Abstract
The neuronal mechanisms that establish 24-hour rhythms in feeding and metabolism remain incompletely understood. In this issue of the JCI, Adlanmerini and colleagues explored the relationship between temporal and homeostatic control of energy balance by focusing on mice that lacked the genes encoding the clock repressor elements REV-ERBα and –β, specifically in the tuberal hypothalamus. Notably, the clock transcription cycle mediated intraneuronal response to the adipostatic hormone leptin. These results show that REV-ERBα and –β in the hypothalamus are necessary for maintaining leptin responsiveness and metabolic homeostasis and lay the foundation to explore how transcriptional changes may link energy-sensing cell types with day/night rhythms. Such information may lead to therapeutics that alleviate the adverse effects of chronic shift work.
Clock genes
In the early 1970s, Ron Konopka and Seymour Benzer proved that genes control behavior in Drosophila melanogaster by isolating progeny of mutagenized flies that exhibited alteration in 24-hour circadian sleep/wake activity (1). Core clock genes were ultimately shown to encode transcription factors that form an autoregulatory feedback loop, including the nuclear receptors REV-ERBα and –β, and to play a critical role in repressing the activating components CLOCK and BMAL1 (2). Clock genes are expressed within pacemaker neurons and many other regions of the brain, and throughout most peripheral cells. Mice with disrupted clock genes that consume high-fat diets feed on a schedule that is misaligned with the light/dark cycle, indicating that both meal timing and macronutrient content contribute to energy homeostasis and metabolic health (3, 4). Understanding how central nervous system (CNS) mechanisms coordinate sleep/wake and appetitive behaviors may ultimately reveal countermeasures to prevent and treat the epidemic of obesity and its comorbidities.
As a dominant appetitive hormone, leptin follows the sleep/wake cycle across mammalian species, and signals to the brain to regulate appetite and energetics (5, 6). First-order leptin-responsive neurons modulate neuroendocrine function according to nutrient state and are distributed within the arcuate (ARC), ventromedial (VMH), and dorsomedial (DMH) hypothalamic nuclei. Accordingly, these brain regions control whole-body energetics and feeding behavior (7–9). Similarly to studies that disrupt the function of the core clock component, Clock, whole-body knockout of Reverbα and -β disrupts metabolic homeostasis, body weight maintenance, and feeding behavior (3, 10, 11).
Hypothalamic functions of REV-ERBs
In this issue of the JCI, Adlanmerini et al. ablated Reverbα and -β clock genes exclusively in the tuberal hypothalamus (HDKO mice). These mice exhibited increased longitudinal weight gain as well as reduced energy expenditure (EE) and body temperature (12). Notably, cells within the ARC, DMH, and VMH are known to regulate rhythms in body temperature (13). Whereas Adlanmerini and colleagues observed lower EE and body temperature, especially during the rest (light) cycle, a previous study observed that ablation of Bmal1 in the VMH (using Sf1-driven CRE recombinase expression) resulted in higher EE and body temperature, especially during the active dark period (9). These results suggest that some effects in the hypothalamus-specific REV-ERBα– and –β–deficient mice may be due to loss of rhythmic inhibition that REV-ERBα and –β normally exert on genes such as Bmal1.
In Adlanmerini et al., HDKO mice also exhibited increased feeding during the light period, and greater rebound food intake following a fasting period (Figure 1). The authors explored the underlying genetic mechanisms in the ARC of HDKO compared with control mice, by performing transcriptional profiling during the time of day when protein expression of REV-ERBα peaked, i.e., in the early rest period. These analyses pointed to changes involving leptin and insulin signaling, and also revealed that genes normally repressed by REV-ERBs exhibited increased expression, whereas genes involved in mediating leptin signaling exhibited decreased expression in the ARC of HDKO mice. The authors also carried out genome-wide chromatin profiling, using immunoprecipitation to capture the DNA segments bound by REV-ERBs, to define the transcriptional targets of these transcription factors within the ARC. Genes that inhibited leptin responsiveness, such as Fabp7 and Socs3, were transcriptionally regulated via REV-ERB binding. The findings of Adlanmerini et al. (12) therefore add to recent work that indicate a role for the molecular clock in energy-sensing AgRP neurons, which are also located within the ARC (5). Notably, Adlanmerini and authors observed that Fabp7 also increased in relative expression in HDKO versus control mice when these groups were administered leptin (12). The physiological relevance of a transcriptional change in leptin pathways was verified by administering leptin; compared with control mice, HDKO mice consumed more, which was paralleled within the ARC by reduced phosphorylation of the leptin-responsive transcription factor STAT3. Altogether, the findings further highlight the important role of the molecular clock within the hypothalamus for maintaining rhythms in leptin responsiveness. Animal models of shift work (recurrent jet lag) and diet-induced obesity also indicate that such paradigms disrupt leptin sensitivity, including at the level of the CNS (5, 6). Given that disrupted sleep/wake cycles and obesity are two intertwined phenomena also in humans, it remains to be determined whether CNS leptin action is altered in subjects who carry out shift work, or in subjects who have had their circadian system intentionally misaligned, which has been tied to disrupted energetics and food intake regulation (14).
