Abstract
As the interface between the gut microbiota and the mucosal immune system, there has been great interest in the maintenance of colonic epithelial integrity through mitochondrial oxidation of butyrate, a short-chain fatty acid produced by the gut microbiota. Herein, we showed that the intestinal epithelium could also oxidize long-chain fatty acids, and that luminally delivered acylcarnitines in bile could be consumed via apical absorption by the intestinal epithelium, resulting in mitochondrial oxidation. Finally, intestinal inflammation led to mitochondrial dysfunction in the apical domain of the surface epithelium that may reduce the consumption of fatty acids, contributing to higher concentrations of fecal acylcarnitines in murine Citrobacter rodentium–induced colitis and human inflammatory bowel disease. These results emphasized the importance of both the gut microbiota and the liver in the delivery of energy substrates for mitochondrial metabolism by the intestinal epithelium.
Keywords: Gastroenterology, Inflammation
Keywords: Fatty acid oxidation, Inflammatory bowel disease, Mitochondria
Introduction
The intestinal epithelial barrier, along with specialized cells responsible for the secretion of mucus, antimicrobial peptides, and enteric hormones, is constantly renewed every 4–5 days, making the intestinal epithelium the most rapidly proliferating cell type in adult animals. This rapid turnover is controlled by an intestinal stem cell population at the base of the intestinal crypts, known as crypt base cells, giving rise to transit amplifying cells that rapidly divide 4–5 times before becoming terminally differentiated (1). The regeneration of the intestinal epithelium is particularly critical in the setting of intestinal inflammation, where hypertrophy of the proliferating crypt compartment is observed. Wnt signaling is a critical pathway that regulates intestinal epithelial progenitor cell proliferation, as well as pathways involved in cellular metabolism that are critical for cell growth and proliferation, where glucose metabolism is essential to support rapid cellular proliferation in both nontransformed and tumor cells (2).
By contrast, fatty acid oxidation (FAO), principally of short-chain fatty acids (SCFAs) produced by the gut microbiota, is widely believed to be the primary source of metabolic fuel for the colonic epithelium. Acetate, propionate, and butyrate are SCFAs produced by gut microbiota through fermentation of undigested carbohydrates. They reach millimolar concentrations in the colonic lumen. After they are absorbed by colonocytes via the monocarboxylate transporter 1 (3) and passive diffusion, SCFAs (particularly butyrate) are oxidized by the colonic epithelium, accounting for 70% to 90% of energy and 70% of oxygen consumption (4–7). The observation that colonocytes in germ-free mice show evidence of autophagy pathway activation relative to colonized mice supports the importance of SCFAs as a source of energy for the colonocyte (8).
Butyrate is not only a substrate for energy production, but it also has epigenetic effects through its ability to inhibit histone deacetylases (HDACs), leading to histone hyperacetylation and alterations in gene expression (9). In the colonic epithelium, these effects are determined by the status of cellular differentiation (10) and whether or not the cells have undergone neoplastic transformation (8). In the former, mitochondrial beta-oxidation of butyrate by the differentiated surface cells of the colonic epithelium prevents the inhibition of HDACs and impairment of cellular proliferation by stem cells in the colonic crypt (10). In the latter, aerobic glycolysis in neoplastic colonic cells via the “Warburg effect” reduces mitochondrial beta-oxidation of butyrate, making it available for HDAC inhibition, leading to epigenetic effects and the inhibition of cellular proliferation (11). Thus, because of the high levels of butyrate production by the gut microbiota, mitochondrial beta-oxidation of this SCFA plays a unique tissue-specific role in the biology of the colonic epithelium.
Alterations in butyrate metabolism by the colonic epithelium are also associated with colonic inflammation. It has been more than 2 decades since Roediger proposed that deficits in the oxidation of SCFAs by the colonic epithelium may play a role in the pathogenesis of inflammatory bowel disease (IBD), particularly ulcerative colitis (UC) (5). Indeed, pharmacological inhibitors of FAO lead to the development of colitis (12), and reduction of colonic SCFAs by diversion of the fecal stream lead to the development of diversion colitis, an inflammatory disease that can be treated by instillation of SCFAs by enema (13). The identification of the SLC22A5 gene encoding OCTN2, a sodium-dependent L-carnitine transporter critical for FAO, as a Crohn’s disease–associated (CD-associated) genomic locus provides support for the notion that mitochondria may play a role in IBD pathogenesis via alterations in FAO (14). Nevertheless, the mechanisms for this deficit remain poorly defined, and various explanations have been proposed, such as reductions in the expression of specific oxidative pathways (5, 15, 16).
Although the regulation of mitochondrial butyrate beta-oxidation by the colonic epithelium is central to all of these biological effects, a systematic characterization of mitochondrial metabolic function in the colonic epithelium has not been previously described. Herein, we showed that the colonic epithelium could oxidize SCFAs and long-chain fatty acids (LCFAs), and the relationship between the two was substrate-dependent and inversely related. Inhibition of SCFA oxidation in short-chain acyl-CoA dehydrogenase–deficient (SCAD-deficient) mice led to greater long-chain FAO, whereas the addition of butyrate to colonic explant cultures led to a suppression of long-chain FAO. However, both long- and short-chain FAO were reduced in the setting of colitis induced by infection with Citrobacter rodentium, providing evidence for a more global mitochondrial dysfunction where both the number and function of colonic mitochondria were reduced. Colonic mitochondria were dysmorphic and reduced in number throughout the surface colonic epithelium in both C. rodentium–induced murine colitis and in human IBD.
Concurrent with our observation that the colonic epithelium can oxidize SCFAs and LCFAs, we showed that high levels of medium- and long-chain acylcarnitines, fatty acid intermediates needed for transport into mitochondria, were delivered at high concentrations into the gut lumen via biliary secretions. Based on in vitro modeling, we showed that they were taken up along the apical side of the colonic epithelium and metabolized via mitochondrial oxidation. Reduction of intestinal epithelial mitochondrial number and function in C. rodentium colitis and in human IBD may contribute to the observed higher levels of fecal acylcarnitines that serve as a biomarker for IBD in humans.
In total, these observations provide evidence for the dynamic regulation of colonocyte mitochondrial FAO in health and how it is disrupted in the setting of colonic inflammation, where there can be a reduction in gut microbiota–produced energy substrates for epithelial metabolism, such as butyrate, and in host-derived metabolites like medium- and long-chain acylcarnitines. These findings provide insights into the growing interest in the multifaceted interaction between the host- and gut microbiota–derived molecules for metabolic homeostasis at the intestinal epithelial interface in general, and the role for epithelial mitochondrial function in the pathogenesis of IBD in particular.
Results
Both long- and short-chain fatty acids are oxidized in the colon, where there is an inverse relationship between the two regulated by the abundance of the SCFA butyrate.
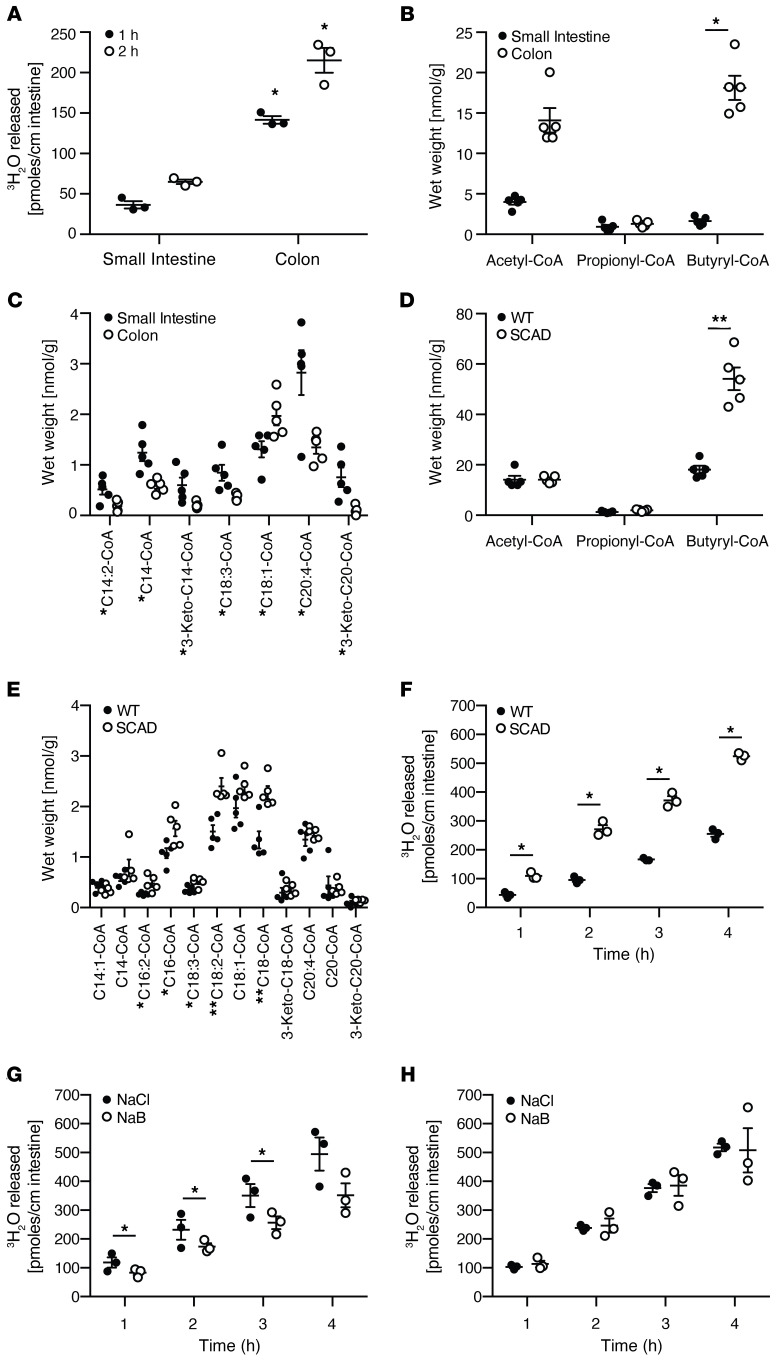
Long-chain FAO was quantified ex vivo using an intestinal explant culture system whereby an intestinal segment was inverted, placing the intestinal epithelium in contact with the assay medium to minimize the metabolic contribution of nonepithelial cells to this assay. The oxidation of tritiated oleic acid led to the time-dependent accumulation of tritiated water that remained linear, indicating the viability of the explant tissues over several hours. FAO of oleic acid was very robust in the colon, where it was approximately 5- to 10-fold greater than in the small intestine (Figure 1A). After transport into the mitochondria, fatty acids are conjugated to coenzyme A–generating acyl-CoAs that serve as substrates for beta-oxidation. Profiling of acyl-CoAs in various tissues by tandem mass spectrometry has been used to provide a static representation of FAO intermediates in the liver (17). To broadly quantify the spectrum of FAO intermediates, acyl-CoAs were quantified in both the small and large intestine of mice. Short-chain acyl-CoAs predominated in the colon relative to the small intestine (Figure 1B), reflecting the abundance of SCFAs produced by the gut microbiota in the colon. By contrast, long-chain acyl-CoAs were lower in the colon compared with the small intestine (Figure 1C).
