Abstract
Osteocytes, the most abundant bone cell type, are derived from osteoblasts through a process in which they are embedded in an osteoid. We previously showed that nutrient restriction promotes the osteocyte transcriptional program and is associated with increased mitochondrial biogenesis. Here, we show that increased mitochondrial biogenesis increase reactive oxygen species (ROS) levels and consequently, NRF2 activity during osteocytogenesis. NRF2 activity promotes osteocyte-specific expression of Dmp1, Mepe, and Sost in IDG-SW3 cells, primary osteocytes, and osteoblasts, and in murine models with Nfe2l2 deficiency in osteocytes or osteoblasts. Moreover, ablation of Nfe2l2 in osteocytes or osteoblasts generates osteopenia and increases osteoclast numbers with marked sexual dimorphism. Finally, treatment with dimethyl fumarate prevented the deleterious effects of ovariectomy in trabecular bone masses of mice and restored osteocytic gene expression. Altogether, we uncovered the role of NRF2 activity in osteocytes during the regulation of osteocyte gene expression and maintenance of bone homeostasis.
Keywords: Bone, Osteocyte, NRF2, Oxidative stress, Osteoporosis, Bone homeostasis
Graphical abstract
Highlights
-
•
ROS levels and NRF2 activity are increased during osteocytogenesis.
-
•
NRF2 drives osteocyte specification and activate the transcription of osteocyte-specific genes.
-
•
NRF2 in osteocytes has a fundamental role in bone homeostasis and its deletion induces osteopenia.
-
•
Activation of NRF2 with dimethyl fumarate prevents osteopenia induced by ovariectomy.
1. Introduction
Bone is a dynamic tissue that undergoes continuous remodeling to adapt to the physiological demands of the whole organism. Of the major bone cell types, osteocytes make up more than 90% of the total bone cells. Presently, it has become clear that osteocytes communicate to osteoblasts and osteoclasts to coordinate bone remodeling, acting as physical mechanosensors and secreting endocrine regulators [[1], [2], [3]]. Lower osteocyte activity has been directly linked to bone fragility, osteopenia associated with aging or diabetes, chronic kidney disease, and atherosclerosis [1,4,5].
Osteocytes are derived from osteoblasts through an active process in which they are embedded in an osteoid matrix. During osteocytogenesis, dramatic morphological changes occur with the acquisition of a unique gene expression profile [6]. During osteocytogenesis, the progressive expression of osteocyte-specific genes can be identified. Dmp1 and Mepe are markers of osteocyte specification, whereas Fgf23 and Sost are indicative of mature osteocytes [7]. However, the exact mechanisms that govern the transition from osteoblasts into osteocytes, as well as how osteocytes exert regulatory functions are not completely defined. One of the driving forces of osteoblast-osteocyte transition is likely to be their encasement within a mineralized bone matrix. The lacuno-canalicular system connects osteocytes and allows those near the surface to interact with bone lining cells and vasculature [8]. Nonetheless, the mechanisms of diffusion between the bone surface and osteocytes are challenged to maintain an adequate supply and exchange of oxygen, nutrients, and metabolic products [[9], [10], [11]]. We recently demonstrated that restriction of the glucose supply promotes the osteocyte transcriptional program and is associated with increased mitochondrial biogenesis [12]. Since mitochondria are the main source of reactive oxygen species (ROS), it is likely that increased electron transport chain activity will also result in increased ROS production during osteocytogenesis. It has been shown that controlled ROS production contributes to cell specification of osteoclasts, whereas excessive ROS production is detrimental for osteoblastogenesis and bone homeostasis [[13], [14], [15]].
Controlled ROS generation is the net effect of a balance between ROS generation and its clearance by cellular antioxidant systems. Upon sensing endogenous or exogenous oxidative stress, the major inducible system to maintain redox homeostasis is NRF2-KEAP1 signaling. Nuclear factor (erythroid-derived 2)-like2 (Nfe2l2, NRF2) is a member of a family of basic leucine zipper transcription factors that binds to antioxidant response elements (AREs) in the regulatory regions of genes encoding detoxifying enzymes and antioxidant proteins [16]. NRF2-KEAP1 is a two-component system that controls NRF2 nuclear levels in the cytoplasm Kelch-like ECH-associated protein 1 (KEAP1), which forms a ubiquitin E3 ligase complex with cullin3 (CUL3) and polyubiquitinates and promotes NRF2 degradation. Increased oxidative/electrophilic stress directly modifies key cysteine residues in KEAP1, reduces ubiquitination and results in stabilization of NRF2. Accumulated nascent NRF2 can then translocate to the nucleus, heterodimerize with sMAF, and activate transcription. The role of NRF2-KEAP1 in bone homeostasis may depend on factors, such as age, sex, or high vs. low activation levels [17,18]. Some studies revealed that deletion of Nfe2l2 in female mice resulted in a deficit in postnatal bone acquisition and increased bone loss [17,19]. However, similar Nfe2l2-deletion approaches in male mice led to controversial results. Some studies reported increased bone acquisition and mass [17,20], whereas others showed reduced bone density, load-driven bone formation, and increased radiation-induced bone loss [21,22]. Similarly, moderate NRF2 activation by heterozygous deletion of Keap1 led to sexual dimorphic results with higher bone mass in male but not female mice [18]. A limitation of these previous studies was that genetic modifications of Nfe2l2 in mice affected all cell types and tissues. Therefore, we cannot exclude the influence of paracrine and endocrine factors; studies with cell-type specific knockout mice of NRF2 would provide more insight into their specific role in bone biology and pathophysiology.
