Graphical abstract
Keywords: Genome-scale metabolic modelling, Microbial community, Optimization, Design, Engineering, Computational methods, Synthetic microbial consortia
Abstract
Microbes do not live in isolation but in microbial communities. The relevance of microbial communities is increasing due to growing awareness of their influence on a huge number of environmental, health and industrial processes. Hence, being able to control and engineer the output of both natural and synthetic communities would be of great interest. However, most of the available methods and biotechnological applications involving microorganisms, both in vivo and in silico, have been developed in the context of isolated microbes. In vivo microbial consortia development is extremely difficult and costly because it implies replicating suitable environments in the wet-lab. Computational approaches are thus a good, cost-effective alternative to study microbial communities, mainly via descriptive modelling, but also via engineering modelling. In this review we provide a detailed compilation of examples of engineered microbial communities and a comprehensive, historical revision of available computational metabolic modelling methods to better understand, and rationally engineer wild and synthetic microbial communities.
1. Introduction
Microbes play a pivotal role in fields as diverse as human health, environmental science and biotechnology. Focus on the latter, microbial production of chemicals has become increasingly attractive across industry due to its role in delivering sustainable manufacturing technology. Microbial biotechnology platforms integrating systems and synthetic biology tools have successfully contributed to delivering a large portfolio of chemical compounds [1], [2], [3], [4], [5], [6], [7]. Early applications of systems metabolic engineering to maximize metabolite production focused on engineering single competitive strains, and thus faced hurdles such as metabolic burden and heterogeneity [8], [9]. Consequently, production of target chemicals is not always cost-effective and great efforts must be made to improve yield and productivity. The use of microbial consortia has thus been promoted as an alternative to overcome these limitations [10], [11] because cooperation among several strains allows microbial communities to function at higher levels of complexity than individual cells. Pathway modularization allows distribution of metabolic reactions among highly specialized strains, thus reducing genetic load requirements per individual. Increased bioproduction performance and efficiency in source transformation can be achieved using different substrates, and/or synthesizing products in parallel, and/or avoiding intermediate metabolite accumulation. In addition, robustness provided by microbial communities avoids environmental stresses [12], [13], [14].
The advantages described have underpinned recent progress in analysing, understanding, designing and developing both natural and synthetic microbial communities. Such progress has been applied to improving health, food and chemical production and to dealing with environmental challenges. Therefore, microbial biotechnology will probably lead to microbial community engineering using species selection, manipulation of strain ratios and/or genetic engineering of community members. However, metabolically engineering microbial communities entails certain unresolved challenges [15], e.g. defining co-culture conditions and growth compatibility, and selecting the cross-feeding metabolites among different strains in the consortium. Microbial community modelling emerged with the need to improve the knowledge and understanding of interactions among heterogeneous cells [16]. Such interactions can be described using metabolite exchange modelling, where the actual metabolites and the extent to which they are exchanged need to be defined. Community interactions include cross-feeding, competition for nutrients, symbiotic relationships (such as plant-microorganism) or parasitism (such as human-pathogen) or multi-tissues. Therefore, these pioneering efforts have highlighted the need for novel computational-system approaches in order to facilitate more rational designs. Here we review pioneering achievements in the field of microbial consortia-based bioprocesses and available computational metabolic modelling tools that provide a better understanding and support rational engineering of microbial communities.
2. Learning from nature: functional and stability-based design of synthetic microbial consortia
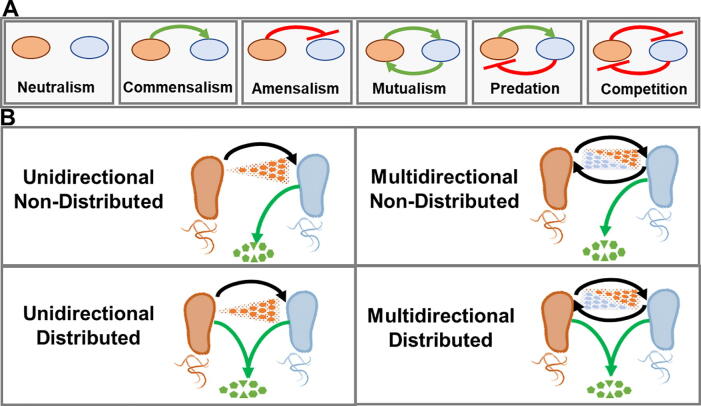
In nature, microorganisms are involved in a large array of complex interactions with other organisms and their environment, thus contributing to stability and functionality. Among others, such complex relationships traditionally include neutralism, commensalism, amensalism, mutualism, predation and competition. These natural relationships have been profusely used as mechanisms for the establishment of synthetic metabolic interactions when designing synthetic microbial communities (SMC) [13], [17], [18] (Fig. 1A). Therefore, the two main questions emerging when designing a SMC are: i) how will the microbial community structure be established to ensure the consortium’s stability? and ii) how will relationships be established within the SMC to drive the community’s output?
Fig. 1.
A, Schematic representation of the basic ecological interactions between the microbial strains in co-culture, green positive and red negative interactions. B, Schematic representation of the SMCs categories. Black arrows stability interactions and green arrows functionality interactions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The stability of the population in a microbial community involves complex interactions among its components. These relationships are usually established through primary interaction mechanisms i.e., interactions that are strictly necessary to deliver the consortium’s output. However the relationships within a given microbial consortium are not exempt from the emergence of secondary interactions which, although contributing to the community’s stability and/or functionability , are not strictly necessary. For instance, these primary and secondary interactions are easily visible in SMCs formed by Synechococcus elongatus and Pseudomonas putida. The production of sucrose by the cyanobacterium provides an organic carbon source to the heterotrophic partner and is the primary interaction within the consortium, while the production of O2 derived from S. elongatus’ photosynthetic activity and the CO2 released by P. putida would be secondary interactions, as additional sources of CO2 and O2 exist in the consortium [19], [20]. In terms of stability, and considering only primary interactions, microbial relationships can be grouped into two main categories: unidirectional and multidirectional. Unidirectional interactions are those in which the population of one of the consortium’s components is regulated (either positively or negatively) by another component. In contrast, multidirectional interactions are those in which all of the community’s microbial components actively interact with each other to support the stability of the entire consortium via positive or negative feedback (Fig. 1B). Similarly, relationships within the community also determine to a great extent the SMC’s functionality and level of complexity. Overall, functionality as a function of complexity level can be categorized as non-distributed or distributed. Non-distributed functionality implies low complexity levels and often only one member of the consortium is responsible for the final community’s output. On the contrary, more complex SMC outputs require the involvement of several community partners within the consortium. Labour is split in a distributed process, thus increasing the consortium’s efficiency. SMC can therefore be classified into four main categories according to the relationships contributing to stability and functionality: Unidirectional Non-Distributed, Multidirectional Non-distributed, Unidirectional Distributed and Multidirectional Distributed (Fig. 1B). In order to define the field’s state-of-the-art, in the following sections we have categorized and contextualized current efforts on synthetic microbial consortia engineering based on the above classification. A detailed review of outstanding examples is summarized in Table 1.
Table 1.
Recent examples of engineering Synthetic Microbial consortia.
| Microorganism | Interaction | Goal to optimize | C-source | Yield | Ref. |
|---|---|---|---|---|---|
| Unidirectional Non-Distributed | |||||
|
Synechoccocus elongates Pseudomonas putida |
S. elongatus produces sucrose from CO2 and light. It was used for P. putida, growing and cleaning 2,4-DNT while produces polyhydroxyalcanoates (PHA) |
|
CO2 |
|
[19] |
|
S. elongatus Escherichia coli |
S. elongatus produces sucrose from CO2 and light. The sucrose is used as C-source for E. coli, producing 3-hydroxypropinoic acid (3-HP) |
|
CO2 |
|
[20] |
|
Klebsiella pneumoniae Shewanella oneidensis |
K. pneumoniae uses glycerol as C-source, producing lactate. S. oneidensis uses the lactate producing electrons. |
|
Glycerol |
|
[139] |
|
Ralstonia eutropha Bacillus subtilis |
B. subtilis hydrolyses sucrose in fructose and glucose, producing propionic acid. They are used by R. eutropha, producing PHA or poly (3-hydroxybutyrate-co-3hydroxyvalerate) [P(3HB-co-3HV]. |
|
Sucrose |
|
[140] |
|
Citrobacter amalonaticus Sporomusa ovata |
C. amalonaticus uses CO as carbon source, producing CO2 and H2 which are used by S. ovata producing acetate |
|
CO |
|
[141] |
|
Trichoderma reesei Rhizopus delemar or T. ressei R. orizae |
T. reesei hydrolyses cellulose into monomeric sugars. R. delemar uses these sugars producing fumaric acid and R. oizae producing lactic acid. |
|
Corn stove |
|
[142] |
|
Clostridium thermocellum C. saccharoperbutylacetonicum |
C. thermocellum hydrolyses cellulose releasing the C-source for butanol production by C. saccharoperbutylacetonicum. |
|
Rice straw |
|
[18] |
|
E. coli Acinetobacter baylyi |
E. coli utilizes glucose as C-source producing acetate. The acetate is used by A. baylyi |
|
Glucose |
|
[143] |
|
T. reesei E. coli |
T. reesei hydrolyses cellulose into monomeric sugars. E. coli uses these sugars producing isobutanol. |
|
Cellulose |
|
[21] |
|
E. coli E. coli |
E. coli E609Y produces xylanase extracellularly, hydrolysing xylan to xylooligosaccharides. they are used by E. coli KO11 producing ethanol. |
|
Xylan |
|
[22] |
|
Rhodotorula glutinis Dwbaryomyces castellii |
D. castelli hydrolyses corn syrup into sugars, which are used by R. glutinis, producing carotenoids. |
|
Corn syrup |
|
[144] |
| Multidirectional Non-Distributed | |||||
|
E. coli Corynebacterium glutamicum |
E. coli (Lys auxotroph) produces amylase extracellularly, hydrolysing starch into glucose, which is used by C. glutamicum, producing cadaverine or L-pipecolic acid (L-PA) and Lys, necessary for E. coli growth. |
|
Starch |
|
[23] |
|
Sacharomyces cerevisiae -Bacillus. Amyloliquefacien or S. cerevisiae - Lactobacillus fermentum |
B. amyloliquefaciens/L. fermentum produces amylase, hydrolysing starch into glucose and oligosaccharides. they are used by S. cerevisiae. Its growth stimulates the production of more amylase for B. amyloliquefaciens/L. fermentum. |
|
Starch |
|
[145] |
| Streptomyces sp. Mg1B. subtilis | In co-culture B. subtilis stimulates Streptomyces sp Mg1 to produce chalcomycin A (macrolide antibiotic). Chalcomycin A inhibits B subtilis growth. |
|
Maltose |
|
[24] |
| P. putida - Bdellovibrio bacteriovorus | P. putida producing PHA and polyhydroxybutyrate (PHB) was mixed with the predatory B. bacteriovorus, that feeds on P. putida, releasing the PHA or PHA to the medium. |
|
octanoate |
|
[146] |
| Pseudomonas aeruginosa -Buskholderia cenocepacia | In co-culture at limited iron P. aeruginosa and B. cenoceparia competed for the iron, limiting the growth of B. cenoceparia. When a P. aeruginosa iron cheater mutant was introduced both strains grew well at limited iron |
|
Casamino acids |
|
[25] |
|
P. aeruginosa Enterobacter aerogenes |
E. aerogenes use glucose, producing 2,3-butanediol which is used by P. aeruginosa producing phenazines, They are used for E. aerogenes as electron acceptor. |
|
Glucose |
|
[147] |
| Unidirectional Distributed | |||||
|
E. coli S. cerevisiae |
Hydrogel compartmentalized E. coli and S. cerevisiae were co-cultured, using glucose as C-source, E. coli produces L-DOPA, that is used by S. cerevisiae to produce betaxhantins |
|
Glucose |
|
[148] |
| Three E. coli strains | The rosmarinic acid biosynthetic pathway was divided in three E. coli strains, one producing caffeic acid, other salvinic acid, and a third strains that use those intermediaries to produce rosmarinic acid. All of them use glucose as carbon source |
|
Glucose |
|
[30] |
|
E. coli E.coli |
The glutarate biosynthetic pathway from Lys was splitted in two E. coli strains. The first one use Lys, producing 5-aminovaleric acid, that is used by the second E. coli strain producing glutarate |
|
Lysine |
|
[149] |
|
E. coli E.coli |
E. coli RES produces resveratrol from p-coumarate. The resveratrol is glycosylated by E. coli RGL. Both strains use glucose as carbon source. |
|
Glucose |
|
[28] |
|
Halomonas sp. HL-48 Marinobacter sp. HL-58 |
When both strains are growing using glucose as carbon source they compete for it. When xylose is used instead of glucose, Halomonas consumes xylose, producing metabolites that are used for Marinobacter growth. |
|
Xylose |
|
[150] |
|
E. coli E. coli |
E. coli P2C produces Tyr and p-coumarate from glucose. Both are used for E. coli BLNA to produce naringenin using glucose as carbon source |
|
Glucose |
|
[27] |
| Four strains of E. coli | The synthetic plants pathway to produce Anthocyanins was divided and inserted in four different E. coli strains. The first produces phenylpropanoic acid, that is used for the second, producing flavonones. A third strain produces flavan-3-ols from flavonones. Finally, the last E. coli strain produces anthocyanins from flavan-3-ols. |
|
Glucose |
|
[31] |
|
E. coli E. coli |
The resveratrol biosynthetic pathway is divided in two E. coli strains. Both strains use glycerol as carbon source. One of them produces P-coumarate, which is used for the other to produce resveratrol. |
|
Glycerol |
|
[29] |
|
E. coli S. cerevisiae |
E. coli utilizes xylose as C-source, producing acetate which is the C-source for S. cerevisiae. In parallel, E. coli is producing taxadiene, that is oxygenated by S. cerevisiae. |
|
Xylose |
|
[26] |
|
E. coli E. coli |
One E. coli strain uses xylose as C-source, producing 3-dehydroshikimic acid (DHS), uses for the other strain to produce muconic acid or 4-hydroxybenzoic acid, using glucose as C-source. |
|
Glucose Xylose |
|
[151] |
| Four strains of S. cerevisiae | The enzymatic pathway to produce ethanol from cellulose was divided in four S. cerevisiae strains. |
|
Cellulose |
|
[17] |
| Multidirectional Distributed | |||||
|
Dietzia sp strain DQ1245-1b Pseudomonas stutzeri SLG510A3-8 |
Dietzia uses hexadecane as C-source, producing hexadecanoid acid, α-ketoglutaric acid and R-3-hydroxybutanoic acid, that are used by P. stutzeri, that in turn produces glutamate and acetate for Dietzia. The consortium increase the diesel degradation |
|
Hexadecane |
|
[152] |
|
E. coli E. coli |
One E. coli strain uses xylose, producing tyrosol. The other consumes glucose and produces salidroside (from tyrosol). The relationship between both strains had been stablished by cross-feeding. The xylose consuming strain is Phe auxotroph, while the glucose consuming is Tyr auxotroph. |
|
Xylose Glucose |
|
[32] |
|
E. coli E. coli |
One E. coli strain uses glucose as C-source, producing lysine. The other E. coli strain intakes the lysine producing cadaverine. This strain use glycerol as carbon source |
|
Glucose Glycerol |
|
[153] |
|
E. coli B. subtilis S. oneidensis |
E. coli utilizes glucose as C-source, producing lactate and an electron donor; B. subtilis uses also glucose producing riboflavin as an electron shuttle. S. oneidensis uses the electron donor and the shuttle generating electricity and oxidizing lactate to acetate, which is used by E. coli and B. subtilis as C-source |
|
Glucose |
|
[154] |
|
S. oneidensis E. coli |
E. coli ferments glucose producing formate, which is used by S. oneidensis, producing flavins, uses by E. coli. Their activity increase the electric current from cathode to anode in a MFC |
|
Glucose |
|
[155] |
|
E. coli E. coli |
E. coli L is Leu auxotroph and E. coli K is Lys. They co-culture provide each other with the necessary amino acids, increasing the growth rate and the biomass. |
|
Glucose |
|
[156] |
2.1. Unidirectional non-distributed
This category includes the simplest community, in which stability is provided by a single unidirectional relationship (e.g., one microbial component is responsible for feeding the other, either directly or by feedstock processing) while the second strain is in charge of the whole consortium’s functionality. Unidirectional Non-Distributed SMC designs provide significant advantages over single cultures by joining complementary metabolic traits of the cognate microbial partners. Therefore, SMC expand the scope of the target bioprocess in terms of either access to new feedstock and/or providing additional biosynthetic properties. In this scenario, a single component of the consortium addresses the catabolism of a complex feedstock (e.g., xylan, cellulose, syrup, etc.) to release low-complexity carbon sources. These easy-to-uptake carbon sources are subsequently used by the other component of the consortium, which is in charge of delivering the non-distributed biotechnological output. Many consortia fitting these criteria have already been constructed, mainly for the production of biofuels. For instance, the fungi Trichoderma reesei was used to hydrolyse cellulose in a co-culture with Escherichia coli, which was in charge of synthetizing isobutanol [21]. Similarly, the co-culture of two clostridium species (C. thermocellum and C. saccharoperbutylacetonicum) were used for the production of butanol [18], while a consortium made up of two specialized strains of E. coli was used to produce ethanol from xylan [25]. Within this category, an interesting group of consortia are those in which one of the components is the producer of the primary carbon source. For instance, the cyanobacteria S. elongatus has been profusely used as sucrose producer in co-cultures with heterotrophic organisms such as P. putida and E. coli for the production of polyhydroxyalkanoates and 3-hydroxypropinoic acid, respectively [19], [20].
