Abstract
Polyglutamine (polyQ) diseases are devastating, slowly progressing neurodegenerative conditions caused by expansion of polyQ-encoding CAG repeats within the coding regions of distinct, unrelated genes. In spinal and bulbar muscular atrophy (SBMA), polyQ expansion within the androgen receptor (AR) causes progressive neuromuscular toxicity, the molecular basis of which is unclear. Using quantitative proteomics, we identified changes in the AR interactome caused by polyQ expansion. We found that the deubiquitinase USP7 preferentially interacts with polyQ-expanded AR and that lowering USP7 levels reduced mutant AR aggregation and cytotoxicity in cell models of SBMA. Moreover, USP7 knockdown suppressed disease phenotypes in SBMA and spinocerebellar ataxia type 3 (SCA3) fly models, and monoallelic knockout of Usp7 ameliorated several motor deficiencies in transgenic SBMA mice. USP7 overexpression resulted in reduced AR ubiquitination, indicating the direct action of USP7 on AR. Using quantitative proteomics, we identified the ubiquitinated lysine residues on mutant AR that are regulated by USP7. Finally, we found that USP7 also differentially interacts with mutant Huntingtin (HTT) protein in striatum and frontal cortex of a knockin mouse model of Huntington’s disease. Taken together, our findings reveal a critical role for USP7 in the pathophysiology of SBMA and suggest a similar role in SCA3 and Huntington’s disease.
Keywords: Neuroscience
Keywords: Neurodegeneration, Neuromuscular disease, Ubiquitin-proteosome system
Introduction
Spinal and bulbar muscular atrophy (SBMA) is 1 of 9 polyQ diseases that are part of a larger family of neurodegenerative diseases characterized by protein misfolding and accumulation; these diseases include Alzheimer’s disease, Huntington’s disease (HD), Parkinson’s disease and amyotrophic lateral sclerosis (ALS). SBMA is caused by a polyglutamine–encoding (polyQ-encoding) CAG triplet repeat expansion within exon 1 of the androgen receptor (AR) gene (1). Unlike the other 8 diseases, which show autosomal dominant inheritance, SBMA is X-linked and affects only males due to a requirement for circulating testosterone (2–4). Key symptoms of SBMA are muscle atrophy, weakness, and fasciculations. The underlying cellular basis of the disease is a loss of brainstem and spinal cord motor neurons along with the associated innervated muscles (5, 6), although a primary role for muscle atrophy has also been demonstrated (7, 8). Surviving motor neurons display intranuclear inclusions that contain predominantly insoluble and truncated AR (9, 10). Such inclusions are also an important feature of the other polyQ diseases.
AR is a 110 kDa steroid/thyroid hormone receptor that regulates androgen-responsive genes to control male sexual differentiation, spermatogenesis, and male gonadotrophin levels. In the absence of androgens, AR resides in the cytoplasm in an aporeceptor complex consisting of the molecular chaperones Hsp90, Hsp70, Hsp40, HIP, and HOP, p23, and the immunophilins FKBP51 and FKBP52 (11–13). Binding of testosterone or 5α-dihydrotestosterone (DHT) results in conformational changes to and concomitant activation of the AR, which then translocates to the nucleus, where it modulates transcription of target genes in association with coregulators. Upon dissociation from DNA, the AR shuttles back into the cytoplasm, thus completing the AR cycle (14, 15). AR metabolism is modulated by posttranslational modifications, such as phosphorylation, acetylation, SUMOylation, and ubiquitination, and several of these have been shown to play a role in SBMA pathogenesis (16–18). The ubiquitination-deubiquitination cycle of AR is complex, with distinct outcomes driven by specific E3 ubiquitin ligases. For example, ubiquitination of AR by CHIP (19, 20) or Mdm2 (21) promotes AR degradation by the 26S proteasome, and the involvement of other E3 ubiquitin ligases, such as Parkin, modulates polyQ-expanded AR levels (20). In contrast, polyubiquitination of AR by RNF6 or E6-AP enhances AR-dependent transcriptional activity (22, 23). Deubiquitination of AR plays an equally important role in AR metabolism. Several deubiquitinases have been reported as interacting with AR and regulating its transcriptional activity; these include USP10, USP26, and USP12 (24–26). In addition, a recent study demonstrated that USP7 interacts with AR and facilitates AR binding to chromatin in prostate cancer cells (27).
PolyQ tract expansion in the AR results in ligand-dependent protein misfolding and aggregation as well as changes in its transcriptional activation function (reviewed in ref. 28). Available evidence supports the hypothesis that neuronal cell death is caused by proteotoxicity of the polyQ-expanded AR rather than by loss of AR function per se. Although the functional significance of inclusion bodies in cell viability in SBMA is yet to be fully appreciated, studies of HD and spinocerebellar ataxia type 7 (SCA7), which are caused by polyQ tract expansions in the Huntingtin (HTT) and Ataxin-7 proteins, respectively, suggest that inclusions may be less toxic than soluble aggregation species and that their formation may be important for cell survival due to their sequestration of toxic intermediates (29–31). Recent proteomics studies, however, revealed inclusion-mediated disruption of a select set of pathways involved in proteostasis (32), suggesting a discrete role for these structures in polyQ toxicity. In contrast, several lines of evidence implicate soluble, aggregated, misfolded polyQ-expanded proteins as likely mediators of neuronal dysfunction and cytotoxicity (33–35). In support of the latter view, our work demonstrated that motor neurons expressing polyQ-expanded AR undergo death at least 1 week before the detection of nuclear inclusions in surviving neurons, suggesting that the biochemical properties of the polyQ-expanded AR before its incorporation into inclusions are likely to be important determinants of toxicity (33). Although the mechanisms are unknown, several models have been proposed to explain the pathogenic nature of polyQ-expanded proteins in neurodegenerative disease (36, 37), including changes in the protein interaction network of polyQ-expanded proteins. For example, prior studies have shown polyQ expansions to alter the interactomes of HTT and Ataxin-1 in HD and SCA1, respectively (32, 38–40). Moreover, dysregulation of the interactomes of mutant proteins has been invoked as a key driver of cellular toxicity in a variety of neurodegenerative disease states (39). Although several proteins have been shown to differentially interact with WT or mutant polyQ-expanded AR (41–43), global unbiased differential interactome studies of the WT and polyQ-expanded AR have not been carried out.
In this study, we hypothesized that an unbiased evaluation of changes in the AR-interaction network due to polyQ expansion would reveal insights into the pathophysiology of SBMA as well as potential therapeutic targets. We identified several proteins that differentially associate with WT and polyQ-expanded AR. One differentially interacting protein identified here is USP7, a preferential interactor of polyQ-expanded AR. Herein, we show that USP7 mediates DHT-dependent mutant AR aggregation and cellular toxicity in SBMA cell models. We also show that lowering USP7 attenuates these effects and ameliorates disease phenotypes in a mouse model of SBMA and in fly models of SBMA and SCA3, implicating this deubiquitinase more broadly in the pathogenic mechanism of polyQ disease.
Results
Unbiased quantitative interaction screen for WT and polyQ-expanded AR.
The expanded polyQ region of the soluble AR is likely to change the overall conformation of the protein (44, 45); thus, we postulated that such structurally altered AR species might interact aberrantly with protein partners and that identifying the differential polyQ-expanded AR interactome could provide insights into pathogenic mechanisms at work in SBMA. Therefore, to characterize the AR interactome and the polyQ expansion–dependent alterations of this network, we employed stable isotope labeling of amino acids in cell culture–based (SILAC-based) quantitative proteomics analysis to identify the binding partners that differentially coimmunoprecipitate (co-IP) with AR10Q or AR112Q in PC12 cells (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI134565DS1). These PC12 cells (grown either in heavy or light media) express either AR10Q or AR112Q under the control of a tetracycline-inducible promoter; addition of doxycycline to these cells induces AR expression. Subsequent treatment with DHT triggers nuclear translocation of AR and, in the case of AR112Q, results in time-dependent AR aggregation and formation of cytologically detectable intranuclear inclusions (17, 18, 33, 46–49). Since we sought to understand the interactome of the soluble, preinclusion state of polyQ-expanded AR, we analyzed cells 48 hours after AR induction and DHT treatment when approximately 10% of cells present with nuclear inclusions and the vast majority of the protein are still in the preinclusion state (ref. 33 and data not shown).
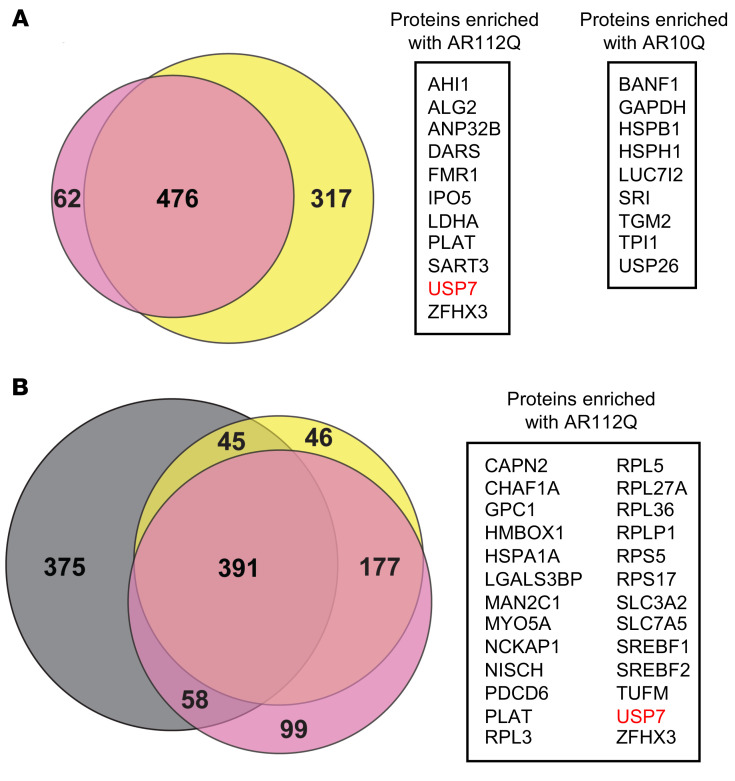
AR was immunoprecipitated with either an anti-AR antibody (which immunoprecipitates both WT and polyQ-expanded AR with near-equal efficiency; Supplemental Figure 1B) or the conformation-specific polyQ antibody 3B5H10 (which selectively recognizes the soluble preinclusion form of polyQ-expanded proteins) (50). Two independent pull-down experiments with the anti-AR antibody yielded 476 common proteins (Figure 1A). Of these, 11 proteins preferentially associated with AR112Q, and 9 proteins were enriched with AR10Q by 1.5-fold or more (Figure 1A and Supplemental Tables 1 and 2).
Figure 1. Unbiased quantitative interaction screen for WT and polyQ-expanded AR.
Three independent SILAC experiments were performed. Experiment 1: PC12 cells expressing AR 112Q or AR10Q were grown in heavy or light SILAC medium, respectively (shown in pink). Anti-AR (A) or 3B5H10 antibody (B) was used for IP. Experiment 2 (label swap): AR112Q- or AR10Q-expressing cells were grown in light or heavy SILAC medium, respectively (shown in yellow). Anti-AR (A) or 3B5H10 antibody (B) was used for IP. Experiment 3: cells expressing AR 112Q or AR10Q were grown in heavy or light SILAC medium, respectively (shown in gray). 3B5H10 antibody (B) was used for IP. Cells were induced with doxycycline to express AR and treated with DHT for 48 hours. (A) Left: Venn diagram comparison of common proteins pulled down with anti-AR antibody in 2 experiments. Right: common proteins that were enriched either with AR112Q or AR10Q by 1.5-fold or more. Fold enrichment is shown in Supplemental Tables 1 and 2. (B) Left: Venn diagram comparison of common proteins pulled down with 3B5H10 antibody in 3 independent experiments. Right: common proteins enriched with AR112Q by 1.5-fold or more in at least 2 experiments. Fold enrichment is shown in Supplemental Table 1.
We also performed 3 independent co-IP experiments using 3B5H10 antibody and identified 671 proteins that were common in at least 2 out of 3 independent experiments (Figure 1B). Because 3B5H10 is specific for expanded polyQ tracts, the amount of AR10Q precipitated by this antibody is 30- to 90-fold lower than AR112Q (Supplemental Figure 1B). Therefore, preferential AR10Q interactors cannot be identified in these co-IPs. Analysis of common AR112Q preferential interactors identified 3 proteins (USP7, Glypican-1, and α-Mannosidase 2C1) across 3 3B5H10 IP experiments, and 23 proteins that were found in 2 experiments. Thus, USP7 was identified as a preferential interactor with polyQ-expanded AR with both antibodies. The limited number of interactors and the modest levels of enrichment observed in our study (Supplemental Tables 1 and 2) suggest that the interactome of AR undergoes subtle, yet potentially physiologically relevant, changes due to polyQ expansion.
