Abstract
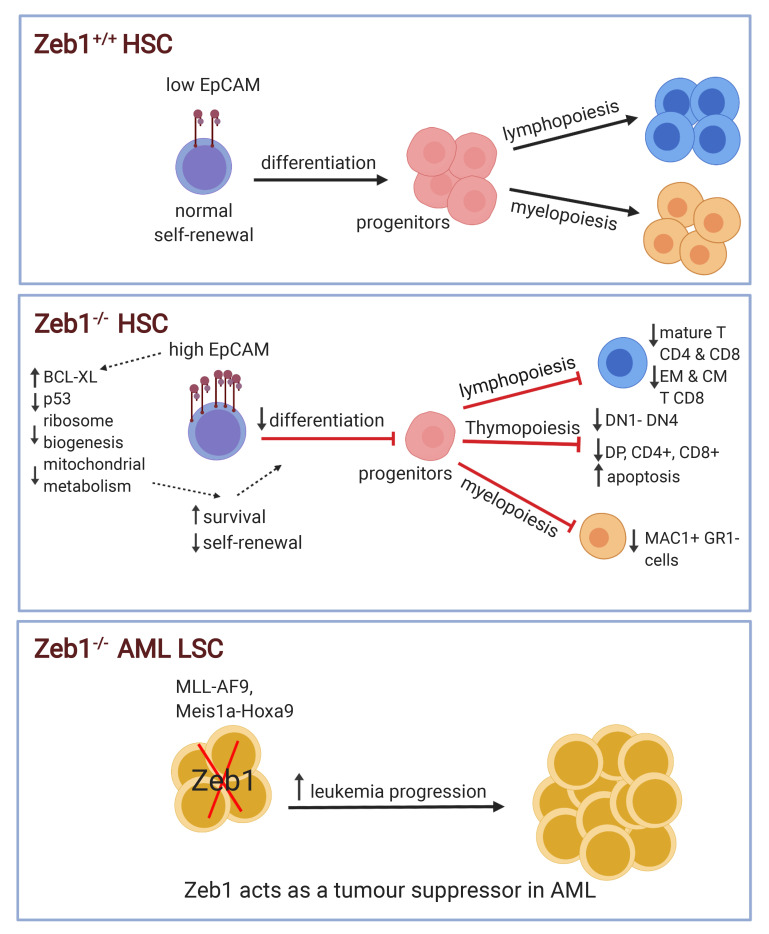
Zeb1, a zinc finger E-box binding homeobox epithelial-mesenchymal transition (EMT) transcription factor, confers properties of “stemness,” such as self-renewal, in cancer. Yet little is known about the function of Zeb1 in adult stem cells. Here, we used the hematopoietic system as a well-established paradigm of stem cell biology to evaluate Zeb1-mediated regulation of adult stem cells. We employed a conditional genetic approach using the Mx1-Cre system to specifically knock out (KO) Zeb1 in adult hematopoietic stem cells (HSCs) and their downstream progeny. Acute genetic deletion of Zeb1 led to rapid-onset thymic atrophy and apoptosis-driven loss of thymocytes and T cells. A profound cell-autonomous self-renewal defect and multilineage differentiation block were observed in Zeb1-KO HSCs. Loss of Zeb1 in HSCs activated transcriptional programs of deregulated HSC maintenance and multilineage differentiation genes and of cell polarity consisting of cytoskeleton-, lipid metabolism/lipid membrane–, and cell adhesion–related genes. Notably, epithelial cell adhesion molecule (EpCAM) expression was prodigiously upregulated in Zeb1-KO HSCs, which correlated with enhanced cell survival, diminished mitochondrial metabolism, ribosome biogenesis, and differentiation capacity and an activated transcriptomic signature associated with acute myeloid leukemia (AML) signaling. ZEB1 expression was downregulated in AML patients, and Zeb1 KO in the malignant counterparts of HSCs — leukemic stem cells (LSCs) — accelerated MLL-AF9– and Meis1a/Hoxa9-driven AML progression, implicating Zeb1 as a tumor suppressor in AML LSCs. Thus, Zeb1 acts as a transcriptional regulator in hematopoiesis, critically coordinating HSC self-renewal, apoptotic, and multilineage differentiation fates required to suppress leukemic potential in AML.
Keywords: Hematology, Stem cells
Keywords: Bone marrow differentiation
Introduction
Epithelial-mesenchymal transition (EMT) is a complex process that organizes specific changes in cellular fate and phenotype and is usually accompanied by loss of cell polarity and adhesion and increased locomotion (1). EMT is an important step in embryonic development and regeneration, which largely promotes a program of cellular plasticity and migration (2). This program is regulated by specific transcription factors (TFs), such as members of the ZEB, SNAI, and TWIST families. To this end, Zeb1, a zinc finger TF that binds to E-box motifs, has been implicated in myogenesis (3–6), neuronal development and differentiation (7–10), postgastrulation embryogenesis (11) and T cell development (12, 13).
It has also been posited that EMT is a critical regulator in cancer pathogenesis and, in particular, cancer stem cell behavior (14–16), which facilitates cancer cells becoming more metastatic, with resultant tumor progression (17). Abundant evidence shows that Zeb1 regulates stem cell properties in cancer, including self-renewal (18–20). While it is established that Zeb1 regulates expression of multiple stem cell–associated TFs, including those with oncogenic potential, such as BMI1, KLF4, and SOX2 (19, 21), and that loss of Zeb1 promotes cellular differentiation during development of the embryonic CNS (9) and skeletal muscle (22), the wider role of Zeb1 in normal stem cell–fate decisions remains unclear.
By exploiting a mouse model engineered to contain conditional alleles of Zeb1 and an inducible Mx-1-Cre (Zeb1fl/fl;Mx1-Cre+) (23), where Zeb1 expression can be deleted in hematopoietic stem cells (HSCs) and their progeny by administering polyinosinic-polycytidylic acid (pIpC), we used the hematopoietic system, as an established stem cell model, to evaluate Zeb1-mediated regulation of somatic stem cells. Herein, we identify Zeb1 as a crucial, indispensable regulator of adult T cell maturation and differentiation. In a broader context, as judged by conditional deletion within the hematopoietic system, we identify Zeb1 as an essential transcriptional repressor balancing adult stem cell self-renewal, apoptotic, and global, multi-lineage differentiation fates of stem cells. Finally, we find that Zeb1-mediated regulation of these stem cell fates is required to suppress malignancy in the context of acute myeloid leukemia (AML).
Results
Acute conditional deletion of Zeb1 reduces the frequency of MAC1+ myeloid cells and CD8+ memory T cells.
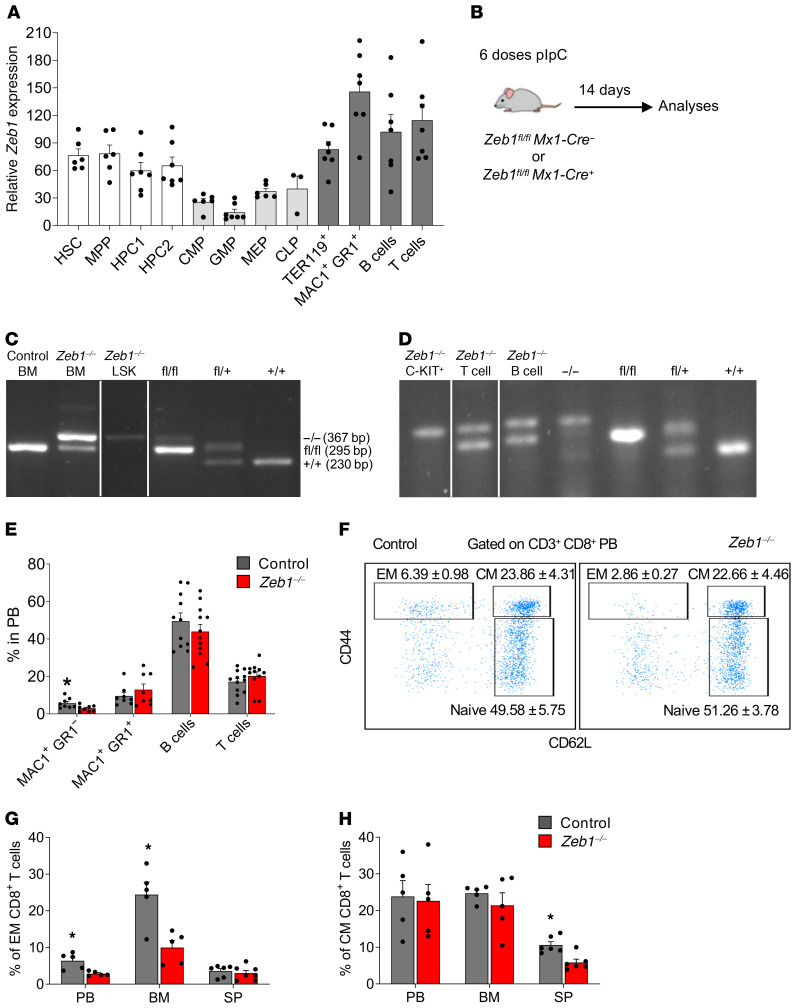
Zeb1 expression has been observed in hematopoietic cells from BM, thymus, spleen, fetal liver, and lymph nodes (12, 24). However, the Zeb1 expression pattern in different subsets of hematopoietic cells, including hematopoietic stem and progenitor cells (HSPCs), remains unclear. We therefore conducted quantitative PCR (Q-PCR) analysis of Zeb1 expression in hematopoietic cell compartments prospectively isolated by FACS. Zeb1 was expressed at high levels in stem and progenitor cells (HSCs, multipotent progenitor [MPP], hematopoietic progenitor cell 1 [HPC1], and HPC2) and in terminally differentiated cells (myeloid, erythroid, and B and T lineages) whereas it was lower in committed myeloid and lymphoid progenitors (common myeloid progenitors [CMP], granulocyte-monocyte progenitors [GMP], megakaryocyte-erythroid progenitors [MEP], common lymphoid progenitors [CLP]) (Figure 1A).
Figure 1. Loss of Zeb1 affects effector and CM CD8+ T cells.
(A) Q-PCR analysis of mRNA Zeb1 expression in different hematopoietic populations (n = 6–7 except CLP n = 3). (B) Schematic of pIpC treatment to delete Zeb1 in Zeb1fl/fl;Mx1-Cre– (control) and Zeb1fl/fl;Mx1-Cre+ (Zeb1–/–) mice and analysis at day 14 after the last pIpC dose. (C) Representative gel electrophoresis analysis confirming Zeb1 deletion in BM cells and LSK population 14 days after the last dose of pIpC. (D) Representative gel electrophoresis analysis of Zeb1 deletion in BM C-KIT+ cells and spleen B (B220+) and T (CD3+) cells 14 days after the last dose of pIpC. (E) Frequency of differentiated cells in PB from control and Zeb1–/– mice 14 days after the last dose of pIpC from 4 independent experiments (n = 8–12 per group). (F) Gating strategy of naive, EM, and CM T cells using CD62L and CD44 markers along with T cell markers CD3, CD4, and CD8 in PB. Frequency of EM T cells (G) and CM T cells (H) within CD3+CD8+ T cells in PB, BM, and SP from control (n = 5 PB and BM, 6 SP) and Zeb1–/– (n = 5 PB and BM, 6–7 SP) mice from 2 independent experiments. Error bars show mean ± SEM. Mann-Whitney U test was used to calculate significance. *P < 0.05.
To evaluate the genetic requirement for Zeb1 in adult HSCs, their progenitors, and fully differentiated blood and immune cells, we bred mice harboring conditional alleles of Zeb1 (Zeb1fl/fl mice) (23) with Mx1-Cre (25) to obtain either Zeb1fl/fl;Mx1-Cre+/– or control (Zeb1fl/fl;Mx1-Cre–/–) mice and administered pIpC on alternate days for 10 days to achieve deletion of Zeb1 (Zeb1–/–). Hematopoiesis in control or Zeb1–/– mice was analyzed 14 days after the last dose of pIpC (Figure 1B). Zeb1 was partially deleted in total BM cells (Figure 1C). To assess whether Zeb1 was completely deleted in HSCs from BM, we prospectively isolated Lin– SCA-1+C-KIT+ (LSK) cells (which contain HSCs) and, by genomic PCR, observed complete deletion of Zeb1 (Figure 1C). Similarly, C-KIT+ cells, which constitute both HSCs and committed myeloid and lymphoid progenitors, were fully deleted for Zeb1 (Figure 1D). In contrast, only partial deletion of Zeb1 was observed in terminally differentiated T and B cells isolated from the spleen (Figure 1D), suggesting that incomplete deletion observed in BM cells may be ascribed to these cell types.