In this study, we addressed the specific role of NRF2 in osteocytes and osteoblasts in vivo. We report that increased mitochondrial biogenesis increased ROS levels and NRF2 activity during osteocytogenesis. NRF2 activity promoted osteocytic specification through transcriptional activation of osteocyte-specific genes. Furthermore, conditional ablation of Nfe2l2 in osteocytes or osteoblasts induces osteopenia and increases the number of osteoclasts with marked sexual dimorphism. Finally, our results showed that treatment with the NRF2 activator dimethyl fumarate (DMF) prevents the deleterious effects of ovariectomy in trabecular bone mass of mice and restored osteocytic gene expression suggesting that might be beneficial for osteopenic bone pathologies.
2. Materials and methods
2.1. Cell cultures
IDG-SW3 cells were obtained from Dr. L. Bonewald and cultured as previously described by Ref. [7]. Briefly, IDG-SW3 were maintained and expanded in proliferative conditions (33 °C in αMEM with 5 mM glucose, 10% FBS, 2 mM glutamine, 1 mM pyruvate, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 50 U/ml IFN-γ) on type I collagen-coated plates. To induce osteogenesis, cells were seeded on type I collagen-coated plates and cultured in osteogenic media (37 °C in αMEM with 10% FBS, 2 mM glutamine, 1 mM pyruvate, 100 U/ml penicillin, and 0.1 mg/ml streptomycin with 50 μg/ml ascorbic acid and 4 mM β-glycerophosphate, in the absence of IFN-γ).
Primary osteocytes were isolated with a protocol derived from Nijweide et al. [23]. Briefly, mice limbs (tibiae, femurs, and humeri) and calvariae from 5 mice were pooled, cut, and serially digested on a rotating shaker at 37 °C. Bone pieces were incubated in a collagenase solution [trypsin (0.025%)/collagenase II (1 mg/ml) in α-MEM] for 25 min. The solution was discarded and the bone pieces were washed in PBS. This was repeated two more times, for a total of three digestions (digestions 1 to 3). Then, bone pieces were incubated with EDTA solution (5 mM EDTA in PBS containing 1% BSA, pH = 7.4) for 25 min. The solution was discarded (digestion 4), and the bone pieces were washed in PBS (digestion 4). Bone pieces were incubated with collagenase solution for 25 min, the solution was kept for osteoblast isolation, and the bone pieces were washed in PBS (digestion 5). Bone pieces were incubated with EDTA solution for 25 min, the solution was kept for osteoblast isolation (digestion 6), and the bone pieces were washed in PBS. Digestion 5 and 6 were combined and spun down at 200×g for 5 min. The cell pellet was resuspended in osteogenic media (αMEM with 10% FBS, 2 mM glutamine, 1 mM pyruvate, 100 U/ml penicillin, and 0.1 mg/ml streptomycin with 50 μg/ml ascorbic acid and 4 mM β-glycerophosphate). Bone pieces were incubated with collagenase solution for 25 min, the solution was kept for cell plating, and the bone pieces were washed in PBS (digestion 7). Bone pieces were incubated with EDTA solution for 25 min, the solution was kept for cell plating, and the bone pieces were washed in PBS (digestion 8). Bone pieces were incubated with collagenase solution for 25 min, the solution was kept for cell plating (digestion 9). Digestions 7 to 9 were combined and spun down at 200×g for 5 min. The cell pellet was resuspended in α-MEM culture media and plated on type-I rat tail collagen coated culture plates. Osteocytes were cultured in α-MEM supplemented with 5% FBS, 5% calf serum, 1 mM pyruvate, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Osteoblasts and osteocytes were maintained at 37 °C and 5% CO2 in a humidified incubator for up to 7 days.
2.2. Animal models
To delete Nfe2l2 specifically in osteocytes, mice carrying loxP sequences flanking exon 5 of Nfe2l2 gene (SN25433 from The Jackson Laboratory) were crossed with Dmp1-cre/ERT2 transgenic line D77 (SN029594 from The Jackson Laboratory) that expressed tamoxifen-inducible Cre recombinase (cre/ERT2) under the control of the mouse dentin matrix protein 1 (DMP1) promoter to generate Nfe2l2; Dmp1-cre/ERT2 mice. Heterozygous Dmp1-cre/ERT2 mice with floxed Nfe2l2 (Nfe2l2f/f) were crossed with Nfe2l2f/f to obtain experimental (Nfe2l2f/f; Dmp1-Cre/Ert2) and control (Nfe2l2f/f) mice. The resulting Nfe2l2f/f; Dmp1-cre/ERT2 mice were fertile and born at the anticipated Mendelian ratio. To induce recombination of floxed Nfe2l2 alleles, 3-week-old Nfe2l2f/f (WT) and Nfe2l2f/f; Dmp1-Cre/Ert2 (KO) mice received a total of 5 intraperitoneal injections of 20 mg/kg tamoxifen free base (Sigma) with 48h in between. Tamoxifen powder was dissolved in ethanol at a concentration of 100 mg/ml and suspended in corn oil at a concentration of 20 mg/ml by shaking for 1 h at 37 °C. Eight-week-old mice were euthanized and samples were collected for μCT and gene expression analysis.
Osteoblast-specific Nfe2l2-deficient mice (Nfe2l2;Col1a1-Cre) were generated by crossing heterozygous Col1a1-Cre mice (Tg(Col1a1-cre)1Kry [24] with Nfe2l2 floxed mice, which was described previously. Nfe2l2f/f;Col1a1-Cre (KO) mice were fertile and born at the Mendelian ratio. Eight-week-old mice were euthanized and samples were collected for μCT and gene expression analysis.