USP7 preferentially interacts with polyQ-expanded AR in vitro and in vivo.
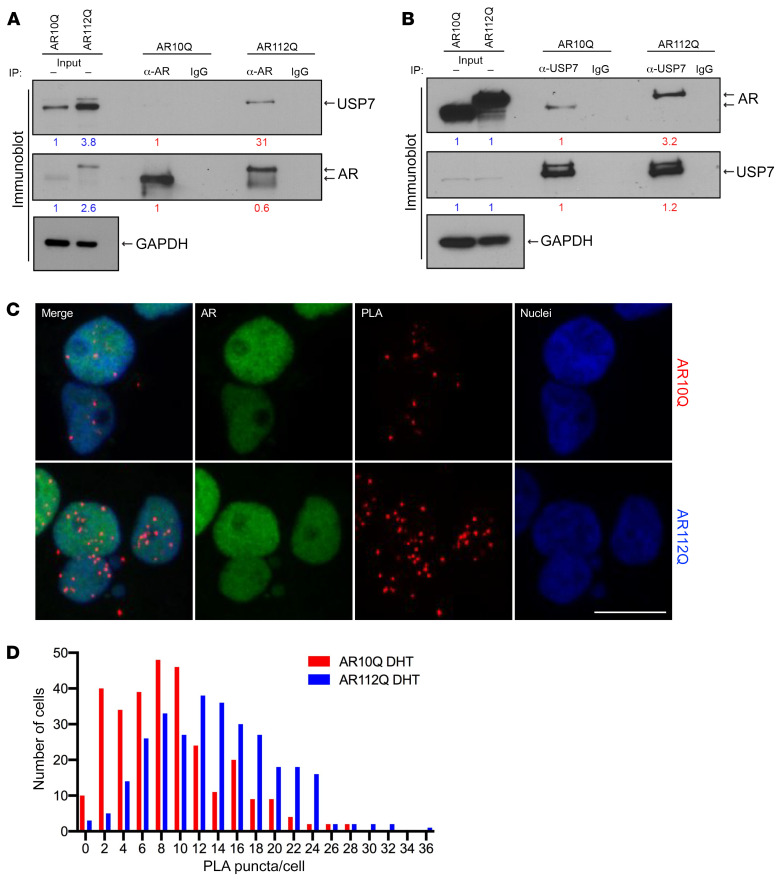
Our analysis of the differential AR interactome identified a robust and reproducible preferential interaction between AR112Q and USP7, as judged by an approximately 2-fold enrichment of USP7 with polyQ-expanded AR using 2 different antibodies (Figure 1 and Supplemental Table 1). We validated this preferential interaction by immunoblot analyses of coimmunoprecipitates from AR10Q- or AR112Q-expressing PC12 cell lysates; these experiments confirmed that USP7 coimmunoprecipitated to a greater extent with AR112Q, compared with AR10Q (Figure 2A). In the reciprocal pull-down assay, endogenous USP7 coprecipitated more AR112Q than AR10Q (Figure 2B). These findings support the quantitative proteomics analyses and confirm that AR-USP7 complex formation in cells is enhanced by the expanded polyQ tract in the AR protein.
Figure 2. USP7 preferentially interacts with AR112Q in cells.
(A) Co-IP of USP7 with AR after pull-down with an anti-AR antibody, followed by immunoblot with anti-AR or anti-USP7 antibodies; GAPDH detection served as loading control. Input levels of AR or USP7 in AR112Q- relative to AR10Q-expressing cells are shown in blue (data normalized to AR10Q cells). Relative amounts of immunoprecipitated AR or USP7 are shown in red, normalized to IP recovery from AR10Q-expressing cells. (B) Co-IP of AR with USP7 following pull-down with an anti-USP7 antibody, followed by immunoblot with anti-AR or anti-USP7 antibodies; GAPDH detection served as loading control. As in A, relative input levels are shown in blue, and amounts of immunoprecipitated proteins are shown in red. (C) USP7-AR interaction was evaluated by PLA (red puncta) in PC12 cells. AR was predominantly nuclear, as judged by subsequent immunostaining with anti-AR antibody (green signal). Scale bar: 10 μm. (D) Quantification of PLA puncta in cells expressing AR10Q and AR112Q, respectively, based on images taken before staining for total AR (see Supplemental Figure 2C), with 100 cells evaluated per condition and carried out in triplicate. P < 0.0001, Kolmogorov-Smirnov test.
To further evaluate the USP7-AR interaction in situ, we performed proximity ligation assay (PLA) using anti-AR and anti-USP7 antibodies. PLA signals result from the 2 labeled antibodies residing in close physical proximity (<40 nm); thus, the detection of such a signal allows the inference of USP7-AR interaction. PC12 cells expressing AR10Q and treated with DHT displayed PLA puncta indicative of the steady-state levels of AR10Q-USP7 interaction in these cells. However, in cells expressing AR112Q under the same conditions, we detected a substantial and statistically significant increase in the fraction of cells harboring a larger number of puncta per cell (Figure 2, C and D). We also observed that USP7 interacts with AR in the absence of DHT, although to a lesser extent, and this interaction is enhanced by the expansion of the polyQ tract as well (Supplemental Figure 2, A and B). Therefore, these PLA data further confirm our biochemical results that USP7 preferentially interacts with AR112Q in PC12 cells and reveal that USP7-AR interaction is enhanced in the presence of DHT.
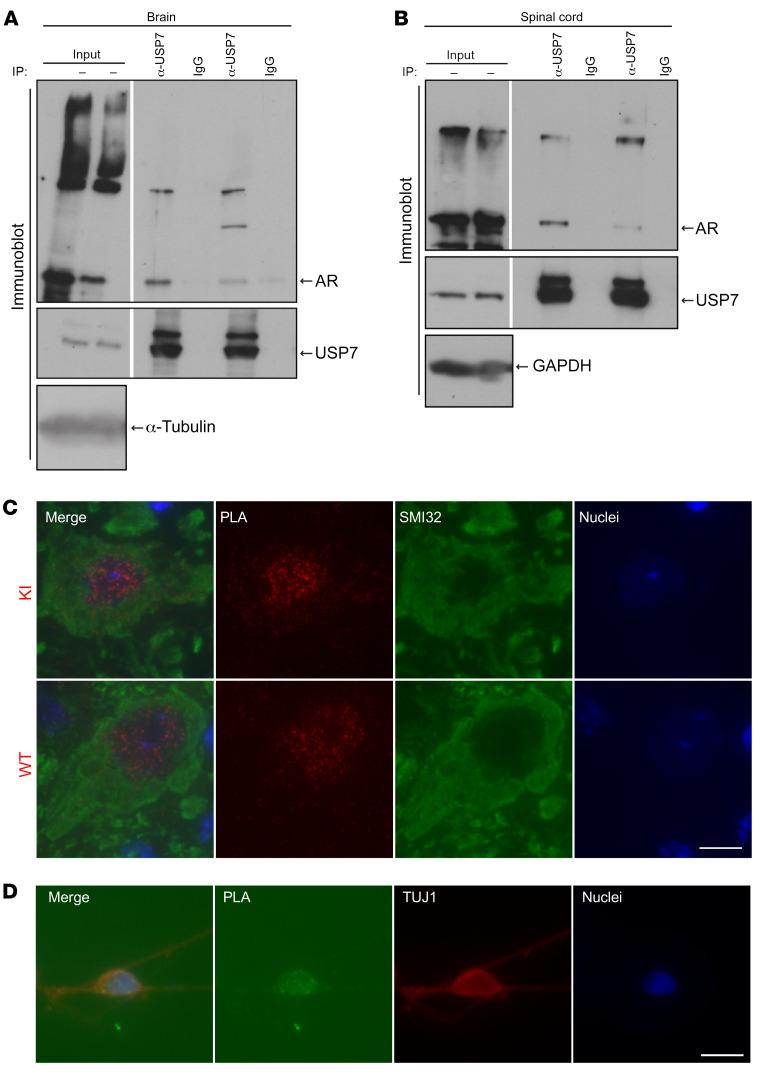
To determine whether USP7 interacts with polyQ-expanded AR in vivo, we used an SBMA transgenic mouse model that expresses human, full-length, polyQ-expanded AR (AR112Q) in the CNS under the control of the prion protein (PrP) promoter (2). This model has been extensively characterized and recapitulates several key aspects of pathology associated with the human disease (2, 46, 49). We immunoprecipitated USP7 from tissue lysates prepared from brains and spinal cords of males of this transgenic line and detected coprecipitation of polyQ-expanded AR by immunoblot (Figure 3, A and B), indicating that these proteins interact in vivo. To further investigate the interaction of USP7 with endogenous mutant AR in vivo, we carried out PLA in spinal cord sections of a knockin (KI) mouse model of SBMA, in which a portion of human Ar exon 1 bearing a CAG repeat of 112 is recombined into the endogenous mouse Ar exon 1; these mice exhibit progressive neuromuscular deficits and pathology (4). PLA puncta were observed in 7-month-old male KI and WT motor neurons (Figure 3C), confirming that the AR/USP7 interaction occurs in motor neurons in vivo with endogenous levels of both AR and USP7 proteins. Analysis of USP7 levels in adjacent spinal cord sections revealed no differences in USP7 protein levels between SBMA and WT male mice (Supplemental Figure 3, C and D). We also evaluated the AR/USP7 interaction using PLA in induced pluripotent stem cell–derived (iPSC-derived) motor neurons from an SBMA patient, further confirming the interaction in patient-derived cells (Figure 3D).
Figure 3. USP 7 interacts with AR in vivo.
(A and B) Western blot analysis of immunoprecipitates from brain and spinal cord of 10-week-old SBMA male transgenic mice. Aggregated AR species can be observed as high molecular weight species at the top of the gel. (C) PLA analysis of AR-USP7 interaction in motor neurons from spinal cord sections of a 7-month-old KI SBMA mouse or a WT mouse, followed by immunostaining for unphosphorylated NF-H (SMI32 Ab) to identify motor neurons and with Hoechst 33258 to mark nuclei. Scale bar: 10 μm. PLA quantification and PLA technical controls are shown in Supplemental Figure 3A and Supplemental Figure 3B, respectively. (D) USP7-AR interaction was evaluated by PLA (green) in iPS-derived motor neurons from SBMA patient, followed by immunostaining with anti-β III-tubulin antibody (TUJ1) (red). Scale bar: 10 μm. PLA technical controls are shown in Supplemental Figure 3E.
USP7 interacts with soluble forms of polyQ-expanded AR.