Figure 1. The reciprocal relationship between short- and long-chain fatty oxidation by the colonic epithelium.
(A) Quantification of oleic acid oxidation using a tritiated water release assay in inverted colonic and small intestinal mouse explant cultures harvested from mice. n = 3, mean ± SEM, *P < 0.05. Representative results of at least 2–3 independent experiments. (B–E) Quantification of the fatty acid oxidation intermediates, acyl-CoAs, by mass spectrometry in mouse intestinal tissues. (B) Short- and (C) long-chain acyl-CoAs in the small intestine and colon. (D) Short- and (E) long-chain acyl-CoAs in the colon of wild-type versus SCAD-deficient mice. Representative results of at least 3 experiments. n = 5, mean ± SEM, *P < 0.05, **P < 0.01. (F) Quantification of oleic acid oxidation using the tritiated water release assay in colonic explants isolated from wild-type versus SCAD-deficient mice. n = 3, mean ± SEM, *P < 0.05. The assays were repeated in the presence or absence of 10 mM sodium butyrate in using colonic explants isolated from (G) wild-type or (H) SCAD-deficient mice. n = 3, mean ± SEM, *P < 0.05. Representative results of 2 independent experiments. P values represent a 2-tailed Student’s t test and paired-sample t test.
The inverse relationship between short-chain and long-chain acyl-CoAs in the small versus large intestine might reflect alternative fuel utilization by the intestinal epithelium due to substrate abundance. For example, such a functional effect would be consistent with the example of the reciprocal relationship between glycolysis versus FAO regulated by cellular energy status via AMPK (18). Because of its juxtaposition with gut microbiota–dependent production of SCFAs, the colonic epithelium may be the only cell type in the body that might demonstrate alternative fuel utilization based on the chain length of fatty acid substrate. To explore this notion, we examined the spectrum of acyl-CoAs in the SCAD-deficient mouse. SCAD deficiency is the result of a mutation in the Acads gene, leading to a block of the first step in the FAO pathway for C4–C6 SCFAs (19, 20). The BALB/cByJ mouse strain, a murine model of the SCAD deficiency (21), has an autosomal recessive mutation of Acads and has recently been shown to have a reduction in crypt cell proliferation (10). We observed a significant increase in butyryl-CoA in the colon of SCAD-deficient mice (Figure 1D) due to a defect in the first step of the FAO pathway, a finding similar to that reported in liver tissue of these mice (17). There was a general increase in long-chain acyl-CoAs in the colon of these mice relative to wild-type mice (Figure 1E). Based on this acyl-CoA profile, we quantified oleic acid oxidation via tritiated water release in the colon of SCAD-deficient mice and their wild-type counterparts. We observed a significant increase in the oxidation of oleic acid in the colonic explants of SCAD-deficient mice (Figure 1F). Since the bioenergetic cellular state regulates the choice of energy substrates through mechanisms that involve AMPK (22), we determined whether SCFA oxidation inhibits long-chain FAO in the colonic epithelium. Indeed, there was an inhibitory effect of butyrate short-chain FAO on oleic acid long-chain FAO in the colon of wild-type mice (Figure 1G) but not in the SCAD-deficient mice (Figure 1H). These results demonstrated that FAO of SCFAs and LCFAs in the colonic epithelium was inversely related.
High concentrations of acylcarnitines are delivered to the gut lumen via biliary secretions, where intestinal epithelial modeling shows that they can be absorbed apically and utilized for mitochondrial oxidative metabolism.
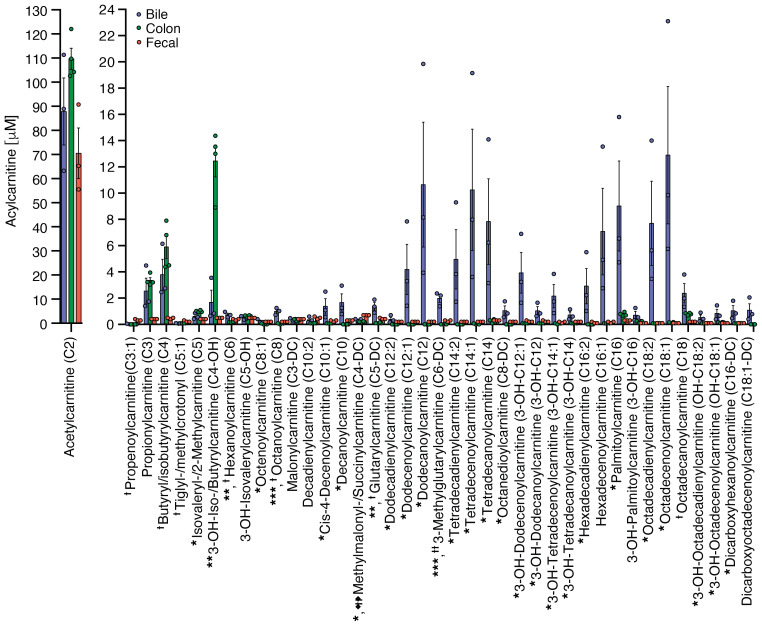
Having provided evidence for long-chain FAO in colonic explants and high levels of acyl-CoAs in colonic tissue (Figure 1), we postulated that the host can produce endogenous LCFAs that could also be utilized by colonic epithelium for energy production. Medium- and long-chain fatty acids are esterified to carnitine to produce acylcarnitines so that they can be transported into the mitochondria (23). The cellular secretion of acylcarnitines may not only be a detoxification mechanism: when excreted into the plasma, growing evidence, reviewed by McCoin et al., suggests that they might also act as signaling molecules that regulate various metabolic and inflammatory pathways (24). The liver can produce acylcarnitines that are exported from hepatocytes into the plasma (25) as well as into the bile (26). Indeed, in mice, biliary levels of medium- and long-chain acylcarnitines were extremely high relative to levels found in either colonic tissue or stool (Figure 2). Although much attention has been paid to plasma levels of acylcarnitines, the levels were significantly higher in human stool (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI133371DS1). Although the gut microbiota played a role in the production of many small-molecule metabolites, it did not play a significant role in the production of acylcarnitines because fecal levels were actually higher in germ-free mice compared with conventionally housed mice (Supplemental Figure 1B). To the contrary, bacterial batch cultures using 3 different culture media showed that palmitoylcarnitine could be consumed by the bacteria in the bacterial batch cultures, where it had a modest growth effect under 2 of the 3 culture conditions (Supplemental Figure 2).
Figure 2. Quantification of acylcarnitine levels in bile (n = 3), colonic tissue (n = 4), and fecal pellets (n = 3) from conventionally housed mice.
*Bile compared to colon, *P < 0.05, **P < 0.01, ***P < 0.001; †Bile compared to fecal, †P < 0.05, ††P < 0.01, †††P < 0.005. P values represent a 2-tailed Student’s t test and paired-sample t test.
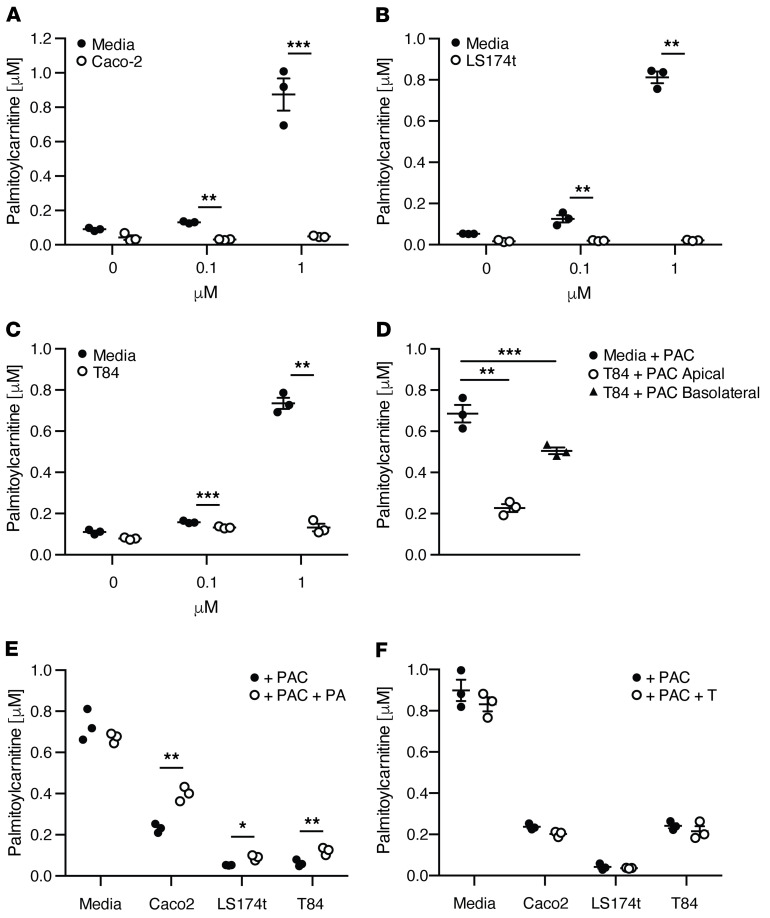
To determine whether acylcarnitines could be consumed by the intestinal epithelium, we examined concentration-dependent consumption of the long-chain acylcarnitine, palmitoylcarnitine (spiked with 0, 0.1, and 1 mM), in 3 model intestinal epithelial cell lines: Caco-2, LS174T, and T84. All 3 showed significant consumption over 24 hours (Figure 3, A–C). Consistent with much higher levels of acylcarnitines in stool than plasma (Supplemental Figure 1A), palmitoylcarnitine consumption was greater when delivered apically than basolaterally to the T84 polarized cell line (Figure 3D). The consumption of palmitoylcarnitine was dependent upon mitochondrial but not peroxisomal oxidation because palmitoylcarnitine uptake was reduced in all 3 cell lines treated with the mitochondrial inhibitor piericidin A, but not with the peroxisomal inhibitor thioridazine (Figure 3, E and F, respectively). These results are consistent with the notion that medium- and long-chain acylcarnitines, delivered to the gut lumen via biliary secretion, can be taken up apically by the intestinal epithelium and used as an alternative source of energy via mitochondrial beta-oxidation.