To analyze the effect of dimethyl fumarate in osteoporosis, 8-week-old female mice underwent a bilateral ovariectomy (OVX) and an equal number of mice underwent SHAM surgery (SHAM). OVX and SHAM were randomly divided into DMF (animals treated with 100 mg/kg of DMF) or vehicle treatment groups. For the treatment, DMF was suspended in 0.8% methylcellulose and sonicated until it was completely dissolved. The DMF or vehicle was given daily by oral gavage for 8 weeks, starting 72 h after surgery. Mice were euthanized 24 h after the last administration and samples were collected for μCT and gene expression analysis.
All animals had ad libitum access to food (Teklad, 2014: Harlan‐Teklad, Indianapolis, IN, USA) and water. Mice were maintained at 22–24 °C on a 12-h light/12-h dark cycle. All procedures were approved by the Ethics Committee for Animal Experimentation of the Generalitat of Catalunya.
2.3. Retroviral transduction
Retroviral pMSCV-NRF2 (generated from pcDNA4-NRF2 from Addgene) and the pMSCV-GFP control virus were used for NRF2 overexpression in primary osteocytes. After infection, cells were cultured for five days in osteogenic media. Retroviral pMSCV-puro-Cre-ERT2, pMSCV-puro, and pMSCV-GFP virus were used for Nfe2l2 deletion assays in primary osteoblasts and osteocytes from Nfe2l2 f/f mice. Puromycin (5 μg/ml) was used for the selection of infected cells. Tamoxifen (1 μM) was added at 48 h for the activation of Cre recombinase. Cells were lysate at least 24 h after the last tamoxifen administration.
2.4. Fluorescence confocal imaging
IDG-SW3 and primary osteoblasts and osteocytes were stained with 250 nM Mitotracker-Deep-red (ThermoFisher), 5 μM CellROX Deep Red (Invitrogen) for 30 min or 2 μM MitoSOX Red (Invitrogen) for 15 min at 37 °C. Cells were examined in a Zeiss LSM 880 laser scanning confocal spectral microscope equipped with an incubation control system (37 °C, 5% CO2). Images corresponding to single confocal sections were taken using a 63x oil immersion objective lens. Images were analyzed using Zeiss software.
2.5. Flow cytometry
IDG-SW3 and osteoblasts were stained with Mitotracker Deep Red (ThermoFisher), CellROX Deep Red (ThermoFisher) or MitoSOX Red (Invitrogen). Then, cells were rinsed twice with PBS. Cells were incubated with 0.25% Trypsin and 0.02% EDTA at 37 °C for 5–10 min until cells became rounded and started to detach. Cell suspensions were collected and centrifuged at 150xg for 5 min. Cell pellets were rinsed three times with PBS and resuspended in PBS. Before flow cytometry analysis, cell suspensions were filtered with a 0.70 μm nylon mesh to remove aggregates. Cell viability was assessed with 7-ADD. Results were analyzed with BD FACSDiva™ Software.
2.6. Intact cell respirometry
Mitochondrial function of intact cells was measured by high-resolution respirometry (Oxygraph-2k, Oroboros Instruments). IDG-SW3 cells were cultured in differentiation media with the indicated times. Cells were trypsined and resuspended again in differentiation media without FBS. 700,000 cells were added to the experimental chamber. Following stabilization of routine respiration, ATP synthesis was inhibited with 1 μg/ml of oligomycin to analyze respiratory uncoupling. Next, carbonylcyanide-4-(trifluoromethoxy)-phenyl-hydrazone 1 μM (FCCP) was titrated to achieve maximum flux through the electron transfer system (ETS). Finally, respiration was inhibited by the sequential addition of rotenone (0.1 μM) and antimycin A (2.5 μM). The remaining O2 flux after inhibition with antimycin A (O2 flux independent of the electron transfer system) was subtracted to calculate the different respiratory states. Oxygen flux values were expressed relative to protein content determined by the BCA method.
2.7. Gene expression
Total RNA was isolated from IDG-SW3, primary osteoblasts, primary osteocytes, and mouse calvaria cells using the TRIsure reagent (Bioline, London, UK). Purified RNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Quantitative PCR was carried out using the ABI Prism 7900 HT Fast Real-Time PCR System and a Taqman 5′-nuclease probe method (Applied Biosystems) with SensiFAST Probe Hi-ROX Mix (Bioline). All transcripts were normalized to TATA binding protein (Tbp) expression.
2.8. Western blot assay
Identification of proteins from cell extracts was performed by immunoblotting against NRF2 (Cell Signaling #12721), KEAP1 (Santa Cruz Biotechnology #514914), OXPHOS complexes (Mitosciences #607300), or α-TUBULIN (Millipore #CP06), diluted at 1:1000. Immuno-reactive bands were detected with horseradish-peroxidase-conjugated secondary antibodies and an EZ-ECL kit (Biological Industries, Cromwell, CT, USA).
2.9. Chromatin immunoprecipitation assay
A ChIP assay was performed as previously described [25]. Primary osteocytes were treated with 5 μM DMF for 48 h. Cells were fixed with 1% formaldehyde for 10 min. ChIP was carried out using 1 μg of anti-NRF2 (Cell Signaling #12721) or anti-IgG (Upstate) and purified with 20 μl Magna ChIP Protein A + G Magnetic Beads (Millipore). Purified DNA fragments were analyzed by qPCR with SYBR Green.
2.10. Determination of mtDNA by qPCR
Two μg of total DNA was used for mitochondrial DNA determination by qPCR. Primers for a mitochondrial DNA gene (tRNA-Glu) and a nuclear DNA gene (Fgf23) were used. The cycling conditions used were 43 cycles of 30 s at 95 °C, 30 s at 70 °C, and 60 s at 72 °C.
