysis of intact A549 and H460 cells: routine respiration (basal), leak respiration (oligomycin), ETS (uncoupled), and ROX (in presence of rotenone and antimycin A). All the parameters were corrected by ROX values. (C) Effect of trimetazidine (TMZ 500 μM; 24 hours), 5-fluorouracil (5-FU 50 μM, 24 hours), doxorubicin (DOX 5 μM; 24 hours), and etoposide (ETO 20 μM, 24 hours) on cell viability. (D) Reduction of BrdU incorporation in A549 cells treated with HADHA shRNA or with 500 μM TMZ for 24 hours in vitro. A doxycycline-induced shRNA targeting HADHA was also used to study the reversibility and the specificity of HADHA inhibition on cell proliferation after 96 hours of doxycycline washout. (E) Total cellular ATP content of A549 cells treated with saline, 500 μM TMZ for 48 hours, or doxycycline-induced shRNA-mediated HADHA knockdown. (F) Principles of the lipidomic analysis of [U-13C]-palmitate oxidative metabolism. (G) The incorporation of [U-13C]-palmitate carbons in citrate was quantified for different isotopomers (M1–M6) after 4 hours in A549 cells treated with scramble-shRNA, shRNA targeting HADHA, or 500 μM TMZ. (H) Lipidomic analysis of A549 cells treated with saline or TMZ. Differences between the treated and the untreated groups were compared using a 2-sided Student’s t test (panels A–E). For panels H and G, 1-way ANOVA with Dunnett’s correction multiple t test comparison was used. Data are expressed as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The on-target effect of MTP inhibition by HADHA-shRNA or TMZ was verified in A549 cells by using a lipidomic approach. HADHA-shRNA and TMZ triggered a significant reduction in the incorporation of [13C]-palmitate in the TCA cycle (Figure 4F), as shown by a reduced content of [13C]-citrate isotopomers after 2 hours of [13C]-palmitate oxidation (Figure 4G). The different citrate isotopomers M1–M5 were generated by successive cycles of the TCA cycle. Furthermore, FAO inhibition by TMZ resulted in the accumulation of unprocessed fatty acids inside A549 cells (Figure 4H). In particular, the TMZ treatment induced the accumulation of lipotoxic long-chain acylcarnitines (C18-OH, C18-1OH, C18-2OH, C16OH, C16:1OH, and C14OH), as specifically found in patients with inherited MTP deficiency (15) or in a transgenic mouse model of VLCAD deficiency (16).
MTP inhibition alters OX+ LUAD tumor growth in vivo.
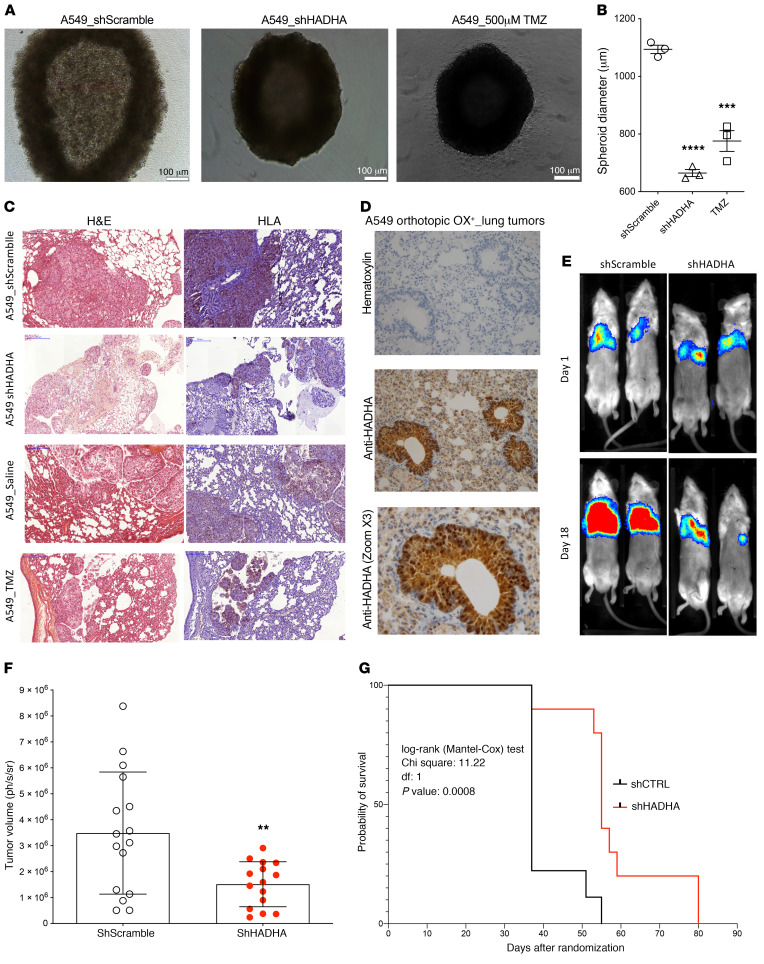
Tumor bioenergetics is strongly dependent on the microenvironment, and the anticancer efficacy of HADHA inhibition had to be verified in vivo in the lung. First, we observed that HADHA knockdown using shRNA reduced A549 cell growth in anchorage-independent 3D conditions (Figure 5, A and B). Then, we developed an orthotopic mouse model of human HADHA+/OX+ LUAD, using A549 cells that stably expressed luciferase and formed orthotopic adenocarcinoma tumors in murine lungs (17, 18). Human A549 cells were detected in the mouse lung parenchyma using an anti-HLA antibody (Figure 5C), and MTP expression in tumors was observed using an antibody against HADHA (Figure 5D). A significant reduction in lung tumor size was observed in vivo when HADHA was inhibited genetically (Figure 5, E and F), and a corresponding and significant gain in animal survival was also measured (Figure 5G). OX+ LUAD mice treatment with TMZ 20 mg/kg administered per os (p.o.), as recommended for the clinical utilization of this drug, led to similar results (Supplemental Figure 5, A and B). However, administration of TMZ 20 mg/kg or 80 mg/kg by i.p. injection also reduced OX+ LUAD tumor size, but the benefits in terms of survival were not significant (Supplemental Figure 5, C and D) because animal weight loss was also observed (Supplemental Figure 5E). Preclinical drug efficacy studies were performed on 3 additional orthotopic human LUAD mouse models generated using the HADHA+/OX+ cells PC-9 and the HADHA–/OX– cells HCC-1395 and H460, as determined from Figure 3G. The results indicated that TMZ 20 mg/kg reduced lung tumor growth in the HADHA+/OX+ PC9 LUAD model (Supplemental Figure 6A), as observed for A549 cells (Supplemental Figure 5F). Of importance to the demonstration of bioenergetic precision medicine, no effect of HADHA inhibition by TMZ was seen on the OX– LUAD orthotopic mouse models obtained using HCC-1395 or H460 human adenocarcinoma cells (Supplemental Figure 6, B–D). Accordingly, little or no accumulation of hydroxylated acylcarnitines was observed in H460 cells treated with TMZ (Supplemental Figure 6E). Lastly, the evaluation of the TMZ oral treatment (20 mg/kg p.o.) in an orthotopic LLC-C57BL/6 immunocompetent LUAD mouse model only showed a limited efficacy that might be explained by the OX– profile of these murine lung cancer cells (19) (Supplemental Figure 5F).
Figure 5. Anticancer effect of HADHA genetic inhibition in OX+ LUAD.
(A) Reduction of OX+ LUAD spheroid growth using HADHA shRNA or 50 μM TMZ on A549 cells cultured in anchorage-independent conditions. (B) Effect of MTP inhibition on A549 OX+ LUAD spheroid diameter. (C) Representative (10×) fields of 2 separate areas of A549 OX+ tumors: H&E staining (left; pink) and anti-human marker HLA (right; blue) staining of A549 cells in excised orthotopic human A549-OX+ LUAD tumors in NSG mice. (D) Immunohistology staining of HADHA in excised orthotopic human A549-OX+ LUAD tumors in NSG mice. H&E staining 20× zoom. Anti-HADHA 40× zoom. (E) Representative evolution of the bioluminescence signal from day 1 to day 18 in 2 groups of NSG mice: (i) orthotopic model of A549 expressing luciferase, (ii) orthotopic model of A549 shHADHA expressing luciferase. (F) Relative tumor volume obtained from the luminescence signal in the groups of mice treated with shRNA scramble and shRNA HADHA (N = 20 animals per group). (G) Animal survival (Kaplan-Meier representation) in the 2 groups of mice treated with shRNA scramble and shRNA HADHA (N = 20 animals per group). Values represent mean ± SEM; N = 4–6 for the in vitro experiments; N = 8–20 for the in vivo experiments. One-way ANOVA with Dunnett’s correction was used to analyze the results of panels A and B. Two-sided unpaired Student’s t test was used to compare the groups of mice in panel F. Log-rank (Mantel-Cox) test was used to compare animal survival in panel G. **P < 0.01, ***P < 0.001, ****P < 0.0001.
