Abstract
Small extracellular vesicles (SEVs) are functional messengers of certain cellular niches that permit noncontact cell communications. Whether niche-specific SEVs fulfill this role in cancer is unclear. Here, we used 7 cell type–specific mouse Cre lines to conditionally knock out Vps33b in Cdh5+ or Tie2+ endothelial cells (ECs), Lepr+ BM perivascular cells, Osx+ osteoprogenitor cells, Pf4+ megakaryocytes, and Tcf21+ spleen stromal cells. We then examined the effects of reduced SEV secretion on progression of MLL-AF9–induced acute myeloid leukemia (AML), as well as normal hematopoiesis. Blocking SEV secretion from ECs, but not perivascular cells, megakaryocytes, or spleen stromal cells, markedly delayed the leukemia progression. Notably, reducing SEV production from ECs had no effect on normal hematopoiesis. Protein analysis showed that EC-derived SEVs contained a high level of ANGPTL2, which accelerated leukemia progression via binding to the LILRB2 receptor. Moreover, ANGPTL2-SEVs released from ECs were governed by VPS33B. Importantly, ANGPTL2-SEVs were also required for primary human AML cell maintenance. These findings demonstrate a role of niche-specific SEVs in cancer development and suggest targeting of ANGPTL2-SEVs from ECs as a potential strategy to interfere with certain types of AML.
Keywords: Hematology, Stem cells
Keywords: Leukemias
Introduction
Hematopoietic stem cells (HSCs) reside in a specialized microenvironment (or niche). The niche ensures hematopoietic homeostasis by controlling the self-renewal, differentiation, and migration of HSCs under steady-state conditions and in response to emergencies and injury (1–3). Data suggest that the bone marrow (BM) niche has a similar role in controlling the behavior of leukemia cells (4, 5); therefore, normal HSCs and leukemia cells reside in and compete for the same niche (6, 7). Moreover, leukemia cells can remodel the local environment to create a unique leukemia-permissive niche that supports leukemia cell survival at the expense of normal hematopoiesis (8–12). Targeting the active, bidirectional crosstalk between leukemia cells and their remodeled niche might be a potential strategy for leukemia therapy (13–19).
Small extracellular vesicles (SEVs) function as important cell-cell messengers (20). After being secreted by one cell, SEVs can induce physiological and pathological changes in recipient cells by transmitting molecular cargo that includes mRNA, microRNA, and proteins (20). The SEVs released by leukemia cells can promote leukemia cell growth in an autocrine manner (21, 22), or can suppress normal hematopoiesis (23, 24) and remodel the niche through a paracrine mechanism (24–30). One study showed that direct injection of leukemia cell–derived SEVs can suppress normal hematopoiesis in vivo, mimicking the effects of leukemia cells (31). These studies suggest that targeting leukemia-derived SEVs might be a potential therapeutic strategy.
Although accumulating studies have demonstrated the impact of leukemia cell–derived SEVs on normal hematopoiesis and the BM niche (27, 32, 33), little is known about how the SEVs from a specific niche cell type regulate normal HSCs and leukemia cells. To our knowledge, most studies have used SEV injection or in vitro coculture when exploring SEV function; however, this approach does not distinguish the origins of specific SEVs secreted by a specific cell type in vivo, which is a current knowledge gap in the field. Furthermore, it is unclear which SEV components contribute to leukemia progression and by which pathways they act. Therefore, the exact roles of SEVs secreted by different niche cells in regulating homeostasis and hematological malignancies in vivo are unknown.
Here, we applied an MLL-AF9–induced acute myeloid leukemia (AML) model and aimed to determine the roles of different niche cell–derived SEVs in HSC function and AML progression. In our previous study, we showed that vacuolar protein sorting 33b (Vps33b) is required for SEV maturation and secretion (21). In the present study, we used 7 cell type–specific mouse Cre lines to conditionally knock out Vps33b in endothelial cells (ECs), BM mesenchymal stem cells (MSCs), osteoprogenitor cells (OPCs), megakaryocytes (Mks), and spleen MSCs. We then examined the effects of reduced SEV secretion on AML progression as well as normal hematopoiesis.
Results
Bone marrow microenvironment–derived SEVs directly bind to leukemia stem cells.
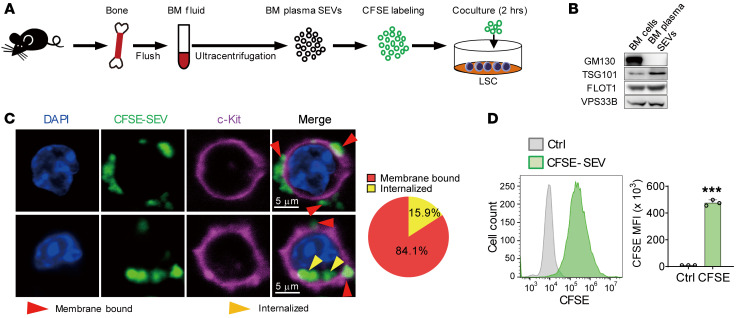
We first wanted to understand whether SEVs existing in the BM microenvironment can physically bind to leukemia cells or even leukemia stem cells (LSCs). To do so, we isolated SEVs from the BM and labeled them with CFSE (a cell-permeable dye) before coculturing with immunophenotypical LSCs (L-GMP cells) (34) or Mac-1+c-Kit+ LSCs for 2 hours (Figure 1A). Western blotting and electron microcopy confirmed the purity and characteristics of the isolated SEVs (Figure 1B and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI138986DS1). After 2 hours of coculture, 79%–85% of SEVs bound the membrane of LSCs, and 15%–21% SEVs were found in the cytoplasm (Figure 1, C and D, and Supplemental Figure 1B). In addition, prolonged coculture time did not obviously change the internalization percentage (Supplemental Figure 1C). These data indicate that BM microenvironment–derived SEVs can indeed target LSCs.
Figure 1. BM fluid SEVs bind to LSCs.
(A) The experimental procedure. BM cells were flushed out, and BM fluid SEVs were isolated by ultracentrifugation. The SEVs (100 μg) were labeled with CFSE and cocultured with 2 × 105 L-GMP cells or Mac-1+c-Kit+ AML cells. (B) Western blot analysis of GM130, TSG101, FLOT1, and VPS33B protein levels in BM cells and BM fluid SEVs. GM130 is a cis-Golgi marker. TSG101 and FLOT1 are SEV markers. (C) Left: Images of LSCs (L-GMP cells) after coculture with CFSE-labeled SEVs. Scale bars: 5 μm. Right: The statistics of membrane-bound and internalized SEVs. In total, 145 AML cells were counted after staining. (D) Flow cytometry (left) and histogram (right) analysis of the CFSE signal of L-GMP cells (n = 3). Ctrl, control. The data represent the means ± SD; ***P < 0.001, Student’s t test. Experiments were conducted 2 times for validation.
Reducing EC-derived SEV secretion prolongs survival in AML mice.
To determine whether Vps33b is required for SEV release by the different niche cells, we knocked down the Vps33b gene with shRNA lentivirus in primary mouse MSCs and ECs, isolated SEVs from supernatant by ultracentrifugation, then quantified the SEVs with nanoparticle tracking analysis and a bicinchoninic acid kit. The data demonstrated that knockdown of Vps33b in MSCs and ECs indeed decreased the number of nanoparticles and protein levels of secreted SEVs (Supplemental Figure 2).
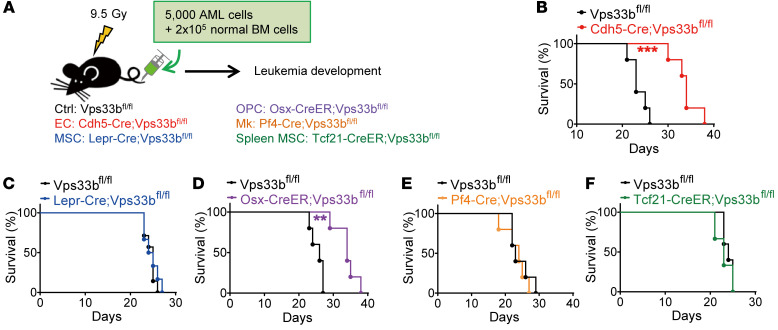
To define which cell sources of the SEVs are important for leukemia progression, we generated Cdh5-Cre;Vps33bfl/fl, Lepr-Cre;Vps33bfl/fl, Osx-CreER;Vps33bfl/fl, Pf4-Cre;Vps33bfl/fl, and Tcf21-CreER;Vps33bfl/fl mice to block SEV secretion from ECs, BM MSCs (35), OPCs (36), Mks (37, 38), and spleen MSCs (39), respectively (Figure 2A). The deletion efficiency of Vps33b in different models was confirmed by real-time quantitative PCR (qPCR) (Supplemental Figure 3). We injected 5000 AML cells together with 2 × 105 whole BM cells into lethally irradiated hosts and then monitored the effects on survival (Figure 2A). Survival was significantly extended up to 12 days in Cdh5-Cre;Vps33bfl/fl and Osx-CreER;Vps33bfl/fl AML mice, in which EC-derived or OPC-derived SEVs were reduced. No effect on AML progression was observed after reducing SEV secretion from BM MSCs, Mks, and spleen MSCs (Figure 2, B–F). We thus focused on EC-derived SEVs (EC-SEVs) in our subsequent experiments.