2.2. Multidirectional non-distributed
These SMC add an additional layer of complexity. In these consortia, the stability of the community is achieved through the establishment of co-dependency relationships between community microorganisms, while only one consortium component is in charge of the labour. Co-dependency relationships allow better control of the stability of microbial populations. An example of a co-dependency interaction is metabolite cross-feeding, where all members of a consortium are responsible for feeding each other. For example, Sgobba et al [23] developed a multidirectional non-distributed consortium for cadaverine production in which a lysine auxotroph E. coli strain released glucose from starch, feeding C. glutamicum that in turn produced lysine for E. coli. Multidirectional relationships can also be established by competition mechanisms, e.g. in a co-culture of Bacillus subtilis and Streptomyces sp. Mg1, the growth of B. subtilis stimulated the production of chalcomycin A by Streptomyces sp Mg1, which is an inhibitor of B subtilis’ growth [24]. An interesting approach to stabilizing SMC by means of competition mechanisms is the introduction of cheaters strains. For example, when Pseudomonas aeruginosa and Bulkholderia cenoceparia were co-cultured under iron-limiting conditions, despite both microorganisms possessing the ability to secrete siderophores, only P. aeruginosa achieved stable growth. When mutant strains of P. aeruginosa unable to secrete siderophores were used instead, both microorganisms grew at the expense of the siderophores secreted by B. cenoceparia [25].
2.3. Unidirectional distributed
This category adds complexity as all members in the SMC contribute to the system’s functionality. Engineering Distributed SMC relies mainly on splitting complex biosynthetic pathways among two or more microbial strains. The division of microbial labour allows resource optimization, thus reducing metabolic burdens and increasing the efficiency of the whole process. In an interesting example E. coli and the yeast S. cerevisiae were used for the production of oxygenated taxanes. Both strains were engineered so that E. coli produced taxadiene, which in turn was used by S. cerevisiae to produce oxygenate taxane [26]. In this SMC, E. coli utilized xylose as carbon source and produced acetate, which in turn was used by S. cerevisiae. Unidirectional distributed consortia have been extensively used for phenylpropanoid production (see Table 1). For example, naringenin was produced by a SMC where an E. coli strain was engineered to produce p-coumarate while another E. coli strain was in charge of synthetizing naringenin from p-coumarate [27]. A similar strategy was used to produce resveratrol and resveratrol glucosides using E. coli-based synthetic consortia [28], [29]. The functional specialization in these kinds of consortia can be addressed by more than two microbial strains. For example, Li et al. [30] used three genetically modified strains of E. coli for the production of rosmarinic acid and four different engineered E. coli strains were used for synthetizing anthocyanins[31] (see Table 1 for details).
2.4. Multidirectional distributed
In this category, all members of the SMC have a role in maintaining both the stability of the system and its functionality. Multidirectional Distributed SMC are significantly more complex and require substantial metabolic engineering of the involved strains to ensure stability and functionality. One example is a consortium comprising two E. coli strains for the production of salidroside [32]. Both strains were engineered to establish cooperative metabolite cross-feeding so that each strain complemented the other’s auxotrophy. One of the strains produced tyrosol, which was used by the other to synthetize salidroside. To avoid competition over the carbon source, both strains were also engineered so that the tyrosol producer used xylose and the salidroside producer used glucose.
Overall it becomes apparent that increasing the complexity of SMC in terms of stability and functionality and using complex feedstocks delivers cost-effective production of increasingly complex metabolites. However, upscaling SMC design from simple Unidirectional Non-Distributed to Multidirectional Distributed is not always straightforward, but is instead a trial and error process. Thus, more holistic approaches to microbial community engineering are needed. In this sense, biotechnological applications using monocultures benefit from simpler construction and application of metabolic models.
Given the clear advantages that modelling contributes to the development of more precise and complex SMC, in the following sections we provide a comprehensive review of classical and recent computational modelling methods developed to describe and engineer natural and synthetic microbial consortia.
3. The long journey of community-based modelling: From ecological to genome-scale models
Multiple community-level modelling approaches have been developed to gain insights in the understanding of complex ecosystems [33], [34]. Overall, they have been classified as ecological, individual-based and metabolic models [35], [36]. Ecological models describe communities using ecological parameters such as pairwise interactions, growth rates, etc. and interactions depend mainly on correlations. Individual-based models focus on the individual rather than the population level. Finally, just as ecological models predict interactions between components of a given system, metabolic models predict interactions in a metabolic context while providing a community dynamics description. The lack of knowledge of kinetic parameters has promoted wide use of stoichiometric, constraint-based rather than kinetic models [37].
3.1. Ecological models
Ecological models focus on representing and/or discovering potential interactions among different species [38], [39]. They include mainly the evolutionary game theory and non-linear dynamics where evolution is driven by stochastic processes [40]. Evolutionary game theory emerges as an adaptation of the classical game theory to biological systems after stating that the assumption that the success of one individual depends on the choices of others does not apply in biology [41]. Thus, in evolutionary game theory, natural selection and mutation are what drive change in biological communities. This theory has been used to explain the behaviour of microbial communities in terms of interactions such as cooperation [42], [43] or competition [44]. A new application of game theory combined with metabolic models has been recently suggested in community modelling to infer evolutionary stable interactions by analysing the behaviour of a pair of microbes with complementary autotrophies and cross-feeding relationships [45].
Lotka-Volterra (LV) are the first non-linear dynamic systems describing biological populations with a mathematical model based on ordinary or partial differential equations. LVs are deterministic and do not consider the randomness present in nature. From a static point of view, they are used to model similarity- or regression- or rule-based networks [46]; while from a dynamic point of view, generalized Lotka-Volterra (gLV) is the main approach [47], [48]. gLV requires knowledge of the growth rates and the strength of the interactions between different components of the community. gLV equations have been used to model a variety of different microbial communities including cheese fermentation communities [49], marine phage communities [50] and the human microbiome [51], [52], [53]. In this last case, gLV was extended to take into account external perturbations over time [51], and successfully applied to predicting species abundances in the community [52]. Finally, it was possible to qualitatively infer interaction types without a dynamic model, quantitative assessment of interaction strengths and growth rates when gLV was considered [53].
3.2. Individual-based modelling.
In individual-based modelling (IBM), otherwise known as agent-based modelling [54], [55] microbes are individually simulated as concentration state variables rather than at population-level. Each cell evolves over time following predefined probabilistic rules that introduce the randomness required to model dynamics. This approach includes genes, transcripts, proteins and metabolites, although usually just a representative subset of them is selected to reduce the complexity. Its main advantage is that they take intra-population heterogeneity into account. For example, IBM has been used to model biofilms with a microbial consortia dynamics simulator set up on a high performance computing platform [56]. A detailed review of IBM models is included in [57].
3.3. Genome-scale metabolic models (GEMs)
GEMs are structured representations of a target organism based on existing genetic, biochemical and physiological information. Therefore, GEMs represent the metabolic capabilities of a particular organism and can be used in combination with algorithms such as Flux Balance Analysis (FBA) to predict phenotype from genotype [58], [59], [60]. Before applying FBA, the metabolic network of a given organism is converted into a numerical stoichiometric matrix (S), where rows describe individual metabolites and columns describe reactions. Cells’ subscripts < i,j > refer to their row (i) and column (j). To predict growth (Z) and the vector v of unsolved individual flux values for each metabolic reaction, a linear programming optimization problem can be solved by maximizing a given objective function Z = sT ·v, subject to a set of constraints (Eq. (1)). FBA assumes the steady-state with mass-balance constraint (S v = 0), where metabolites’ change in concentration as a function of time equals zero. Flux is constrained for each reaction by defining its lower (lb) and upper (ub) bounds in mmol/g of dry weight * h) units. Z is usually set to maximize biomass (g/L), although any metabolic reaction could be selected as a target for flux optimization. A good example is setting the bounds for a particular metabolite of interest’s exchange reaction to maximize its production rate.
| (1) |
An important advantage of metabolic models is that they provide accuracy without requiring kinetic information [60]. Therefore, it is not surprising that this modelling approach has started to be used successfully irrespective of the level of complexity, i.e. from individual organisms right up to microbial communities. Therefore, GEMs are seen by many as optimal computational tools for optimizing SMC-based biotechnological endeavours. The GEM design procedure includes a posteriori experimental validation of the model, including a performance assessment using different carbon sources, gene essentiality and flux prediction in known given conditions. Flux prediction could be validated with experimental techniques that retrieve in-vivo fluxomics data, i.e. 13C metabolic flux analysis (MFA), based on stable isotope tracing studies [61]. For instance, 13C-MFA was extended to measure metabolic fluxes at the microbial community level [62] and further improved by avoiding cell separation. This allows quantification of microbe-specific fluxes and metabolite cross-feeding rates and has been applied successfully with E. coli biofilms [63].
Community models based on GEMs have been used at microbial scale, but also to create multi-tissue models such as the human liver and to predict the effect of kinetically-modelled drugs [64]. Interestingly, this approach has also been applied to the construction of whole-plant models including the leaves, the stem, the seeds and the roots of barley [65]. The combination of several genome-scale models in a community, however, entails several challenges [66]. For instance, defining a community’s target function is a tricky point both in biological and mathematical terms. On the other hand, setting exchange rates and metabolic fluxes to constrain the models in the context of a community is a new topic. Current techniques measure the in vivo data for individual organisms and are thus not applicable to the community. Instead, the individual contribution of each organism must be measured. An additional challenge is to define the composition of the medium for combined culture taking into account the exchange of metabolites within the SMC.
Due to the increasing interest and applicability of GEMs, we focus here on the different approaches available so far to model microbial communities using these metabolic models. Two different stages can be distinguished in microbial community modelling with GEMs: The first stage includes descriptive methods to understand and describe communities. The second level of development, which took off recently, focuses on building methods to describe and engineer these microbial communities.