Figure 3. Consumption of palmitoylcarnitine by intestinal cell lines.
Supernatant concentrations of palmitoylcarnitine 24 hours after incubation with the intestinal cell lines (A) Caco2, (B) LS174t, and (C) T84. (D) Basolateral versus apical uptake of palmitoylcarnitine after 24-hour incubation with T84 cell monolayers grown on Transwell plates to a high transepithelial resistance (TER). (E) Palmitoylcarnitine uptake in intestinal cell lines 24 hours after incubation with the mitochondrial inhibitor piericidin A or (F) the peroxisomal inhibitor thioridazine. n = 3, mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001. P values represent a 2-tailed Student’s t test and paired-sample t test.
Based on this evidence, we then performed a comprehensive characterization of acylcarnitine metabolism using primary human intestinal epithelial cells in culture. We showed that colonoids generated from a healthy human subject grown as high resistance 2D monolayers could consume palmitoylcarnitine apically and basolaterally (Supplemental Figure 3A). In fact, palmitoylcarnitine (C16) was consumed apically and basolaterally to a greater degree than all other long-chain acylcarnitines (see apical and basolateral controls, shown as CT, in Supplemental Table 1). We therefore examined the impact of apical and basolateral supplementation of palmitoylcarnitine (C16) on apical and basolateral consumption and production of a spectrum of acylcarnitines in these 2D colonoid cultures (Supplemental Table 1). In general, acylcarnitines were consumed from the basolateral surface and secreted apically. A notable exception to this was the robust consumption of acetylcarnitine (C2) from the apical and basolateral surfaces, where a reversal to secretion was induced specifically into basolateral compartment by supplementation of palmitoylcarnitine (C16) at either the apical or basolateral surface. This may be a reflection of cellular energy homeostasis, whereby excess nutrient is secreted rather than converted into triglyceride or glycogen for storage. Consistent with this notion, basolateral palmitoylcarnitine (C16) supplementation led to a generalized increase in apical secretion of several acylcarnitines (see CT+BPAC in Apical Fraction, Supplemental Table 1). Thus, in addition to being used for oxidative metabolism, acylcarnitines may also act as a signaling molecule that may alter cellular metabolic homeostasis, as has been proposed in other tissues (24). Despite this effect, the addition of palmitoylcarnitine had minimal effects on transepithelial resistance relative to control conditions (Supplemental Figure 3B).
These studies also showed that the metabolism of acylcarnitines by the intestinal epithelium may be selective, with the greatest levels of consumption being observed for C16, C14:2, C10:2, C3, C3:1, and C2 acylcarnitines both apically and basolaterally (see controls, CT, in Supplemental Table 1). Since we only stimulated the colonoids with palmitoylcarnitine, additional pharmacokinetic studies will be needed to examine the capacity of the intestinal epithelium to consume other acylcarnitines.
To examine the dynamics of palmitoylcarnitine metabolism ex vivo, similar to our studies quantifying oleic acid oxidation in explant cultures (Figure 1, G and H), colonic explants from wild-type and SCAD mice were incubated with palmitoylcarnitine in the presence of either sodium butyrate or NaCl (Supplemental Figure 4). Consistent with our observation that 2D colonoids were able to secrete acylcarnitines apically under certain conditions (Supplemental Table 1), palmitoylcarnitine levels increased during the first 2 hours of incubation. However, after 4 hours in culture, the level of palmitoylcarnitine in the culture supernatant was significantly higher in the wild-type colonic tissue supplemented with sodium butyrate relative to the other 3 conditions (Supplemental Figure 4), consistent with the notion that oxidation of SCFAs could reduce the oxidation of long-chain fatty acids, as we described in Figure 1, E–H.
Colonic inflammation induced by C. rodentium infection inhibits both SCFA and LCFA oxidation.
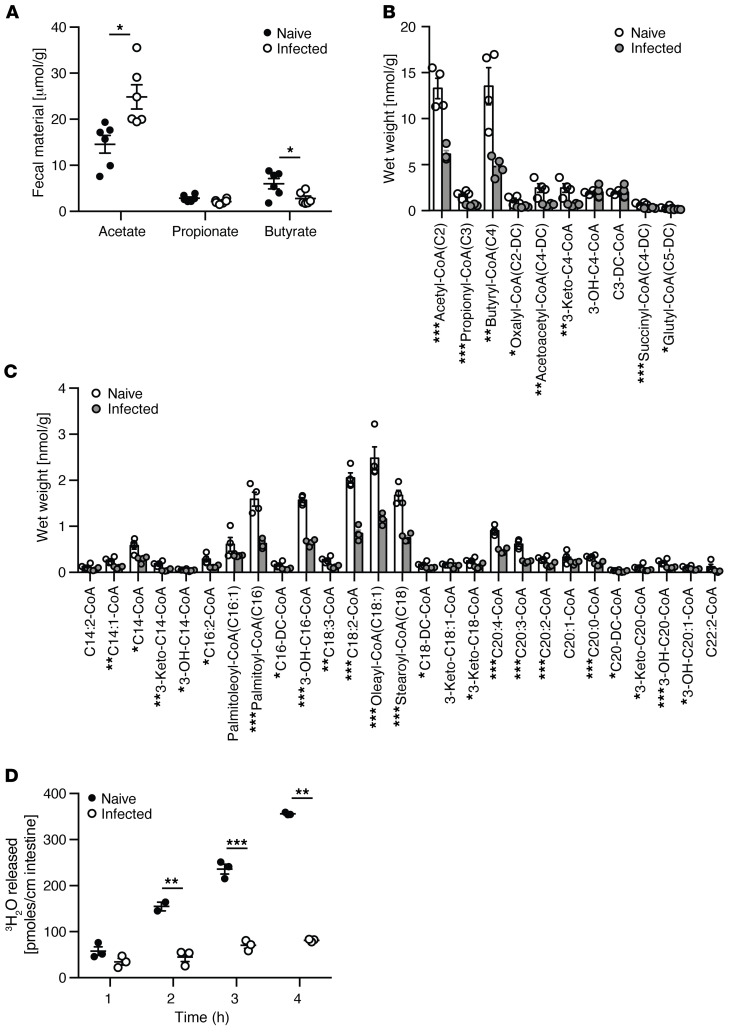
Multiple studies have shown that FAO of SCFAs is reduced in the setting of colonic inflammation in patients with UC, where this metabolic alteration has been proposed to play a role in disease pathogenesis (3, 27). Based on the reciprocal functionality of short-chain versus long-chain FAO (Figure 1), we hypothesized that colonic inflammation would reduce short-chain but increase long-chain FAO. We examined this notion using the C. rodentium model of mouse colitis in which the colonic epithelium remains relatively intact with crypt hypertrophy and epithelial hyperproliferation (28). Fecal SCFAs, quantified by proton nuclear magnetic resonance (1H NMR), showed that C. rodentium infection led to a reduction in butyrate but increase in acetate (Figure 4A). Consistent with this reduction in substrate and previous studies in human IBD (29, 30), acyl-CoA profiling in the colon of C. rodentium–infected mice showed a significant reduction in short-chain acyl-CoAs (Figure 4B). Unexpectedly and inconsistent with our observations in healthy wild-type mice (Figure 1), a significant decrease in long-chain acyl-CoAs in the colon of C. rodentium–infected mice was also observed (Figure 4C). We confirmed that long-chain FAO was functionally reduced in C. rodentium infection using the tritiated oleic acid colonic explant assay (Figure 4D). Collectively, these results suggest that C. rodentium–induced colitis led to a global alteration in FAO.
Figure 4. Quantification of the fatty acid oxidation intermediates, acyl-CoAs, in the colon of mice infected with C.
rodentium. (A) 1H NMR quantification of SCFAs from fecal water of naive and C. rodentium–infected mice. Quantification of (B) short- and (C) long-chain acyl-CoAs extracted from the colon of naive and C. rodentium–infected mice. (D) Oleic acid oxidation quantified by tritiated water release assay in colonic explants of naive and C. rodentium–infected mice. All experiments n = 5, mean ± SEM, *P < 0.05, **P < 0.01. Representative results of at least 2 independent experiments. P values represent a 2-tailed Student’s t test and paired-sample t test.