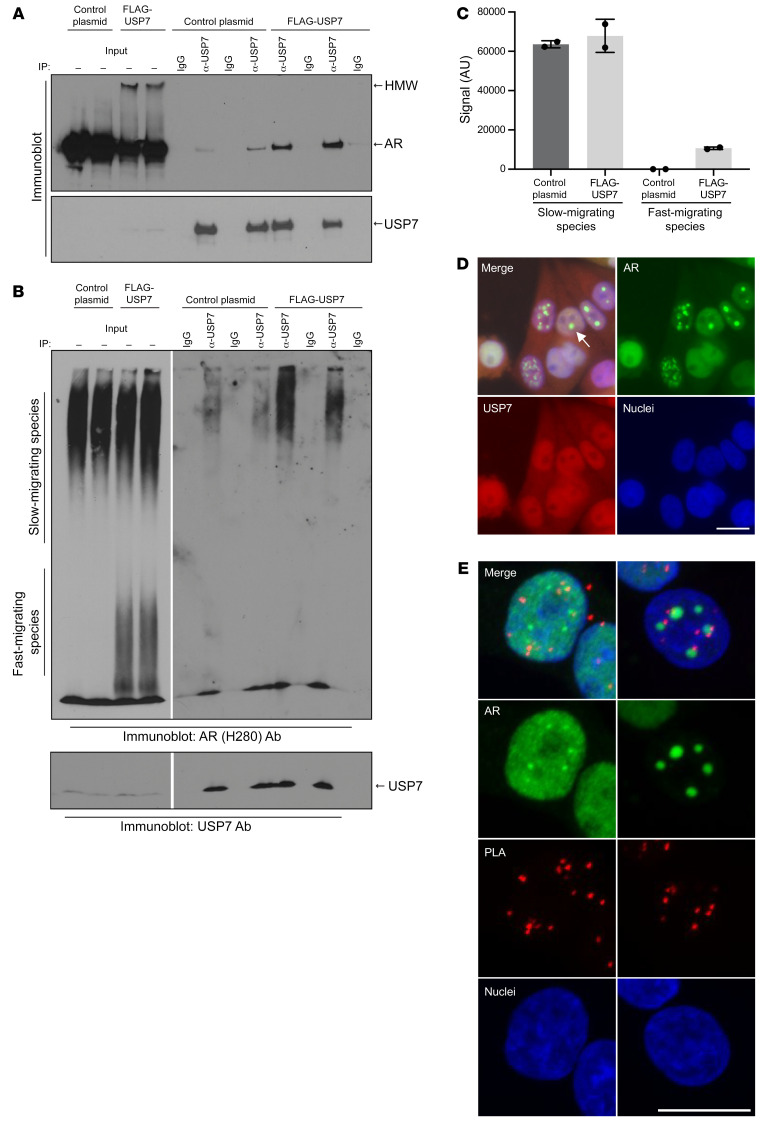
Previous studies have shown that AR112Q can be resolved by SDS-agarose gel electrophoresis into a soluble, 3B5H10-reactive, slow-migrating aggregate species composed of full-length protein and an insoluble, fast-migrating species consisting of both full-length AR and N-terminal AR fragments (33, 51). To determine which of these species associates with USP7, lysates of AR112Q-expressing PC12 cells with or without constitutive coexpression of FLAG-USP7 were subjected to IP with an anti-USP7 antibody. Immunoblot analysis by SDS-PAGE (Figure 4A) or SDS-agarose (Figure 4, B and C) of the same USP7 immunoprecipitates revealed that the USP7-associated AR112Q migrates in monomeric form by SDS-PAGE and as a soluble, slow-migrating species on SDS-agarose. Additionally, immunofluorescence analysis revealed that USP7 does not colocalize with nuclear inclusions of aggregated AR112Q (Figure 4D). Furthermore, PLA experiments establish that USP7-interacting AR puncta do not overlap with AR inclusions, despite frequent close proximity (Figure 4E), further confirming that USP7 preferentially interacts with soluble AR112Q.
Figure 4. USP7 preferentially interacts with soluble polyQ-expanded AR and does not colocalize with nuclear inclusions.
IP of AR112Q-expressing PC12 cell protein lysates with anti-USP7 antibody, resolved by SDS-PAGE (A) or SDS-agarose (B). (C) Quantification of slow- and fast-migrating species from input lanes of B. (D) Immunofluorescence images of PC12 cells expressing AR112Q. Arrow points to a nuclear inclusion. Scale bar: 10 μm. (E) AR immunofluorescence images of cells examined by PLA in Figure 2C. Approximately 10% (9.5 ± 1.1) of AR112Q-expressing cells contained nuclear inclusions. Scale bar: 10 μm.
Knockdown of USP7 rescues polyQ-expanded AR toxicity and aggregation.
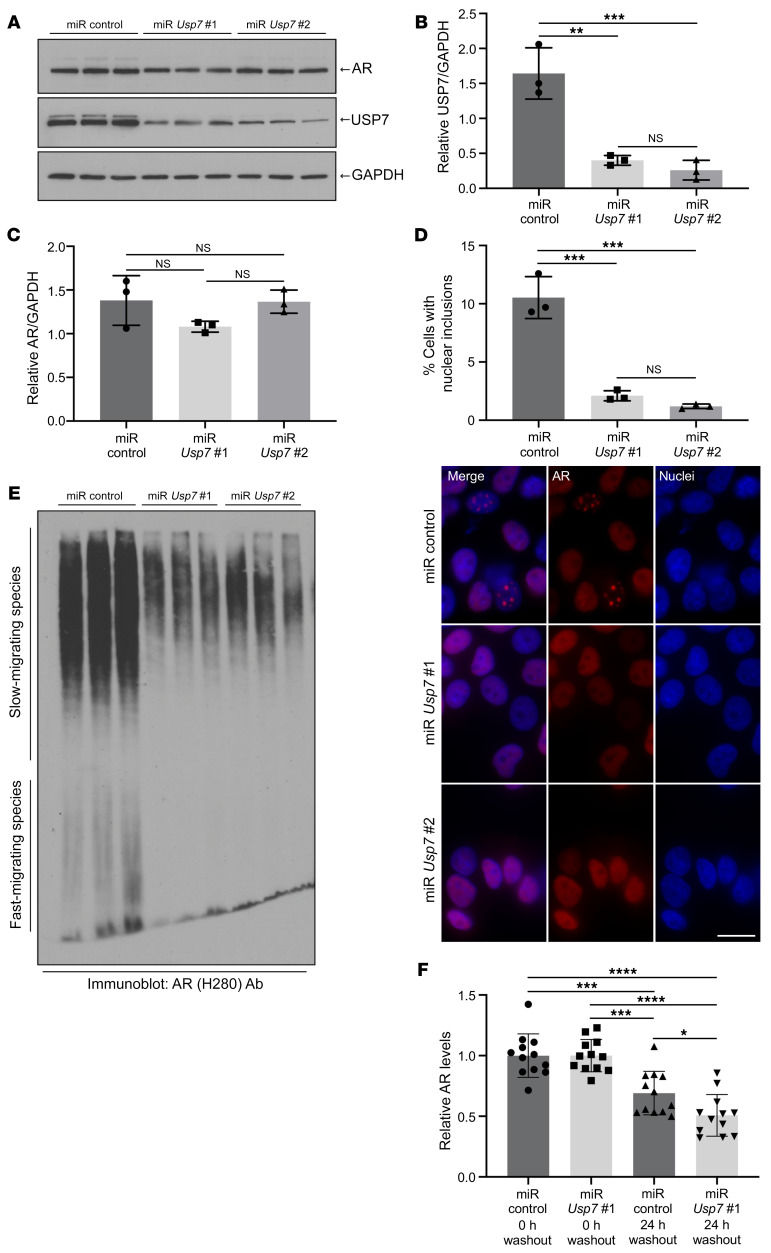
In order to evaluate whether the preferential interaction between USP7 and polyQ-expanded AR plays a role in cellular dysfunction, we determined the effect of USP7 knockdown on AR112Q aggregation and cytotoxicity. AR112Q-expressing PC12 cells were stably transfected with miRNA-expressing constructs aimed at silencing Usp7 (miR Usp7) or with a control nontargeting miRNA-expressing construct (miR control). Expression and activation of AR112Q were induced by addition of doxycycline to the culture medium in the presence of DHT. Two different miRs for Usp7 (miR Usp7 no. 1 and miR Usp7 no. 2) were able to reduce USP7 protein levels by 75% and 84%, respectively (Figure 5, A and B). Lowering USP7 levels resulted in a dramatic decrease (5- to 8-fold) in the number of cells harboring nuclear inclusions (Figure 5D), without altering the steady-state levels of the AR monomer (Figure 5, A and C). Despite the lack of effect of USP7 lowering on AR steady-state levels, the half-life of AR under the condition of USP7 knockdown was modestly but statistically significantly reduced compared with that of AR112Q under the condition of endogenous USP7 levels (Figure 5F and Supplemental Figure 4G), suggesting that USP7 knockdown modestly increases AR turnover. In addition, SDS-agarose gel analysis revealed that partial knockdown of USP7 reduced the amount of the fast migrating species (which are temporally correlated with inclusion formation; refs. 33, 51) relative to control (Figure 5E). Importantly, the amount of the soluble slow migrating species was also reduced by over 50% (Figure 5E and Supplemental Figure 4F).
Figure 5. Knockdown of USP7 decreases polyQ-expanded AR aggregation and increases AR turnover in a cell model of SBMA.
PC12 cells inducibly expressing AR112Q and constitutively expressing miRNAs targeting different regions of Usp7 mRNA (miR Usp7 no. 1 and miR Usp7 no. 2) or nontargeting miRNA (miR control) were (A) analyzed for USP7 and AR protein levels by immunoblot. (B and C) Quantification of USP7 and AR protein levels from A. (D) Percentage of cells with nuclear inclusions upon USP7 knockdown (top); representative immunofluorescence images (bottom). Scale bar: 10 μm. For each condition, 500 cells were counted in triplicate. Experiment was repeated 3 times. (E) Cell lysates from A were resolved on SDS-agarose followed by immunoblot analysis with an anti-AR antibody. (F) AR turnover upon USP7 knockdown. AR levels were determined by Western blot analysis (images shown in Supplemental Figure 4G). Graph represents an average of 4 independent experiments, with each experiment performed in triplicate. Data were normalized to the 0-hour washout time point. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, 1-way ANOVA with post hoc Tukey’s test. Error bars represent SD.
Although the lowering of USP7 in cells substantially rescued polyQ-dependent AR phenotypes, we also observed that the near-complete (>90%) loss of USP7 had limited effect on AR112Q nuclear inclusion formation (Supplemental Figure 4). The reasons for this result remain under investigation; however, it is noteworthy that a similar observation was documented in normal human fibroblast cells, in which partial reduction of USP7 levels by RNAi resulted in destabilization of p53 protein, but near-complete loss of USP7 stabilized p53 (52). We postulate that this phenomenon may be related to the feedback regulation of USP7 and its role in proteostasis in dividing cells.
Haploinsufficiency of Usp7 rescues DHT-induced cell toxicity.
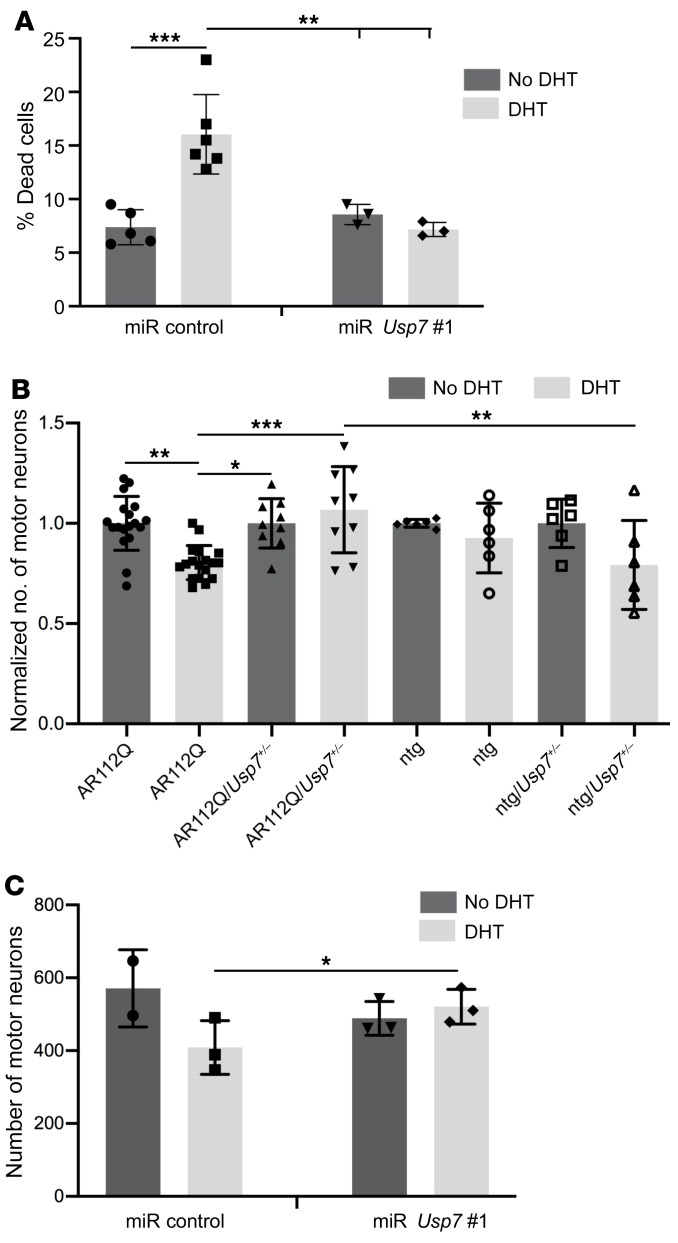
Reducing USP7 levels rescued DHT-induced cytotoxicity in polyQ-expanded AR-expressing cells (Figure 6A). These results suggest that partial lowering of USP7 levels in polyQ-expanded AR-expressing cells rescues polyQ-dependent phenotypes. Because DHT-induced cell death is a characteristic feature of cultured primary motor neurons derived from SBMA mice (46, 49), we also evaluated the effect of Usp7 haploinsufficiency on this in vitro phenotype. As shown in Figure 6B (and Supplemental Figure 5A), primary motor neurons derived from E13.5 embryos of AR112Q mice underwent cell death upon treatment with DHT, whereas neurons derived from AR112Q-expressing transgenic mice that are haploinsufficient for Usp7 (AR112Q/Usp7+/–) were refractory to this effect. Similarly, primary motor neurons derived from E13.5 embryos of AR113Q KI mice, in which the mutant AR is expressed from the endogenous AR promoter, exhibited DHT-dependent toxicity, which was abrogated by AAV delivery of miR Usp7 (Figure 6C). Note that expression of miR Usp7 resulted in an approximately 50% USP7 knockdown (Supplemental Figure 5B). These data confirm the important role of USP7 in SBMA motor neuron toxicity.