At 14 days after ablation of Zeb1, no significant changes were observed in BM and spleen cellularity or spleen size (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI129115DS1). Immunophenotyping of peripheral blood (PB) revealed a significant reduction in the proportion of MAC1+GR1– monocytic cells in Zeb1–/– mice, while no significant changes were observed in MAC1+GR1+ cells, which contain granulocytes, or in T cells and B cells in PB or BM (Figure 1E and Supplemental Figure 1B) (26, 27). Intriguingly, despite incomplete deletion of Zeb1 in lymphoid cells from Zeb1–/– mice, we observed a selective reduction in CD8+ effector memory (CD8+ EM) (CD44hiCD62L–) T cells in PB and BM of Zeb1–/– mice (Figure 1, F and G). We also found a reduction in CD8+ central memory (CD8+ CM) (CD44hiCD62Lhi) T cells in spleen of Zeb1–/– mice, collectively demonstrating a critical role of Zeb1 in CD8+ T cell function (Figure 1H).
Acute loss of Zeb1 results in a cell-survival defect during thymocyte differentiation and a cell-autonomous T cell differentiation defect.
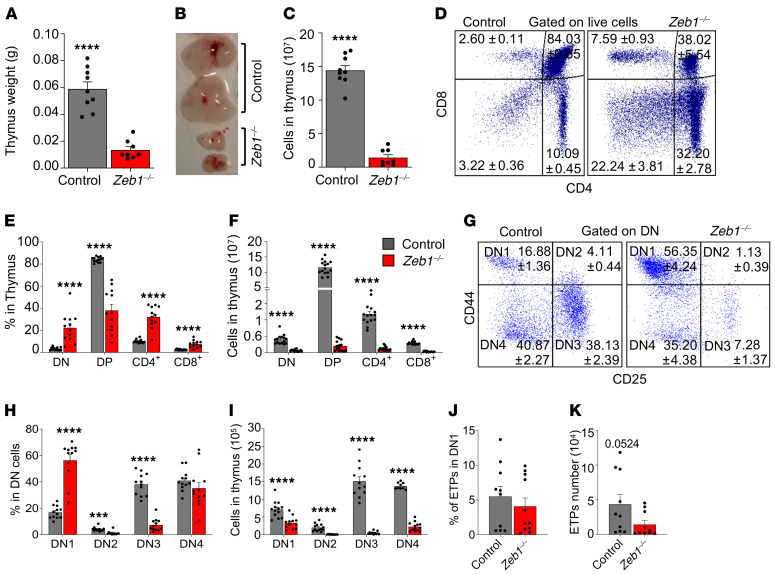
Having shown a defect in CD8+ T cells in Zeb1–/– mice and given that germline KO of Zeb1 results in a developmental defect in the T cell lineage (12, 13), we opted to assess T cell development in the thymus of adult Zeb1–/– mice. Fourteen days after the last dose of pIpC, Zeb1–/– mice displayed diminutive thymi coupled with a dramatic reduction in cellularity (Figure 2, A–C). Immunophenotypic analysis of T cell subsets in the thymus revealed an increased frequency of immature double-negative (DN) CD4–CD8– cells and mature single-positive (SP) CD4 (CD4+) and SP CD8 (CD8+) T cells, contrasting with a significant reduction in the proportion of double positive (DP) CD4+CD8+ cells in Zeb1–/– mice (Figure 2, D and E). Normalizing for reduced thymic cellularity in Zeb1–/– mice led to a significant reduction in total cell numbers observed in DN, DP, CD4+, and CD8+ cells from Zeb1–/– mice (Figure 2F). This correlated with increased apoptosis in DP, CD4+, and CD8+ populations, but surprisingly, not in DN cells from Zeb1–/– mice (Supplemental Figure 1C).
Figure 2. Loss of Zeb1 results in T cell reduction in thymus associated with early differentiation defects in thymus.
Thymus weight (A), representative photograph (B), and total thymus cellularity (C) from control (n = 9) and Zeb1–/– (n = 8) mice from 5 independent experiments at day 14 after the last pIpC dose. (D) Representative FACS plots of T cell analysis in thymus based on CD4 and CD8 cell-surface markers (DN: CD4–CD8–, DP: CD4+CD8+, CD4+, CD8+). (E) Frequency of T cell subsets in thymus from control (n = 13) and Zeb1–/– (n = 12) mice from 6 independent experiments at day 14 after the last pIpC dose. (F) Total cell count of T cell subsets in thymus from control (n = 14–15) and Zeb1–/– (n = 14–15) mice from 7 independent experiments at day 14 after the last pIpC dose. (G) Representative FACS plots showing gating strategy of early stages within CD4+CD8+ DN population using CD25 and CD44 (DN1: CD44+CD25–, DN2: CD44+CD25+, DN3: CD44–CD25+, DN4: CD44–CD25–) between control and Zeb1–/– at day 14 after the last pIpC dose. (H) Frequency of DN populations (DN1, DN2, DN3, DN4) in DN cells from control (n = 12) and Zeb1–/– (n = 12) mice from 5 independent experiments at day 14 after the last pIpC dose. (I) Total cell count of DN populations in thymus from control (n = 9-13) and Zeb1–/– (n = 11–13) mice from 4 independent experiments at day 14 after the last pIpC dose. (J) Frequency and (K) total count of ETPs (DN1 cKithi) from control (n = 10) and Zeb1–/– (n = 10–11) mice from 4 independent experiments at day 14 after last pIpC dose. Error bars show mean ± SEM. Mann-Whitney U test was used to calculate significance. ***P < 0.001; ****P < 0.0001.
Given that the earliest stages of T cell development were affected by Zeb1 ablation, we further evaluated the DN cell compartment, which represents the initial stage of thymocyte selection (28, 29). Using CD44 and CD25, the DN population can be subdivided chronologically into 4 populations: DN1 (CD44+CD25–), DN2 (CD44+CD25+), DN3 (CD44–CD25+), and DN4 (CD44–CD25–) (29), before they become DP cells and ultimately CD4+ or CD8+ mature cells (29). Immunophenotypic analysis showed increased frequency of DN1 cells and a reduction in DN2 and DN3, while no change was observed in the frequency of DN4 in Zeb1–/– mice, suggestive of a Zeb1-mediated differentiation block in the transition between DN1 and DN2/DN3 (Figure 2, G and H). Analysis of apoptosis in DN subsets revealed increased apoptotic levels in DN2 and DN3 after Zeb1 deletion, accounting for the differentiation block observed in these compartments, whereas DN1 and DN4 displayed comparable levels of apoptosis between genotypes (Supplemental Figure 1D). When the absolute number of these populations was quantified to account for reduced thymic cellularity of Zeb1–/– mice, a significant decrease was found across all DN populations following Zeb1 deletion (Figure 2I).
Next, we evaluated whether the defect observed within immature DN cells was caused by a failure of thymocyte survival preceding selection. We therefore assessed early thymic progenitors (ETPs), characterized as CD4–CD8–CD44+CD25–C-KIThi (30–33). Fourteen days after the last dose of pIpC, we found a comparable frequency of ETPs between control and Zeb1–/– mice (Figure 2J) with the absolute count of ETPs in Zeb1–/– thymus showing a near significant reduction compared with control due to reduced thymic cellularity in Zeb1–/– mice (Figure 2K).
Since the Mx1-Cre system deletes genes in nonhematopoietic tissues, such as BM niche cells (25, 34), we assessed whether Zeb1-mediated regulation of T cell development was cell autonomous by competitively transplanting CD45.2+ BM cells from Zeb1fl/fl;Mx1-Cre+ or control mice, admixed with unfractionated CD45.1+ BM cells (Supplemental Figure 2A). Six weeks after transplantation, Zeb1 deletion was induced by injection of pIpC, and 14 days after the last pIpC injection, analysis of donor engraftment in the thymus revealed a dramatic reduction in the Zeb1–/– genotype compared with control (Supplemental Figure 2B). Consistent with this, a substantial attenuation in donor contribution to DN, DP, CD4+, and CD8+ cell populations was detected in the Zeb1–/– genotype (Supplemental Figure 2C). With the exception of ETPs, a significant reduction in donor contribution was found across nearly all DN populations following cell-autonomous Zeb1 deletion, confirming that Zeb1 mediates T cell maturation in a cell-autonomous manner (Supplemental Figure 2D). Since mature T cell frequency did not change 14 days after Zeb1 loss during steady state due to incomplete Zeb1 deletion (Figure 1D), we also analyzed the donor contribution to peripheral T cells 14 days after Zeb1 ablation in a cell-autonomous manner, which revealed complete deletion of Zeb1 and a significant reduction in mature T cells in PB, BM, and spleen (Supplemental Figure 2, E and F). Further, we confirmed that Zeb1 mediates cell-autonomous reduction in EM CD8+ T cells in PB (Supplemental Figure 2G). Together, these data suggest that Zeb1 is critical for cell survival at the earliest stages of thymocyte differentiation as well as for T cell maturation and maintenance in the thymus. Thus, Zeb1 is required for cell-autonomous T cell development in the thymus.
Acute conditional deletion of Zeb1 results in a reduction of lymphoid lineage commitment in BM.
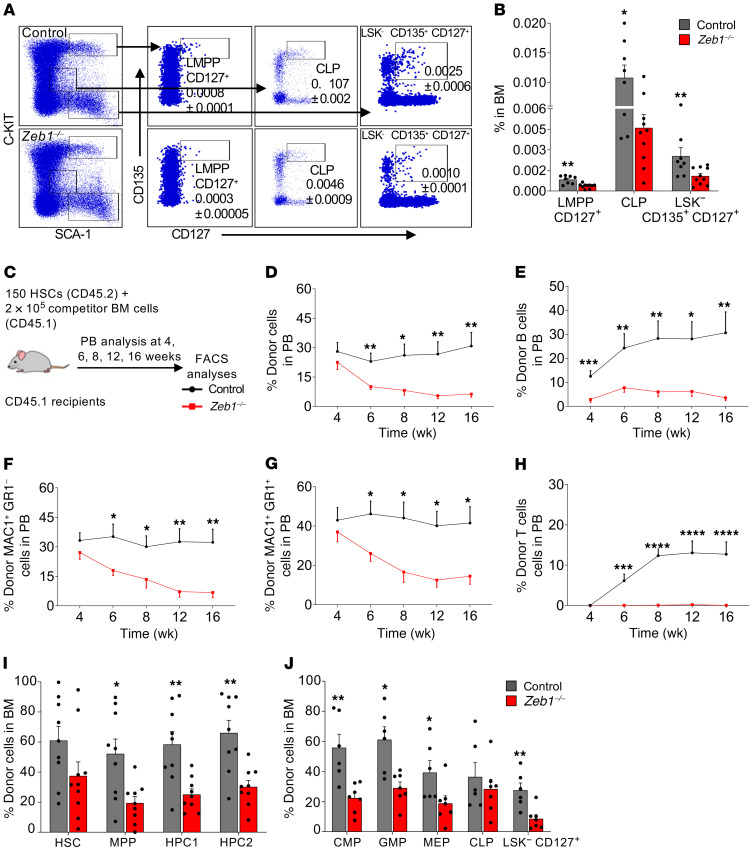
We next gauged the impact of Zeb1 on early T lymphoid lineage commitment in the BM. LSK CD135hiCD127hi lympho-myeloid MPPs (LMPPs CD127+, nonconventional LMPP) rapidly and efficiently generate T and innate lymphoid cells (35) compared with conventional LMPP (LSK CD34+CD135hi) (36) or HPC1 (LSK CD150–CD48+) that overlap functionally with conventional LMPP by 80% (37–39). Interestingly, we found a significant reduction in the proportion of T cell lineage–primed LMPP CD127+, but not conventional LMPP, which showed a statistically insignificant trend toward reduction after acute Zeb1 ablation (Figure 3, A and B, and Supplemental Figure 3A). We assessed other BM lymphoid progenitor compartments, including CLP (LIN–SCA-1loC-KITloCD127hiCD135hi) (35, 40) and LIN– SCA-1+C-KIT– (LSK–CD135+CD127+) (41) that were reduced in Zeb1–/– mice (Figure 3, A and B). Together, these data suggest that Zeb1 acts as a critical modulator of incipient lymphoid progenitor commitment from HSCs.