2.11. Mitochondrial isolation
Mitochondria were isolated as described by Ref. [26]. Briefly, cells were trypsinizate and centrifuged at 300xg for 5 min. Cell pellet was resuspended in 1 ml MIB Buffer (200 mM sucrose, 10 mM TRIS/MOPS, 1 mM EGTA/Tris, pH 7.4). Cell suspension was homogenized using a glass potter on ice. Homogenate was centrifuged at 600xg at 4 °C for 10 min to remove cell debris. Supernatant was collected and mitochondria were pellet by centrifugation at 1,0000xg, 4 °C for 10–15 min. Pellet was washed once with MIB buffer. The final pellet was resuspended in MIB buffer or the appropriate buffer for each assay.
2.12. Determination of cellular GSSG and GSH
The GSSG/GSH ratio was determined as described by Senft et al. [27]. Briefly, 2 × 105 cells were lysed with 600 μl of ice-cold PCA-RQB buffer [20 mM HCl, 5 mM diethylenetriaminepentaacetic acid (DTPA)], 10 mM ascorbic acid, and 6% perchloric acid. Samples were centrifuged at 12,000g for 5 min at 4 °C. Supernatants were collected for GSSG/GSH ratio determination while the pellet was kept for protein quantification and normalization. The acidic pH of deproteinized samples was neutralized with 1 mM phosphate buffer at pH 7. For the determination of GSH, 45 μl of neutralized sample was incubated with 2 μl of RQB buffer (20 mM HCl, 5 mM DTPA, 10 mM ascorbic acid) or NEM (7.5 mM N‐ethylmaleimide in RQB buffer) for 5 min at room temperature. Then, 100 μl of 0.1 M phosphate buffer and 15 μl of O-PA (5 mg of ortho‐phthaldialdehyde in ethanol) were added to each tube. The reaction was incubated for 30 min at room temperature in the dark. Samples were loaded in a black 96-well Costar plate with a flat clear bottom. Fluorescence was measured in a FLUOstar plate (excitation 365 ± 5 nm, emission 430 ± 20 nm) and GSH concentration was calculated relative to a GSH standard curve. For the determination of GSSG, 45 μl of neutralized sample were incubated with 2 μl NEM (7.5 mM N‐ethylmaleimide in RQB buffer) for 5 min at room temperature. Then, 3 μl of RQB buffer or 100 mM Na2S2O3 were added to each tube. The reaction was incubated for 1 h at room temperature in the dark. Then, 100 μl of 0.1 M phosphate buffer and 15 μl of O-PA (5 mg of ortho‐phthaldialdehyde in ethanol) were added to each tube. The reaction was incubated for 30 min at room temperature in the dark. Samples were loaded in a black 96-well Costar plate with a flat clear bottom. Fluorescence was measure in a FLUOstar plate (excitation 365 nm, emission 430 nm) and GSSG concentration was calculated relative to a GSH standard curve.
2.13. Determination of mitochondrial GSSG and GSH
Mitochondrial glutathione levels were determined using the Quantification kit for oxidized and reduced glutathione (Sigma-Aldrich) based on the 5,5′-dithio-bis(2-nitro-benzoic acid; DTNB) assay. Briefly, Mitochondria were isolated as described previously. Mitochondrial pellets were resuspended in 10 mM HCl, and lysed by freezing and thawingtwice. 5-Sulfosalicylic acid was added to a final concentration of 0.5%. GSSG and GSH were determined following manufacture instructions. Color changes were monitored by absorbance at 405 nm. GSH and GSSG levels were quantified using standard curves.
2.14. Hydrogen peroxidase assay
The generation of Mitochondrial H2O2 was determined using Amplex™ Red Hydrogen Peroxide/Peroxidase Assay Kit (Thermofisher) following manufacturer's instructions. Briefly, mitochondria were isolated as described previously. Mitochondrial pellets were diluted in 1x Reaction Buffer and incubated in presence of 100 μM Amplex® Red reagent and 0.2 U/mL HRP for 30 min at room temperature and protected from light. Fluorescence was measured in a fluorimeter (Ex: 550 nm/Em: 590 nm). H2O2 levels were quantified using a standard curve.
2.15. MicroCT analysis
Femurs from mice were dissected, cleaned of soft tissue, and fixed in 4% paraformaldehyde (PFA) for 24 h. High‐resolution images from the femur were acquired using a microCT imaging system (Skyscan 1272, Bruker microCT, Kontich, Belgium) in accordance with recommendations from the American Society of Bone and Mineral Research (ASBMR). Samples were scanned in air at 60 kV and 166 μA with an exposure time of 5600 ms, using a 1 mm aluminum filter and an isotropic voxel size of 11 μm. Two‐dimensional images were obtained every 1° of a 180° rotation and subsequently reconstructed using NRecon reconstruction software and analyzed with CT‐Analyzer (SkyScan). For trabecular measurements, manual VOI was employed, starting at 100 slices from the distal growth plate of the femur and extending to the diaphysis for 150 slices. Cortical measurements were performed by delineating the femur medial cortex for 100 slices around the femoral midshaft. A Gaussian noise filter was applied for reconstruction and a global binary threshold was manually established at 15 for trabecular analysis and at 50 for cortical analysis.
2.16. Histological analysis
For histological preparations, samples were fixed in 4% paraformaldehyde for 24 h at 4 °C, decalcified in 14% EDTA (pH 7.4) for 6 weeks, and embedded in paraffin. Samples were cut into 7 μm sections and stained with Hematoxylin/Eosin or TRAP.
2.17. Statistical analysis
Statistical analyses were performed using Student's t-test or two-way analysis of variance (ANOVA). Quantitative data are presented as the mean ± standard error of the mean (SEM). Differences were considered significant at *p < 0.05, **p < 0.01, and ***p < 0.001.