4. Dynamics-based classification of descriptive approaches to metabolic modelling
Descriptive methods for metabolic modelling of microbial communities are useful to describe how consortia work, to understand them and to identify relationships within the community. However, they cannot be used to engineer consortia for reasons explained below. Several attempts have been made to classify and categorize different approaches to modelling microbial communities. The first classification was based on available knowledge about the community and its complexity [67], [68]. An alternative categorization was based on the scope of the community, as defined by Bosi et al [69]. A most recent classification is based on the definition of the target function (simplified linear, multi-level or non-linear function) [70]. In order to complement current approaches, we propose a classification based on microbial community dynamics (see Table 2). Thus, we classified available descriptive methods as static/unified, static/multi-part and dynamic.
Table 2.
Descriptive microbial community modelling methods classification. The ‘In-vivo consortia categories’ defines the most complex category from those defined in Fig. 1 that could be modelled with the descriptive computational approach (both unidirectional and multidirectional could be modelled in all computational categories). There are additional multiple ad-hoc algorithms or methods not listed in the ‘Tool’ column but collected in Table 3.
| In-vivo consortia categories | Properties | Tool | |
|---|---|---|---|
| Static/Unified | Uni/Multidirectional Non-Distributed |
|
MO-FBA/FVA, Kbase |
| Static/Multi-part | Uni/Multidirectional Distributed |
|
OptCom, cFBA, Mminte, SteadyCom, Microbiome modeling toolbox, CarveMe, MICOM |
| Dynamic | Uni/Multidirectional Distributed |
|
DMMM, d-OptCom, COMETS, MCM, evoFBA, BacArena, Daphne, MMODES |
4.1. Static/Unified methods
This approach considers all strains unified in a common metabolic model, with only one copy of the shared reactions and metabolites. The model is completed by adding strain-specific metabolic content and a combined community-based biomass target function. This approach, also called ‘lumped network’ or ‘enzyme soup’, is the simplest and, although only useful to have a general perspective of how the community works, it allows high scalability (Table 3). In network-based models, the unified approach would be the closest as it considers all reactions in a single graph, irrespective of stoichiometry, which is ignored in favour of topology. Network-based models consider the metabolic reactions of each of the strains in the community to plot a graph where metabolites are represented by nodes connected to each other following the direction of metabolic reactions, i.e. from substrates to products. Reactions are in turn represented by edges. This approach could be applied to poor-quality GEM reconstructions because the main source of data is the reaction sequence. Some tools or algorithms that follow this unified approach are:
-
•
Borenstein’s group uses a graph or network-based community model representation that does not consider stoichiometry. With this unified static approach, they mainly study relationships among different microbes [71].
-
•
Kbase is a community data-driven network reconstruction [72]. It builds a single community model rather than aggregating individual models and is focused on predicting interactions between species in a community. In the absence of data for certain species due to lack of individual cultivability, this approach uses relevant community-level data as input. Single and community modelling is carried out using the Kbase software platform, including automatic gap-filling analysis by providing a particular community-based growth condition (www.kbase.us) [73].
-
•
MO-FBA and MO-FVA, multi-objective FBA and FVA (Flux Variability Analysis) algorithm extensions to community level [74]. These methods model microbial consortia by grouping several constraint-based individual models in a large, combined stoichiometric matrix. The multi-objective feature allows weighted combination of each strain’s individual objective.
Table 3.
Applications descriptive microbial community modelling approaches. There are three blocks corresponding to the descriptive modelling approach category described in Table 2. The ‘tool’ column includes the name of the algorithm or method defined in that application to describe the communities, and link to the software if it is available. ‘In vivo validation’ column indicates if the application has been validated with in vivo data or they are in silico-based results.
| Modelled Species | Application | In vivo validation | Tool | Ref. |
|---|---|---|---|---|
| Static/unified | ||||
| Several anaerobic fermentative strains | Description of product formation in fermentative conditions, from glucose depending on pH and substrate concentration. | No | ad hoc | [157] |
|
− 478 species − 154 human microbiome species |
Large-scale studies based on integration of metabolic capabilities in a common network with multiple species, with nodes representing metabolites and edges connecting substrates to products. Phylogenetic analysis and prediction of interactions based on that metabolic network. | No | ad hoc | [158], [159] |
| Assorted 113 bacterial species | Study of metabolic variability and cohabitation categorize interactions versus growth rate. | No | ad hoc | [160] |
| Synechococcus spp, Chloroflexus spp, and sulfate reducing bacteria | The first microbial consortia modelling classification, representing the consortium with different approaches. Description of relative abundances, biomass productivity and generation of toxic by-products. | No | ad hoc | [67] |
| Clostridium cellulolyticum, Desulfovibrio vulgaris Hildenborough, and Geobacter sulfurreducens | Study of trophic and electron accepting interactions of subsurface anaerobic environments. | Yes | ad hoc | [161] |
| 2 naphthalene-contaminated soil communities; with 13 and 12 species, including: Achromobacter, Azospirillum, Comamonas, Achromobacter and Pseudoxanthomonas[162]. | Description of common metabolic network of naphthalene-degrading bacterial communities based on metaproteomic and taxonomic data. | Yes | ad hoc | [163] |
| 261 assorted species of diverse habitats, such as soil, water and the human gut. | Study the extent of resource competition and metabolic exchanges in over 800 microbial communities. | No | ad hoc | [164] |
| Microbialites and microbial mats (structures similar to corals and stromatolites) | Study of autotrophic capabilities (identification of pathways for C and N assimilation) with a metabolic network based on metagenomics data. | No | ad hoc | [165] |
| Thermosynechococcus elongatus BP-1 and Meiothermus ruber Strain A | Study of photoautotrophic cyanobacterium-heterotroph consortium. | No | KBase | [72] |
| The same as Taffs et al., 2009[67](see above) | Description of ecosystem of hot spring microbial mats, with different behaviour between day and night. | No | MO-FBA/FVA | [74] |
| Static/multi-part | ||||
| D. vulgaris and Methanococcus maripaludis | Study of mutualistic interactions between sulphate-reducing bacteria and methanogens, predicting fluxes (intracellular and exchange between species). | Yes | ad hoc | [75] |
| Clostridium butyricum and Methanosarcina mazei | Studying a syntrophic interaction to increase methane production in anaerobic conditions, with an efficient consumption of by-products. | No | ad hoc | [166] |
|
- Hepatocyte (liver), adipocyte (fat) and myocyte (skeletal muscle) human cells - leaf, stem and root of Arabidopsis thaliana cells |
Defining multi-tissue models, to study diabetes in human (including gene expression data) or analysing how Arabidopsis minimizes energy usage for plant growth. | No | ad hoc | [79], [167] |
| Plasmodium falciparum and the host red blood cell (erythrocyte) | Study of the metabolism of malaria infection, over different life cycle stages of the pathogen. | No | ad hoc | [168] |
| E. coli, Bacillus subtilis, Helicobacter pylori, Salmonella typhimurium, Methanosarcina barkeri, S. oneidensis and Methylobacterium extorquens | Estimation of medium composition to allow symbiosis between binary pairs of species. | No | ad hoc | [77] |
| 46 pairs of auxotroph E. coli | Description of synthetic mutualism interactions in auxotrophic E.coli. | Yes | ad hoc | [169] |
| The same as Stolyar,2007, Taffs et al., 2009 and Miller et al.,2010[67], [75], [161](see above) | Quantifying a syntrophic association; assessing the level of sub-optimal growth in phototrophic microbial mats depending on community composition; and evaluating the direction of inter-species metabolite and electron transfer. | No | OptCom | [80] |
|
- Two imaginary species ‘i’ consuming glucose and ammonium and producing succinate and species ‘j’ consuming succinate, fixing nitrogen gas and excreting ammonium. - E. coli polymorphism in Long Term Experimental Evolution experiment[170]. |
Analysis community parameters (relative biomass abundances, etc) at balanced growth. | No | cFBA | [78] |
| Geobacter metallireducens and G. sulfurreducens | Study of interspecies electron transfer mechanisms in syntrophic associations, in genomic and transcriptomics. | Yes | ad hoc | [171] |
| Bacteroides thetaiotamicron, Eubacterium rectale and Methanobrevibacter smithii | Prediction of interactions between 3 key representative bacteria in the human gut, and analysing their individual contributions to secrete SCFA. | Yes | ad hoc | [100] |
| Bifidobacterium adolescentis L2-32 and Faecalibacterium prausnitzii A2-165 | Predicting demand for acetate and production of butyrate, in 2 gut strains related to Chron’s disease, using OptCom tool. | No | ad hoc | [172] |
| Ketogulonicigenium vulgare and Bacillus megaterium | Understanding of vitamin C production by an artificial consortium, study of subsystems and other possible metabolites to secrete. | No | ad hoc | [173] |
| 11 representative gut microbes (E. coli, H. pylori, Salmonella enterica, S. thermophilus, etc.) | Study of interactions between gut microbes and human small intestinal enterocytes, under anoxic and normoxic conditions. | No | ad hoc | [103] |
| Leptospirillum ferriphilum and Ferroplasma acidiphilum | Study of bioleaching (oxidizing iron) in a bacteria-archaea consortium presents in natural environment, with chemo-mixotrophic growth. | No | ad hoc | [174] |
| AOB, ammonia oxidizing bacteria: Nitrosomonas europaea, Nitrosomonas eutropha, Nitrosospira multiformis, and Nitrosococcus oceani. And NOB, nitrite oxidizing bacteria: Candidatus Nitrospira defluvii, Nitrobacter winogradskyi, Nitrobacter hamburgensis, Nitrospina gracilis. | Assessment of NO redox reactions contributes to N2O formation during nitrification, in 9 different consortia with variable composition selected among 4 AOB and 4 NOB. | Yes | ad hoc | [175] |
|
- Human Microbiome Project data - Desulfovibrio piger, B. thetaiotaomicron, Bacteroides caccae, Bacteroides ovatus, E. rectale, Marvinbryantia formatexigens, Collinsella aerofaciens, E. coli and Clostridium symbiosium |
Exploring pairwise microbial metabolic interactions, using 16S data from microbiome studies. Evaluating a sulphate-reducing bacteria growth in gut microbiome with different diets with data from [176].. | No | MMinte | [82] |
| − 4 E. coli auxotrophic for amino acids- Gut microbiome | Maximizing community stability (common growth). | No | SteadyCom | [83] |
|
− 74 human gut bacterial strains from AGORA collection − 5587 species from NCBI RefSeq in groups of 20 strains per community |
Automatic reconstruction of single strain models (from 238 to 2472 reactions per model) with the possibility to merge in a community one, analysing the number of compounds that can be exchanged. | No | CarveMe | [84] |
| Human gut strains from AGORA collection[102]and human cells (Brunk et al., 2018)[177] | Analysis of pairwise interactions (microbe-microbe and host-microbe) of different types (competition, parasitism, etc.) with a join matrix of GEMs, and modelling of microbial communities given the relative abundances, used to personalize community biomass reaction and simulating under different diets. | No | Microbiome modelling toolbox | [85] |
| Human gut strains from AGORA collection | Predicting growth rates and metabolic fluxes from microbe abundances as input. Using an heuristic optimization approach based on L2 regularization to allow different growth rates per strain. | No | MICOM | [86] |
| Dynamic | ||||
| E. coli | Exploring the metabolic variability among bacterial strains and identifying interactions, across different single-carbon-source conditions. They use a combination of a graph-theoretic approach together with a metabolic model. | No | ad hoc | [178] |
| Clostridium acetobutylicum and Clostridium cellulolyticum | Improving bioprocessing of cellulose with a clostridial consortia, with DMMM. | No | ad hoc | [179] |
| G. sulfurreducens and Rhodoferax ferrireducens | Designing of uranium bioremediation scenarios with two competing heterogeneous species | No | DMMM | [88], [180] |
|
- E. coli auxotrophs - G. sulfurreducens, R. ferrireducens, and S. oneidensis |
Study of impact of lactate vs acetate addition on the composition of uranium-reducing community. In-vivo validation of E. coli auxotrophs with Wintermute and Silver, 2010 [181] results. | Yes | d-OptCom | [81] |
| E. coli, S. enterica and Methylobacterium extorquens | Simulation of spatiotemporal dynamics of microbial communities, predicting species ratios and investigating the influence of spatial structure on competition in mutualistic systems, and with a competitor between the cross-feeding pair. | Yes | COMETS | [89] |
| Homogeneous E.coli consortia | Combining metabolic model with statistical analysis and calibration to experimental data, in this case related to Lenski’s experiment LTEE. | Yes | MCM | [91] |
|
- E. coli and S. enterica[87] - B. fragilis, B. longum, C. difficile, E. coli, H. pylori and L. acidophilus |
Visualization of metabolic interaction networks between microbes in a community. | No | VisANT | [90] |
| E. coli (E. coli B, not K12) | Analysis of evolution. LTEE: divergence in glucose-limited conditions, with daily transfers. | No | evoFBA | [182] |
|
- Clostridium beijerinckii and M. barkeri - Anaerostipes caccae, B. thetaiotaomicron, Bifidobacterium longum, Blautia producta, Clostridium ramosum, E. coli and Lactobacillus plantarum |
Analysis of interactions and spatial and temporal distributions of microbes in communities using individual-based metabolic modelling. | No | BacArena | [94] |
| L. plantarum | Study of cross-feeding with short-chain fatty acids from glucose in the human gut microbiome, using DMMM with spatial addition. The L. plantarum GEM is converted in a ‘supra-model’ increased by pathways crucial in carbohydrate fermentation in the colon. | No | ad hoc | [183] |
| N.s europaea and N. winogradskyi | Study of the dynamics of nitrification-derived N oxide production, with aerobic ammonia- and nitrite-oxidizing bacteria, using DMMM. | Yes | ad hoc | [184] |
| E.coli | Analysis of diauxic shift in two homogeneous subpopulations, combining ordinary differential equations (ODE) with GEMs. | No | Daphne | [95] |
|
- F. prausnitzii and B. adolescentis - P. aurescens, H. stevensii, Halobacillus sp. |
Simulation of heterogeneous microbial communities behaviour over time with ODE and GEMs under perturbations, i.e. changes in availability of metabolites and biomass of different strains. | No | MMODES | [96] |
4.2. Static/Multi-part methods
This category of models preserves the individual metabolic matrices and introduces a pool of metabolites, which could be defined by pre-fixed reactions (guild compartment in other classifications) or by new stoichiometric reactions after an initial optimization step (bi-level optimization). Single strain models are directly connected by exchange reactions, assuming no change in the concentrations of extracellular metabolites and no accumulation in the medium. This approach has been profusely applied to describe microbial communities (Table 3). Several algorithms that fit this category are summarized below:
-
•
The method described by Stolyar et al [75] provided the first metabolic model of a microbial consortium and the distribution of its associated metabolic fluxes. This method has since been used in applications pursuing different objectives, such as categorizing interactions [76], estimating medium composition [77], predicting relative biomass abundances [78] and defining a host-pathogen interaction between the human alveolar macrophage and M. tuberculosis in a multi-tissue model [79].
