Abstract
Human herpes simplex virus 1 (HSV-1) encephalitis can be caused by inborn errors of the TLR3 pathway, resulting in impairment of CNS cell-intrinsic antiviral immunity. Deficiencies of the TLR3 pathway impair cell-intrinsic immunity to vesicular stomatitis virus (VSV) and HSV-1 in fibroblasts, and to HSV-1 in cortical but not trigeminal neurons. The underlying molecular mechanism is thought to involve impaired IFN-α/β induction by the TLR3 recognition of dsRNA viral intermediates or by-products. However, we show here that human TLR3 controls constitutive levels of IFNB mRNA and secreted bioactive IFN-β protein, and thereby also controls constitutive mRNA levels for IFN-stimulated genes (ISGs) in fibroblasts. Tlr3–/– mouse embryonic fibroblasts also have lower basal ISG levels. Moreover, human TLR3 controls basal levels of IFN-β secretion and ISG mRNA in induced pluripotent stem cell–derived cortical neurons. Consistently, TLR3-deficient human fibroblasts and cortical neurons are vulnerable not only to both VSV and HSV-1, but also to several other families of viruses. The mechanism by which TLR3 restricts viral growth in human fibroblasts and cortical neurons in vitro and, by inference, by which the human CNS prevents infection by HSV-1 in vivo, is therefore based on the control of early viral infection by basal IFN-β immunity.
Keywords: Immunology, Infectious disease
Keywords: Innate immunity
Introduction
TLR3 on endosomes recognizes double-stranded RNA (dsRNA) intermediates or by-products generated during viral infection. TLR3 signaling leads to the activation of IFN regulatory factor 3 (IRF3), NF-κB, and ATF/c-jun, promoting the induction of antiviral IFNs and downstream IFN-stimulated genes (ISGs) (1–4). The discovery of inborn errors of human TLR3 and its pathway in children with herpes simplex virus 1 (HSV-1) encephalitis (HSE) led to the suggestion that TLR3 serves as a key sensor for HSV-1 replication in the CNS (5–7). Childhood HSE is a rare, sporadic, and life-threatening complication of primary infection with HSV-1 in which the virus replicates in the CNS. HSV-1 infection is ubiquitous in the general population. The virus resides in the trigeminal (TG) ganglion, where it remains latent, but can later reactivate to cause benign herpes labialis or other rare complications, including HSE (8). The pathogenesis of HSE remained unexplained until our description of the first genetic etiologies for this disease (9, 10). Germline HSE-causing mutations have since been reported in 7 genes of the TLR3 pathway (TLR3, UNC93B1, TRIF, TRAF3, TBK1, IRF3, NEMO) and 2 genes of the IFN-α/β receptor pathway (IFNAR1, STAT1) (9–17). UNC-93B is a membrane-bound molecule that regulates the signaling of endosomal TLR3, TLR7, TLR8, and TLR9 by binding to their transmembrane domains and maintaining TLR3 expression (18–21). TRIF is the sole adaptor of TLR3, whereas TRAF3, TBK1, IRF3, and NEMO are key molecules required for the TLR3-TRIF–dependent induction of antiviral IFNs (7). Candidate mutations in other genes of the pathway have also been reported (22). Mutations of TLR3 pathway genes have been shown to impair the TLR3-dependent induction of antiviral IFNs, whereas IFNAR1 and STAT1 mutations impair cellular responses to type I IFNs. Collectively, these findings suggest that HSE can result from the impaired production of IFN-α/β and/or IFN-λ in response to TLR3 stimulation by HSV-1 in the CNS.
This hypothesis was initially supported by experiments conducted in dermal fibroblasts infected with HSV-1 and another neurotropic virus, vesicular stomatitis virus (VSV), an ssRNA virus typically innocuous in humans, chosen for these studies because it is highly cytopathic and induces IFN more effectively than HSV-1 in these cells (23). Fibroblasts with TLR3 signaling deficiencies display impaired responses to the TLR3 agonist polyinosinic-polycytidylic acid (poly(I:C)), a synthetic analog that mimics dsRNA by-products and intermediates of viral replication. Following infection with HSV-1 or VSV, TLR3 pathway–deficient fibroblasts produce less IFN-β and -λ than control cells, and display higher rates of viral replication and virus-induced cell death (9–11, 13–16, 24). Moreover, the viral phenotype of fibroblasts from patients with mutations of TLR3 pathway genes is rescued by pretreatment with IFN-α or -β, but not IFN-λ (9–11, 13–16, 24). The viral phenotype of fibroblasts from patients not responding to IFN-α/β and -λ due to STAT1 deficiency is not rescued by any type of IFN (10). We have also shown that cortical neurons and oligodendrocytes derived from TLR3-deficient induced pluripotent stem cells (iPSCs) display impaired IFN responses to poly(I:C) and HSV-1, and do not control HSV-1, with this viral phenotype being rescued by pretreatment with IFN-α or -β but not IFN-λ (25). Collectively, these findings suggest that the viral (HSV-1 and VSV) and IFN-β phenotypes of TLR3-deficient fibroblasts observed in vitro are a surrogate for that of iPSC-derived cortical neurons in vitro and predisposition to HSE in vivo.
Unlike iPSC-derived cortical neurons, iPSC-derived peripheral TG neurons from healthy donors control HSV-1 as poorly as poly(I:C)-unresponsive TLR3-deficient TG neurons, in terms of viral growth (26). Pretreatment with IFN-α or -β, but not IFN-λ, rescues susceptibility to viral infections in both types of TG neurons, whereas pretreatment with poly(I:C) rescues only control TG neurons, in which IFNs and ISGs are induced in response to TLR3-dependent poly(I:C) stimulation. These data indicate that TG neurons are vulnerable to HSV-1 in the absence of preemptive stimulation via TLR3 or IFN-α/β receptors, whereas control cortical neurons display TLR3-dependent constitutive resistance that is sufficiently strong to block incoming HSV-1 in the absence of prior antiviral signals. This experimental observation in vitro is consistent with HSV-1 infecting TG neurons and establishing latency in these cells, but not in cortical neurons in vivo in most individuals (27). Overall, these findings suggest a cellular model of HSE with a TLR3-dependent IFN-mediated phenotype in fibroblasts and iPSC-derived cortical but not TG neurons. However, the molecular basis of these 2 cellular phenotypes in vitro and of HSE in vivo remained unexplained. Indeed, although both HSV-1 and VSV produce dsRNAs (28, 29), HSV-1 recognition depends largely on the cGAS DNA sensor in mouse fibroblasts and myeloid cells (30), whereas VSV recognition in mouse fibroblasts is dependent on RIG-I (31). It therefore remained unclear whether TLR3 actually recognizes dsRNA intermediates or by-products generated during the infection of fibroblasts and cortical neurons with HSV-1 and VSV, or whether it controls the IFN-mediated immunity of these cells against these viruses by other mechanisms. As a first step toward addressing this question, we performed a comprehensive analysis of the connection between IFN induction and VSV infection in human fibroblasts. We then investigated whether our findings also applied to HSV-1 infection in iPSC-derived cortical and TG neurons.
Results
RIG-I–dependent overproduction of IFN-β and -λ in response to VSV-M51R in TLR3-deficient fibroblasts.
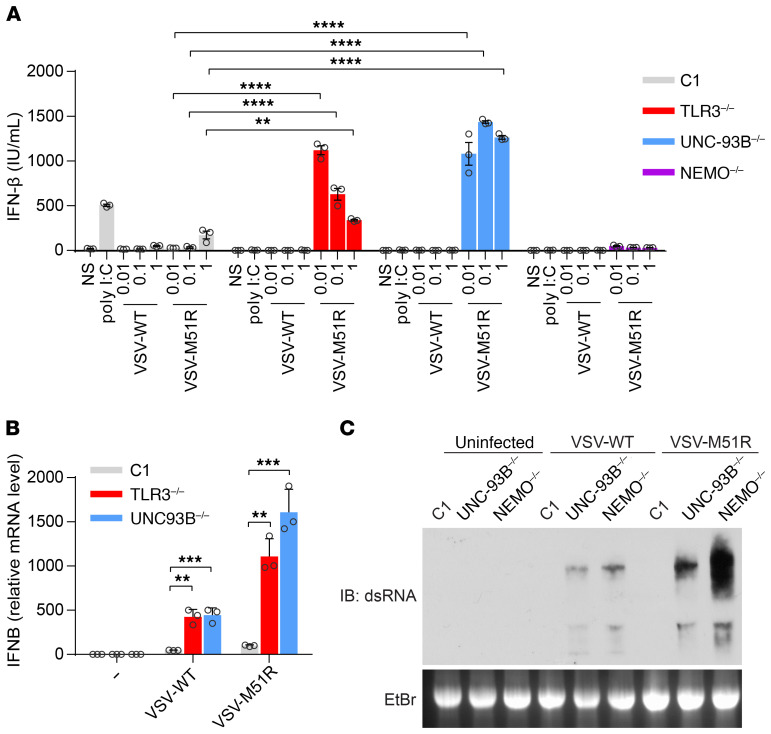
In human fibroblasts, high levels of mRNA for IFN-β and IFN-λ (including all 3 types of IFN-λ, IFNL1, IFNL2, and IFNL3), but not other subtypes of antiviral IFNs, can be induced by extracellular poly(I:C) stimulation, which activates TLR3 in endosomes (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI134529DS1), or intracellular poly(I:C) stimulation, which activates RIG-I and MAD5 in the cytosol (Supplemental Figure 1) (28). We hypothesized that, if the higher levels of viral growth and cell death observed in TLR3-deficient cells were due to an impairment of virus-induced IFN production, then a potent IFN-inducing stimulus would rescue viral susceptibility. We made use of a natural mutant of VSV, VSV-M51R, which induces IFN very strongly in most of the cells tested, much more so than VSV-WT (32, 33). We first assessed the production of IFN-β and -λ following infection with VSV-WT and -M51R at various MOIs for 24 hours, in simian virus 40 (SV-40) T antigen–transformed fibroblasts (SV-40 fibroblasts) from a healthy control, a patient with HSE with autosomal recessive (AR) UNC-93B deficiency (UNC-93B–/–), and a patient with HSE with AR TLR3 deficiency (TLR3–/–). Surprisingly, UNC-93B–/– and TLR3–/– cells produced about 30 times more IFN-β than healthy control cells after 24 hours of infection with VSV-M51R at a MOI of 0.01 (Figure 1A). Similar results were obtained for IFN-λ (as measured by ELISA, which recognizes all IFN-λ1/2/3 in a nonspecific manner, Supplemental Figure 2, A and B). NEMO–/– fibroblasts displayed no activation of the transcription factors IRF3 and NF-κB in response to viral infection (11, 32), and were thus included as a negative control (Figure 1A). Consistently, IFNB and IFNL1 mRNA levels were higher in UNC-93B–/– and TLR3–/– cells than in healthy control cells 24 hours after infection with VSV-M51R or -WT (Figure 1B, Supplemental Figure 2C), although no IFN-β or IFN-λ was detected by ELISA in UNC-93B–/– and TLR3–/– cells following VSV-WT infection (Figure 1A, Supplemental Figure 2A). In UNC-93B–/– and TLR3–/– fibroblasts, the response to TLR3 stimulation with extracellularly added poly(I:C) was completely abolished, whereas the responses to transfection with poly(I:C) (a nonspecific agonist of RIG-I and MDA5) and 7sk-as (a specific agonist of RIG-I) were intact (Figure 1A, Supplemental Figure 3, A–C) (9, 10). IFN induction in response to VSV may be RIG-I dependent, as suggested by previous studies on mouse embryonic fibroblasts (MEFs) (28, 31, 33). Indeed, following the short-hairpin RNA–mediated (shRNA-mediated) knockdown of RIG-I or its downstream signaling molecule MAVS (34–37), the production of IFN-β in response to VSV-M51R infection, or to transfected poly(I:C) and 7sk-as, was much weaker in RIG-I or MAVS-knockdown UNC-93B–/– cells than in those transduced with a control scrambled RNA, indicating the essential role of RIG-I in sensing VSV-M51R (Supplemental Figure 3, A and B). Thus, VSV-M51R can induce IFN-β or IFN-λ via RIG-I in UNC-93B–/– and TLR3–/– fibroblasts, and, paradoxically, UNC-93B–/– and TLR3–/– cells respond to VSV-M51R by producing markedly larger amounts of IFN-β and -λ than control cells.