Figure 3. Loss of Zeb1 results in a reduction of lymphoid progenitors in BM and a multilineage hematopoietic differentiation defect after HSC transplantation.
(A) Representative FACS plots of the analysis of LMPP CD127+ (nonconventional LMPP: LSK CD135+CD127+), CLP (LIN– SCA-1loC-KITloCD135+CD127+), and LSK–CD127+CD135+ 14 days after the last dose of pIpC. (B) Frequency of LMPP CD127+, CLP, and LSK–CD 127+CD135+ in the BM from control (n = 8) and Zeb1–/– (n = 10) mice from 4 independent experiments at day 14 after the last pIpC dose. (C) Schematic of competitive HSC transplantation. 150 HSCs from control or Zeb1–/– mice (donor CD45.2) mixed with 2 × 105 BM competitor cells (CD45.1) were transplanted into lethally irradiated recipients (CD45.1), and the mice were monitored by bleeding the tail vein at different time points until week 16. (D) Percentage of donor cells in PB at different time points after HSC transplantation from control (n = 10) and Zeb1–/– (n = 10) mice from 3 independent experiments. Analysis of PB donor contribution to B cells (B220+) (E), MAC1+GR1– myeloid cells (F), MAC1+GR1+ myeloid cells (G), and T cells (CD4+CD8+) (H) from control (n = 10) and Zeb1–/– (n = 8–10) mice from 3 independent experiments. Donor contribution to BM HSPCs (I) (HSC: LSK CD150+CD48–, MPP: LSK CD150–CD48–, HPC1: LSK CD150–CD48+, HPC2: LSK CD150+CD48+) from control (n = 9) and Zeb1–/– (n = 9) from 3 independent experiments and BM committed progenitors (J) (CMP: LK CD34+CD16/32–, GMP: CD34+CD16/32+, MEP: CD34–CD16/32–, CLP: LIN– SCA-1loC-KITloCD127+, and LSK–CD127+ from control (n = 6) and Zeb1–/– (n = 7) from 2 independent experiments. Error bars show mean ± SEM. Mann-Whitney U test was used to calculate significance. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Acute conditional deletion of Zeb1 in HSCs results in a profound self-renewal and multilineage hematopoietic differentiation defect.
To directly assess the impact of acute deletion of Zeb1 in HSCs, we performed flow cytometry analysis on immunophenotypically defined HSCs and all MPP populations from control or Zeb1–/– mice. The frequency of HSCs (LSK CD150+CD48–) and MPPs (LSK CD150–CD48–) was comparable between control and Zeb1–/– genotypes (Supplemental Figure 3B). HPC1 (LSK CD150–CD48+) showed a nonsignificant reduction after Zeb1 deletion similar to that observed in conventional LMPP (LSK CD34+CD135+) (Supplemental Figure 3, A and B). Zeb1–/– HPC2 (LSK CD150+CD48+), which possesses both myeloid and lymphoid potential (38), showed a significant reduction in the frequency in total BM compared with control (Supplemental Figure 3B). These data demonstrate that Zeb1 regulates the abundance of select MPP populations.
Having observed a reduction of MAC1+GR1– myeloid cells in PB following acute deletion of Zeb1 in the hematopoietic system, we asked whether this was due to defects in committed myeloid progenitors from BM Zeb1–/–. No significant difference in CMP (LK CD34+CD16/32–), GMP (LK CD34+CD16/32+), and MEP (LK CD34–CD16/32–) populations was noted between control and Zeb1–/– mice (Supplemental Figure 3C). Thus, Zeb1-mediated regulation of terminal MAC1+GR1– myeloid cell differentiation appears to be independent of committed myeloid progenitor maturation from BM.
To stringently test the functionality and differentiation capacity of HSCs from Zeb1–/– mice, we prospectively isolated 150 HSCs (CD45.2) from control or Zeb1–/– mice at 14 days following deletion, mixed them with 2 × 105 BM competitor cells (CD45.1), and transplanted this cell preparation into lethally irradiated recipients (CD45.1) (Figure 3C). The engraftment capacity of transplant recipients in PB was monitored until week 16 (Figure 3C). Significant engraftment failure was observed by week 6 and continued to decrease progressively until week 16 (Figure 3D). To test the donor contribution to PB of specific hematopoietic lineages, we analyzed PB for CD45.2 (donor) and CD45.1 (competitor) in conjunction with MAC1+GR1– myeloid, MAC1+GR1+ myeloid, B220+ B cells, and CD4+CD8+ T cells. A profound reduction in donor contribution to B cells (Figure 3E), MAC1+GR1– myeloid cells (Figure 3F), and MAC1+GR1+ myeloid cells (Figure 3G) was observed in recipients of Zeb1–/– HSCs. No engrafted T cells were derived from recipients transplanted with Zeb1–/– HSCs (Figure 3H).
Having observed multilineage defects in terminally differentiated blood cells in recipients of Zeb1–/– HSCs, we asked whether these defects originated in parental HSPCs or lineage-committed progenitors. Within LSK compartments, the donor contribution to HSC was equal between recipients of control or Zeb1–/– HSCs (Figure 3I). However, there was a significant reduction in the donor contribution to MPP, HPC1, and HPC2 in Zeb1–/– compared with control (Figure 3I). We also analyzed committed progenitors downstream of HSPCs and found a dramatic reduction in donor contribution to CMP, GMP, MEP, and LSK–CD127+, but no change was observed in donor contribution to CLP (Figure 3J). These data directly link the functional defects observed after transplantation of Zeb1-deficient HSCs to alterations in specific HSPC and lineage committed progenitor compartments.
An integral part of successful engraftment after BM transplantation is homing of i.v. infused HSPCs to the BM niche, the main home of adult hematopoiesis. To assess whether the Zeb1–/– engraftment defect was due to abnormal homing of Zeb1–/– HSPCs to the BM, we transplanted 7 × 106 total BM cells (CD45.2) from control or Zeb1–/– mice into lethally irradiated recipients (CD45.1) and analyzed donor cell presence in recipients at 18 hours after transplantation (Supplemental Figure 4A). Relative parity was observed in the homing capacity of total BM cells or LSK populations in the 2 genotypes (Supplemental Figure 4B). Similarly, homing of donor cells to the spleen and thymus was comparable between control and Zeb1–/– genotypes (Supplemental Figure 4C). Thus, acute deletion of Zeb1 does not impact the homing ability of hematopoietic cells in vivo.
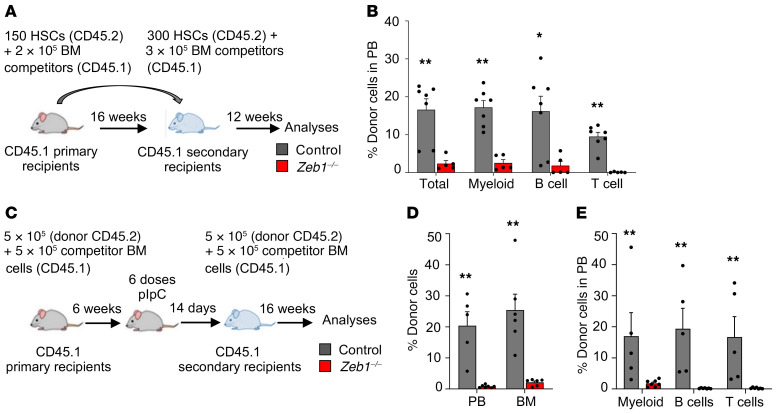
To directly test the impact of Zeb1 deletion on the self-renewal capacity of HSCs, we performed secondary transplantation of Zeb1–/– HSCs. We sorted 300 HSCs (CD45.2) from control or Zeb1–/– primary recipients and admixed them with 3 × 105 competitor BM cells before transplanting them into lethally irradiated recipients. We observed a strong defect in PB engraftment associated with multilineage hematopoietic impairment in secondary transplant recipients by week 12 (Figure 4, A and B), indicative of a self-renewal defect in Zeb1–/– HSCs.
Figure 4. Zeb1 regulates HSC self-renewal and differentiation in a cell-autonomous manner.
(A) Schematic of secondary HSC transplantation. 300 CD45.2+ HSCs from primary recipients from control or Zeb1–/– mice mixed with 3 × 105 BM competitor cells (CD45.1) were transplanted into lethally irradiated recipients (CD45.1), and the mice were analyzed at week 12. (B) Percentage of donor cells in PB and donor contribution to myeloid (MAC1+), B (B220+), and T (CD4+/CD8+) cells at week 12 after secondary HSC transplantation from control (n = 7) and Zeb1–/– (n = 5) from 2 independent experiments. (C) Schematic of the secondary total BM transplantation in cell-autonomous manner. 5 × 105 CD45.2+ BM cells from primary recipients 14 days after the last pIpC dose from control or Zeb1–/– mice mixed with 5 × 105 BM competitor cells (CD45.1) were transplanted into lethally irradiated recipients (CD45.1), and the mice were analyzed at week 16. (D) Percentage of donor cells in PB and BM at week 16 after secondary cell autonomous total BM transplantation from control (PB n = 5, BM = 6) and Zeb1–/– (PB n = 7, BM = 6) mice from 1 experiment. (E) Donor contribution to PB MAC1+ myeloid cells, B220+ B cells, and CD4+/CD8+ T cells at week 16 after secondary cell-autonomous total BM transplantation from control (n = 5) and Zeb1–/– (n = 7) mice from 1 experiment. Error bars show mean ± SEM. Mann-Whitney U test was used to calculate significance. *P < 0.05; **P < 0.01.
Zeb1 is required for cell-autonomous HSC function.
To assess whether the acute requirement for Zeb1 in maintaining HSC function was cell autonomous, we performed a competitive BM transplantation by transplanting 5 × 105 BM cells from control and Zeb1fl/fl;Mx1-Cre+ (CD45.2) mice admixed with equal number of WT competitor cells (CD45.1) into lethally irradiated recipients (CD45.1). Six weeks later, Zeb1 deletion was induced by administering recipients with pIpC,and 14 days after the last dose of pIpC, mice were sacrificed and 5 × 105 donor BM cells (CD45.2) from primary recipients (14 days after Zeb1 ablation) mixed with 5 × 105 competitor BM cells (CD45.1) were transplanted into lethally irradiated recipients (Figure 4C). Sixteen weeks after transplantation, we found a dramatic reduction in donor engraftment in PB and BM (Figure 4D). Further analysis of donor contribution to PB lineages revealed a marked reduction in myeloid cells and near loss of B and T cells (Figure 4E).
To determine whether the Zeb1–/– BM microenvironment plays a role in Zeb1-mediated HSC regulation, we transplanted 1 × 106 WT BM cells (CD45.1) into lethally irradiated control or Zeb1fl/fl;Mx1-Cre+ (CD45.2) mice. Six weeks later, we injected the recipients with pIpC to delete Zeb1 and analyzed the mice at week 16 after the last dose of pIpC (Supplemental Figure 4D). PB analyses of myeloid cells (MAC1+), B cells (B220+), and T cells (CD4+/CD8+) showed no significant difference between control and Zeb1–/– (Supplemental Figure 4E). Next, we asked whether the altered BM niche would affect the composition of HSPC and committed progenitors. The data showed that the frequency of these populations was comparable between control and Zeb1–/– (Supplemental Figure 4, F and G). Together, these data demonstrate that Zeb1 is required for cell-autonomous HSC functionality.
Zeb1–/– HSCs display deregulated cell polarity and hematopoietic differentiation transcriptional signatures.