C. rodentium infection reduces both the number and function of colonic mitochondria.
The reduction of both short- and long-chain FAO in C. rodentium–induced colitis may be due to a reduction in colonic epithelial mitochondria, functional abnormalities, or both. To address this issue, isolated colonic epithelial cells from naive and C. rodentium–infected mice were evaluated genomically, biochemically, and functionally (Figure 4 and Supplemental Figure 5). Quantitative PCR of mitochondrial DNA and mitochondrial citrate synthase assays (Supplemental Figure 5, A and B) showed that C. rodentium–induced colitis reduced both the number and function of colonic mitochondria, respectively. These results were confirmed by flow cytometry of mitochondria stained with MitoTracker Green and Deep Red (Supplemental Figure 5, C–F). MitoTracker probes contain chloromethyl moieties that diffuse across the plasma membrane into mitochondria and bind to sulfhydryls. MitoTracker Green is not dependent on membrane potential and is therefore an indicator of mitochondrial mass, whereas MitoTracker Deep Red is a positively charged rosamine-containing probe dependent on negative mitochondrial membrane potential (31, 32). The decrease in both MitoTracker Green and Deep Red staining indicates a decrease in mitochondrial content as well as function in isolated epithelial cells from the colon of C. rodentium–infected mice compared with naive mice.
C. rodentium infection and IBD lead to an alteration in number, structure, and position of colonic epithelial mitochondria.
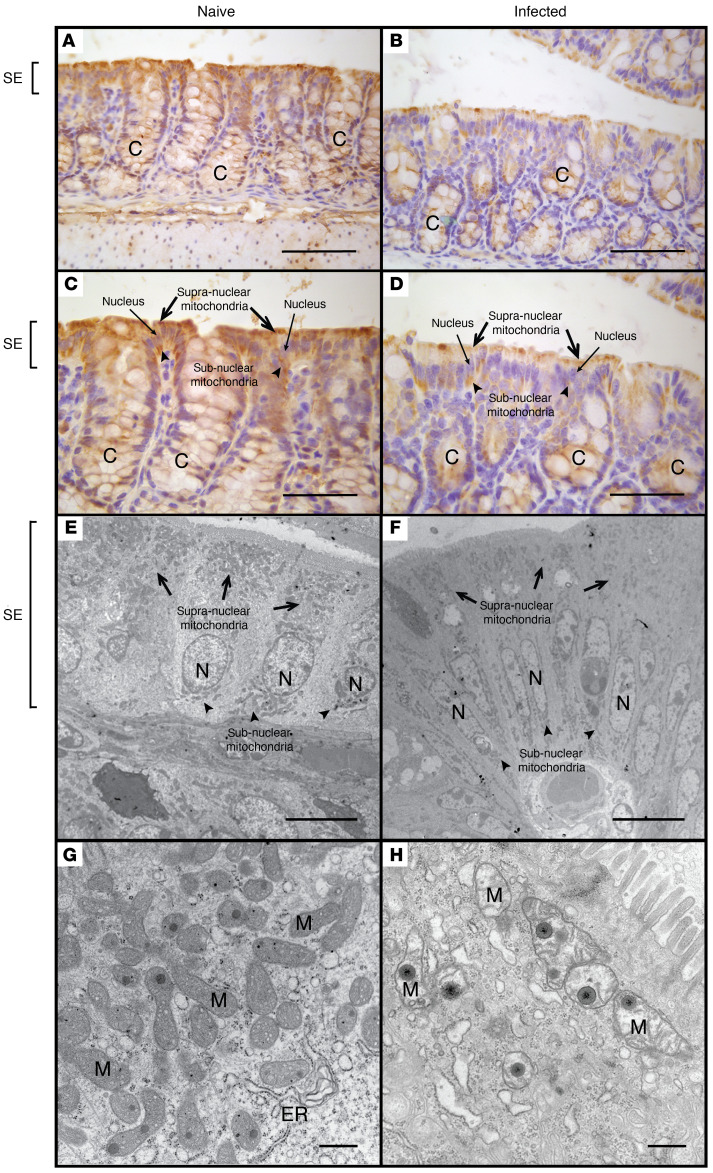
MTCO1 (mitochondrially encoded cytochrome c oxidase I), also known as COX-1 (cytochrome c oxidase), is one of 3 mitochondrial DNA-encoded subunits of respiratory complex IV, the final enzyme of the electron transport chain of mitochondrial oxidative phosphorylation (33). MTCO1 staining can serve as a biomarker for both the presence of mitochondria and functional mitochondrial respiration (33). In colonic tissue of naive mice, MTCO1 was most prominent in epithelial cells, where a diffuse pattern of cytoplasmic staining was observed (Figure 5, A and C). By contrast, there was a decrease in MTCO1 staining in the colonic epithelium of C. rodentium–infected mice that was most pronounced in the superficial epithelium, where staining was more preserved in the subnuclear than the supranuclear compartment (Figure 5, B and D). These observations were confirmed by electron microscopy comparing the number of mitochondria and their morphological characteristics in the superficial colonic epithelium of naive mice (Figure 5, E and G) with C. rodentium–infected mice (Figure 5, F and H), where the latter showed dysmorphic mitochondria (i.e., swelling with loss of cristae abundance and organization) and a reduction in the number of mitochondria that were most striking in the supranuclear compartment of the colonic epithelium.
Figure 5. Comparative morphological analysis of colonic epithelial mitochondria in naive and C.
rodentium–infected mice. MTCO1 staining of colonic tissue isolated from naive (A and C) and C. rodentium–infected mice (B and D). Electron microscopy of the superficial colonic epithelium in naive (E and G) and C. rodentium–infected mice (F and H). Scale bars: 200 μm (A and B), 150 μm (C and D), 10 μm (E and F), 500 nm (G and H). Symbols: crypt (C), nucleus (N), surface epithelium (SE), mitochondria (M), endoplasmic reticulum (ER), supranuclear mitochondria (black arrows), subnuclear mitochondria (black arrowheads).
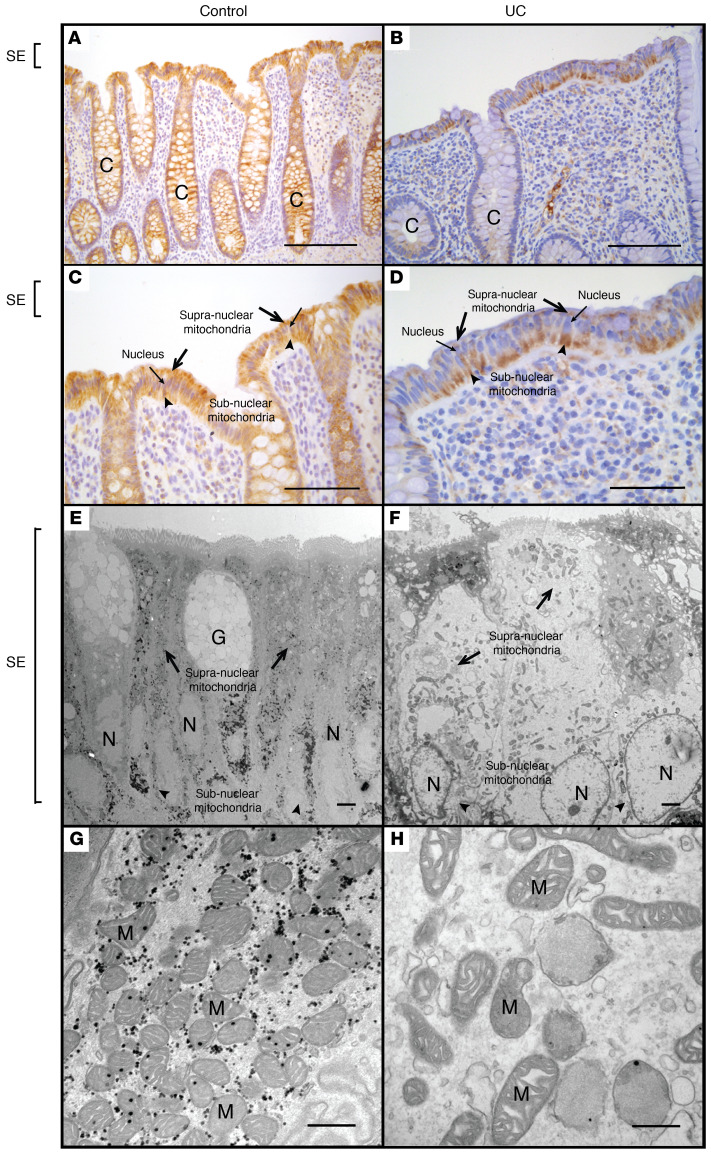
To determine whether these alterations are also associated with colonic inflammation in human disease, we examined MTCO1 staining in surgical tissue specimens obtained from healthy subjects versus patients with IBD (Supplemental Table 2). In healthy subjects, we observed diffuse prominent staining throughout the colonic epithelium, where staining was uniformly distributed throughout the cytoplasm (Figure 6, A and C). By contrast, IBD samples showed a marked decrease in MTCO1 staining in the supranuclear region of the superficial epithelium relative to the subnuclear region in 56% of patients with CD and 60% patients with UC (Figure 6, B and D). We found no differences in the rate of subnuclear staining between IBD types (CD vs. UC, Fisher’s exact test P value 1.00). Electron microscopy also confirmed a reduction in supranuclear mitochondria in IBD as well as dysmorphic mitochondrial characteristics in the superficial epithelium relative to the images of normal colonic tissue (Figure 6, E–H), similar to those characteristics observed in C. rodentium–induced colitis (Figure 5, E–H). In total, these results support the notion of C. rodentium–induced colitis as a model for mitochondrial dysfunction in human IBD, where both show similar morphological alterations in colonic epithelial mitochondria. The biochemical and functional characterization of mitochondrial dysfunction in C. rodentium–induced colitis (Figure 4, Figure 5, and Supplemental Figure 5) therefore provides an explanation for the reduction in butyrate oxidation observed in patients with IBD (5).
Figure 6. Comparative morphological analysis of colonic epithelial mitochondria in healthy human subjects versus patients with IBD.