TMZ inhibits respiratory chain complex I and reprograms the mitochondrial proteome.
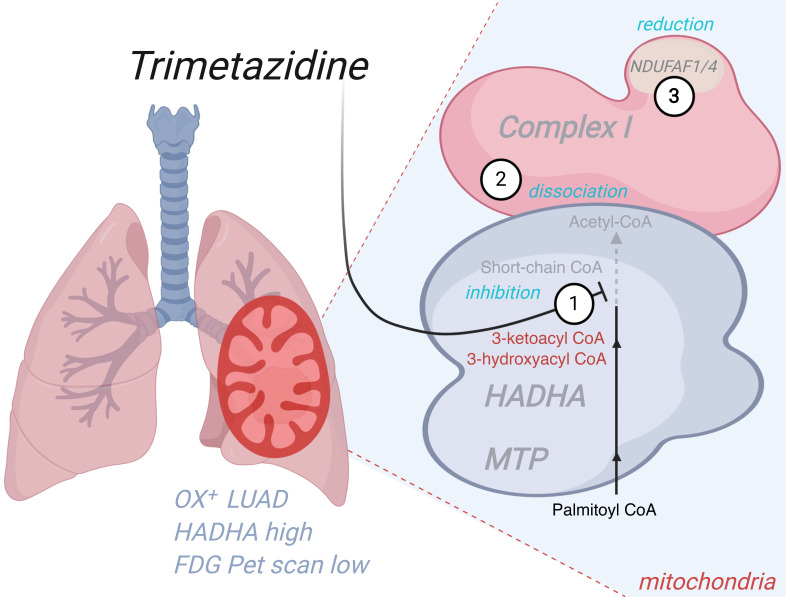
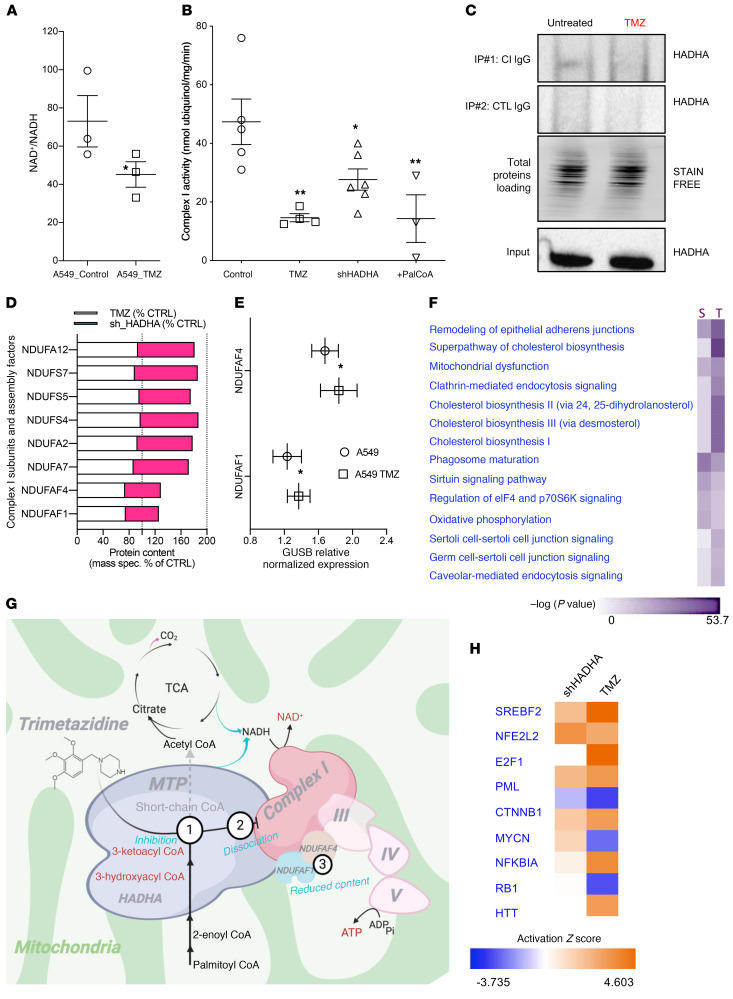
Our findings indicate that TMZ triggers the accumulation of unprocessed lipotoxic fatty acids in OX+ lung cancer cells (Figure 4H). Likewise, secondary inhibition of the mitochondrial respiratory chain by lipotoxicity has been proposed in the pathophysiology mechanisms of MTP genetic deficiencies. Accordingly, we observed that TMZ treatment of A549 cells triggered the accumulation of NADH (Figure 6A) as explained by indirect inhibition of complex I activity by 48 hours of TMZ treatment, by shHADHA, or by direct interaction with palmitoyl-CoA (Figure 6B). To clarify the link between HADHA and complex I, we considered the numerous biochemical studies that have demonstrated a physical association between HADHA and complex I to form a supercomplex (20–26). In particular, IP studies revealed that HADHA physically interacts with complex I subunits (25), and genetic defects in HADHA are associated with complex I deficiency (27). Using IP and Western blot studies, we also found a physical link between HADHA and complex I, and we discovered that HADHA inhibition by TMZ dissociates this link (Figure 6C). Moreover, we found a reduction in the protein content of NDUFAF1 and NDUFAF4, 2 essential complex I assembly factors, in A549 cells treated with TMZ or shHADHA (Figure 6D). No change in the mRNA level of these factors was observed (Figure 6E), suggesting that protein degradation may have occurred. The unbiased proteomic study of these cells further revealed that HADHA inhibition also stimulated cholestero-genesis (Figure 6F and Supplemental Table 3), as typically observed in conditions of FAO inhibition.
Figure 6. Trimetazidine disrupts the HADHA–complex I interaction.
(A) NAD+/NADH ratio in A549 (OX+) cells treated with 500 μM trimetazidine (TMZ) for 48 hours. (B) NADH-oxidizing complex I enzymatic activity on A549 cells treated with 500 μM TMZ for 48 hours, on A549 cells expressing a shRNA against HADHA, or on A549 cell homogenate treated with 100 μM palmitoyl-CoA. (C) Impact of HADHA downregulation using shRNA on complex I–HADHA physical association. HADHA detection was performed by Western blot. A negative control IgG was also used to verify the specificity of the capture. (D) Changes in complex I composition in A549 cells expressing the shRNA targeting HADHA or in cells treated with TMZ. Complex I subunit content was quantified using mass spectrometry. The data are expressed as a percentage of the corresponding control. (E) NDUFAF1 and NDUFAF4 subunit mRNA content in A549 cells treated with TMZ. (F) Differential proteomic analysis performed on A549 cells treated with TMZ versus untreated cells, or between A549 cells expressing an shRNA against HADHA and cells expressing a scramble shRNA. The proteome modifications were compared using the “comparative analysis module” of IPA (QIAGEN). Only proteins with changes greater than 15% and P < 0.05 were kept for this analysis. The IPA canonical pathways altered in the 2 conditions are shown as well as the –log P value (purple color scale). (G) The mechanism of action of TMZ in lung cancer cells. Three related effects are proposed: HADHA inhibition, dissociation from complex I, and reduced content of NDUFAF1 and NDUFAF4. (H) Transcription factors potentially activated or inhibited by the shHADHA or TMZ treatments in A549 cells, as predicted using the upstream analysis module of IPA. The activation z score is shown as a color code (orange = activated and blue = inhibited). Data are expressed as mean ± SEM. *P < 0.05, **P < 0.01.