-
•
OptCom [80] and d-OptCom [81]: are two closely related methods focusing mainly on engineering microbial communities (see a longer description in section 5), although descriptive versions are also available.
-
•
cFBA [78]: this method assumes a balanced and fixed growth rate for all microbes in the consortium. Subsequently, cFBA (community FBA) maximizes this growth rate using a non-linear multi-objective function. This approach implies constant species abundance ratios in the community and is applicable to cells grown in chemostat or in waste-water microbial community scenarios [66].
-
•
MMinte [82]: this method supports the assessment of the pairwise microbial metabolic interactions that occur in a community model limited to two strains. Metabolic models are automatically reconstructed using ModelSEED and metagenomics data as input (16S rRNA sequences).
-
•
SteadyCom [83]: this system maximizes community stability, i.e. constant growth rate across all microbes in the community, with an iterative linear programming approach. Additionally, it applies FVA to predict microbial abundances under changing uptake rates.
-
•
CarveMe [84]: this focuses mainly on automatic reconstruction of single strains. In addition, it allows to automatically merge several single-species models into a single community model with a common or individual extracellular compartments.
-
•
Microbiome modelling toolbox [85]: a COBRA Toolbox extension to analyze microbial communities and study interactions (intra- or with the host).
-
•
MICOM [86]: this static approach predicts growth rates and fluxes from in to vivo data such as species abundances in a microbiome sample. Consequently, it infers metabolic interactions among the microbiota.
4.3. Dynamic methods
Static approaches ignore temporal events. The dynamic or hybrid approaches are based on dynamic Flux Balance Analysis (dFBA) [87], which allows representations of the community’s temporal evolution, including variations of metabolite concentrations and cell densities over time. This is the preferred approach to simulate microbial interactions because shared metabolites vary dynamically. It is however limited by the fact that dynamic approaches entail kinetic parameter configuration and require higher computational resources, running FBA multiple time points per strain and thus limiting the analysis to smaller sized communities.
-
•
DMMM [88]: this was the first method that used dFBA at community level. DMMM optimizes growth rates for each strain.
-
•
COMETS [89]: in addition to dynamic simulation of communities using dFBA, this algorithm considers the cells’ spatial distribution. The biomasses and fluxes per time points reported as output can be visualized using the VisANT tool [90].
-
•
MCM [91]: this framework simulates dynamic community models and adds statistical evaluation and parameter calibration based on experimental data. It was initially tested with a homogeneous E. coli community and subsequently with species assemblages in nitrifying and methanogenic bioreactors [92], [93].
-
•
BacArena [94]: this combines metabolic modelling with individual-based modelling instead of using population-based modelling (one model per strain with a certain amount of biomass). Therefore, BacArena supports modelling of metabolically heterogeneous populations where each individual cell is represented by a unique metabolic model depending on its spatial resource allocation. In BacArena, metabolite diffusion (implemented with partial differential equations) produces gradient concentrations resulting in spatial niches where different metabolic pathways are activated. It is also able to predict novel cross-feeding interactions through fermentation products. COMETS and BacArena allow spatial resolution taking diffusion parameters into account.
-
•
Daphne [95]: Daphne combines two different modelling strategies: GEM (metabolism) and ODE (Ordinary Differential Equations). ODE supports modelling the strain’s growth kinetics and the medium metabolite consumption and production dynamically. It is underpinned by a set of equations that can be solved mathematically.
-
•
MMODES [96]: this also integrates GEMs and ODEs to simulate biomass and metabolite dynamics over time. In addition, it is possible to add perturbations using external longitudinal interventions such as changes in the medium and/or strain ratio (e.g. increasing a metabolite concentration and/or biomass of a species).
4.4. Practical applications of descriptive modelling methods
Overall, each of the three categories of descriptive methods described above are suitable for modelling different microbial scenarios and their interactions. The unified approach is appropriate for multiple strain systems and/or where knowledge is limited, e.g unknown details of the individual assignment of reactions and metabolites, such as in metagenomics. Quantifying metabolic fluxes and representing inter-species interactions requires more complex approaches, such as the multi-part or dynamic models. However, the dynamic approach is the only suitable one to model medium composition and predict metabolite concentration because the multi-part feature transfers metabolites from one model strain to another. Therefore, the dynamic approach is better able to represent complex situations in microbial communities. Dynamic approaches consider all time-dependent elements, although they only can be applied to small communities because they require describing individual strains in great detail and are more time consuming than the other approaches. Nevertheless, they are recommended for engineered or synthetic microbial communities used in biotechnology applications, i.e. scenarios where species richness is generally low. Computing requirements tend to increase from Static/Unified to Static/Multi-part to Dynamic approaches. This is particularly true for Dynamic approaches because dFBA requires solving each GEM for every time slot in the time-series. Final requirements will depend on the size of the strain models (number of reactions and number of metabolites) and any additional factors, such as the length of the time-series.
Static/Unified, Static/Multi-part and Dynamic approaches have been used to model a large number of microbial communities (see ‘modelled species’ column in Table 3) in a range of scenarios; such as food biotechnology [97], human health (including GEMs for microbes, tissues and organs) [98] and marine microbiome [99]. Table 3 collates applications to microbial consortia from a descriptive point of view. Applications have been grouped according to the categories defined in Table 2. The most extended approach is the static/multi-part one, which has around twice the number of applications of static/unified and dynamic approaches. In some cases, the study defines a new computational method, while in others the methods are re-used. Sometimes the corresponding in vivo consortium has been deployed, although this is generally not the case. Microbial consortia applications are sorted according to: i) species richness (some are monoclonal populations, others are consortia comprising less than 10 strains or hundreds of heterogeneous strains, such as those present in the gut microbiota), ii) species diversity (ranging from only one cell per strain to large consortia with hundreds of cells per organism) and iii) environment, industrial bio-transformations, human health and plants.
The application of these methods to human gut microbiome modelling has received special attention [70]. Despite improvements in sequencing having broadened our knowledge of the components of the gut microbial community, the relationships among them and between them and the human cells remain mostly unknown. Hence, microbial community modelling techniques contribute to improving our understanding of the complex behaviour of the gut microbiome and its associations with human diseases. In many cases, only a small (less than 10) and simplified subset of representative species from the microbiome have been taken into account [100], [101]. However, in recent studies the size of the modelled microbiomes has been expanded to tens or even hundreds of species. Static/multi-part applications often focus on this scenario. It is noteworthy that Thiele’s group has modelled the metabolism of the entire human gut microbiota communities using constraint-based models [102]. This approach to modelling has been used to study the interactions between gut microbes and human intestinal enterocytes under anoxic and normoxic conditions [103], to predict levels of short chain fatty acids used to treat Crohn’s disease [104] and to determine whether metformin treatment increases agmatine production by gut microbiota, explaining changes throughout the host’s lifespan [105]. More recently, microbiome modelling has been used to develop human organ models [106]. Finally, in multi-omics modelling, metabolome data have begun to be combined with microbiome data [107], [108], [109]. In general, integration of different omics data, such as metagenomics, proteomics, metabolomics and fluxomics with GEMs offer wider modeling scenarios [110], [111], [112].
5. Engineering metabolic modelling: Design and optimization
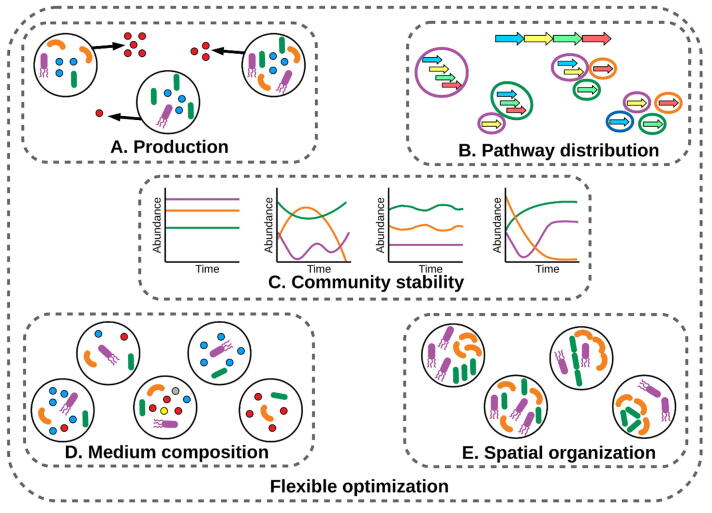
All of the descriptive approaches discussed in the previous section are non-optimizing modelling methods. In the context individual cell modelling, following the development of descriptive methods, new tools to design and engineer high performance strains were profusely developed [113], including strain designing algorithms such as OptKnock [114], OptStrain [115], OptGene [116] and GDLS [117]. Unfortunately, progress at community level has not caught up, i.e. most of the current approaches support neither design nor optimization of natural and/or synthetic microbial communities. However, pioneering efforts in this field pave the way for future development of community-based design and optimization methods. A set goal is to be able to optimize SMC based on their final application. Key parameters/goals to be optimized include, but are not limited to, production, pathway distribution, community stability, medium composition, spatial cell organization and a combination of goals, i.e. a flexible objective. Therefore, beyond methodological classifications [59], the applications of microbial community engineering can be also grouped according to their optimization goal (see Fig. 2). In this context, there are a few tools that can be considered generic, i.e. they could be used to optimize several applications (see Table 4). However, multiple applications have been developed as ad hoc systems to optimize very particular tasks (Table 4). In the following sections we classify and describe in detail the first GEM-based attempts to design microbial communities for biotechnological applications.
Fig. 2.
Microbial community optimization/design goal categories. Section 5 describes each category in detail. Table 4 shows detailed applications and methods of these different categories. A. Optimize production of a metabolite of interest (red circle) depending on community parameters. B. Optimize distribution of the reactions within a metabolic pathway among different strains. C. Optimize individual strain growth to reach a stable community over time. D. Optimize concentration of nutrients (circles) available in the microbial community medium. E. Optimize physical distribution of the strains in the community. The flexible optimization category covers all the optimization goals. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 4.
Engineering modelling applications. Grouped by the optimization community goal. Focus on optimization/engineering topics. ‘Production’ group includes to optimize different community parameters (strains ratio, carbon source ratio, initial biomass, etc). GR = Growth Rate. Output means the configuration parameters that are predicted. If there is a software available, it is referred to and linked in the column ‘references’ too.
| Specific goal of optimization | Output | Strains | Results and additional details | Ref. |
|---|---|---|---|---|
| Production | ||||
| Maximizing ethanol production |
|
|
|
[118], [185], [186] |
| Maximizing flavonoids production |
|
E. coli strains (flavonoid pathway fragmented in 2 strains) |
|
[119] |
2 maximization goals:
|
|
- D. vulgaris - M. maripaludis - M. barkeri |
|
[131] |
| Maximizing yield |
|
|
|
[120], [188] |
Maximizing (together):
|
|
- D. vulgaris - M. maripaludis |
|
OptCom [80] |
Maximizing (together):
|
|
|
|
OptCom [80] |
Maximizing (together):
|
|
|
|
OptCom [80] |
| Maximizing uranium reduction |
|
|
|
OptCom [81] |
2 cases of study:
|
Interventions in medium composition or biomass of strains |
|
|
MDPbiomeGEM [96] |
| Pathway distribution | ||||
| Optimizing metabolite secretion Secondary goal: medium composition |
|
122 strains (6 from [77]) and 116 from [76] combined in > 6500 different consortia of 2 members |
|
[189] |
| Maximizing growth or compound yield | Allocated reactions per strain | 2 generic bacteria with reduced central carbon metabolism |
|
[122] |
| Minimizing number of species | Selected species to combine in the community | Human gut microbiome |
|
CoMiDA [121] |
2 cases of study:
|
All reactions to include and their distribution among strains |
- Streptomyces cattleya and M. barkeri (selected from 4 strains) - K. pneumoniae and M. mazei |
|
MultiPlus [124] |
| Optimizing metabolic exchange rates |
|
|
|
[190] |
| Surviving under constraints | Cross-feeding partnerships and division of labor | E. coli (2–3 strains) |
|
DOLMN [125] |
| Maximizing ethanol yield | KO in strains |
S. cerevisiae E. coli |
|
BioLEGO 2 [126] |
| Stability | ||||
Maximizing (together):
|
strains ratio | Auxotrophic E. coli pairs: (argH-lysA) (lysA, trpC) (metA, ilvE) |
argH-lysA: 0.8:0.2 in vivo and 0.97:0.03 in silicolysA-trpC: 0.9:0.1 in vivo and 0.98:0.02 in silicometA-ilvE: 0.15:0.85 in vivo and 0.15:0.85 in silico | dOptCom [81] |
| GR in auxotroph evolution | strains ratio | E. coli lysine and leucine KOs long-term |
|
[156] |
| Common growth Secondary goal: spatial distribution |
|
|
|
COMETS [89] |
| GR with optimum distribution of resources |
|
E. rectale or F. prausnitzii, B. thetaiotaomicron, B. adolescentis and R. bromii |
|
CASINO [101] |
| Common growth |
|
|
|
SteadyCom [83] |
| Medium composition | ||||
| Minimizing the cost of metabolic cooperation | Combination of nutrients allowing synergistic growth | E. coli arginine and leucine KOs |
|
[127] |
| Spatial organization | ||||
| Spatial Partitioning |
|
P. aeruginosa S. aureus (chronic wound biofilm) |
|
[129] |
| Spatial Partitioning |
|
2 case of study (reduced models):
|
|
IndiMeSH [128] |
| Flexible | ||||
| Optimizing PHA accumulated Secondary goal: production |
|
- S. elongatus - P. putida |
|
FLYCOP [59] |
| Stability maximization (common growth) |
|
4 E. coli auxotrophic for amino acids |
|
FLYCOP [59] |
| Several optimization goals: maximizing yield or biomass or GR, and minimizing time | Uptake rates per strain (glucose, acetate, oxygen) | 2 E. coli polymorphism:
|
|
FLYCOP [59] |
5.1. Production
The optimization goal in this group of applications is often maximizing production parameters in terms of either production rates, yields or titers of a specific metabolite of industrial, health or environmental interest. The outputs are parameters used to design the community that fulfils this aim, e.g., strain ratio, C-source ratio, metabolite uptakes, initial biomasses, cross-feeding rates, etc.