Figure 1. Paradoxical IFN response to VSV-M51R infection in fibroblasts with TLR3 signaling deficiencies.
(A) IFN-β production in SV-40–transformed dermal fibroblasts (SV-40 fibroblasts) left nonstimulated (NS), treated with poly(I:C), or infected with VSV-WT (WT) or VSV-M51R mutant at various MOIs (0.01, 0.1, 1) for 24 hours, as measured by ELISA. C1 is a healthy control. (B) IFN-β mRNA levels in fibroblasts left NS or infected for 24 hours with VSV-WT or -M51R at a MOI of 1. β-glucuronidase mRNA levels were used for normalization. The error bars indicate SD of biological triplicates from 3 independent experiments. P values were obtained for 1-way ANOVA and subsequent Tukey’s multiple comparison tests. (C) dsRNA from VSV-WT– and VSV-M51R–infected fibroblasts, visualized by electrophoresis in 1.5% agar gels, blotting onto nylon membranes and incubation with a monoclonal antibody against dsRNA. Ethidium bromide (EtBr) staining of the agar gel is shown as a loading control. Data from 3 independent experiments are shown. **P < 0.01, ***P < 0.001, ****P < 0.0001.
The hyper-IFN response to VSV-M51R in TLR3-deficient fibroblasts is triggered by enhanced viral replication.
We then investigated whether RIG-I was hyperactive in UNC-93B–/– fibroblasts, which would account for IFN overinduction as a means of compensating for the lack of TLR3 signaling. We overcame the problem of the confounding effect of a larger viral stimulus in UNC-93B–/– cells by transfecting the fibroblasts with total cellular RNA isolated from VSV-WT– or VSV-M51R–infected Vero cells (vRNA). IFN-β production levels were almost identical between UNC-93B–/– and healthy control cells stimulated by transfection with vRNA (Supplemental Figure 3C). As no viral proteins antagonistic to IFN were produced, VSV-WT and VSV-M51R RNA induced IFN-β to similar levels (Supplemental Figure 3C). This result suggests that the RIG-I pathway is equally active in the cells of patients with UNC-93B deficiency and healthy controls. Moreover, when we coinfected fibroblasts with both VSV-WT and VSV-M51R, the high levels of IFN-β production induced by the M51R virus were completely abolished by coinfection with VSV-WT (Supplemental Figure 3D) (38). This suggests that the inability to block mRNA export from the nucleus, and thus, to abolish IFN production by VSV-WT, is responsible for this phenomenon rather than the stimulation, by VSV-M51R, of an additional signaling pathway different from that stimulated by the WT virus (39). However, consistent with the high levels of virus-triggered IFN-β and IFN-λ production in UNC-93B–/– and TLR3–/– fibroblasts, we detected substantially more dsRNA in the cells of patients with UNC-93B and NEMO deficiency than in control fibroblasts (Figure 1C), 8 hours after VSV-WT or -M51R infection, by Western blotting with anti–dsRNA antibody (28). The high levels of dsRNA are also consistent with the previously known rapid VSV-WT replication in UNC-93B–/– and TLR3–/– fibroblasts (9, 10, 13), which was confirmed by determining VSV glycoprotein (VSV-G) mRNA levels by quantitative real-time PCR (RT-qPCR) (Supplemental Figure 3E). Interestingly, the induction of IFNB mRNA by VSV-WT was detectable only at late time points (not at the 6 hour time point) (Supplemental Figure 3F), suggesting that viral replication is required for IFN induction. Indeed, this enhanced production of IFN in UNC-93B–/– and TLR3–/– cells required an actively replicating virus, as the ultraviolet irradiation of VSV-WT and -M51R blocked their stimulatory effect (Supplemental Figure 3G). Thus, the potent IFN production observed after VSV-M51R infection in cells with TLR3 pathway deficiencies was due to the presence of large amounts of dsRNA upon infection, rather than hyperactive RIG-I signaling.
Uncontrolled VSV-M51R replication and virus-induced cell death in TLR3-deficient fibroblasts despite high levels of virus-induced IFN production.
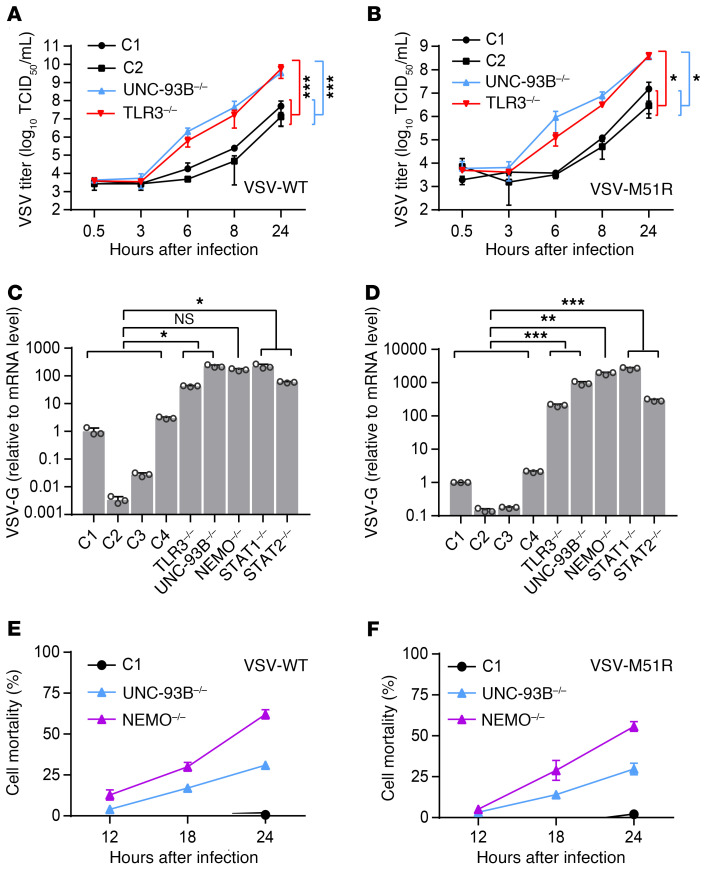
We then investigated the second cellular phenotype related to inborn errors of TLR3 immunity — viral susceptibility — in TLR3–/– and UNC-93B–/– fibroblasts. We measured VSV growth kinetics and virus-induced cell death. Surprisingly, although VSV-M51R complemented the IFN phenotype, with IFN levels even exceeding those observed in controls, this potent induction of IFN production by the virus did not decrease viral replication and cell mortality in TLR3–/– or UNC-93B–/– SV-40 fibroblasts upon infection with VSV-M51R at a MOI of 1, from 0.5 hour to 24 hours after infection. Both the WT and M51R viruses replicated much more rapidly in UNC-93B–/– and TLR3–/– cells than in control cells, and, by 6 hours after infection, there was at least a 100-fold difference in titer (Figure 2, A and B), whereas IFN induction was not readily detectable at this time point (Supplemental Figure 3F). We then measured VSV replication, by assessing viral VSV-G RNA levels in fibroblasts from patients with inborn errors of TLR3 (TLR3, UNC93B1, NEMO) or IFN (STAT1, STAT2) immunity (40, 41), comparing these levels with those in WT control cells. VSV levels were markedly higher in the cells of all patients 16 hours after infection (Figure 2, C and D; and Supplemental Figure 4, A and B). Moreover, cell death rates were greater in UNC-93B–/– cells than in control cells after 24 hours of VSV-WT or -M51R infection (Figure 2, E and F), despite the extremely high levels of IFN production following infection with VSV-M51R (Figure 1, A and B; Supplemental Figure 2, A–C). Thus, although the production of larger amounts of immunostimulatory viral replication intermediates in TLR3 pathway-deficient fibroblasts, such as dsRNA (Figure 1C), led to enhanced IFN production by the patients’ cells upon infection with VSV-M51R, this enhanced IFN production was not sufficient to protect these cells against viral replication, which reached very high levels before IFN production was induced by the virus.
Figure 2. Unrestricted virus growth and cytotoxicity in fibroblasts with TLR3 signaling deficiencies.
VSV-WT (A) and VSV-M51R (B) single-cycle replication curves for fibroblasts from healthy controls (C1 and C2) and patients with UNC-93B or TLR3 deficiency at a MOI of 1 over 24 hours. Control fibroblasts (C1–C4) and TLR3–/–, UNC93B–/–, NEMO–/–, STAT1–/–, and STAT2–/– fibroblasts were infected with VSV-WT (C) and VSV-M51R (D) at a MOI of 0.01 for 16 hours. Viral VSV-G RNA levels were then determined by RT-qPCR and normalized against C1. Cell mortality following infection with VSV-WT (E) and VSV-M51R (F) at a MOI of 1 for C1, UNC-93B–/–, and NEMO–/– fibroblasts, as measured by the release of lactate dehydrogenase (LDH) at the times indicated, in hours postinfection (hpi). Values are expressed relative to those for uninfected cells. Triplicate measurements from 3 independent experiments (A–B, E–F) or representative results from 3 independent experiments (C–D) are shown. The error bars indicate SD of biological triplicates. P values were obtained through log transformation followed by 1-way ANOVA and subsequent Tukey’s multiple comparison tests (A–B) or likelihood ratio tests (C–D), by comparing each patient’s fibroblasts with control fibroblasts, and the respective P value is indicated. *P < 0.05, **P < 0.01, ***P < 0.001.
General antiviral defect in fibroblasts with deficiencies of the TLR3 and IFN signaling pathways.