Therefore, our findings demonstrated that TMZ targeted the multifunctional FAO-CI supercomplex (20–26) with a mechanism of action that involved 3 effects (Figure 6G): (i) on-target HADHA catalytic inhibition; (ii) dissociation of the link between HADHA and complex I; and (iii) reduction in the content of NDUFAF1 and NDUFAF4, 2 essential complex I assembly factors. Likewise, biochemical studies performed in patients’ cells with mutations in HADHA also showed a reduction in NDUFAF1 protein content (27), in full agreement with our findings. Last, the direct inhibition of complex I with rotenone could stratify the OX+ and OX– cells (Supplemental Figure 4D), although TMZ showed higher specificity and potency, suggesting differences in the mechanism of action (Figure 6G). To further delineate the mode of action of TMZ, we compared the perturbome of TMZ and shHADHA treatments on the proteome of A549 cells. The corresponding pathway analysis (IPA) revealed conserved changes in epithelial adherens junctions, cholesterol biosynthesis, and mitochondrial function (Figure 6F). The observed upregulation of cholesterol synthesizing enzymes is a common consequence of FAO inhibition, and the computational analysis of the proteome changes predicted activation of the sterol regulatory element binding transcription factor 2 (SREBF2) and of the nuclear factor erythroid–derived 2 related factor 2 (NRF2) (Figure 6H and Supplemental Figure 7). Altogether, these findings delineate the impact of MTP inhibition on mitochondrial physiology and cell composition in lung cancer.
Discussion
Current developments in cancer biology indicate that peculiarities in tumor metabolism can eventually be targeted for anticancer therapy, but interpatient tumor heterogeneity must be considered in the early design of bioenergetic therapeutic approaches. In our study, we deciphered the functional status of mitochondria in situ on freshly excised surgical pieces of human LUAD using high-resolution respirometry, and this approach may eventually be considered for the bioenergetic profiling of lung tumors in the clinical setting.
More specifically, we performed 2 comparative bioenergetic and proteomic analyses: (i) OX+ versus OX– LUAD tumors and (ii) OX+ LUAD tumors versus the paired, histologically normal, cancer-adjacent tissue. The first observation (OX+/OX– tumors) revealed that half of the LUAD surgical pieces obtained from the University Hospital of Bordeaux or from the TCGA cohort were of the oxidative (OX+) bioenergetic subtype. Proteomic analysis showed that OX+ LUAD tumors were enriched in mitochondrial proteins of the OXPHOS system and enriched in components of the protein translation machinery as ribosome subunits. This proteomic observation was verified on the TCGA LUAD oxidative tumors transcriptome and suggests that LUADs with intense biosynthetic activity could have a predominant mitochondrial metabolism. The mechanisms involved in coordinating OXPHOS and protein translation still remain unknown, and a recent investigation revealed a role for the translation initiation factor EIF3F in the stimulation of OXPHOS in human LUAD (18). Our work also showed that OX+ LUADs were further enriched in components of the antioxidant systems, and a similar observation was reported in oxidative ovarian tumors (28). The main genetic driver of the observed oxidative, FAO-dependent phenotype of OX+ LUAD was activation of β-catenin. Yet, although OX+ LUADs have higher levels of β-catenin protein, they were not enriched in β-catenin mutations, suggesting that noncanonical activation of this pathway may have occurred. The second bioenergetic class of human LUAD tumors (OX–) showed enrichment in proteins involved in glycolysis associated with an increased expression of cell-adhesion molecular systems. The mechanical regulation of glycolysis via cytoskeleton architecture was recently shown in NSCLC cells (29), suggesting that the OX– phenotype of LUADs could be linked to cell-adhesion remodeling processes. Moreover, we discovered enrichment in components of the PI3K-Akt signaling in OX– TCGA LUADs, and previous studies showed that this pathway coordinates glycolysis with cytoskeleton architecture remodeling (30). Two key differences between OX+ and OX– LUADs were (i) the reduced patient survival in the former group and (ii) an increased amount of copy number alterations in the genome. Additional copy number alteration comparative profiling studies between OX+ and OX– LUADs will be required to better understand the genetic determinism, if any, of the bioenergetic heterogeneity of these tumors. More specifically, deletions in the S-methyl-5′-thioadenosine phosphorylase (MTAP) were enriched 35-fold in OX+ LUADS. This enzyme is responsible for the first step in the methionine salvage pathway and could be related to the enrichment in protein translation machineries discovered in the OX+ tumor proteome.
A key finding of our bioenergetic study was that respiration of the paired, cancer-adjacent tissue systematically opposed respiration of the tumor region, supporting the potentially novel concept of bioenergetic mispairing. Yet, a significantly higher respiratory rate exists between OX+ and OX– lung tumors (PZ), even without normalization to the adjacent tissue (HT). The normalization to the paired, cancer-adjacent tissue aimed at considering 2 points: (i) the eventual preexisting differences in lung tissue bioenergetics observed between patients. Such differences could result from variable exposure of the lung to chemicals (exposome) but also from interpatient disparity in nutrition, exercise training, or disease history. (ii) changes in the adjacent tissue proteome and bioenergetics, which could also result from the mutual interaction with the tumor. In our work, consideration of the adjacent lung tissue background respiration led to a stronger stratification between the OX+ and OX– tumor groups. Accordingly, recent transcriptomic studies showed that intrinsic tumor subtypes are reflected in histologically normal, cancer-adjacent tissue, suggesting that biomarkers and targets derived from the knowledge of the coupled tumor-adjacent tissue may be helpful in defining personalized therapeutic strategies or in predicting prognosis and recurrence risk (31, 32). In our work, the HADHA target was identified in OX+ LUADs after comparison of the tumor proteome (OX+ PZ) to that of the adjacent paired lung tissue (OX+ HT), which brings consistency to our approach of tumor bioenergetic stratification. The molecular basis of mitochondrial activation was analyzed in OX+ LUAD tumors (PZOX+) by comparative proteomics and revealed enrichment in 7 FAO proteins and deprivation in 9 glycolytic enzymes compared with the paired adjacent tissue (HTOX+). We demonstrated that HADHA expression as determined by tumor histology scoring, RNA expression, or Western blot could stratify a subgroup (OX+/HADHA+ LUAD) of human lung tumors with predominant FAO linked to oxidative phosphorylation. Our findings also demonstrated that PET imaging of glucose uptake, a well-established oncology staging technique, in contrast to respirometry, could be used to stratify the OX+/HADHA+ LUAD patients. Therefore, we suggest that the population of LUAD OX+ patients for targeting with TMZ should present with elevated HADHA tumor expression and low 18-FDG–PET scan SUVmax value. More generally, different criteria used for selecting the tumors eligible for OXPHOS inhibition therapy have been proposed in different studies: the Pten-null genetic status, Akt activation status, resistance to BRAF inhibitors, resistance to Ibrutinib, enrichment in cancer-initiating cells, or glycolysis deficiency (33–39).
The identification of MTP (HADHA subunit) as a primary target for HADHA+/OX+ human lung tumors must be discussed in the context of the current understanding of lung tumor bioenergetics. Hensley et al. (3) showed that the oxidative subgroup of lung tumors, defined as incorporating [13C]-labeled glucose carbons in TCA cycle intermediates, relied predominantly on a carbon source other than glucose and suggested the existence of alternative fuels. In our work, we identified that i) fatty acid is one source of carbon contributing to OX+ LUAD bioenergetics and ii) these tumors enhanced the expression of mitochondrial fatty acid oxidizing enzymes as the MTP subunits HADHA and HADHB. We also showed that cellular energy homeostasis monitored through steady-state ATP levels was reduced, and that cell proliferation was altered when targeting HADHA. A similar anticancer bioenergetic mechanism, reduction of cellular ATP levels, was proposed when targeting fatty acid metabolism through ACSL3 (4) or CPT1 (40, 41) inhibition. Furthermore, our work demonstrated that MTP inhibition using genetic or pharmacological means altered HADHA+/OX+ LUAD tumor growth in vivo, with no effect on HADHA–/OX– LUAD orthotopic tumors. These findings provide proof-of-concept data for bioenergetic precision medicine. The proteomic comparative analysis of OX+ PZ and OX+ HT also revealed upregulation of the bifunctional enzyme acting on the peroxisomal β-oxidation (HSD17B4), suggesting a possible metabolic vulnerability at this level. Likewise, the observed upregulation of the nonspecific lipid-transfer protein (SCP2); 2,4-dienoyl-CoA reductase (DECR); probable phospholipid-transporting ATPase IH (AT11A); and fatty acid-binding protein 4 (FABP4) in OX+ PZ could contribute to the OX+/FAO-dependent phenotype.