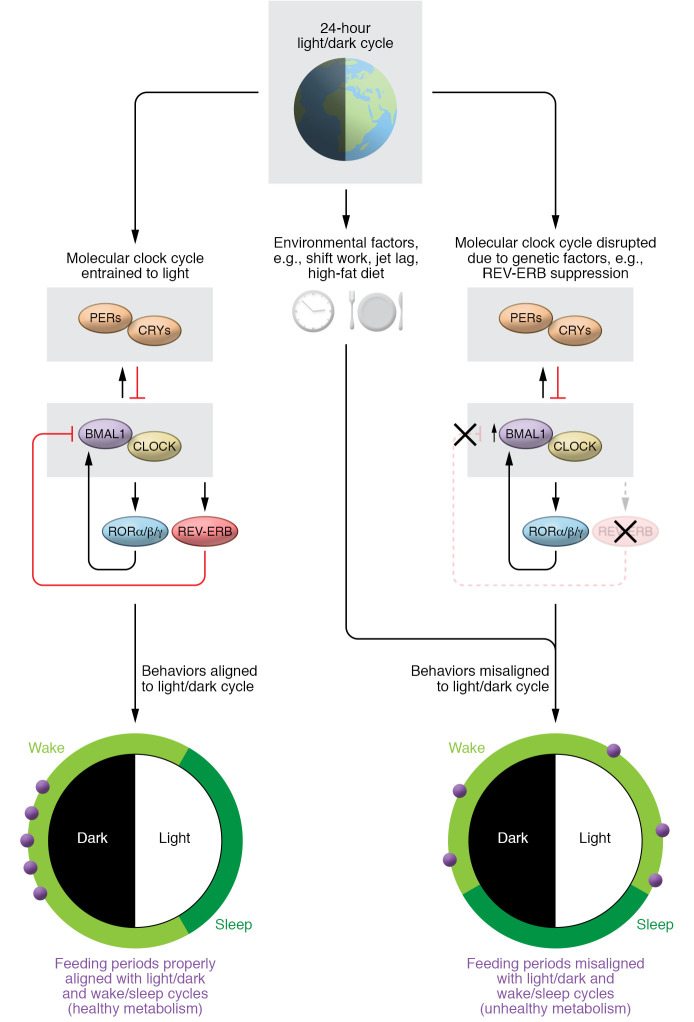
Figure 1. The molecular clock aligns behavior and physiology with the 24-hour light/dark cycle.
The circadian system synchronizes sleep/wake and fasting/feeding cycles with the light/dark cycle. Core clock components including CLOCK and BMAL1 activate REV-ERBα and –β, which repress BMAL1 transcription in an autoregulatory feedback loop (left). Disruption of the circadian system can occur through environmental factors such as shift work and jet lag, resulting in misaligned behavior (including feeding) and metabolic rhythms (right). The work by Adlanmerini et al. (12) indicates that core clock components in energy-sensing neurons of the hypothalamus coordinate behavior and physiology via transcription to coordinate hunger with activity cycles. Disrupting REV-ERBα and –β in the hypothalamus increased Bmal1 mRNA, altered leptin signaling, and changed feeding behavior (right). PERs, period proteins; CRYs, cryptochrome proteins; ROR, retinoic acid receptor–related orphan receptor.
Critical intertissue differences
The circadian system functions in a hierarchical manner, wherein light-modulated neurons within the hypothalamic suprachiasmatic nucleus (SCN) constitute the central pacemaker, to synchronize physiological and behavioral rhythms with the external light/dark cycle. An important aspect of the work by Adlanmerini et al. (12) is that the employed mouse model spared ablation of REV-ERBα and –β from the SCN, a region that is often targeted in similar studies, for instance in models that also result in disrupted temporal regulation of feeding (3, 5). By maintaining clock function within the SCN, the researchers avoided the potential confounder that this brain region sits at the top of the hierarchy, for instance being critical for maintenance of rhythmic activity behavior (15). The present work (12) therefore adds to other studies that indicate that the molecular clock, even outside the SCN, plays a critical role in maintenance of energy homeostasis (5, 9).