MTCO1 staining of colonic tissue obtained from a representative healthy (control) human subject (A and C) and a representative patient with IBD (UC) (B and D). Electron microscopy of the superficial colonic epithelium obtained from a representative healthy human subject (E and G) and a representative patient with IBD (UC) (F and H). Scale bars: 400 μm (A and B), 200 μm (C and D), 2 μm (E and F), 500 nm (G and H). Symbols: crypt (C), goblet cell (G), nucleus (N), surface epithelium (SE), mitochondria (M), supranuclear mitochondria (black arrows), subnuclear mitochondria (black arrowheads).
Elevation of fecal acylcarnitines is associated with IBD in patients and C. rodentium colitis in mice.
The inhibition of palmitoylcarnitine uptake by an intestinal epithelial cell line in vitro by pharmacological mitochondrial inhibitors (Figure 3E), together with functional and histological evidence of mitochondrial dysfunction in both C. rodentium–induced colitis and human IBD, suggests that the decrease in mitochondrial FAO might lead to an elevation of fecal acylcarnitine levels in intestinal inflammatory disease. Indeed, the levels of a wide spectrum of acylcarnitines were elevated in mice with C. rodentium–induced colitis (Supplemental Figure 7).
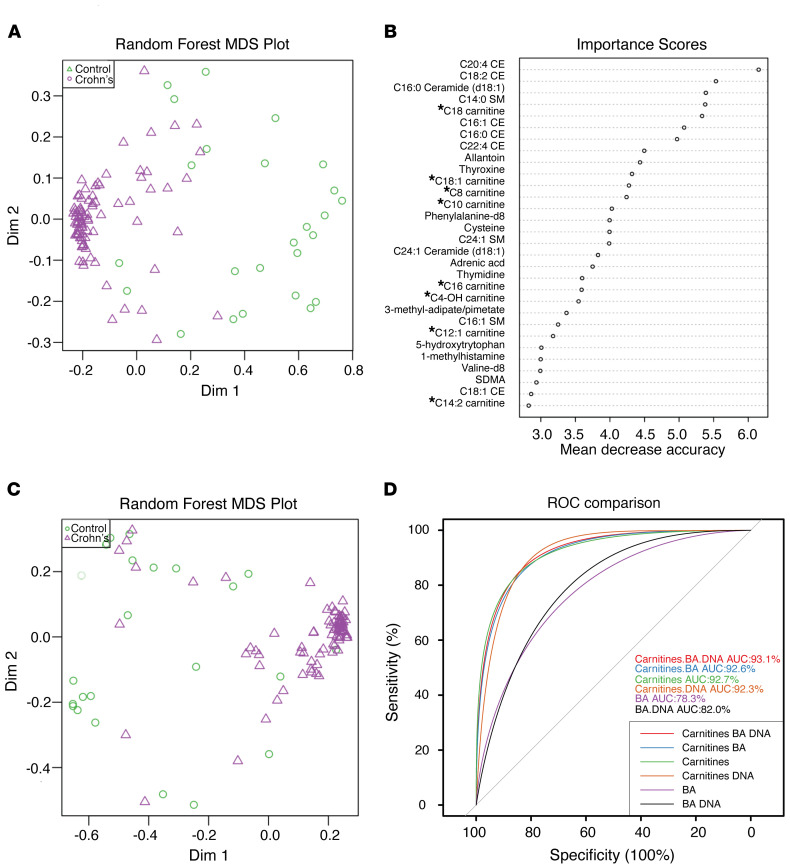
This was also the case for patients with CD, a form of IBD where elevations were associated with increased disease activity (quantified by fecal calprotectin levels), an increase in human DNA in the feces, and dysbiosis, defined as “cluster 2” (34) (Supplemental Figure 6). Two acylcarnitines, C3 carnitine and C20:4 carnitine, showed the opposite pattern with a decrease in abundance, the former being consistent with the known reduction in fecal propionate levels associated with intestinal inflammation (Figure 1B) (35). The association between dysbiosis in IBD and bile acids as well as acylcarnitines in the feces has recently been reported (30), where a number of associations were described with bacterial taxa. An analysis of our cohort revealed that there is a very robust separation that distinguishes the fecal metabolome of healthy subjects from those with CD in a multidimensional scaling (MDS) plot (Figure 7A), where a random forest analysis showed that acylcarnitines were among the most important features responsible for this separation (Figure 7B). Using only the previously described metabolomic features used as biomarkers for IBD and dysbiosis, bile acids (30) and human DNA (34), together with acylcarnitines, showed good separation on the MDS plot between healthy subjects and patients with IBD, where the IBD subjects were more tightly clustered together on the right of the plot than the more widely dispersed healthy control subjects (Figure 7C). Among these 3 features, alone or in combination, acylcarnitines alone were the single strongest predictor of the presence of IBD, having an AUC of 92.7% on a receiver operating characteristic curve (ROC) analysis (Figure 7D). In total, these results support the notion that intestinal epithelial mitochondrial function plays a role in defining fecal acylcarnitines as a robust biomarker for IBD.
Figure 7. Fecal acylcarnitines as a biomarker for IBD.
(A) MDS plot of fecal metabolites from a previously described cohort of healthy pediatric subjects and those with CD (34, 47). (B) Random forest analysis of fecal metabolites stratified by importance in distinguishing healthy subjects from patients with IBD. * indicates acylcarnitines. (C) MDS plot of the same cohort in A focused on human DNA, bile acids, and acylcarnitines. (D) A receiver operating characteristic curve (ROC) showing the discriminative ability of human DNA, bile acids, and acylcarnitines to individually or in combination predict a condition of health versus IBD.
Discussion
It is well known that mitochondria play a critical role in the regulation of many cellular processes, such as energy generation, apoptosis, the unfolded protein response, and autophagy (15). Recently, it has become apparent that there are functional features of mitochondria that are unique to the colonic epithelium because of its juxtaposition with the gut microbiota and its production of metabolites such as SCFAs by bacterial fermentation of indigestible carbohydrates (10). Specifically, the oxidation of butyrate by the colonic epithelium prevents epigenetic alterations of the genome marked by histone hyperacetylation via the inhibition of HDACs (11). The consequence of such alterations leads to the inhibition of cellular proliferation, a phenotype that could reduce colonic epithelial wound healing (10). The regulation of FAO by the colonic epithelium may therefore play a critical role in disease states where the epithelial barrier is wounded, such as IBD, as well as in colonic neoplasia. A few reports in the literature have documented a reduction of butyrate oxidation by the colonic epithelium in the setting of UC (12, 13). However, the mechanisms have remained poorly defined. Indeed, there are currently no studies that systematically characterize the overall regulation of FAO by the colonic epithelium in either health or disease.
Herein, we used a combination of technologies to characterize the FAO of a spectrum of fatty acids in the colonic epithelium both functionally using explant cultures and biochemically via tandem mass spectrometry. The results demonstrated that the colonic epithelium could oxidize not only SCFAs, as have been previously described, but also LCFAs at levels actually exceeding that observed in the small intestine. By studying the impact of butyryl-CoA oxidation on long-chain FAO using SCAD-deficient mice, we showed that butyrate inhibited long-chain FAO. Although additional studies will be needed to determine the mechanism(s) involved in the reciprocal relationship between short- and long-chain FAO, it is plausible that AMPK might be involved, similar to the reciprocal relationship between glycolysis and beta-oxidation (36), given the primary role of mitochondria in maintaining cellular energy homeostasis. Regardless of the mechanism, the high levels of butyrate produced by the gut microbiota as an energy substrate for the colonic epithelium would suppress long-chain FAO, supporting the conclusion that the SCFAs are the primary source of metabolic fuel for colonocytes (4–7).
The ability of the colonic epithelium to use both SCFAs and LCFAs for oxidative metabolism suggests that, in addition to microbiota-based substrates in the form of the SCFA butyrate, the host might also contribute substrates for FAO in the form of medium- to long-chain fatty acids. Indeed, we provide evidence that acylcarnitines delivered to the gut lumen in bile can be consumed apically in a polarized intestinal epithelial model system, where they undergo mitochondrial but not peroxisomal oxidation. This previously unappreciated mechanism by which biliary secretions may have a physiological impact on the intestinal tract is consistent with other interactions between the liver and the gut, such as the enterohepatic circulation of bile acids, as well as bile acid–mediated activation of the farnesoid X nuclear hormone receptor in the terminal ileal epithelium (37). The mechanism by which luminal acylcarnitines are transported apically into the intestinal epithelium is currently unknown, but may involve solute carrier (SLC) membrane transport proteins, such as SLC22A1, which is responsible for the efflux of acylcarnitine from the liver into the circulation (25) and has been shown to be expressed on the apical domain of the intestinal epithelium (38). Indeed, we showed that human 2D colonoids could be stimulated to secrete acylcarnitines apically when stimulated by palmitoylcarnitine basolaterally. Although the impact of polarized secretion in the setting of disease remains to be determined, our result may suggest an additional mechanism by which intestinal epithelial mitochondrial dysfunction might lead to greater levels of fecal acylcarnitine levels, namely enhanced apical secretion due to reduced mitochondrial consumption. Indeed, elevated plasma acylcarnitine levels are often monitored clinically in patients with mitochondrial disorders (39). Interestingly, our data in the 2D colonoid model also suggests that both apical and basolateral consumption may be restricted to acylcarnitines, a number of which are found at higher levels in human stool than plasma. Ultimately, it will be of interest to determine the impact of host-derived acylcarnitines relative to microbiota-generated SCFAs on the metabolic function of the colonic epithelium, especially since we provide evidence that these are reciprocally related.
We determined the impact of disease on epithelial mitochondrial structure and function through a series of genomic, biochemical, and FACS-based analyses, revealing that a reduction in both short- and long-chain acyl-CoAs in colonic tissue of mice infected with C. rodentium was due to a decrease in the number and function of mitochondria in the colonic epithelium. MTCO1 immunohistochemistry and electron microscopy confirmed and extended this finding, showing not only a dysmorphic appearance of mitochondria, but also a reduction in number that was particularly severe in the superficial surface epithelium of the colon, consistent with previously reported observations by Ma et al. (40). These mitochondrial alterations are in part due to a C. rodentium effector protein known as mitochondrial-associated protein (40). Similar findings were observed in patients with IBD, providing an explanation for previous studies reporting a reduction in SCFA metabolism by the colonic epithelium (29).