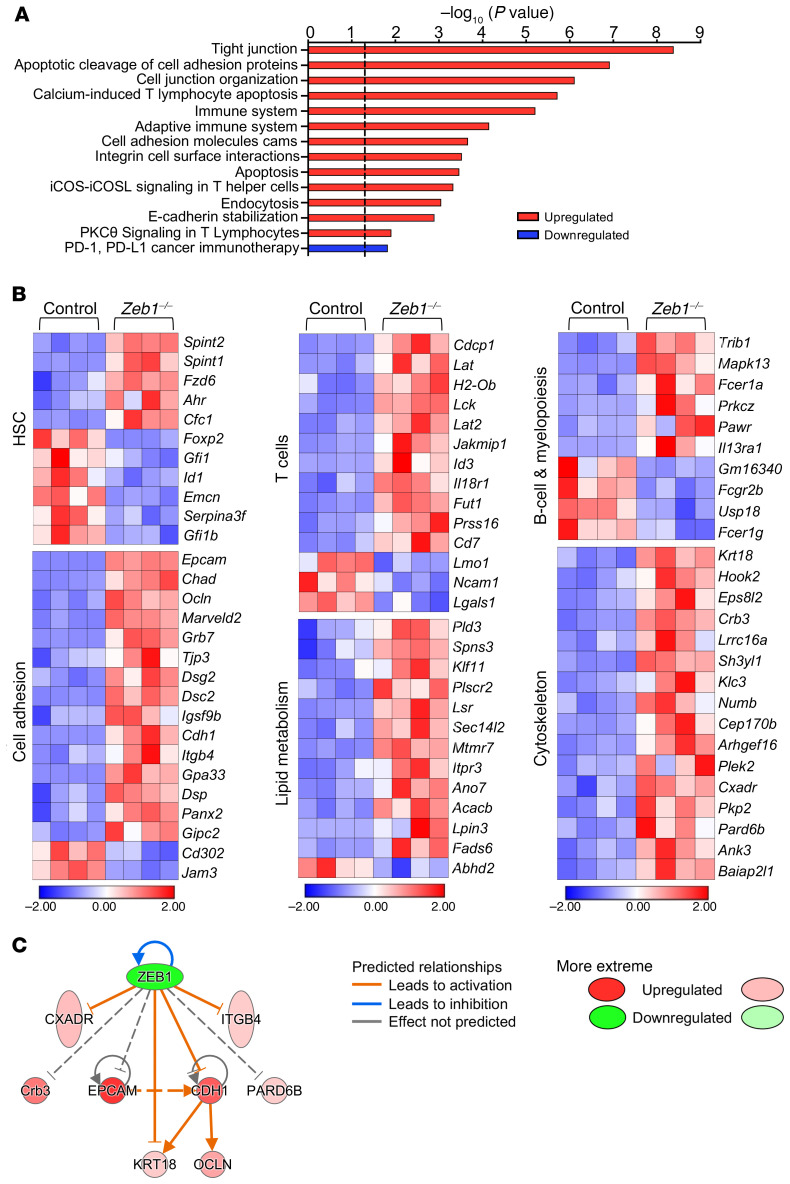
In order to understand the transcriptional signature underpinning Zeb1-mediated regulation in HSCs, we performed RNA-Seq on purified HSCs (LSK CD150+CD48–) from control or Zeb1–/– mice 14 days after the last dose of pIpC. Of 222 differentially expressed genes (DEG), 47 genes (21%) were downregulated and 175 upregulated (79%) from Zeb1–/– HSCs. These data are largely consistent with the notion that Zeb1 functions as a transcriptional repressor (42). Biological pathway analysis confirmed that the most enriched pathways were upregulated and included tight junction, cell adhesion, cell junction organization, immune system, and endocytosis pathways (Figure 5A). Zeb1 appears to regulate a transcriptional signature related to cell polarity, consisting of genes related to cytoskeleton, cell adhesion, and lipid metabolism/lipid membrane biology (Figure 5B), congruent with the idea that Zeb1 acts as a potent inducer of the EMT process, involving Zeb1-mediated repression of cell polarity genes (43–45). Using Ingenuity Pathway Analysis software (IPA), we generated a gene-interaction network showing the direct regulation of Zeb1-specific target genes related to cell polarity, cytoskeleton, and cell adhesion that included a regulatory node involving epithelial cell adhesion molecule (EpCAM), CRB3, PARD6b, ITGB4, CDH1, KRT18, and OCLN (10, 43–50) (Figure 5C). In agreement with this transcriptional network, EpCAM, CDH1, and ITGB4 upregulation in Zeb1–/– HSPCs was confirmed at the protein level by flow cytometry (Figure 6, A and B, and Supplemental Figure 5, A and B). Reflecting the global, multilineage differentiation functional defects of Zeb1–/– HSCs, we observed a broad, robust pattern of deregulated HSC maintenance and hematopoietic lineage affiliated from both myeloid and lymphoid lineages (Figure 5B).
Figure 5. Zeb1–/– HSCs display deregulation of hematopoietic function and cell polarity transcriptional programming.
RNA-Seq was performed in sorted control and Zeb1–/– HSCs (LSK CD150+CD48–) 14 days after last pIpC dose (n = 4 for each genotype). (A) Biological pathway analysis shows the top enriched pathways in Zeb1–/– HSCs compared with control. Data are shown as –log10 (P value), and the dashed black line indicates P value of 0.05. (B) Heatmaps of the DEGs after Zeb1 deletion related to HSC function, T cells, and B cells as well as cytoskeleton, lipid metabolism, and cell adhesion. Heatmap scale represents z score. (C) A network of Zeb1 interaction with several target genes related to polarity, cytoskeleton, and cell adhesion using IPA software. Due to their confirmed binding to ZEB1 in the literature, Epcam, Pard6b, and Crb3 were added manually.
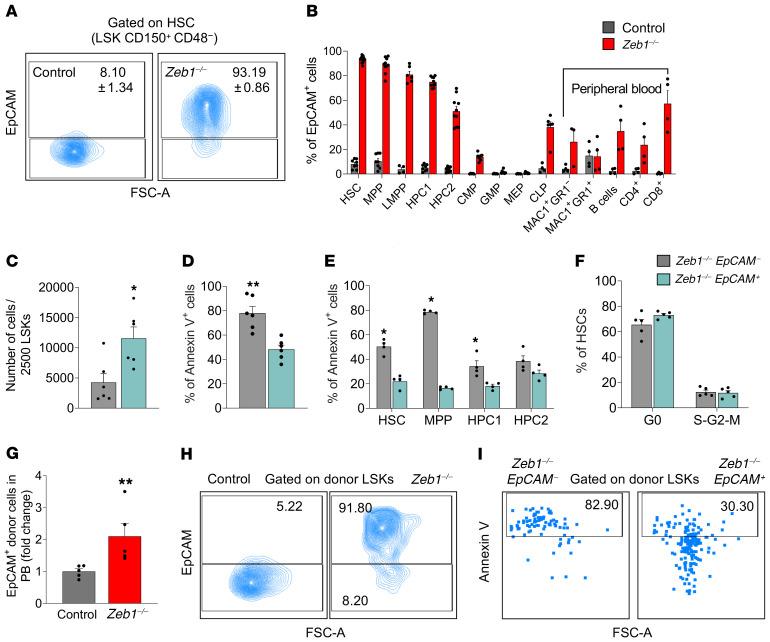
Figure 6. Increased EpCAM expression confers survival advantage and differentiation block in Zeb1–/– HSCs.
(A) Representative flow cytometry plots of EpCAM expression in HSCs 14 days after pIpC injection. (B) Analysis of EpCAM expression in BM subpopulations and PB mature cells 14 days after pIpC injection from control (n = 8 for HSC, MPP, HPC1, and HPC2; n = 4 for LMPP, CLP and mature PB populations; n = 5 for CMP, GMP, and MEP) and Zeb1–/– (n = 10 for HSC, MPP, HPC1, and HPC2; n = 6 for LMPP and CLP; n= 4 for mature PB populations except MAC1+GR1– n = 3; n = 7 for CMP, GMP, and MEP). (C) Cell number after culturing 2500 LSKs from Zeb1–/– EpCAM– (n = 6) and Zeb1–/– EpCAM+ (n = 6) from 3 independent experiments. (D) Analysis of apoptosis in LSKs after culture from Zeb1–/– EpCAM– (n = 6) and Zeb1–/– EpCAM+ (n = 6) from 3 independent experiments. (E) Analysis of apoptosis in fresh BM HSPCs 14 days after pIpC injection from Zeb1–/– EpCAM– (n = 4) and Zeb1–/– EpCAM+ (n = 4) from 2 independent experiments. (F) Cell cycle analysis of HSCs using Ki67 and DAPI 14 days after pIpC injection from Zeb1–/– EpCAM– (n = 5) and Zeb1–/– EpCAM+ (n = 5) from 1 experiment. (G) Analysis of EpCAM expression in donor PB at week 16 after primary HSC transplantation from control (n = 5) and Zeb1–/– (n = 5) from 1 experiment represented as fold change. (H) Representative FACS plots of the analysis of EpCAM expression in LSKs 16 weeks after primary HSC transplantation from control (n = 2) and Zeb1–/– (n = 1) from 1 experiment. (I) Representative FACS plots of the analysis of apoptosis using annexin V in EpCAM-negative and -positive fractions within donor LSKs 16 weeks after primary HSC transplantation from control (n = 2) and Zeb1–/– (n = 1) from 1 experiment. Error bars show mean ± SEM. Mann-Whitney U test was used to calculate significance. *P < 0.05; **P < 0.01.
With prominent transcriptional deregulation of T cell pathways being observed in Zeb1–/– HSCs (Figure 5A), we next asked whether the defects in Zeb1–/– HSCs could be associated with those observed in Zeb1–/– T cells. To address this question at the transcriptional level, we conducted RNA-Seq from Zeb1–/– or control CLPs, which have T lymphoid but not myeloid potential, and compared their transcriptional signatures with that from Zeb1–/– HSCs. Zeb1–/– CLPs displayed gene expression pathways comparable to those of Zeb1–/– HSCs, including deregulated cell-cell junction, tight junction, cell adhesion, cytoskeleton, and T cell pathways (Supplemental Figure 5, C–E). Remarkably, of the 47 DEGs in Zeb1–/– CLPs, 27 genes (57%) were also differentially expressed in Zeb1–/– HSCs (Supplemental Figure 5F). Other biological pathways reflecting T cell function were deregulated in Zeb1–/– HSCs but not Zeb1–/– CLPs (e.g., calcium-induced T lymphocyte apoptosis, iCOS-iCOSL signaling in T helper cells) (Figure 5A). Transcriptional signatures relating to the CtBP1 pathway (51, 52) were observed only in Zeb1–/– CLPs (Supplemental Figure 5C). However, the majority of transcriptional programing mediating the differentiation defect of Zeb1–/– T cells was instigated in HSCs and transmitted to CLPs. Overall, in its capacity as a transcriptional repressor, Zeb1 acts as a potent regulator of cell polarity and differentiation-affiliated transcriptional signatures in HSCs.
Increased EpCAM expression confers a cell-survival advantage in Zeb1–/– HSCs that alters self-renewal and differentiation fates.
EpCAM, a glycoprotein mediating cell adhesion in epithelia (53), was the most highly upregulated gene in Zeb1–/– HSCs (Figure 5B). In other types of stem cells, EpCAM has been established as a crucial regulator of stem cell maintenance and differentiation (46). We therefore elected to examine the impact of EpCAM expression in the context of Zeb1-mediated regulation of HSC fate. We first confirmed enhanced expression of EpCAM at the protein level by flow cytometry of Zeb1–/– HSC, MPP, HPC1, and HPC2 populations (Figure 6, A and B). While EpCAM expression was nearly extinguished during differentiation to CMP, GMP, and MEP committed progenitors, it was upregulated in terminally differentiated cells (myeloid, B and T lineages) from PB (Figure 6B). EpCAM-positive HSPCs from Zeb1–/– LSKs expanded more than their EpCAM-negative counterparts in vitro (Figure 6C), suggesting that EpCAM expression confers a cell-survival signal in Zeb1–/– HSCs. We directly addressed whether EpCAM expression mediates cell survival in freshly isolated Zeb1–/– HSCs and in vitro and observed reduced apoptosis in Zeb1–/– EpCAM+ HSCs and MPP subsets in both settings (Figure 6, D and E). Cell cycle status based on EpCAM expression was unperturbed in Zeb1–/– HSCs (Figure 6F). By evaluating the impact of EpCAM expression in Zeb1–/– HSC survival and differentiation in vivo, we showed at 16 weeks after HSC transplantation that Zeb1–/– cells in PB displayed a 2-fold increase in EpCAM expression compared with controls (Figure 6G). Thus, EpCAM expression in Zeb1–/– HSC correlates with the multilineage differentiation block observed during transplantation (Figure 3 and Figure 4). Yet high expression of EpCAM was preserved in Zeb1–/– HSPCs at 16 weeks after transplant and these EpCAM+ Zeb1–/– HSPCs had a lower propensity for apoptosis (Figure 6, H and I). Together, these data suggest that augmented EpCAM expression confers a cell survival advantage in Zeb1–/– HSCs that causes an imbalance between self-renewal and differentiation fates.