Figure 2. Blocking SEV secretion from ECs delays AML progression.
(A) The experimental design. (B–F) Survival curves of AML recipients with different genotypes. Cdh5-Cre;Vps33bfl/fl (B), Lepr-Cre;Vps33bfl/fl (C), Osx-CreER;Vps33bfl/fl (D), Pf4-Cre;Vps33bfl/fl (E), and Tcf21-CreER;Vps33bfl/fl (F) are shown (n = 5–8; **P < 0.01, ***P < 0.001, log-rank test). Experiments were conducted 3 times for validation.
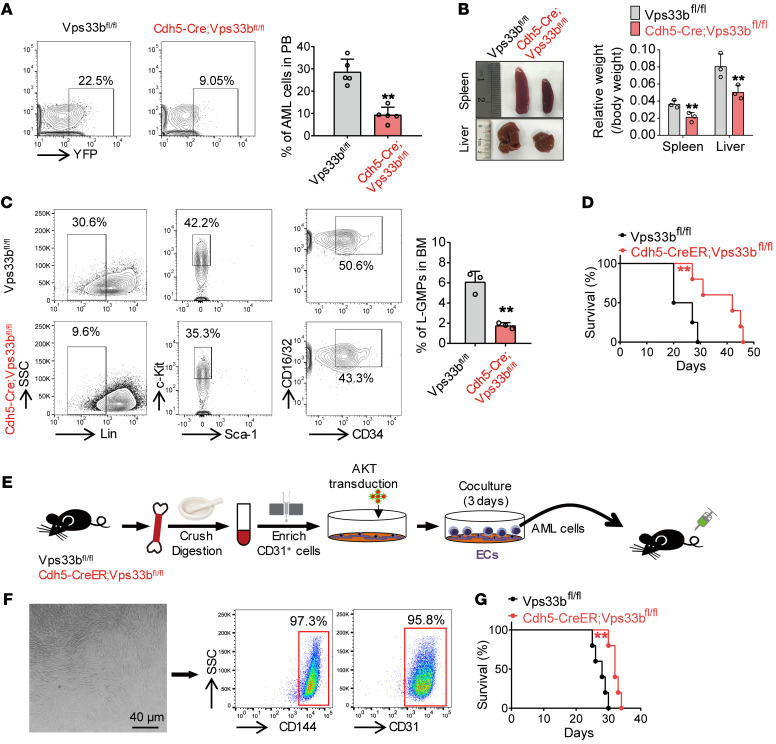
We next examined leukemia cells in peripheral blood (PB) from recipient mice 3 weeks after injection and found that the percentage of yellow fluorescent protein–positive (YFP+) AML cells was significantly decreased in Cdh5-Cre;Vps33bfl/fl mice compared with their control counterparts (22.5% vs. 9.05%, respectively; Figure 3A). To exclude the possibility that the delay in leukemia progression in Cdh5-Cre;Vps33bfl/fl mice was due to a homing defect of AML cells, we measured the homing efficiency of AML cells in Vps33bfl/fl and Cdh5-Cre;Vps33bfl/fl mice. We did not observe any significant changes in the percentage of homed AML cells in either the BM or the spleen (Supplemental Figure 4A). However, because the transplanted leukemia cell number was much more than that used for the survival analysis (2.5 × 105 vs. 5000), we have to admit that we cannot completely exclude the homing effect of Vps33b deletion on the AML progression unless other solid evidence is provided.
Figure 3. EC-SEVs support leukemia development.
(A) Flow cytometry (left) and histogram (right) analysis of the percentage of YFP+ leukemia cells in the peripheral blood (PB) of recipients 20 days after transplantation (n = 5; the data represent the means ± SD; **P < 0.01, Student’s t test). (B) Recipient spleen and liver size (left) and weight (right) 20 days after transplantation (n = 3; the data represent the means ± SD; **P < 0.01, Student’s t test). (C) Flow cytometry (left) and histogram (right) analysis shows the percentages of L-GMP cells in the BM of recipients (n = 3; the data represent the means ± SD; **P < 0.01, Student’s t test). (D) Survival curves of Cdh5-CreER;Vps33bfl/fl mice and Vps33bfl/fl control mice after AML cell injection (n = 4–5; **P < 0.01, log-rank test). (E) The experimental procedure for BM EC isolation and coculture with AML cells. In brief, BM from Cdh5-CreER;Vps33bfl/fl mice or Vps33bfl/fl control mice was crushed and digested, and enriched using anti-CD31 beads. The enriched ECs were transduced with AKT lentiviruses and cocultured with AML cells for 3 days. After coculture, the AML cells were injected into C57BL/6J recipients. (F) Left: The morphology of the enriched and cultured ECs. Scale bar: 40 μm. Right: Flow cytometry analysis to determine the purity of cultured ECs. (G) Survival curves of C57BL/6J recipients injected with AML cells cocultured with WT ECs or Vps33b-null ECs (n = 5; **P < 0.01, log-rank test). Experiments were conducted 2–4 times for validation.
LSCs are thought to be a key cellular element in leukemia initiation, maintenance, and relapse (40, 41). We next evaluated the frequency of Mac-1+Gr-1– leukemia cells (enriched for LSCs). The proportion of these cells was markedly reduced in Cdh5-Cre;Vps33bfl/fl mice (44.9% vs. 18.5%; Supplemental Figure 4B). The sizes of spleens and livers from Cdh5-Cre;Vps33bfl/fl recipients were much smaller compared with those in control recipients, indicating reduced infiltration in these organs (Figure 3B). More importantly, the number of L-GMP cells in Cdh5-Cre;Vps33bfl/fl recipients was lower than that in control recipients (Figure 3C). In addition, we obtained similar results in Tie2-Cre;Vps33bfl/fl mice, in which SEV secretion was also reduced from ECs (Supplemental Figure 4, C–F). Taken together, with 2 independent EC reporter mouse strains, our data strongly suggest that reducing EC-derived SEVs delays leukemia onset, decreases LSC numbers, and impairs LSC infiltration.
Because neither Cdh5-Cre nor Tie2-Cre is absolutely specific to ECs, we further generated a new Cdh5-CreER knockin mouse strain. We first generated Cdh5-CreER;R26-tdTomato mice to verify the utility of this new knockin mouse strain (Supplemental Figure 4, G and H). Consistent with our previous findings, Cdh5-CreER;Vps33bfl/fl recipients also exhibited longer survival than control mice when injected with AML cells (Figure 3D). To further demonstrate the importance of EC-SEVs to AML cells, we isolated ECs from Cdh5-CreER;Vps33bfl/fl and control mice (42, 43) and cocultured them with the same number of AML cells (Figure 3E). We confirmed the purity of the isolated ECs by flow cytometry (Figure 3F). We also characterized the SEVs secreted from the cultured ECs (Supplemental Figure 4, I–K). After 3 days of coculture, AML cells were collected and transplanted into WT mice. We found that exposure to AML cells cocultured with Vps33b-knockout ECs prolonged the survival of AML mice (Figure 3G). Collectively, our data demonstrate that EC-SEVs are of bona fide importance to leukemia progression.
Reducing EC-derived SEV production has no effect on hematopoiesis.