The main generic method in this group has been developed using OptCom’s optimization capabilities [80]. OptCom implemented two-level optimization for single strains and communities. By default, OptCom optimizes the community biomass by assuming fixed single strain growth, and returns strain ratio, substrate uptakes and secretion rates. OptCom has been applied to different microbial communities: a syntrophic association through hydrogen between D. vulgaris and M. maripaludis [80], a phototrophic microbial community based on Synechococcus spp. in daylight metabolism as the primary feeder [80], sub-surface anaerobic environments with electron accepting interactions [80] and communities involved in uranium reduction [81].
Apart from OptCom, some ad-hoc methods have been developed to optimize production: to maximize ethanol production with S. cerevisiae and E. coli [118]; to maximize flavonoid production with E.coli strains using a scaled-Gaussian model [119] and to maximize yield with three E.coli mutants in a chemostat model of competition for a simple sugar (glucose limited conditions) [120].
5.2. Pathway distribution
Methods and applications in this category support consortia engineering by fragmenting and distributing a given complex metabolic pathway between the components of a consortium. This allows division of labour through metabolite exchange, i.e. intermediate metabolites are secreted by one strain and then used by another. These are mainly graph-based methods [39] and thus allow easy identification of i) species responsible for producing a certain metabolite and ii) metabolite trafficking among strains. For example, CoMiDA [121] identifies putative sub-pathways responsible for synthetizing a target product from a series of given substrates while minimizing the number of necessary species in the community. Non-stoichiometry methods are the most usual among graph-based methods even though they are not independent and require post-processing steps (where stoichiometry is taken into account) to verify whether designs are plausible. Some approaches based on MILP (Mixed Integer Linear Programming) rather than FBA, consider stoichiometry to allocate reactions among the community’s single-strain metabolic models. This supports optimization of specific community goals (growth rate or uptake of one compound) [122]. Other approaches have been designed to expand the network with an agglomerative algorithm that adds reactions iteratively instead of fragmenting the network [123].
Generic methods have been also developed to optimize pathway distribution, including: MultiPlus (static/unified), DOLMN (static/multi-part) and BioLEGO 2 (static/multi-part). MultiPlus [124] starts with a hypergraph that integrates several GEM models and has two fixed objectives: minimizing the number of reactions and minimizing exchanged metabolites in a de novo synthesis pathway. Following a MILP optimization approach, DOLMN [125] identifies communities able to survive under constraints (e.g. limited number of reactions) that are difficult to identify manually. BioLEGO 2 [126] allows large-scale simulations of several simultaneous knockouts (KO) and runs comprehensive searches to identify the KOs maximizing ethanol yield.
5.3. Community stability
These applications predict optimal individual growth parameters resulting in stable communities over time. Two generic methods have been developed to optimize this goal, d-OptCom [81] and SteadyCom [83].
d-OptCom includes both descriptive (dynamic) and engineering approaches that are similar to those of its static version, OptCom. d-OptCom is a highly complex method, e.g. it requires a bilinear FBA solver. Optimization is based on a global search feature (BARON), kinetics parameters and additional LP constraints that need to be defined in order to configure a MILP problem with new reactions for new interactions between strains. In addition, d-OptCom is defined as a ‘comprehensive computational framework’ and does not provide any software that supports neither reproducibility nor the development of new applications. It was used to predict the optimal strain ratios in several auxotrophic pairs of E. coli consortia [81]. SteadyCom focuses on predicting a common growth rate for all members in a community and then expecting it to be stable. Contrary to other multi-objective methods such as d-OptCom and the flexible methods, it entails a fixed objective. SteadyCom requires linear FBA solver complexity and iterative LP-based optimization. Apart from FBA, SteadyCom is compatible with FVA. The method was applied to a multi-E. coli community with amino acid auxotrophy as proof of concept and a simplified human gut microbiome community that was reduced to 9 species to analyse the influence of fibre content from diet [83].
On the other hand, the ad-hoc CASINO toolbox [101] is a computational platform focusing on the human gut microbiome. It is designed to study metabolic interactions among microbial species and the host metabolism. From a static point of view, CASINO predicted alterations of amino-acid metabolism due to dietary interventions. This method follows a two-level optimization approach, similar to d-OptCom. It begins by maximizing growth rate at the individual species level to determine uptakes and subsequently optimizes growth rate and resource distribution at the community level. Contrary to other previous methods, CASINO requires experimental data (strain abundances, etc.) as input to configure models.
5.4. Medium composition
Applications in this category aim to predict the optimal concentration of metabolites in a given medium that deliver maximal community performance in terms of growth, production, decontamination, etc. A study by Zampieri and Sauer [127] describes an application to meet this optimization. It is based on a model that returns ideal medium composition to minimize the cost of metabolic cooperation in microbial ecosystems. The system maximized metabolite concentration of medium inputs to minimize the cost of shared essential metabolites while guaranteeing that growth was only possible within the consortium, not as individual strains. This is a comprehensive optimization approach that solves a two-level MILP problem with high computational complexity. A descriptive static/multi-part approach has also been used to optimize medium composition [77]. This used a pair of metabolic models to initially define a minimum medium containing all the metabolites required to sustain growth. Subsequently, they authors iteratively removed carbon sources to hamper growth and added new metabolites to recover growth. It was concluded that medium composition makes symbiotic relationships possible between binary pairs of 7 different strains.
5.5. Spatial organization
Methods developed to meet this objective predict the physical distribution of the community’s strains along a 2D/3D space. The only generic method in this group is IndiMeSH [128], which dynamically modelled bacterial dispersion and nutrient diffusion in a 2D pore network based on pore size and nutrient gradients. Modelling space implies simplifying other issues, such as using a simplified versions of GEMs (reducing numbers of reactions and metabolites from thousands to hundreds) or integrating all bacterial biomass per spatial unit in a single reaction regardless of intra-species variation. IndiMeSH was applied to soil habitats using two different consortia: a syntrophic community of E. coli with S. enterica, and a multi-strain community comprising the obligate aerobic P. putida and the facultative anaerobic P. stutzeri. BacArena and MMODES, listed above in the descriptive methods, include some spatial features, although optimizing spatial organization is not their principal goal. In an ad hoc application, a combination of GEMs and partial differential equations to describe metabolite diffusion resulted in a dynamic model that was able to predict biofilm thickness [129].
Spatially Linked Microbial Consortia (SLMC) is a conceptual design to engineer consortia. Spatial distribution is optimized using isolated modules and bespoke growth media to improve control and facilitate new strain combinations. SLMC is reviewed by Sala [130] by including GEMs in the process of designing compatible synthetic communities.
5.6. Flexible optimization
This section describes the usage of methods for goal-agnostic optimization, i.e. the optimization goal and the consortium-parameters-to-be-predicted can be independently defined and remain different in each case. They support design and engineering of microbial communities by selecting the consortium configuration that best optimizes a given goal. To the best of our knowledge, FLYCOP [59] is the only system with the ability to do this. The goal can be defined flexibly depending on the consortium’s functionality and a particular interest, e.g. community growth rate, stability, medium composition, etc. For example, FLYCOP can be configured to optimize medium composition by selecting metabolites and their initial concentrations from a finite list. FLYCOP’s flexible approach could contribute to improving metabolic modelling of microbial communities in ways that go beyond multiple optimization goals not limited to maximizing growth rate. One example of this are applications where we would seek to maximize yields of certain a product of interest when its synthesis pathway has been split between different strains. Another example would be the comparison optimization approaches for different products as is done experimentally in [26] with different fitness functions. Another advantage of FLYCOP’s flexible approach is that multiple parameters can be optimized at once rather than having to implement independent optimization processes for each parameter [118]. Besides, FLYCOP lends itself to applications with obligatory mutualistic communities where other engineering approaches do not, e.g. d-OptCom [81]. While production optimization methods are able to maximize yield at the individual cell level, FLYCOP optimizes reaction fluxes within the metabolic model. FLYCOP can manage applications involving GEMs with thousands of reactions where other methods are limited to small models. FLYCOP’s flexibility can also applied to single-strain models where each individual strain has a different growth rate, thus not requiring a single growth rate for all strains in the model as other applications do [78], [131]. Additionally, FLYCOP supports automatic search optimization versus the systematic assessment of different configurations. Regarding technical features, FLYCOP has linear FBA solver complexity and optimizes using a local search approach (SMAC).
Among the different categories of applications designed to engineer microbial communities using GEMs (see Fig. 2), the most widespread optimization goal is production followed by pathway distribution. Fewer examples focus on the optimization of other parameters, e.g., stability, medium composition, spatial organization and even a flexible goal. The most common consortia are two-strain (see ‘strains’ column in Table 4), although there are some with a higher number of strains, both homogeneous and heterogeneous.
6. Summary and outlook
As we realize that microbes are rarely found alone but in the context of complex communities, the need for computational tools able to provide mechanistic knowledge of how these communities work and evolve over time becomes critically important. Microbial communities are already recognized as key players in human health and they have begun to be seen as promising biocatalysts in biotechnology applications. Following the development of individual-cell modelling approaches and pioneering efforts on community-level modelling, it is largely expected that methods for the efficient analysis and engineering of such communities will spring up in the coming years. Combining GEMs with Machine Learning or Artificial Intelligence techniques have been suggested as promising developments for metabolic modeling [132], [133] and its application to metabolic engineering [134].
In this context, the debate over the real objective function at community level is a long running one [135]. The general assumption is that the microbial community's goal is to maximize growth under a natural selection scenario. However, optimizing biomass might not be the right microbial goal with genetically engineered organisms or when the environment is different from that where its evolution can occur [91]. Thus, alternative community configurations implying alternative goals are ignored by most of the available methods. It would therefore be interesting to have methods that support optimization of different community goals.
Current dynamic methods rely on the analysis of a few species and only static/unified methods can be used to analyse complex communities, thus hampering a deeper understanding of such communities. An important challenge to address in the near future is the development of tools to support dynamic analyses of large microbial communities in the context of high-quality GEMs [70]. This will require not just a larger GEM portfolio but also collecting large sets of kinetic parameters and developing novel computational methods to reduce the very time-consuming dFBA-based solving stage.
Beyond increasing the complexity of microbial community engineering using GEMs, it is important to note that the components of a given community often operate under different sets of conditions. Differences involving nutrient preference can be easily taken into account, so model-based analysis becomes very useful when defining a common medium that supports growth for all the strains in the consortium. However, GEMs cannot be used directly to model many other environmental conditions such as pH and temperature. Approaches based on the inclusion of omics data to constrain the models have shown to be an interesting alternative. In any case, applications of computational modelling to synthetic has only been tested with a set of microbes living in physiologically compatible environmental conditions.
Another important challenge in microbial community modeling is validation, i.e. proving model usability beyond the computational context. Experimental validation requires a very controlled environment to reduce microbial communities’ high complexity [136]. Validating community dynamics approaches is even more complex. Thus, experimental validation is currently viable when working with small size communities, i.e. two or three components, or synthetic communities. Validation becomes much more difficult when working with larger or natural communities (soil & gut microbiomes, etc.). In vitro simulators could be a suitable alternative [137], [138].
In the long term, model-guided microbial community engineering should trend towards the development of technologies capable of predicting potential genetic modifications at the community-level (similar to what the individual-level OptKnock or GDLS algorithms are capable of). The same applies to all the useful and comprehensive COBRA-related algorithms currently used to engineer individual strains.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the European Union's Horizon 2020 Research and Innovation Programme under Grant Agreements no. 814650 (SynBio4Flav) and 870294 (MixUp) and the “Severo Ochoa Program for Centres of Excellence in R&D” from the Agencia Estatal de Investigación of Spain (grant SEV-2016-0672 (2017-2021)) to the CBGP. BGJ was supported by a Postdoctoral contract associated to the Severo Ochoa Program. The authors thank Clive A. Dove for critical reading of the manuscript.
References
- 1.Park S.Y., Yang D., Ha S.H., Lee S.Y. Metabolic Engineering of Microorganisms for the Production of Natural Compounds. Adv Biosys. 2018;2(1):1700190. doi: 10.1002/adbi.201700190. [DOI] [Google Scholar]
- 2.Khalil A.S., Collins J.J. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11(5):367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar V., Baweja M., Singh P.K., Shukla P. Recent developments in systems biology and metabolic engineering of plant-microbe interactions. Front Plant Sci. 2016;7:1421. doi: 10.3389/fpls.2016.01421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Revuelta J.L., Buey R.M., Ledesma‐Amaro R., Vandamme E.J. Microbial biotechnology for the synthesis of (pro)vitamins, biopigments and antioxidants: challenges and opportunities. Microb Biotechnol. 2016;9(5):564–567. doi: 10.1111/1751-7915.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallero DA. Environmental biotechnology : a biosystems approach. 2015.