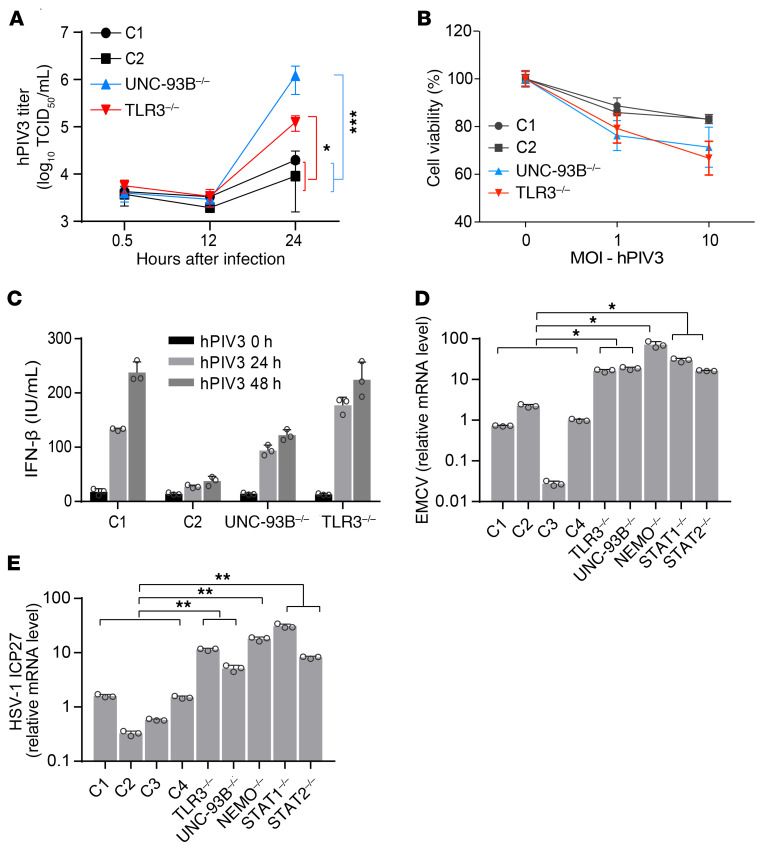
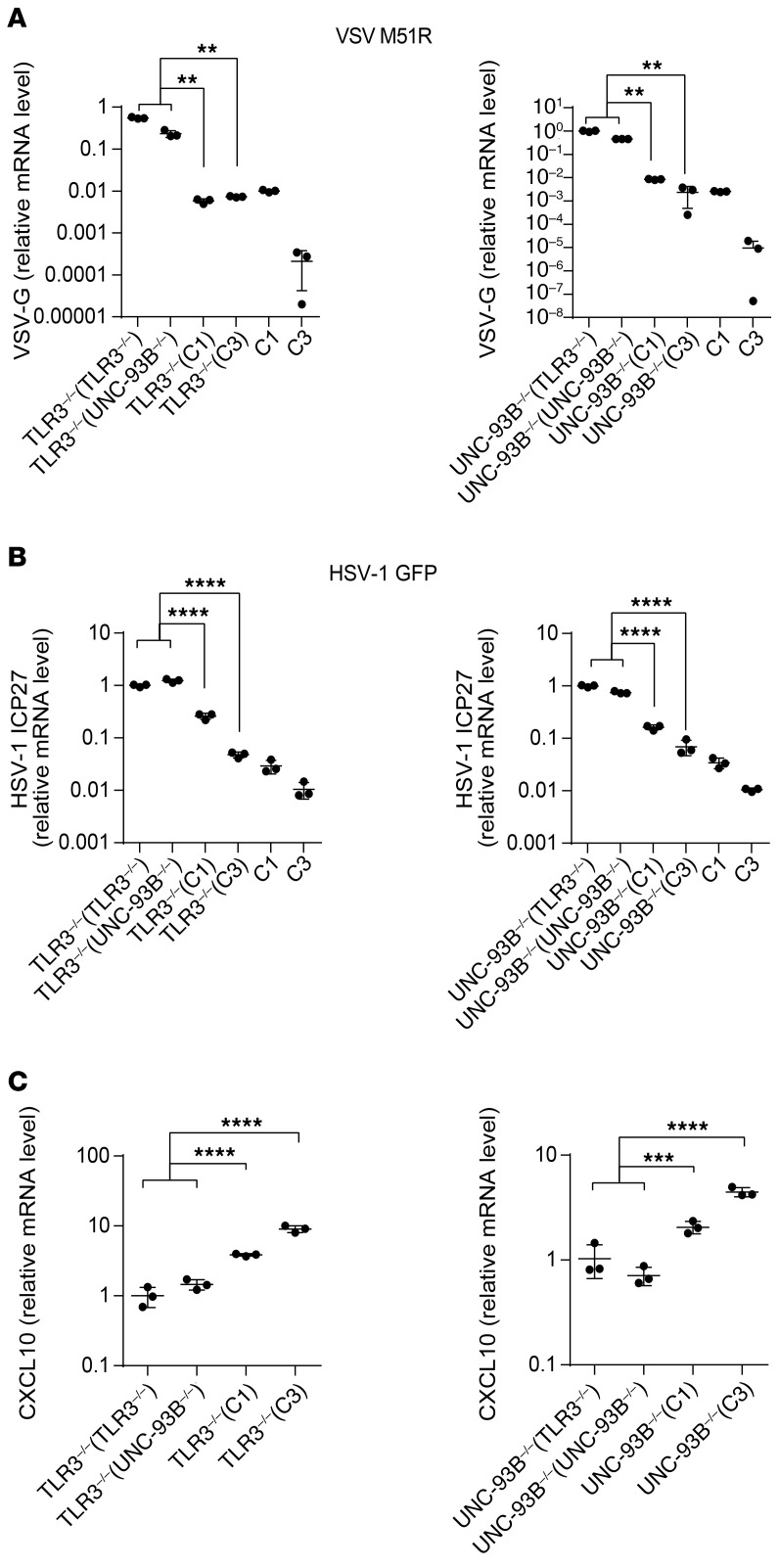
We then assessed the susceptibility of UNC-93B–/– and TLR3–/– fibroblasts to other viruses, which may or may not rely on TLR3 for virus-triggered IFN induction, by evaluating viral replication and virus-induced cell death. We found that another ssRNA virus, human parainfluenza virus 3 (hPIV3), like VSV, replicated faster and to higher titers in UNC-93B–/– and TLR3–/– cells than in control cells (Figure 3A). As a result, viral cytotoxicity was also higher in UNC-93B–/– and TLR3–/– fibroblasts (Figure 3B). IFN-β production was similar in control and TLR3- or UNC-93B–deficient fibroblasts infected with hPIV3 (Figure 3C), probably reflecting intact IFN induction via TLR3-independent pathways. We subsequently challenged fibroblasts with encephalomyocarditis virus (EMCV), another RNA virus that has been shown to induce IFN production in an MDA5-dependent manner in MEFs (31, 42), and that, like hPIV3, induces normal levels of IFN production in TLR3-deficient human fibroblasts (10, 13). EMCV also replicated to high levels in cells with TLR3 pathway deficiencies (TLR3, UNC93B1, NEMO) or IFN pathway deficiencies (STAT1, STAT2) (Figure 3D). Finally, consistent with our previous reports of enhanced HSV-1 replication in TLR3 pathway–deficient fibroblasts (9, 10), we detected higher levels of HSV-1 viral ICP27 RNA in TLR3 pathway–deficient or IFN pathway–deficient cells than in control cells (Figure 3E; and Supplemental Figure 4C), 16 hours after infection, whereas IFNB and IFNL mRNA induction was detected 24 hours after infection (Supplemental Figure 4, D and E). Overall, these data indicate that fibroblasts with deficiencies of the TLR3 and IFN signaling pathways are highly susceptible to infection with at least 3 RNA viruses (VSV, hPIV3, and EMCV) and one DNA virus (HSV-1), despite a high level of IFN production induced by VSV-M51R and the normal induction of IFN production by hPIV3 and EMCV in TLR3 pathway–deficient cells. This is paradoxical, as poor IFN production by these cells upon infection with VSV-WT and HSV-1 had been thought to underlie the cellular vulnerability to both viruses.
Figure 3. High susceptibility to different viruses in TLR3 signaling–deficient cells.
(A) hPIV3 single-cycle replication curves in fibroblasts from healthy controls (C1 and C2) and patients with UNC-93B or TLR3 deficiency over 24 hours. (B) Viability of fibroblasts 48 hours after infection with hPIV3 at the indicated MOIs. (C) IFN-β production, measured by ELISA, after 24 or 48 hours of infection with hPIV3 in C1, C2, UNC-93B–/–, and TLR3–/– fibroblasts. Control fibroblasts (C1–C4) and TLR3–/–, UNC93B–/–, NEMO–/–, STAT1–/–, and STAT2–/– fibroblasts were infected with EMCV (D) or HSV-1-GFP (E) at a MOI of 0.01 for 16 hours. Viral RNA levels were then quantified by RT-qPCR, with normalization against the values for C1. Triplicate measurements from 3 independent experiments (A–C) or representative results from 3 independent experiments (D–E) are shown. The error bars indicate SD of biological triplicates. P values were obtained through log transformation followed by 1-way ANOVA and subsequent Tukey’s multiple comparison tests (A) or likelihood ratio tests, by comparing each patient’s fibroblasts with control fibroblasts (D, E), and the respective P values are indicated. *P < 0.05, **P < 0.01 ***P < 0.001.
Low basal levels of IFN and ISG expression in fibroblasts with TLR3-IFN signaling deficiencies.
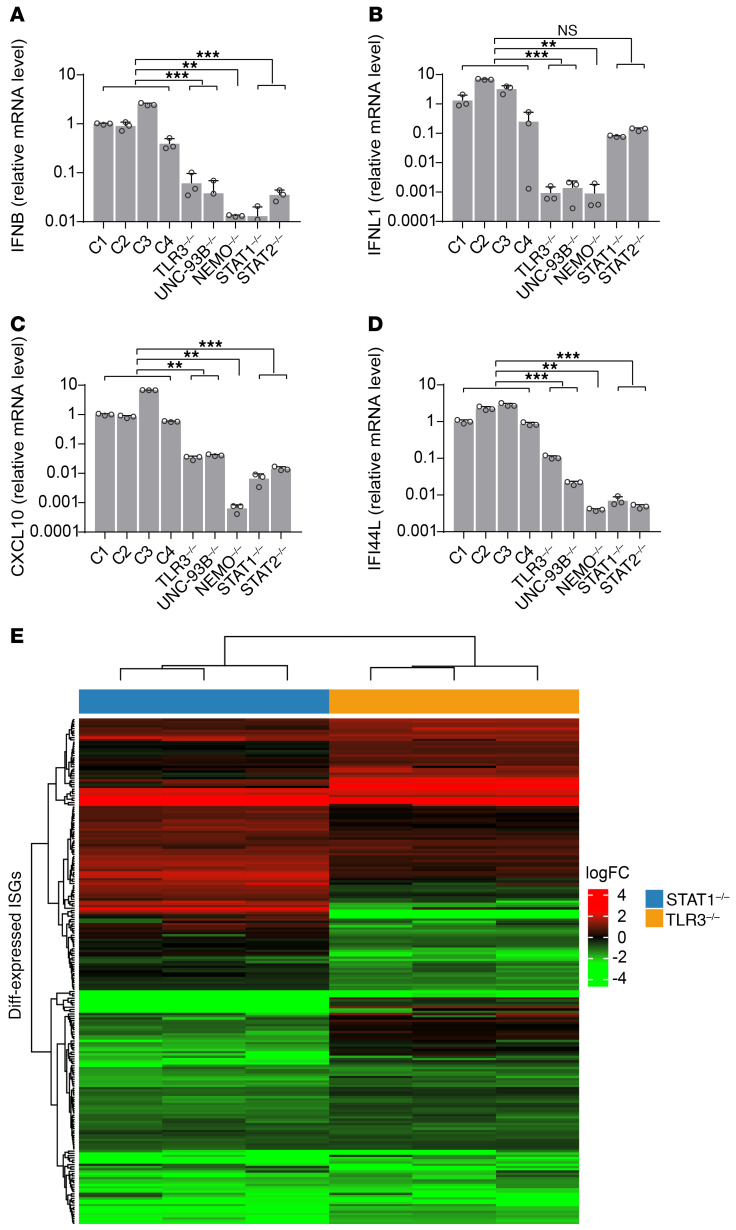
Our previous and current data show that the prior treatment (but not treatment at the time of infection) of TLR3 signaling–deficient fibroblasts with recombinant IFN-α2b or -β, but not IFN-λ, protects them against VSV-WT and -M51R, or HSV-1 replication and virus-induced cell death (Supplemental Figure 5A) (10). In this study, we also found that TLR3 signaling–deficient cells sustained very high levels of virus replication before the induction of IFN production in response to viral infection (Figure 3E; and Supplemental Figure 4, C–E). We therefore hypothesized that viral replication might be limited in control cells by basal levels of IFNs constitutively expressed in a TLR3-dependent manner. We tested this hypothesis, first by assessing basal levels of IFN production in unstimulated SV-40 fibroblasts. Control fibroblasts secreted higher basal levels of IFN-β than TLR3–/– or UNC-93B–/– fibroblasts (Supplemental Figure 5B). We further determined basal mRNA levels for IFNB, IFNL1 (IL29), and downstream ISGs, including CXCL10 and IFI44L, in unstimulated fibroblasts. Control cells contained larger amounts of mRNA for these genes than SV-40 fibroblasts from patients with TLR3, UNC93B, or NEMO deficiency (Figure 4, A–D) whose TLR3 response signaling was impaired (9, 11, 13). Fibroblasts from previously reported patients with IRAK4 or MYD88 deficiencies, in which responses to all TLRs except TLR3 were impaired, displayed normal levels of IFNs and ISGs mRNA (Supplemental Figure 5C). Basal levels of mRNA for IFNB, IFNL1, CXCL10, and IFI44L were also lower in SV-40 fibroblasts from patients with STAT1 and STAT2 deficiency whose IFN response signaling is impaired than in control cells (Figure 4, A–D) (40, 41). The immortalization of fibroblasts with SV-40 T antigen has been reported to affect IFN immunity (43). We then analyzed the transcriptomes of control, TLR3–/–, and STAT1–/– primary fibroblasts by RNA sequencing (RNA-Seq) to rule out the possibility that the apparently TLR3-IFN signaling–related basal levels of IFN and ISG were an SV-40 fibroblast–specific phenomenon. We found that mRNA for 716 fibroblastic ISGs, but not IFNs, was detectable in control primary fibroblasts in basal conditions. mRNA levels were significantly lower in both TLR3–/– and STAT1–/– primary fibroblasts than in control cells for 43 of the 225 differentially expressed ISGs, whereas 13 ISGs were upregulated in these cells relative to control cells (Figure 4E, Supplemental Figure 5D, Supplemental Table 1). An analysis, with DAVID software (44), of the 43 ISGs with low basal expression levels further confirmed the significant downregulation of type I IFN signaling pathway genes and antiviral immune genes in TLR3–/– and STAT1–/– primary fibroblasts (Supplemental Table 2). Thus, TLR3 and IFN signaling deficiencies have a profound impact on basal IFN-β production and ISG expression in SV-40–transformed and primary fibroblasts in the absence of exogenous stimulation, demonstrating the role of TLR3 in maintaining constitutive antiviral gene expression.