3. Results
3.1. Increased OXPHOS activity and ROS generation are associated with osteocytogenesis
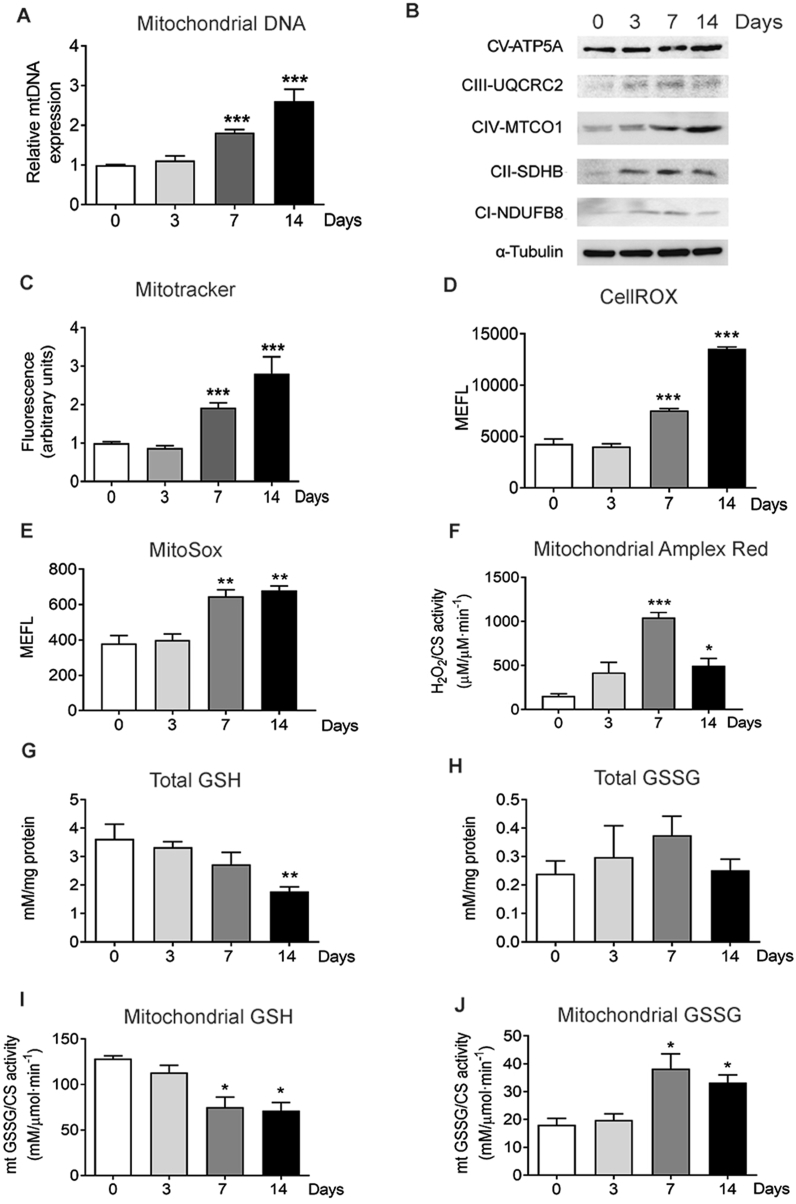
Confluent IDG-SW3 cells differentiated into osteocytes after 14–21 days in culture, constituting a well-established model of osteocytogenesis [7]. Previous reports from our group suggested metabolic reprogramming towards oxidative phosphorylation during osteocyte differentiation [12]. Therefore, using this cell model, we investigated the mitochondrial content during osteocyte specification. Measurement of mitochondrial DNA (mtDNA) revealed that differentiation resulted in a progressive increase in the number of mitochondria per cell (Fig. 1A). This increase in mtDNA was consistent with increased protein levels of different oxidative phosphorylation (OXPHOS) complexes and higher signal of the mitochondrial membrane potential probe MitoTracker, suggesting that the newly formed mitochondria were metabolically active (Fig. 1B and C). Moreover, we also measured mitochondrial respiration in intact, differentiated IDG-SW3 cells. We determined routine O2 consumption, leak state (uncoupled respiration after addition of oligomycin), electron transfer capacity (after FCCP addition, (ETS)). Differences in respiratory parameters were not significant between groups although slightly increased electron transfer capacity was observed in cells differentiated for 14 days (Suppl. Figure 1A). To determine whether osteocytic differentiation also resulted in altered ROS levels, we used confocal microscopy and FACS analysis to assess oxidative stress in these cells. Enhanced levels of ROS measured by CellRox were observed in parallel with higher mitochondrial content during osteocytogenesis in intact IDG-SW3 cells (Fig. 1D and E and Suppl. Figure 1B). Determination of mitochondrial peroxides by MitoSOX and AmplexRed in isolated mitochondria from differentiated IDG-SW3 cells showed increased levels of mitochondrial peroxides (Fig. 1F). Reduced glutathione (GSH) is one of the most important scavengers of ROS and its ratio with oxidized glutathione (GSSG) is used as a marker of increased ROS levels. During osteocytogenesis of IDG-SW3 cells, we found a progressive decrease in GSH levels and an increase in GSSG levels in both intact cells and isolated mitochondria (Fig. 1G–J). The mitochondrial content of murine primary osteoblasts and osteocytes was compared. Osteocytes displayed a 30–40% increase in the mitochondrial number per cell compared to osteoblasts (Suppl. Figure 2). Moreover, confocal images showed that primary osteoblast and osteocytes arranged their mitochondria in elongated tubules, forming networks that were shown to be more efficient at producing ATP [28]. These networks follow a similar pattern as that of ROS staining, suggesting that mitochondrial activity is the major source of ROS in these cells (Suppl. Figure 2).