The mode-of-action study detailed in our work showed that when TMZ or shHADHA inhibited the MTP, they altered in turn the catalytic activity of respiratory chain complex I. TMZ and shHADHA trigger a decrease in the cellular content of NDUFAF1 and of NDUFAF4, 2 proteins required for complex I assembly and function (42, 43). More generally, a functional and molecular link between FAO and complex I structure and function has been suggested in previous studies showing that mutations in ACAD9, a complex I–bound acyl-CoA dehydrogenase catalyzing the initial rate-limiting step of the fatty acid β-oxidation, leads to complex I disassembly and consecutive altered function (26). Moreover, biochemical studies showed that the MTP physically associates with the respiratory chain to form the FAO-supramolecular complex (20–26), and genetic defects in HADHA trigger complex I deficiency (27). Using biochemical IP methods, we discovered that HADHA pharmacological inhibition altered the physical and functional interaction between the MTP and complex I. We also discovered in 3 types of cancer that HADHA expression was positively correlated with that of COX7A2L and MICOS60, 2 proteins involved in the assembly of mitochondrial respiratory supercomplexes. Therefore, further evaluation of the interaction between HADHA, NDUFAF1, NDUFAF4, COX7A2L, and MICOS60 could contribute to a better understanding of the regulation of respiratory chain supercomplex assembly and function in lung cancer.
Our findings indicate that targeting MTP involved destabilization of complex I, and the link between complex I inhibition and a decrease of tumor growth was observed in previous preclinical investigations using small-molecule chemical inhibitors (36, 38, 44–47). However, the clinical efficacy and the lack of toxicity of these compounds in humans have not been demonstrated yet. We observed that the genetic inhibition of HADHA induced a highly significant gain in animal survival and a 60% reduction of tumor size. The pharmacological treatment with TMZ showed less efficacy, although significant effects were measured on tumor size and animal survival. Therefore, we argue that HADHA is a target of valuable interest, as validated by genetic means, but also that TMZ is not the ideal drug inhibitor for metabolic cancer monotherapy because of a rather modest preclinical efficacy. Yet, we also raise the argument that more potent electron transport chain inhibitors could have limitations, as recently reported for the complex I inhibitor IACS-01075, which triggered peripheral neuropathy, myalgia, and fatigue during the clinical trial (48). Limitations were also reported for the complex I inhibitor Bay 2243 (49, 50). More generally, complex I inhibitors can induce neurotoxicity, weight loss, and hypocaloric state (51, 52). Moreover, the degree of complex I inhibition is a critical parameter to assess in order to avoid protumorigenic effects (53–55). Interestingly, mild complex I inhibitors such as mubritinib (35), TMZ, or metformin have anticancer properties, although the clinical trials on metformin showed a poor efficacy in pancreatic tumors (56) and melanoma (57). Regarding use of FAO inhibitors for cancer treatment, etomoxir and perhexiline have been discouraged in clinical practice because of hepatotoxicity and cardiotoxicity. These findings suggest that potent complex I or FAO blockers, even with strong tumor reduction effects at the preclinical stage, are associated with stronger toxicity during clinical safety evaluations. This observation is key for the preclinical and clinical evaluation of our approach.
In contrast with the above-discussed complex I–targeting anticancer strategies, TMZ is a safe HADHA drug inhibitor approved by the European Medicines Agency in 2009. Seventeen clinical trials have been performed with TMZ, and it is estimated that 532,544,831 patient-months have been exposed to this drug between 1964 and 2011 (https://www.ema.europa.eu/en/documents/referral/trimetazidine-article-31-referral-assessment-report_en.pdf). Hence, the repurposing of TMZ for cancer therapy in OX+ LUAD patients could be attractive because of the good safety profile of this drug, which is already approved for the treatment of angina.
The on-target effect of this drug was previously demonstrated in biochemical studies showing that TMZ is a 3-ketoacylcoenzyme A thiolase inhibitor (58). In our study, we verified this effect and further discovered that TMZ mode of action involved inhibition of respiratory chain complex I, leading to a mitochondrial bioenergetic and REDOX crisis. The low level of glycolytic enzyme expression observed in the proteome of OX+ tumors might indicate the incapacity to perform a reverse Pasteur effect, i.e., compensatory glycolysis in a condition of OXPHOS inhibition, leading to cellular energy crisis.
Methods
NSCLC subjects and lung tumor sampling.
Nineteen surgical pieces of human LUAD (see Supplemental Table 1, sheet 1 for patient descriptions) were obtained from the University Hospital of Bordeaux, departments of thoracic surgery (M. Thumerel) and oncology and pathology (H. Bégueret). The surgeon (MT) performed tumor resection and the pathologist (HB) immediately collected the 2 areas of interest for the study: the PZ and the tumor-adjacent HT. We used the terms PZOX+ and PZOX– to designate the 2 bioenergetic types of lung tumors, and HTOX+ and HTOX– to designate their corresponding cancer-adjacent tissues. Immediately after tumor excision and sample preparation by the pathologist, the fresh tissues (PZ and HT) were divided in 3 parts. One part was analyzed for mitochondrial respiration by high-resolution respirometry (Oroboros), one part was fixed in formol for histopathology investigations, and the last part was dissected and snap-frozen in liquid nitrogen for the proteomic analysis, as detailed below.
In vivo assessment of lung tumor glucose uptake by 18-FDG–PET scan.
As part of the NSCLC diagnostic procedure performed at the University Hospital of Bordeaux (CHU Bordeaux; Haut-Lévèque), a 4D acquisition was performed in the radiotherapy treatment position, 50 minutes after i.v. injection of 3.7 MBq/kg 18-FDG, on a PET/CT integrated system (Discovery RX, General Electric Medical System) using the Real-time Position Management (RPM) device (Varian) and “Motion Free” software (General Electric Medical System). The respiratory cycle was rebinned in 6 phases. The lung tumors’ glucose uptake was calculated as the SUVmax normalized to tumor area.
IHC and image analysis.
Surgical specimens, including the tumor (PZ) and adjacent tissue (HT), were fixed in 10% buffered formaldehyde, and tissue sections from tumoral (PZ) and nontumoral (HT) parenchyma were routinely processed for the purpose of diagnosis. Selected representative slides, including tumoral and nontumoral cancer-associated tissue (HT), were processed for HADHA IHC. The 2.5-μm-thick sections were dewaxed and rehydrated, and antigen retrieval was performed in a 1 mM Tris-EDTA pH 9 solution. All staining procedures were performed in an automated auto-stainer (Dako-Agilent) using standard reagents provided by the manufacturer. The sections were incubated with the HADHA antibody (Abcam, 203114) at a 1:100 dilution for 45 minutes at room temperature. EnVision Flex/HRP (Dako-Agilent) was used for signal amplification for 20 minutes and 3,3′-diamino-benzidine (DAB, Dako) development was used for detecting the primary antibodies. The slides were counterstained with hematoxylin, dehydrated, and mounted. Each IHC assay contained a negative control (buffer, no primary antibody). Sections were visualized using a Nikon Eclipse 501 microscope and scanned. HADHA immunopositivity was analyzed under pathologist supervision using an automated computer system with positive pixel count (Panoramic VIWER, 3D Histech). The quantification of HADHA staining was performed using a color deconvolution algorithm (ImageJ [NIH]) to isolate individual stains for quantification: hematoxylin or DAB. HADHA intensity level was determined by calculating the ratio between the intensity of the DAB color staining and the number of cells measured by the area of hematoxylin labeling. Then, the HADHA immunostaining score was obtained by calculating the ratio between HADHA intensity level in the cancer region (PZ) to that determined in the noncancer (HT) region.
High-resolution respirometry analysis of human lung tumors and cancer-associated tissue and NSLC cell lines.