Figure 4. Impact of TLR3 signaling deficiencies on basal IFN-related gene expression.
mRNA levels of IFNB (A), IFNL1 (B), CXCL10 (C), and IFI44L (D) (relative to GAPDH) in unstimulated fibroblasts from healthy controls (C1–C4) and individuals deficient for TLR3-IFN signaling, as quantified by RT-qPCR with normalization against C1. Representative data from 3 independent experiments are shown. The error bars indicate SDs of triplicate measurements. (E) Gene expression profile of the ISGs differentially expressed in patients with STAT1 (blue bar) and TLR3 (orange bar) deficiencies, relative to mean expression levels in controls, as assessed by RNA-Seq. The heatmap shows the log fold-change in ISG expression, with red indicating upregulation and green downregulation. P values were obtained for likelihood ratio tests by comparing each patient’s fibroblasts with control fibroblasts (A–D), and the respective P values are indicated. **P < 0.01, ***P < 0.001.
Low basal levels of IFN production underlie the enhanced viral growth in TLR3-deficient fibroblasts.
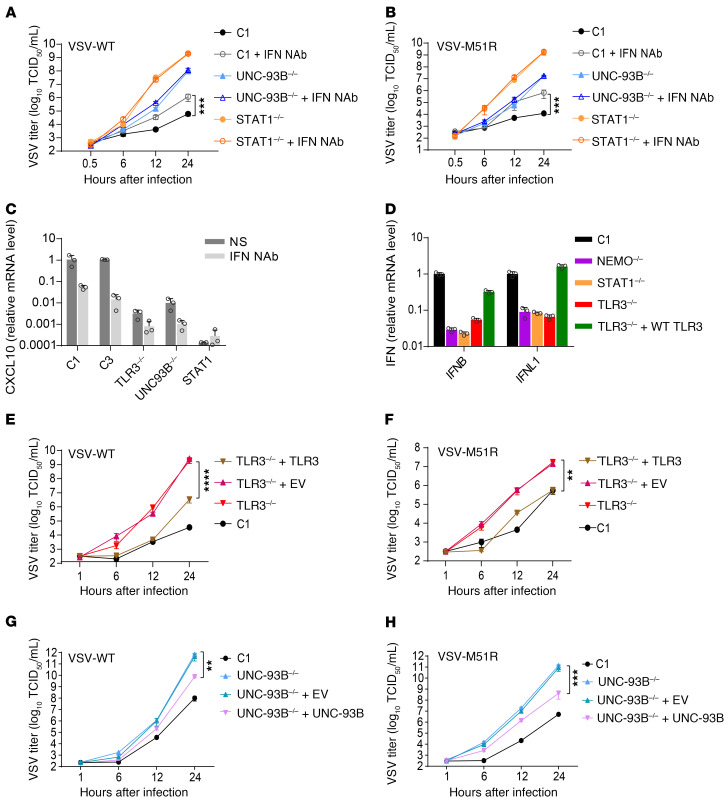
We assessed the importance of basal levels of IFN production in control cells, by neutralizing the IFNs and assessing the impact of this intervention on VSV replication. We cultured UNC-93B–/–, STAT1–/–, and control SV-40 fibroblasts in the presence or absence of neutralizing polyclonal antibodies (NAbs) raised against IFN-α, -β and -λ, for 72 hours. We measured VSV-WT and VSV-M51R growth kinetics over 24 hours. Viral titers were similar in UNC-93B–/– and STAT1–/– fibroblasts with and without NAbs. By contrast, the growth of VSV in control fibroblasts was greater at 8 and 24 hours after infection in the presence of anti-IFN NAbs than in their absence (Figure 5, A and B), suggesting a substantial contribution of basal IFN production, which was below the limit of detection for ELISA, in limiting viral growth. Consistent with this interpretation, basal ISG CXCL10 mRNA levels were strongly decreased by NAb treatment (Figure 5C). We then rescued TLR3 signaling in TLR3–/– SV-40 fibroblasts by ectopically expressing WT TLR3 to demonstrate the requirement of TLR3 signaling for this constitutive antiviral response limiting viral replication. Basal levels of IFNB and IL29 mRNA were restored to levels similar to those in a healthy control following the expression of WT TLR3 (Figure 5D). In TLR3–/– cells complemented with WT TLR3, replication rates for VSV-WT and VSV-M51R were lower than those in nontransfected TLR3–/– cells or in TLR3–/– cells transfected with an empty vector (Figure 5, E and F). A similar rescue was observed in the growth curve of VSV-WT and VSV-M51R in WT UNC-93B–expressing UNC-93B–/– fibroblasts (Figure 5, G and H). Collectively, these experiments demonstrate that a lack of TLR3 signaling results in a deficiency of cell-intrinsic, constitutive antiviral IFN responses in fibroblasts, leading to early viral replication that may overwhelm the activity of RIG-I–dependent IFN production, which is induced later in response to viral replication.
Figure 5. Constitutive IFN-β production in fibroblasts is TLR3 dependent.
Replication of VSV-WT (A) and VSV-M51R (B) in fibroblasts from a healthy control (C1) or patients with UNC-93B or STAT1 deficiency, cultured in the presence or absence of neutralizing antibodies against IFN-α, -β, and -λ (IFN Nab). (C) CXCL10 mRNA levels in unstimulated fibroblasts after treatment with IFN NAb for 24 hours. (D) IFN-β and IFN-λ1 (IL-29) mRNA levels in TLR3–/– fibroblasts transfected with WT TLR3, measured by RT-qPCR and normalized against GUS expression. Replication of VSV-WT (E) and VSV-M51R (F) in TLR3–/– fibroblasts stably transfected with empty vector (+EV) or WT TLR3 (+TLR3). Replication of VSV-WT (G) and VSV-M51R (H) in UNC-93B–/– fibroblasts stably transfected with empty vector or WT UNC-93B (+UNC-93B). Representative results are shown for 3 (A, B, E–H) or 2 (C–D) independent experiments. The error bars indicate SDs of biological triplicates (C, D) or the SEM of biological triplicates (A, B, E–H). P values were obtained through log transformation followed by 1-way ANOVA and subsequent Tukey’s multiple comparison tests, and the respective P values are indicated. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Basal IFN-β production by control cells restricts viral growth.
In human fibroblasts, IFNB was the only interferon gene strongly expressed in basal conditions, and only IFNB, IFNL1, IFNL2, and IFNL2 displayed a strong induction of expression upon activation of the TLR3 or MAVS pathway (Supplemental Figure 1). However, these cells did not express the receptor for type III IFNs (Supplemental Figure 6A). We hypothesized that the basal production of small amounts of IFN-β by healthy control cells can rescue the TLR3-deficient cell phenotype. We evaluated the effects of basal IFN, by stimulating patients’ SV-40 fibroblasts with conditioned medium from healthy control unstimulated cell cultures. This medium effectively decreased the growth of VSV-M51R (Figure 6A) and HSV-1 (Figure 6B, Supplemental Figure 6B) at 24 hours in TLR3–/– and UNC-93B–/– cells from patients, to levels below those in cells treated with medium from TLR3–/– and UNC-93B–/– fibroblasts, with virus titers close to those measured in control cells. Moreover, mRNA levels for ISGs, including CXCL10, MxA, and RIG-I, were upregulated in the patients’ fibroblasts by stimulation with conditioned medium from healthy controls, but not with conditioned medium from TLR3–/– and UNC-93B–/– fibroblasts (Figure 6C, Supplemental Figure 6C). Fibroblasts are unable to respond to IFN-λ, probably because they lack IFNLR expression (Supplemental Figure 6A) (45). However, the pretreatment of TLR3 signaling–deficient fibroblasts with either IFN-β or IFN-α2b can rescue resistance to viral infection, consistent with the relatively high levels of IFNAR expression (Supplemental Figure 6A). We found that IFN-β NAbs abolished both the protection against VSV-M51R growth and ISG induction conferred by the conditioned culture medium from control fibroblasts (Supplemental Figure 6, D and E), demonstrating that IFN-β is the protective cytokine in this context. STAT1–/– fibroblasts failed to upregulate ISGs when treated with conditioned medium from healthy controls (Supplemental Figure 6E), suggesting that the IFN signaling pathway is essential for the maintenance of basal ISG expression by basal IFN levels. Thus, TLR3-dependent, constitutive IFN-β production governs intrinsic antiviral immunity in human fibroblasts.
Figure 6. TLR3-dependent constitutive IFN-β production restricts VSV growth.
(A) TLR3–/– (left panel) and UNC93B–/– (right panel) fibroblasts were subjected to pretreatment with conditioned medium from unstimulated cell cultures (as indicated in parentheses) for 18 hours and infected with VSV-M51R for 24 hours. VSV-G mRNA levels were then assessed by RT-qPCR, with normalization against GAPDH. (B) Similar to A, except that HSV-1 GFP was used to infect cells and ICP27 mRNA levels were measured by RT-qPCR. (C) TLR3–/– (left panel) and UNC93B–/– (right panel) fibroblasts were treated with conditioned medium, as indicated in parentheses, for 24 hours and CXCL10 expression was then quantified by RT-qPCR. The error bars indicate SDs of biological triplicates from 3 independent experiments. P values were obtained for likelihood ratio tests, by comparing each patient’s fibroblasts treated with conditioned medium from control or patient fibroblasts, and the respective P values are indicated. **P < 0.01, ***P < 0.001, ****P < 0.0001.
TLR3 also controls constitutive antiviral IFN immunity in mouse fibroblasts.