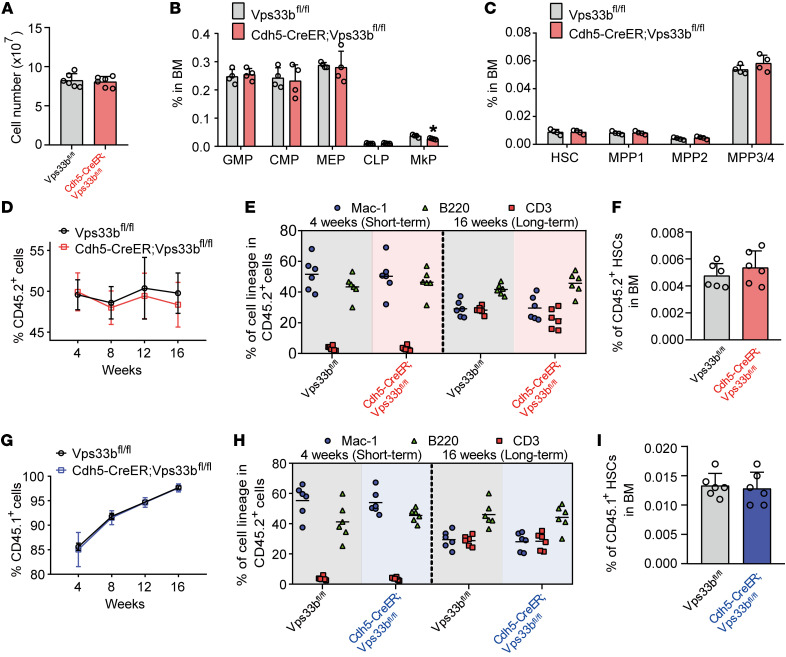
To test whether reducing EC-SEVs in vivo affects normal hematopoiesis, we treated 4-week-old Cdh5-CreER;Vps33bfl/fl and littermate control mice with tamoxifen for 5 days. After 1 month, we measured the cellularity of the BM and the frequencies of hematopoietic stem cells and progenitor cells. We found similar results between Cdh5-CreER;Vps33bfl/fl mice and littermate controls (Figure 4, A–C, and Supplemental Figure 5A), indicating that reducing EC-SEVs does not affect normal hematopoiesis in hemostasis. We confirmed this result by using spectral cytometry (Supplemental Figure 5, B and C). We also found no changes in EC numbers in the bone, BM, or spleen (Supplemental Figure 5, D and E), suggesting that EC-SEVs are not required for EC survival under steady state.
Figure 4. EC-SEVs have no effect on normal hematopoiesis.
(A–C) Cellularity analysis of Cdh5-CreER;Vps33bfl/fl mice and Vps33bfl/fl littermate control mice. The total BM cell numbers (A), frequencies of hematopoietic progenitor cells (B), and frequencies of HSCs/multipotent progenitors (MPPs) (C) are shown (n = 4–6; the data represent the means ± SD, Student’s t test). (D) The percentage of CD45.2+ donor cells in the PB of recipient mice at the indicated time points after competitive BM transplantation. The donor cells from Cdh5-CreER;Vps33bfl/fl mice and Vps33bfl/fl littermate control mice were CD45.2+ (n = 10; the data represent the means ± SEM, Student’s t test). (E) The short-term and long-term multilineage reconstitution capacities of donor-derived (CD45.2) PB cells in recipients (n = 6; Student’s t test). (F) The percentage of CD45.2+ donor-derived HSCs in the BM of recipients (n = 6; the data represent the means ± SD, Student’s t test). (G) The percentage of CD45.1+ donor cells in the PB of recipient mice at the indicated time points after BM transplantation. The donor cells were CD45.1+ (n = 10; the data represent the means ± SEM, Student’s t test). (H) The short-term and long-term multilineage reconstitution capacities of donor-derived (CD45.1) PB cells in recipients (n = 6; Student’s t test). (I) The percentage of CD45.1+ donor-derived HSCs in the BM of recipients (n = 6; the data represent the means ± SD, Student’s t test). Experiments were conducted 2–3 times for validation. GMP, granulocyte-macrophage progenitor; CMP, common myeloid progenitor; MEP, megakaryocyte-erythrocyte progenitor; CLP, common lymphoid progenitor; MkP, megakaryocyte progenitor.
To stringently test whether reducing EC-SEVs affects HSC function in vivo, we performed a competitive BM transplantation assay. We found that the repopulation ability of BM cells from Cdh5-CreER;Vps33bfl/fl mice was similar to that of BM cells from control mice (Figure 4D). The donors (CD45.2+) from the 2 groups had the same capacity to generate myeloid (Mac-1+) and lymphoid (CD3+ and B220+) lineages (Figure 4E) and to output HSCs (Figure 4F). These results indicate that reducing EC-SEVs has no effect on the reconstitution potential of HSCs from Cdh5-CreER;Vps33bfl/fl mice.
We then performed a reciprocal transplantation experiment in which we transplanted WT BM cells (CD45.1+) into Cdh5-CreER;Vps33bfl/fl and littermate control mice. The reconstitution ability of the donor cells in Cdh5-CreER;Vps33bfl/fl mice did not differ from that in controls with respect to repopulation efficiency (Figure 4G), lineage regeneration (Figure 4H), and HSC output (Figure 4I). We confirmed this finding in another independent transplantation experiment in which we transplanted WT CD45.1+ cells into Tie2-Cre;Vps33bfl/fl or Vps33bfl/fl mice. After 16 weeks of reconstitution, we sorted CD45.1+ BM cells from the primary recipients and cotransplanted them with CD45.2+ competitor cells at a 1:1 ratio into secondary recipients (Supplemental Figure 6A). We found that CD45.1+ donor cells from Tie2-Cre;Vps33bfl/fl mice had the same reconstitution capacity as cells from Vps33bfl/fl mice (Supplemental Figure 6, B–D). Together, these data demonstrate that reducing EC-SEVs has no effect on HSC function in vivo. We thus conclude that EC-SEVs are dispensable for normal hematopoiesis.
EC-derived SEVs contain high ANGPTL2 protein levels.
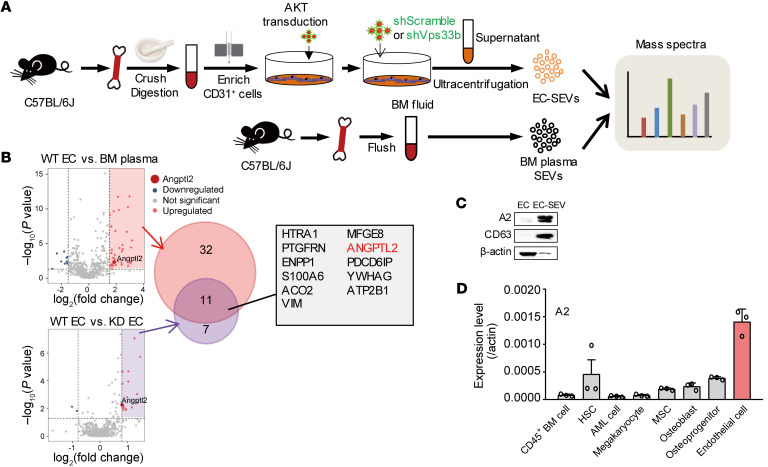
Given that SEVs can transmit molecular cargos into recipient cells, we presumed that EC-SEVs might deliver proteins to support AML development. To address this hypothesis, we undertook a global comparison of the protein cargos contained within SEVs from WT ECs, Vps33b-knockdown ECs (Supplemental Figure 7A), and BM fluid by mass spectrometry (Figure 5A). Compared with the BM fluid and Vps33b-knockdown ECs, the WT EC-SEVs contained a very high level of angiopoietin-like protein 2 (ANGPTL2) (Figure 5B). Western blot analysis confirmed this observation (Figure 5C).
Figure 5. EC-SEVs contain high levels of ANGPTL2 protein.
(A) The experimental procedure for mass spectra analysis. The SEVs were collected from the culture supernatant of WT ECs, Vps33b-knockdown ECs, and normal BM fluid. The proteins were extracted from the SEVs for mass spectrometry. (B) Mass spectra results. The overlap in the proteins enriched in WT EC-SEVs versus BM-SEVs and WT EC-SEVs versus Vps33b-knockdown (KD) EC-SEVs is shown. (C) Western blot analysis of ANGPTL2 (A2), β-actin, and CD63 protein levels in BM ECs and EC-SEVs. (D) qRT-PCR detection of Angptl2 mRNA in the indicated cells (n = 3; the data represent the means ± SD). β-Actin was used as an internal control. Experiments were conducted 2–3 times for validation.