Zeb1–/– EpCAM+ HSPCs display enhanced cell survival and diminished mitochondrial metabolism, RNA biogenesis, and differentiation transcriptional signatures.
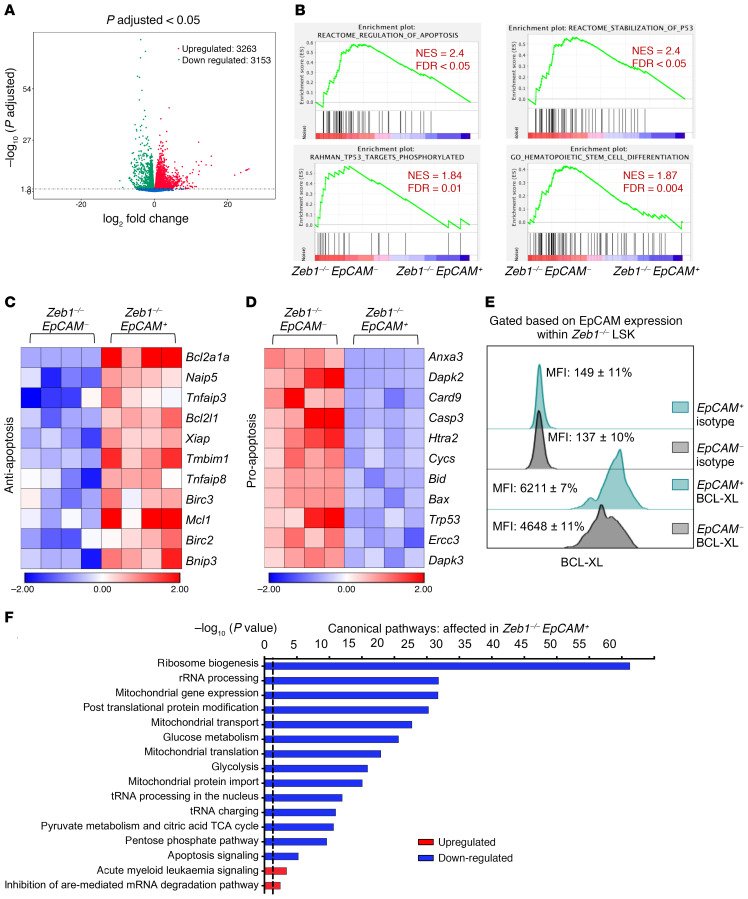
To evaluate the transcriptomic signature demarcating Zeb1–/– EpCAM+ HSPCs from Zeb1–/– EpCAM– HSPCs, we performed RNA-Seq on Zeb1–/– EpCAM+ LSK cells or Zeb1–/– EpCAM– LSK cells 14 days after the last dose of pIpC. In Zeb1–/– EpCAM+ HSPCs, 3263 genes were upregulated and 3153 genes were downregulated (Figure 7A). In agreement with enhanced cell survival and a functional block in differentiation associated with Zeb1–/– EpCAM+ HSCs (Figure 6), biological pathway analysis revealed a robust p53-mediated prosurvival signature and an antihematopoietic differentiation signature in Zeb1–/– EpCAM+ HSPCs (Figure 7, B–D). Furthermore, we observed augmented expression of antiapoptotic BCL-XL (54) in Zeb1–/– EpCAM+ HSPCs (Figure 7, C and E) and an EpCAM-p53-BCL-XL (BCL2L1) interacting gene network of apoptotic regulation in Zeb1–/– HSPCs (Supplemental Figure 6).
Figure 7. Zeb1–/– EpCAM+ HSPCs display enhanced cell survival and diminished mitochondrial metabolism, RNA biogenesis, and differentiation transcriptional signatures.
(A) Volcano plot showing the relationship between magnitude of gene expression change (log2 of fold-change; x axis) and statistical significance of this change (–log10 of adjusted P value; y axis) in a comparison of Zeb1–/– EpCAM+ to Zeb1–/– EpCAM– LSK cells. Colored points represent DEGs (cutoff FDR < 0.05) that are either overexpressed (red) or underexpressed (green) in Zeb1–/– EpCAM+ compared with Zeb1–/– EpCAM–. (B) GSEA plots of regulation of apoptosis, stabilization of P53, TP53 targets phosphorylated, and HSC differentiation. Heatmaps of the DEGs within EpCAM+ and EpCAM– LSK after Zeb1 deletion related to antiapoptosis (C) and proapoptosis (D). (E) Representative histogram of BCL-XL levels in EpCAM fractions within Zeb1–/– LSK. (F) Canonical pathways that were mostly enriched in Zeb1–/– EpCAM+ LSK cells derived from the IPA, BioCarta, KEGG, PID, and Reactome pathway databases. Data are shown as –log10 (P value), and the dashed black line indicates P value of 0.05. Analysis was performed using the GSEA software and IPA.
Mitochondria play crucial regulatory roles in fundamental cellular processes, such as apoptosis and bioenergetic provisions (55), and in the context of HSCs, act as a gatekeeper limiting HSC self-renewal ability (56). Zeb1–/– EpCAM+ HSPCs displayed reduced mitochondrial gene expression, transport, translation, and protein import as well as reduced associated mitochondrial metabolic pathways (e.g., pyruvate metabolism and TCA cycle) critical to HSC fate (57) (Figure 7F). Further highlighting the relatively low bioenergetic state of Zeb1–/– EpCAM+ HSPCs, ribosome biogenesis and ribosome-associated pathways, such as rRNA processing, whose reduction has previously been associated with conferral of stress resistance in preleukemic HPSCs (58), were similarly downregulated in Zeb1–/– EpCAM+ HSPCs (Figure 7F). Consistent with this, Zeb1–/– EpCAM+ HSPCs also displayed augmented AML signaling (Figure 7F). Therefore, in addition to control of cell survival, Zeb1-mediated repression of EpCAM appears to be critical in regulating mitochondrial metabolism and ribosomal pathways associated with normal HSC maintenance and prevention of preleukemic and leukemic signaling.
ZEB1 expression is downregulated in AML patients, and acute deletion of Zeb1 in leukemic stem cells enhances disease progression in MLL-AF9 and Meis1a/Hoxa9 driven AML.
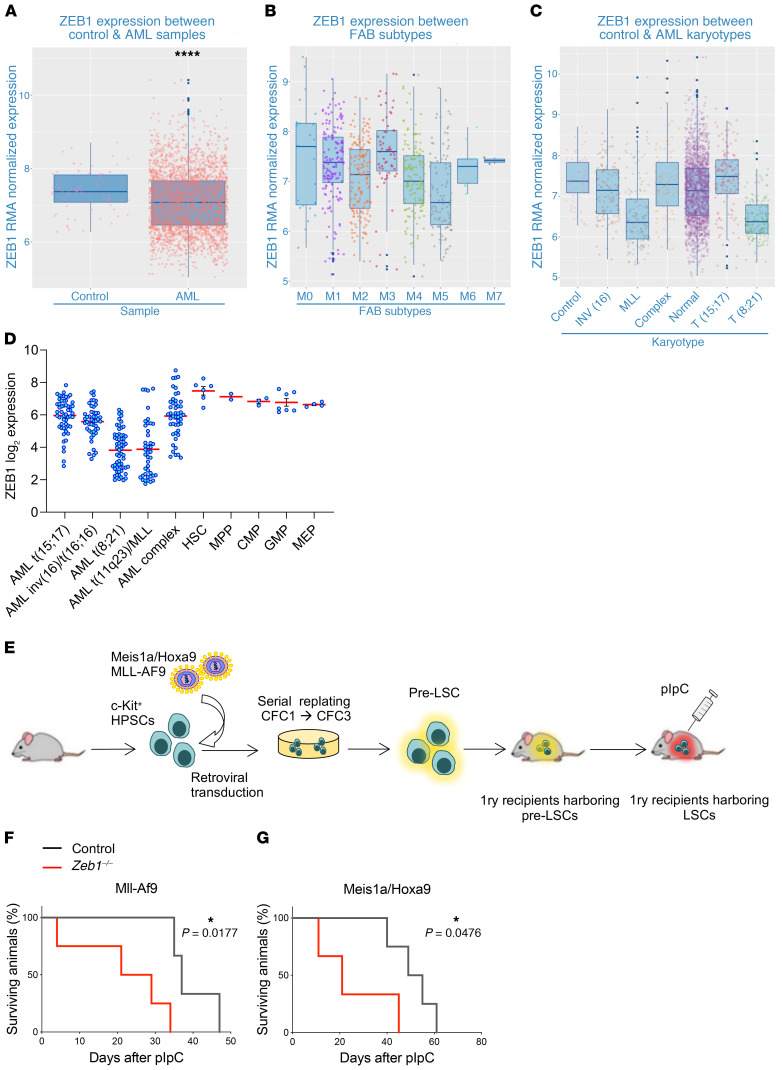
Subversion of HSC fates may cause hematologic neoplasia, including leukemia (59). Having found that Zeb1 deficiency leads to critical impairments in HSC self-renewal, apoptotic, and differentiation fates and because increased EpCAM expression in Zeb1-deficient HSCs enhanced AML signaling, we assessed the role of Zeb1 in AML. We initially evaluated ZEB1 expression in a large cohort of AML patients. AML (n = 2611) and control (n = 77) patient data sets were obtained from NCBI’s Gene Expression Omnibus (GEO) to assemble a case/control cohort hybridized to Affymetrix Human Genome U133 Plus 2.0 GeneChip array and analyzed through R using bioconductor packages, where data was normalized using Robust Multi-Array Average (RMA). We observed that ZEB1 expression was lower in AML patients compared with healthy controls (Figure 8A). Attenuated expression of ZEB1 was particularly prevalent in M4 and M5 FAB subtypes and also in AML patients with t(8;21) and MLL chromosomal translocations (60, 61) (Figure 8, B and C). Using an independent AML patient database, BloodSpot (http://www.bloodspot.eu), we validated lower ZEB1 expression in patients harboring these chromosomal translocations (Figure 8D).
Figure 8. Zeb1 is downregulated in AML patient samples and acts as a tumor suppressor in MLL-AF9 and Meis1a/Hoxa9-driven AML.
(A–C) ZEB1 RMA–normalized expression from combined published data sets in (A) control and AML, (B) across FAB subtypes and, (C) karyotypes. (D) ZEB1 log2 expression data in human HSPC and AML karyotypes. Data from BloodSpot. Error bars show mean ± SEM. Student’s t test was used unless otherwise indicated. ****P < 0.0001. (E) C-KIT+ cells from control and Zeb1fl/fl;Mx1-Cre+ mice were transduced with retroviruses expressing MLL-AF9 or Meis1a/Hoxa9 oncogenes and plated into CFC assays. After CFC3 (6 days each CFC), pre-LSCs (CD45.2+C-KIT+) were sorted and transplanted into lethally irradiated recipients together with CD45.1+ unfractionated BM cells. Three weeks later, mice were administered pIpC to induce gene deletion and monitored for AML development. (F and G) Kaplan-Meier survival curve of primary recipients transplanted with (F) MLL-AF9 (n = 4) or (G) Meis1a/Hoxa9 (n = 4) pre-LSCs. Mantel-Cox test. *P < 0.05.