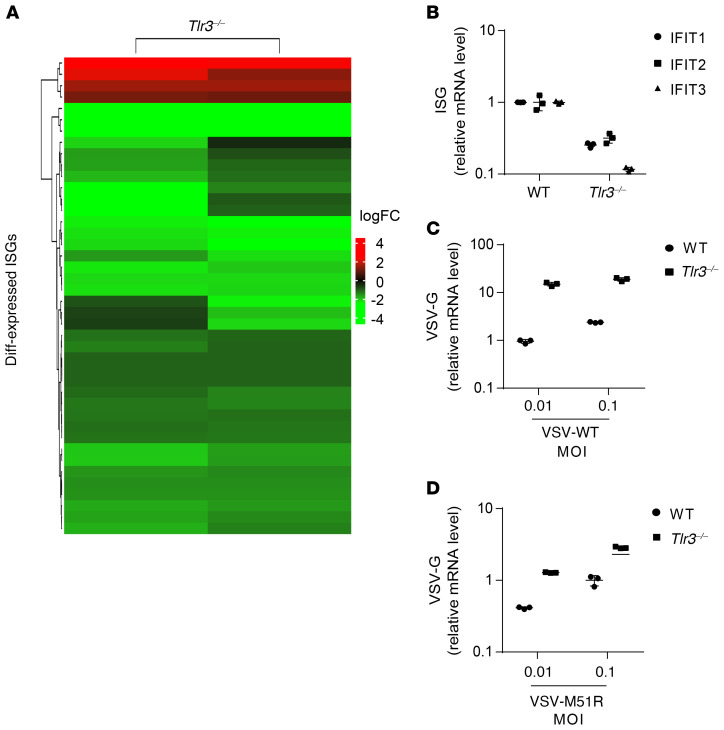
We investigated whether TLR3-dependent constitutive IFN responses also governed cell-intrinsic constitutive antiviral immunity in MEFs. In mice, spontaneous IFN-α/β production in vivo in the absence of viral infection primes and enhances the immune response (46–48). We performed RNA-Seq on WT primary MEFs and Tlr3–/– MEFs to determine whether TLR3 plays a critical role in controlling cell-intrinsic basal IFN and antiviral immunity in mouse cells. Basal mRNA levels were significantly lower in Tlr3–/– MEFs than in Tlr3 WT MEFs for 38 of the 42 ISGs differentially expressed in WT and Tlr3–/– MEFs, whereas 4 ISGs were more strongly expressed in these cells than in Tlr3 WT MEFs (Figure 7A, Supplemental Figure 7A, Supplemental Table 3). Seven of the 38 ISGs downregulated in Tlr3–/– MEFs were also downregulated in TLR3-deficient human fibroblasts, but different ISGs were upregulated in mouse and human TLR3-deficient fibroblasts (Supplemental Figure 7B, Supplemental Table 4). As in human fibroblasts, an analysis of the 38 ISGs downregulated in Tlr3–/– MEFs with DAVID software (44) confirmed a significant downregulation of type I IFN signaling pathway genes and antiviral immune genes (Supplemental Table 5). We confirmed these findings by measuring basal levels of expression for several ISGs by RT-qPCR. Consistent with the human data, Tlr3–/– MEFs had lower levels of mRNA for various ISGs, including Ifit1, Ifit2, and Ifit3 (Figure 7B). Finally, we assessed the viral susceptibility of MEFs by measuring viral RNA levels after infection with VSV-WT and -M51R. Tlr3–/– MEFs were more susceptible to both VSV-WT and -M51R than Tlr3 WT MEFs, as high levels of VSV RNA were detected in Tlr3–/– MEFs as early as 5 hours after infection (Figure 7, C and D; Supplemental Figure 7, C and D). These data suggest that TLR3 signaling controls constitutive antiviral IFN immunity in both mouse and human fibroblasts, which use this mechanism to restrict infection with various viruses.
Figure 7. TLR3 ablation decreases ISG expression and increases vulnerability to viruses in MEFs.
(A) Gene expression profile of all differentially expressed ISGs in Tlr3–/– MEFs relative to mean levels in WT mice, as assessed by RNA-Seq. The heatmap shows the log fold-changes of ISG gene expression, with red indicating upregulation and green downregulation. (B) ISG expression was assessed in unstimulated WT and Tlr3–/– MEFs, by RT-qPCR with normalization against RPL19. WT and Tlr3–/– MEFs were infected with VSV-WT (C) and VSV-M51R (D) for 24 hours. Viral RNA levels were then quantified by RT-qPCR, with normalization against RPL19. The error bars indicate SDs of technical triplicates.
Impaired basal IFN-β and intrinsic antiviral immunity in TLR3-deficient iPSC-derived cortical but not TG neurons.
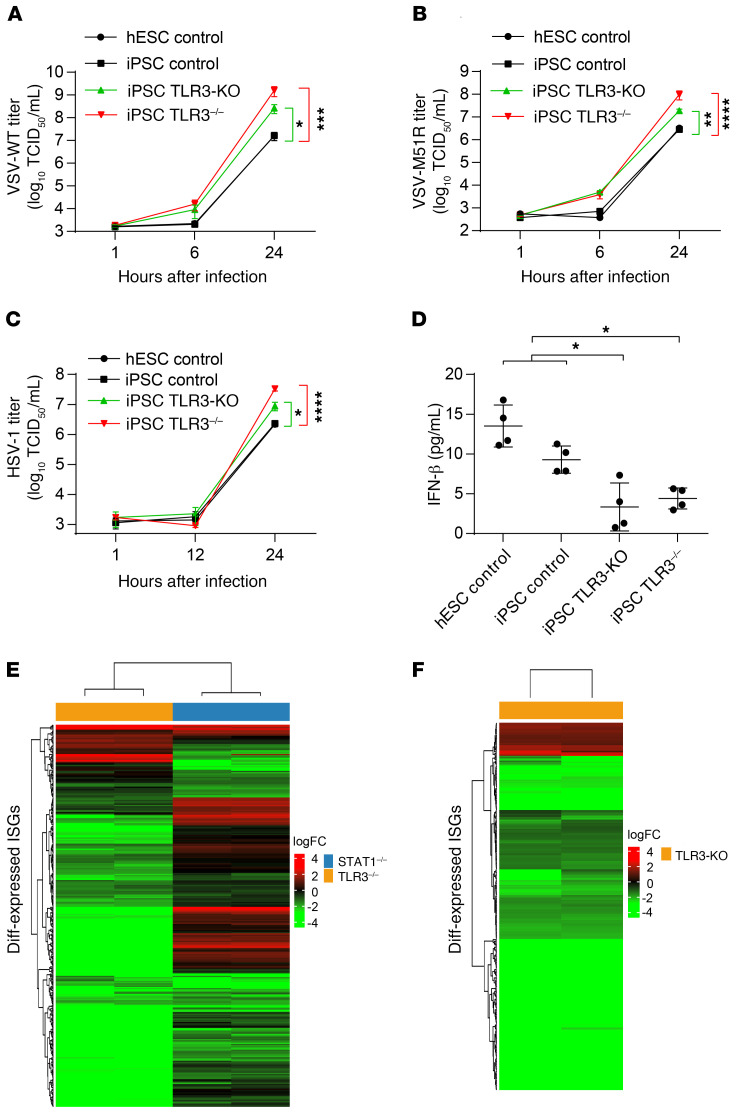
We previously showed that TLR3 deficiency impairs cell-autonomous defense against HSV-1 infection in iPSC-derived cortical neurons and oligodendrocytes, but not astrocytes, neural stem cells, and TG neurons (25, 26). As with fibroblasts, prior treatment (but not treatment at the time of infection) of TLR3 pathway–deficient iPSC-derived cortical neurons and oligodendrocytes with recombinant IFN-α2b or -β, but not IFN-λ, protected them against HSV-1 replication (25, 26). We investigated whether our findings for fibroblasts also applied to cortical and TG neurons. We derived cortical and TG neurons from one healthy control iPSC line, one healthy hESC line, an iPSC line from a patient with AR complete TLR3 deficiency (26), and an isogenic TLR3-KO iPSC line that we generated by CRISPR/Cas9-mediated gene editing (Supplemental Figure 8, A and B) (49). As in SV-40 fibroblasts, virus-induced IFNB and IFNL1 induction occurred late, 16 and 24 hours after HSV-1 infection (Supplemental Figure 8, C and D). When cortical neurons were infected with VSV-WT, VSV-M51R, and HSV-1, higher levels of viral replication were observed as early as 6 hours after infection in TLR3-deficient cells, confirming that TLR3 is essential for the control of HSV-1 in these cells, and showing that this receptor is also essential for control of the 2 types of VSV tested (Figure 8, A–C). By contrast, the replication of the 3 viruses was not higher in TG neurons lacking TLR3 than in the other cells (Supplemental Figure 8, E–G). TLR3 protein levels were undetectable in both patient-specific and isogenic TLR3-deficient iPSC-derived cortical neurons, as shown by Western blotting (Supplemental Figure 8A). We also assessed basal IFN-β protein secretion by cortical neurons in the Simoa assay (50). We showed that basal levels of IFN-β production were low in TLR3-deficient cells (Figure 8D). We further analyzed the transcriptomes of control iPSC-derived cortical neurons, comparing them with those of TLR3-deficient patient and isogenic TLR3-KO iPSC-derived cortical neurons. Consistent with our results for fibroblasts, TLR3-deficient cortical neurons had low levels of ISG mRNAs (Figure 8, E and F; and Supplemental Figure 9, A and B). The IFN mRNAs themselves were not detected by RNA-Seq. In total, we detected the expression of 734 ISGs in control cortical neurons in basal conditions. The mRNA levels for these genes were significantly lower in both TLR3-deficient patient and TLR3-KO cortical neurons than in control cells, for 243 of 265 and 283 of 311 differentially expressed ISGs, respectively, whereas 22 and 28 ISGs were upregulated in these cells relative to control cells (Supplemental Figure 9, C and D; and Supplemental Table 6). As many as 196 of the downregulated ISGs and 9 of the upregulated ISGs were common to TLR3-KO and TLR3-deficient patient neurons. An analysis with DAVID software (44) of the 196 ISGs with low basal levels of expression confirmed the significant downregulation of the genes controlled by the IFN-α/β–responsive pathway, and of antiviral genes in both TLR3-KO and TLR3-deficient cortical neurons (Supplemental Table 7). Collectively, these findings strongly suggest that TLR3 also controls constitutive levels of IFN-β, and thus, cell-intrinsic antiviral immunity, in human hPSC-derived cortical neurons. This process is crucial for infection control, at least for VSV and HSV-1 in vitro and, by inference, probably HSV-1 in vivo.
Figure 8. TLR3 controls basal IFN-β and antiviral immunity in cortical neurons.