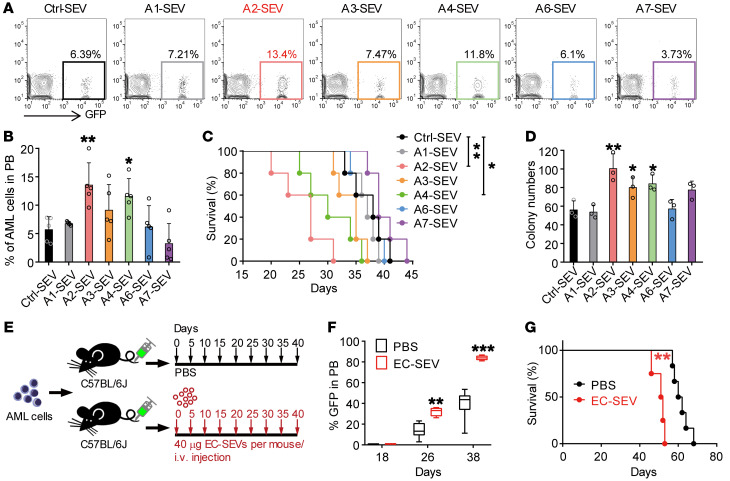
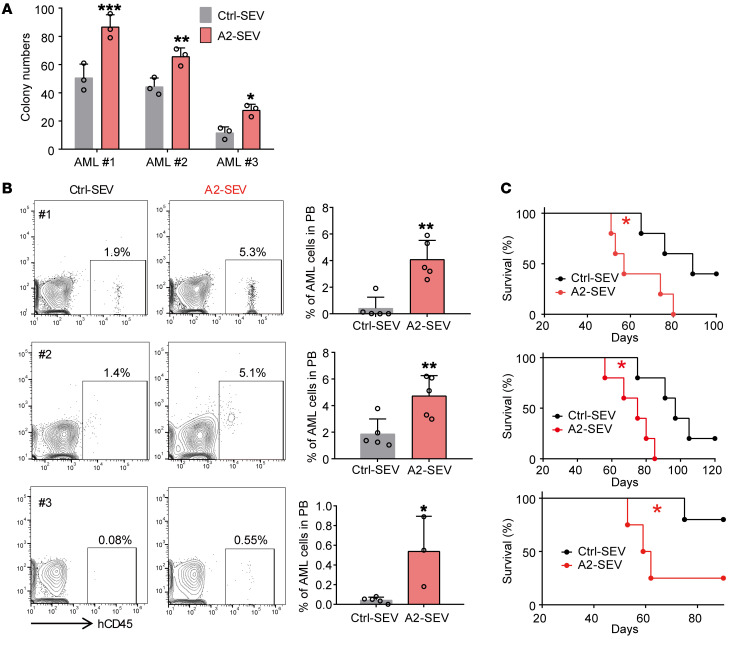
There are 6 murine ANGPTL family members: ANGPTL1, 2, 3, 4, 6, and 7. We next isolated CD45+ cells, HSCs, AML cells, Mks, MSCs, osteoblasts (OBs), OPCs, and ECs by flow cytometry (Supplemental Figure 7, B–I) and performed real-time qPCR to examine the ANGPTL expression levels in these cells. Among the 6 members, Angptl2 exhibited the highest mRNA expression in ECs (Figure 5D and Supplemental Figure 7J). These data suggest that ANGPTL2 expression in EC-SEVs might have important roles in supporting AML development. To test this hypothesis, we conducted a functional assay whereby we transduced 293T cells with an empty vector (Ctrl) or ANGPTL1, ANGPTL2, ANGPTL3, ANGPTL4, ANGPTL6, or ANGPTL7 vector and collected the supernatant. Then we purified the SEVs via ultracentrifugation (Supplemental Figure 7K) and confirmed ANGPTL protein expression by Western blot (Supplemental Figure 7L). We cocultured 1 × 105 AML cells with each set of transduced SEVs (3 μg) for 3 days, then collected the AML cells for in vivo transplantation and in vitro colony assay (Supplemental Figure 7K). Compared with Ctrl-SEVs, ANGPTL2-SEVs and ANGPTL4-SEVs accelerated AML progression and reduced the survival of AML mice, whereas ANGPTL1-, ANGPTL3-, ANGPTL6-, or ANGPTL7-containing SEVs did not affect AML progression (Figure 6, A–C). In addition, ANGPTL2-SEV and ANGPTL4-SEV exposure resulted in an increase in the colony numbers of AML cells after coculture (Figure 6D). Notably, ANGPTL2-SEVs had a stronger effect on leukemia progression than ANGPTL4-SEVs. Because the EC-SEVs contained a very high level of ANGPTL2 (Figure 5C), we took ANGPTL2 forward for further analysis. We collected SEVs from the EC culture medium and injected them into AML mice every 5 days for 40 days (Figure 6E). We found that EC-SEVs could accelerate leukemia progression (Figure 6, F and G). We thus conclude that ANGPTL2-SEVs secreted by ECs support AML development.
Figure 6. ANGPTL2-SEVs from ECs support leukemia development.
(A and B) Flow cytometry (A) and histogram (B) analysis of the percentages of GFP+ leukemia cells in the PB of recipients 16 days after transplantation (n = 5; the data represent the means ± SD; *P < 0.05, **P < 0.01, 1-way ANOVA followed by Dunnett’s test). Ctrl, control. (C) Survival analysis of recipients injected with AML cells cocultured with the indicated SEVs (n = 5; *P < 0.05, **P < 0.01, log-rank test). (D) The colony-forming ability of AML cells cocultured with the indicated SEVs (n = 3; the data represent the means ± SD; *P < 0.05, **P < 0.01, 1-way ANOVA followed by Dunnett’s test). (E) The in vivo administration of EC-SEVs. Equal volumes of PBS or EC-SEVs (40 μg) were intravenously injected every 5 days for 40 days. (F) The percentages of GFP+ leukemia cells in the PB of the recipients at indicated time points (n = 4–6; **P < 0.01, ***P < 0.001, Student’s t test). (G) Survival curves of AML recipients injected with PBS or EC-SEVs (n = 4–6; **P < 0.01, log-rank test). Experiments were conducted 2–3 times for validation.
VPS33B regulates ANGPLT2-SEV release from ECs to support AML development.
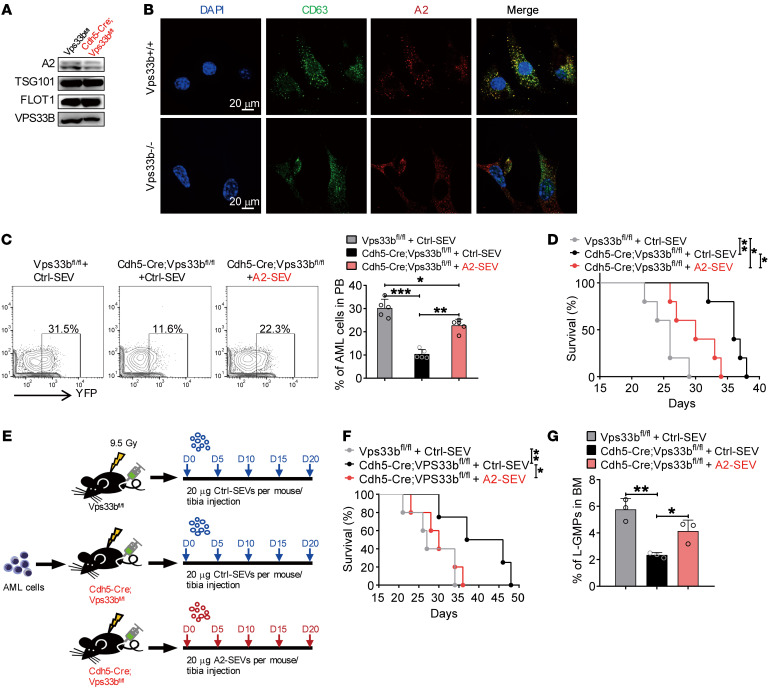
Because SEV maturation and release is regulated by VPS33B (21), we attempted to investigate whether ANGPTL2-SEVs from ECs are also controlled by VPS33B. Western blot analyses showed decreased ANGPTL2 protein level in BM fluid from Cdh5-CreER;Vps33bfl/fl mice compared with the expression levels in control mice (Figure 7A). Vps33b knockout in ECs or VPBS33B knockdown in 293T cells impaired ANGPTL2-SEV formation (Figure 7B and Supplemental Figure 8A), indicating that VPS33B is a key regulator of ANGPTL2-SEV maturation. Our protein analyses also confirmed that the ANGPLT2 protein level was reduced in EC-SEVs from Vps33b-knockdown ECs (Figure 5B).
Figure 7. VPS33B regulates ANGPTL2-SEV release.