Figure 6. Knockdown of USP7 rescues DHT-induced toxicity in cell models of SBMA.
(A) DHT-induced cell death in PC12 cells expressing AR112Q and either miR Usp7 no. 1 or miR control. Two hundred cells were counted in triplicate per condition (except for miR control no DHT, n = 5; and miR control DHT, n = 6). The experiment was repeated 3 times. (B) Dissociated spinal cord cultures from mouse embryos with the genotypes AR112Q, AR112Q/Usp7+/–, ntg/Usp7+/–, or nontransgenic (ntg) were treated with DHT or vehicle (ethanol) for 7 days and motor neurons from 10 random fields were counted per experimental condition. Three independent experiments were performed for AR112Q (n = 6) and AR112Q/Usp7+/– (n = 3), and 2 independent experiments were performed in triplicate for ntg/Usp7+/– and ntg. Due to experimental variability in total motor neuron number, relative motor neuron number in DHT vs. vehicle treatment conditions is presented. An example of the raw data is presented in Supplemental Figure 5A. *P < 0.05; **P < 0.01; ***P < 0.001, 1-way ANOVA with post hoc Tukey’s. Error bars represent SD. (C) Dissociated spinal cord cultures from mouse embryos of KI SBMA mice were infected with AAV1 expressing either miR control or miR Usp7 no. 1 for 5 days, followed by DHT or ethanol treatment for an additional 7 days. Motor neurons were counted from the entire coverslip per experimental condition in triplicate (except for the miR control ethanol condition, where the experiment was done in duplicate). *P < 0.05, 1-tailed t test. Error bars represent SD.
USP7 knockdown rescues SBMA phenotypes in a fly model.
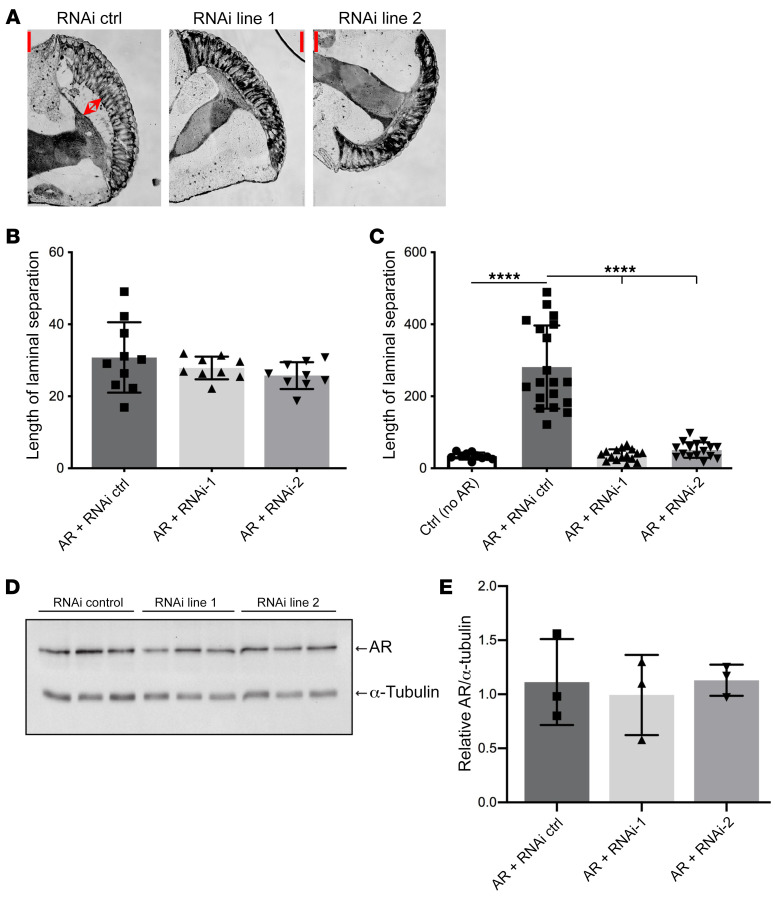
Because lowering the levels of USP7 had a protective effect in cell models of SBMA, we next sought to evaluate the role of USP7 in SBMA pathophysiology in vivo. Therefore, we used a Drosophila melanogaster model of SBMA, in which full-length human, polyQ-expanded AR (AR52Q) is expressed under the control of the glass multimer reporter driver (GMR-GAL4). The use of this driver results in AR expression in fly eyes (53–55) and retinal degeneration in flies expressing AR52Q, but not AR12Q, in the presence of DHT (55). Flies expressing AR52Q display an eye phenotype characterized by detachment of the photoreceptor array from the underlying lamina when raised on a DHT-containing diet and aged as adults for 10 days (Figure 7, A and C, RNAi control). Analysis of the flies at day 1 of DHT treatment did not reveal clear signs of eye pathology (Figure 7B), establishing that the observed effects at day 10 of DHT treatment are not a result of developmental deficiency, but rather the result of DHT-induced AR52Q pathology. To determine the role of USP7 in this phenotype, we knocked down USP7 with 2 different RNAi lines (Figure 7, RNAi line 1, RNAi line 2, each of which leads to ≥60% Usp7 mRNA reduction in the intact fly; ref. 56). Both RNAi lines resulted in rescue of the AR52Q-dependent and DHT-induced eye phenotype (Figure 7, A and C) to a level comparable to that of flies that do not express AR52Q (Figure 7C). No changes in steady-state AR monomer levels were observed upon USP7 knockdown (Figure 7, D and E), in agreement with what we observed upon USP7 knockdown in PC12 cells. These data establish a functional role for USP7 in mediating polyQ-expanded AR-mediated pathology in an in vivo model of SBMA.
Figure 7. Knockdown of USP7 rescues DHT-induced toxicity in a fly model of SBMA.
(A) Histological sections of fly eyes expressing pathogenic AR52Q either with RNAi control or 2 independent USP7-directed RNAi; red arrows in the left panel mark detachment of array from the lamina. Scale bar: 50 μm. (B) Flies expressing AR52Q were sacrificed at day 1 of DHT treatment as adults. Eyes from 9–10 flies per group were sectioned, with 3–6 sections stained and imaged; the length of the detachment of the array from the lamina was measured and averaged for each fly. (C) Adult flies expressing AR52Q were sacrificed at 10 days of DHT treatment. Eyes from 10–19 flies per experimental group were sectioned and analyzed as in B. (D) Western blots from fly heads expressing AR52Q. (E) Quantification of AR levels in each experimental group from D. ****P < 0.0001, 1-way ANOVA with post hoc Tukey’s test. Error bars represent SD.
USP7 knockdown rescues SCA3 phenotype in a fly model.
In order to determine whether USP7 plays a role in the pathogenicity in other polyQ diseases, we evaluated the effect of USP7 knockdown in a fly model of SCA3. In this model, full-length Ataxin-3 harboring 77 glutamines is expressed along with, but independently of, membrane-targeted GFP in the fly eye (57, 58). Expression of full-length Ataxin-3 77Q in fly eyes leads to degeneration of internal eye structures without clear impact on the external eye, necessitating the use of histological assays to assess degeneration (57, 58), which would be a time-consuming endeavor for screening purposes. Therefore, we turned to the CD8-GFP–based technique, which enables the visualization of internal eye degeneration without the need for histological assays. As photoreceptors die, GFP signal is diminished, leading to reduced overall fluorescence as well as increased mosaicism. As we have shown before, the CD8-GFP–based technique is a sensitive and reliably quantifiable reporter of internal retinal degeneration (57, 59–61). We elected to use this technique to examine the role of USP7 and other DUBs and related proteins on toxicity caused by pathogenic Ataxin-3 77Q.
As shown in Supplemental Figure 6A, expression of Ataxin-3 77Q resulted in loss of GFP expression, indicative of eye degeneration (57). In a targeted assay for suppressors of toxicity from pathogenic Ataxin-3, we found that knocking down USP7 with the same RNAi lines used for AR52Q (Figure 7) resulted in rescue of the degeneration phenotype (Supplemental Figure 6B). In comparison, knockdown of other deubiquitinases, such as Rnp11 and its partner Rpn8, exacerbated the toxicity, and the lowering of CYLD deubiquitinase levels was without effect. Thus, these data suggest that phenotypic rescue of a fly model of SCA3 is not due to a general loss of deubiquitination function, but rather to a specific consequence of reduced USP7. In addition, we observed that knockdown of USP7 decreased Ataxin-3 77Q aggregation in flies (Supplemental Figure 6C), further supporting the idea that USP7 plays a functional role in polyQ-expanded Ataxin-3 toxicity. Our findings therefore suggest that USP7 might play a broad role in the pathophysiology of polyQ diseases.
USP7 interacts with full-length HTT protein.
Previous studies revealed the interaction between mutant exon 1–encoded HTT protein and USP7 (32). We wished to evaluate the interaction of USP7 with mutant polyQ-expanded full-length HTT protein from a KI mouse model of HD (62). In these KI mice (zQ175), 1 allele of the mouse Htt exon 1 was replaced by the human HTT exon 1 sequence harboring a 175 CAG repeat tract. Immunoblot analyses of USP7 pull-downs from lysates prepared from striata and cortex of these mice indicate that USP7 associates with both WT and mutant HTT, but preferentially interacts with polyQ-expanded HTT (Supplemental Figure 7, A–C). Moreover, PLA analysis in iPS cells derived from an HD patient and a control individual revealed interaction between HTT and USP7 in these human-derived cells (Supplemental Figure 7, D and E). The heterozygosity of the Htt alleles in these patient-derived cells likely obscured potential USP7 differential interaction with mutant HTT. Nonetheless, these combined findings lend further support to the notion that polyQ-expanded proteins preferentially associate with USP7, and this association may potentially underlie the pathogenicity of multiple polyQ expansion diseases.
USP7 modifies SBMA disease phenotype in a transgenic mouse model.
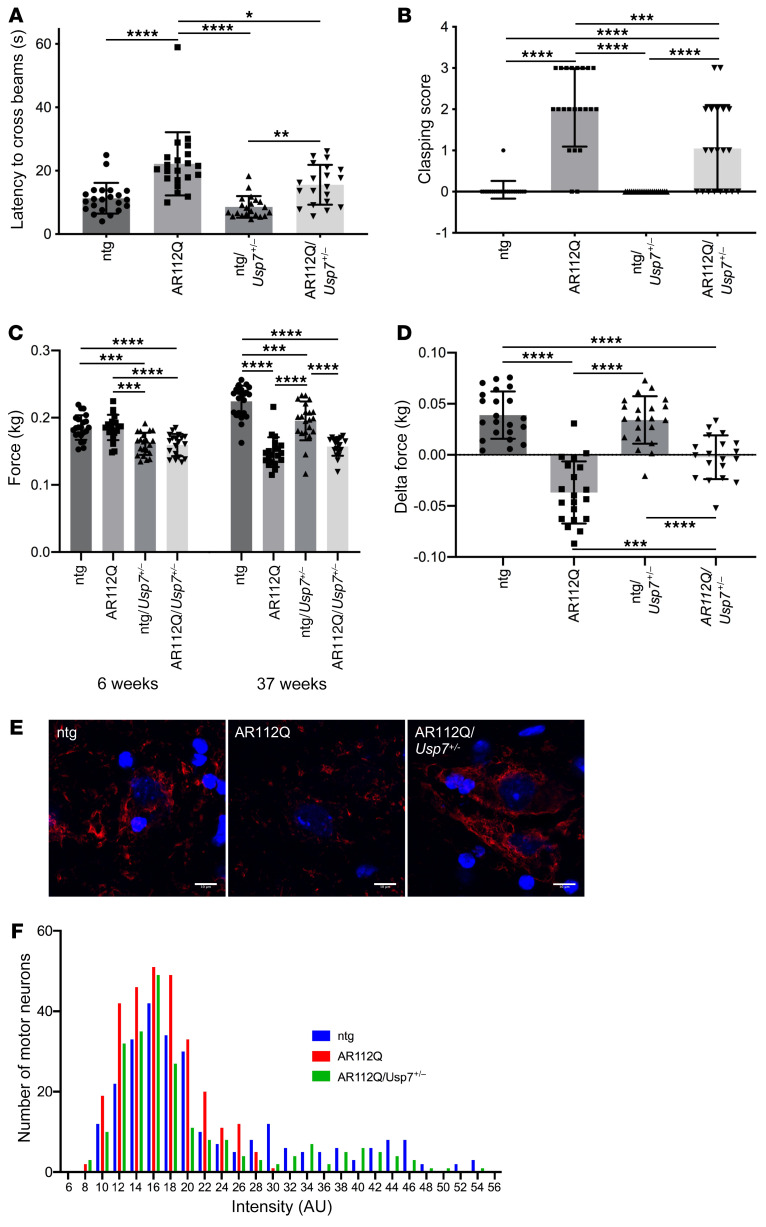
The observation of cytoprotective effects of USP7 lowering in cell and fly models of SBMA suggested the possibility that reducing USP7 levels may ameliorate disease phenotypes and/or slow disease progression in a transgenic mouse model of SBMA. Although complete genetic ablation of Usp7 results in embryonic lethality in mice, Usp7+/– heterozygous mice are nevertheless viable and do not manifest adverse disease pathology (63). Therefore, SBMA transgenic mice that are haploinsufficient for Usp7 (AR112Q/Usp7+/–) were compared with corresponding SBMA mice that were WT for Usp7 (AR112Q), and 2 control groups: (a) nontransgenic (ntg, for SBMA) but haploinsufficient for Usp7 (ntg/Usp7+/–) and (b) WT for both Usp7 and SBMA (ntg).
