Abstract
Obesity occurs when energy expenditure is outweighed by energy intake. Tuberal hypothalamic nuclei, including the arcuate nucleus (ARC), ventromedial nucleus (VMH), and dorsomedial nucleus (DMH), control food intake and energy expenditure. Here we report that, in contrast with females, male mice lacking circadian nuclear receptors REV-ERBα and –β in the tuberal hypothalamus (HDKO mice) gained excessive weight on an obesogenic high-fat diet due to both decreased energy expenditure and increased food intake during the light phase. Moreover, rebound food intake after fasting was markedly increased in HDKO mice. Integrative transcriptomic and cistromic analyses revealed that such disruption in feeding behavior was due to perturbed REV-ERB–dependent leptin signaling in the ARC. Indeed, in vivo leptin sensitivity was impaired in HDKO mice on an obesogenic diet in a diurnal manner. Thus, REV-ERBs play a crucial role in hypothalamic control of food intake and diurnal leptin sensitivity in diet-induced obesity.
Keywords: Endocrinology, Metabolism
Keywords: Diabetes, Leptin, Neuroendocrine regulation
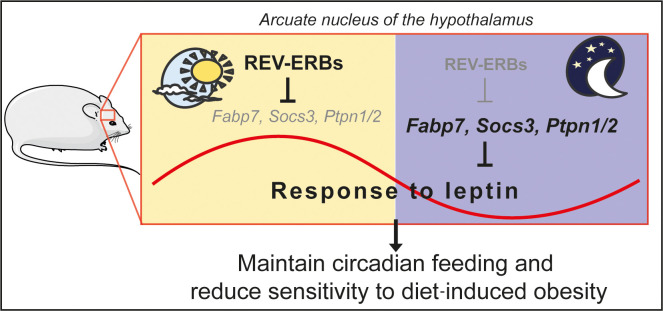
REV-ERB nuclear receptors, which link circadian rhythms and metabolism in the periphery, coordinately regulate hypothalamic gene expression to control diurnal food intake and leptin sensitivity.
Introduction
In humans and mice, circadian regulation and metabolic homeostasis are coupled. Circadian disruption is a risk factor for metabolic diseases including obesity, diabetes, and cardiovascular diseases, with epidemiological studies revealing that shift and night workers have higher body mass index and increased incidence of metabolic syndrome (1, 2). Several studies have supported the possibility that hypothalamic deregulation, through derangement of brain clock-gene expression, contributes to the pathogenesis of metabolic disorders (3–8). The mammalian circadian system is hierarchically organized with a master clock conductor in the suprachiasmatic nucleus (SCN) that controls peripheral clocks throughout the body.
Each cell contains a molecular clock composed of transcription factors (TFs) regulating one another in transcriptional-translational feedback loops that generate near-24-hour oscillations. REV-ERBα and –β nuclear receptors comprise a negative loop by repressing the expression of Bmal1, a positive regulator of clock output genes. In addition to their role in the cell-autonomous clock, REV-ERBs regulate many tissue-specific genes and metabolic functions (9). Our understanding of the physiological functions of REV-ERBs has mainly been derived from phenotypic and molecular analysis of mice lacking REV-ERBα in all tissues, as REV-ERBβ appears to be less important but provides redundancy for metabolic and behavioral rhythms (10, 11). REV-ERBα whole-body-KO mice exhibit metabolic disorders including abnormal blood glucose level, increased adiposity, steatosis, and increased thermogenic capacity (10, 12, 13), as well as circadian behavioral changes such as reduction in locomotor activity period (14), alteration of sleep homeostasis (15), mood disorders (16, 17), and altered feeding behaviors (18, 19).
Feeding behaviors and metabolism are highly dependent on the function of the hypothalamus. In particular, the paraventricular hypothalamus (PVH), the dorsomedial hypothalamus (DMH), the ventromedial hypothalamus (VMH), and the arcuate nucleus (ARC) regulate several diurnal physiological functions including energy expenditure, thermogenesis, glucose and lipid metabolism, and feeding behaviors (4–6, 20). Although the SCN is the master clock driving these rhythms, these non-SCN nuclei are suggested to have circadian functions independent of the SCN (21, 22).
Although the REV-ERBα total-body KO has been informative, the specific role of REV-ERBs in the non-SCN hypothalamic nuclei cannot be inferred through this model. To address this, we deleted both REV-ERBα and –β in the tuberal hypothalamic nuclei, including the ARC, VMH, and DMH. Our studies demonstrate that hypothalamic REV-ERBs are critical for body weight homeostasis due to their regulation of diurnal food intake and leptin sensitivity.
Results
Specific deletion of REV-ERBα and –β in non-SCN hypothalamic nuclei.
We first determined the diurnal expression of REV-ERBα in non-SCN hypothalamic nuclei of mice with epitope-tagged endogenous REV-ERBα (HA-REV-ERBα mice, previously described in ref. 23) to specifically visualize the protein by immunofluorescence. Peak expression was observed between zeitgeber time 1 (ZT1) and ZT7 (Figure 1A), and therefore studies of REV-ERBs’ function in the hypothalamus were conducted at ZT1–ZT4.
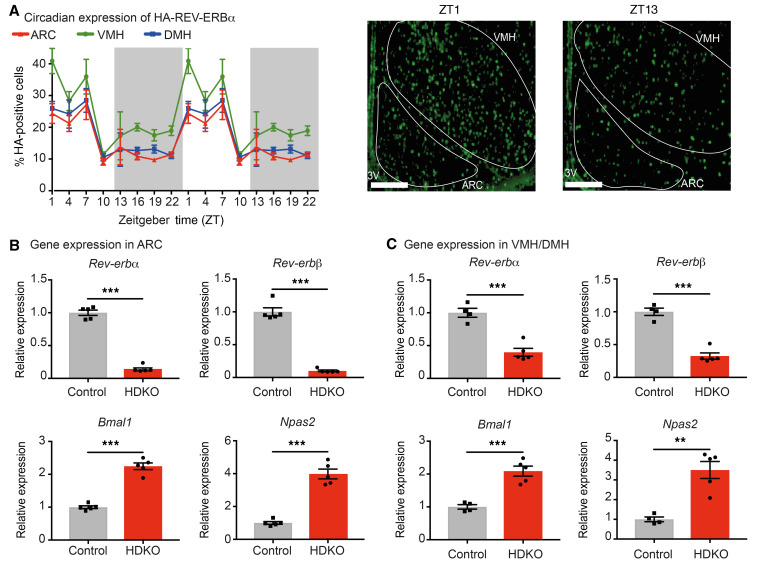
Figure 1. Generation of mice with specific deletion of both REV-ERBα and –β in non-SCN hypothalamic nuclei.
(A) Diurnal expression of HA-REV-ERBα in ARC, VMH, and DMH from HA-REV-ERBα mice housed in regular 12-hour light/12-hour dark cycles. Percentage of HA-positive cells was determined over total number of cells detected by DAPI staining (n = 6–10, mean ± SEM, double-plotted). Representative pictures of HA-REV-ERBα staining in the ARC and VMH at the peak (ZT1) and trough (ZT13) are presented. Scale bars: 100 μm. 3V, third ventricle. (B and C). Reverbα, Reverbβ, and canonical REV-ERB target gene expression in the ARC (B) and VMH/DMH (C) from Reverbαfl/fl-Reverbβfl/fl-Nkx2.1-Cre+ mice (named HDKO mice) compared with their control littermates (Reverbαfl/fl-Reverbβfl/fl-Nkx2.1-Cre–) at ZT4 on NCD (n = 4–5, mean ± SEM). Results were compared by unpaired t test. **P < 0.01, ***P < 0.001.
To delineate hypothalamus-specific functions of REV-ERBs, mice homozygous for floxed alleles of REV-ERBα and REV-ERBβ (23, 24) were crossed with Nkx2.1-Cre mice, which express Cre in the ventromedial tuberal hypothalamic nuclei but, importantly, not in the SCN (25). This Cre driver is also expressed in the lung and thyroid gland but not in any other peripheral metabolic organs (25, 26). In these Reverbαfl/fl-Reverbβfl/fl-Nkx2.1-Cre+ mice (hereafter referred to as HDKO mice), Reverbα and Reverbβ mRNA levels were markedly depleted in the ARC (80%–90%, Figure 1B) and in the VMH/DMH (V-DMH) (60%–70%, Figure 1C) relative to control littermates (Reverbαfl/fl-Reverbβfl/fl-Nkx2.1-Cre–). Consistent with loss of REV-ERB–mediated repression, Bmal1 and Npas2 mRNAs were increased in HDKO mice in both the ARC (Figure 1B) and V-DMH (Figure 1C). Importantly, expression of Reverbα and Reverbβ mRNA was not affected in the SCN and liver from HDKO mice (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI140424DS1), allowing the dissociation of REV-ERB functions in non-SCN hypothalamic nuclei independently of its functions in the SCN master clock.
Deletion of REV-ERBs in non-SCN hypothalamic nuclei exacerbates diet-induced obesity.