- 6.P. Dvořák P.I. Nikel J. Damborský V. de Lorenzo Bioremediation 3. 0: Engineering pollutant-removing bacteria in the times of systemic biology Biotechnol Adv 2017;35:845–66. 10.1016/j.biotechadv.2017.08.001. [DOI] [PubMed]
- 7.Yuan S.-F., Alper H.S. Metabolic engineering of microbial cell factories for production of nutraceuticals. Microb Cell Fact. 2019;18(1) doi: 10.1186/s12934-019-1096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H., Ding M.-Z., Jia X.-Q., Ma Q., Yuan Y.-J. Synthetic microbial consortia: from systematic analysis to construction and applications. Chem Soc Rev. 2014;43(20):6954–6981. doi: 10.1039/C4CS00114A. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein H.C., Carlson R.P. Microbial consortia engineering for cellular factories: In vitro to in silico systems. Comput Struct Biotechnol J. 2012;3(4):e201210017. doi: 10.5936/csbj.201210017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R., Zhao S., Wang Z., Koffas M.AG. Recent advances in modular co-culture engineering for synthesis of natural products. Curr Opin Biotechnol. 2020;62:65–71. doi: 10.1016/j.copbio.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Sgobba E., Wendisch V.F. Synthetic microbial consortia for small molecule production. Curr Opin Biotechnol. 2020;62:72–79. doi: 10.1016/j.copbio.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Amor DR, Bello MD. Bottom-up approaches to synthetic cooperation in microbial communities. Life 2019. https://doi.org/10.3390/life9010022. [DOI] [PMC free article] [PubMed]
- 13.Che S., Men Y. Synthetic microbial consortia for biosynthesis and biodegradation: promises and challenges. J Ind Microbiol Biotechnol. 2019;46(9-10):1343–1358. doi: 10.1007/s10295-019-02211-4. [DOI] [PubMed] [Google Scholar]
- 14.Qian X., Chen L., Sui Y., Chen C., Zhang W., Zhou J., Dong W., Jiang M., Xin F., Ochsenreither K. Biotechnological potential and applications of microbial consortia. Biotechnol Adv. 2020;40:107500. doi: 10.1016/j.biotechadv.2019.107500. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H., Wang X. Modular co-culture engineering, a new approach for metabolic engineering. Metab Eng. 2016;37:114–121. doi: 10.1016/j.ymben.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Antoniewicz M.R. A guide to deciphering microbial interactions and metabolic fluxes in microbiome communities. Curr Opin Biotechnol. 2020;64:230–237. doi: 10.1016/j.copbio.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Goyal G., Tsai S.-L., Madan B., DaSilva N.A., Chen W. Simultaneous cell growth and ethanol production from cellulose by an engineered yeast consortium displaying a functional mini-cellulosome. Microb Cell Fact. 2011;10(1):89. doi: 10.1186/1475-2859-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiyoshi K., Furukawa M., Seyama T., Kadokura T., Nakazato A., Nakayama S. Butanol production from alkali-pretreated rice straw by co-culture of Clostridium thermocellum and Clostridium saccharoperbutylacetonicum. Bioresour Technol. 2015;186:325–328. doi: 10.1016/j.biortech.2015.03.061. [DOI] [PubMed] [Google Scholar]
- 19.Fedeson D.T., Saake P., Calero P., Nikel P.I., Ducat D.C. Biotransformation of 2,4‐dinitrotoluene in a phototrophic co‐culture of engineered Synechococcus elongatus and Pseudomonas putida. Microb Biotechnol. 2020;13(4):997–1011. doi: 10.1111/1751-7915.13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L.i., Chen L., Diao J., Song X., Shi M., Zhang W. Construction and analysis of an artificial consortium based on the fast-growing cyanobacterium synechococcus elongatus UTEX 2973 to produce the platform chemical 3-hydroxypropionic acid from CO2. Biotechnol Biofuels. 2020;13(1) doi: 10.1186/s13068-020-01720-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minty J.J., Singer M.E., Scholz S.A., Bae C.-H., Ahn J.-H., Foster C.E., Liao J.C., Lin X.N. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc Natl Acad Sci USA. 2013;110(36):14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin H.-D., McClendon S., Vo T., Chen R.R. Escherichia coli binary culture engineered for direct fermentation of hemicellulose to a biofuel. AEM. 2010;76(24):8150–8159. doi: 10.1128/AEM.00908-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sgobba E., Stumpf A.K., Vortmann M., Jagmann N., Krehenbrink M., Dirks-Hofmeister M.E., Moerschbacher B., Philipp B., Wendisch V.F. Synthetic escherichia coli-corynebacterium glutamicum consortia for l-lysine production from starch and sucrose. Bioresour Technol. 2018;260:302–310. doi: 10.1016/j.biortech.2018.03.113. [DOI] [PubMed] [Google Scholar]
- 24.Barger S.R., Hoefler B.C., Cubillos-Ruiz A., Russell W.K., Russell D.H., Straight P.D. Imaging secondary metabolism of Streptomyces sp. Mg1 during cellular lysis and colony degradation of competing Bacillus subtilis. Antonie Van Leeuwenhoek. 2012;102(3):435–445. doi: 10.1007/s10482-012-9769-0. [DOI] [PubMed] [Google Scholar]
- 25.Leinweber A., Fredrik Inglis R., Kümmerli R. Cheating fosters species co-existence in well-mixed bacterial communities. ISME J. 2017;11(5):1179–1188. doi: 10.1038/ismej.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou K., Qiao K., Edgar S., Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33(4):377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganesan V., Li Z., Wang X., Zhang H. Heterologous biosynthesis of natural product naringenin by co-culture engineering. Synth Syst Biotechnol. 2017;2(3):236–242. doi: 10.1016/j.synbio.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thuan N.H., Trung N.T., Cuong N.X., Van Cuong D., Van Quyen D., Malla S. Escherichia coli modular coculture system for resveratrol glucosides production. World J Microbiol Biotechnol. 2018;34(6) doi: 10.1007/s11274-018-2458-z. [DOI] [PubMed] [Google Scholar]
- 29.Camacho-Zaragoza J.M., Hernández-Chávez G., Moreno-Avitia F., Ramírez-Iñiguez R., Martínez A., Bolívar F., Gosset G. Engineering of a microbial coculture of Escherichia coli strains for the biosynthesis of resveratrol. Microb Cell Fact. 2016;15(1) doi: 10.1186/s12934-016-0562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z., Wang X., Zhang H. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metab Eng. 2019;54:1–11. doi: 10.1016/j.ymben.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Jones J.A., Vernacchio V.R., Collins S.M., Shirke A.N., Xiu Y.u., Englaender J.A., Cress B.F., McCutcheon C.C., Linhardt R.J., Gross R.A., Koffas M.A.G., Lee S.Y. Complete biosynthesis of anthocyanins using E. coli polycultures. mBio. 2017;8(3) doi: 10.1128/mBio.00621-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Li X.-B., Jiang J., Liu Z.-N., Qiao B., Li F.-F., Cheng J.-S., Sun X., Yuan Y.-J., Qiao J., Zhao G.-R. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides. Metab Eng. 2018;47:243–253. doi: 10.1016/j.ymben.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Succurro A., Moejes F.W., Ebenhöh O., O'Toole G. A diverse community to study communities: integration of experiments and mathematical models to study microbial consortia. J Bacteriol. 2017;199(15) doi: 10.1128/JB.00865-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eng A., Borenstein E. Microbial community design: methods, applications, and opportunities. Curr Opin Biotechnol. 2019;58:117–128. doi: 10.1016/j.copbio.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendes-Soares H., Chia N. Community metabolic modeling approaches to understanding the gut microbiome: bridging biochemistry and ecology. Free Radical Biol Med. 2017;105:102–109. doi: 10.1016/j.freeradbiomed.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Succurro A, Ebenhöh O. Review and perspective on mathematical modeling of microbial ecosystems. Biochem Soc Trans 2018;46:403–12. https://doi.org/10.1042/BST20170265. [DOI] [PMC free article] [PubMed]
- 37.Conde P do RM, Sauter T, Pfau T. Constraint based modeling going multicellular. Front Mol Biosci 2016;3. https://doi.org/10.3389/fmolb.2016.00003. [DOI] [PMC free article] [PubMed]
- 38.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol 2014;5. https://doi.org/10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed]
- 39.Cardona C., Weisenhorn P., Henry C., Gilbert J.A. Network-based metabolic analysis and microbial community modeling. Curr Opin Microbiol. 2016;31:124–131. doi: 10.1016/j.mib.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Frey E. Evolutionary game theory: Theoretical concepts and applications to microbial communities. Physica A. 2010;389(20):4265–4298. doi: 10.1016/j.physa:2010.02.047. [DOI] [Google Scholar]
- 41.S.J. Maynard Evolution and the Theory of Games 1982 Cambridge University Press
- 42.West S.A., Griffin A.S., Gardner A., Diggle S.P. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 43.Nadell C.D., Foster K.R., Xavier J.B. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehmann L., Keller L. The evolution of cooperation and altruism – A general framework and a classification of models. J Evol Biol. 2006;19:1365–1376. doi: 10.1111/j.1420-9101.2006.01119.x. [DOI] [PubMed] [Google Scholar]
- 45.Zomorrodi A.R., Segrè D. Genome-driven evolutionary game theory helps understand the rise of metabolic interdependencies in microbial communities. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faust K., Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 47.Hofbauer J., Sigmund K. Cambridge University Press; 1998. Evolutionary Games and Population Dynamics. [Google Scholar]
- 48.May RM. Stability and Complexity in Model Ecosystems. vol. 1. Princeton University Press; 1974. https://doi.org/10.2307/j.ctvs32rq4.
- 49.Mounier J., Monnet C., Vallaeys T., Arditi R., Sarthou A.S., Hélias A. Microbial interactions within a cheese microbial community. Appl Environ Microbiol. 2008;74:172–181. doi: 10.1128/AEM.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffmann K.H., Rodriguez-Brito B., Breitbart M., Bangor D., Angly F., Felts B. Power law rank-abundance models for marine phage communities. FEMS Microbiol Lett. 2007;273:224–228. doi: 10.1111/j.1574-6968.2007.00790.x. [DOI] [PubMed] [Google Scholar]
- 51.Stein R.R., Bucci V., Toussaint N.C., Buffie C.G., Rätsch G., Pamer E.G. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science (80-) 2015;350:663–6. https://doi.org/10.1126/science.aad2602. [DOI] [PubMed]
- 53.Xiao Y., Angulo M.T., Friedman J., Waldor M.K., Weiss S.T., Liu Y.Y. Mapping the ecological networks of microbial communities. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Judson O.P. The rise of the individual-based model in ecology. Trends Ecol Evol. 1994;9:9–14. doi: 10.1016/0169-5347(94)90225-9. [DOI] [PubMed] [Google Scholar]
- 55.Hellweger F.L., Clegg R.J., Clark J.R., Plugge C.M., Kreft J.U. Advancing microbial sciences by individual-based modelling. Nat Rev Microbiol. 2016;14:461–471. doi: 10.1038/nrmicro.2016.62. [DOI] [PubMed] [Google Scholar]
- 56.Li B., Taniguchi D., Gedara J.P., Gogulancea V., Gonzalez-Cabaleiro R., Chen J. NuFeb: a massively parallel simulator for individual-based modelling of microbial communities. PLoS Comput Biol. 2019 doi: 10.1371/journal.pcbi.1007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kreft J.-U., Plugge C.M., Prats C., Leveau J.H.J., Zhang W., Hellweger F.L. From genes to ecosystems in microbiology: modeling approaches and the importance of individuality. Front Microbiol. 2017;8:2299. doi: 10.3389/fmicb.2017.02299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu C., Kim G.B., Kim W.J., Kim H.U., Lee S.Y. Current status and applications of genome-scale metabolic models. Genome Biol. 2019 doi: 10.1186/s13059-019-1730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.García-Jiménez B, García JL, Nogales J. FLYCOP: Metabolic modeling-based analysis and engineering microbial communities. Bioinformatics, vol. 34, Oxford University Press; 2018, p. i954–63. https://doi.org/10.1093/bioinformatics/bty561. [DOI] [PMC free article] [PubMed]