HPSC-derived cortical neurons from 2 control lines (hESC control, iPSC control), a CRISPR/Cas9-mediated TLR3 knockout (TLR3-KO) line, and a TLR3–/– patient line were infected with VSV-WT (A), VSV-M51R (B), or HSV-1 (C) for the indicated times. Viral replication was assessed in 50% TCID50 assays. (D) IFN-β levels were measured in the culture supernatants of hPSC-derived cortical neurons, by Simoa digital ELISA. (E) Gene expression profile of the ISGs differentially expressed in patients with TLR3 (orange bar) and STAT1 (blue bar) deficiencies, relative to controls, as assessed by RNA-Seq. (F) Gene expression profile of the ISGs differentially expressed in TLR3-KO iPSC-derived cortical neurons, relative to parental control iPSC-derived cortical neurons. The heatmap shows the log fold-change in ISG expression, as assessed from 2 technical duplicates of the RNA-Seq, with red indicating upregulation and green downregulation. The error bars indicate the SEM (A–C) or SDs (D) of biological triplicates from 3 independent experiments. P values were obtained with (A–C) or without (D) log transformation followed by 1-way ANOVA and subsequent Tukey’s multiple comparison tests, by comparing TLR3–/– or TLR3-KO cortical neurons with controls, and the respective P values are indicated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
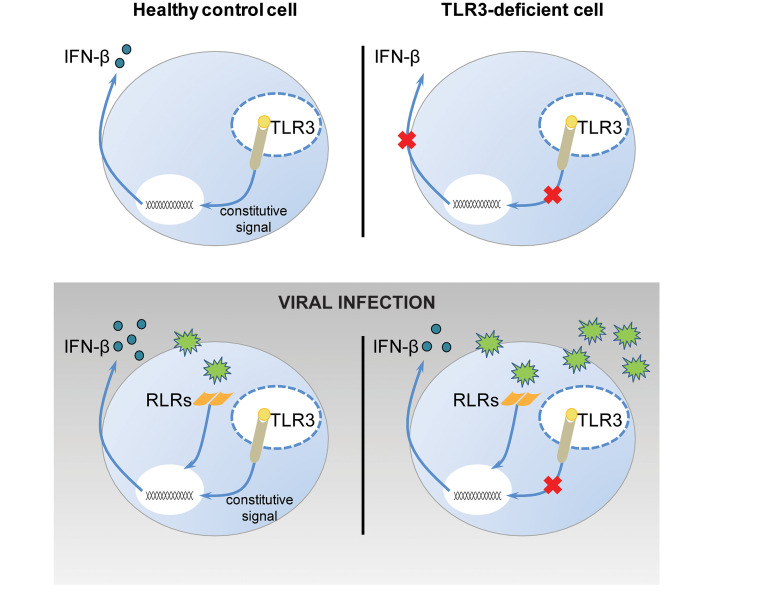
We describe an unexpected role of TLR3 signaling in antiviral immunity: as a rheostat controlling constitutive low-level production of IFN-β, which is crucial for limiting viral replication at early stages in the viral infection of dermal fibroblasts and cortical neurons. IFN-β was previously thought to be the fibroblastic IFN because it was discovered in these cells, in which no other type I IFNs were detectable (51). In most other cells, it is the first IFN to be induced after viral infection, triggering the amplification of other IFNs via IRF7 (52). We discovered that the restriction of VSV growth in human fibroblasts was heavily dependent on the TLR3-dependent basal IFN-β production of these cells, rather than their recognition of VSV and the subsequent RIG-I–dependent induction of IFN-β. Basal IFN-β production is impaired in TLR3–/– and UNC-93B–/– fibroblasts, and these cells are therefore overwhelmed by VSV-WT and even M51R replication within as little as 6 hours after infection, before the RIG-I–dependent induction of IFN-β production in response to viral infection. This situation contrasts with that in healthy control fibroblasts, in which VSV replication and cell death are limited by basal IFN-β levels, which are higher not only than those in UNC-93B–/– and TLR3–/– cells, but also than those in NEMO–/–, STAT1–/–, and STAT2–/– cells. In human fibroblasts lacking TLR3 signaling (e.g., UNC-93B–/–, TLR3–/–, NEMO–/–), the impairment of constitutive low-level TLR3-dependent IFN-β production may reduce the responsiveness and intrinsic defenses of uninfected cells against viral infection, by reducing the basal level of expression of specific ISGs. In cells lacking IFN signaling responses (e.g., STAT1–/–, STAT2–/–), impaired responses to IFN-β result in lower levels of basal expression for ISGs, including TLR3 and STAT1. As a more general consequence, fibroblasts lacking TLR3 and IFN signaling are highly susceptible to infection with the 3 RNA viruses (VSV, hPIV3, EMCV) and the DNA virus (HSV-1) tested, regardless of the levels of IFN-β induction by these viruses. Similarly, TLR3-deficient iPSC-derived cortical neurons are highly susceptible to infection with VSV and HSV-1. Collectively, our data indicate that inborn errors of the TLR3 pathway impair basal and protective IFN-β antiviral immunity in human fibroblasts and cortical neurons.
We have shown that this TLR3 rheostat also operates in mouse fibroblasts. Previous studies have shown that Tlr3–/– mice are no more susceptible than their WT littermates to VSV, lymphocytic choriomeningitis virus, or reovirus (53), and that they are even more resistant to Punta Toro virus (54), influenza virus (55), and vaccinia virus (56). Mouse fibroblasts may not have been infected in the course of such experiments, or their contribution to host defense may be compensated by other cells. However, mouse TLR3 is required for antiviral immunity to mouse cytomegalovirus (57), respiratory syncytial virus (58), coxsackievirus group B serotype 3 (CVB3) (59), and coxsackievirus B4 (CVB4) viruses (60). The contribution of fibroblasts in these models is unknown. Other cell types in mice may also control basal IFN-β immunity through TLR3. Interestingly, the DNA sensor cGAS controls basal levels of IFNs and ISGs in mouse bone marrow–derived macrophages (61). It is therefore possible that TLR3 and cGAS control basal ISG levels in different cell types. In certain cell types, both may be required for basal antiviral immunity. Mouse cGAS is essential for defense against various infections, including HSV-1 infection (30). The genetic ablation of cGAS revealed that this protein was required for antiviral responses to both DNA viruses and RNA viruses in vivo (61). Future studies with single and double knockouts of TLR3 and cGAS in mice and humans may delineate the respective contributions of TLR3 and cGAS, as viral sensors versus rheostats of IFN immunity in host defense. Virus-induced IFN-α/β and IFN-λ production could be protective against viral infection in vivo, probably following the earliest stage of viral infection. The study of MDA5 and RIG-I in this context is also warranted by the identification of patients with MDA5 deficiency displaying selective vulnerable to respiratory viruses (62, 63). The identification of patients with cGAS, STING, or RIG-I deficiency could provide new insight into this aspect.
The natural TLR3 stimuli controlling basal IFN-β production in fibroblasts and cortical neurons remain unknown. In the absence of exogenous stimulation, endogenous ligands such as mRNAs or debris from necrotic cells can trigger TLR3 signaling in human dendritic cells (64) and murine macrophages (65), respectively. Self-noncoding RNAs generated during cell damage and the microtubule regulator stathmin have been identified as endogenous agonists of TLR3 in human keratinocytes and in human astrocytes and microglia cells, respectively (66, 67). One or more of these known cell-endogenous agonists may activate TLR3 in fibroblasts, cortical neurons, and other cells, thereby contributing to basal IFN and ISG production. However, it remains possible that TLR3 can sustain basal activation in the absence of stimulation with any ligand. In any event, our study clearly demonstrates that TLR3 is essential for the maintenance of constitutive IFN-β immunity in human fibroblasts and cortical neurons, and this observation also applies to mouse fibroblasts. It may also operate in other CNS-resident cells, such as oligodendrocytes, in which the control of HSV-1 is also dependent on TLR3, contrasting with the situation in astrocytes, neural stem cells, and TG neurons (25, 26). Constitutive TLR3 activity in the brain may also be important in the prevention of other neurological conditions via mechanisms that may or may not be related to IFN-mediated immunity (68, 69). While we cannot yet discern the respective roles of constitutive and virus-induced cell-intrinsic IFN immunity in the course of natural infection with HSV-1 in TLR3-deficient individuals, our findings provide a plausible molecular and cellular basis for HSE in patients with inborn errors of TLR3 immunity. They also suggest that IFN-α/β, rather than IFN-λ, are critical for protective immunity to HSV-1 in the CNS.
Our findings also suggest that TLR3-dependent cell-intrinsic constitutive IFN immunity may be a critical first line of defense against HSV-1 and perhaps other viruses in certain tissues, and its disruption may result in tissue-specific severe viral infections in various organs. Patients with TLR3 deficiency may also be more susceptible to diseases other than HSE. This was confirmed by our recent identification of 3 unrelated patients with TLR3 mutations and severe influenza pneumonia (70), a finding that is consistent with TLR3 being a major IFN-β– and IFN-λ–inducing receptor in human and mouse pulmonary epithelium cells (PECs), in which large amounts of IFN-λ may be produced with potent antiviral activity (70, 71). Furthermore, a deleterious TLR3 mutation has also been found in a patient with varicella zoster virus ophthalmic neuritis (72). Overall, our findings indicate that TLR3 controls baseline tone of IFN-β and IFN-λ in dermal fibroblasts and of IFN-β in cortical neurons (both cell types respond to IFN-β but not IFN-λ). They also suggest that TLR3 may control baseline levels of IFN-β and/or IFN-λ in PECs (which respond to both IFN-β and IFN-λ). The genetic dissection of various viral diseases, including coronavirus disease 2019 (COVID-19) (73–75), may help to delineate the roles of human TLR3 and other IFN-inducing sensors in host defense. It also seems likely that deficiencies of other type I IFN responsiveness circuit genes (e.g., IRF7, IRF9, STAT1, STAT2, IFNAR1, IFNAR2) render patients prone to a broad spectrum of viral diseases, other than the previously reported phenotypes including severe influenza pneumonia (IRF7, IRF9), HSE (IFNAR1, STAT1), and severe adverse reactions to live attenuated viral vaccines (IRF9, STAT1, STAT2, IFNAR1, IFNAR2) (12, 17, 40, 76–79), partly due to the low basal levels of IFN-β, IFN-λ, and ISG in tissue-specific cells. Our findings highlight the importance of host cell–intrinsic and constitutive, as opposed to pathogen-induced, IFN and ISG immunity in antiviral defenses, particularly during early stages of viral infection (80–82). Our findings also suggest that TLR3 may govern the first line of broad antiviral responses in some cell types, such as fibroblasts, cortical neurons, and perhaps PECs, or even more broadly in certain tissues or organs, such as the CNS and lungs in particular, as opposed to governing immunity to specific viruses, such as HSV-1, in various cells, tissues, and organs.
Methods
Cells and viruses.
Primary fibroblasts were isolated from skin biopsy specimens from patients with AR UNC-93B, AR STAT1, XR NEMO, and AR TLR3 deficiencies and healthy controls, as previously described (9, 11, 13, 41). The fibroblasts were immortalized by transfection with the SV-40 large T antigen and maintained in DMEM supplemented with 10% FBS.
Cells stably expressing shRNAs against RIG-I and MAVS were generated by transduction with shRNA-expressing retroviral particles (Santa Cruz Biotechnology). Stably transfected cell lines were generated by transfecting TLR3–/– or UNC-93B–/– fibroblasts with TLR3 or UNC-93B WT or empty vector, with Lipofectamine 2000 (Invitrogen). Transfectants were selected in DMEM containing 5 μg/mL blasticidin (Invitrogen).
VSV, Indiana strain, WT, and M51R mutant (harboring a substitution of an arginine residue in place of the methionine in position 51 of the matrix (M) protein) were provided by Pierre Lebon and have been described elsewhere (10, 39, 83–85). MEFs were generated from 14-day-old WT and Tlr3−/− embryos (86), and maintained in DMEM supplemented with 10% FBS.
Cell culture of iPSCs and hESCs (together referred to as hPSCs), and neuronal induction and differentiation.
Control MRC5 and TLR3–/– patient-specific iPSC lines were reprogrammed with a nonintegrating Sendai viral vector, as previously described (26). All hPSC lines were used at passage 20–50, and were maintained on vitronectin with Essential 8 medium (Fisher Scientific), with twice-weekly passaging with EDTA (87). The HPSC lines used here were as follows: TLR3–/– patient-specific iPSCs, control iPSCs MRC5, isogenic MRC5 TLR3 KO iPSCs, and a control hESC line H9 (WA-09) (26). All cell lines were karyotyped to ensure genomic integrity and weekly tests for mycoplasma were performed. TG neuron and cortical neuron differentiation was performed as previously described (26).
Cell stimulation.
SV-40 fibroblasts were plated at a density of 105 cells per well in a 24-well plate and were incubated overnight. Polyinosinic-polycytidylic acid (poly(I:C); Amersham) was added to the culture medium at a concentration of 25 μg/mL, transfected with the RIG-I agonist, 7sk-as at a concentration of 0.45 ng, or with 25 μg of poly(I:C), with Lipofectamine 2000 (Invitrogen), or infection with VSV at the indicated MOIs, and the culture medium was harvested after 24 hours of stimulation. Cytokine concentrations in culture medium were determined by ELISA.
Measurement of cytokine production.
IFN-β production was assessed in an enzyme-linked immunosorbent assay (ELISA; TFB Fujirebio). For IFN-λ determinations, culture supernatants were incubated for 2 hours in plates coated with 1 μg/mL monoclonal anti-human IL29 antibody (R&D Systems). A biotinylated monoclonal secondary antibody directed against human IL-29 (R&D Systems) and streptavidin peroxidase were added, together with TMB (3,3′,5,5′-tetramethylbenzidine). The signal at 450 nm was then read with a plate fluorescence reader.
Immunoblotting.