We evaluated the in vivo effects of Usp7 haploinsufficiency in SBMA transgenic male mice on key disease phenotypic readouts of motor function using 4 assays: (a) balance beam walking; (b) clasping; (c) grip strength; and (d) ability to stay on a rotating rod (rotarod activity). An objective assay to assess motor function is the balance beam–crossing assay, which is a useful measure of fine motor coordination and balance capabilities and has been used to assess motor skills of mouse models of SBMA, HD, and other neurodegenerative diseases (49, 64, 65). AR112Q males crossed the 12 mm beam significantly more slowly than both ntg and ntg/Usp7+/– males (Figure 8A). Notably, the loss of 1 allele of Usp7 in AR112Q mice (AR112Q/Usp7+/–) resulted in significantly improved performance on this task (Figure 8A).
Figure 8. Haploinsufficiency of Usp7 rescues several motor deficits and restores levels of unphosphorylated NF-H in spinal motor neurons in a mouse model of SBMA.
(A) Effect of monoallelic knockout of Usp7 on balance beam deficits of AR112Q male mice. ntg (n = 22), AR112Q (n = 21), ntg/Usp7+/– (n = 21), and AR112Q/Usp7+/– (n = 20) mice were evaluated at 33 weeks of age. (B) ntg (n= 22), AR112Q (n = 22), ntg/Usp7+/– (n = 21), and AR112Q/Usp7+/– (n = 20) male mice were evaluated for clasping at 30 weeks of age. (C) Effect of monoallelic knockout of Usp7 on grip strength of 6-week-old (n = 25 per cohort) and 37-week-old mice. ntg (n = 22), AR112Q (n = 20), ntg/Usp7+/– (n = 21) and AR112Q/Usp7+/– (n = 19); average relative change in grip strength between 6 and 37 weeks is represented in D. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, 1-way ANOVA with post hoc Tukey’s test. Error bars represent SD. (E) Unphosphorylated NF-H (SMI32 Ab) immunofluorescence (red) in spinal cords from 37-week-old mice (3 mice per experimental group), with Hoechst 33258 to identify nuclei. Scale bars: 10 μm. (F) Intensity of SMI32 staining was evaluated from at least 230 motor neurons per experimental group. For comparison of distributions, statistical significance was determined by the Kolmogorov-Smirnov test (ntg vs. AR112Q P < 0.0001; AR112Q/Usp7+/– vs. ntg P < 0.01; AR112Q/Usp7+/– vs. AR112Q P < 0.001).
To further evaluate the effect of Usp7 haploinsufficiency on motor impairments, we compared the cohorts for limb-clasping tendency and grip strength. Although the pathology underlying clasping is not well understood, this behavior has been documented in several neurodegenerative disease mouse models, including SBMA transgenic animals (49, 65). AR112Q/Usp7+/– displayed reduced clasping phenotype compared with AR112Q mice at 30 weeks of age (Figure 8B), suggesting that a partial loss of USP7 results in significant improvement of this behavior.
AR112Q males are known to develop a progressive decrease in grip strength (2, 46, 49); therefore, we examined the effect of Usp7 haploinsufficiency on grip strength of AR112Q mice. At 6 weeks of age, the grip strengths of AR112Q and ntg mice were indistinguishable. However, over the next 31 weeks, while the SBMA mice displayed a significant decline in grip strength, the ntg cohort showed significant improvement in this parameter, consistent with previous observations (2, 46, 49). In contrast, while AR112Q/Usp7+/– mice performed worse than ntg mice, their grip strength did not decline with time and was significantly improved relative to that of the AR112Q mice (Figure 8, C and D). Although our findings suggest that haploinsufficiency of Usp7 can rescue motor function defects in SBMA mice, as judged by the parameters discussed above, we did not observe improved rotarod performance (Supplemental Figure 8A). However, it is noteworthy in this regard that ntg/Usp7+/– mice and AR112Q/Usp7+/– mice were significantly smaller than AR112Q mice starting at 6 weeks of age and remained smaller throughout the course of the experiment (Supplemental Figure 8B). Although the reasons for the size differences between these animals is unclear, it is possible that they reflect aspects of USP7 function in early development that could potentially influence the outcome of the rotarod performance (66). Nevertheless, taken together, our data clearly indicate that USP7 lowering has beneficial effects on behavioral phenotypes in SBMA transgenic mice.
Haploinsufficiency of Usp7 alters several pathologies in SBMA mice.
Our finding that 70%–80% knockdown of USP7 significantly decreased the aggregation of AR112Q in PC12 cells (Figure 5D) led us to investigate whether lowering USP7 levels decreased AR aggregation in spinal cord motor neurons of SBMA transgenic mice. Motor neurons of the spinal cord ventral horn at 37 weeks of age did not display any differences in nuclear inclusion load between AR112Q and AR112Q/Usp7+/– mice (data not shown). The reason for this lack of effect could be that 40%–50% decrease of USP7 levels (Supplemental Figure 8, C and D) may not be sufficient to affect polyQ-expanded AR aggregation in this mouse model. It should also be noted that reduction of USP7 levels in these mice had no effect on the steady-state levels of AR monomer (Supplemental Figure 8, C and E).
Previous studies have documented a correlation between SBMA pathology and a decrease in unphosphorylated neurofilament heavy chain (NF-H) in spinal cord ventral horn motor neurons (2, 49). Given that Usp7 haploinsufficiency delayed some motor deficits in SBMA male mice, we next determined whether lowering USP7 levels would alter the phosphorylation state of NF-H as judged by the intensity of SMI32 immunofluorescence in spinal cord ventral horn motor neurons at 37 weeks of age. SMI32 immunofluorescence was significantly reduced in AR112Q male mice relative to ntg animals (P < 0.0001), in agreement with previous studies (2, 49). Monoallelic knockout of Usp7 largely restored SMI32 immunofluorescence to ntg levels (Figure 8, E and F). AR112Q/Usp7+/– SMI32 staining was significantly different from both AR112Q animals (P < 0.001) and ntg animals (P < 0.01), indicating that Usp7 haploinsufficiency substantially rescues a key molecular feature associated with disease pathology.
USP7 catalytic activity promotes AR aggregation.
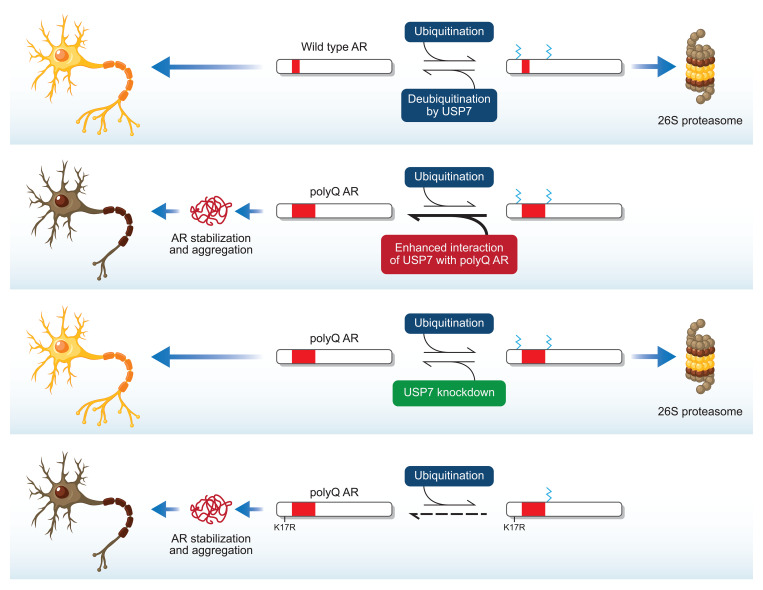
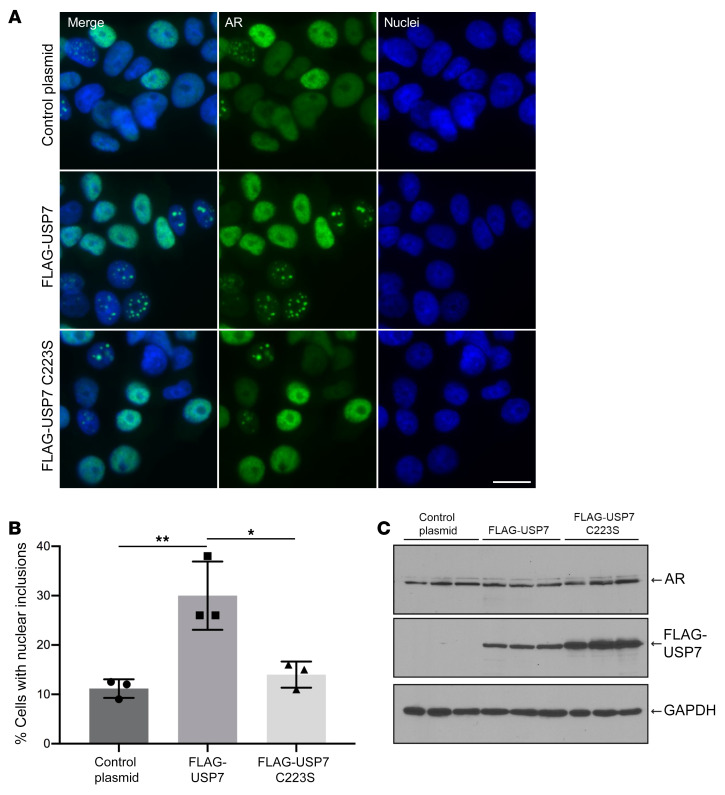
USP7 is a ubiquitously expressed protein that possesses ubiquitin-specific protease activity and has been implicated in the modulation of steady-state levels of several proteins, including Mdm2 and p53 (52). Although our genetic knockout and RNA-based gene-silencing approaches indicate that reduction of USP7 protein levels rescues SBMA-associated phenotypes in vitro and in vivo, the role of the deubiquitinase function of the protein in SBMA pathophysiology was uncertain. Therefore, we evaluated the effect of a catalytically inactive form of USP7 on polyQ-expanded AR aggregation in PC12 cells. To this end, we generated AR112Q-expressing PC12 cell lines that constitutively overexpress either FLAG-tagged WT USP7 or a catalytically inactive (C223S) form of the protein (67) (Figure 9). Corresponding control lines that harbor stably transfected vector plasmid were also generated. USP7 overexpression resulted in a substantial increase in the fraction of cells harboring AR inclusions (relative to vector-transfected cells) (Figure 9, A and B), an effect that is consistent with our finding that USP7 lowering reduced polyQ-expanded AR aggregation (Figure 5D). We observed that expression of USP7 C223S resulted in a markedly lower level of AR112Q-induced inclusions (relative to WT USP7) (Figure 9, A and B). These findings implicate the USP7 deubiquitinase activity in polyQ-expanded AR aggregation.
Figure 9. USP7 catalytic activity promotes AR aggregation.
(A) Immunofluorescence images of PC12 cells expressing AR112Q and FLAG-USP7, the catalytic mutant FLAG-USP7 C223S, or a control vector. Scale bar: 10 μm. (B) Quantification of the number of cells containing nuclear inclusions from A. For each condition, 500 cells were counted in triplicate. Experiment was repeated 3 times. *P < 0.05; **P < 0.01, 1-way ANOVA with post hoc Tukey’s test. Error bars represent SD. (C) Expression levels of AR, FLAG-USP7, and FLAG-USP7 C223S were evaluated by immunoblot.