While these data imply that ZEB1 acts as a tumor suppressor in AML, the functional requirement of Zeb1 in AML disease progression remains unknown. To directly assess this, we employed an assay in which leukemic transformation of murine C-KIT+ HSPCs is mediated by retroviral overexpression of either MLL-AF9 or Meis1a/Hoxa9 AML oncogenes (62–64). MLL-AF9– or Meis1a/Hoxa9-transduced cells were serially passaged for 3 rounds in colony-forming cell (CFC) assays to generate pre–leukemic stem cells (pre-LSCs), which, on i.v. injection into primary lethally irradiated mice, become LSCs — the malignant counterparts of HSCs that drive disease progression in AML (65) (Figure 8E). We cotransduced HSPCs from noninduced Zeb1fl/fl;Mx1-Cre+ or control mice with retroviruses expressing either MLL-AF9 or Meis1a/Hoxa9, collected pre-LSCs, and transplanted them into primary recipients alongside unfractionated BM support cells (Figure 8E). By flow cytometry, we assessed the PB of recipients for engrafting leukemic cells and induced Zeb1 deletion with pIpC after disease onset, when 20% engraftment of leukemic cells was observed in the PB (66) (data not shown). In both MLL-AF9 and Meis1a/Hoxa9 leukemic models, recipients of Zeb1-KO LSCs succumbed to AML with enhanced rapidity compared with recipients receiving control LSCs, indicating that Zeb1 deletion accelerates LSC-mediated disease progression (Figure 8, F and G). Thus, Zeb1 acts as a tumor suppressor in MLL-AF9 and Meis1a/Hoxa9 AML LSCs.
Discussion
Zeb1, in its capacity as a critical EMT regulator, controls myriad processes in embryonic development and, through the agency of tissue-specific stem cells, acts as a critical regulator of adult tissue homeostasis (18). Deregulation of Zeb1 activity has been implicated in multiple cancer types and, in these settings, Zeb1 acts as an instigator of the activity of cancer stem cells, a subset of cancer cells driving therapy resistance and metastasis, which ultimately cause fatality (42, 43). Understanding of the cellular and molecular mechanisms underpinning Zeb1-mediated regulation of stem cell self-renewal, lineage fate, and differentiation in normal and cancer stem cells remains incomplete. Here, in stem cells of the hematopoietic system, we find that acute conditional deletion of Zeb1 causes a profound cell-autonomous self-renewal defect and differentiation block across all lineages after transplantation and deregulates a transcriptional program associated with cell polarity. Strikingly, acute conditional deletion of Zeb1 in HSCs and their progeny affects the lineage fate and cell survival of T cells, leading to a rapid loss of thymocytes and CD8+ T cell subsets.
While it is known that Zeb1 and other TFs, such as zinc finger TF Gata3, are essential in T cell development (67, 68), the process in adults is less clear. Here, we identify Zeb1 as an indispensable regulator of transcriptional programming for the entire adult T cell repertoire, during initial T cell commitment from HSPC BM progenitor subsets through to cell survival during positive and negative selection in the thymus. In spite of incomplete gene deletion using the Mx-Cre system, we also identified Zeb1 as a regulator of CD8+ EM in BM and PB and CD8+CM T cells from spleen, supporting previous observations that Zeb1 is critical to CD8+ T cell function during infection (69). Our data are also consistent with the notion that the ZEB family member Zeb2 plays reciprocal roles in CD8+ T cell biology (69) and that it does not compensate for the absence of Zeb1. Further understanding Zeb1-mediated control of adult T cell differentiation may have implications for immunosurveillance, a naturally occurring immune mechanism involving CD8+ T cells and other immune subsets that eradicate tumor cells (70, 71). In particular, the complex interplay between the necessity for Zeb1 in immune cell subsets involved in immunosurveillance and the tumor microenvironment, where paradoxically, ZEB1 expression can drive metastasis by interfering with immune checkpoints, requires further exploration to negate possible toxic effects associated with targeting ZEB1 or ZEB1 target genes therapeutically in cancer. Nonetheless, the benefits of modulating the Zeb1 transcriptional/epigenetic network in cancer immunotherapy have been clearly illustrated in the blockade of CD47, a direct transcriptional target of Zeb1, that enhances phagocytosis of breast cancer cells undergoing EMT (72).
Zeb1–/– HSCs were functionally defective in their capacity to generate other blood lineages in transplantation, suggesting that Zeb1 modulates the ability of HSCs to differentiate correctly in vivo through repression of lineage-commitment–affiliated gene programs in HSCs. In keeping with the notion of lineage-specific transcriptional repression licensed by Zeb1, we observed upregulation of 79% of genes in Zeb1–/– HSCs together with a robust gene expression signature associated with deregulated multilineage differentiation. Relatively few transcriptional repressors, including Gfi1 and Gfi1b, have been shown to regulate HSC self-renewal and differentiation function (73). Gfi1–/– HSCs have a phenotype resembling that observed in Zeb1–/– HSCs (74, 75), and notably, both Gfi1 and Gfi1b were transcriptionally repressed in Zeb1–/– HSCs, suggesting positive regulation by ZEB1. Zeb1 therefore likely acts as a transcriptional repressor that regulates HSC self-renewal and global differentiation via a transcriptional repressor network that includes both Gfi1 and Gfi1b.
Zeb1 regulates HSC self-renewal and differentiation in association with a transcriptional program of cell polarity, which relates to the structural and cellular changes that occur to a cell, facilitating specialized function, such as cell division, adhesion, or migration (76). Several studies in Drosophila melanogaster male germline stem cells (77, 78) support the longstanding hypothesis that cell polarity acts as a critical mechanism that asserts control of symmetric versus asymmetric stem cell division and therefore stem cell fate, simply put, striking a balance between self-renewal and differentiation fates in tissue homeostasis and under conditions of physiologic stress (76, 79). Notably, Numb, a marker of asymmetric division in Drosophila melanogaster neuroblasts (80), and Crb3, which asymmetrically distributes polarity proteins in mouse preimplantation embryos (81), were deregulated in Zeb1–/– HSCs. Taken together with the observation that Zeb1–/– HSCs have decreased self-renewal potential, these data suggest that Zeb1–/– HSCs favor symmetric, differentiating divisions over asymmetric divisions. Other regulators of cell polarity, including genes associated with apical-basal polarity (82), such as tight junctions (Ocln, Marveld2, Tjp3), adherens junctions (Cdh1) and desmosomes (Krt8, Dsc2, Dsg2), were upregulated in Zeb1–/– HSCs. We provide evidence that derepression of EpCAM in Zeb1–/– HSCs correlates with enhanced HSC survival in homeostasis and transplantation. Yet enhanced HSC survival and relative upregulation of EpCAM in PB correlate with a block in engraftment in transplantation of Zeb1–/– HSCs, suggesting that enhanced survival of HSCs mediated by EpCAM disturbs the delicate balance of self-renewal versus differentiation fates.
EpCAM expression in Zeb1–/– HSCs reduced mitochondrial metabolism, which is also important for HSC self-renewal, cell survival,and differentiation fates (57). For example, reduced pyruvate metabolism was observed in EpCAM+ Zeb1–/– HSPCs, which is consistent with evidence that ablating aspects of pyruvate metabolism causes HSC exhaustion and a block in HSC differentiation (83, 84). Associated with a low metabolic state, EpCAM+ Zeb1–/– HSPCs also exhibited reduced ribosome biogenesis, which may reflect enhanced cell survival mediated by the combinatorial lack of p53 target gene phosphorylation, stabilization of p53, and reduced rRNA observed in EpCAM+ Zeb1–/– HSPCs (58, 85). In future work, it will be of interest to further evaluate the genetic requirement for EpCAM in Zeb1-mediated HSC function in the context of mitochondrial metabolism and ribosome biogenesis.
Deregulation of cell adhesion molecules or other cell polarity genes, such as EpCAM, which are a normal feature of epithelial cells, could also be incompatible with the predominantly mesenchymal milieu of the BM environment and may facilitate aberrant, cell-autonomous–driven interactions of HSCs with components of the BM niche (86). This could restrain the motility or alter survival and quiescence of HSCs (or HSPC subsets) within the BM. Studies should be conducted using in vivo imaging of Zeb1–/– HSCs in their BM habitat to define the broader role of cell polarity–related molecules in vascular and osteoblastic BM niches and how they might influence HSC fate.
Perturbation of cell polarity is also a hallmark of cancer development, (87) and given our findings of Zeb1’s impact on myeloid differentiation — which is blocked in AML — we explored the function of Zeb1 in AML. In hematologic malignancies, Zeb1 has variably been reported as either a tumor suppressor (in Sézary syndrome and adult T cell leukemia/lymphoma) (88–90) or an oncogene (in mantle cell lymphoma) (91). We found ZEB1 expression was reduced in select AML patient subtypes, including those involving MLL chromosomal translocations that confer poor prognosis in AML (92). Induction of Zeb1 KO in LSC mouse models of MLL-AF9– and Meis1a/Hoxa9-driven AML accelerated disease progression, implying that Zeb1 acts as a tumor suppressor in AML LSCs. In support of this, ablating prooncogenic Gata2 in AML LSCs caused upregulation of Zeb1 expression (66). Taken together with analysis of Zeb1–/– HSCs, these data suggest that Zeb1-mediated control of HSC self-renewal, apoptosis, and differentiation fates is integral to suppressing the vulnerability of HSCs to leukemic transformation and disease progression in AML. This view is in agreement with the likely preleukemic selective advantage provided by decreased ribosome biogenesis (58) and enhanced AML signaling in Zeb1–/– EpCAM+ HSPCs. In contrast, however, and consistent with an oncogenic function for Zeb1 in AML, high expression of Zeb1 has been found to drive dissemination of AML LSCs and leukemic cells to extramedullary sites and other organs (92). Yet the cell context–dependent requirement for Zeb1 in initiating and propagating AML (93) remains ambiguous and requires further in-depth experimentation.
Methods
Mice.
We generated Zeb1fl/fl mice (23) which were bred with Mx1-Cre+/– mice (25) to generate an experimental cohort of Zeb1fl/fl;Mx1-Cre–/– (control) and Zeb1fl/fl;Mx1-Cre+/– (Zeb1–/–). Zeb1 was deleted after i.p. administration of pIpC (6 doses every alternate day, 0.3 mg per dose, GE Healthcare). Genotyping is described in Supplemental Methods.
Flow cytometry analysis.
Bones (femurs, tibias, iliac bones) were crushed using a pestle and mortar in PBS supplemented with 2% FBS, and the BM cell suspension was filtered through a 70 μm cell strainer (Miltenyi Biotec). Spleen and thymi were minced through a 70 μm cell strainer to obtain a homogeneous cell suspension. PB was obtained from the tail vein in EDTA-treated tubes (Starstedt). Red blood cells were lysed by ammonium chloride solution (STEMCELL Technologies). For the immunophenotypic analysis, cells were stained as follows: HSPCs (LSK SLAM): lineage cocktail was prepared from a pool of biotinylated antibodies of differentiated cell markers in PBS 2% FBS (MAC1 and GR1 for myeloid cells, TER119 for erythroid lineage, B220 for B cells, CD3e, CD4, CD8a for T cells), SCA-1-APCCy7, C-KIT-APC, CD150-PECy7, and CD48-FITC to study HSC, MPP, HPC1, and HPC2; for the committed progenitors (LK), LIN cocktail as in LSK SLAM, SCA-1-APCCy7, CKIT-APC, CD34-FITC, CD16/32-PECy7, CD135-PE, and CD127-BV650 to study LMPP, CMP, GMP, MEP, and CLP. The lineage cocktail was detected by adding streptavidin as a secondary antibody. Lineage-positive cells from the BM and spleen were stained for GR1-PECy7 and MAC1-APC (myeloid cells), CD3-APC, CD4-PE, CD8-APCCy7 (T cells), B220-FITC (B cells), CD62L-PECy7, and CD44-APC (naive, effector, and memory T cells). For thymocytes, cells were stained for CD4 and CD8, CD44, CD25, and C-KIT to study early and late stages of T cell development in thymus. For apoptosis assay, after staining the cells for cell-surface markers, they were stained with annexin V–PE antibody (BioLegend) for 30 minutes in the dark at room temperature (RT), and DAPI (1 μg/mL) (Molecular Probes) was added before running the samples. Ki67 for cell-cycle analysis in HSCs and intracellular staining were done after the extracellular staining, cells were fixed in 1% paraformaldehyde (PFA) (Thermo Fisher) for 20 minutes at 4°C, permeabilized using PBS containing 0.1% Saponin (MilliporeSigma) for 30 minutes at 4°C, and then stained with the antibodies for 30 minutes at 4°C in the dark. For cell cycle, cells were incubated with DAPI at a final concentration of 5 μg/mL in the dark for 5 minutes before running the samples. Samples were analyzed using BD LSRFortessa (BD Biosciences). Data were analyzed using FlowJo, version 10.0.8 (Tree Star). A full list of antibodies used is shown in Supplemental Table 1.