Cells were lysed in NP40 lysis buffer (280 mM NaCl, 50 mM Tris pH 8, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 0.5% NP40) supplemented with 1 mM DTT, 5 mM Na3VO4, and Complete protease inhibitor cocktail (Roche). The protein lysate was subjected to SDS-PAGE and the bands obtained were transferred to a PVDF membrane, which was probed with unconjugated rabbit anti–RIG-I (Cell Signaling, catalog 3743), goat anti-MAVS (Santa Cruz Biotechnology, catalog sc-365333), TLR3 (R&D Systems, catalog 1487) and HRP-conjugated secondary antibodies. A β-tubulin antibody (MilliporeSigma, catalog T4026) and a GAPDH (Santa Cruz Biotechnology, catalog sc-365062) antibody were used as loading controls.
For the detection of dsRNA in VSV-infected cells, SV-40 fibroblasts were infected as described above, with VSV at a MOI of 10 for 8 hours. RNA was isolated with TRIzol reagent (Invitrogen), according to the manufacturer’s protocol. We subjected 30 μg total RNA to electrophoresis in a 1.5% agarose/TBE gel, and the bands were then transferred to a nylon membrane (Hybond N+, GE Healthcare). The RNA was fixed by ultraviolet irradiation, probed with the anti–dsRNA antibody K1, and the blot was developed for enhanced chemiluminescence (Pico ECL, Pierce).
Viral assays.
SV-40–transformed fibroblasts were plated at a density of 105 cells per well in 24-well plates. For one-step growth curves, the cells were incubated with VSV-WT and -M51R, hPIV3, EMCV, or HSV-1 at the indicated MOI for 30 minutes or 1 hour, washed twice in PBS, and then transferred to fresh DMEM. Virus samples were collected at the indicated times and the viral titer was determined by 50% tissue culture infective dose (TCID50) assays on Vero E6 cells (for VSV and hPIV3), according to the Reed and Muench calculation (13), or by qPCR methods for EMCV (88), determinations of ICP27 mRNA levels for HSV-1, and determinations of VSV-G mRNA levels for VSV.
For coinfection experiments, cells were infected with VSV-WT at a MOI of 3 and with VSV-M51R at a MOI of 1 in DMEM supplemented with 10% FBS for 24 hours. VSV-WT and -M51R were inactivated by exposure to ultraviolet light for 10 minutes at a distance of 15 cm. Cytokine production was measured in the culture medium by ELISA.
For IFN neutralization experiments, cells were cultured in the presence of neutralizing polyclonal antibodies against IFN-α and -β (PBL), replaced daily for 3 days. They were then infected with VSV-WT or -M51R at a MOI of 3, as described above. Samples were harvested at the times indicated and titered on Vero cells.
Cytotoxicity assays.
Cells were plated at a density of 2 × 104 cells per well in 96-well plates, in DMEM with or without IFNα2b (105 IU/mL, Schering-Plough) treatment for 18 hours. The medium was replaced with medium containing VSV at the indicated MOIs. Mortality was assessed 24 hours after infection, with the LDH Cytoxicity Detection KitPLUS (Roche). The death of cells is expressed relative to that of uninfected cells. For hPIV3 infection, fibroblasts were plated as above and infected at the indicated MOIs. Viability was assessed in a resazurin-based viability assay (MilliporeSigma). The viability of infected cells is expressed relative to that of uninfected cells.
Determination of mRNA levels by RT-qPCR.
Total RNA was extracted from cells with TRIzol (Invitrogen) or the RNeasy kit (QIAGEN). Samples were treated with DNase at 37°C for 1 hour and cleaned by passage through an RNeasy column (QIAGEN). Reverse transcriptase-PCR was performed with random hexamers (Applied Biosystems). RT-qPCR was performed with Applied Biosystems TaqMan assays, using the β-glucuronidase (GUS) housekeeping gene for normalization, or in SYBR Green assays with the GAPDH (human) or RPL19 (mouse) housekeeping gene for normalization. Results are expressed according to the ΔΔCt method, as described by the manufacturer.
RNA sequencing and analysis.
Total RNA was extracted from human primary fibroblasts, C57BL/6 mouse primary fibroblasts, or hPSC-derived cortical neurons. RNA samples were treated with DNase (Ambion) to remove residual genomic DNA. RNA-Seq libraries were prepared with the Illumina RiboZero TruSeq Stranded Total RNA Library Prep Kit (Illumina) and sequenced on the Illumina NextSeq platform in the 150 nt with paired-end configuration. We sequenced 3 (human samples) or 2 (mouse samples) technical replicates for each sample. The raw sequencing data is available under National Center for Biotechnology Information Sequence Read Archive (NCBI-SRA) accession number SRP288648.
The RNA-Seq FASTQ raw data were inspected with multiQC v1.6 to ensure that they were of high quality (89). The sequencing reads of human and mouse samples were mapped onto the UCSC human reference genome GRCh37/hg19 and the UCSC mouse reference genome GRCm38/mm10, respectively (90), with STAR aligner v2.6 (91). The quality of each mapped alignment in BAM files was evaluated with RSeQC (92). Reads were quantified to determine the number of gene-level read counts forming the read alignment, with featureCounts v1.6.0 and GENCODE GRCh37.p13 human gene annotation V19 and GENCODE GRCm38.p6 mouse gene annotation M19, respectively (93, 94). The gene-level read counts were normalized and log2-transformed by DESeq2 to obtain the gene expression value for all genes and all samples (95).
We extracted expression data for 905 ISGs based on our previous IFN-stimulated microarray data analysis (13). Differential ISG expression was analyzed by applying TMM normalization and gene-wise generalized linear model regression in edgeR (96). The ISGs displaying significant differential expression were selected on the basis of a FDR < 0.05. We used ComplexHeatmap (97) to plot a heatmap of the fold-change in gene expression. Gene set enrichment analysis was performed with Ingenuity Pathway Analysis (IPA) (98) and DAVID (44) software.
Conditioned medium experiments.
Conditioned culture medium was harvested from healthy control, TLR3–/–, or UNC-93B–/– unstimulated cell cultures after 2 days of culture. Cell debris was removed by centrifugation. Healthy control, TLR3–/–, UNC-93B–/–, or STAT1–/– fibroblasts were then cultured in fresh DMEM. The conditioned culture supplemented with 100 U neutralizing antibodies against IFN-β or -α or normal sheep serum (isotype control) was added to the culture medium of the cells, and was maintained in the medium for 18 hours. RNA was isolated for RT-qPCR, and VSV growth was measured at the time points indicated.
Gene editing.
Gene-editing experiments were performed as previously described (99). Briefly, guide RNA (gRNA) sequences were generated with the CRISPR design tool (http://crispr.mit.edu/). GTCATCGAATCAAATTAAAG was selected as the guide sequence. Forward and reverse oligonucleotides for each gRNA were then inserted into the MLM3636 vector (Addgene, 43860). We electroporated 2 million hPSCs with 20 μg of Cas9-GFP plasmid and 5 μg gRNA plasmid mixed in electroporation buffer (BTX, catalog 45-0805). Green cells were sorted by FACS 48 hours after electroporation. About 50,000 cells with a moderate GFP fluorescence intensity were cultured, plated at clonal density in 96-well plates, and amplified. Genomic DNA was then extracted from each clone and Sanger sequencing for TLR3 was performed. Forward: 5′-CAACACTCCACCTCACTATC-3′; reverse: 5′-CATTGAAGAGAAATGTTCCCAGAC-3′. The TLR3-KO line used in this study carries a homozygous insertion that leads to a frameshift at amino acid position 210 and a premature stop codon 17 amino acids later (c. 628A>AA; p. K210fs17*).
Simoa assay.
IFN-β protein levels were quantified in a digital ELISA (Simoa, Quanterix) developed with the Quanterix Homebrew kit, as previously described (50).
Statistics.
Data are expressed as the mean of at least 3 biological replicates ± SD unless otherwise indicated, or are representative of 3 independent experiments. For most data, linear mixed models were used for log-transformed relative values, to account for repeated measurements. The difference between cases and controls was assessed with a likelihood ratio test. Analysis was performed with the nlme package of R software. Where indicated, 1-way ANOVA tests or log-transformations followed by 1-way ANOVA tests were used for statistical analysis. Data were analyzed in PRISM7. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Study approval.
All experiments were conducted in accordance with local regulations and with the approval of the institutional review boards of The Rockefeller University and Institut National de la Santé et de la Recherche Médicale (INSERM).
Author contributions
DG, MJC, OH and JC performed the experiments. DG, MJC, PZ, and JC analyzed the data. VB, MH, XM, YI, AC, VSS, BB, LL, GC, JM, EA, EJ, DC, IM, LA, SH, GAS, LN, DD, LS helped design the study and performed some of the experiments. MSD provided MEF cells. JLC and SYZ supervised the study. DG, MJC, JLC and SYZ wrote the manuscript. All authors contributed and edited the manuscript.
Supplementary Material
Acknowledgments
We thank the patients and their families for their participation. We thank the members of both branches of the St. Giles Laboratory of Human Genetics of Infectious Diseases, in particular Tatiana Kochetkov, for expertise and assistance with cell culture, Dusan Bogunovic for invaluable discussions and advice, and Dominick Papandrea, Cécile Patissier, and Yelena Nemirovskaya for administrative assistance. We thank Pierre Lebon (Laboratory of Virology, Paris University, Assistance Publique-Hôpitaux de Paris, Cochin Hospital) for providing us with VSV and technical advice. This work was funded in part by the National Center for Advancing Translational Sciences, NIH Clinical and Translational Science Award program (UL1TR001866), NIH (R01NS072381, R01AI088364, and R21AI151663), the French National Research Agency (ANR) under the “Investments for the future” program (ANR-10-IAHU-01), Integrative Biology of Emerging Infectious Diseases Laboratoire d’Excellence (ANR-10-LABX-62-IBEID), and grants ANR-14-CE14-0008-01 and ANR-18-CE15-0020-02, The Rockefeller University, Institut National de la Santé et de la Recherche Médicale (INSERM), Paris Descartes University, and the St. Giles Foundation. DG is supported by the Charles H. Revson Senior Fellowship in Biomedical Sciences and the National Natural Science Foundation of China (grant 319708550). IM is supported by KU Leuven C1 grant C16/18/007 and Fonds Wetenschappelijk Onderzoek Vlaanderen grant G0C8517N.
Version 1. 11/18/2020
In-Press Preview
Version 2. 01/04/2021
Print issue publication
Footnotes
Authorship note: MJC, PZ, and OH contributed equally to this work. VB, MH, and JC contributed equally to this work. JLC and SYZ contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e134529. https://doi.org/10.1172/JCI134529.
Contributor Information
Daxing Gao, Email: daxing@ustc.edu.cn.
Michael J. Ciancanelli, Email: michael.ciancanelli@gmail.com.
Peng Zhang, Email: pzhang@mail.rockefeller.edu.
Oliver Harschnitz, Email: harschno@mskcc.org.
Vincent Bondet, Email: vincent.bondet@pasteur.fr.
Mary Hasek, Email: mhasek@mail.rockefeller.edu.
Jie Chen, Email: jchen03@mail.rockefeller.edu.
Xin Mu, Email: xin_mu@tju.edu.cn.
Yuval Itan, Email: yitan@rockefeller.edu.
Aurélie Cobat, Email: aurelie.cobat@inserm.fr.
Vanessa Sancho-Shimizu, Email: v.sancho-shimizu@imperial.ac.uk.
Benedetta Bigio, Email: bbigio@rockefeller.edu.
Lazaro Lorenzo, Email: lazaro.lorenzo-diaz@inserm.fr.
Gabriele Ciceri, Email: cicerig@mskcc.org.
Esperanza Anguiano, Email: espy.anguiano@gmail.com.
Emmanuelle Jouanguy, Email: emmanuelle.jouanguy@inserm.fr.
Damien Chaussabel, Email: dchaussabel@sidra.org.
Isabelle Meyts, Email: isabelle.meyts@uzleuven.be.
Michael S. Diamond, Email: diamond@borcim.wustl.edu.
Laurent Abel, Email: laurent.abel@inserm.fr.
Sun Hur, Email: Sun.Hur@childrens.harvard.edu.
Gregory A. Smith, Email: g-smith3@northwestern.edu.
Darragh Duffy, Email: darragh.duffy@pasteur.fr.