On normal chow diet (NCD), HDKO mice did not exhibit significant changes in body weight (Supplemental Figure 2A), adipose tissue mass (Supplemental Figure 2, B and C), activity (Supplemental Figure 2, D and E), energy expenditure (Supplemental Figure 2, F–H), body temperature (Supplemental Figure 2I), and food consumption (Supplemental Figure 2, J–L). Remarkably, however, high-fat diet (HFD) feeding resulted in HDKO male mice gaining more weight than controls (Figure 2A), with increased adipose tissue mass (Figure 2, B and C). This effect seemed to be sex-specific, as female mice lacking REV-ERBs in non-SCN hypothalamic nuclei did not exhibit exacerbation of the diet-induced obesity (DIO) effects (Figure 2, D–F). In the males, physical activity (Figure 2, G and H) and respiratory exchange ratio (Figure 2I) were not affected, but energy expenditure was decreased in both light and dark phases (Figure 2, J–K).
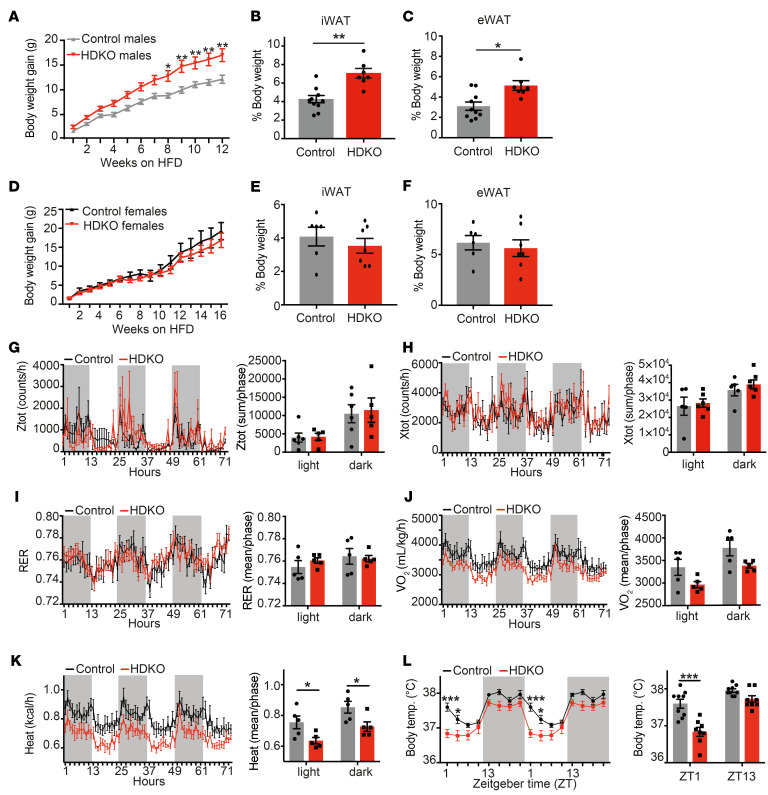
Figure 2. Deletion of REV-ERBs in non-SCN hypothalamic nuclei exacerbates DIO only in males.
(A) Body weight gain of HDKO male mice and their control littermates on HFD (n = 9–14, mean ± SEM). Results were compared by repeated measures (RM) 2-way ANOVA and Holm-Šidák multiple-comparison test. A representative experiment is shown (of 3 separate experiments with similar results). (B) Inguinal and (C) epididymal white adipose tissue (iWAT and eWAT) weights from HDKO male mice and their control littermates after 12 weeks of HFD (n = 6–8, mean ± SEM). Results were compared by Mann-Whitney test. (D) Body weight gain of HDKO female mice and their control littermates on HFD (n = 7–9, mean ± SEM). Results were compared by Mann-Whitney test. (E) iWAT and (F) eWAT weights from HDKO female mice and their control littermates after 16 weeks of HFD (n = 6–7, mean ± SEM). Results were compared by Mann-Whitney test. (G and H) Vertical (Ztot) and horizontal (Xtot) diurnal activity, (I) respiratory exchange ratio (RER), (J) oxygen consumption (VO2), and (K) heat production in HDKO male mice and their control littermates after 12 weeks of HFD. (D–H) Diurnal recording for 3 days is displayed (left panel), as well as corresponding quantification during the light and dark phases (right panel). Results were compared by RM 2-way ANOVA and Holm-Šidák multiple-comparison test (n = 5, mean ± SEM). (L) Body temperature in HDKO male mice and their control littermates after 12 weeks of HFD (n = 8–9, mean ± SEM). Diurnal recording is displayed (left panel, 24-hour recording duplicated), as well as ZT1 and ZT13 (right panel). Results were compared by RM 2-way ANOVA and Holm-Šidák multiple-comparison test. *P < 0.05, **P < 0.01, ***P < 0.001.
Interestingly, body temperature was decreased in HDKO mice at ZT1–ZT4 (Figure 2L), corresponding to the peak of REV-ERB expression in the hypothalamus and suggesting a direct effect of hypothalamic REV-ERB signaling on core temperature rather than a consequence of increased body weight. Reverbα, Reverbβ, and Ucp1 expression was not affected in brown adipose tissue from HDKO mice on HFD at ZT4 and ZT16 compared with control mice (Supplemental Figure 3A). Moreover, in contrast with Reverbα whole-body-KO mice (12), HDKO mice on HFD did not display an altered thermogenic capacity in response to a norepinephrine challenge (Supplemental Figure 3B). These results suggest that the phenotype observed in whole-body Reverbα-KO mice was independent of REV-ERB expression in non-SCN hypothalamic nuclei. Importantly, triiodothyronine (T3) and thyroxine (T4) circulating levels were also comparable in control and HDKO mice on HFD (Supplemental Figure 3, C and D), suggesting that thyroid signaling is preserved in HDKO mice despite potential Cre expression in the thyroid gland (25, 26). Also, despite the antisense relationship between Reverbα and thyroid hormone receptor α2 (Thrα2) (27, 28), Thrα2 expression was unchanged, as were genes involved in thyroid hormone action in the ARC from HDKO mice (Supplemental Figure 3, E and F). Thrα1 expression was modestly decreased in HDKO mice compared with control littermates (Supplemental Figure 3E), but similar changes were seen in mice lacking only REV-ERBα in non-SCN hypothalamic nuclei (HSKO, Reverbαfl/fl-Nkx2.1-Cre+) (Supplemental Figure 3, G and H), even though these mice do not exhibit a metabolic phenotype on HFD (Supplemental Figure 3I). Together, these results highly suggest that both central and peripheral thyroid signaling are preserved in HDKO mice and that the observed phenotype directly results from both REV-ERBs’ deletion and their redundant impact on gene expression in non-SCN hypothalamic nuclei on HFD.
Hypothalamic REV-ERBs regulate diurnal feeding behavior.
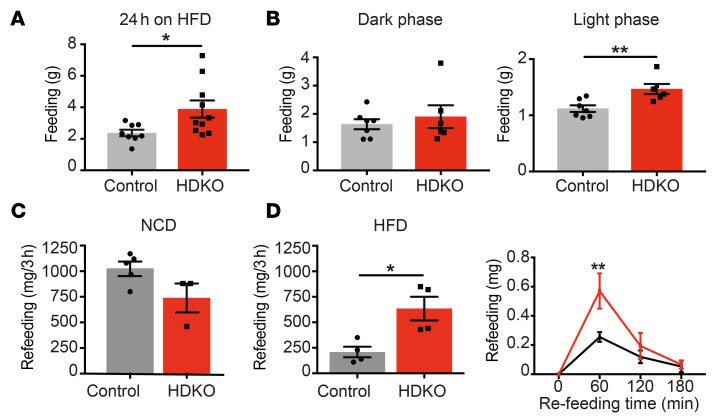
To understand how hypothalamic REV-ERBs control the propensity to develop DIO, food intake was measured in control and HDKO mice on HFD. Twenty-four-hour food intake was increased in HDKO mice (Figure 3A) only during the light phase (Figure 3B), corresponding to the physiological expression of hypothalamic REV-ERBs. Of particular note, after a 24-hour fast, rebound feeding at ZT1–ZT4 was massively increased in HDKO mice only in the setting of DIO, with the maximal effect observed 1 hour after refeeding (Figure 3, C and D). Interestingly, whereas control mice adapted to calorie-dense HFD by eating less food than when on NCD, this phenomenon was not observed in HDKO mice (compare Figure 3, C and D), suggesting an absence of homeostatic control of food intake in HDKO mice.
Figure 3. Hypothalamic REV-ERBs regulate diurnal eating behaviors on HFD.
(A and B) Food consumption in HDKO mice and their control littermates on HFD. (A) Twenty-four-hour food consumption calculated from 8 days of records are presented (n = 8–10, mean ± SEM). (B) Light phase and dark phase food consumption calculated from 48 hours of records are presented (n = 6–7, mean ± SEM). Results were compared by Mann-Whitney test. (C and D) Rebound feeding at ZT1–ZT4 after 24 hours of fasting in HDKO mice and their control littermates on NCD (C) and HFD (D) (n = 3–5, mean ± SEM). Results were compared by Mann-Whitney test. Three-hour time course of rebound feeding on HFD is also displayed (n = 3, mean ± SEM). A representative experiment is shown (of 2 separate experiments with similar results). Results were compared by repeated measures 2-way ANOVA and Holm-Šidák multiple-comparison test. *P < 0.05, **P < 0.01.