Fig. 1.
Increased OXPHOS activity and ROS generation are associated with osteocytic differentiation. (A) Quantification of mtDNA in undifferentiated IDG-SW3 after 3, 7, and 14 days of differentiation. Results are plotted as expression relative to undifferentiated IDG-SW3 (mean ± SEM; n = 7). (B) Analysis of the levels of mitochondrial complexes in undifferentiated IDG-SW3 after 3, 7, and 14 days of differentiation (C) Flow cytometry analysis of mitochondrial membrane potential using Mitotracker Deep Red in undifferentiated IDG-SW3 and after 3, 7, and 14 days of differentiation. Results are plotted as expression relative to undifferentiated IDG-SW3 (mean ± SEM; n = 8). (D) Flow cytometry analysis of total ROS stained with CellROX Deep Red in undifferentiated IDG-SW3 and after 3, 7, and 14 days of differentiation. Results are plotted as Molecules of Equivalent Fluorescein (mean ± SEM; n = 4). (E) Flow cytometry analysis of mitochondrial superoxide stained with MitoSOX Red in undifferentiated IDG-SW3 and after 3, 7, and 14 days of differentiation. Results are plotted as Molecules of Equivalent Fluorescein (mean ± SEM; n = 4). (F) Determination of superoxide produced in isolated mitochondria using Amplex Red. Results were normalized to citrate synthase (CS) activity. (mean ± SEM; n = 3). (G and H) Total GSH (G) and GSSG (H) in undifferentiated IDG-SW3 and after 3, 7, and 14 days of differentiation. Results are plotted as mean ± SEM (n = 5). (I, and J) GSH (I) and GSSG (J) levels in isolated mitochondria from undifferentiated IDG-SW3 and after 3, 7, and 14 days of differentiation. Results were normalized to citrate synthase (CS) activity (mean ± SEM; n = 4 *p < 0.05, **p < 0.01, and ***p < 0.001 using Student's t-test.
3.2. Increased NRF2 activity during osteocytogenesis
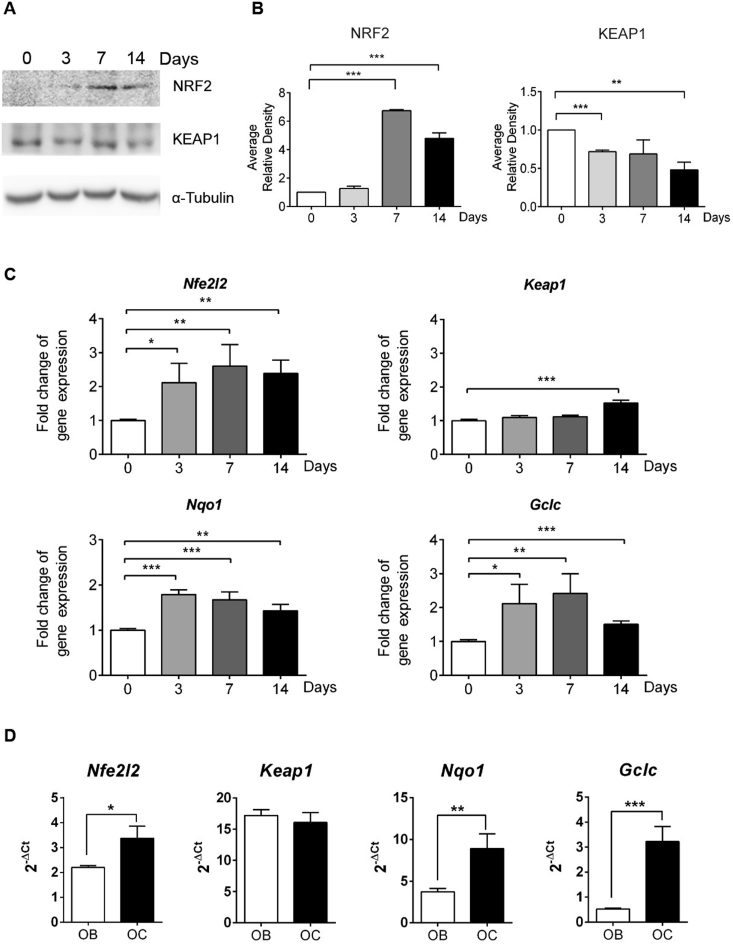
In view of the fact that NRF2 is a major oxidative stress sensor and inducer of the antioxidant response, we hypothesized an increase in NRF2 levels and activity in response to the increase in ROS levels and GSSG/GSH ratio. We found a significant increase in total NRF2 protein levels in IDG-SW3 cells after 7 and 14 days of differentiation, whereas KEAP1 protein levels slightly lowered over time (Fig. 2A and B). We confirmed this observation of higher NRF2 activity, as the NRF2 target genes NAD(P)H dehydrogenase quinone 1 (Nqo1) and glutamate-cysteine ligase catalytic subunit (Gclc) were transcriptionally activated during IDG-SW3 differentiation (Fig. 2C). Similarly, transcription of Nfe2l2 itself was also higher throughout osteocytogenesis, whereas Keap1 mRNA levels remained unaltered until the 14th day when they were also increased (Fig. 2C). We confirmed that isolated primary murine osteocytes displayed higher expression of Nfe2l2 mRNA and NRF2 targets Nqo1 and Gclc than primary osteoblasts, whereas Keap1 mRNA levels were not altered (Fig. 2D). Altogether, these findings demonstrate elevated NRF2 activity in osteocytes and during differentiation of IDG-SW3 cells and suggest that NRF2 may be required to facilitate osteocytogenesis.
Fig. 2.