(A) Western blot analysis of ANGPTL2, TSG101, FLOT1, and VPS33B protein levels in BM fluid SEVs from Vps33bfl/fl mice and Cdh5-Cre;Vps33bfl/fl mice. (B) Colocalization of CD63 and ANGPTL2 in WT and VPS33B-null ECs. (C) Flow cytometry (left) and histogram (right) analysis of the percentages of YFP+ AML cells in the PB of the indicated recipients injected with AML cells cocultured with Ctrl-SEVs or ANGPTL2-SEVs (n = 5; the data represent the means ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, 1-way ANOVA with Tukey’s multiple-comparison test). (D) Survival analysis of the indicated recipients injected with AML cells cocultured with Ctrl-SEVs or ANGPTL2-SEVs (3 μg per 1 × 105 AML cells) (n = 5; *P < 0.05, **P < 0.01, log-rank test). (E) The in vivo administration of ANGPTL2-SEVs into Cdh5-Cre;Vps33bfl/fl mice and Vps33bfl/fl control recipients. Equal volumes of Ctrl- or ANGPTL2-SEVs (20 μg) were intratibially injected every 5 days for 20 days. (F) Survival analysis of AML recipients injected with Ctrl- or ANGPTL2-SEVs (n = 5; *P < 0.05, **P < 0.01, log-rank test). (G) The percentage of L-GMP cells in BM of indicated recipients (n = 5; the data represent the means ± SD; *P < 0.05, **P < 0.01, 1-way ANOVA with Tukey’s multiple-comparison test). Experiments were conducted 2 times for validation.
Next, we wanted to determine whether the SEVs from the 293T cells after VPBS33B knockdown were deficient in supporting AML cells. Here, we infected 293T cells with shScramble (Ctrl, #1) or shVPS33B (#2), then transduced the Ctrl or knockdown cells with an ANGPTL2 vector. We purified SEVs from the supernatant (#1 and #2) and then incubated them with AML cells. After 3 days of culture, we injected the AML cells into recipient mice to induce leukemia (Supplemental Figure 8B, #1 and #2). We found that the SEVs from shVPS33B-knockdown 293T cells impaired AML cell function and that this effect resulted in extended survival of the recipient mice (Supplemental Figure 8, C and D). We next established a rescue experiment whereby we added normal ANGPTL2-SEVs to the SEVs collected from shVPS33B-transduced 293T cells (#3) and cocultured these with AML cells. This coculture restored AML cell function, as injection of these AML cells into recipients could accelerate leukemia progression (Supplemental Figure 8, C and D). This finding indicates that VPS33B controls ANGPTL2-SEV release to support leukemia development.
To further demonstrate the role of VPS33B in regulating ANGPTL2-SEV release in ECs, we injected AML cells cocultured with Ctrl-SEVs or ANGPTL2-SEVs (Supplemental Figure 7K) into Cdh5-CreER;Vps33bfl/fl mice. We found that including ANGPTL2-SEVs in the culture medium enhanced the function of AML cells, as exhibited by the accelerated leukemia progression in injected mice (Figure 7C) and the shortened survival time of leukemia recipients (Figure 7D). In an independent experiment, we directly injected ANGPTL2-SEVs into the tibia of recipients every 5 days for 20 days after AML cell injection (Figure 7E). Cdh5-CreER;Vps33bfl/fl recipient mice injected with ANGPTL2-SEVs showed a survival profile similar to that seen in Vps33bfl/fl control recipients (Figure 7F), indicating that ANGPTL2-SEVs alone can rescue delayed AML progression caused by impaired ANGPTL2-SEV release from ECs. In addition, ANGPTL2-SEV injection increased the LSC number to the same level as in Vps33bfl/fl controls (Figure 7G). We also cocultured Angptl2-knockdown BM ECs (Supplemental Figure 9, A and B) with AML cells for 3 days in Transwell. AML cells were then collected, counted (Supplemental Figure 9C), and transplanted into WT mice. The dynamics of AML cells in peripheral blood (Supplemental Figure 9D) and the survival of recipient mice (Supplemental Figure 9E) were examined, which showed that knockdown of Angptl2 in ECs indeed reduced the protein level of ANGTPL2 in EC-SEVs and delayed the AML progression. The results demonstrate that VPS33B regulates ANGPTL2-SEV release by ECs to support AML development.
ANGPTL2-SEVs bind to PIRB/LILRB2 to support AML development.
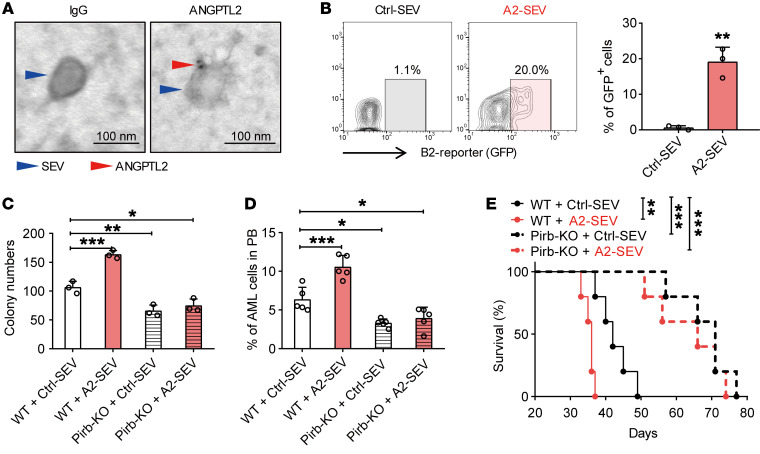
The data thus far suggested that ANGPTL2-SEVs support AML development, but the underlying mechanisms of this regulation were unclear. To fill this gap, we transduced 293T with an ANGPTL2-mCherry or Ctrl vector and collected the SEVs from the 293T culture medium. We then cocultured SEVs with LSCs to determine whether ANGPTL2-SEVs can bind to LSCs. Flow cytometric analyses showed increased mCherry expression in LSCs after coculture with ANGPTL2-mCherry-SEVs (Supplemental Figure 10A). We next used CFSE to label the SEVs from the supernatant of 293T cells transduced with ANGPTL2 and also found that the ANGPTL2-SEVs could bind to the surface of Mac-1+c-Kit+ LSCs (Supplemental Figure 10B). Moreover, consistent with our previous study showing that ANGPTL2-SEVs can bind to LILRB2 on HSCs (21), we observed by immunoelectron microscopy that ANGPTL2 anchored on the SEV surface (Figure 8A). A reporter-based system further verified that ANGPTL2-SEVs could bind to LILRB2 on leukemia cells (Figure 8B). These results indicate that ANGPTL2-SEVs bind to LSCs via its receptor LILRB2.
Figure 8. ANGPTL2-SEVs bind to LILRB2.
(A) Immunoelectron microscopy showing the location of ANGPTL2 protein in SEVs. Scale bars: 100 nm. (B) Flow cytometry (left) and histogram (right) analysis of ANGPTL2-SEVs binding to LILRB2-chimeric reporter cells. In this experiment, LILRB2-chimeric reporter cells will express GFP when ANGPTL2-SEVs bind to LILRB2 on the cell surface (n = 3; the data represent the means ± SD; **P < 0.01, Student’s t test). The stable chimeric receptor reporter cell system has been previously described (49). The extracellular domain of LILRB2 was chimerically fused with the intracellular domain of paired immunoglobulin-like receptor (PILR). Upon binding with ANGPTL2-SEVs, the receptor can further recruit the adaptor DAP-12 to activate the NFAT promoter, followed by the increase in GFP expression in reporter cells. (C) Colony-forming ability of Pirb+/+ and Pirb–/– AML cells cocultured with control- or ANGPTL2-SEVs (10 μg per 50,000 AML cells) (n = 3; the data represent the means ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, 1-way ANOVA followed by Dunnett’s test). (D) The percentage of AML cells in the PB of recipients injected with Pirb+/+ and Pirb–/– AML cells cocultured with control- or ANGPTL2-SEVs (n = 5; the data represent the means ± SD; *P < 0.05, ***P < 0.001, 1-way ANOVA followed by Dunnett’s test). (E) Survival curves of recipients injected with Pirb+/+ and Pirb–/– AML cells cocultured with control- or ANGPTL2-SEVs (n = 5; **P < 0.01, ***P < 0.001, log-rank test). Experiments were conducted 2–4 times for validation.
Pirb is the murine homolog gene for human LILRB2. Thus, to demonstrate that ANGPTL2-SEVs support AML development via LILRB2, we transformed Pirb–/– c-Kit+ BM cells into AML cells and cocultured the resulting cells with Ctrl-SEVs or ANGPTL2-SEVs. After coculture, we conducted colony assays and transplantation experiments. First, ANGPTL2-SEV coculture improved the colony-forming ability of Pirb+/+ AML cells 1.5-fold, but ANGPTL2-SEVs were unable to stimulate Pirb–/– AML cells to produce more colonies (Figure 8C). Importantly, Pirb–/– AML cells cocultured with Ctrl-SEVs formed fewer colonies than Pirb+/+ AML cells (Figure 8C), indicating that ANGPTL2-SEVs exert their effects on AML cells via PIRB in vitro.