High-resolution respirometry using the Oroboros O2k apparatus was used for precisely measuring mitochondrial respiration on intact lung tumors (PZ) and paired, cancer-adjacent tissue (HT). Each piece of lung tumor (PZOX+ or PZOX–) or paired-adjacent tissue (HTOX+ or HTOX–) was divided in 3 to 5 samples of 10–20 mg and mitochondrial respiration was evaluated at 37°C using the O2k system (Oroboros Instruments). The respiratory medium consisted of 5 mM glucose DMEM supplemented with 10% FCS. To assess the respiratory coupling ratio (RCR), 20 μg/μL oligomycin (Sigma-Aldrich) was used. To exclude nonmitochondrial respiration, 2 mM KCN was used. The rate of respiration was normalized to the dry weight of the tissue, and the mean of 3–5 pieces was calculated for each sample. The respiratory rates were expressed as nmol O2/min/mg dry tissue.
For the A549 high-resolution respirometry study, the FAO OXPHOS capacity was determined using 20 μM palmitoyl-carnitine as an energy substrate. DatLab software (Oroboros Instruments) was used for data acquisition and analysis.
Human lung tumors, adjacent tissue, and NSCLC cell lines in label-free proteomic analysis.
Immediately after tumor excision and dissection by the pathologist, lung tumor samples (PZ and HT) were snap-frozen in liquid nitrogen and stored at –80°C. Tissue lysis was performed in Eppendorf tubes using RIPA buffer (150 mM sodium chloride, 1.0% NP-40 or Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) under agitation for 2 hours at 4°C. The lysate was centrifuged for 20 minutes at 18,000g and 4°C, and the supernatant was used for the proteomic analysis at the mass spectrometry facility of the University of Bordeaux, as recently described (17, 18).
Computational analysis of human LUAD samples from the GDC-TCGA database using the proteomic OX+/OX– signature. Level 3 released gene level expression data for RNA-Seq were retrieved for 586 LUADs from TCGA. The data processing, quality control, and normalization were done by TCGA workgroup. Normalized RSEM (RNA-Seq by expectation-maximization) values were used for the analysis. A hierarchical clustering was applied using the Euclidean distance and the Ward method for aggregation using the R package EMA. Kaplan-Meier curves were performed using https://xenabrowser.net Differential expression analysis was performed using DESeq2 R package. Pathways enrichment analysis on genes differentially expressed between OX+ and OX– groups (adjusted P < 0.05 and fold-change > 1.5) was conducted using DAVID Bioinformatics Resources version 6.8 (https://david.ncifcrf.gov/). GO Biological Process and KEGG pathways were used. For survival analysis, log-rank test was performed to test the significant difference between 2 curves.
Computational analysis of human LUAD samples from TCGA using the HADHA z score.
In silico analysis of MTP α-subunit (HADHA) expression in the TCGA cohort of human LUAD was performed using the HADHA z score expression value that took into account the difference in the mean expression of HADHA between the tumor and the normal tissue and the standard deviation of these measurements. The z score indicates the number of standard deviations away from the mean of expression in the reference and was calculated by TCGA as follows: z = (expression in tumor sample – mean expression in reference sample)/standard deviation of expression in reference sample. Comparison of HADHA tumor expression was also performed directly using the RNA-Seq log2 FPKM values. Comparison of the transcriptome of HADHA+ LUADs and HADHA– LUADs (as segregated using cBioPortal according to the HADHA z score value) was then performed using DESeq2 and Metascape (adjusted P < 0.05). The network was visualized with Cytoscape (v3.1.2) with “force-directed” layout and with edge bundled for clarity. Lastly, a coexpression analysis was performed on the entire TCGA LUAD population using cBioPortal.
Cell culture conditions and lentiviral constructions and transductions.
The 12 human lung cancer cells listed in Supplemental Table 1, sheet 2 were obtained from ATCC (A549, H1975, H460, LLC, H1299, H596, H1770, H1437) and from D. Santamaria’s group (INSERM U1218, Bordeaux, France) (H23, H358, PC9, H2279, H2935, HCC4006, HCC364, and HCC1395). The cells were maintained in 5 mM glucose DMEM medium (Gibco) supplemented with 10% FBS, pH 7.4 at 37°C in a humidified incubation chamber with 5% CO2. Cells were subcultured every 2 days and used on experiments when they reached 85% confluency. Also, A549, H460, PC9, 1395, and LLC cells were stably transfected with luciferase (pRRLsin-MND-Luc-WPRE). The clones were tested as luciferase-positive using direct application of 2 mL of a 150 mg/mL solution of luciferine D-luciferin (Interchim), followed by immediate bioluminescent imaging using the photonIMAGER optima system. Specific suppression of human HADHA mRNA was achieved by transduction of lentiviral particles expressing oligonucleotides bearing a short hairpin structure (OriGene, TL312528V). A549 cells grown in a 24-well plate at confluence of 50% were incubated with control or HADHA shRNA lentiviral particles. Cells were grown in culture media for 48 hours. After washing with PBS, cells were cultured in fresh media for an additional period of 24 hours. Cells were then diluted in a 1:5 suspension and cultured for 10 days in the presence of 1 μg/mL of puromycin dihydrochloride (Sigma-Aldrich) to select resistant cells (shHADHA). Transfected cells with scramble vector were used as negative control and denominated shScramble. The efficacy of shHADHA was determined by real-time qPCR using predesigned primers (Sigma-Aldrich) and Western blot assay using a specific antibody anti-HADHA (Abcam, 203114). The lentiviral shRNAmiR construction for generating the doxycycline-inducible HADHA knockdown in A549 cells was obtained as follows. shRNAmiR target-specific sequences were designed from the shRNA sequence and the shRNA targeting HADHA described above (Origene, TL312528A, TL312528D) and shRNA control sequence (SCR negative control used in pLKO1) (63) according to the technical manual (Thermo Fisher Scientific Open Biosystems). The hairpin stem consisted of 23 nt of dsRNA and a 19 nt loop from human miR30. The single-strain 114-mers were designed, synthesized by Eurofins, and annealed and phosphorylated. Double-strand oligos were then cloned into pTRIPZ plasmid linearized by EcoRI/XhoI and checked by sequencing. Lentiviral vector production was done by the service platform for lentiviral vector production Vect′UB′ of the TMB-Core of the University of Bordeaux. Cell transduction was performed as described above for the shRNA.
The following is the primer shRNAmiR sense XhoI overhang: TCGAGAAGGTATATTGCTGTTGACAGTGAGCGAXXXXXXXXXXXXXXXXXXXXXTAGTGAAGCCACAGATGTAXXXXXXXXXXXXXXXXX XXXXGTGCCTACTGCCTCGG. The following is the sequence primer shRNAmiR reverse EcoRI overhang: AATTccgaggcagtaggcaCXXXXXXXXXXXXXXXXXXXXXtacatctgtggcttcactaXXXXXXXXXXXXXXX XXXXXXTcgctcactgtcaacagcaatataccttc.
The following is the overhang for ligation mir30 context (stem): Mismatch for processing — Target sequence 21 nt (second reverse-complement to first, sense versus antisense) loop — mirHADHA1 CCTATTGTGGCTGCCATCAATGG; mirHADHA TAGAAGCGGTGATTCCAGATCAC; mirSCR (NON-TARGETING) CCTAAGGTTAAGTCGCCCTCG.
Multicellular tumor spheroids and cell viability and proliferation assays.
3D cell cultures were essentially prepared as described by Ho et al. (64). Briefly, spheroids were produced by seeding U-bottom wells previously coated with 1% agarose with 2 × 104 cells suspended in 200 μL DMEM/F-12 medium containing 10% FBS. With this setup, cells do not adhere to the plastic matrix and are thus allowed to associate, forming a multicellular 3D structure. The size of the aggregates was monitored every 24 hours in the inverted microscope for a total of 72 hours. The sulforhodamine B (SRB) cell viability assay and BrdU proliferation assay were performed as recently described (17).
ATP measurements.
The steady-state intracellular ATP content was measured using the CellTiter-Glo luminescent cell viability assay (Promega) following the manufacturer’s instructions. Luminescence was read on a Clariostar plate reader (Promega), and values were calculated based on an ATP standard curve. The assay was normalized to cell number.
Complex I enzymatic activity assay.