NRF2 activity during osteocytogenesis. (A) Undifferentiated IDG-SW3 cells after 3, 7, and 14 days of differentiation were analyzed for NRF2 and KEAP1 protein expression. Tubulin levels were used as a protein load control. (B) NRF2 and KEAP1 levels were quantified and normalized by tubulin levels. Results are plotted as mean ± SEM (n = 3). (C) mRNA expression of Keap1, Nfe2l2, and target genes in undifferentiated IDG-SW3 after 3, 7, and 14 days of differentiation. mRNA levels were quantified by qRT-PCR, normalized by Tbp, and plotted as expression relative to undifferentiated IDG-SW3 (mean ± SEM; n = 8) (D) mRNA expression of Keap1, Nfe2l2, and target genes in primary cultures of osteoblasts and osteocytes. mRNA expression was quantified by RT-qPCR and normalized to Tbp expression. Results are plotted as 2-ΔΔCt (mean ± SEM; n = 6). *p < 0.05, **p < 0.01, and ***p < 0.001 using Student's t-test.
3.3. Nfe2l2 deletion in osteocytes causes osteopenia and impairs osteocyte-specific gene expression
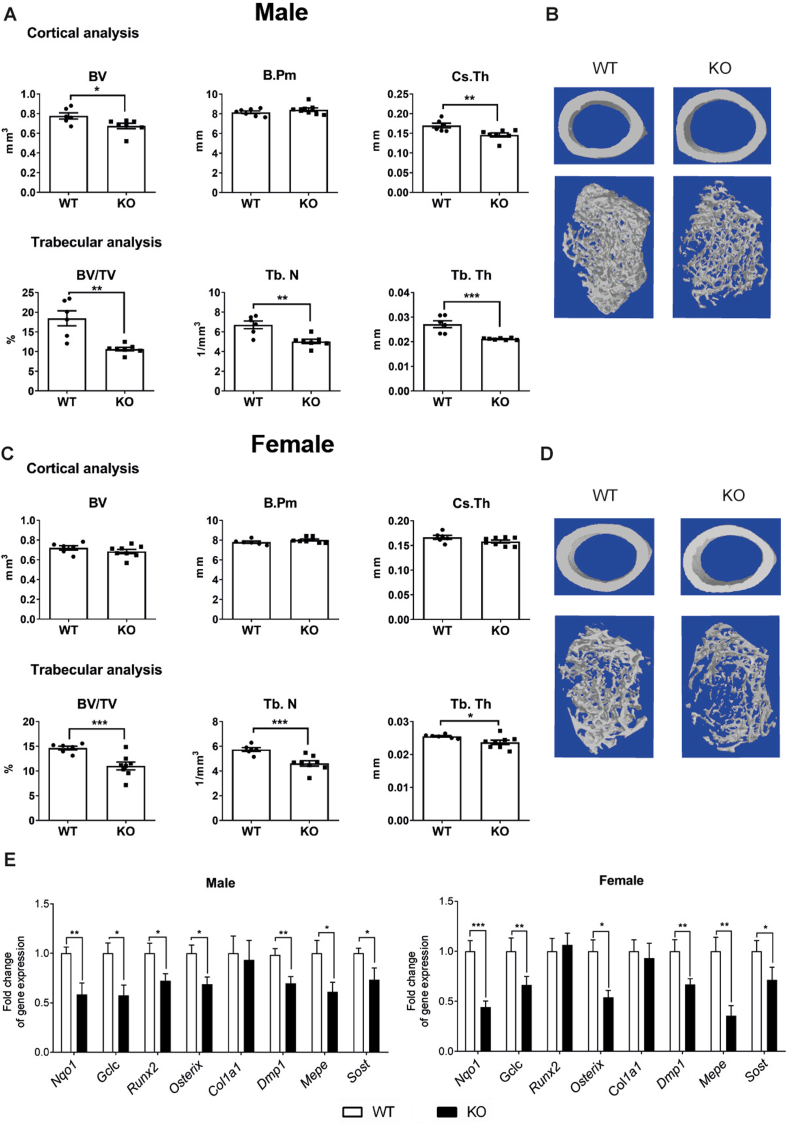
Whereas all previous studies reported a consistent sexual dimorphism, they also reported conflicting results on the effects on bone mass after whole-body deletion of Nfe2l2 in male mice. Moreover, information about the role of NRF2 in specific bone cell types is missing. To determine the potential role of NRF2 in the osteocytic cell lineage, we first deleted its expression in mature osteoblasts and osteocytes by breeding Dmp1-CreERt2 mice with Nfe2l2fl/fl mice [29]. In these mice, Cre recombinase fused to a triple mutant form of the human estrogen receptor does not bind its natural ligand (17β-estradiol) under physiological concentrations, but is active upon tamoxifen induction. Deletion was performed with low doses of tamoxifen (20 mg/kg) in three-week-old mice, therefore avoiding the effects of deletion at the embryonic or neonatal period. As previously shown using this tamoxifen dose, we minimized the effects of tamoxifen on bone turnover in mice and a robust and restricted activation of the transgene in osteocytes and mature osteoblasts was achieved [[29], [30], [31], [32]]. Four weeks after deletion, we visualized bones in male and female mice in distal femurs by micro‐computed tomography scanning (μCT) and histological analysis. Deletion of Nfe2l2 led to a decrease in both trabecular and cortical bone architecture. Dmp1-CreERt2:Nf2l2fl/fl (hereafter Dmp1/Nfe2l2-KO) male mice presented lower cortical bone volume (BV) associated with reduced cortical thickness (Cs.Th) while the bone perimeter (B.Pm) around the midshaft was not affected (Fig. 3A and B). Moreover, distal femurs in males also presented less trabecular bone volume (BV/TV) resulting from a significantly lower trabecular number (Tb.N) and thickness (Tb.Th) (Fig. 3A and B). Although lower in magnitude, trabecular analysis of distal femurs in Dmp1/Nfe2l2-KO females also showed a significant reduction in trabecular bone volume (BV/TV), trabecular number (Tb.N), and thickness (Tb.Th). However, reduction in cortical bone parameters only reached significant differences for cortical thickness (Fig. 3C and D). To clarify the underlying reason for the osteopenic phenotype of Dmp1/Nfe2l2-KO mice, we analyzed the expression of osteoblast and osteocyte genes. As expected, expression of the NRF2 targets genes Nqo1 and Gclc were reduced in both male and female bones. Dmp1/Nfe2l2-KO male and female mice also displayed reduced expression of osteocyte genes, including Dmp1, Mepe, and Sost, and additional bone-cell genes, such as Runx2 or Osx (Fig. 3E). Reduced osteocyte gene expression occurred without significant changes in the density of osteocytes per bone area (N.Oc/B.Ar) (Suppl. Figure 3). Notably, the number of osteoclasts was significantly higher in male Dmp1/Nfe2l2-KO mice, whereas this increase was not significant in female mice (Suppl. Figure 2). These effects in osteoclasts took place without altered Rankl/Opg expression (Suppl. Figure 3).