The reduction of mitochondria specifically in the supranuclear compartment of the superficial colonic epithelium is particularly intriguing — not only may this histological characteristic be a biomarker for the intestinal inflammatory state, but it may have functional implications for the cellular biology of the colonic epithelium. Differential mitochondrial positioning has been described in polarized cell types, such as budding yeast, neurons, and pancreatic acinar cells (41–43). Studies have shown in these model systems that mitochondria localize to areas of the cell needed for metabolically intensive processes. This phenomenon has not been previously described in the intestinal or colonic epithelium. We hypothesize that the polarized colonocyte would have a large number of mitochondria at the apical side of the cell, where butyrate concentrations are high and a high level of FAO is occurring. The loss of mitochondria would be consistent with the observed reduction in SCFA oxidation associated with IBD (3, 27), where it may have an impact on the cellular proliferation of stem cells located in the colonic crypt via the inhibition of HDACs (10). It also provides an alternative explanation by which colonic inflammation, induced by C. rodentium in mice or associated with IBD in humans, might lead to the expansion of aerotolerant, facultative anaerobic organisms at the mucosal interface. C. rodentium might increase the diffusion of oxygen into the luminal environment, not only by reducing the consumption of oxygen via mitochondrial beta-oxidation by the superficial colonic epithelium due to a reduction in gut microbiota butyrate production as proposed by Lopez et al. (44), but as our results suggest, a similar phenomenon might also occur because of the impaired function and/or number of mitochondria in this same cellular compartment.
Finally, we showed that alteration of epithelial mitochondrial function, effectively reducing FAO, may have an impact on substrate availability of fecal acylcarnitines, which were elevated in both C. rodentium–induced murine colitis and in patients with CD. Although there are bacterial taxonomic associations with the elevation of fecal acylcarnitines in human IBD, leading to the notion that the gut microbiota might be important for their production (30), our results in germ-free mice showing high levels of fecal acylcarnitines strongly indicates that they are primarily of host and not of microbial origin. Nevertheless, we provide preliminary evidence that palmitoylcarnitine can be consumed by the human gut microbiota in culture, where it has a modest effect on growth. Previous studies have shown that acylcarnitines as well as carnitine itself can serve as a nitrogen and carbon source for bacteria (45). Indeed, antibiotic treatment of mice can increase fecal acylcarnitine levels (46), suggesting the interesting possibility that the IBD-associated dysbiosis may play a role in the elevation of fecal acylcarnitines due to reduced consumption. This would be consistent with our observation that elevated fecal acylcarnitines in CD were positively correlated with dysbiosis (“cluster 2” in Supplemental Figure 6). The performance of fecal acylcarnitine as a predictor of IBD was impressive, having an AUC of over 92% in the ROC analysis, which was better than previous fecal biomarkers for IBD, such as human DNA and bile acid composition. In addition to providing support for the notion that acylcarnitines are consumed by the intestinal epithelium for mitochondrial oxidation, the elevation of these lipids may be a useful marker of disease in patients with intestinal inflammation.
Using FAO as a biomarker of mitochondrial function, we provide mechanistic insight into the use of energy substrates provided by the gut microbiota in the form of butyrate, as well as the host in the form of acylcarnitines — the latter being a previously unappreciated modality of enterohepatic communication. The most significant reduction in mitochondrial number and morphology restricted to the superficial colonic epithelium in the supranuclear apical subcellular compartment may have significant effects on the host via the inhibition of stem cell proliferation, as well as the gut microbiota with the expansion of facultative anaerobes driven by aerobic respiration. The unique environment in which the superficial colonic epithelium resides, juxtaposed with an anaerobic environment high in fatty acid substrates, emphasizes the importance of mitochondrial function via FAO in this cellular compartment.
Methods
Animals.
All experiments were approved by the IACUC at the University of Pennsylvania Perelman School of Medicine. C57BL/6J, BALB/cByJ (SCAD deficient) and BALB/cJ (wild-type) were obtained from the Jackson Laboratory. Germ-free mice were obtained from the University of Pennsylvania Gnotobiotic Mouse Facility. All animals used in experiments were 7–10-week-old female mice. Animals were fed a standard chow diet and were kept on 12-hour light/12-hour dark cycles. For the C. rodentium model, conventionally housed, female, 8–10-week-old, C57BL/6J mice were inoculated by oral gavage with 6 × 108 CFU of C. rodentium (strain DBS100, gift from David Artis, Weill Cornell Medicine at Cornell University, New York, New York). Mice were euthanized by CO2 asphyxiation followed by cervical dislocation on day 11 after infection.
Human studies.
Plasma and fecal samples assayed for acylcarnitine levels as well as rectal biopsies for colonoid primary cell line generation were obtained from the Food and Resulting Microbial Metabolite (FARMM) study (47). The FARMM study recruited healthy volunteers ages 18–60. Stool, urine, blood, and rectal biopsies were collected throughout the 15-day inpatient study designed to determine the impact of diet on the gut microbiome, fecal, and plasma metabolome. The samples used for this study were obtained at the baseline timepoint at the beginning of the study. The fecal metabolomic analysis in pediatric CD, shown in Figure 7 and Supplemental Figure 6, was performed as part of the PLEASE study that has been previously described (34). Briefly, stool samples from pediatric patients with CD and healthy controls were used for a prospective cohort study to assay metabolites such as acylcarnitines and measure calprotectin and human DNA. Both the FARMM and PLEASE studies received approval from the University of Pennsylvania IRB.
Bacterial batch culture.
Bacteria were grown in lysogeny broth (LB, Thermo Fisher Scientific); brain heart infusion (BHI, Anaerobe Systems); gut microbiota medium (GMM, Anaerobe Systems); or yeast extract, casitone, and fatty acid (YCFA, Anaerobe Systems) culture media shaken at 37°C. A fecal sample from a healthy 24-year-old male subject (47) was grown in BHI, GMM, or YCFA with or without palmitoylcarnitine, final concentration 1 μM. OD (600) was measured to determine growth rate in the presence or absence of palmitoylcarnitine. Culture media supernatants were taken at 0 and 24 hours for acylcarnitine analysis by mass spectrometry. Control samples consisted of media with or without palmitoylcarnitine.
Human colon cancer cell lines.
Human colon cancer cell lines T84, Caco-2, and LS174T (ATCC) were maintained at 37°C in 5% CO2 in a humidified incubator. LS174T and Caco-2 cells were maintained in DMEM supplemented with penicillin-streptomycin and 10% FBS. T84 cells were maintained in DMEM/Ham’s F12 supplemented with penicillin-streptomycin and 10% FBS. Cultures were used for experiments when grown to confluent monolayers on 6-well plates unless otherwise noted.
Healthy human colonoids.
Healthy human colonoids (from FARMM study) were generated and propagated in Matrigel (Corning, 356231) on 24-well polystyrene plates as previously described (48) with full growth medium comprising basal medium (advanced DMEM supplemented with 10% FBS, 1× penicillin/streptomycin, 1× GlutaMAX, 10 mM HEPES, 1× N2 supplement, 1× B27 supplement, all from Invitrogen), 1 mM N-acetyl-cysteine, 10 nM leu-gastrin (all from MilliporeSigma) and WRN cell conditioned medium (WRN cells from ATCC; basal medium and WRN conditioned medium mixed at 1:1 ratio), and supplements (500 nM A83-01 [Tocris], 10 μM SB-20210, 10 mM nicotinamide, 50 ng/mL human EGF [all from MilliporeSigma], and 2 μL/mL 1× Primocin [InvivoGen]). For the first 48 hours of colonoid culture, growth medium was supplemented with 10 μM Y27632, 2.5 μM CHIR99021 (both from MilliporeSigma), and 2.5 μM thiazovivin (Reprocell). Media was replaced every 48 hours.
Transepithelial electrical resistance assay.
Transepithelial electrical resistance (TER) was measured by an epithelial volt/ohm meter (World Precision Instruments, EVOM2 300523) with colon cancer cell lines and healthy human colonoid-derived cells grown as monolayers on Transwell permeable supports (Costar, 3470). To grow colonoid-derived cells on the Transwell cell culture insert, established colonoids were dislodged and resuspended in TrypLE (Invitrogen, 12605010) for 5–10 minutes in a 37oC water bath to disrupt colonoid formation and generate single cells. The cell suspension was supplemented with 10% v/v of FBS and pipetted using a 100 μL pipette tip by gentle strokes of approximately 50 times. Cells were counted using an automated Corning cell counter. Next, 100 μL of single-cell suspension containing 2 × 105 cells in growth medium were plated on 0.4 μm pore, 12-mm diameter polycarbonate membrane inserts (Corning, 3413) coated with 33.3 μg/mL human collagen IV (Millipore, C954D93). After 24 hours, the lower chamber medium was replaced with differentiation medium (growth medium supplemented with 2 μg/mL hBMP4 [MilliporeSigma]) and the upper chamber medium was supplemented with 100 μL of differentiation medium. Fresh media were replaced every 48 hours. Cells were maintained at 37°C, 5% CO2. TER was evaluated when filter-grown monolayers reached full resistance (at least 800 Ω-cm2 of TER).
Chemical reagents.
Palmitoylcarnitine (MilliporeSigma) was used at 1 μM. Piericidin A (Cayman Chemical) was used at 100 nM. Thioridazine (Cayman Chemical) was used at 200 nM.
Isolation of colonocytes.