Lorenz Studer, Email: studerl@mskcc.org.
Jean-Laurent Casanova, Email: Jean-Laurent.Casanova@rockefeller.edu.
Shen-Ying Zhang, Email: shzh289@rockefeller.edu.
References
- 1.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 2.Beutler BA. TLRs and innate immunity. Blood. 2009;113(7):1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 4.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 5.De Tiège X, Rozenberg F, Héron B. The spectrum of herpes simplex encephalitis in children. Eur J Paediatr Neurol. 2008;12(2):72–81. doi: 10.1016/j.ejpn.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Sancho-Shimizu V, et al. Genetic susceptibility to herpes simplex virus 1 encephalitis in mice and humans. Curr Opin Allergy Clin Immunol. 2007;7(6):495–505. doi: 10.1097/ACI.0b013e3282f151d2. [DOI] [PubMed] [Google Scholar]
- 7.Zhang SY, et al. TLR3 immunity to infection in mice and humans. Curr Opin Immunol. 2013;25(1):19–33. doi: 10.1016/j.coi.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. doi: 10.3402/jom.v5i0.22766. Grinde B. Herpesviruses: latency and reactivation - viral strategies and host response. J Oral Microbiol. [Published online Oct 25, 2013] [DOI] [PMC free article] [PubMed]
- 9.Casrouge A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314(5797):308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S-Y, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317(5844):1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 11.Audry M, et al. NEMO is a key component of NF-κB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J Allergy Clin Immunol. 2011;128(3):610–7.e1. doi: 10.1016/j.jaci.2011.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupuis S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33(3):388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 13.Guo Y, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208(10):2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman M, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med. 2012;209(9):1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez de Diego R, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33(3):400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancho-Shimizu V, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121(12):4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. doi: 10.1172/JCI139980. Bastard P, et al. Herpes simplex encephalitis in a patient with a distinctive form of inherited IFNAR1 deficiency [published ahead of print September 22, 2020]. J Clin Invest . [DOI] [PMC free article] [PubMed]
- 18.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177(2):265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y-M, Brinkmann MM, Paquet M-E, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452(7184):234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 20.Tabeta K, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7(2):156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 21.Pelka K, et al. The chaperone UNC93B1 regulates Toll-like receptor stability independently of endosomal TLR transport. Immunity. 2018;48(5):911–922.e7. doi: 10.1016/j.immuni.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mørk N, et al. Mutations in the TLR3 signaling pathway and beyond in adult patients with herpes simplex encephalitis. Genes Immun. 2015;16(8):552–566. doi: 10.1038/gene.2015.46. [DOI] [PubMed] [Google Scholar]
- 23.Rieder M, Conzelmann KK. Rhabdovirus evasion of the interferon system. J Interferon Cytokine Res. 2009;29(9):499–509. doi: 10.1089/jir.2009.0068. [DOI] [PubMed] [Google Scholar]
- 24.Lim HK, et al. TLR3 deficiency in herpes simplex encephalitis: high allelic heterogeneity and recurrence risk. Neurology. 2014;83(21):1888–1897. doi: 10.1212/WNL.0000000000000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lafaille FG, et al. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature. 2012;491(7426):769–773. doi: 10.1038/nature11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmer B, et al. Human iPSC-derived trigeminal neurons lack constitutive TLR3-dependent immunity that protects cortical neurons from HSV-1 infection. Proc Natl Acad Sci U S A. 2018;115(37):E8775–E8782. doi: 10.1073/pnas.1809853115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roizman B, Whitley RJ. An inquiry into the molecular basis of HSV latency and reactivation. Annu Rev Microbiol. 2013;67:355–374. doi: 10.1146/annurev-micro-092412-155654. [DOI] [PubMed] [Google Scholar]
- 28.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80(10):5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341(6152):1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 32.Smahi A, et al. Genomic rearrangement in NEMO impairs NF-kappaB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature. 2000;405(6785):466–472. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- 33.Kato H, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23(1):19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 35.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437(7062):1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 36.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19(6):727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Marcus PI, Sekellick MJ. Interferon induction by viruses. XIII. Detection and assay of interferon induction-suppressing particles. Virology. 1985;142(2):411–415. doi: 10.1016/0042-6822(85)90349-6. [DOI] [PubMed] [Google Scholar]
- 39.von Kobbe C, et al. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell. 2000;6(5):1243–1252. doi: 10.1016/S1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 40.Moens L, et al. A novel kindred with inherited STAT2 deficiency and severe viral illness. J Allergy Clin Immunol. 2017;139(6):1995–1997.e9. doi: 10.1016/j.jaci.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 41.Chapgier A, et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol. 2006;176(8):5078–5083. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 42.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103(22):8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rathi AV, Cantalupo PG, Sarkar SN, Pipas JM. Induction of interferon-stimulated genes by Simian virus 40 T antigens. Virology. 2010;406(2):202–211. doi: 10.1016/j.virol.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 45.Lasfar A, et al. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 2006;66(8):4468–4477. doi: 10.1158/0008-5472.CAN-05-3653. [DOI] [PubMed] [Google Scholar]
- 46.Hida S, et al. CD8(+) T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-alpha/beta signaling. Immunity. 2000;13(5):643–655. doi: 10.1016/S1074-7613(00)00064-9. [DOI] [PubMed] [Google Scholar]
- 47.Sato M, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13(4):539–548. doi: 10.1016/S1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 48.Takaoka A, et al. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science. 2000;288(5475):2357–2360. doi: 10.1126/science.288.5475.2357. [DOI] [PubMed] [Google Scholar]
- 49.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodero MP, et al. Detection of interferon alpha protein reveals differential levels and cellular sources in disease. J Exp Med. 2017;214(5):1547–1555. doi: 10.1084/jem.20161451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stewart WE. The Interferon System. Springer Vienna; 1979. [Google Scholar]
- 52.Sato M, et al. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441(1):106–110. doi: 10.1016/S0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 53.Edelmann KH, et al. Does Toll-like receptor 3 play a biological role in virus infections? Virology. 2004;322(2):231–238. doi: 10.1016/j.virol.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 54.Gowen BB, et al. TLR3 deletion limits mortality and disease severity due to Phlebovirus infection. J Immunol. 2006;177(9):6301–6307. doi: 10.4049/jimmunol.177.9.6301. [DOI] [PubMed] [Google Scholar]
- 55.Goffic RL, et al. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2006;2(6):e53. doi: 10.1371/journal.ppat.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hutchens M, et al. TLR3 increases disease morbidity and mortality from vaccinia infection. J Immunol. 2008;180(1):483–491. doi: 10.4049/jimmunol.180.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabeta K, et al. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci U S A. 2004;101(10):3516–3521. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rudd BD, et al. Deletion of TLR3 alters the pulmonary immune environment and mucus production during respiratory syncytial virus infection. J Immunol. 2006;176(3):1937–1942. doi: 10.4049/jimmunol.176.3.1937. [DOI] [PubMed] [Google Scholar]
- 59.Negishi H, et al. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc Natl Acad Sci U S A. 2008;105(51):20446–20451. doi: 10.1073/pnas.0810372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richer MJ, Lavallée DJ, Shanina I, Horwitz MS. Toll-like receptor 3 signaling on macrophages is required for survival following coxsackievirus B4 infection. PLoS One. 2009;4(1):e4127. doi: 10.1371/journal.pone.0004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schoggins JW, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505(7485):691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamborn IT, et al. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J Exp Med. 2017;214(7):1949–1972. doi: 10.1084/jem.20161759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asgari S, et al. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc Natl Acad Sci U S A. 2017;114(31):8342–8347. doi: 10.1073/pnas.1704259114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karikó K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem. 2004;279(13):12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 65.Cavassani KA, et al. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. J Exp Med. 2008;205(11):2609–2621. doi: 10.1084/jem.20081370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernard JJ, et al. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat Med. 2012;18(8):1286–1290. doi: 10.1038/nm.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amor S, et al. The microtubule regulator stathmin is an endogenous protein agonist for TLR3. J Immunol. 2018;184(12):6929–6937. doi: 10.4049/jimmunol.0902419. [DOI] [PubMed] [Google Scholar]
- 68.Okun E, et al. Toll-like receptor 3 inhibits memory retention and constrains adult hippocampal neurogenesis. Proc Natl Acad Sci U S A. 2010;107(35):15625–15630. doi: 10.1073/pnas.1005807107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee-Kirsch MA. The type I interferonopathies. Annu Rev Med. 2017;68:297–315. doi: 10.1146/annurev-med-050715-104506. [DOI] [PubMed] [Google Scholar]
- 70.Lim HK, et al. Severe influenza pneumonitis in children with inherited TLR3 deficiency. J Exp Med. 2019;216(9):2038–2056. doi: 10.1084/jem.20181621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galani IE, et al. Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46(5):875–890.e6. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 72.Sironi M, et al. TLR3 mutations in adult patients with herpes simplex virus and varicella-zoster virus encephalitis. J Infect Dis. 2017;215(9):1430–1434. doi: 10.1093/infdis/jix166. [DOI] [PubMed] [Google Scholar]
- 73.Casanova JL, Su HC, COVID human genetic effort a global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020;181(6):1194–1199. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Q, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bastard P, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hernandez N, et al. Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J Exp Med. 2018;215(10):2567–2585. doi: 10.1084/jem.20180628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ciancanelli MJ, et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348(6233):448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duncan CJ, et al. Human IFNAR2 deficiency: lessons for antiviral immunity. Sci Transl Med. 2015;7(307):307ra154. doi: 10.1126/scitranslmed.aac4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hernandez N, et al. Inherited IFNAR1 deficiency in otherwise healthy patients with adverse reaction to measles and yellow fever live vaccines. J Exp Med. 2019;216(9):2057–2070. doi: 10.1084/jem.20182295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu BC, Sarhan J, Poltorak A. Host-intrinsic interferon status in infection and immunity. Trends Mol Med. 2018;24(8):658–668. doi: 10.1016/j.molmed.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Belardelli F, Vignaux F, Proietti E, Gresser I. Injection of mice with antibody to interferon renders peritoneal macrophages permissive for vesicular stomatitis virus and encephalomyocarditis virus. Proc Natl Acad Sci U S A. 1984;81(2):602–606. doi: 10.1073/pnas.81.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gresser I. Wherefore interferon? J Leukoc Biol. 1997;61(5):567–574. doi: 10.1002/jlb.61.5.567. [DOI] [PubMed] [Google Scholar]
- 83.Faria PA, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17(1):93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 84.Desforges M, et al. Different host-cell shutoff strategies related to the matrix protein lead to persistence of vesicular stomatitis virus mutants on fibroblast cells. Virus Res. 2001;76(1):87–102. doi: 10.1016/S0168-1702(01)00251-9. [DOI] [PubMed] [Google Scholar]
- 85.Ferran MC, Lucas-Lenard JM. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol. 1997;71(1):371–377. doi: 10.1128/JVI.71.1.371-377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Daffis S, Samuel MA, Suthar MS, Gale M, Diamond MS. Toll-like receptor 3 has a protective role against West Nile virus infection. J Virol. 2008;82(21):10349–10358. doi: 10.1128/JVI.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen G. Splitting hESC/hiPSC lines with EDTA in feeder free conditions. Harvard Stem Cell Institute; 2008. [PubMed] [Google Scholar]
- 88.Wang Z, Liu Y, Lin W, Cui S. A real-time PCR to detect and analyze virulent EMCV loads in sows and piglets. Mol Biol Rep. 2012;39(12):10013–10017. doi: 10.1007/s11033-012-1870-y. [DOI] [PubMed] [Google Scholar]
- 89.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Porcaro F, Corsello G, Pajno GB. SLIT’s prevention of the allergic march. Curr Allergy Asthma Rep. 2018;18(5):31. doi: 10.1007/s11882-018-0785-7. [DOI] [PubMed] [Google Scholar]
- 91.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 93.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 94.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 98.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2013;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paquet D, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533(7601):125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.