AR is a substrate for the deubiquitinase function of USP7.
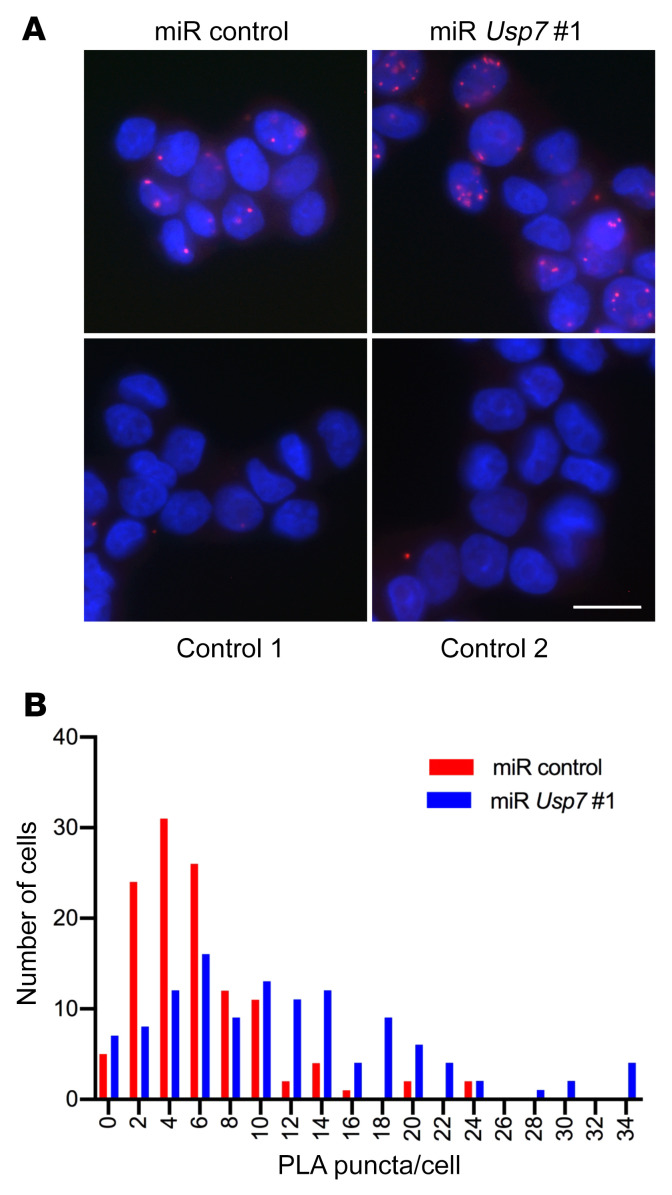
The identification of a catalytic role for USP7 in AR aggregation raised the possibility that AR itself is a substrate for this deubiquitinase, especially given that ubiquitination of AR has been documented previously (19–21). Therefore, to determine whether USP7 knockdown alters the ubiquitination status of polyQ-expanded AR in the cell, we performed PLAs using anti-Ub and anti-AR antibodies. PC12 cells expressing AR112Q displayed PLA puncta (Figure 10A) indicative of the steady-state levels of AR ubiquitination in these cells. Upon USP7 knockdown by stable expression of miR Usp7 no. 1 (Figure 5A), we observed a statistically significant increase in the fraction of cells harboring a larger number of puncta per cell (Figure 10, A and B). This finding suggests that the deubiquitinase function of USP7 modulates the ubiquitination state of AR.
Figure 10. USP7 knockdown enhances polyQ-expanded AR ubiquitination.
AR ubiquitination in PC12 cells expressing AR112Q and miR Usp7 no. 1 (or a miR control) was evaluated by PLA, using antibodies to AR and ubiquitin. (A) Fluorescence images of PLA puncta. Control 1, anti-AR antibody was omitted; control 2, PC12 cells that do not express AR. Scale bar: 10 μm. (B) Quantification of the number of PLA puncta in miR control–expressing cells or miR Usp7 no. 1–expressing cells, respectively; n = 120 cells per condition. Experiment was repeated 3 times. For comparison of distributions, statistical significance was determined by the Kolmogorov-Smirnov test. P < 0.0001.
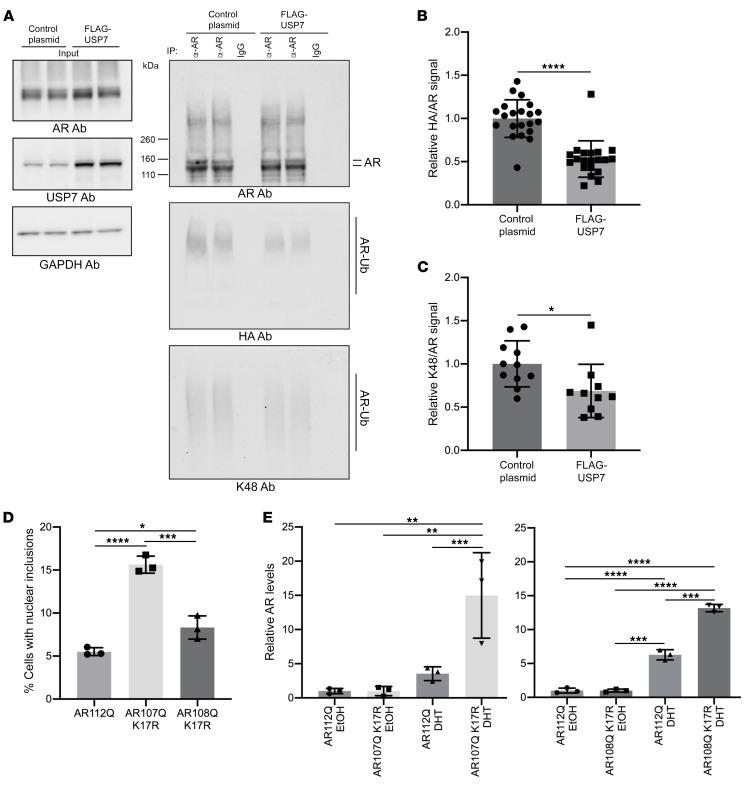
To investigate the impact of USP7 deubiquitinase function on the AR ubiquitination state biochemically, we also evaluated USP7-dependent AR ubiquitination by transiently transfecting HEK293T cells with plasmids expressing (a) AR111Q (with an N-terminal nuclear localization signal), (b) HA-tagged Ub, and (c) FLAG-tagged USP7 or GFP control. Ubiquitin modifications were preserved by the treatment of cells with proteasome inhibitor (10 μM MG132) for 6 hours before harvest. Transient coexpression of HA-Ub with AR111Q resulted in robust ubiquitination of AR111Q as judged by immunoblot analyses of the stringently immunoprecipitated AR using an anti-AR antibody (Figure 11, A and B). This effect was attenuated upon overexpressing USP7 in these cells, suggesting that USP7 directly deubiquitinates the AR. To further characterize the nature of the ubiquitin lysine linkages on AR, we analyzed the immunoprecipitated AR using a K48-specific antibody. This experiment revealed that AR ubiquitination in HEK293T cells occurs (at least in part) via K48 linkages; moreover, USP7-dependent AR deubiquitination substantially reduced such linkages (Figure 11, A and C). These findings have implications for the functional role of USP7 in modulating the species-specific steady-state levels of the AR.
Figure 11. AR is a substrate for the deubiquitinase function of USP7.
(A) Immunoprecipitated AR from HEK293T cells transiently expressing AR111Q, HA-tagged Ub, and FLAG-USP7 (or GFP control) was analyzed by immunoblotting with anti-AR antibody (top), HA-specific antibody (middle) and K48-linked polyubiquitin-specific antibody (bottom). Input levels are shown on the left. (B) Quantitation (from 8 independent experiments performed in duplicate or triplicate) of the relative change in HA to AR monomer signals upon FLAG-USP7 overexpression. (C) Quantitation of the relative change in K48 to AR monomer signals upon FLAG-USP7 overexpression from 4 independent experiments (each experiment performed in duplicate or triplicate). *P < 0.05; ****P < 0.0001, 2-tailed Student’s t test. Error bars represent SD. (D) Percentage of cells with nuclear inclusions in cells expressing AR112Q, AR107Q K17R, or AR108Q K17R. *P < 0.05; ***P < 0.001; ****P < 0.0001, 1-way ANOVA with post hoc Tukey’s test. Error bars represent SD. (E) To evaluate AR turnover, AR112Q, AR107Q K17R, or AR108Q K17R expression was induced with DOX for 48 hours. Following DOX washout, cells were treated with cycloheximide and either DHT or vehicle (EtOH) for an additional 24 hours. AR levels were analyzed by Western blotting (example images shown in Supplemental Figure 10E). Data were normalized to the vehicle (EtOH) treatment condition. Three independent experiments (each one in triplicate) were performed. **P < 0.01; ***P < 0.001; ****P < 0.0001, 1-way ANOVA with post hoc Tukey’s test. Error bars represent SD.
In order to identify the lysine residues on AR112Q that are subject to ubiquitination by USP7, we characterized the whole cell ubiquitinome from cells in which USP7 was knocked down. Thus, PC12 cells inducibly expressing AR112Q and constitutively expressing Usp7-directed miRNA (miR Usp7 no. 1) or nontargeting miRNA (miR control) were treated with DHT and the proteasome inhibitor MG132 and subjected to SILAC ubiquitinome analysis. These experiments revealed 8 lysine residues on AR112Q that showed ubiquitination changes of greater than 1.5-fold upon knockdown of Usp7 (relative to nontargeting miRNA control) in 2 ubiquitinome experiments (Supplemental Table 3). Although knockdown of the USP7 deubiquitinase may have nominally been expected to cause higher levels of ubiquitination, we observed a reduction in the steady-state ubiquitination levels of these 8 lysine residues. One explanation for these findings is that cells lacking USP7 were depleted of a subset of highly ubiquitinated AR112 polypeptides due to proteasomal degradation. In fact, this scenario is supported by the observation that knockdown of USP7 in cells expressing AR112Q results in a reduction of soluble aggregates that migrate on SDS-agarose as a slow migrating species even upon treatment with a proteasome inhibitor (Figures 5E and Supplemental Figure 9C). To determine whether prevention of AR ubiquitination on lysine residues regulated by USP7 (Supplemental Table 3) affects its stability and aggregation, we mutated 1 of the 8 identified lysines (Lys-17, which resides in a region of the mutant AR that plays an important role in disease pathogenesis) to arginine. We created clonal PC12 cells expressing AR107Q K17R and AR108Q K17R and evaluated the role of K17R mutation on polyQ-expanded AR aggregation. Cells expressing polyQ-expanded AR-K17R exhibited a higher frequency of cells bearing aggregated AR (Figure 11D and Supplemental Figure 10, A and D). Moreover, these cells exhibited enhanced DHT-dependent AR stabilization (Figure 11E), suggesting an important role for ubiquitylation of AR at lysine 17 in its degradation. Although lysines can be modified by numerous posttranslational modifications, most of which would be prevented by the K17R mutation, the results shown here, combined with our ubiquitinome analysis revealing lysine 17 as a USP7 substrate, support the importance of USP7 deubiquitylation of polyQ-expanded AR at lysine 17.
Discussion
The causative mutation of SBMA was identified nearly 3 decades ago as an expansion of a CAG triplet repeat within the AR gene. Since then, tremendous progress has been made toward understanding the molecular mechanisms underlying disease pathophysiology, with particular emphasis on the role of the CAG-encoded expanded polyQ tract in cellular dysfunction. There is wide consensus that polyQ expansion leads to AR misfolding and aggregation as well as proteotoxicity. Nevertheless, the pathways that mediate expanded polyQ-AR–dependent toxicity are not well understood. Hence, there is limited information on putative molecular targets that can be modulated pharmacologically for modification of SBMA disease onset or progression.