Animals were euthanized by CO2 asphyxiation followed by cervical dislocation. Colons were dissected, flushed with sterile PBS, and splayed open longitudinally and washed again with PBS. Tissue was incubated on ice in PBS containing 30 mM EDTA and 1.5 mM DTT for 20 minutes with occasional inverting. This was followed by shaking every 2 minutes in PBS containing 30 mM EDTA for 10 minutes to remove crypts from the submucosa. The mesenchyme was removed, and cells were pelleted by centrifugation at 400 g for 5 minutes at 4°C and washed in PBS.
Quantification of mitochondrial DNA.
Mitochondrial DNA was isolated using Qiagen Blood and Tissue Kit following the manufacturer’s protocol (Qiagen). Primers for mitochondria D-loop (encoded by mitochondrial DNA) and Ikb-β (encoded by nuclear DNA) were utilized to determine relative mitochondrial DNA level, and β-actin was used as an internal control; primers are shown in the format of gene, forward primer (F), reverse primer (R): mtD-loop, (F) ACTATCCCCTTCCCCATTTG, (R) TGTTGGTCATGGGCTGATTA; Ikb-β, (F) GCTGGTGTCTGGGGTACAGT, (R) ATCCTTGGGGAGGCATCTAC; β-actin, (F) TGTTACCAACTGGGACGACA, (R) ACCAGAGGCATACAGGGACA.
Quantitative mass spectrometry.
Acyl-CoAs from small intestinal (duodenum, 5 cm from the stomach) or colonic tissue were extracted with chloroform and methanol (2:1, v/v), together with stable isotope-labeled and unlabeled standards, were measured by flow injection tandem mass spectrometry as previously described (17) and normalized to the wet tissue weight.
Mass spectrometry for acylcarnitines.
Acylcarnitines were measured as their butylated derivatives using tandem mass spectrometry. Stable isotope-labeled internal standards (Cambridge Isotope Laboratories) were added to samples. Ethanol was added and the samples were dried under a stream of nitrogen at 60°C. After adding butanolic hydrochloric acid (Regis Technologies), the samples were heated to 65°C for 15 minutes and dried under a stream of nitrogen. The dried samples were reconstituted with acetonitrile/water (80:20) and injected into a Xevo TQ-XS tandem mass spectrometer (Waters Corporation). Data were acquired to collect the parent compounds of mass m/z 85. Quantitation was against the nearest chain-length stable isotope labeled internal standard. The mass spectrometer was operated in positive ion mode with resolving power of 140,000 at m/z 200, mass scanning range being m/z 140–600, and 5 μL of sample was delivered to mass spectrometer through direct flow injection. Mobile phase was acetonitrile/water (80:20, v/v). Flow rate was 0.1 mL/minute. The source temperature was 150°C and desolvation temperature was 200°C, with gas flow of 550 L/hour. The cone and capillary voltage was 50 V and 3.2 kV, respectively. Data were acquired in multiple reaction monitoring (MRM). The mass spectrometer was operated in positive ion mode with resolving power of 140,000 at m/z 200, mass scanning range being m/z 140–600.
NMR metabolomics.
SCFAs were extracted and analyzed as described previously (49). An NMR spectrum for each fecal sample was acquired using NOESY-presaturation on a 4-channel Bruker Ascend 700 MHz spectrometer (Bruker Biospin) with a TXI 700 MHz 3 mm probe with triple-axis gradients. All spectra were acquired with a 14 ppm sweep width, 4-second acquisition time, 4 dummy scans, and 384 transients. All spectra were zero filled to 128,000 data points, Fourier transformed with 0.1 Hz line broadening applied, manually phased, and baseline-corrected using TopSpin NMR software. The SCFA concentrations were quantitatively determined using the 700 MHz library from Chenomx NMR Suite 7.1 after acquiring an NMR spectrum from each fecal sample. The resulting quantified metabolites were used as input variables into subsequent statistical analysis and normalized by fecal weight.
FAO assay.
FAO was measured with 3H-oleic acid (American Radiolabeled Chemicals) and quantified using tritiated water released per centimeter length of intestine. Briefly, the colon or duodenum (first 5 cm from the stomach) was isolated and then flushed with oxygenated PBS to remove waste. The intestinal segment was everted on a gavage needle and filled with PBS and ligated at both ends to form a sac. The explant was incubated in a buffer containing 3H-oleic acid and 100 mM carnitine for 4 hours at 37°C. At each hour, a 400 μL aliquot of assay buffer was removed from each sample and replaced with 400 μL of fresh assay buffer. The collected aliquot was quenched with 400 μL of 10% (w/v) trichloroacetic acid and centrifuged at 6800 g for 10 minutes. The supernatant was neutralized with 6 N sodium hydroxide solution and applied to columns containing AGX-1 resin (BioRad). The flow-through containing the unretained sample and the fraction eluted with 2 mL of water were pooled. Then, 10 mL of scintillation fluid was added and radioactivity measured on a scintillation counter. For the butyrate studies, explants were incubated with either 10 mM sodium chloride or 10 mM sodium butyrate.
Flow cytometry.
Flow cytometry was performed using FACSCanto or LSR II cytometers (BD Biosciences) and FlowJo software (Tree Star) for cells suspended in DPBS containing 1% BSA (Sigma-Aldrich) to assess mitochondrial mass and membrane potential (50). Single cell suspensions generated from murine intestinal crypts were incubated with 5 nM MitoTracker Green (Molecular Probes; membrane potential insensitive) and 1 nM MitoTracker Deep Red (Molecular Probes; membrane potential sensitive) in DPBS containing 10% FBS at 37°C for 30 minutes. DAPI (Molecular Probes) was used to determine cell viability. A suspension of unstained cells pooled from all samples was utilized to establish background fluorescence in each experiment. Geometric MFIs for MitoTracker Green and MitoTracker Deep Red were determined in the live-cell fraction (DAPI–) for each specimen after subtraction of background fluorescence.
Histology.
Paraffin-embedded 4% paraformaldehyde-fixed surgical colonic tissue sections from the Cooperative Human Tissue Network and anonymized archival tissue from the Department of Pathology, Penn Presbyterian Medical Center (expedited IRB approval) were stained with H&E. Immunohistochemistry was performed with MTCO1 (Abcam, ab14705, clone 1D6E1A8). Slides were analyzed and scored by a blinded pathologist for disease activity, localization, and intensity of MTCO1 staining. In Supplemental Table 2, if any biopsy section from an individual patient demonstrated a loss of supranuclear staining in the superficial epithelium spanning at least 5 crypts, the patient was classified as having “Sub-nuclear Staining.” If no sections for each patient showed a loss of supranuclear staining in the superficial epithelium, the patient was classified as having “Diffuse Cytoplasmic Staining.”
Transmission electron microscopy.
Tissue samples were fixed in cacodylate-buffered 2.5% (w/v) glutaraldehyde, postfixed in 2.0% osmium tetroxide, then embedded in epoxy resin. Ultrathin sections were poststained with uranyl acetate and lead citrate. Imaging was performed using a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera and AMT Advantage imaging software. Micrographs were assessed by 2 investigators blinded to clinical diagnosis.
Statistics.
Results are expressed as the mean ± SEM. A 2-tailed Student’s t test and paired-sample t test were used for direct comparisons between 2 groups and within groups, respectively. A P value of less than 0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001. A Fisher’s exact test was used to calculate P values for the MTCO1 scoring of subnuclear versus supranuclear staining. A 2-way ANOVA with Holm-Sidak’s post hoc test for multiple comparisons was used for direct comparisons between palmitoylcarnitine-treated and untreated groups.
Metabolites with more than 80% missing values across all samples were removed from the analysis. For each metabolite, samples with the missing values were imputed with its minimum abundance across samples. For each sample, the metabolite abundances were further normalized by dividing 90% cumulative sum of the abundances of all metabolites. The 90% cumulative sum, instead of total abundance, was chosen to avoid potential large outliers in the samples. The normalized abundances were then log transformed and used in all analyses. The metabolite annotation was obtained from the Human Metabolome Database (HMDB). Two group comparisons were performed using Wilcoxon rank-sum test. P values were adjusted for multiple comparisons with FDR control.
Study approval.
All animal studies were performed under approval by the IACUC of the University of Pennsylvania. All study protocols involving human samples were collected according to IRB-approved protocols at the University of Pennsylvania for which written informed consent was obtained and there was no access to identifiable information.
Author contributions
SAS, LC, SAO, KAW, KH, JC, BS, CBC, RJX, JB, and SAK performed experiments. FF and MB provided human colonic tissues and analyzed them by electron microscopy and immunohistochemistry. AMW, MB, HN, FDB, JDL, and GDW designed the experiments. EC and HL performed statistical analyses. SAS, LC, and GDW generated the figures and wrote the manuscript. The order of the co–first authors, SAS, SAO, and LC, was based upon the amount of experiments performed as well as contributions to data analysis and drafting of the text for this manuscript.
Supplementary Material
Acknowledgments
We acknowledge the following funding sources: K01DK103953 (to KAW), R03DK114463 (to KH), and R01DK114436 (to HN) from NIH/NIDDK; R01AA026297 (to HN) from NIAAA; R01GM103591 (to GW) from NIGMS; Crohn’s and Colitis Foundation and P30DK050306 for the Human-Microbial Analytic and Repository Core of the Center for Molecular Studies in Digestive and Liver Disease (to GDW) from NIH/NIDDK. The authors acknowledge the PennCHOP Microbiome Program (to GDW) for assistance in metabolomic analyses and bacterial culture as well as Raymond Meade and Biao Zou for assistance with electron microscopy at the Electron Microscopy Resource Laboratory at the University of Pennsylvania
Version 1. 11/03/2020
In-Press Preview
Version 2. 01/04/2021
Print issue publication
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e133371.https://doi.org/10.1172/JCI133371.
Contributor Information
Sarah A. Smith, Email: sarahsa@pennmedicine.upenn.edu.
Sayaka A. Ogawa, Email: saogawa@pennmedicine.upenn.edu.
Lillian Chau, Email: lcwang04@gmail.com.
Kelly A. Whelan, Email: Kelly.whelan@temple.edu.
Kathryn E. Hamilton, Email: hamiltonk1@email.chop.edu.