Food consumption is regulated by REV-ERB–dependent leptin signaling in the ARC.
To understand the molecular mechanisms by which REV-ERBs regulate food consumption to exacerbate DIO, transcriptomic analysis was performed at ZT4 in control and HDKO mice fed ad libitum with NCD or HFD. Gene expression profiling revealed that the majority of HFD-induced transcriptional changes occurred in the ARC and not the V-DMH (results not shown). As expected, the expression of several canonical clock genes was dysregulated in the ARC of HDKO mice on both NCD and HFD (Supplemental Figure 4A).
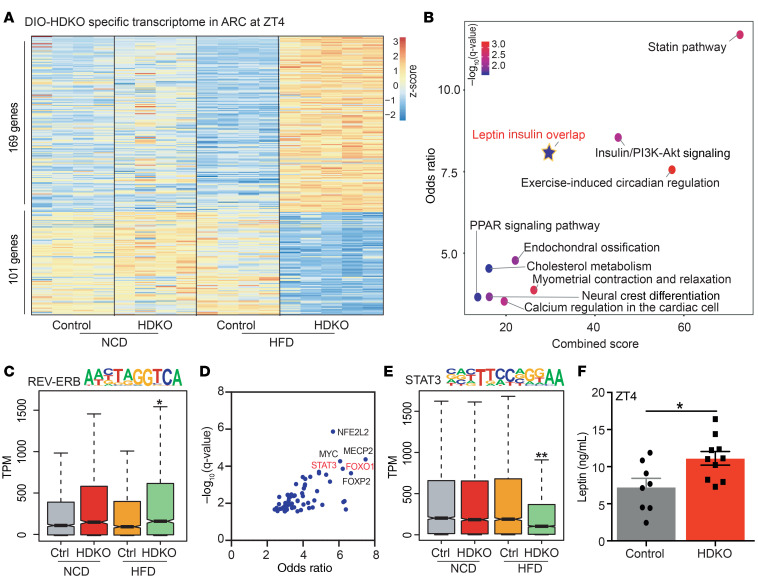
The deletion of REV-ERBs specifically altered the expression of numerous other genes in the ARC of HFD-fed mice (169 genes up-, 101 genes downregulated) (Figure 4A and Supplemental Figure 4B). Interestingly, pathway analysis revealed an enrichment for leptin and insulin signaling (Figure 4B), which are known to regulate feeding behaviors, in part through activation of cellular substrates in the ARC (29, 30). To note, although loss of REV-ERBα has been shown to alter dopamine signaling in the hippocampus and midbrain (16, 17), this was not observed in gene set enrichment analysis (GSEA) of ARC gene expression on HFD in the HDKO mice (Supplemental Figure 4C). Consistent with the deletion of repressive REV-ERBs in the HDKO model, genes that were upregulated in the ARC of these mice were highly enriched for the REV-ERB DNA-binding motif (Figure 4C).
Figure 4. Food consumption is regulated by REV-ERB–dependent leptin signaling in the ARC on HFD.
(A) Heatmap of the genes differentially expressed in HDKO mice in the ARC only on HFD. Gene expression analysis at ZT4 from HDKO mice and their control littermates on NCD and HFD in the ARC (TPM > 0.1, FDR < 0.05). Two to 4 independent hypothalamic nuclei punches were pooled together per biological replicate (n = 4–5). (B) Pathway analysis of genes differentially expressed in HDKO mice on HFD in the ARC (Enrichr q value < 0.05). (C) Box-and-whisker plot showing minimum, maximum, median, first quartile, and third quartile from quantification of enrichment of the REV-ERB motif in the transcriptome from HDKO mice and their control littermates on NCD or HFD in the ARC. P values were calculated with the Mann-Whitney test. (D) Enrichr transcription factor analysis on the genes downregulated in HDKO mice on HFD in the ARC. (E) Box-and-whisker plot showing minimum, maximum, median, first quartile and third quartile from quantification of enrichment of the STAT3 motif in the transcriptome from HDKO mice and their control littermates on NCD or HFD in the ARC. P values were calculated with the Mann-Whitney test. (F) Leptin circulating levels at ZT4 in control and HDKO mice on HFD (n = 8–10, mean ± SEM). Results were compared by Mann-Whitney test. *P < 0.05, **P < 0.01.
To predict the TFs controlling downregulated genes, we applied the Enrichr TF algorithm (31, 32). Interestingly, motifs for STAT3 and FOXO1, both known to mediate leptin transcriptional responses (29, 30, 33), were enriched near genes downregulated in the ARC of HDKO mice (Figure 4D). In accordance with this prediction, the STAT3 motif was enriched at genes whose expression decreased in the HDKO ARC (Figure 4E), providing further evidence of impaired of leptin signaling in the absence of REV-ERBs. Moreover, circulating leptin levels were upregulated in HDKO mice on HFD (Figure 4F), consistent with a trend toward increased leptin gene expression in inguinal white adipose tissue (iWAT) (Supplemental Figure 4D). Interesting, circulating leptin levels already tended to be elevated at ZT4–ZT10 in HDKO mice on NCD (Supplemental Figure 4E), corresponding to the rhythmic expression of hypothalamic REV-ERBs. As expected, both control and HDKO leptin levels largely increased after maintenance on HFD, but circulating leptin levels increased even further in HDKO mice compared with control mice (Supplemental Figure 4F), suggesting an acceleration of diet-induced leptin resistance in HDKO mice. In contrast with the circulating levels of leptin, serum insulin levels were not dysregulated in HDKO mice maintained on either NCD or HFD (Supplemental Figure 4, G and H).
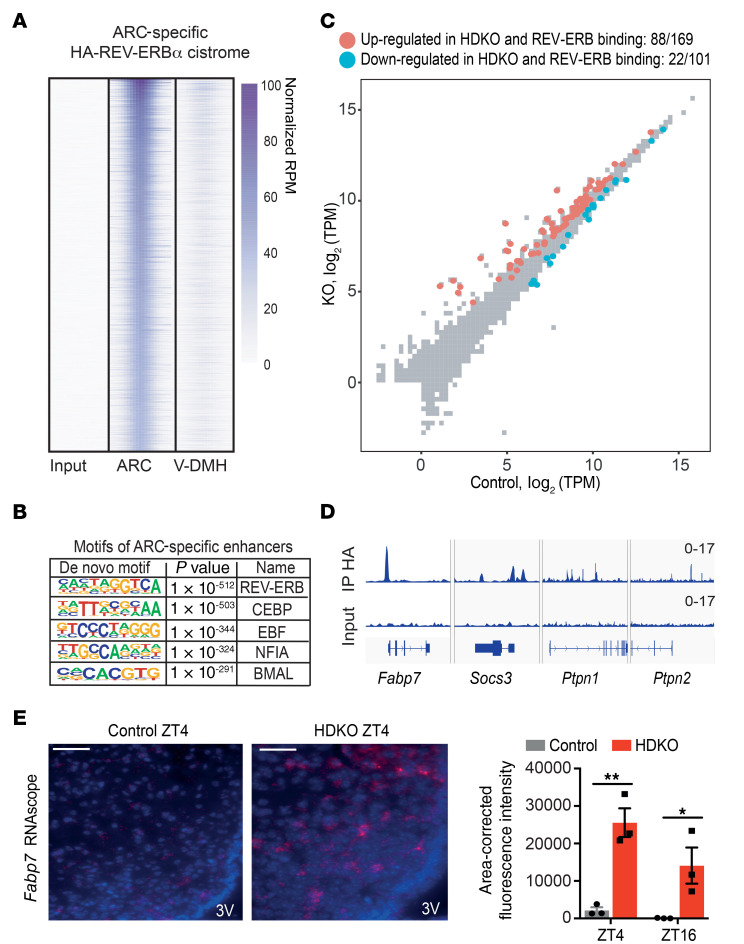
We next performed chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) to determine direct genomic targets of REV-ERBs in the ARC, taking advantage of the HA-REV-ERBα mice in which the endogenous protein is epitope tagged. Given the small size of hypothalamic nuclei and the low abundance of TFs, ARC or V-DMH punches from 10 independent HFD-treated HA-REV-ERBα mice were harvested at ZT4 and pooled for study. A number of REV-ERBα binding sites were common to the ARC and V-DMH (Supplemental Figure 5A), and de novo motif analysis revealed marked enrichment for the canonical REV-ERB motif at these sites (Supplemental Figure 5B). However, a greater number of binding sites were specific to either the ARC (Figure 5A) or V-DMH (Supplemental Figure 5C). ARC-specific binding sites were highly enriched for the REV-ERB motif (Figure 5B). Approximately 52% (88 of 169) of genes upregulated in the HDKO ARC had a REV-ERB binding site within 100 kb, suggesting direct regulation by REV-ERBα in the ARC of DIO mice (Figure 5C). Only 22% (22 of 101) of downregulated genes also had a REV-ERB binding site within 100 kb. Importantly, direct REV-ERB binding sites were noted near a number of genes that have been described to downregulate leptin signaling including Fabp7, Socs3, Ptpn1 (PTP1B), and Ptpn2 (TCPTP) (29, 34–37) (Figure 5D). Fabp7 upregulation in HDKO mice maintained on an HFD was confirmed by RNAscope fluorescent in situ hybridization (FISH) in the ARC at both ZT4 and ZT16 (Figure 5E). Together, these results further suggest that REV-ERBα directly represses gene expression of factors known to downregulate leptin signaling.