- 60.O’Brien E.J., Monk J.M., Palsson B.O. Using genome-scale models to predict biological capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiechert W. 13C metabolic flux analysis. Metab Eng. 2001 doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- 62.Rühl M., Hardt W.D., Sauer U. Subpopulation-specific metabolic pathway usage in mixed cultures as revealed by reporter protein-based 13C analysis. Appl Environ Microbiol. 2011 doi: 10.1128/AEM.02696-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolfsberg E., Long C.P., Antoniewicz M.R. Metabolism in dense microbial colonies: 13 C metabolic flux analysis of E. coli grown on agar identifies two distinct cell populations with acetate cross-feeding. Metab Eng. 2018 doi: 10.1016/j.ymben.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 64.Krauss M., Schaller S., Borchers S., Findeisen R., Lippert J., Kuepfer L. Integrating Cellular Metabolism into a Multiscale Whole-Body Model. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grafahrend-Belau E., Junker A., Eschenröder A., Müller J., Schreiber F., Junker B.H. Multiscale metabolic modeling: dynamic flux balance analysis on a whole-plant scale. Plant Physiol. 2013;163:637–647. doi: 10.1104/pp.113.224006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gottstein W., Olivier B.G., Bruggeman F.J., Teusink B. Constraint-based stoichiometric modelling from single organisms to microbial communities. J R Soc Interface. 2016;13:20160627. doi: 10.1098/rsif.2016.0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taffs R., Aston J.E., Brileya K., Jay Z., Klatt C.G., McGlynn S. In Silico approaches to study mass and energy flows in microbial consortia: a syntrophic case study. BMC Syst Biol. 2009;3 doi: 10.1186/1752-0509-3-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Garcia O., Lear G., Singhal N. Metabolic network modeling of microbial interactions in natural and engineered environmental systems. Front Microbiol. 2016;7:673. doi: 10.3389/fmicb.2016.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bosi E., Bacci G., Mengoni A., Fondi M. Perspectives and challenges in microbial communities metabolic modeling. Front Genet. 2017;8 doi: 10.3389/fgene.2017.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar M., Ji B., Zengler K., Nielsen J. Modelling approaches for studying the microbiome. Nat Microbiol. 2019;4:1253–1267. doi: 10.1038/s41564-019-0491-9. [DOI] [PubMed] [Google Scholar]
- 71.Borenstein E., Kupiec M., Feldman M.W., Ruppin E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci USA. 2008;105:14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Henry C.S., Bernstein H.C., Weisenhorn P., Taylor R.C., Lee J.Y., Zucker J. Microbial community metabolic modeling: a community data-driven network reconstruction. J Cell Physiol. 2016;231:2339–2345. doi: 10.1002/jcp.25428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S. KBase: the united states department of energy systems biology knowledgebase. Nat Biotechnol. 2018;36:566–569. doi: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Budinich M., Bourdon J., Larhlimi A., Eveillard D. A multi-objective constraint-based approach for modeling genome-scale microbial ecosystems. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0171744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stolyar S., Van Dien S., Hillesland K.L., Pinel N., Lie T.J., Leigh J.A. Metabolic modeling of a mutualistic microbial community. Mol Syst Biol. 2007;3:1–14. doi: 10.1038/msb4100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Freilich S., Zarecki R., Eilam O., Segal E.S., Henry C.S., Kupiec M. Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun. 2011;2:1–7. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- 77.Klitgord N., Segrè D. Environments that induce synthetic microbial ecosystems. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khandelwal R.A., Olivier B.G., Röling W.F.M., Teusink B., Bruggeman F.J. Community flux balance analysis for microbial consortia at balanced growth. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0064567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bordbar A., Feist A.M., Usaite-Black R., Woodcock J., Palsson B.O., Famili I. A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology. BMC Syst Biol. 2011;5 doi: 10.1186/1752-0509-5-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zomorrodi A.R., Maranas C.D. OptCom: a multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zomorrodi A.R., Islam M.M., Maranas C.D. d-OptCom: dynamic multi-level and multi-objective metabolic modeling of microbial communities. ACS Synth Biol. 2014;3:247–257. doi: 10.1021/sb4001307. [DOI] [PubMed] [Google Scholar]
- 82.Mendes-Soares H., Mundy M., Soares L.M., Chia N. MMinte: an application for predicting metabolic interactions among the microbial species in a community. BMC Bioinf. 2016;17:343. doi: 10.1186/s12859-016-1230-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chan S.H.J., Simons M.N., Maranas C.D. SteadyCom: predicting microbial abundances while ensuring community stability. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Machado D., Andrejev S., Tramontano M., Patil K.R. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018;46:7542–7553. doi: 10.1093/nar/gky537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baldini F., Heinken A., Heirendt L., Magnusdottir S., Fleming R.M.T., Thiele I. The microbiome modeling toolbox: from microbial interactions to personalized microbial communities. Bioinformatics. 2019;35:2332–2334. doi: 10.1093/bioinformatics/bty941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Diener C, Gibbons SM, Resendis-Antonio O. MICOM: Metagenome-Scale Modeling To Infer Metabolic Interactions in the Gut Microbiota. MSystems 2020;5. https://doi.org/10.1128/mSystems.00606-19. [DOI] [PMC free article] [PubMed]
- 87.Mahadevan R., Edwards J.S., Doyle F.J. Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys J. 2002;83:1331–1340. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhuang K., Izallalen M., Mouser P., Richter H., Risso C., Mahadevan R. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J. 2011;5:305–316. doi: 10.1038/ismej.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harcombe W.R., Riehl W.J., Dukovski I., Granger B.R., Betts A., Lang A.H. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Granger BR, Chang Y-C, Wang Y, DeLisi C, Segrè D, Hu Z. Visualization of Metabolic Interaction Networks in Microbial Communities Using VisANT 5.0. PLOS Comput Biol 2016;12:e1004875. https://doi.org/10.1371/journal.pcbi.1004875. [DOI] [PMC free article] [PubMed]
- 91.Louca S., Doebeli M. Calibration and analysis of genome-based models for microbial ecology. Elife. 2015;4 doi: 10.7554/eLife.08208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Louca S., Doebeli M. Taxonomic variability and functional stability in microbial communities infected by phages. Environ Microbiol. 2017;19:3863–3878. doi: 10.1111/1462-2920.13743. [DOI] [PubMed] [Google Scholar]
- 93.Louca S., Doebeli M. Reaction-centric modeling of microbial ecosystems. Ecol Modell. 2016;335:74–86. doi: 10.1016/j.ecolmodel.2016.05.011. [DOI] [Google Scholar]
- 94.Bauer E., Zimmermann J., Baldini F., Thiele I., Kaleta C. BacArena: individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Succurro A., Segrè D., Ebenhöh O. Emergent subpopulation behavior uncovered with a community dynamic metabolic model of escherichia coli diauxic growth. MSystems. 2019;4 doi: 10.1128/msystems.00230-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.García-Jiménez B., Carrasco J., Medina J., Wilkinson M.D. Dynamic simulations of microbial communities under perturbations: opportunities for microbiome engineering. Res Sq - Prepr. 2020 [Google Scholar]
- 97.Rau M.H., Zeidan A.A. Constraint-based modeling in microbial food biotechnology. Biochem Soc Trans. 2018;46:249–260. doi: 10.1042/BST20170268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cook DJ, Nielsen J. Genome-scale metabolic models applied to human health and disease. Wiley Interdiscip Rev Syst Biol Med 2017;9. https://doi.org/10.1002/wsbm.1393. [DOI] [PubMed]
- 99.Fondi M., Fani R. Constraint-based metabolic modelling of marine microbes and communities. Mar Genomics. 2017;34:1–10. doi: 10.1016/j.margen.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 100.Shoaie S., Karlsson F., Mardinoglu A., Nookaew I., Bordel S., Nielsen J. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci Rep. 2013;3 doi: 10.1038/srep02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shoaie S., Ghaffari P., Kovatcheva-Datchary P., Mardinoglu A., Sen P., Pujos-Guillot E. Quantifying diet-induced metabolic changes of the human gut microbiome. Cell Metab. 2015;22:320–331. doi: 10.1016/j.cmet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 102.Magnúsdóttir S., Heinken A., Kutt L., Ravcheev D.A., Bauer E., Noronha A. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017;35:81–89. doi: 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- 103.Heinken A., Thiele I. Anoxic conditions promote species-specific mutualism between gut microbes In Silico. Appl Environ Microbiol. 2015;81:4049–4061. doi: 10.1128/AEM.00101-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bauer E., Thiele I. From metagenomic data to personalized in silico microbiotas: predicting dietary supplements for Crohn’s disease. Npj Syst Biol Appl. 2018;4:27. doi: 10.1038/s41540-018-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pryor R., Norvaisas P., Marinos G., Best L., Thingholm L.B., Quintaneiro L.M. Host-microbe-drug-nutrient screen identifies bacterial effectors of metformin therapy. Cell. 2019;178 doi: 10.1016/j.cell.2019.08.003. 1299–1312.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thiele I., Sahoo S., Heinken A., Hertel J., Heirendt L., Aurich M.K. Personalized whole-body models integrate metabolism, physiology, and the gut microbiome. Mol Syst Biol. 2020;16 doi: 10.15252/msb.20198982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chong J., Xia J. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites. 2017;7 doi: 10.3390/metabo7040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Garza D.R., Van Verk M.C., Huynen M.A., Dutilh B.E. Towards predicting the environmental metabolome from metagenomics with a mechanistic model. Nat Microbiol. 2018;3:456–460. doi: 10.1038/s41564-018-0124-8. [DOI] [PubMed] [Google Scholar]
- 109.Noecker C., Chiu H.-C., McNally C.P., Borenstein E. Defining and evaluating microbial contributions to metabolite variation in microbiome-metabolome association studies. MSystems. 2019;4 doi: 10.1128/msystems.00579-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vijayakumar S., Conway M., Lió P., Angione C. Optimization of multi-omic genome-scale models: Methodologies, hands-on tutorial, and perspectives. Methods Mol Biol. 2018 doi: 10.1007/978-1-4939-7528-0_18. [DOI] [PubMed] [Google Scholar]
- 111.Ramon C., Gollub M.G., Stelling J. Integrating -omics data into genome-scale metabolic network models: principles and challenges. Essays Biochem. 2018 doi: 10.1042/EBC20180011. [DOI] [PubMed] [Google Scholar]
- 112.Hyduke D.R., Lewis N.E., Palsson B.O. Analysis of omics data with genome-scale models of metabolism. Mol Biosyst. 2013 doi: 10.1039/c2mb25453k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Machado D., Herrgård M.J. Co-evolution of strain design methods based on flux balance and elementary mode analysis. Metab Eng Commun. 2015;2:85–92. doi: 10.1016/j.meteno.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burgard A.P., Pharkya P., Maranas C.D. OptKnock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 115.Pharkya P., Burgard A.P., Maranas C.D. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 2004;14:2367–2376. doi: 10.1101/gr.2872004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Patil K.R., Rocha I., Förster J., Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinf. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lun D.S., Rockwell G., Guido N.J., Baym M., Kelner J.A., Berger B. Large-scale identification of genetic design strategies using local search. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Henson M.A., Hanly T.J. Dynamic flux balance analysis for synthetic microbial communities. IET Syst Biol. 2014;8:214–229. doi: 10.1049/iet-syb.2013.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jones J.A., Vernacchio V.R., Sinkoe A.L., Collins S.M., Ibrahim M.H.A., Lachance D.M. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab Eng. 2016;35:55–63. doi: 10.1016/j.ymben.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 120.Gudelj I., Kinnersley M., Rashkov P., Schmidt K., Rosenzweig F. Stability of cross-feeding polymorphisms in microbial communities. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Eng A., Borenstein E. An algorithm for designing minimal microbial communities with desired metabolic capacities. Bioinformatics. 2016;32:2008–2016. doi: 10.1093/bioinformatics/btw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhao Q, Segre D, Paschalidisy IC. Optimal allocation of metabolic functions among organisms in a microbial ecosystem. 2016 IEEE 55th Conf. Decis. Control. CDC 2016, Institute of Electrical and Electronics Engineers Inc.; 2016, p. 7063–8. https://doi.org/10.1109/CDC.2016.7799357.