Because dysregulation of protein interaction networks has been suggested to be at least partly responsible for the cellular dysfunction observed in several polyQ expansion diseases, we reasoned that identification of pathologically relevant protein interactions might yield tractable disease-modifying targets. Therefore, we sought to identify protein partners of AR whose interactions are altered by the polyQ expansion by evaluating the differential protein interaction networks of polyQ-expanded and WT AR in PC12 cells. As shown in Figure 1 and Supplemental Tables 1 and 2, we identified several proteins that preferentially interact either with WT or polyQ-expanded AR. Although these AR interactors do not cluster into defined pathways, they include key regulators of vital cellular functions, such as ubiquitination, protein folding, translation, and nuclear-cytoplasmic shuttling. The ubiquitin-specific proteases USP7 and USP26, identified here as preferred partners of AR112Q and AR10Q, respectively, have been previously reported to interact with AR (25, 27). However, the polyQ length–dependent differences in binding suggest a role for protein ubiquitination-deubiquitination in AR toxicity in SBMA, an aspect that we have explored further in this study. We have also found that the heat shock proteins, HSPB1 and HSP105 are enriched with AR10Q, whereas HSP70 preferentially binds AR112Q. Since these factors play an important role in the cellular stress response and protein folding, our findings may have implications for the involvement of these mechanisms in cellular toxicity associated with expanded polyQ-containing AR. It is especially noteworthy that increasing HSP70 expression or activity has been shown to enhance the degradation of polyQ-expanded AR in a cellular model and to rescue toxicity in a fly model of SBMA, respectively (68, 69). Several ribosomal proteins were also found to associate with AR (Figure 1 and Supplemental Table 1), in agreement with a previous study (70). Although these are primarily cytoplasmic proteins, it has been suggested that their association with AR is reflective of acute hormone-induced ribosome biogenesis (70, 71). In addition, other preferential interactors include Glypican1, Importin 5, FMR1, and α-Mannosidase 2C1, the functional significance of which warrants further investigation.
Using co-IP assays combined with SILAC-based quantitative proteomics, we identified USP7 as a preferential physical interactor with polyQ-expanded AR. The impact of such a preferential interaction between these 2 proteins can be the consequence of either (a) enhanced USP7 deubiquitination of polyQ-expanded AR due to recruitment of USP7 by polyQ-expanded AR or (b) attenuation of normal USP7 function due to its sequestration by polyQ-expanded AR. To test these hypotheses, we determined the effects of genetic knockdown of USP7 or overexpression of WT or catalytically inactive USP7 in SBMA cell models. These experiments established that not only does loss of USP7 result in rescue of polyQ-expanded AR-associated phenotypes, but also that overexpression of USP7 exacerbates polyQ-expanded AR aggregation. The latter effect is not observed upon overexpression of a catalytically inactive mutant of USP7, indicating that the role of USP7 in cellular dysfunction requires its enzymatic function.
USP7 is a ubiquitin-specific protease that predominantly resides in the nucleus (72, 73), although a minor fraction exists in the cytosol (66). The deubiquitinase activity of USP7 regulates protein function as well as the stability of proteins involved in several cellular pathways including the DNA damage response, apoptosis, transcription, and DNA replication (reviewed in refs. 74, 75). Because ubiquitination can mark proteins for proteasomal degradation, the deubiquitinase function of USP7 may serve to increase the half-life of its polypeptide substrates. The best characterized role of USP7 is in the proteasomal regulation of the steady-state levels of the tumor suppressor p53 and its E3 ubiquitin ligase Mdm2. Thus, the deubiquitinase function of USP7 stabilizes p53 under conditions of DNA damage stress and promotes p53 degradation by stabilizing Mdm2 in normally dividing cells (52, 76).
The role of USP7 in the modulation of AR function is only just being unraveled. A recent study (27) showed that USP7 interacts with AR, mediates its deubiquitination, and facilitates AR binding to chromatin in prostate cancer cells. Additionally, overexpression of USP7 in this system resulted in stabilization of AR, suggesting a role for USP7 in regulating AR half-life. Our work has further revealed that polyQ-expanded AR is a likely substrate for the deubiquitinase function of USP7 and that this activity results in reduction of K48-linked polyubiquitin chains from AR. Because K48-linked polyubiquitination usually targets a polypeptide for proteasomal degradation, we hypothesized that USP7 regulates polyQ-expanded AR stability. A prediction of this model is that USP7 modulates steady-state levels of AR. However, as shown in Figure 5C, Figure 7E, and Supplemental Figure 8E, we did not observe significant changes in steady-state levels of AR monomer upon lowering of USP7 in cell, fly, and mouse models of SBMA nor upon overexpression of USP7 in cells (Figure 9C). Nevertheless, USP7 knockdown resulted in a substantial (>50%) decrease in polyQ-expanded AR soluble oligomers (slow migrating species revealed by SDS-agarose) (Figure 5E and Supplemental Figure 4, E and F). Further evaluation revealed a modest but statistically significant decrease in mutant AR half-life following USP7 knockdown (Figure 5F). These results suggest that, although the overall levels of monomeric AR appear relatively refractory to changes in USP7 expression, a subpopulation of soluble monomeric and oligomeric AR is sensitive to the activity of USP7; reducing USP7 levels or activity may enhance the turnover of these soluble, toxic species.
It is also possible that because USP7 deubiquitinates polyQ-expanded AR, the polyubiquitin chains left on AR (upon USP7 knockdown) pose steric barriers to AR oligomer formation, accounting for our observation of reduced amounts of slow-migrating polyQ-expanded AR species in the absence of USP7. Support for this view comes from a recent study by Dao et al. (77) in which the proteasomal shuttle protein UBQLN2 was shown to undergo liquid-liquid phase separation (LLPS) into protein-rich droplets. Noncovalent binding of ubiquitin or K48-linked polyubiquitin chains to UBQLN2 eliminates LLPS, serving as a switch between droplet and dispersed phases.
Our in vivo studies have demonstrated that knockdown of USP7 in a fly model of SBMA has a protective effect on the degeneration of photoreceptor neurons and accessory pigment cells. Because total knockout of Usp7 is embryonic lethal in mice, we were limited to evaluating the effects of germline haploinsufficiency of Usp7 in SBMA transgenic mice. These studies showed that even a modest reduction in USP7 improves motor functions, such as grip strength and balance beam walking. The beneficial effects of USP7 lowering in animal models of SBMA suggests that pharmacological suppression of USP7 levels and/or activity by antisense technology or small molecules could be therapeutically beneficial in the treatment of SBMA. That USP7 is a druggable target is attested to by the fact that small molecule inhibitors of USP7 activity have been developed and are being evaluated for the treatment of several types of cancers (78–80). These approaches are predicated on the notion that, while loss of USP7 may be deleterious during embryonic development (63, 66), inhibition of USP7 in the postdevelopment state (as we have achieved in cultured motor neurons) may be a therapeutically viable strategy. We are currently pursuing these ideas.
The notion that USP7 modulates polyQ-expanded AR function raises the question, Does USP7 also play a role in the pathophysiology of other polyQ-expansion diseases? Indeed, we have shown that knockdown of USP7 has both neuroprotective and antiaggregation effects in a fly model of SCA3. Moreover, differential interaction of USP7 with polyQ-expanded or WT Ataxin-1 has been documented in mammalian cells (81). It is also noteworthy that USP7 preferentially interacts with soluble HTT exon1 harboring expanded polyQ tracts in human cells, raising the possibility that USP7 may play a role in HD as well (32). In fact, our validation that USP7 interacts with endogenous HTT in iPS cells, coupled with our observation that USP7 preferentially interacts with polyQ-expanded HTT in a KI mouse model of HD (Supplemental Figure 7), further supports this view.
In summary, our studies have revealed what we believe is a hitherto unknown requirement for USP7 in the pathogenic mechanisms underlying SBMA and reveal the potential of manipulating this role as a possible therapeutic approach for the treatment of SBMA patients.
Methods
Statistics.
For multiple comparisons, statistical significance was determined by 1-way ANOVA with post hoc Tukey’s test. Comparisons between 2 experimental conditions were done by 1-tailed Student’s t test, unless otherwise indicated. For comparison of distributions, statistical significance was determined by the Kolmogorov-Smirnov test. A P value of less than 0.05 was considered significant. Data were analyzed using Prism 7 (GraphPad).
Study approval.
All animal procedures were performed following the guidelines of the Office of Laboratory Animal Welfare (NIH) and with the approval of the Thomas Jefferson University Institutional Animal Care and Use Committee.
For further information, see Supplemental Methods.
Author contributions
The experiments were conceived and analyzed by AP, DEM, and SVT. AP, YL, EM, FJA, GBM, and BR conducted assays, analyzed data, and prepared figures. SSC and BD dissected and contributed HTT mouse samples. APL, NP, and WG provided Ar113+/– mice, iPS cell lines, and Usp7+/– mice, respectively, and contributed to the experimental design. AP, DEM, SVT, SSC, and BD wrote the manuscript.
Supplementary Material
Acknowledgments
We thank S. Finkbeiner (Gladstone Institute of Neurological Disease and UCSF) for providing 3B5H10 antibody, H.-Y. Tang from the Wistar Institute for proteomics analyses, T. Berger for initial discussions, and G. Ristic (University of Michigan) for assistance with initial assays of AR52Q in Drosophila. We thank K. Fischbeck for SBMA iPS cells and the Cedars Sinai Board of Governors Regenerative Medicine Institute for HD iPS cells. Research reported in this publication used the Bioimaging Facility, with the support of Maria Covarrubias, at Sidney Kimmel Cancer Center at Thomas Jefferson University, which was supported by the National Cancer Institute of the NIH under award P30CA056036. This work was supported by NIH grants R01NS090335 and R01NS108114 (to DEM), R03NS103060 (to AP), R01NS086778 (to SVT), R01NS055746 (to APL), and a grant from the Kennedy’s Disease Association (to AP).
Version 1. 11/10/2020
In-Press Preview
Version 2. 01/04/2021
Print issue publication
Funding Statement
to Diane Merry
to Diane Merry
to Anna Pluciennik
to Sokol Todi
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e134565.https://doi.org/10.1172/JCI134565.
Contributor Information
Anna Pluciennik, Email: anna.pluciennik@jefferson.edu.
Yuhong Liu, Email: yuhong.liu@jefferson.edu.
Elana Molotsky, Email: elana.molotsky@jefferson.edu.
Gregory B. Marsh, Email: gregory.marsh@wayne.edu.
Bedri Ranxhi, Email: fi2905@wayne.edu.
Frederick J. Arnold, Email: frederick.arnold@duke.edu.
Naemeh Pourshafie, Email: naemeh.pourshafie@nih.gov.
Andrew P. Lieberman, Email: liebermn@umich.edu.
Wei Gu, Email: wg8@cumc.columbia.edu.
Sokol V. Todi, Email: stodi@med.wayne.edu.