Jie Chen, Email: chenji@email.chop.edu.
Lu Tan, Email: TANL@EMAIL.CHOP.EDU.
Eric Z. Chen, Email: chvlyl@gmail.com.
Sue Keilbaugh, Email: sakeilbaugh@gmail.com.
Franz Fogt, Email: franzfogt@gmail.com.
Meenakshi Bewtra, Email: mbewtra@pennmedicine.upenn.edu.
Jonathan Braun, Email: Jonathan.Braun2@cshs.org.
Ramnik J. Xavier, Email: xavier@molbio.mgh.harvard.edu.
Clary B. Clish, Email: clary@broadinstitute.org.
Barry Slaff, Email: barry.slaff@gmail.com.
Aalim M. Weljie, Email: aalim@upenn.edu.
Frederic D. Bushman, Email: bushman@mail.med.upenn.edu.
James D. Lewis, Email: lewisjd@mail.med.upenn.edu.
Hongzhe Li, Email: hongzhe@mail.med.upenn.edu.
Stephen R. Master, Email: masters@email.chop.edu.
Michael J. Bennett, Email: bennettmi@email.chop.edu.
Hiroshi Nakagawa, Email: nakagawh@mail.med.upenn.edu.
Gary D. Wu, Email: gdwu@mail.med.upenn.edu.
References
- 1.Crosnier C, Stamataki D, Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet. 2006;7(5):349–359. doi: 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346(6205):1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 3.Kles KA, Chang EB. Short-chain fatty acids impact on intestinal adaptation, inflammation, carcinoma, and failure. Gastroenterology. 2006;130(2 suppl 1):S100–S105. doi: 10.1053/j.gastro.2005.11.048. [DOI] [PubMed] [Google Scholar]
- 4.Hadjiagapiou C, Schmidt L, Dudeja PK, Layden TJ, Ramaswamy K. Mechanism(s) of butyrate transport in Caco-2 cells: role of monocarboxylate transporter 1. Am J Physiol Gastrointest Liver Physiol. 2000;279(4):G775–G780. doi: 10.1152/ajpgi.2000.279.4.G775. [DOI] [PubMed] [Google Scholar]
- 5.Roediger WE. The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet. 1980;2(8197):712–715. doi: 10.1016/s0140-6736(80)91934-0. [DOI] [PubMed] [Google Scholar]
- 6.Roediger WE, Rae DA. Trophic effect of short chain fatty acids on mucosal handling of ions by the defunctioned colon. Br J Surg. 1982;69(1):23–25. doi: 10.1002/bjs.1800690108. [DOI] [PubMed] [Google Scholar]
- 7.Scheppach W, et al. Effect of short-chain fatty acids on the human colonic mucosa in vitro. JPEN J Parenter Enteral Nutr. 1992;16(1):43–48. doi: 10.1177/014860719201600143. [DOI] [PubMed] [Google Scholar]
- 8.Donohoe DR, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133(suppl 7):2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 10.Kaiko GE, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;167(4):1137. doi: 10.1016/j.cell.2016.10.034. [DOI] [PubMed] [Google Scholar]
- 11.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48(4):612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roediger WE, Nance S. Metabolic induction of experimental ulcerative colitis by inhibition of fatty acid oxidation. Br J Exp Pathol. 1986;67(6):773–782. [PMC free article] [PubMed] [Google Scholar]
- 13.Harig JM, Soergel KH, Komorowski RA, Wood CM. Treatment of diversion colitis with short-chain-fatty acid irrigation. N Engl J Med. 1989;320(1):23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- 14.Peltekova VD, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36(5):471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 15.Rath E, Haller D. Mitochondria at the interface between danger signaling and metabolism: role of unfolded protein responses in chronic inflammation. Inflamm Bowel Dis. 2012;18(7):1364–1377. doi: 10.1002/ibd.21944. [DOI] [PubMed] [Google Scholar]
- 16.Novak EA, Mollen KP. Mitochondrial dysfunction in inflammatory bowel disease. Front Cell Dev Biol. 2015;3:62. doi: 10.3389/fcell.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palladino AA, Chen J, Kallish S, Stanley CA, Bennett MJ. Measurement of tissue acyl-CoAs using flow-injection tandem mass spectrometry: acyl-CoA profiles in short-chain fatty acid oxidation defects. Mol Genet Metab. 2012;107(4):679–683. doi: 10.1016/j.ymgme.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbull DM, et al. Short-chain acyl-CoA dehydrogenase deficiency associated with a lipid-storage myopathy and secondary carnitine deficiency. N Engl J Med. 1984;311(19):1232–1236. doi: 10.1056/NEJM198411083111906. [DOI] [PubMed] [Google Scholar]
- 20.Amendt BA, et al. Short-chain acyl-coenzyme A dehydrogenase deficiency. Clinical and biochemical studies in two patients. J Clin Invest. 1987;79(5):1303–1309. doi: 10.1172/JCI112953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood PA, Amendt BA, Rhead WJ, Millington DS, Inoue F, Armstrong D. Short-chain acyl-coenzyme A dehydrogenase deficiency in mice. Pediatr Res. 1989;25(1):38–43. doi: 10.1203/00006450-198901000-00010. [DOI] [PubMed] [Google Scholar]
- 22.Hardie DG. Sensing of energy and nutrients by AMP-activated protein kinase. Am J Clin Nutr. 2011;93(4):891S–8916. doi: 10.3945/ajcn.110.001925. [DOI] [PubMed] [Google Scholar]
- 23.Eaton S, Bartlett K, Pourfarzam M. Mammalian mitochondrial beta-oxidation. Biochem J. 1996;320(pt 2):345–357. doi: 10.1042/bj3200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCoin CS, Knotts TA, Adams SH. Acylcarnitines--old actors auditioning for new roles in metabolic physiology. Nat Rev Endocrinol. 2015;11(10):617–625. doi: 10.1038/nrendo.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HI, et al. Fine mapping and functional analysis reveal a role of SLC22A1 in acylcarnitine transport. Am J Hum Genet. 2017;101(4):489–502. doi: 10.1016/j.ajhg.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton JJ, Hahn P. Carnitine and carnitine esters in rat bile and human duodenal fluid. Can J Physiol Pharmacol. 1987;65(9):1816–1820. doi: 10.1139/y87-283. [DOI] [PubMed] [Google Scholar]
- 27.Cook SI, Sellin JH. Review article: short chain fatty acids in health and disease. Aliment Pharmacol Ther. 1998;12(6):499–507. doi: 10.1046/j.1365-2036.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 28.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell Microbiol. 2005;7(12):1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 29.Huda-Faujan N, et al. The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem J. 2010;4:53–58. doi: 10.2174/1874091X01004010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd-Price J, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poot M, et al. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J Histochem Cytochem. 1996;44(12):1363–1372. doi: 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- 32.Poot M, Pierce RH. Detection of changes in mitochondrial function during apoptosis by simultaneous staining with multiple fluorescent dyes and correlated multiparameter flow cytometry. Cytometry. 1999;35(4):311–317. doi: 10.1002/(SICI)1097-0320(19990401)35:4<311::AID-CYTO3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 33.Dennerlein S, Rehling P. Human mitochondrial COX1 assembly into cytochrome c oxidase at a glance. J Cell Sci. 2015;128(5):833–837. doi: 10.1242/jcs.161729. [DOI] [PubMed] [Google Scholar]
- 34.Lewis JD, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18(4):489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152(2):327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Randle PJ. Regulatory interactions between lipids and carbohydrates: the glucose fatty acid cycle after 35 years. Diabetes Metab Rev. 1998;14(4):263–283. doi: 10.1002/(SICI)1099-0895(199812)14:4<263::AID-DMR233>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 37.Inagaki T, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103(10):3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han TK, et al. Organic cation transporter 1 (OCT1/mOct1) is localized in the apical membrane of Caco-2 cell monolayers and enterocytes. Mol Pharmacol. 2013;84(2):182–189. doi: 10.1124/mol.112.084517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rinaldo P, Cowan TM, Matern D. Acylcarnitine profile analysis. Genet Med. 2008;10(2):151–156. doi: 10.1097/GIM.0b013e3181614289. [DOI] [PubMed] [Google Scholar]
- 40.Ma C, et al. Citrobacter rodentium infection causes both mitochondrial dysfunction and intestinal epithelial barrier disruption in vivo: role of mitochondrial associated protein (Map) Cell Microbiol. 2006;8(10):1669–1686. doi: 10.1111/j.1462-5822.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 41.Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8(12):1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller KE, Sheetz MP. Axonal mitochondrial transport and potential are correlated. J Cell Sci. 2004;117(pt 13):2791–2804. doi: 10.1242/jcs.01130. [DOI] [PubMed] [Google Scholar]
- 43.Tinel H, et al. Active mitochondria surrounding the pancreatic acinar granule region prevent spreading of inositol trisphosphate-evoked local cytosolic Ca(2+) signals. EMBO J. 1999;18(18):4999–5008. doi: 10.1093/emboj/18.18.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez CA, et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science. 2016;353(6305):1249–1253. doi: 10.1126/science.aag3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meadows JA, Wargo MJ. Carnitine in bacterial physiology and metabolism. Microbiology (Reading) 2015;161(6):1161–1174. doi: 10.1099/mic.0.000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujisaka S, et al. Diet, genetics, and the gut microbiome drive dynamic changes in plasma metabolites. Cell Rep. 2018;22(11):3072–3086. doi: 10.1016/j.celrep.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ni J, et al. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci Transl Med. 2017;9(416):eaah6888. doi: 10.1126/scitranslmed.aah6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahe MM, Sundaram N, Watson CL, Shroyer NF, Helmrath MA. Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J Vis Exp. 2015;(97):52483. doi: 10.3791/52483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu GD, et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut. 2016;65(1):63–72. doi: 10.1136/gutjnl-2014-308209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whelan KA, et al. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene. 2017;36(34):4843–4858. doi: 10.1038/onc.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.