For HSC sorting, BM cell suspension was obtained and red blood cells were lysed by ammonium chloride solution (STEMCELL Technologies). Cells were enriched for CKIT by MACS (Miltenyi Biotec) using anti-CKIT magnetic beads (Miltenyi Biotec). CKIT+ cells were stained as described earlier, and HSCs were sorted using a BD FACSAria Fusion (BD Biosciences).
Transplantation experiments.
C57BL/6 SJL mice (CD45.1) were used as recipients for all the transplantations, except that the niche transplantation C57BL/6 (CD45.2) mice (The Jackson Laboratory) were used as recipients. The mice were lethally irradiated at 9 Gy (split dose). For primary transplantation, 150 HSCs from Zeb1–/– and control cells mixed with 2 × 105 whole BM (CD45.1) (supporting cells) were i.v. transplanted into lethally irradiated mice (CD45.1). To monitor the engraftment, tail-vein bleeding was performed at different time points after transplant. To further assess the capacity of Zeb1-deficient HSCs to repopulate secondary recipients, 300 HSCs from Zeb1–/– and control mixed with 3 × 105 whole BM (CD45.1) (supporting cells) were i.v. transplanted into lethally irradiated mice (CD45.1). The engraftment ability was monitored via tail-vein bleeding as was done with the primary recipients.
For cell-autonomous transplantation, Zeb1 was deleted specifically in hematopoietic cells (but not in BM niche cells) after transplanting 5 × 105 whole BM (CD45.2) from Zeb1fl/fl;Mx1-Cre+ and Zeb1fl/fl;Mx1-Cre– along with 5 × 105 whole BM (CD45.1) (supporting cells) into lethally irradiated recipients (CD45.1). Six weeks later, 6 doses of pIpC (every alternate day, 0.3 mg per dose) were i.p. injected to delete Zeb1. Mice were dissected at day 14 after the last dose of pIpC and analyzed. For cell-autonomous secondary transplantation, 5 × 105 CD45.2 donor BM cells were sorted from control and Zeb1–/– primary recipients and mixed with competitor cells and retransplanted into lethally irradiated recipients.
For niche transplantation, 1 × 106 total BM cells from WT CD45.1+ mice were transplanted into lethally irradiated Zeb1fl/fl;Mx1-Cre+ and Zeb1fl/fl;Mx1-Cr– mice. Six weeks later, 6 doses of pIpC (every alternate day, 0.3 mg per dose) were i.p. injected to delete Zeb1. Mice were dissected at week 16 after the last dose of pIpC.
Leukemia transformation assay.
1 × 106 CD45.2+C-KIT+ cells were obtained from control and Zeb1fl/fl;Mx1-Cre–/+ mice and cultured in IMDM 10% FBS supplemented with 40 ng/mL SCF, 20 ng/mL IL-3, and 20 ng/mL IL-6. The next day, the cells were transduced with retroviral vectors encoding Meis1a/Hoxa9 and MLL-AF9 using retronectin-coated plates (TaKaRa) as described previously (64, 66). After 72 hours, 5000 cells were plated in colony-forming units assay 1 (CFC1) using MethoCult M3231 (STEMCELL Technologies) semi-solid media for 6 days, and this process was repeated for up to 3 rounds of CFCs. At the end of CFC3, pre-LSCs were harvested and sorted according to C-KIT expression and transplanted into lethally irradiated primary recipients. Three weeks later, Zeb1 was deleted after i.p. administration of pIpC (8 doses every alternate day, 0.3 mg per dose, GE Healthcare). Mice were monitored for AML development.
RNA-Seq.
RNA from HSCs (LSK CD150+CD48–) and CLPs (LIN– SCA-1loC-KITloCD127+) from control and Zeb1–/– mice 14 days after the last dose of pIpC injection was extracted using the RNAeasy Micro Kit (QIAGEN). Total RNA quality and quantity were assessed using Agilent 2100 Bioanalyzer and the RNA Nano 6000 Kit (Agilent Technologies). The library was prepared using the NEB Ultra II Directional RNA Library Prep Kit for Illumina. The libraries then were sequenced using a 75 base paired end (2 × 75bp PE) dual index read format on the Illumina HiSeq4000 according to the manufacturer’s instructions. Further details on sequencing and bioinformatics were described previously (66).
The heatmap was created using Morpheus, an online tool, (Broad Institute). DEGs with an FDR of less than 0.05 were used for heatmaps. The biological pathway analysis was performed using BioCarta, KEGG, and Reactome pathway databases run on GSEA software (94) as well as IPA software (QIAGEN). IPA was used to create a prediction network of Zeb1 interactions with its target genes.
RNA-Seq for EPCAM+ and EPCAM– from Zeb1–/– mice is described in Supplemental Methods.
All RNA-Seq data are available in the NCBI’s GEO database (GSE153664, GSE154615).
AML patients bioinformatic analysis.
ZEB1 expression data were analyzed from a cohort of 2611 AML patients and 77 controls obtained from the GEO database (GSE14468, GSE22845, GSE10358, GSE12417, GSE13159, GSE14062, GSE15434, GSE16015, GSE38987, GSE22056, GSE33223, GSE17855, GSE15389) (95) and ArrayExpress (E-MTAB-3444) (96). R software was used to analyze and produce data. Data processing, normalization, and analysis were described previously (97).
Statistics.
Figures were prepared using Prism (GraphPad Software). Statistical analyses were done using Mann-Whitney U test to calculate significance, which was defined as P < 0.05\.
Study approval.
All animal experiments were performed according to protocols ratified by the UK Home Office and carried out at Cardiff University animal facility under project number 30/3380.
Author contributions
A Almotiri designed and performed experiments, analyzed and interpreted data, prepared the figures, and contributed to writing the manuscript. HA contributed to experimental design, performed experiments, analyzed and interpreted data, analyzed RNA-Seq data, and reviewed the manuscript. JBMG contributed to experimental design, performed experiments, analyzed and interpreted data, and reviewed the manuscript. A Abdelfattah and BA performed experiments, analyzed data, and reviewed the manuscript. LS, A Greene, MG, A Gibbs, A Alsayari, ST, LAT, and DS performed experiments and contributed to data analysis. SE and PG performed bioinformatics analyses. MPS, SB, and TB contributed to experimental design and analysis and reviewed the manuscript. ASB and FAS contributed significantly to experimental design, data analysis and interpretation, and manuscript preparation. NPR conceived and supervised the project, designed experiments, analyzed and interpreted the data, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work is supported by the British Society for Hematology, Life Science Research Network Wales, the Leukemia Cancer Society, Leukemia Research Appeal for Wales, the Lady Tata Memorial Trust (to NPR), and Shaqra University (to A Almotiri). We thank Shelley Rundle for assistance with RNA-Seq and Robert Andrews for assistance with bioinformatics. The graphical abstract was drawn using BioRender.
Version 1. 10/27/2020
In-Press Preview
Version 2. 01/04/2021
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021 Almotiri et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2021;131(1):e129115.https://doi.org/10.1172/JCI129115.
Contributor Information
Alhomidi Almotiri, Email: hamidi.salem@gmail.com.
Juan Bautista Menendez-Gonzalez, Email: MenendezGonzalezJ@cardiff.ac.uk.
Ali Abdelfattah, Email: AbdelfattahAM@cardiff.ac.uk.
Badi Alotaibi, Email: AlotaibiB@cardiff.ac.uk.
Lubaid Saleh, Email: lubaid.saleh@sheffield.ac.uk.
Adelle Greene, Email: adellegreene@hotmail.co.uk.
Alex Gibbs, Email: gibbsa@cardiff.ac.uk.
Sarab Taha, Email: tahasn@cardiff.ac.uk.
Leigh-anne Thomas, Email: thomasl85@cardiff.ac.uk.
Dhruv Shah, Email: dhruvkat675@gmail.com.
Sarah Edkins, Email: edkinss@cardiff.ac.uk.
Marc P. Stemmler, Email: marc.stemmler@fau.de.
Simone Brabletz, Email: simone.brabletz@fau.de.
Thomas Brabletz, Email: thomas.brabletz@fau.de.
Ashleigh S. Boyd, Email: a.boyd@ucl.ac.uk.
Florian A. Siebzehnrubl, Email: fas@cardiff.ac.uk.
Neil P. Rodrigues, Email: RodriguesN@cardiff.ac.uk.