For the in vivo transplantation experiments, we also found that Pirb–/– AML cells cocultured with ANGPTL2-SEVs had no effect on leukemia progression, while Pirb+/+ AML cells did (Figure 8, D and E). Pirb+/+ AML cells cocultured with ANGPTL2-SEVs aggravated splenomegaly in the recipients compared with Pirb+/+ AML cells cocultured with Ctrl-SEVs (Supplemental Figure 10, C and D). However, the sizes of spleens from recipients injected with Pirb–/– AML cells cocultured with Ctrl-SEVs and ANGPTL2-SEVs were similar, and much smaller than the sizes in recipients injected with Pirb+/+ AML cells (Supplemental Figure 10, C and D). These results indicate that loss of LILRB2 eliminates the effect of ANGPTL2-SEVs on AML cells. Moreover, after 24 hours of ANGPTL2-SEV treatment, both phosphorylated SHP2 (p-SHP2) and p-CREB levels were significantly increased in WT AML cells, but not in Pirb–/– AML cells, suggesting that Pirb depletion increases the survival of AML cells through the p-SHP2/p-CREB pathway, independently of ANGPTL2 presence (Supplemental Figure 10E). The results indicate that ANGPTL2-SEVs must bind to LILRB2 to support leukemia development in vivo.
ANGPTL2-SEVs maintain self-renewal of primary human AML cells.
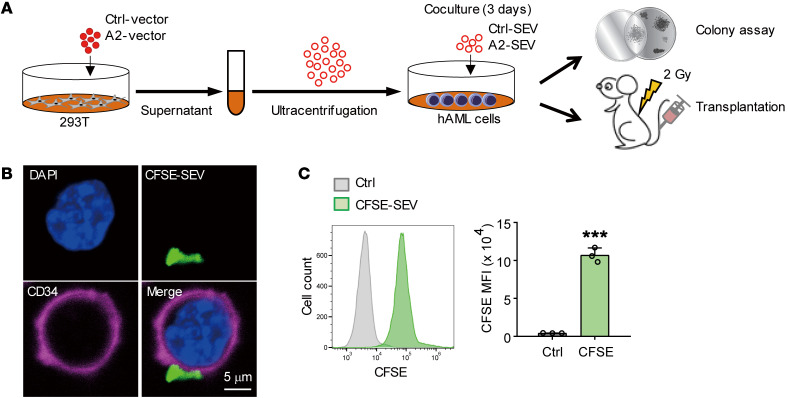
In our final analyses, we investigated whether ANGPTL2-SEVs could also support human leukemia cell growth. To this aim, we cocultured primary human AML cells with Ctrl-SEVs or ANGPTL2-SEVs, then performed colony-forming and xenotransplantation assays (Figure 9A). Similar to our findings in the mouse AML model, CFSE-labeled ANGPTL2-SEVs could target CD34+-enriched LSCs (Figure 9, B and C). AML cells cocultured with ANGPTL2-SEVs formed more colonies than those cocultured with Ctrl-SEVs (Figure 10A). Our xenotransplantation assay showed that ANGPTL2-SEVs accelerated AML progression (Figure 10B) and shortened the survival of recipient mice (Figure 10C). These data demonstrate that ANGPTL2-SEVs maintain the function of human AML cells.
Figure 9. ANGPTL2-SEVs can target human LSCs.
(A) The experimental procedure for studying the effect of ANGPTL2-SEVs on human AML cells. 293T cells were transduced with Ctrl vectors or ANGPTL2 vectors. Ctrl-SEVs or ANGPTL2-SEVs were collected from the supernatant of the transduced 293T cells. Human AML cells (1 × 106) were cocultured with 10 μg Ctrl-SEVs or ANGPTL2-SEVs, and then colony assay and transplantation into NOD/SCID mice were performed. (B) Images of CD34+ human AML cells captured after coculture with CFSE-labeled SEVs. Scale bar: 5 μm. (C) Flow cytometry (left) and histogram (right) analysis of the CFSE signals of CD34+ human AML cells (n = 3; the data represent the means ± SD; ***P < 0.001, Student’s t test). Experiments were conducted 3 times for validation.
Figure 10. ANGPTL2-SEVs support human AML development.
(A) The colony-forming ability of human AML cells after coculture with Ctrl-SEVs or ANGPTL2-SEVs (n = 3; the data represent the means ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test). (B) Flow cytometry (left) and histogram (right) analysis of the percentages of human CD45+ AML cells in the PB of recipients injected with AML cells after coculture with Ctrl-SEVs or ANGPTL2-SEVs (n = 3–5; the data represent the means ± SD; *P < 0.05, **P < 0.01, Student’s t test). (C) Survival analysis of recipients injected with AML cells after coculture with Ctrl-SEVs or ANGPTL2-SEVs (n = 5; *P < 0.05, log-rank test). Experiments were conducted 2–3 times for validation.
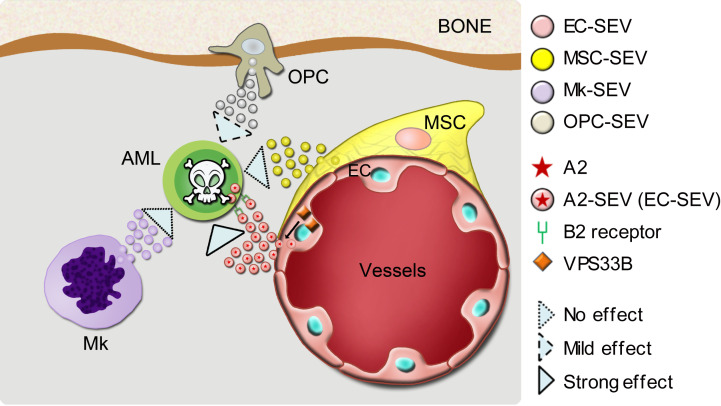
To evaluate the supportive roles of human EC-derived SEVs in human AML cells, we isolated SEVs from primary HUVEC cells (HUVEC-SEVs) and cocultured them with human AML cells for 60 hours. We used normal culture medium without SEVs as a control. First, the HUVEC-SEVs maintained CD34+ AML cells in the culture. We then observed that the percentage of CD34+ AML cells was higher when the cells were cocultured with HUVEC-SEVs than when they were cultured without SEVs (Supplemental Figure 11A). Importantly, both HUVEC cells and HUVEC-SEVs contained a high level of ANGPTL2 protein (Supplemental Figure 11B), suggesting that HUVEC-SEVs might also maintain CD34+ AML cells through ANGPTL2. In addition, supplementing the culture medium with HUVEC-SEVs enhanced the function of AML cells: these cells exhibited an increased colony-forming ability (Supplemental Figure 11, C and D) and accelerated leukemia progression (Supplemental Figure 11, E and F). We demonstrate that VPS33B controls the homeostasis of ANGPTL2-SEVs of ECs, which further enhances leukemia development via its binding to the LILRB2/PIRB receptor. These data provide a paradigm as to how niche cell–derived SEVs support AML development (Figure 11).
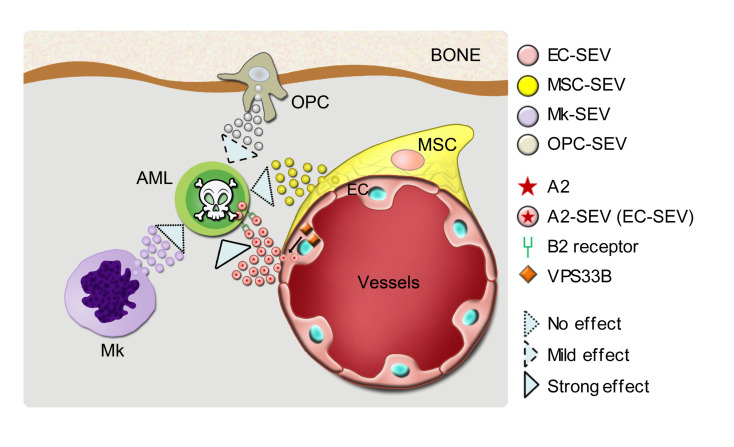
Figure 11. A model showing how EC-derived SEVs regulate AML development.
VPS33B controls the release of ANGPTL2-SEVs from ECs, which further enhances leukemogenesis via its binding to the receptor LILRB2.