Figure 5. REV-ERBα directly regulates ARC transcriptome on HFD.
(A) HA-REV-ERBα cistrome specific to the ARC on HFD. ChIP-seq analysis of HA immunoprecipitation in the ARC from HA-REV-ERBα mice on HFD at ZT4 (reads per million mapped reads [RPM] > 1, >7-fold over input, FDR < 0.0001). Ten independent hypothalamic nuclei punches were pooled together (n = 1). (B) Motif analysis of ARC-specific cistrome of HA-REV-ERBα on HFD with default HOMER parameters. (C) Scatterplot analysis of the HFD-HDKO–specific ARC transcriptome at ZT4. The direct regulome was identified by the integration of the HA-REV-ERBα cistrome in the ARC on HFD (TPM > 0.1, FDR < 0.05 for differentially expressed genes; RPM > 1, >7-fold over input for peaks within 100 kb). (D) Genome browser view of HA-REV-ERBα ChIP-seq peaks in the ARC. (E) Fabp7 staining by RNAscope in the ARC from control and HDKO mice on HFD. Representative pictures at ZT4 and quantification at ZT4 and ZT16 are presented. Scale bars: 50 μm. 3V, third ventricle. Results were compared by 2-way ANOVA (interaction P = 0.1705) and Holm-Šidák multiple-comparison test. *P < 0.05, **P < 0.01.
Hypothalamic REV-ERBs regulate diurnal leptin sensitivity on HFD.
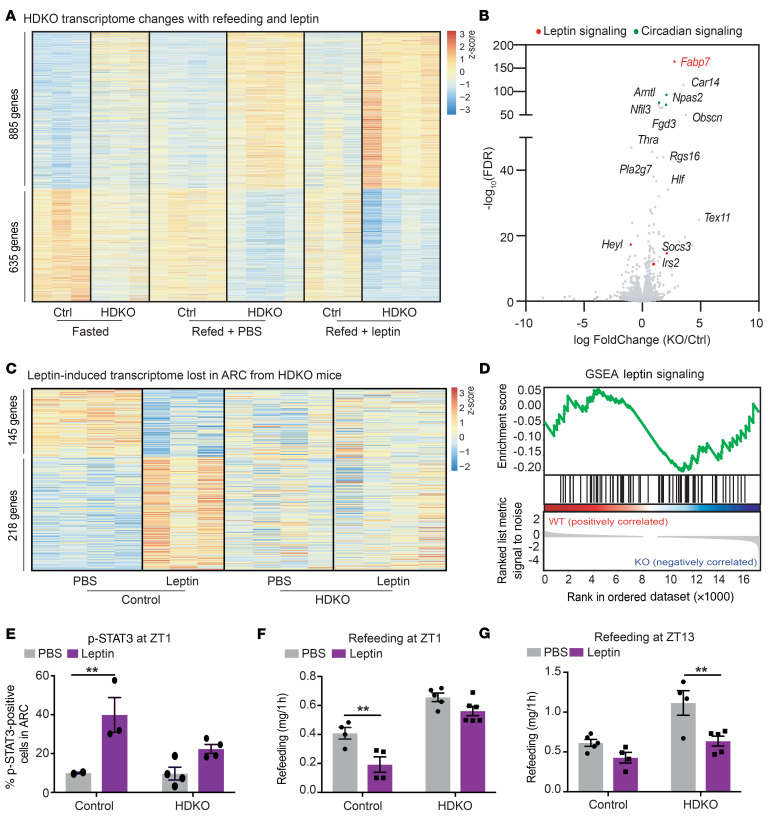
To directly test the hypothesis that REV-ERBs regulate leptin responsiveness in the ARC of DIO mice, we performed transcriptomic profiling on ARC punches from 24-hour-fasted control and HDKO mice injected with PBS or leptin 15 minutes prior to an hour-long refeeding with HFD. To understand the mechanism by which deletion of REV-ERBs impairs leptin sensitivity, we initially looked at genes differentially expressed in HDKO mice in the basal condition (fasted), whose differential expression was exacerbated by refeeding and leptin treatment (Figure 6A). A total of 1520 genes met these criteria (885 genes upregulated, 635 genes downregulated). Interestingly, among these genes, we observed several involved in leptin signaling (Figure 6B), including Fabp7 and Socs3, which contain strong REV-ERBα binding sites in the ARC and were upregulated in HDKO mice. Indeed, bioinformatic analysis revealed that 363 genes were differentially expressed (218 genes upregulated, 145 genes downregulated) in response to leptin in control mice but not in HDKO mice (Figure 6C and Supplemental Figure 6A). GSEA confirmed the enrichment of the leptin signaling pathway in control mice only (Figure 6D). Furthermore, transcriptomic profiling between 24-hour fasting and 1 hour after refeeding revealed 805 dysregulated genes in HDKO compared with control ARC after refeeding (645 genes up-, 160 genes downregulated) (Supplemental Figure 6, B and C), and GSEA confirmed the impairment of leptin signaling in HDKO ARC during refeeding (Supplemental Figure 6D).
Figure 6. Hypothalamic REV-ERBs regulate diurnal leptin sensitivity on HFD.
(A) Heatmap of the genes differentially expressed in HDKO mice in the ARC in basal condition (fasted) and exacerbated during refeeding and leptin treatments. Gene expression analysis at ZT4 from HDKO mice and their control littermates after 24 hours of fasting and refeeding with HFD after injection of PBS or leptin (TPM > 0.1, FDR < 0.05). Two independent hypothalamic nuclei punches were pooled together per biological replicate (n = 3–4). (B) Volcano plot of the genes differentially expressed in HDKO mice identified in panel A, highlighting genes involved in circadian regulation (in green) or leptin signaling (in red). (C) Heatmap of the genes differentially expressed in HDKO mice in response to leptin. Gene expression analysis at ZT4 from HDKO mice and their control littermates after 24 hours of fasting and refeeding with HFD after injection of PBS or leptin (TPM > 0.1, FDR < 0.05). Two independent hypothalamic nuclei punches were pooled together per biological replicate (n = 3–4). (D) GSEA of leptin signaling during refeeding in response to leptin in the ARC from HDKO mice and their control littermates. (E) Percentage of p-STAT3–positive cells in the ARC after 24 hours of fasting, and PBS or leptin injection at ZT1 in HDKO mice and their control littermates on HFD (n = 2–4, mean ± SEM). Results were compared by 2-way ANOVA (interaction P = 0.1187) and Holm-Šidák multiple-comparison test. (F and G) Rebound feeding on HFD at ZT1 (F) and ZT13 (G) after 24 hours of fasting and injection of PBS or leptin in HDKO mice and their control littermates (n = 4-6, mean ± SEM). A representative experiment is shown at ZT1 (of 2 separate experiments with similar results). Results were compared by 2-way ANOVA and Holm-Šidák multiple-comparison test. **P < 0.01.
To test whether the absence of a transcriptional response to leptin resulted in physiological leptin resistance, leptin sensitivity was assessed in vivo. Phosphorylated STAT3 (p-STAT3) was evaluated by immunofluorescence detection in the ARC from HDKO and control mice that had been fasted for 24 hours and treated at ZT1 with leptin (or PBS) 1 hour prior to harvest. As expected, leptin increased the percentage of p-STAT3–positive cells in the ARC in control mice, but the effect of leptin was markedly attenuated in HDKO mice (Figure 6E). In addition, rebound feeding was evaluated at ZT1 after 24 hours of fasting in control and HDKO mice injected with PBS or leptin 15 minutes prior to refeeding with HFD for 1 hour (Figure 6F). Remarkably, while leptin decreased rebound feeding in control mice, as expected, no such response was noted in HDKO mice. Importantly, this leptin insensitivity of the HDKO mice was diurnal, as administration of leptin did attenuate rebound feeding at ZT13, when expression of REV-ERBs is at its trough (Figure 6G). Taken together, these findings implicate REV-ERBs as direct regulators of the ARC leptin signaling during DIO.