References
- 1.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Postigo AA, Dean DC. ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. EMBO J. 1997;16(13):3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Postigo AA, Ward E, Skeath JB, Dean DC. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol Cell Biol. 1999;19(10):7255–7263. doi: 10.1128/MCB.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jethanandani P, Kramer RH. Alpha7 integrin expression is negatively regulated by deltaEF1 during skeletal myogenesis. J Biol Chem. 2005;280(43):36037–36046. doi: 10.1074/jbc.M508698200. [DOI] [PubMed] [Google Scholar]
- 6.Postigo AA, Dean DC. Independent repressor domains in ZEB regulate muscle and T-cell differentiation. Mol Cell Biol. 1999;19(12):7961–7971. doi: 10.1128/MCB.19.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark SG, Chiu C. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development. 2003;130(16):3781–3794. doi: 10.1242/dev.00571. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, et al. Zinc finger E-box-binding homeobox 1 (ZEB1) is required for neural differentiation of human embryonic stem cells. J Biol Chem. 2018;293(50):19317–19329. doi: 10.1074/jbc.RA118.005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh S, et al. Zeb1 controls neuron differentiation and germinal zone exit by a mesenchymal-epithelial-like transition. Elife. 2016;5:e12717. doi: 10.7554/eLife.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, et al. ZEB1 represses neural differentiation and cooperates with CTBP2 to dynamically regulate cell migration during neocortex development. Cell Rep. 2019;27(8):2335–2353.e6. doi: 10.1016/j.celrep.2019.04.081. [DOI] [PubMed] [Google Scholar]
- 11.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. Delta-crystallin enhancer binding protein delta EF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119(2):433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 12.Higashi Y, et al. Impairment of T cell development in deltaEF1 mutant mice. J Exp Med. 1997;185(8):1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagi T, Moribe H, Kondoh H, Higashi Y. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125(1):21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 14.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells — an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5(9):744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 15.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta Rev Cancer. 2017;1868(2):584–591. doi: 10.1016/j.bbcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Berx G, Raspe E, Christofori G, Thiery JP, Sleeman JP. Pre-EMTing metastasis? Recapitulation of morphogenetic processes in cancer. Clin Exp Metastasis. 2007;24(8):587–597. doi: 10.1007/s10585-007-9114-6. [DOI] [PubMed] [Google Scholar]
- 18.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop — a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11(9):670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 20.Siebzehnrubl FA, et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5(8):1196–1212. doi: 10.1002/emmm.201302827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh DK, et al. Oncogenes activate an autonomous transcriptional regulatory circuit that drives glioblastoma. Cell Rep. 2017;18(4):961–976. doi: 10.1016/j.celrep.2016.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G, et al. miRNA-223 upregulated by MYOD inhibits myoblast proliferation by repressing IGF2 and facilitates myoblast differentiation by inhibiting ZEB1. Cell Death Dis. 2017;8(10):e3094. doi: 10.1038/cddis.2017.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brabletz S, et al. Generation and characterization of mice for conditional inactivation of Zeb1. Genesis. 2017;55(4):e23024. doi: 10.1002/dvg.23024. [DOI] [PubMed] [Google Scholar]
- 24.Postigo AA, Dean DC. Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc Natl Acad Sci U S A. 2000;97(12):6391–6396. doi: 10.1073/pnas.97.12.6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 26.Lagasse E, Weissman IL. Flow cytometric identification of murine neutrophils and monocytes. J Immunol Methods. 1996;197(1–2):139–150. doi: 10.1016/0022-1759(96)00138-x. [DOI] [PubMed] [Google Scholar]
- 27.Sunderkotter C, et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172(7):4410–4417. doi: 10.4049/jimmunol.172.7.4410. [DOI] [PubMed] [Google Scholar]
- 28.Bhandoola A, Sambandam A. From stem cell to T cell: one route or many? Nat Rev Immunol. 2006;6(2):117–126. doi: 10.1038/nri1778. [DOI] [PubMed] [Google Scholar]
- 29.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150(10):4244–4252. [PubMed] [Google Scholar]
- 30.Matsuzaki Y, et al. Characterization of c-kit positive intrathymic stem cells that are restricted to lymphoid differentiation. J Exp Med. 1993;178(4):1283–1292. doi: 10.1084/jem.178.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ceredig R, Rolink T. A positive look at double-negative thymocytes. Nature Reviews Immunology. 2002;2(11):888–897. doi: 10.1038/nri937. [DOI] [PubMed] [Google Scholar]
- 32.Allman D, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4(2):168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 33.Bhandoola A, Sambandam A, Allman D, Meraz A, Schwarz B. Early T lineage progenitors: new insights, but old questions remain. J Immunol. 2003;171(111):5653–5658. doi: 10.4049/jimmunol.171.11.5653. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425(6960):836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 35.Ghaedi M, Steer CA, Martinez-Gonzalez I, Halim TYF, Abraham N, Takei F. Common-lymphoid-progenitor-independent pathways of innate and T lymphocyte development. Cell Rep. 2016;15(3):471–480. doi: 10.1016/j.celrep.2016.03.039. [DOI] [PubMed] [Google Scholar]
- 36.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Pietras EM, et al. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell. 2015;17(1):35–46. doi: 10.1016/j.stem.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13(1):102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mooney CJ, Cunningham A, Tsapogas P, Toellner KM, Brown G. Selective expression of Flt3 within the mouse hematopoietic stem cell compartment. Int J Mol Sci. 2017;18(5):1037. doi: 10.3390/ijms18051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sitnicka E, Bryder D, Theilgaard-Monch K, Buza-Vidas N, Adolfsson J, Jacobsen SE. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 2002;17(4):463–472. doi: 10.1016/S1074-7613(02)00419-3. [DOI] [PubMed] [Google Scholar]
- 41.Kumar R, Fossati V, Israel M, Snoeck HW. Lin-Sca1+kit- bone marrow cells contain early lymphoid-committed precursors that are distinct from common lymphoid progenitors. J Immunol. 2008;181(11):7507–7513. doi: 10.4049/jimmunol.181.11.7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14(4):481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aigner K, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26(49):6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sánchez-Tilló E, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29(24):3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 45.Vannier C, Mock K, Brabletz T, Driever W. Zeb1 regulates E-cadherin and Epcam (epithelial cell adhesion molecule) expression to control cell behavior in early zebrafish development. J Biol Chem. 2013;288(26):18643–18659. doi: 10.1074/jbc.M113.467787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.González B, Denzel S, Mack B, Conrad M, Gires O. EpCAM is involved in maintenance of the murine embryonic stem cell phenotype. Stem Cells. 2009;27(8):1782–1791. doi: 10.1002/stem.97. [DOI] [PubMed] [Google Scholar]
- 47.Chapple RH, et al. Lineage tracing of murine adult hematopoietic stem cells reveals active contribution to steady-state hematopoiesis. Blood Adv. 2018;2(11):1220–1228. doi: 10.1182/bloodadvances.2018016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bendriem RM, Singh S, Aleem AA, Antonetti DA, Ross ME. Tight junction protein occludin regulates progenitor self-renewal and survival in developing cortex. Elife. 2019;8:e49376. doi: 10.7554/eLife.49376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bierie B, et al. Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc Natl Acad Sci U S A. 2017;114(12):E2337–E2346. doi: 10.1073/pnas.1618298114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips JL, et al. Distinct mechanisms of regulation of the ITGA6 and ITGB4 genes by RUNX1 in myeloid cells. J Cell Physiol. 2018;233(4):3439–3453. doi: 10.1002/jcp.26197. [DOI] [PubMed] [Google Scholar]
- 51.Postigo AA, Dean DC. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci U S A. 1999;96(12):6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purbey PK, Singh S, Notani D, Kumar PP, Limaye AS, Galande S. Acetylation-dependent interaction of SATB1 and CtBP1 mediates transcriptional repression by SATB1. Mol Cell Biol. 2009;29(5):1321–1337. doi: 10.1128/MCB.00822-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schnell U, Cirulli V, Giepmans BN. EpCAM: structure and function in health and disease. Biochim Biophys Acta. 2013;1828(8):1989–2001. doi: 10.1016/j.bbamem.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 54.Rodrigues NP, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106(2):477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 55.Osellame LD, Blacker TS, Duchen MR. Cellular and molecular mechanisms of mitochondrial function. Best Pract Res Clin Endocrinol Metab. 2012;26(6):711–723. doi: 10.1016/j.beem.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hinge A, et al. Asymmetrically segregated mitochondria provide cellular memory of hematopoietic stem cell replicative history and drive HSC attrition. Cell Stem Cell. 2020;26(3):420–430.e6. doi: 10.1016/j.stem.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Filippi MD, Ghaffari S. Mitochondria in the maintenance of hematopoietic stem cells: new perspectives and opportunities. Blood. 2019;133(18):1943–1952. doi: 10.1182/blood-2018-10-808873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai X, et al. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell. 2015;17(2):165–177. doi: 10.1016/j.stem.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corces MR, Chang HY, Majeti R. Preleukemic hematopoietic stem cells in human acute myeloid leukemia. Front Oncol. 2017;7:263. doi: 10.3389/fonc.2017.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reikvam H, Hatfield KJ, Kittang AO, Hovland R, Bruserud Ø. Acute myeloid leukemia with the t(8;21) translocation: clinical consequences and biological implications. J Biomed Biotechnol. 2011;2011:104631. doi: 10.1155/2011/104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winters AC, Bernt KM. MLL-rearranged leukemias — an update on science and clinical approaches. Front Pediatr. 2017;5:4. doi: 10.3389/fped.2017.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lawrence HJ, et al. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13(12):1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 63.Dorsam ST, et al. The transcriptome of the leukemogenic homeoprotein HOXA9 in human hematopoietic cells. Blood. 2004;103(5):1676–1684. doi: 10.1182/blood-2003-07-2202. [DOI] [PubMed] [Google Scholar]
- 64.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10(4):257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 65.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 66.Menendez-Gonzalez JB, et al. Gata2 as a crucial regulator of stem cells in adult hematopoiesis and acute myeloid leukemia. Stem Cell Reports. 2019;13(2):291–306. doi: 10.1016/j.stemcr.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grégoire JM, Roméo PH. T-cell expression of the human GATA-3 gene is regulated by a non-lineage-specific silencer. J Biol Chem. 1999;274(10):6567–6578. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- 68.Hosoya T, et al. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206(13):2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guan T, et al. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+ T cell fates. J Exp Med. 2018;215(4):1153–1168. doi: 10.1084/jem.20171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jansen CS, et al. An intra-tumoral niche maintains differentiates stem-like CD8 T cells. Nature. 2019;576(7787):465–470. doi: 10.1038/s41586-019-1836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boyd AS, Rodrigues NP. Stem cells cycle toward immune surveillance. Immunity. 2018;48(2):187–190. doi: 10.1016/j.immuni.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 72.Noman MZ, et al. CD47 is a direct target of SNAI1 and ZEB1 and its blockade activates the phagocytosis of breast cancer cells undergoing EMT. Oncoimmunology. 2018;7(4):e1345415. doi: 10.1080/2162402X.2017.1345415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teitell MA, Mikkola HK. Transcriptional activators, repressors, and epigenetic modifiers controlling hematopoietic stem cell development. Pediatr Res. 2006;59(4 Pt 2):33R–39R. doi: 10.1203/01.pdr.0000205155.26315.c7. [DOI] [PubMed] [Google Scholar]
- 74.Hock H, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431(7011):1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 75.Zeng H, Yücel R, Kosan C, Klein-Hitpass L, Möröy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23(20):4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Florian MC, et al. Aging alters the epigenetic asymmetry of HSC division. PLoS Biol. 2018;16(9):e2003389. doi: 10.1371/journal.pbio.2003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Voog J, D’Alterio C, Jones DL. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature. 2008;454(7208):1132–1136. doi: 10.1038/nature07173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boyle M, Wong C, Rocha M, Jones DL. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1(4):470–478. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 79.Wu M, et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1(5):541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cayouette M, Raff M. Asymmetric segregation of Numb: a mechanism for neural specification from Drosophila to mammals. Nat Neurosci. 2002;5(12):1265–1269. doi: 10.1038/nn1202-1265. [DOI] [PubMed] [Google Scholar]
- 81.Jiang X, et al. Asymmetric distribution of CRUMBS polarity complex proteins from compacted 8-cell to blastocyst stage during mouse preimplantation development. Gene Expr Patterns. 2018;27:93–98. doi: 10.1016/j.gep.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 82.Roignot J, Peng X, Mostov K. Polarity in mammalian epithelial morphogenesis. Cold Spring Harb Perspect Biol. 2013;5(2):a013789. doi: 10.1101/cshperspect.a013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takubo K, et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12(1):49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu WM, et al. Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell. 2013;12(1):62–74. doi: 10.1016/j.stem.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Donati G, et al. The balance between rRNA and ribosomal protein synthesis up- and downregulates the tumour suppressor p53 in mammalian cells. Oncogene. 2011;30(29):3274–3288. doi: 10.1038/onc.2011.48. [DOI] [PubMed] [Google Scholar]
- 86.Schreck C, et al. Niche WNT5A regulates the actin cytoskeleton during regeneration of hematopoietic stem cells. J Exp Med. 2017;214(1):165–181. doi: 10.1084/jem.20151414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halaoui R, McCaffrey L. Rewiring cell polarity signaling in cancer. Oncogene. 2015;34(8):939–950. doi: 10.1038/onc.2014.59. [DOI] [PubMed] [Google Scholar]
- 88.Vermeer MH, et al. Novel and highly recurrent chromosomal alterations in Sézary syndrome. Cancer Res. 2008;68(8):2689–2698. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]
- 89.Hidaka T, et al. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood. 2008;112(2):383–393. doi: 10.1182/blood-2008-01-131185. [DOI] [PubMed] [Google Scholar]
- 90.Nakahata S, Yamazaki S, Nakauchi H, Morishita K. Downregulation of ZEB1 and overexpression of Smad7 contribute to resistance to TGF-beta1-mediated growth suppression in adult T-cell leukemia/lymphoma. Oncogene. 2010;29(29):4157–4169. doi: 10.1038/onc.2010.172. [DOI] [PubMed] [Google Scholar]
- 91.Sánchez-Tilló E, et al. The EMT activator ZEB1 promotes tumor growth and determines differential response to chemotherapy in mantle cell lymphoma. Cell Death Differ. 2014;21(2):247–257. doi: 10.1038/cdd.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stavropoulou V, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell. 2016;30(1):43–58. doi: 10.1016/j.ccell.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 93.Basheer F, et al. Contrasting requirements during disease evolution identify EZH2 as a therapeutic target in AML. J Exp Med. 2019;216(4):966–981. doi: 10.1084/jem.20181276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kolesnikov N, et al. ArrayExpress update — simplifying data submissions. Nucleic Acids Res. 2015;43(Database issue):D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Menendez-Gonzalez JB, et al. Inhibition of GATA2 restrains cell proliferation and enhances apoptosis and chemotherapy mediated apoptosis in human GATA2 overexpressing AML cells. Sci Rep. 2019;9(1):12212. doi: 10.1038/s41598-019-48589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.