The recent work by Adlanmerini et al. (12) ablated REV-ERBα and –β by driving CRE expression from the gene Nkx2.1, disrupting genes across multiple brain regions that independently are implicated in a wide range of behaviors and metabolic functions. It therefore remains to be determined whether the role of REV-ERBα and –β differs depending on where these core clock elements are anatomically inactivated. In the context of the circadian system, genomic regulation of the chromatin landscape by core clock elements has so far mostly been explored in peripheral tissues. Such studies do, however, indicate that there are critical intertissue differences in how these genes may confer physiological rhythms specific to a given tissue (16). Separate studies indicate that there exists marked heterogeneity in the chromatin landscape across neuronal populations (17). As such, Adlanmerini and colleagues lay the foundation to explore whether there also exist critical differences in how energy-sensing cell types are rhythmically regulated at the epigenetic level, and whether such mechanisms may be involved in day/night rhythms in physiology and behavior, such as food intake (13).
Notably, using the gene Nkx2.1 targets both glia and neurons, and there is an increasing focus on molecular clock function specifically within glial cells (18). Thus far, this relevance has largely been explored in the context of the SCN, even though glial cells also are recognized as essential in metabolic homeostasis throughout multiple hypothalamic nuclei, including for leptin responsiveness (19). Further studies may indeed shed light on whether the behavioral and physiological changes observed by Adlanmerini and colleagues (12) depend on specific cell populations within each of these hypothalamic centers, given that many such regions may be composed of cells with opposing functions on behaviors such as food intake, such as the POMC and AgRP cells within the ARC (5, 13). Given that circadian disruption, as occurs in shift work, may also produce adverse metabolic effects by disrupting the circadian interplay across brain regions (20), a related question is whether there are cells in which restoration of clock function can serve a more critical role in reestablishing metabolic homeostasis. Such knowledge may hopefully aid the development of therapeutics targeted toward the numerous adverse effects of chronic shift work. Clock systems originally were identified based on studies of sleep/wake behavior and Adlanmerini and authors add to the growing evidence (12) that this system is also integral to the regulation of metabolism at the level of the brain.
Acknowledgments
We acknowledge Biliana Marcheva for help with the figure and support from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants R01DK090625, R01DK100814, 1R01DK113011-01A1, and K99DK124682, the National Institute on Aging (NIA) grants P01AG011412 and R01AG065988 (to JB), the Swedish Research Council grant 2014-6888, and the Swedish Society for Medical Research (to JC).
Version 1. 01/04/2021
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e144655. https://doi.org/10.1172/JCI144655.
Contributor Information
Jonathan Cedernaes, Email: jonathan.cedernaes@neuro.uu.se.
Joseph Bass, Email: j-bass@northwestern.edu.
References
- 1.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68(9):2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354(6315):994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 3.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Cedernaes J, et al. Transcriptional basis for rhythmic control of hunger and metabolism within the AgRP neuron. Cell Metab. 2019;29(5):1078–1091.e5. doi: 10.1016/j.cmet.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kettner NM, et al. Circadian dysfunction induces leptin resistance in mice. Cell Metab. 2015;22(3):448–459. doi: 10.1016/j.cmet.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balthasar N, et al. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42(6):983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Garfield AS, et al. Dynamic GABAergic afferent modulation of AgRP neurons. Nat Neurosci. 2016;19(12):1628–1635. doi: 10.1038/nn.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orozco-Solis R, et al. The circadian clock in the ventromedial hypothalamus controls cyclic energy expenditure. Cell Metab. 2016;23(3):467–478. doi: 10.1016/j.cmet.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerhart-Hines Z, et al. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature. 2013;503(7476):410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feillet CA, et al. Rev-erbα modulates the hypothalamic orexinergic system to influence pleasurable feeding behaviour in mice. Addict Biol. 2017;22(2):411–422. doi: 10.1111/adb.12339. [DOI] [PubMed] [Google Scholar]
- 12.Adlanmerini M, et al. Hypothalamic REV-ERB nuclear receptors control diurnal food intake and leptin sensitivity in diet-induced obese mice. J Clin Invest. 2021;131(1):e140424. doi: 10.1172/JCI140424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cedernaes J, et al. Neurogenetic basis for circadian regulation of metabolism by the hypothalamus. Genes Dev. 2019;33(17–18):1136–1158. doi: 10.1101/gad.328633.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHill AW, et al. Impact of circadian misalignment on energy metabolism during simulated nightshift work. Proc Natl Acad Sci U S A. 2014;111(48):17302–17307. doi: 10.1073/pnas.1412021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDearmon EL, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314(5803):1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perelis M, et al. Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science. 2015;350(6261):aac4250. doi: 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fullard JF, et al. An atlas of chromatin accessibility in the adult human brain. Genome Res. 2018;28(8):1243–1252. doi: 10.1101/gr.232488.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brancaccio M, et al. Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science. 2019;363(6423):187–192. doi: 10.1126/science.aat4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuente-Martin E, et al. Leptin regulates glutamate and glucose transporters in hypothalamic astrocytes. J Clin Invest. 2012;122(11):3900–3913. doi: 10.1172/JCI64102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288(5466):682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]