Discussion
We have demonstrated that REV-ERBs in hypothalamic nuclei associated with feeding and metabolism play an important role in controlling DIO. Both ad libitum feeding during the light phase and rebound feeding after 24 hours of fasting are increased in HDKO mice maintained on HFD. Moreover, feeding adaptation from NCD to HFD is impaired in HDKO mice, suggesting an impairment of homeostatic feeding regulation. Integration of transcriptomic and cistromic analyses of REV-ERBs’ functions in non-SCN hypothalamic nuclei upon ad libitum feeding and feeding challenges revealed the impairment of leptin signaling in the ARC in the absence of REV-ERB expression. In strong support of this mechanism, the transcriptomic response to leptin in the ARC is disturbed by deletion of REV-ERBs, and leptin responsiveness in vivo is impaired in a diurnal manner in mice lacking hypothalamic REV-ERBs.
Food intake is temporally organized; the feeding period is restricted to the active phase, while the resting phase corresponds to a fasting period and mobilization of energy stores. The circadian variations in metabolism and feeding behavior are anticipated and coordinated by the circadian clocks (38), which are synchronized by environmental cues. Although the SCN master clock is mainly synchronized by light cycles, feeding rhythms are a powerful time giver for non-SCN central and peripheral clocks (39). Thus, misalignment of the SCN master clock with these food-entrainable clocks uncouples central and peripheral clocks and has deleterious effects on metabolic health (40). It is thus important to understand circadian regulation of food intake to understand the etiology of metabolic disorders.
Leptin is an adipokine that exhibits a circadian rhythm in terms of both synthesis by adipose tissue and central signaling (41, 42). Moreover, several studies demonstrate that circadian disruption induces leptin resistance, even without changes in feeding rhythms (41, 43–45), suggesting that leptin signaling is highly dynamic in response to both external feeding cues and internal circadian timing to maintain long-term energy balance. Indeed, central leptin signaling has been demonstrated to be diurnal using a Bmal1-KO model that completely impaired cell-autonomous clocks (42), which does not occur following deletion of REV-ERBs (14, 46, 47). As Reverb expression is BMAL1 dependent (48), the present work suggests a direct role for REV-ERBs in the effect of BMAL1 on leptin signaling.
Interestingly, hypothalamic REV-ERBα–single-KO (HSKO) mice did not exhibit the metabolic phenotype of the HDKO mice, suggesting a role for REV-ERBβ. In the liver, loss of REV-ERBβ has minor impact on the clock or metabolism except when REV-ERBα is also deleted (10, 11), but our studies do not rule out a more important role of REV-ERBβ in the non-SCN hypothalamus. We also note that the metabolic dysfunction induced by deletion of REV-ERBs in non-SCN hypothalamic nuclei was observed only in males. This is consistent with other studies reporting strong sex effects in sensitivity to circadian and metabolic disruptions (49–51). Estrogen and leptin effects overlap in their regulation of hypothalamic p-STAT3 (52), which could also contribute to the observed resistance of females to the loss of hypothalamic REV-ERBs.
Here we showed that deletion of REV-ERBs impaired diurnal leptin signaling in males and presented strong genomic and transcriptomic evidence in favor of a direct mechanism in the ARC. Interestingly, HA-REV-ERBα occupied genomic regions near a number of genes that negatively regulated leptin signaling. One of these, Fabp7, has been proposed to function in glial cells within the ARC to impair leptin signaling especially on HFD (34). Several studies demonstrated a key role of glial cells in leptin and feeding regulation by the hypothalamus (53–58). It is critical to note that the Nkx2.1-Cre model used here is expressed in both neuronal and glial cells (26, 59–61), allowing for the delineation of the global effect of hypothalamic REV-ERB signaling. Thus, we cannot exclude hypothetical cancelling effects of expression in different cell types, which could theoretically mask some of the potential metabolic phenotypes. Interestingly, SOCS3, which was also directly regulated by REV-ERBα, has also been described to inhibit leptin signaling and to be especially induced by HFD (62). Moreover, the responsiveness of HDKO mice to leptin at ZT13 suggests that leptin sensitivity was phase shifted rather than impaired only at ZT1–ZT4 in HDKO mice. This is consistent with a recent study demonstrating that DIO shifts circadian leptin sensitivity from the beginning of the light phase to the beginning of the dark phase (63). Both Socs3 and Fabp7 deletions prevent DIO (34, 64), suggesting that their circadian repression by REV-ERBs may be an adaptative mechanism to maintain circadian leptin sensitivity on HFD. Together, these results reveal that hypothalamic REV-ERBs are necessary for the homeostatic adaptation of feeding and metabolism to prevent DIO.
Methods
Experimental models
Reverbαfl/fl-Reverbβfl/fl mice (24) were bred with C57BL/6J-Tg(Nkx2.1-Cre)2Sand/J mice maintained on a C57BL/6J background (The Jackson Laboratory, stock 008661). HA-REV-ERBα mice were generated and maintained on a C57BL/6J genetic background (23). Except for some specific experiments specified below, mice were bred and group housed in a specific pathogen–free animal facility at 22°C and under a 12-hour light/12-hour dark cycle (lights on at 7 am, lights off at 7 pm), with free access to water and NCD (LabDiet, 5010) or HFD composed of 60:20:20 kcal percentage of fat/carbohydrate/protein (Research Diets, D12492i) starting at 5–6 weeks old. Unless otherwise specified in the figure legends, all experiments were carried out on 11- to 14-week-old male littermates. Control mice are Reverbαfl/fl-Reverbβfl/fll-Nkx2.1-Cre–, while Reverbαfl/fl-Reverbβfl/fl-Nkx2.1-Cre+ are named HDKO mice. For HSKO mice (Reverbαfl/fl-Nkx2.1-Cre+), control littermates used are Reverbαfl/fl-Nkx2.1-Cre–.
Method details
Diurnal HA-REV-ERBα staining in hypothalamus.
HA-REV-ERBα mice were sacrificed at 8 time points in 3-hour intervals (8 am = ZT1, 11 am = ZT4, 2 pm = ZT7, 5 pm = ZT10, 8 pm = ZT13, 11 pm = ZT16, 2 am = ZT19, and 5 am = ZT22) (n = 5 at each time point). After cervical dislocation, mouse brains were removed and placed in a brain matrix for coronal sectioning. Sections containing the hypothalamic region were taken and fixed in 4% paraformaldehyde (PFA) in 1× PBS at 4°C for 12–18 hours and then washed in PBS and incubated in 30% sucrose in PBS for 1–2 days. Samples were then incubated in 50:50 30% sucrose plus 1× PBS/OCT for 2 hours and embedded in OCT. Sections (14 μm) were made on a cryostat (Leica) and collected on Superfrost Plus slides (VWR) in 3 series of slides, each slide containing 4 brain sections. To identify the slices of interest, Nissl staining was performed on serial section 1. Slides were rehydrated for 1 minute in distilled water and stained 4 minutes with warm 0.5% cresyl violet stain solution (MilliporeSigma, C5042) in distilled water and 0.3% glacial acetic acid (MilliporeSigma, ARK2183). Slides were washed in distilled water 3 times (1 minute each), 50% ethanol (EtOH) (30 seconds), and 70% EtOH/1% glacial acid acetic (30 seconds) before dehydration with 95% EtOH (30 seconds), 100% EtOH (1 minute), and 2 xylene (MilliporeSigma, 534056) baths (3 minutes each). Slides were mounted with Cytoseal (Thermo Fisher Scientific, 8312-4) and visualized using a Nikon Eclipse E200 microscope.
Selected slides from serial section 2 were used to perform immunofluorescent staining for HA (C29F4, Cell Signaling Technology, 3724S) and counterstain with DAPI. Briefly, slides were washed twice with distilled water for 2 minutes each and antigen retrieval was performed using citrate buffer pH 6.0 (120°C for 30 seconds) in a Decloaking Chamber (Biocare, DC2002), before blocking the endogenous peroxidase with 3% hydrogen peroxide in distilled water for 15 minutes and washing the slides 3 times with distilled water (1 minute each). Slices were then washed with 0.1% Tween 20 in PBS and blocked with 2.5% horse serum in 0.5% Tween 20 in PBS for 1 hour at room temperature before incubation with the primary antibody overnight at 4°C (C29F4, 1:1000 in 0.1% Tween 20 in PBS). After 4 washes with 0.1% Tween 20 in PBS (5 minutes each), slides were incubated with undiluted ImmPRESS secondary antibody (Vector Laboratories, MP-7401) for 60 minutes at room temperature. After 4 washes with 0.1% Tween 20 in PBS (5 minutes each), slides were incubated with TSA Fluorescence System working solution for 4 minutes at room temperature (PerkinElmer, SAT701001EA). After 4 washes with 0.1% Tween 20 in PBS (5 minutes each), slides were counterstained with DAPI (4 μg/mL) and mounted with Vectashield mounting medium (Vector Laboratories, H-1000). Images were taken using an inverted Olympus IX81 microscope and MetaMorph software, set to grayscale and the fluorescence intensity at a background-excluding threshold using ImageJ (NIH). HA-REV-ERBα–positive cells were quantified with the Image-based Tool for Counting Nuclei (ITCN) plugin in ImageJ on each of area of interest and normalized to DAPI-positive nuclei. During image capture and analysis, the investigator was blinded to genotype and time point.
FISH of ARC Fabp7.