The enzymatic activity of complex I was assessed by monitoring the oxidation of NADH. First, a cell homogenate was prepared as follows: 10 million cells were detached from the plate using 0.25% trypsin-EDTA solution, and a cell pellet was obtained by centrifugation 10 minutes at 300g. The pellet was frozen at –80°C and thawed at room temperature on ice. Then, mechanical cell lysis was performed using a glass/glass potter at 4°C (10 strokes in complex I enzyme assay buffer [35 mM KH2PO4, 5 mM MgCl2, 2 mM KCN, pH 7.2]). A cell homogenate was obtained and protein concentration was determined using the BCA assay (Thermo Fisher Scientific). Then, the rate of NADH oxidation by complex I was recorded using the ubiquinone analogue decylubiquinone (Sigma-Aldrich, D-7911) as electron acceptor as described previously (59).
NAD+/NADH determination.
The NAD+/NADH ratio was measured using the NAD+/NADH colorimetric assay kit (Abcam), according to the manufacturer’s instructions.
Immunopurification and Western blot studies.
The complex I pulldown assay was performed using Dynabeads protein A IP kit from Invitrogen. Dynabeads were coated with 1 μg anti–complex I immunocapture antibody from Abcam (ab109798, 18G12BC2) or 1 μg of the negative control IgG (Sigma-Aldrich). Total proteins from A549 cells were extracted with a RIPA buffer. IP experiments were realized with positive control such as purified IgG and negative control using beads without antibody coating. Western blot was performed as previously described (18). Proteins were detected using primary antibodies against HADHA (Abcam, 203114).
Oxygen consumption of intact lung cancer cells by Seahorse.
The oxygen consumption rate was determined on 12 human lung cancer cell lines described in Supplemental Table 1, sheet 2 using the Seahorse XF96 equipment as detailed in Gentric et al. (28). The OCR was normalized to cell number as determined using the SRB assay described above.
RNA extraction and cDNA synthesis.
Total RNA was isolated from A549 cells using QiaZOL reagent (QIAGEN) according to the manufacturer’s instructions. Total RNA was quantified spectrophotometrically and 1 μg was treated with 1 U of RNase-free DNase (Bio-Rad). cDNA synthesis was performed using the DNase-treated RNA according to the iScript cDNA Synthesis kit (Bio-Rad).
Real-time PCR.
Gene expression analysis was performed using CFX96 real-time PCR (Bio-Rad) and Power SYBR Green PCR Master Mix (Applied Biosystems). For this test, predesigned primer pairs were purchased from Sigma-Aldrich (KiCqStart SYBR Green predesigned primers for gene expression analysis). The comparative Ct method was used to compare changes in gene expression levels. GUSB and RPLP0 were used as an endogenous control.
Acylcarnitine measurements by liquid chromatography/mass spectrometry.
Extracted samples (plasma, cells, and medium) were dissolved in 100 μL CHCl3, and 15 μL was used for the liquid chromatography/mass spectrometry analysis. Liquid chromatography separation was achieved using a Bio-Bond 5U C4 column (Dikma). The liquid chromatography solvents were as follows: buffer A, 95:5 H2O/methanol plus 0.03% NH4OH; buffer B, 60:35:5 isopropanol/methanol/H2O plus 0.03% NH4OH. A typical liquid chromatography run consisted of the following for 70 minutes after injection: 0.1 mL/min 100% buffer A for 5 minutes, 0.4 mL/min linear gradient from 20% buffer B to 100% buffer B over 50 minutes, 0.5 mL/min 100% buffer B for 8 minutes, and equilibration with 0.5 mL/min 100% buffer A for 7 minutes. Free fatty acid analysis was performed using a Thermo Fisher Scientific Q Exactive Plus fitted with a heated electrospray ionization source. The mass spectrometry source parameters were 4 kV spray voltage, with a probe temperature of 437.5°C and capillary temperature of 268.75°C. Full-scan mass spectrometry data were collected with a resolution of 70 k, AGC target 1 × 106, maximum injection time of 100 ms, and a scan range of 150–2,000 m/z. Data-dependent mass spectrometry (top 5 mode) was acquired with a resolution of 35 k; AGC target 1 × 105; maximum injection time of 50 ms; isolation window 1 m/z; scan range 200 to 2,000 m/z; and stepped normalized collision energy (NCE) of 20, 30, and 40. Extracted ion chromatograms for each free fatty acid were generated using an m/z ± 0.01 mass window around the calculated exact mass (i.e., palmitic acid, calculated exact mass for M–H was 255.2330, and the extracted ion chromatogram was 255.22–255.24). Quantification of the free fatty acids was performed by measuring the area under the peak.
Analysis of [U-13C]palmitateoxidation by ionic-exchange chromatography and tandem mass spectrometry. A549 cells grown in low-glucose DMEM were washed twice with PBS and resuspended in PBS containing 2 mM [U-13C]palmitate with 10 mM glucose. The cells were incubated for 2 hours at 27°C before fast filtration preparation of the samples for mass spectrometry analysis. Metabolites were analyzed by ionic-exchange chromatography coupled with tandem mass spectrometry (IC-MS/MS) using the method described by Heuillet et al. (60). Retention time on the column and multiple reaction monitoring transition of each analyzed metabolite were determined. The 13C mass isotopomer distribution of intracellular metabolites was determined from relevant isotopic clusters in the IC-MS/MS analysis according to the method of Kiefer et al. (61). 13C mass isotopomer distribution measurements were performed using a triple quadrupole mass spectrometer (4000 QTRAP, Applied Biosystems). To obtain 13C-labeling patterns (13C isotopologs), isotopic clusters were corrected for the natural abundance of isotopes other than 13C using the in-house software IsoCor (available at MetaSys).
Generation of OX+ and OX– LUAD orthotopic mouse models.
The NOD/LtSz-scid/IL-2Rγchainnull (NSG) mice were produced and housed in the University of Bordeaux Segalen animal facility, according to the rules and regulations governed and enforced by the IACUC. Animals between 6 and 8 weeks old were included in protocols. Mice were monitored weekly for body weight and examined for aspect and behavior during the time course of the experiments. No changes were noticed except otherwise indicated. The orthotopic mouse models of OX+ or OX– human LUADs were obtained by using luciferase-expressing clones of A549, H460, PC9, and 1395 human LUAD cell lines, as well as the LLC murine LUAD cell line (18).
In vivo TMZ treatment.
TMZ was purchased from Sigma-Aldrich and solubilized in saline solution. Two types of treatment were considered in our study: p.o. and i.p. In the p.o. administration, a dose of 20 mg/kg was added to the drinking water and replaced every day. For the i.p. injections, 20 mg/kg or 80 mg/kg were administered daily to the treated group of mice.
Statistics.
Results are expressed as mean ± SEM and analyzed using GraphPad Prism 8.1 software. The unpaired Student’s t test was used to compare 2 independent groups, and the paired t test was used for sequential measurements. For multiple comparison analyses, the ANOVA test with Dunnett’s correction was used. Evaluation of the gaussian distribution of the data was performed prior to the t test or ANOVA. Normal distribution of the data and variance similarity were verified using GraphPad Prism. A P value less than 0.05 was considered statistically significant. All experiments were performed with a minimum of N = 3 biological replicates and n = 3 technical replicates. For mouse studies, sample size was determined using the methods described by Berndtson et al. (62). Predetermined exclusion criteria included the absence of signal at the start of the experiment.
Study approval.
All experiments involving the use of human tissues for research were approved by the ethics committee (Comité de Protection des Personnes Sud-Ouest et Outre Mer III, Université de Bordeaux, Bordeaux, France). All procedures using animals were approved by the Animal Care and Use Ethics Committee of the University of Bordeaux (Comité d’éthique et d’expérimentation animale de l’université de Bordeaux, Université de Bordeaux, Bordeaux, France). The animal facility institutional agreement number is A33063916.