- 123.Handorf T., Ebenhöh O., Heinrich R. Expanding metabolic networks: scopes of compounds, robustness, and evolution. J Mol Evol. 2005;61:498–512. doi: 10.1007/s00239-005-0027-1. [DOI] [PubMed] [Google Scholar]
- 124.Julien-Laferrière A., Bulteau L., Parrot D., Marchetti-Spaccamela A., Stougie L., Vinga S. A combinatorial algorithm for microbial consortia synthetic design. Sci Rep. 2016;6:29182. doi: 10.1038/srep29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Thommes M., Wang T., Zhao Q., Paschalidis I.C., Segrè D. Designing metabolic division of labor in microbial communities. MSystems. 2019;4 doi: 10.1128/msystems.00263-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vitkin E., Gillis A., Polikovsky M., Bender B., Golberg A., Yakhini Z. Distributed flux balance analysis simulations of serial biomass fermentation by two organisms. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0227363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zampieri M., Sauer U. Model-based media selection to minimize the cost of metabolic cooperation in microbial ecosystems. Bioinformatics. 2016;32:1733–1739. doi: 10.1093/bioinformatics/btw062. [DOI] [PubMed] [Google Scholar]
- 128.Borer B., Ataman M., Hatzimanikatis V., Or D. Modeling metabolic networks of individual bacterial agents in heterogeneous and dynamic soil habitats (IndiMeSH) PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1007127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Phalak P., Chen J., Carlson R.P., Henson M.A. Metabolic modeling of a chronic wound biofilm consortium predicts spatial partitioning of bacterial species. BMC Syst Biol. 2016;10 doi: 10.1186/s12918-016-0334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ben S.S., Or D., Ben Said S., Or D. Synthetic microbial ecology: engineering habitats for modular consortia. Front Microbiol. 2017;8:1125. doi: 10.3389/fmicb.2017.01125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koch S., Benndorf D., Fronk K., Reichl U., Klamt S. Predicting compositions of microbial communities from stoichiometric models with applications for the biogas process. Biotechnol Biofuels. 2016;9:17. doi: 10.1186/s13068-016-0429-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rana P., Berry C., Ghosh P., Fong S.S. Recent advances on constraint-based models by integrating machine learning. Curr Opin Biotechnol. 2020 doi: 10.1016/j.copbio.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 133.Zampieri G., Vijayakumar S., Yaneske E., Angione C. Machine and deep learning meet genome-scale metabolic modeling. PLoS Comput Biol. 2019 doi: 10.1371/journal.pcbi.1007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim G.B., Kim W.J., Kim H.U., Lee S.Y. Machine learning applications in systems metabolic engineering. Curr Opin Biotechnol. 2020 doi: 10.1016/j.copbio.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 135.Liao X., Rong S., Relatedness Q.DC. Conflict, and the Evolution of Eusociality. PLoS Biol. 2015;13 doi: 10.1371/journal.pbio.1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lawson C.E., Harcombe W.R., Hatzenpichler R., Lindemann S.R., Löffler F.E., O’Malley M.A. Common principles and best practices for engineering microbiomes. Nat Rev Microbiol. 2019 doi: 10.1038/s41579-019-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gil-Sánchez I., Cueva C., Tamargo A., Quintela J.C., de la Fuente E., Walker A.W. Application of the dynamic gastrointestinal simulator (simgi®) to assess the impact of probiotic supplementation in the metabolism of grape polyphenols. Food Res Int. 2020 doi: 10.1016/j.foodres.2019.108790. [DOI] [PubMed] [Google Scholar]
- 138.Zengler K., Hofmockel K., Baliga N.S., Behie S.W., Bernstein H.C., Brown J.B. EcoFABs: advancing microbiome science through standardized fabricated ecosystems. Nat Methods. 2019 doi: 10.1038/s41592-019-0465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li F., Yin C., Sun L., Li Y., Guo X., Song H. Synthetic klebsiella pneumoniae-shewanella oneidensis consortium enables glycerol-fed high-performance microbial fuel cells. Biotechnol J. 2018;13 doi: 10.1002/biot.201700491. [DOI] [PubMed] [Google Scholar]
- 140.Bhatia S.K., Yoon J.J., Kim H.J., Hong J.W., Gi Hong Y., Song H.S. Engineering of artificial microbial consortia of Ralstonia eutropha and Bacillus subtilis for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production from sugarcane sugar without precursor feeding. Bioresour Technol. 2018;257:92–101. doi: 10.1016/j.biortech.2018.02.056. [DOI] [PubMed] [Google Scholar]
- 141.Lee C.R., Kim C., Song Y.E., Im H., Oh Y.K., Park S. Co-culture-based biological carbon monoxide conversion by Citrobacter amalonaticus Y19 and Sporomusa ovata via a reducing-equivalent transfer mediator. Bioresour Technol. 2018;259:128–135. doi: 10.1016/j.biortech.2018.02.129. [DOI] [PubMed] [Google Scholar]
- 142.Scholz S.A., Graves I., Minty J.J., Lin X.N. Production of cellulosic organic acids via synthetic fungal consortia. Biotechnol Bioeng. 2018;115:1096–1100. doi: 10.1002/bit.26509. [DOI] [PubMed] [Google Scholar]
- 143.Santala S., Karp M., Santala V. Rationally engineered synthetic coculture for improved biomass and product formation. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0113786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Buzzini P. Batch and fed-batch carotenoid production by rhodotorula glutinis-debaryomyces castellii co-cultures in corn syrup. J Appl Microbiol. 2001;90:843–847. doi: 10.1046/j.1365-2672.2001.01319.x. [DOI] [PubMed] [Google Scholar]
- 145.Fossi B.T., Tavea F., Fontem L.A., Ndjouenkeu R., Wanji S. Microbial interactions for enhancement of alpha-amylase production by Bacillus amyloliquefaciens 04BBA15 and Lactobacillus fermentum 04BBA19. Biotechnol Rep. 2014;4:99–106. doi: 10.1016/j.btre.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Martínez V., Herencias C., Jurkevitch E., Prieto M.A. Engineering a predatory bacterium as a proficient killer agent for intracellular bio-products recovery: The case of the polyhydroxyalkanoates. Sci Rep. 2016 doi: 10.1038/srep24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Venkataraman A., Rosenbaum M.A., Perkins S.D., Werner J.J., Angenent L.T. Metabolite-based mutualism between Pseudomonas aeruginosaPA14 and Enterobacter aerogenes enhances current generation in bioelectrochemical systems. Energy Environ Sci. 2011;4:4550–4559. doi: 10.1039/C1EE01377G. [DOI] [Google Scholar]
- 148.Johnston T.G., Yuan S.F., Wagner J.M., Yi X., Saha A., Smith P. Compartmentalized microbes and co-cultures in hydrogels for on-demand bioproduction and preservation. Nat Commun. 2020;11:563. doi: 10.1038/s41467-020-14371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang X., Su R., Chen K., Xu S., Feng J., Ouyang P. Engineering a microbial consortium based whole-cell system for efficient production of glutarate from L-lysine. Front Microbiol. 2019;10:341. doi: 10.3389/fmicb.2019.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Khan N., Maezato Y., McClure R.S., Brislawn C.J., Mobberley J.M., Isern N. Phenotypic responses to interspecies competition and commensalism in a naturally-derived microbial co-culture. Sci Rep. 2018;8:297. doi: 10.1038/s41598-017-18630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang H., Pereira B., Li Z., Stephanopoulos G. Engineering Escherichia coli coculture systems for the production of biochemical products. Proc Natl Acad Sci U S A. 2015;112:8266–8271. doi: 10.1073/pnas.1506781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hu B., Wang M., Geng S., Wen L., Wu M., Nie Y. Metabolic exchange with non-alkane-consuming pseudomonas stutzeri SLG510A3-8 improves n-alkane biodegradation by the alkane degrader Dietzia sp. strain DQ12-45-1b. Appl Env Microbiol. 2020;86 doi: 10.1128/AEM.02931-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang J., Lu X., Ying H., Ma W., Xu S., Wang X. A novel process for cadaverine bio-production using a consortium of two engineered Escherichia coli. Front Microbiol. 2018;9:1312. doi: 10.3389/fmicb.2018.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu Y., Ding M., Ling W., Yang Y., Zhou X., Li B.-Z. A three-species microbial consortium for power generation. Energy Env Sci. 2017;10:1600–1609. doi: 10.1039/C6EE03705D. [DOI] [Google Scholar]
- 155.Wang V.B., Sivakumar K., Yang L., Zhang Q., Kjelleberg S., Loo S.C. Metabolite-enabled mutualistic interaction between shewanella oneidensis and escherichia coli in a co-culture using an electrode as electron acceptor. Sci Rep. 2015;5:11222. doi: 10.1038/srep11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang X., Reed J.L. Adaptive evolution of synthetic cooperating communities improves growth performance. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0108297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rodríguez J., Kleerebezem R., Lema J.M., Van Loosdrecht M.C.M. Modeling product formation in anaerobic mixed culture fermentations. Biotechnol Bioeng. 2006;93:592–606. doi: 10.1002/bit.20765. [DOI] [PubMed] [Google Scholar]
- 158.La Rosa R., Nogales J., Rojo F. The Crc/CrcZ-CrcY global regulatory system helps the integration of gluconeogenic and glycolytic metabolism in Pseudomonas putida. Environ Microbiol. 2015;17:3362–3378. doi: 10.1111/1462-2920.12812. [DOI] [PubMed] [Google Scholar]
- 159.Levy R., Borenstein E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci U S A. 2013;110:12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Freilich S., Kreimer A., Borenstein E., Yosef N., Sharan R., Gophna U. Metabolic-network-driven analysis of bacterial ecological strategies. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Miller L.D., Mosher J.J., Venkateswaran A., Yang Z.K., Palumbo A.V., Phelps T.J. Establishment and metabolic analysis of a model microbial community for understanding trophic and electron accepting interactions of subsurface anaerobic environments. BMC Microbiol. 2010;10 doi: 10.1186/1471-2180-10-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Guazzaroni M.E., Herbst F.A., Lores I., Tamames J., Peláez A.I., López-Cortés N. Metaproteogenomic insights beyond bacterial response to naphthalene exposure and bio-stimulation. ISME J. 2013 doi: 10.1038/ismej.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tobalina L., Bargiela R., Pey J., Herbst F.-A., Lores I., Rojo D. Context-specific metabolic network reconstruction of a naphthalene-degrading bacterial community guided by metaproteomic data. Bioinformatics. 2015;31:1771–1779. doi: 10.1093/bioinformatics/btv036. [DOI] [PubMed] [Google Scholar]
- 164.Zelezniak A., Andrejev S., Ponomarova O., Mende D.R., Bork P., Patil K.R. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci U S A. 2015;112:6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cerqueda-García D., Falcón L.I. Metabolic potential of microbial mats and microbialites: Autotrophic capabilities described by an in silico stoichiometric approach from shared genomic resources. J Bioinform Comput Biol. 2016;14 doi: 10.1142/S0219720016500207. [DOI] [PubMed] [Google Scholar]
- 166.Bizukojc M., Dietz D., Sun J., Zeng A.P. Metabolic modelling of syntrophic-like growth of a 1,3-propanediol producer, Clostridium butyricum, and a methanogenic archeon, Methanosarcina mazei, under anaerobic conditions. Bioprocess Biosyst Eng. 2010;33:507–523. doi: 10.1007/s00449-009-0359-0. [DOI] [PubMed] [Google Scholar]
- 167.de Oliveira Dal’Molin CG, Quek LE, Saa PA, Nielsen LK. A multi-tissue genome-scale metabolic modeling framework for the analysis of whole plant systems. Front Plant Sci 2015;6:1–12. https://doi.org/10.3389/fpls.2015.00004. [DOI] [PMC free article] [PubMed]
- 168.Huthmacher C, Hoppe A, Bulik S, Holzhütter HG. Antimalarial drug targets in Plasmodium falciparum predicted by stage-specific metabolic network analysis. BMC Syst Biol 2010;4. https://doi.org/10.1186/1752-0509-4-120. [DOI] [PMC free article] [PubMed]
- 169.Wintermute E.H., Silver P.A. Emergent cooperation in microbial metabolism. Mol Syst Biol. 2010;6. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: Genetic differentiation in Escherichia coli. Genetics 1994. [DOI] [PMC free article] [PubMed]
- 171.Nagarajan H., Embree M., Rotaru A.E., Shrestha P.M., Feist A.M., Palsson B. Characterization and modelling of interspecies electron transfer mechanisms and microbial community dynamics of a syntrophic association. Nat Commun. 2013;4 doi: 10.1038/ncomms3809. [DOI] [PubMed] [Google Scholar]
- 172.El-Semman I.E., Karlsson F.H., Shoaie S., Nookaew I., Soliman T.H., Nielsen J. Genome-scale metabolic reconstructions of Bifidobacterium adolescentis L2–32 and Faecalibacterium prausnitzii A2–165 and their interaction. BMC Syst Biol. 2014;8 doi: 10.1186/1752-0509-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ye C., Zou W., Xu N., Liu L. Metabolic model reconstruction and analysis of an artificial microbial ecosystem for vitamin C production. J Biotechnol. 2014;182–183:61–67. doi: 10.1016/j.jbiotec.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 174.Merino M.P., Andrews B.A., Asenjo J.A. Stoichiometric model and flux balance analysis for a mixed culture of Leptospirillum ferriphilum and Ferroplasma acidiphilum. Biotechnol Prog. 2015;31:307–315. doi: 10.1002/btpr.2028. [DOI] [PubMed] [Google Scholar]
- 175.Perez-Garcia O., Chandran K., Villas-Boas S.G., Singhal N. Assessment of nitric oxide (NO) redox reactions contribution to nitrous oxide (N2O) formation during nitrification using a multispecies metabolic network model. Biotechnol Bioeng. 2016;113:1124–1136. doi: 10.1002/bit.25880. [DOI] [PubMed] [Google Scholar]
- 176.Rey F.E., Gonzalez M.D., Cheng J., Wu M., Ahern P.P., Gordon J.I. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1312524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Brunk E., Sahoo S., Zielinski D.C., Altunkaya A., Dräger A., Mih N. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018 doi: 10.1038/nbt.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tzamali E., Poirazi P., Tollis I.G., Reczko M. A computational exploration of bacterial metabolic diversity identifying metabolic interactions and growth-efficient strain communities. BMC Syst Biol. 2011;5 doi: 10.1186/1752-0509-5-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Salimi F., Zhuang K., Mahadevan R. Genome-scale metabolic modeling of a clostridial co-culture for consolidated bioprocessing. Biotechnol J. 2010;5:726–738. doi: 10.1002/biot.201000159. [DOI] [PubMed] [Google Scholar]
- 180.Zhuang K., Ma E., Lovley D.R., Mahadevan R. The design of long-term effective uranium bioremediation strategy using a community metabolic model. Biotechnol Bioeng. 2012;109:2475–2483. doi: 10.1002/bit.24528. [DOI] [PubMed] [Google Scholar]
- 181.Wintermute E.H., Silver P.A. Dynamics in the mixed microbial concourse. Genes Dev. 2010;24:2603–2614. doi: 10.1101/gad.1985210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Großkopf T., Consuegra J., Gaffé J., Willison J.C., Lenski R.E., Soyer O.S. Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long-term evolution experiment. BMC Evol Biol. 2016;16 doi: 10.1186/s12862-016-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hoek MJA va., Merks RMH. Emergence of microbial diversity due to cross-feeding interactions in a spatial model of gut microbial metabolism. BMC Syst Biol 2017;11. https://doi.org/10.1186/s12918-017-0430-4. [DOI] [PMC free article] [PubMed]
- 184.Mellbye BL, Giguere AT, Murthy GS, Bottomley PJ, Sayavedra-Soto LA, Chaplen FWR. Genome-Scale, Constraint-Based Modeling of Nitrogen Oxide Fluxes during Coculture of Nitrosomonas europaea and Nitrobacter winogradskyi . MSystems 2018;3. https://doi.org/10.1128/msystems.00170-17. [DOI] [PMC free article] [PubMed]
- 185.Hanly T.J., Henson M.A. Dynamic metabolic modeling of a microaerobic yeast co-culture: Predicting and optimizing ethanol production from glucose/xylose mixtures. Biotechnol Biofuels. 2013;6 doi: 10.1186/1754-6834-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hanly T.J., Henson M.A. Dynamic flux balance modeling of microbial co-cultures for efficient batch fermentation of glucose and xylose mixtures. Biotechnol Bioeng. 2011;108:376–385. doi: 10.1002/bit.22954. [DOI] [PubMed] [Google Scholar]
- 187.Tatton M.J., Archer D.B., Powell G.E., Parker M.L. Methanogenesis from ethanol by defined mixed continuous cultures. Appl Environ Microbiol. 1989 doi: 10.1128/aem.55.2.440-445.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kinnersley M., Wenger J., Kroll E., Adams J., Sherlock G., Rosenzweig F. Ex uno plures: clonal reinforcement drives evolution of a simple microbial community. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chiu H.C., Levy R., Borenstein E. Emergent biosynthetic capacity in simple microbial communities. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hanemaaijer M., Olivier B.G., Röling W.F.M., Bruggeman F.J., Teusink B. Model-based quantification of metabolic interactions from dynamic microbial-community data. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0173183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Harcombe W. Novel cooperation experimentally evolved between species. Evolution (N Y) 2010 doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]