For quantification of ARC Fabp7 mRNA expression, brains were rapidly removed and fixed in 4% PFA in 0.1 M PBS (pH 7.4) overnight at 4°C and then transferred to a 30% sucrose solution in 0.1 M PBS. Subsequently, brains were sectioned on a cryostat in the coronal plane at 14 μm thickness and collected on Superfrost Plus slides (VWR) in 3 series of slides, each slide containing 4 brain sections. To identify the slices of interest, Nissl staining was performed on serial section 1, as described for HA staining. Sections taken through the rostral-caudal extent of the ARC were stored at –80°C until further analysis. FISH was performed using the RNAscope Multiplex Fluorescent Reagent kit v2 (ACDBio, 323110) per the manufacturer’s instructions. Detection was carried out using probes designed by ACDBio for Fabp7 mRNA (Mm-Fabp7-C2, 414651-C2). Following a series of amplification steps, sections were mounted with Fluorogel DAPI-containing mounting media (Thermo Fischer Scientific). Sections were visualized using a Keyence BZ-X800 fluorescence microscope under a 40× objective. Quantification was done using ImageJ software. Images were set to grayscale and the fluorescence intensity at a background-excluding threshold within selections of the ARC was analyzed. The left and right ARC from at least 2 sections per sample were quantified and averaged. Data are presented as raw integrated density/area (mm2). During image capture and analysis, the investigator was blinded to genotype and time point.
In vivo metabolic phenotyping.
Mice were housed individually in a Comprehensive Lab Animal Monitoring System (CLAMS) (Columbus Instruments) under 12-hour light/12-hour dark conditions for 5 consecutive days. Experimental data were obtained for 3 light/dark cycles after 2 days of adaptation period. The rate of O2 (VO2) and CO2 consumption, respiratory-exchange ratio, heat production, and locomotor activity of each mouse was recorded every 15 minutes sequentially and the mean/hour is presented. Data acquisition and analysis were performed with the Oxymax (Columbus Instruments) and GraphPad Prism software.
Whole-animal energy expenditure in response to norepinephrine.
Oxygen-consumption rates were measured using the CLAMS. As previously described (65), mice were anesthetized with 75 mg/kg pentobarbital (Nembutal) and placed into CLAMS cages preacclimated to 30°C. A subcutaneous injection of 1 mg/kg L-(−)-norepinephrine (+)-bitartrate salt monohydrate (MilliporeSigma, A9512) was performed in the dorsal nuchal region and oxygen consumption rates were recorded until rates began to decline.
Core-body temperature measurements.
Mice were singly housed in climate-controlled rodent incubators (Powers Scientific) maintained at 22°C with free access to food and water. Core-body temperatures were recorded using a rectal thermocouple probe (Physitemp, RET-3 probe) and a digital thermometer (Oakton Instruments, Temp 10T Thermocouple).
Hormonal measurements.
Hormonal measurements were performed in mouse serum using an ultrasensitive ELISA kit for insulin (Crystal Chem, 90080), leptin (MilliporeSigma, EZML-82K), T3 (Alpha Diagnostic, 1700), and T4 (Alpha Diagnostic, 1100) according to the manufacturers’ instructions.
Feeding records.
Mice were singly housed in climate-controlled rodent incubators (Powers Scientific) maintained at 22°C with free access to food and water for 1 week for acclimation. Food intake was measured, including any crumbled food present at the bottom of the cages. If the amount of crumbled food was too great (>1 g/12 hours), the measurement was not included in food intake analyses.
Refeeding and leptin experiments.
For refeeding experiments, mice were singly housed in climate-controlled rodent incubators (Powers Scientific) maintained at 22°C with free access to food and water for 1 week for acclimation. Twenty-four-hour fasting was used to be able to compare refeeding during the light phase at ZT1 and the dark phase at ZT13, as previously described (42). After 24-hour fasting (from ZT0 or ZT12), mice were refed at ZT1 or ZT13, with NCD or HFD, and food consumption was measured for 3 hours. For leptin experiments, mice were injected with PBS or recombinant mouse leptin (National Hormone and Peptide Program) by intraperitoneal injection at 10 mg/kg 10 minutes before refeeding and food consumption was measured after 1 hour. For gene expression analysis, mice were harvested 1 hour after refeeding. All the experiments were carried out in time windows of 3 hours from ZT1 to ZT4 or ZT13 to ZT16.
p-STAT3 staining in the ARC in response to leptin.
For quantification of ARC p-STAT3, the same protocol described for HA staining was used. Sections taken through the rostral-caudal extent of the ARC were stained for p-STAT3 (Cell Signaling Technology, D3A7; 1:100) and then visualized using a Keyence BZ-X710 fluorescent microscope under a 10× objective. Quantification was done using ImageJ software. Images were set to grayscale and the fluorescence intensity at a background-excluding threshold. p-STAT3–positive cells were quantified in each area of interest and normalized to DAPI-positive nuclei. During image capture and analysis, the investigator was blinded to genotype and treatment.
Hypothalamic nuclei microdissection.
This technique was performed as previously described (66) with modifications. All tools used for microdissection were treated with DEPC solution (MilliporeSigma, D5758) overnight at 37°C (0.1 mL/100 mL distilled H2O). To maintain tissue viability during the dissection, Earle’s balanced salt solution (EBSS) (Thermo Fisher Scientific, 14155063) supplemented with sodium bicarbonate (0.44 g/100 mL EBSS) and glucose (0.884 g/100 mL EBSS) was used. EBSS was treated with DEPC overnight at 37°C (0.1 mL/100 mL EBSS) and maintained at 37°C with 95% O2/5% CO2 during the experiment. The whole brain was removed, rinsed with oxygenated EBSS medium, and placed ventral side up on a TC-1 Tissue Chopper (Electron Microscopy Sciences, 93100) to facilitate visual orientation of the structures. The frontal end of the hypothalamus was set as the anatomical origin, and the Allen Brain Atlas (https://portal.brain-map.org/) was used to determine the coordinate of the structures of interest and their spatial relations to the anatomical origin. The sections were placed in oxygenated EBSS in a Petri dish and micropunches of the SCN, ARC, and V-DMH were performed with 1-mm and 2-mm tissue corers (Fine Science Tools, 18035). Importantly, to preserve gene expression, the procedure was performed in less than 5 minutes. To validate the separate dissections of individual hypothalamic nuclei, the expression of VIP (SCN-specific), Rfrp (V-DMH–specific), and Pomc1 (ARC-specific) was confirmed by RT-qPCR (Supplemental Figure 1B).
RNA isolation.
Two to 4 independent hypothalamic nuclei punches were pooled together per biological replicate. Hypothalamic punches were homogenized manually using a Pellet-Pestle (Kimble, 749515) for 1 minute with 1 mL QIAzol Lysis Reagent (QIAGEN). Total RNA was purified and collected from RNeasy Mini spin columns according to the RNeasy Lipid Mini Kit (QIAGEN, 74804). Total RNA was isolated from snap-frozen brown adipose tissue, WAT, and liver, which were mechanically homogenized in TRIzol (Life Technologies) using a TissueLyser (QIAGEN) for 2 minutes. Following RNA extraction with chloroform, mRNA was further purified with RNeasy Mini spin columns and on-column DNase digestion (QIAGEN).
Gene expression analysis (RT-qPCR).
Complementary DNA was made from purified RNA with a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, 43-688-14). cDNA was used for quantitative PCR with a Power SYBR Green PCR Master Mix (Applied Biosystems) on the QuantStudio 6 Flex Real-Time PCR software and system. Results were analyzed by standard curve and normalized to Tbp or 36B4 (Arbp) expression. Primer sequences are listed in Supplemental Table 1.
RNA-seq library preparation and data analysis.
For RNA-seq, RNA integrity was examined using Agilent High Sensitivity RNA ScreenTape. RNA samples (1 μg) with RNA integrity number greater than 7 were used for RNA cleanup and library preparation with an Illumina TruSeq Stranded Total RNA Library Prep kit according to the manufacturer’s instructions. All barcoded libraries were quantified by KAPA Library Quantification Kit (Roche), and equimolarly pooled for subsequent sequencing. All RNA-seq experiments were done with at least 3 biological replicates per condition. High-throughput sequencing data were generated through either the Functional Genomics Core at the University of Pennsylvania or Novagene. Sequencing reads were aligned to UCSC mm10 genome using STAR v 2.6 (67). Read counts were then obtained with featureCounts per the manual’s instructions (68) (Supplemental Table 2). Differentially expressed genes (cutoff defined as FDR < 0.05, transcripts per million reads [TPM] > 0.1) were identified using DESeq2 (69). Heatmaps were generated in R with package pheatmap by (i) identifying differentially expressed genes with the aforementioned cutoffs and (ii) mapping their corresponding z-transformed TPM or log2(fold change) values across all biological replicates and treatment conditions. The heatmap color scheme was scaled accordingly for optimal graphical illustrations. Gene ontology analyses and TF enrichment were performed by Enrichr webserver (32). Gene set enrichment analyses were performed with GSEA software (70) with the mouse leptin gene signaling pathway curated from WikiPathways (https://www.wikipathways.org/index.php/WikiPathways).
ChIP.