References
- 1.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 2.Chevalier-Larsen ES, et al. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J Neurosci. 2004;24(20):4778–4786. doi: 10.1523/JNEUROSCI.0808-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katsuno M, et al. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35(5):843–854. doi: 10.1016/S0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 4.Yu Z, et al. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J Clin Invest. 2006;116(10):2663–2672. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy WR, Alter M, Sung JH. Progressive proximal spinal and bulbar muscular atrophy of late onset. A sex-linked recessive trait. Neurology. 1968;18(7):671–680. doi: 10.1212/WNL.18.7.671. [DOI] [PubMed] [Google Scholar]
- 6.Sobue G, Hashizume Y, Mukai E, Hirayama M, Mitsuma T, Takahashi A. X-linked recessive bulbospinal neuronopathy. A clinicopathological study. Brain. 1989;112(Pt 1):209–232. doi: 10.1093/brain/112.1.209. [DOI] [PubMed] [Google Scholar]
- 7.Lieberman AP, et al. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Rep. 2014;7(3):774–784. doi: 10.1016/j.celrep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, et al. Defects in neuromuscular transmission may underlie motor dysfunction in spinal and bulbar muscular atrophy. J Neurosci. 2016;36(18):5094–5106. doi: 10.1523/JNEUROSCI.3485-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, et al. Nuclear inclusions of the androgen receptor protein in spinal and bulbar muscular atrophy. Ann Neurol. 1998;44(2):249–254. doi: 10.1002/ana.410440216. [DOI] [PubMed] [Google Scholar]
- 10.Li M, et al. Nonneural nuclear inclusions of androgen receptor protein in spinal and bulbar muscular atrophy. Am J Pathol. 1998;153(3):695–701. doi: 10.1016/S0002-9440(10)65612-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohen SP, Kralli A, Yamamoto KR. Hold ‘em and fold ‘em: chaperones and signal transduction. Science. 1995;268(5215):1303–1304. doi: 10.1126/science.7761850. [DOI] [PubMed] [Google Scholar]
- 12.Cano LQ, Lavery DN, Bevan CL. Mini-review: Foldosome regulation of androgen receptor action in prostate cancer. Mol Cell Endocrinol. 2013;369(1–2):52–62. doi: 10.1016/j.mce.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Cato L, Neeb A, Brown M, Cato AC. Control of steroid receptor dynamics and function by genomic actions of the cochaperones p23 and Bag-1L. Nucl Recept Signal. 2014;12:e005. doi: 10.1621/nrs.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black BE, Holaska JM, Rastinejad F, Paschal BM. DNA binding domains in diverse nuclear receptors function as nuclear export signals. Curr Biol. 2001;11(22):1749–1758. doi: 10.1016/S0960-9822(01)00537-1. [DOI] [PubMed] [Google Scholar]
- 15.Shank LC, et al. Activation of the DNA-dependent protein kinase stimulates nuclear export of the androgen receptor in vitro. J Biol Chem. 2008;283(16):10568–10580. doi: 10.1074/jbc.M800810200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basso M, Pennuto M. Serine phosphorylation and arginine methylation at the crossroads to neurodegeneration. Exp Neurol. 2015;271:77–83. doi: 10.1016/j.expneurol.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Chua JP, et al. Disrupting SUMOylation enhances transcriptional function and ameliorates polyglutamine androgen receptor-mediated disease. J Clin Invest. 2015;125(2):831–845. doi: 10.1172/JCI73214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montie HL, Pestell RG, Merry DE. SIRT1 modulates aggregation and toxicity through deacetylation of the androgen receptor in cell models of SBMA. J Neurosci. 2011;31(48):17425–17436. doi: 10.1523/JNEUROSCI.3958-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He B, et al. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP) J Biol Chem. 2004;279(29):30643–30653. doi: 10.1074/jbc.M403117200. [DOI] [PubMed] [Google Scholar]
- 20.Morishima Y, Wang AM, Yu Z, Pratt WB, Osawa Y, Lieberman AP. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum Mol Genet. 2008;17(24):3942–3952. doi: 10.1093/hmg/ddn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21(15):4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nawaz Z, et al. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19(2):1182–1189. doi: 10.1128/MCB.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu K, et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell. 2009;15(4):270–282. doi: 10.1016/j.ccr.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burska UL, et al. Deubiquitinating enzyme Usp12 is a novel co-activator of the androgen receptor. J Biol Chem. 2013;288(45):32641–32650. doi: 10.1074/jbc.M113.485912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dirac AM, Bernards R. The deubiquitinating enzyme USP26 is a regulator of androgen receptor signaling. Mol Cancer Res. 2010;8(6):844–854. doi: 10.1158/1541-7786.MCR-09-0424. [DOI] [PubMed] [Google Scholar]
- 26.Faus H, Meyer HA, Huber M, Bahr I, Haendler B. The ubiquitin-specific protease USP10 modulates androgen receptor function. Mol Cell Endocrinol. 2005;245(1–2):138–146. doi: 10.1016/j.mce.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Chen ST, Okada M, Nakato R, Izumi K, Bando M, Shirahige K. The deubiquitinating enzyme USP7 regulates androgen receptor activity by modulating its binding to chromatin. J Biol Chem. 2015;290(35):21713–21723. doi: 10.1074/jbc.M114.628255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giorgetti E, Lieberman AP. Polyglutamine androgen receptor-mediated neuromuscular disease. Cell Mol Life Sci. 2016;73(21):3991–3999. doi: 10.1007/s00018-016-2275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 30.Bowman AB, Yoo SY, Dantuma NP, Zoghbi HY. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum Mol Genet. 2005;14(5):679–691. doi: 10.1093/hmg/ddi064. [DOI] [PubMed] [Google Scholar]
- 31.Ortega Z, Diaz-Hernandez M, Maynard CJ, Hernandez F, Dantuma NP, Lucas JJ. Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J Neurosci. 2010;30(10):3675–3688. doi: 10.1523/JNEUROSCI.5673-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YE, et al. Soluble oligomers of polyQ-expanded Huntingtin target a multiplicity of key cellular factors. Mol Cell. 2016;63(6):951–964. doi: 10.1016/j.molcel.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 33.Heine EM, Berger TR, Pluciennik A, Orr CR, Zboray L, Merry DE. Proteasome-mediated proteolysis of the polyglutamine-expanded androgen receptor is a late event in spinal and bulbar muscular atrophy (SBMA) pathogenesis. J Biol Chem. 2015;290(20):12572–12584. doi: 10.1074/jbc.M114.617894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95(1):55–66. doi: 10.1016/S0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 35.Simeoni S, et al. Motoneuronal cell death is not correlated with aggregate formation of androgen receptors containing an elongated polyglutamine tract. Hum Mol Genet. 2000;9(1):133–144. doi: 10.1093/hmg/9.1.133. [DOI] [PubMed] [Google Scholar]
- 36.Paulson HL, Bonini NM, Roth KA. Polyglutamine disease and neuronal cell death. Proc Natl Acad Sci U S A. 2000;97(24):12957–12958. doi: 10.1073/pnas.210395797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson AL, Bottomley SP. Towards the treatment of polyglutamine diseases: the modulatory role of protein context. Curr Med Chem. 2010;17(27):3058–3068. doi: 10.2174/092986710791959800. [DOI] [PubMed] [Google Scholar]
- 38.Culver BP, et al. Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J Biol Chem. 2012;287(26):21599–21614. doi: 10.1074/jbc.M112.359307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosp F, et al. Quantitative interaction proteomics of neurodegenerative disease proteins. Cell Rep. 2015;11(7):1134–1146. doi: 10.1016/j.celrep.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim J, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452(7188):713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beauchemin AM, Gottlieb B, Beitel LK, Elhaji YA, Pinsky L, Trifiro MA. Cytochrome c oxidase subunit Vb interacts with human androgen receptor: a potential mechanism for neurotoxicity in spinobulbar muscular atrophy. Brain Res Bull. 2001;56(3–4):285–297. doi: 10.1016/S0361-9230(01)00583-4. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki E, et al. Aberrant E2F activation by polyglutamine expansion of androgen receptor in SBMA neurotoxicity. Proc Natl Acad Sci U S A. 2009;106(10):3818–3822. doi: 10.1073/pnas.0809819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao H, et al. A polyglutamine expansion disease protein sequesters PTIP to attenuate DNA repair and increase genomic instability. Hum Mol Genet. 2012;21(19):4225–4236. doi: 10.1093/hmg/dds246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies P, Watt K, Kelly SM, Clark C, Price NC, McEwan IJ. Consequences of poly-glutamine repeat length for the conformation and folding of the androgen receptor amino-terminal domain. J Mol Endocrinol. 2008;41(5):301–314. doi: 10.1677/JME-08-0042. [DOI] [PubMed] [Google Scholar]
- 45.Eftekharzadeh B, et al. Sequence context influences the structure and aggregation behavior of a polyQ tract. Biophys J. 2016;110(11):2361–2366. doi: 10.1016/j.bpj.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montie HL, et al. Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2009;18(11):1937–1950. doi: 10.1093/hmg/ddp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orr CR, et al. An interdomain interaction of the androgen receptor is required for its aggregation and toxicity in spinal and bulbar muscular atrophy. J Biol Chem. 2010;285(46):35567–35577. doi: 10.1074/jbc.M110.146845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walcott JL, Merry DE. Ligand promotes intranuclear inclusions in a novel cell model of spinal and bulbar muscular atrophy. J Biol Chem. 2002;277(52):50855–50859. doi: 10.1074/jbc.M209466200. [DOI] [PubMed] [Google Scholar]
- 49.Zboray L, et al. Preventing the androgen receptor N/C interaction delays disease onset in a mouse model of SBMA. Cell Rep. 2015;13(10):2312–2323. doi: 10.1016/j.celrep.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller J, et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat Chem Biol. 2011;7(12):925–934. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger TR, Montie HL, Jain P, Legleiter J, Merry DE. Identification of novel polyglutamine-expanded aggregation species in spinal and bulbar muscular atrophy. Brain Res. 2015;1628(Pt B):254–264. doi: 10.1016/j.brainres.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13(6):879–886. doi: 10.1016/S1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 53.Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5(4):583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- 54.Nedelsky NB, et al. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67(6):936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447(7146):859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 56.Tsou WL, et al. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS One. 2012;7(8):e43112. doi: 10.1371/journal.pone.0043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burr AA, Tsou WL, Ristic G, Todi SV. Using membrane-targeted green fluorescent protein to monitor neurotoxic protein-dependent degeneration of Drosophila eyes. J Neurosci Res. 2014;92(9):1100–1109. doi: 10.1002/jnr.23395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutton JR, et al. Interaction of the polyglutamine protein ataxin-3 with Rad23 regulates toxicity in Drosophila models of spinocerebellar ataxia type 3. Hum Mol Genet. 2017;26(8):1419–1431. doi: 10.1093/hmg/ddx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das B, et al. A novel iron (II) preferring dopamine agonist chelator D-607 significantly suppresses α-syn- and MPTP-induced toxicities in vivo. Neuropharmacology. 2017;123:88–99. doi: 10.1016/j.neuropharm.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsou WL, et al. DnaJ-1 and karyopherin α3 suppress degeneration in a new Drosophila model of Spinocerebellar Ataxia Type 6. Hum Mol Genet. 2015;24(15):4385–4396. doi: 10.1093/hmg/ddv174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yedlapudi D, Joshi GS, Luo D, Todi SV, Dutta AK. Inhibition of alpha-synuclein aggregation by multifunctional dopamine agonists assessed by a novel in vitro assay and an in vivo Drosophila synucleinopathy model. Sci Rep. 2016;6:38510. doi: 10.1038/srep38510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Menalled LB, et al. Comprehensive behavioral and molecular characterization of a new knock-in mouse model of Huntington’s disease: zQ175. PLoS One. 2012;7(12):e49838. doi: 10.1371/journal.pone.0049838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29(9):1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carter RJ, et al. Characterization of progressive motor deficits in mice transgenic for the human Huntington’s disease mutation. J Neurosci. 1999;19(8):3248–3257. doi: 10.1523/JNEUROSCI.19-08-03248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Southwell AL, Ko J, Patterson PH. Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington’s disease. J Neurosci. 2009;29(43):13589–13602. doi: 10.1523/JNEUROSCI.4286-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hao YH, et al. USP7 acts as a molecular rheostat to promote WASH-dependent endosomal protein recycling and is mutated in a human neurodevelopmental disorder. Mol Cell. 2015;59(6):956–969. doi: 10.1016/j.molcel.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li M, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416(6881):648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 68.Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2002;11(5):515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 69.Wang AM, et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat Chem Biol. 2013;9(2):112–118. doi: 10.1038/nchembio.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsiao JJ, Smits MM, Ng BH, Lee J, Wright ME. Discovery Proteomics Identifies a Molecular Link between the Coatomer Protein Complex I and Androgen Receptor-dependent Transcription. J Biol Chem. 2016;291(36):18818–18842. doi: 10.1074/jbc.M116.732313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang T, Liao S. A very rapid effect of androgen on initiation of protein synthesis in prostate. Proc Natl Acad Sci U S A. 1975;72(2):706–709. doi: 10.1073/pnas.72.2.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernández-Montalván A, et al. Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. FEBS J. 2007;274(16):4256–4270. doi: 10.1111/j.1742-4658.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- 73.Zapata JM, Pawlowski K, Haas E, Ware CF, Godzik A, Reed JC. A diverse family of proteins containing tumor necrosis factor receptor-associated factor domains. J Biol Chem. 2001;276(26):24242–24252. doi: 10.1074/jbc.M100354200. [DOI] [PubMed] [Google Scholar]
- 74.Bhattacharya S, Chakraborty D, Basu M, Ghosh MK. Emerging insights into HAUSP (USP7) in physiology, cancer and other diseases. Signal Transduct Target Ther. 2018;3:17. doi: 10.1038/s41392-018-0012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim RQ, Sixma TK. Regulation of USP7: a high incidence of E3 complexes. J Mol Biol. 2017;429(22):3395–3408. doi: 10.1016/j.jmb.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 76.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428(6982):1–2. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 77.Dao TP, et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol Cell. 2018;69(6):965–978.e6. doi: 10.1016/j.molcel.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.An T, et al. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem Pharmacol. 2017;131:29–39. doi: 10.1016/j.bcp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 79.Carra G, et al. Therapeutic inhibition of USP7-PTEN network in chronic lymphocytic leukemia: a strategy to overcome TP53 mutated/deleted clones. Oncotarget. 2017;8(22):35508–35522. doi: 10.18632/oncotarget.16348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morra F, et al. The combined effect of USP7 inhibitors and PARP inhibitors in hormone-sensitive and castration-resistant prostate cancer cells. Oncotarget. 2017;8(19):31815–31829. doi: 10.18632/oncotarget.16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hong S, Kim SJ, Ka S, Choi I, Kang S. USP7, a ubiquitin-specific protease, interacts with ataxin-1, the SCA1 gene product. Mol Cell Neurosci. 2002;20(2):298–306. doi: 10.1006/mcne.2002.1103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.