Discussion
This study provides the first evidence to our knowledge that SEVs from a specific niche cell type support cancer development in the multicellular tissue microenvironment. Specifically, we show that EC-derived SEVs contained high levels of ANGPTL2 and support AML development by binding to LILRB2; ANGPTL2-SEVs released from ECs were controlled by VPS33B. Importantly, ANGPTL2 SEVs were required for primary human AML cell maintenance. A previous study showed that T cell acute lymphoblastic leukemia (T-ALL) cells directly contact CXCL12-producing ECs, and that a Cxcl12 deletion in ECs suppresses T-ALL progression (19). Our imaging analysis also showed that during AML progression, AML cells prefer to localize around ECs rather than being stochastically distributed (data not shown), indicating the importance of ECs in supporting different types of leukemia. In our previous study, we demonstrated that VPS33B regulates SEV autocrine signaling to mediate leukemogenesis (21), while this study dissected different contributions of niche cell–derived SEVs to leukemia progression and uncovered the mechanism by which VPS33B regulates SEVs paracrine from ECs to mediate leukemia development.
BM ECs are heterogeneous and have different subtypes, including sinusoidal ECs, arterial ECs, and arteriolar/capillary ECs (44–47). These 3 EC subtypes have different locations and secrete different levels of cytokines (45, 47), which likely underlies their different contributions to HSC maintenance in the BM. Future studies should use specific Cre mice to dissect the contributions of SEVs from different ECs to AML progression and explore the common mechanisms underlying how EC-derived SEVs regulate different types of leukemia.
The BM niche compartment includes a complex network of MSC, OPCs, osteoblasts, ECs, sympathetic nerves, nonmyelinating Schwann cells, Mks, and adipocytes (2, 3, 8, 11, 12). In this study, we only included ECs, MSCs, OPCs, and Mks. The importance of the SEVs derived from other niche cell types deserves further investigation, as they might provide additional insight into AML progression. To our surprise, blocking SEV secretion from MSCs had no effect on AML progression. One possibility to explain this finding is that VPS33B might regulate the maturation and release of only certain types of SEV present in different cell types (21). This possibility might lead us to ignore the effects of MSC-derived SEVs on leukemia. To address this critical question, different genes that are essential for SEV maturation and secretion can be knocked out in MSCs. Although we have shown that Vps33b regulates the SEV maturation and release, and deletion of Vps33b in ECs inhibits leukemia progression, we have to admit that Vps33b may regulate leukemia progression through other pathways in addition to SEV effects. Meanwhile, as described in our previous study (21), not all of the ANGPTL2 protein exists in the SEVs, and the non-SEV form of ANGPTL2 may also be required for the leukemia progression. Moreover, we found that leukemia progression was delayed when SEV secretion was blocked from OPCs; we will study the OPC-associated mechanisms in an independent study.
Interestingly, we noticed that ANGPTL2 could be localized on the SEV surface. This finding is consistent with a recent report showing that PD-L1 can be sorted to the SEV surface to mediate its immunosuppressive effects via binding to the receptor PD-1 (48). This finding reveals the unique pathway by which SEV components exert their pathological function in leukemia progression and might constitute a general SEV-mediated mechanism regarding the interactions between niche components and cancer cells. Although we showed that VPS33B is critical for ANGPTL2 sorting into SEVs, whether other vascular regulator proteins can also coordinate the precise localization of ANGPTL2 to the SEV membrane is unclear. ANGTPL2 is involved in inflammation, lipid metabolism, vascular remodeling, and tumorigenesis. Whether ANGPTL2-SEVs have major roles in these physiological and pathological situations needs further investigation.
In summary, our results in murine and xenograft models demonstrate that ANGPTL2-SEVs or EC-SEVs have cross-species effects. Both murine and human AML cells can be targeted and regulated by EC-derived ANGPTL2-SEVs. Moreover, ECs express much higher Vps33b and Angptl2 in leukemic BM than in healthy BM (Supplemental Figure 12A), indicating that leukemia cells require ANGPTL2-SEVs to survive, and that they therefore stimulate ECs to produce more ANGPTL2-SEVs. Importantly, the expression of LILRB3 was higher in AML than in normal HSCs and myeloid progenitors (Supplemental Figure 12B), which may explain why ANGPTL2-SEVs can affect AML cells but not normal hematopoiesis. This clinically relevant finding suggests that targeting of ANGPTL2-SEVs alone, or more likely in combination with established chemotherapy regiments, might be a potential treatment. Consistent with a previous study (13, 14, 19), our data provide definitive evidence that targeting the leukemia microenvironment is a powerful strategy for leukemia therapy. Although this study is in the context of leukemia, it is reasonable to believe that SEVs might be relevant to other cancers as well.
Methods
Supplemental Methods are provided online with this article.
The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD020946.
Animals.
To specifically delete Vps33b in murine ECs, BM MSCs, Mks, OPCs, or spleen stromal cells, Vps33bfl/fl mice were crossed with Cdh5-Cre, Tie2-Cre, Cdh5-CreER, Lepr-Cre, Pf4-Cre, Osx-CreER, or Tcf21-CreER, respectively. Cre–Vps33bfl/fl littermate mice served as controls. Tcf21-CreER transgenic mice and Cdh5-CreER knockin mice were generated by Beijing Biocytogen Co. Ltd. Six- to eight-week-old C57BL/6 CD45.2 mice, CD45.1 mice, and NOD/SCID mice were purchased from the Shanghai SLAC Laboratory Animal Co. Ltd. or Beijing Vital River Laboratories. Pirb–/– and Col2.3-GFP mice were kept in our institutes. All mice used in this study were 8–10 weeks old and female. To induce deletion by CreER, 4- to 6-week-old mice were given 1 mg tamoxifen (MilliporeSigma) in 100 μL corn oil (MilliporeSigma) daily by intraperitoneal injections for 5 consecutive days. All animals were maintained at the Animal Core Facility of Shanghai Jiao Tong University School of Medicine and the State Key Laboratory of Experimental Hematology.
Statistics.
Statistical analysis was performed using GraphPad Prism v8.0 and SPSS v19.0 (IBM). Significance was calculated using an unpaired 2-tailed Student’s t test for data with 2 groups. For data with more than 2 groups, a 1-way ANOVA with Tukey’s multiple-comparison test or a 1-way ANOVA followed by Dunnett’s test was used. The statistical significance and details of the technical or biological replicates are reported in the respective figure legends. P values less than 0.05 were considered statistically significant. Statistical significance is indicated by asterisks: *P < 0.05, **P < 0.01, ***P < 0.001.
Study approval.
Animal experiments were approved by our institutes and complied with the Guideline for Animal Care at the Shanghai Jiao Tong University School of Medicine and the State Key Laboratory of Experimental Hematology. Human samples were supplied by the Department of Hematology at the First People’s Hospital, Xinhua Hospital, or Tongren Hospital, Shanghai Jiao Tong University School of Medicine. Written informed consent was obtained from the patients and approved by the Ethics Committee for Medical Research (IRB) at Shanghai Jiao Tong University School of Medicine.
Author contributions
DH, GS, X. Hao, and X. He designed and performed the experiments, analyzed the data, and wrote the paper. ZZ, CC, ZY, LX, and SM helped with mouse experiments and flow cytometry. LL provided human samples. BOZ helped with manuscript preparation. HC, JZ, and TC proposed the study, designed the experiments, interpreted the results, wrote the paper, and oversaw the research project.
Supplementary Material
Acknowledgments
We thank our lab members for their assistance with the experiments, and Linzhao Cheng and Timm Schroeder for their suggestions on the work. We thank Hongmei Shen for her help with spectral cytometry. We also appreciate Junling Liu at Shanghai Jiaotong University School of Medicine for his kind help in providing us with the Vps33bfl/fl mice for this study. This work was supported by grants from the Ministry of Science and Technology of China (2020YFE0203000, 2019YFA0801800, 2016YFA0100600, 2017YFA0103400, and 2018YFA0107000), the National Natural Science Foundation of China (81730006, 81825001, 81922002, 81421002, 81721004, 81890990, 81861148029, 81870086, and 81670106), the Chinese Academy of Medical Sciences (CAMS) Initiative for Innovative Medicine (2017-I2M-3-009 and 2016-I2M-1-017), the Natural Science Foundation of Shanghai (17ZR1415500), the Shanghai Science and Technology Commission (19XD1422100), and the CAMS Fundamental Research Funds for Central Research Institutes (2019RC310003); an SKLEH-Pilot Research Grant (Z20-04); and Distinguished Young Scholars of Tianjin (19JCJQJC63400).
Version 1. 10/27/2020
In-Press Preview
Version 2. 01/04/2021
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e138986.https://doi.org/10.1172/JCI138986.
Contributor Information
Dan Huang, Email: 451465668@qq.com.
Guohuan Sun, Email: sunguohuan@ihcams.ac.cn.