Ten independent hypothalamic nuclei punches were pooled together. Hypothalamic nuclei punches were directly thawed in cross-linking solution for 20 minutes at room temperature (1% formaldehyde in 1× PBS), quenched with 0.125 M glycine for 5 minutes, and washed 3 times with 1× PBS at 4°C. Cross-linked hypothalamic nuclei punches were resuspended in ChIP dilution buffer (50 mM HEPES pH 7.5, 155 mM NaCl, 1 mM EDTA, 1.1% Triton X-100, 0.11% sodium deoxycholate, 0.1% SDS) on ice and resuspended during chromatin fragmentation with a Sonic Dismembrator (Thermo Fischer Scientific, FB705) at 4°C: 3 cycles at 10% amplitude, 3 cycles at 15% amplitude. Lipid-cleared samples were resuspended in 1 mL of ChIP dilution buffer and input was saved before incubation with BSA-blocked HA magnetic beads (Pierce, 888367) overnight at 4°C with rotation. Immunoprecipitates were washed with rotation at 4°C with ChIP dilution buffer twice (1 and 5 minutes each), ChIP dilution buffer with 500 mM NaCl (5 minutes), ChIP wash buffer (5 minutes; 10 mM Tris-HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA) and Tris-EDTA buffer (5 minutes; 10 mM Tris-HCl pH 8.0, 1 mM EDTA). Cross-linking was reversed overnight at 65°C in elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) and DNA was isolated using phenol/chloroform and precipitated by NaCl/EtOH overnight at –80°C with 20 μg of glycogen carrier (Thermo Fisher Scientific, AM9515).
ChIP-seq library preparation and data analysis.
ChIP DNA was prepared for sequencing according to the amplification protocol provided by New England Biolabs. Next-generation sequencing of ChIP-seq libraries was performed by the Functional Genomics Core at the University of Pennsylvania. Sequencing reads were aligned to the UCSC mm10 genome using Bowtie2 (71) with the -N 1 option to remove PCR duplicates (Supplemental Table 2). All samples were normalized to 2 × 107 reads. Downstream peak calling and processing were done with HOMER v.4.6. Peaks are defined as RPM greater than 1, fold change IP/input greater than 7, and FDR less than 0.0001. De novo motif analyses were performed with HOMER function findMotifsGenome.pl. To identify REV-ERB binding sites near target genes, bedtools window (72) was used with default parameters. REV-ERB–regulated genes with either REV-ERB or STAT3 motifs were identified with HOMER function annotatepeak.pl with -m option.
Statistics
Data are presented as mean ± SEM. Graphing and statistical analysis were performed using GraphPad Prism. No statistical test was performed to predetermine sample size. As described in figure legends, statistical analyses were performed using the Mann-Whitney test for comparisons between 2 groups, and 1-way analysis of variance (ANOVA) and Holm-Šidák multiple-comparison test for comparisons between more than 2 groups. Repeated measures 2-way ANOVA or 2-way ANOVA were performed for assessment of variable effects (time, diet, treatment, genotype) followed by Holm-Šidák multiple-comparison test (*P < 0.05, **P < 0.01, ***P < 0.001). For repeated measures 2-way ANOVA or 2-way ANOVA analysis, the interaction P value is reported in the figure legend when not significant, as well as the genotype-effect P value when relevant. RStudio (v1.0.153) software was used for graphing and statistical analysis of sequence data.
Data availability
The data sets generated during this study are available at GSE148644 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148644).
Study approval
All animal studies were approved by the University of Pennsylvania Perelman School of Medicine Institutional Animal Care and Use Committee.
Author contributions
MA and MAL designed the research. MA, HCBN, BMK, CWT, CEG, LCP, and BJC performed the research. HCBN performed bioinformatics analysis. MA, HCBN, CEG, MRH, and MAL analyzed dat. MA, HCBN, and MAL wrote the manuscript.
Supplementary Material
Acknowledgments
We gratefully acknowledge L. Woodie for critically reading the manuscript. We thank L. Cheng from the Penn Molecular Pathology and Imaging Core for help with histology, as well as the Gene Expression and Functional Genomics Core of the Penn Diabetes Research Center (DK19525). This work was supported by NIH R01DK45586, the JPB Foundation, and the Cox Institute for Medical Research (to MAL). MA was supported by an American Diabetes Association Training grant (1-18-PDF-126). MRH was supported by NIH grant R01DKDK021397.
Version 1. 10/06/2020
In-Press Preview
Version 2. 01/04/2021
Print issue publication
Funding Statement
to M.A.L.
to M.A.L.
to M.A.L.
to M.A.
to M.R.H.
Footnotes
Conflict of interest: MAL is an advisory board member for Eli Lilly and Pfizer Inc., consultant to Novartis, and receives research support from Pfizer. MRH has received research support from investigator-initiated sponsored proposals from Eli Lilly & Co. and Boehringer-Ingelheim.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e140424.https://doi.org/10.1172/JCI140424.
See the related Commentary at You are when you eat: on circadian timing and energy balance.
Contributor Information
Marine Adlanmerini, Email: marinea@pennmedicine.upenn.edu.
Brianna M. Krusen, Email: brianna.krusen@pennmedicine.upenn.edu.
Clare W. Teng, Email: wanming.teng@pennmedicine.upenn.edu.
Caroline E. Geisler, Email: Caroline.Geisler@pennmedicine.upenn.edu.
Lindsey C. Peed, Email: lindseycpeed@gmail.com.
Bryce J. Carpenter, Email: bryce.carpenter@pennmedicine.upenn.edu.
Matthew R. Hayes, Email: hayesmr@pennmedicine.upenn.edu.
Mitchell A. Lazar, Email: lazar@mail.med.upenn.edu.
References
- 1.Biggi N, Consonni D, Galluzzo V, Sogliani M, Costa G. Metabolic syndrome in permanent night workers. Chronobiol Int. 2008;25(2):443–454. doi: 10.1080/07420520802114193. [DOI] [PubMed] [Google Scholar]
- 2.Peplonska B, Bukowska A, Sobala W. Association of rotating night shift work with BMI and abdominal obesity among nurses and midwives. PLoS One. 2015;10(7):e0133761. doi: 10.1371/journal.pone.0133761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koshiyama H, Hamamoto Y, Honjo S, Wada Y, Lkeda H. Hypothalamic pathogenesis of type 2 diabetes. Med Hypotheses. 2006;67(2):307–310. doi: 10.1016/j.mehy.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 4.Cedernaes J, Waldeck N, Bass J. Neurogenetic basis for circadian regulation of metabolism by the hypothalamus. Genes Dev. 2019;33(17–18):1136–1158. doi: 10.1101/gad.328633.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greco CM, Sassone-Corsi P. Circadian blueprint of metabolic pathways in the brain. Nat Rev Neurosci. 2019;20(2):71–82. doi: 10.1038/s41583-018-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myers MG, Jr , Olson DP. Central nervous system control of metabolism. Nature. 2012;491(7424):357–363. doi: 10.1038/nature11705. [DOI] [PubMed] [Google Scholar]
- 7.Timper K, Brüning JC. Hypothalamic circuits regulating appetite and energy homeostasis: pathways to obesity. Dis Model Mech. 2017;10(6):679–689. doi: 10.1242/dmm.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roh E, Song DK, Kim MS. Emerging role of the brain in the homeostatic regulation of energy and glucose metabolism. Exp Mol Med. 2016;48:e216. doi: 10.1038/emm.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett LJ, Lazar MA. Nuclear receptor Rev-erbα: up, down, and all around. Trends Endocrinol Metab. 2014;25(11):586–592. doi: 10.1016/j.tem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bugge A, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26(7):657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerhart-Hines Z, et al. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature. 2013;503(7476):410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delezie J, et al. The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26(8):3321–3335. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- 14.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 15.Mang GM, et al. Altered sleep homeostasis in rev-erbα knockout mice. Sleep. 2016;39(3):589–601. doi: 10.5665/sleep.5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jager J, et al. Behavioral changes and dopaminergic dysregulation in mice lacking the nuclear receptor Rev-erbα. Mol Endocrinol. 2014;28(4):490–498. doi: 10.1210/me.2013-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung S, et al. Impact of circadian nuclear receptor REV-ERBα on midbrain dopamine production and mood regulation. Cell. 2014;157(4):858–868. doi: 10.1016/j.cell.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 18.Amador A, Wang Y, Banerjee S, Kameneka TM, Solt LA, Burris TP. Pharmacological and genetic modulation of rev-erb activity and expression affects orexigenic gene expression. PLoS One. 2016;11(3):e0151014. doi: 10.1371/journal.pone.0151014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feillet CA, et al. Rev-erbα modulates the hypothalamic orexinergic system to influence pleasurable feeding behaviour in mice. Addict Biol. 2017;22(2):411–422. doi: 10.1111/adb.12339. [DOI] [PubMed] [Google Scholar]