Author contributions
NDA designed and conducted all of the experiments, except the bioenergetic investigations performed on LUAD surgical specimens by EO and NB, and wrote the manuscript. BR, JI, and NDA performed the in vivo studies using the orthotopic LUAD OX+ and OX– mice and provided intellectual input and valuable discussion. LG, TP, and FB performed the 13C-palmitate metabolomic analyses (METATOOL) and IRV performed the lipidomic studies along with GP. MF performed the analysis on the LUAD spheroid tumor model. SC performed the proteomic analyses. YK and AD performed the in silico analysis of the TCGA LUAD cohort. CLP performed the modeling of the complex I structure. WM and HRR performed the lentiviral transductions and provided valuable discussion. VGD generated the doxycycline-inducible shRNA system for HADHA reversible downregulation. MT and JMB performed surgery and prepared lung tumor samples; HB performed the pathology analysis. RR supervised all of the studies and provided valuable discussion and intellectual input. SS participated in the in vivo measurement of lung tumor size in mouse models. PE performed the immunoprecipitation studies. LD participated in the bioenergetic analyses of LUAD cell lines. DE performed enzyme activity assays for complex I. NDS performed the LUAD IHC staining for HADHA expression scoring. DL participated in project management. FMG participated in bioinformatic analyses with YK. All authors edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported, in part, by the French National Institute for Scientific and Medical Research (INSERM), the Brazilian National Council for Scientific and Technological Development (CnPq), Plan Cancer, Fondation ARC, French National Institute against cancer (Inca), Ligue Régionale contre le cancer, ITN Marie Curie TRANSMIT (H2020-MSCA-ITN-2016 N°722605), SIRIC-Brio2 (Project PRIME/IMS/COMMUCAN), and CAPES-COFECUB. The authors thank the Cancéropôle GSO (Club Metabo-Cancer) for guidance and support in the use of metabolomics. We would also like to thank S.R. Cassia for stimulating discussions. The authors also thank Violaine Moreau for providing the plasmid used for T41A β-catenin ectopic expression and D. Santamaria for providing selected NSCLC cell lines. Last, we would like to thank Franklin Rumjanek for his guidance throughout this project, which involved a collaboration between the University of Rio de Janeiro (UFRJ) and the University of Bordeaux. We dedicate this work to Franklin, who passed away during the revision of this manuscript.
Version 1. 01/04/2021
Print issue publication
Footnotes
Conflict of interest: RR and NA declare a patent (PCT/EP2018/086278) entitled “Mitochondrial trifunctional protein (MTP) inhibitors for use in the treatment of high mitochondrial respiration (OX+) cancers.” Institution: AST Aquitaine — University of Bordeaux.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e133081.https://doi.org/10.1172/JCI133081.
Contributor Information
Saharnaz Sarlak, Email: saharnaz.sarlak@u-bordeaux.fr.
Emilie Obre, Email: emilie.obre@elysia-bioscience.com.
Pauline Esteves, Email: pauline.esteves@u-bordeaux.fr.
Yann Kieffer, Email: yann.kieffer@curie.fr.
Alexis Dupis, Email: alexis.dupis@gmail.com.
Julien Izotte, Email: julien.izotte@u-bordeaux.fr.
Laetitia Dard, Email: laetitia.dard@u-bordeaux.fr.
Isabelle Redonnet-Vernhet, Email: isabelle.redonnet-vernhet@chu-bordeaux.fr.
Giuseppe Punzi, Email: peppepunzi@gmail.com.
Elodie Dumon, Email: elodie.dumon32@gmail.com.
Lara Gales, Email: lara.gales@insa-toulouse.fr.
Floriant Bellvert, Email: bellvert@insa-toulouse.fr.
Jean-Marc Baste, Email: Jean-Marc.Baste@chu-rouen.fr.
Didier Lacombe, Email: didier.lacombe@chu-bordeaux.fr.
Ciro Leonardo Pierri, Email: ciroleopierri@gmail.com.
Fatima Mechta-Grigoriou, Email: fatima.mechta-grigoriou@curie.fr.
Matthieu Thumerel, Email: matthieu.thumerel@chu-bordeaux.fr.
Rodrigue Rossignol, Email: rodrigue.rossignol@u-bordeaux.fr.
References
- 1.Garon EB, et al. Five-year overall survival for patients with advanced non–small-cell lung cancer treated with pembrolizumab: results from the phase I KEYNOTE-001 study. J Clin Oncol. 2019;37(28):2518–2527. doi: 10.1200/JCO.19.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong DS, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383(13):1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hensley CT, et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;164(4):681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padanad MS, et al. Fatty acid oxidation mediated by acyl-CoA synthetase long chain 3 is required for mutant KRAS lung tumorigenesis. Cell Rep. 2016;16(6):1614–1628. doi: 10.1016/j.celrep.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kageyama T, et al. HADHA is a potential predictor of response to platinum-based chemotherapy for lung cancer. Asian Pac J Cancer Prev. 2011;12(12):3457–3463. [PubMed] [Google Scholar]
- 7.Chen PH, et al. Metabolic diversity in human non-small cell lung cancer cells. Mol Cell. 2019;76(5):838–851.e5. doi: 10.1016/j.molcel.2019.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vergara D, et al. β-Catenin knockdown affects mitochondrial biogenesis and lipid metabolism in breast cancer cells. Front Physiol. 2017;8:544. doi: 10.3389/fphys.2017.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao F, et al. Paracrine Wnt5a-β-catenin signaling triggers a metabolic program that drives dendritic cell tolerization. Immunity. 2018;48(1):147–160.e7. doi: 10.1016/j.immuni.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senni N, et al. β-Catenin-activated hepatocellular carcinomas are addicted to fatty acids. Gut. 2019;68(2):322–334. doi: 10.1136/gutjnl-2017-315448. [DOI] [PubMed] [Google Scholar]
- 11.Lehwald N, et al. β-Catenin regulates hepatic mitochondrial function and energy balance in mice. Gastroenterology. 2012;143(3):754–764. doi: 10.1053/j.gastro.2012.05.048. [DOI] [PubMed] [Google Scholar]
- 12.Frey JL, et al. β-Catenin directs long-chain fatty acid catabolism in the osteoblasts of male mice. Endocrinology. 2018;159(1):272–284. doi: 10.1210/en.2017-00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu M, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292(1):C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 14.Peng S, et al. The efficacy of trimetazidine on stable angina pectoris: a meta-analysis of randomized clinical trials. Int J Cardiol. 2014;177(3):780–785. doi: 10.1016/j.ijcard.2014.10.149. [DOI] [PubMed] [Google Scholar]
- 15.Sander J, et al. Neonatal screening for defects of the mitochondrial trifunctional protein. Mol Genet Metab. 2005;85(2):108–114. doi: 10.1016/j.ymgme.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Primassin S, et al. Carnitine supplementation induces acylcarnitine production in tissues of very long-chain acyl-CoA dehydrogenase-deficient mice, without replenishing low free carnitine. Pediatr Res. 2008;63(6):632–637. doi: 10.1203/PDR.0b013e31816ff6f0. [DOI] [PubMed] [Google Scholar]
- 17.Dias Amoedo N, et al. Targeting human lung adenocarcinoma with a suppressor of mitochondrial superoxide production. Antioxid Redox Signal. 2020;33(13):883–902. doi: 10.1089/ars.2019.7892. [DOI] [PubMed] [Google Scholar]
- 18.Esteves P, et al. Nuclear control of lung cancer cells migration, invasion and bioenergetics by eukaryotic translation initiation factor 3F. Oncogene. 2020;39(3):617–636. doi: 10.1038/s41388-019-1009-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miralpeix M, et al. The impairment of respiration by glycolysis in the Lewis lung carcinoma. Cancer Lett. 1990;50(3):173–178. doi: 10.1016/0304-3835(90)90261-U. [DOI] [PubMed] [Google Scholar]
- 20.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fulgencio JP, et al. Troglitazone inhibits fatty acid oxidation and esterification, and gluconeogenesis in isolated hepatocytes from starved rats. Diabetes. 1996;45(11):1556–1562. doi: 10.2337/diab.45.11.1556. [DOI] [PubMed] [Google Scholar]