Xiaoxin Hao, Email: haoxiaoxin2011@163.com.
Xiaoxiao He, Email: xxhe0702@163.com.
Zhaofeng Zheng, Email: zhengzhaofeng@ihcams.ac.cn.
Chiqi Chen, Email: Chenchiqi@sjtu.edu.cn.
Zhuo Yu, Email: yuzhuo78@aliyun.com.
Li Xie, Email: xieli100@126.com.
Shihui Ma, Email: mashihui@ihcams.ac.cn.
Ligen Liu, Email: liuligen@shsmu.edu.cn.
Bo O. Zhou, Email: bo.zhou@sibcb.ac.cn.
Hui Cheng, Email: chenghui@ihcams.ac.cn.
Junke Zheng, Email: zhengjunke@shsmu.edu.cn.
Tao Cheng, Email: chengtao@ihcams.ac.cn.
References
- 1.Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol. 2019;20(5):303–320. doi: 10.1038/s41580-019-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crane GM, Jeffery E, Morrison SJ. Adult haematopoietic stem cell niches. Nat Rev Immunol. 2017;17(9):573–590. doi: 10.1038/nri.2017.53. [DOI] [PubMed] [Google Scholar]
- 3.Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014;20(8):833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane SW, Scadden DT, Gilliland DG. The leukemic stem cell niche: current concepts and therapeutic opportunities. Blood. 2009;114(6):1150–1157. doi: 10.1182/blood-2009-01-202606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krause DS, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat Med. 2013;19(11):1513–1517. doi: 10.1038/nm.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glait-Santar C, et al. Functional niche competition between normal hematopoietic stem and progenitor cells and myeloid leukemia cells. Stem Cells. 2015;33(12):3635–3642. doi: 10.1002/stem.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng H, et al. Leukemic marrow infiltration reveals a novel role for Egr3 as a potent inhibitor of normal hematopoietic stem cell proliferation. Blood. 2015;126(11):1302–1313. doi: 10.1182/blood-2015-01-623645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoggatt J, Kfoury Y, Scadden DT. Hematopoietic stem cell niche in health and disease. Annu Rev Pathol. 2016;11:555–581. doi: 10.1146/annurev-pathol-012615-044414. [DOI] [PubMed] [Google Scholar]
- 9.Cheng H, Cheng T. ‘Waterloo’: when normal blood cells meet leukemia. Current Opin Hematol. 2016;23(4):304–310. doi: 10.1097/MOH.0000000000000253. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Aguilera A, Mendez-Ferrer S. The hematopoietic stem-cell niche in health leukemia. Cell Mol Life Sci. 2017;74(4):579–590. doi: 10.1007/s00018-016-2306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schepers K, Campbell TB, Passegué E. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell. 2015;16(3):254–267. doi: 10.1016/j.stem.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passaro D, et al. Increased vascular permeability in the bone marrow microenvironment contributes to disease progression and drug response in acute myeloid leukemia. Cancer Cell. 2017;32(3):324–341.e6. doi: 10.1016/j.ccell.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duarte D, et al. Inhibition of endosteal vascular niche remodeling rescues hematopoietic stem cell loss in AML. Cell Stem Cell. 2018;22(1):64–77.e6. doi: 10.1016/j.stem.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welner RS, et al. Treatment of chronic myelogenous leukemia by blocking cytokine alterations found in normal stem and progenitor cells. Cancer Cell. 2015;27(5):671–681. doi: 10.1016/j.ccell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanoun M, et al. Acute myelogenous leukemia-induced sympathetic neuropathy promotes malignancy in an altered hematopoietic stem cell niche. Cell Stem Cell. 2014;15(3):365–375. doi: 10.1016/j.stem.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao A, et al. Bone marrow endothelial cell-derived interleukin-4 contributes to thrombocytopenia in acute myeloid leukemia. Haematologica. 2019;104(10):1950–1961. doi: 10.3324/haematol.2018.214593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Leukemia cell infiltration causes defective erythropoiesis partially through MIP-1α/CCL3. Leukemia. 2016;30(9):1897–1908. doi: 10.1038/leu.2016.81. [DOI] [PubMed] [Google Scholar]
- 19.Pitt LA, et al. CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance. Cancer Cell. 2015;27(6):755–768. doi: 10.1016/j.ccell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 21.Gu H, et al. Sorting protein VPS33B regulates exosomal autocrine signaling to mediate hematopoiesis and leukemogenesis. J Clin Invest. 2016;126(12):4537–4553. doi: 10.1172/JCI87105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raimondo S, et al. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun Signal. 2015;13:8. doi: 10.1186/s12964-015-0086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornick NI, et al. AML suppresses hematopoiesis by releasing exosomes that contain microRNAs targeting c-MYB. Sci Signal. 2016;9(444):ra88. doi: 10.1126/scisignal.aaf2797. [DOI] [PubMed] [Google Scholar]
- 24.Huan J, et al. Coordinate regulation of residual bone marrow function by paracrine trafficking of AML exosomes. Leukemia. 2015;29(12):2285–2295. doi: 10.1038/leu.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corrado C, Raimondo S, Saieva L, Flugy AM, De Leo G, Alessandro R. Exosome-mediated crosstalk between chronic myelogenous leukemia cells and human bone marrow stromal cells triggers an interleukin 8-dependent survival of leukemia cells. Cancer Lett. 2014;348(1–2):71–76. doi: 10.1016/j.canlet.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Kumar B, Garcia M, Murakami JL, Chen CC. Exosome-mediated microenvironment dysregulation in leukemia. Biochim Biophys Acta. 2016;1863(3):464–470. doi: 10.1016/j.bbamcr.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Kumar B, et al. Acute myeloid leukemia transforms the bone marrow niche into a leukemia-permissive microenvironment through exosome secretion. Leukemia. 2018;32(3):575–587. doi: 10.1038/leu.2017.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tadokoro H, Umezu T, Ohyashiki K, Hirano T, Ohyashiki JH. Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J Biol Chem. 2013;288(48):34343–34351. doi: 10.1074/jbc.M113.480822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paggetti J, et al. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood. 2015;126(9):1106–1117. doi: 10.1182/blood-2014-12-618025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdelhamed S, et al. Extracellular vesicles impose quiescence on residual hematopoietic stem cells in the leukemic niche. EMBO Rep. 2019;20(7):e47546. doi: 10.15252/embr.201847546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyiadzis M, Whiteside TL. Exosomes in acute myeloid leukemia inhibit hematopoiesis. Curr Opin Hematol. 2018;25(4):279–284. doi: 10.1097/MOH.0000000000000439. [DOI] [PubMed] [Google Scholar]
- 33.Boyiadzis M, Whiteside TL. The emerging roles of tumor-derived exosomes in hematological malignancies. Leukemia. 2017;31(6):1259–1268. doi: 10.1038/leu.2017.91. [DOI] [PubMed] [Google Scholar]
- 34.Krivtsov AV, Feng Z, Armstrong SA. Transformation from committed progenitor to leukemia stem cells. Ann N Y Acad Sci. 2009;1176:144–149. doi: 10.1111/j.1749-6632.2009.04966.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15(2):154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizoguchi T, et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev Cell. 2014;29(3):340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao M, et al. Megakaryocytes maintain homeostatic quiescence promote post-injury regeneration of hematopoietic stem cells. Nat Med. 2014;20(11):1321–1326. doi: 10.1038/nm.3706. [DOI] [PubMed] [Google Scholar]
- 38.Bruns I, et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat Med. 2014;20(11):1315–1320. doi: 10.1038/nm.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inra CN, et al. A perisinusoidal niche for extramedullary haematopoiesis in the spleen. Nature. 2015;527(7579):466–471. doi: 10.1038/nature15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 41.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 42.Zhang F, et al. Adenovirus E4 gene promotes selective endothelial cell survival and angiogenesis via activation of the vascular endothelial-cadherin/Akt signaling pathway. J Biol Chem. 2004;279(12):11760–11766. doi: 10.1074/jbc.M312221200. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi H, et al. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nat Cell Biol. 2010;12(11):1046–1056. doi: 10.1038/ncb2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Itkin T, et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 2016;532(7599):323–328. doi: 10.1038/nature17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baryawno N, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177(7):1915–1932.e16. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acar M, et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526(7571):126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu C, et al. Stem cell factor is selectively secreted by arterial endothelial cells in bone marrow. Nat Commun. 2018;9(1):2449. doi: 10.1038/s41467-018-04726-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen G, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng M, et al. A motif in LILRB2 critical for Angptl2 binding and activation. Blood. 2014;124(6):924–935. doi: 10.1182/blood-2014-01-549162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.