- 20.Orozco-Solis R, Ramadori G, Coppari R, Sassone-Corsi P. SIRT1 relays nutritional inputs to the circadian clock through the Sf1 neurons of the ventromedial hypothalamus. Endocrinology. 2015;156(6):2174–2184. doi: 10.1210/en.2014-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe M, et al. Circadian rhythms in isolated brain regions. J Neurosci. 2002;22(1):350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilding C, Hughes AT, Brown TM, Namvar S, Piggins HD. A riot of rhythms: neuronal and glial circadian oscillators in the mediobasal hypothalamus. Mol Brain. 2009;2:28. doi: 10.1186/1756-6606-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adlanmerini M, et al. Circadian lipid synthesis in brown fat maintains murine body temperature during chronic cold. Proc Natl Acad Sci U S A. 2019;116(37):18691–18699. doi: 10.1073/pnas.1909883116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dierickx P, et al. SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc Natl Acad Sci U S A. 2019;116(25):12147–12152. doi: 10.1073/pnas.1904226116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mieda M, Hasegawa E, Kessaris N, Sakurai T. Fine-tuning circadian rhythms: the importance of Bmal1 expression in the ventral forebrain. Front Neurosci. 2017;11:55. doi: 10.3389/fnins.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilbertz TM, Meier S, Perner S. NKX2-1 (NK2 homeobox 1) Atlas Genet Cytogenet Oncol Haematol. 2011;15(1):19–28. [Google Scholar]
- 27.Lazar MA, Hodin RA, Darling DS, Chin WW. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol Cell Biol. 1989;9(3):1128–1136. doi: 10.1128/MCB.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rindfleisch BC, Brown MS, VandeBerg JL, Munroe SH. Structure and expression of two nuclear receptor genes in marsupials: insights into the evolution of the antisense overlap between the α-thyroid hormone receptor and Rev-erbα. BMC Mol Biol. 2010;11:97. doi: 10.1186/1471-2199-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park HK, Ahima RS. Leptin signaling. F1000Prime Rep. 2014;6:73. doi: 10.12703/P6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Rui L. Leptin signaling and leptin resistance. Front Med. 2013;7(2):207–222. doi: 10.1007/s11684-013-0263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghilardi N, Ziegler S, Wiestner A, Stoffel R, Heim MH, Skoda RC. Defective STAT signaling by the leptin receptor in diabetic mice. Proc Natl Acad Sci U S A. 1996;93(13):6231–6235. doi: 10.1073/pnas.93.13.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasumoto Y, et al. Glial fatty acid-binding protein 7 (FABP7) regulates neuronal leptin sensitivity in the hypothalamic arcuate nucleus. Mol Neurobiol. 2018;55(12):9016–9028. doi: 10.1007/s12035-018-1033-9. [DOI] [PubMed] [Google Scholar]
- 35.Bjorbak C, et al. SOCS3 mediates feedback inhibition of the leptin receptor via Tyr985. J Biol Chem. 2000;275(51):40649–40657. doi: 10.1074/jbc.M007577200. [DOI] [PubMed] [Google Scholar]
- 36.Zabolotny JM, et al. PTP1B regulates leptin signal transduction in vivo. Dev Cell. 2002;2(4):489–495. doi: 10.1016/S1534-5807(02)00148-X. [DOI] [PubMed] [Google Scholar]
- 37.Dodd GT, et al. Intranasal targeting of hypothalamic PTP1B and TCPTP reinstates leptin and insulin sensitivity and promotes weight loss in obesity. Cell Rep. 2019;28(11):2905–2922.e5. doi: 10.1016/j.celrep.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Challet E. The circadian regulation of food intake. Nat Rev Endocrinol. 2019;15(7):393–405. doi: 10.1038/s41574-019-0210-x. [DOI] [PubMed] [Google Scholar]
- 39.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherji A, et al. Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc Natl Acad Sci U S A. 2015;112(48):E6691–E6698. doi: 10.1073/pnas.1519807112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kettner NM, Mayo SA, Hua J, Lee C, Moore DD, Fu L. Circadian dysfunction induces leptin resistance in mice. Cell Metab. 2015;22(3):448–459. doi: 10.1016/j.cmet.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cedernaes J, et al. Transcriptional basis for rhythmic control of hunger and metabolism within the AgRP neuron. Cell Metab. 2019;29(5):1078–1091.e5. doi: 10.1016/j.cmet.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shea SA, Hilton MF, Orlova C, Ayers RT, Mantzoros CS. Independent circadian and sleep/wake regulation of adipokines and glucose in humans. J Clin Endocrinol Metab. 2005;90(5):2537–2544. doi: 10.1210/jc.2004-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalsbeek A, et al. The suprachiasmatic nucleus generates the diurnal changes in plasma leptin levels. Endocrinology. 2001;142(6):2677–2685. doi: 10.1210/endo.142.6.8197. [DOI] [PubMed] [Google Scholar]
- 46.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4(2):e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikeda R, et al. REV-ERBα and REV-ERBβ function as key factors regulating mammalian circadian output. Sci Rep. 2019;9(1):10171. doi: 10.1038/s41598-019-46656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibbs JE, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. 2012;109(2):582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu L, et al. Estrogens prevent metabolic dysfunctions induced by circadian disruptions in female mice. Endocrinology. 2015;156(6):2114–2123. doi: 10.1210/en.2014-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmisano BT, Stafford JM, Pendergast JS. High-fat feeding does not disrupt daily rhythms in female mice because of protection by ovarian hormones. Front Endocrinol (Lausanne) 2017;8:44. doi: 10.3389/fendo.2017.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Omotola O, Legan S, Slade E, Adekunle A, Pendergast JS. Estradiol regulates daily rhythms underlying diet-induced obesity in female mice. Am J Physiol Endocrinol Metab. 2019;317(6):E1172–E1181. doi: 10.1152/ajpendo.00365.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Q, Horvath TL. Cross-talk between estrogen and leptin signaling in the hypothalamus. Am J Physiol Endocrinol Metab. 2008;294(5):E817–E826. doi: 10.1152/ajpendo.00733.2007. [DOI] [PubMed] [Google Scholar]
- 53.Chen N, et al. Direct modulation of GFAP-expressing glia in the arcuate nucleus bi-directionally regulates feeding. Elife. 2016;5:e18716. doi: 10.7554/eLife.18716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JG, et al. Leptin signaling in astrocytes regulates hypothalamic neuronal circuits and feeding. Nat Neurosci. 2014;17(7):908–910. doi: 10.1038/nn.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.García-Cáceres C, Fuente-Martín E, Argente J, Chowen JA. Emerging role of glial cells in the control of body weight. Mol Metab. 2012;1(1–2):37–46. doi: 10.1016/j.molmet.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stein LM, et al. Dorsal vagal complex and hypothalamic glia differentially respond to leptin and energy balance dysregulation. Transl Psychiatry. 2020;10(1):90. doi: 10.1038/s41398-020-0767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bouyakdan K, et al. The gliotransmitter ACBP controls feeding and energy homeostasis via the melanocortin system. J Clin Invest. 2019;129(6):2417–2430. doi: 10.1172/JCI123454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Djogo T, et al. Adult NG2-glia are required for median eminence-mediated leptin sensing and body weight control. Cell Metab. 2016;23(5):797–810. doi: 10.1016/j.cmet.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 59.Minocha S, et al. Nkx2.1-derived astrocytes and neurons together with Slit2 are indispensable for anterior commissure formation. Nat Commun. 2015;6:6887. doi: 10.1038/ncomms7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minocha S, et al. Nkx2.1 regulates the generation of telencephalic astrocytes during embryonic development. Sci Rep. 2017;7:43093. doi: 10.1038/srep43093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orquera DP, Tavella MB, de Souza FSJ, Nasif S, Low MJ, Rubinstein M. The homeodomain transcription factor NKX2.1 is essential for the early specification of melanocortin neuron identity and activates Pomc expression in the developing hypothalamus. J Neurosci. 2019;39(21):4023–4035. doi: 10.1523/JNEUROSCI.2924-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olofsson LE, Unger EK, Cheung CC, Xu AW. Modulation of AgRP-neuronal function by SOCS3 as an initiating event in diet-induced hypothalamic leptin resistance. Proc Natl Acad Sci U S A. 2013;110(8):E697–E706. doi: 10.1073/pnas.1218284110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boucsein A, Rizwan MZ, Tups A. Hypothalamic leptin sensitivity and health benefits of time-restricted feeding are dependent on the time of day in male mice. FASEB J. 2019;33(11):12175–12187. doi: 10.1096/fj.201901004R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mori H, et al. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nat Med. 2004;10(7):739–743. doi: 10.1038/nm1071. [DOI] [PubMed] [Google Scholar]
- 65.Cannon B, Nedergaard J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. J Exp Biol. 2011;214(pt 2):242–253. doi: 10.1242/jeb.050989. [DOI] [PubMed] [Google Scholar]
- 66.Atkins N, Miller CM, Owens JR, Turek FW. Non-laser capture microscopy approach for the microdissection of discrete mouse brain regions for total RNA isolation and downstream next-generation sequencing and gene expression profiling. J Vis Exp. 2011;2011(57):3125. doi: 10.3791/3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated during this study are available at GSE148644 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148644).