- 22.Protasoni M, et al. Respiratory supercomplexes act as a platform for complex III-mediated maturation of human mitochondrial complexes I and IV. EMBO J. 2020;39(3):e102817. doi: 10.15252/embj.2019102817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Evidence for physical association of mitochondrial fatty acid oxidation and oxidative phosphorylation complexes. J Biol Chem. 2010;285(39):29834–29841. doi: 10.1074/jbc.M110.139493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Mitochondrial fatty acid oxidation and the electron transport chain comprise a multifunctional mitochondrial protein complex. J Biol Chem. 2019;294(33):12380–12391. doi: 10.1074/jbc.RA119.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narayan SB, et al. Short-chain 3-hydroxyacyl-coenzyme A dehydrogenase associates with a protein super-complex integrating multiple metabolic pathways. PLoS One. 2012;7(4):e35048. doi: 10.1371/journal.pone.0035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiff M, et al. Complex I assembly function and fatty acid oxidation enzyme activity of ACAD9 both contribute to disease severity in ACAD9 deficiency. Hum Mol Genet. 2015;24(11):3238–3247. doi: 10.1093/hmg/ddv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyni T, et al. Pathology of skeletal muscle and impaired respiratory chain function in long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency with the G1528C mutation. Neuromuscul Disord. 1996;6(5):327–337. doi: 10.1016/0960-8966(96)00352-5. [DOI] [PubMed] [Google Scholar]
- 28.Gentric G, et al. PML-regulated mitochondrial metabolism enhances chemosensitivity in human ovarian cancers. Cell Metab. 2019;29(1):156–173. doi: 10.1016/j.cmet.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JS, et al. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature. 2020;578(7796):621–626. doi: 10.1038/s41586-020-1998-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu H, et al. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell. 2016;164(3):433–446. doi: 10.1016/j.cell.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casbas-Hernandez P, et al. Tumor intrinsic subtype is reflected in cancer-adjacent tissue. Cancer Epidemiol Biomarkers Prev. 2015;24(2):406–414. doi: 10.1158/1055-9965.EPI-14-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aran D, et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun. 2017;8(1):1077. doi: 10.1038/s41467-017-01027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baran N, et al. Inhibition of oxphos with Iacs-010759 s\depletes AML leukemia initiating cells (LIC) in vitro and improves survival in pre-clinical AML models. Blood. 2017;130(suppl 1):1350. doi: 10.1182/blood.V130.Suppl_1.1350.1350. [DOI] [Google Scholar]
- 34.Chiche J, et al. GAPDH expression predicts the response to r-chop, the tumor metabolic status, and the response of DLBCL patients to metabolic inhibitors. Cell Metab. 2019;29(6):1243–1257.e10. doi: 10.1016/j.cmet.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Baccelli I, et al. Mubritinib targets the electron transport chain complex I and reveals the landscape of OXPHOS dependency in acute myeloid leukemia. Cancer Cell. 2019;36(1):84–99.e8. doi: 10.1016/j.ccell.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Schöckel L, et al. Targeting mitochondrial complex I using BAY 87-2243 reduces melanoma tumor growth. Cancer Metab. 2015;3:11. doi: 10.1186/s40170-015-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, et al. Metabolic reprogramming toward oxidative phosphorylation identifies a therapeutic target for mantle cell lymphoma. Sci Transl Med. 2019;11(491):eaau1167. doi: 10.1126/scitranslmed.aau1167. [DOI] [PubMed] [Google Scholar]
- 38.Naguib A, et al. Mitochondrial complex I inhibitors expose a vulnerability for selective killing of Pten-null cells. Cell Rep. 2018;23(1):58–67. doi: 10.1016/j.celrep.2018.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scotland S, et al. Mitochondrial energetic and AKT status mediate metabolic effects and apoptosis of metformin in human leukemic cells. Leukemia. 2013;27(11):2129–2138. doi: 10.1038/leu.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samudio I, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120(1):142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu PP, et al. Elimination of chronic lymphocytic leukemia cells in stromal microenvironment by targeting CPT with an antiangina drug perhexiline. Oncogene. 2016;35(43):5663–5673. doi: 10.1038/onc.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stroud DA, et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538(7623):123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 43.Heide H, et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16(4):538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 44.Bastian A, et al. AG311, a small molecule inhibitor of complex I and hypoxia-induced HIF-1α stabilization. Cancer Lett. 2017;388:149–157. doi: 10.1016/j.canlet.2016.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sica V, et al. Lethal poisoning of cancer cells by respiratory chain inhibition plus dimethyl α-ketoglutarate. Cell Rep. 2019;27(3):820–834.e9. doi: 10.1016/j.celrep.2019.03.058. [DOI] [PubMed] [Google Scholar]
- 46.Ellinghaus P, et al. BAY 87-2243, a highly potent and selective inhibitor of hypoxia-induced gene activation has antitumor activities by inhibition of mitochondrial complex I. Cancer Med. 2013;2(5):611–624. doi: 10.1002/cam4.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molina JR, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med. 2018;24(7):1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- 48.Yap TA, et al. Phase I trial of IACS-010759 (IACS), a potent, selective inhibitor of complex I of the mitochondrial electron transport chain, in patients (pts) with advanced solid tumors. J Clin Oncol. 2019;37(15 suppl):3014. doi: 10.1200/JCO.2019.37.15_suppl.3014. [DOI] [Google Scholar]
- 49. Bayer Healthcare. Clinical Trial Study Synopsis [BAY 87-2243]. Study 15044. http://trialfinder.bayerscheringpharma.de/html/pdf/15044 Study Synopsis CTP.pdf Created June 26, 2013. Accessed October 22, 2020.
- 50.Kirkpatrick DL, Powis G. Clinically evaluated cancer drugs inhibiting redox signaling. Antioxid Redox Signal. 2017;26(6):262–273. doi: 10.1089/ars.2016.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schapira AHV. Complex I: inhibitors, inhibition and neurodegeneration. Exp Neurol. 2010;224(2):331–335. doi: 10.1016/j.expneurol.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 52.Heinz S, et al. Mechanistic investigations of the mitochondrial complex I inhibitor rotenone in the context of pharmacological and safety evaluation. Sci Rep. 2017;7:45465. doi: 10.1038/srep45465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gasparre G, et al. A mutation threshold distinguishes the antitumorigenic effects of the mitochondrial gene MTND1, an oncojanus function. Cancer Res. 2011;71(19):6220–6229. doi: 10.1158/0008-5472.CAN-11-1042. [DOI] [PubMed] [Google Scholar]
- 54.Kurelac I, et al. Inducing cancer indolence by targeting mitochondrial complex I is potentiated by blocking macrophage-mediated adaptive responses. Nat Commun. 2019;10(1):903. doi: 10.1038/s41467-019-08839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossignol R, et al. Threshold effect and tissue specificity. Implication for mitochondrial cytopathies. J Biol Chem. 1999;274(47):33426–33432. doi: 10.1074/jbc.274.47.33426. [DOI] [PubMed] [Google Scholar]
- 56.Kordes S, et al. Metformin in patients with advanced pancreatic cancer: a double-blind, randomised, placebo-controlled phase 2 trial. Lancet Oncol. 2015;16(7):839–847. doi: 10.1016/S1470-2045(15)00027-3. [DOI] [PubMed] [Google Scholar]
- 57.Montaudié H, et al. Metformin monotherapy in melanoma: a pilot, open-label, prospective, and multicentric study indicates no benefit. Pigment Cell Melanoma Res. 2017;30(3):378–380. doi: 10.1111/pcmr.12576. [DOI] [PubMed] [Google Scholar]
- 58.Aubert A, et al. A cellular anti-ischemic agent, trimetazidine prevents the deleterious effects of oxygen free-radicals on the internal ear. Article in French. Ann Otolaryngol Chir Cervicofac. 1990;107(suppl 1):28–35. [PubMed] [Google Scholar]
- 59.Jose C, et al. Redox mechanism of levobupivacaine cytostatic effect on human prostate cancer cells. Redox Biol. 2018;18:33–42. doi: 10.1016/j.redox.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heuillet M, et al. Methodology for the validation of isotopic analyses by mass spectrometry in stable-isotope labeling experiments. Anal Chem. 2018;90(3):1852–1860. doi: 10.1021/acs.analchem.7b03886. [DOI] [PubMed] [Google Scholar]
- 61.Kiefer P, et al. Quantitative metabolome analysis using liquid chromatography-high-resolution mass spectrometry. Anal Biochem. 2008;382(2):94–100. doi: 10.1016/j.ab.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 62.Berndtson WE, et al. A simple, rapid and reliable method for selecting or assessing the number of replicates for animal experiments. J Anim Sci. 1991;69(1):67–76. doi: 10.2527/1991.69167x. [DOI] [PubMed] [Google Scholar]
- 63.Sarbassov DD, et al. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 64.Ho WY, et al. Development of multicellular tumor spheroid (MCTS) culture from breast cancer cell and a high throughput screening method using the MTT assay. PLoS One. 2012;7(9):e44640. doi: 10.1371/journal.pone.0044640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.