Fig. 3.
Bone phenotype of mice with Nfe2l2 conditional deletion in osteocytes. (A–D) MicroCT analysis and representative images of femurs obtained from male (A and B) or female (C and D) Nfe2l2f/f;Dmp1-Cre-Ert2 and control (Nfe2l2f/f) mice. (E) mRNA levels in calvaria obtained from NRF2 conditional knock-out (Nfe2l2f/f;Dmp1-Cre-Ert2) and control (Nfe2l2f/f) mice. mRNA expression levels were measured by RT-qPCR and normalized to Tbp expression. Results are plotted as mean ± SEM of seven to eleven independent animals. *p < 0.05, **p < 0.01, and ***p < 0.001 using Student's t-test.
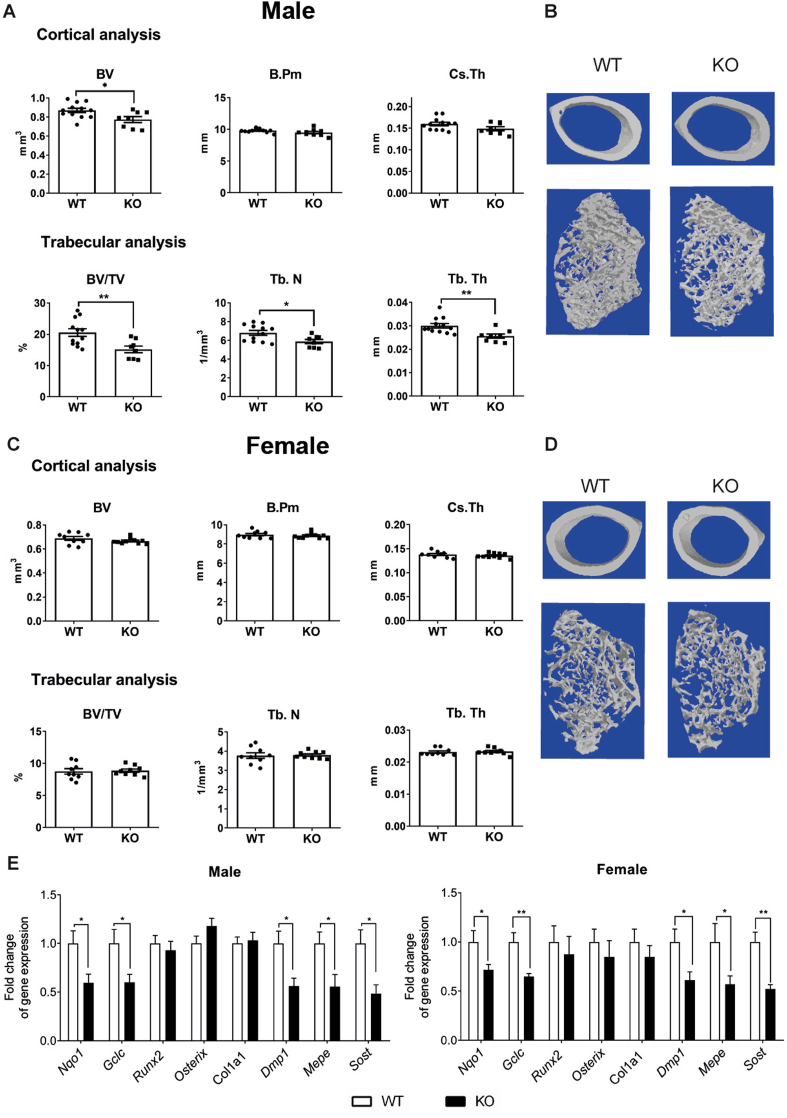
Given the effect of Nfe2l2 deletion in osteocytes we aimed to establish the impact of Nfe2l2 deletion in the previous steps of osteoblast differentiation. Osteoblast-specific Nfe2l2 mutant mice were generated by means of a Cre recombinase adjacent to the 2.3-Col1a1 promoter Nfe2l2;Col1a1-Cre (hereafter Col1a1/Nfe2l2-KO), which drives constitutive expression of Cre exclusively in osteoblasts from E14 [24]. MicroCT analysis of distal femurs in male mice demonstrated less trabecular bone volume (BV/TV) and lower trabecular number (Tb.N) and thickness (Tb.Th), similar to Dmp1/Nfe2l2-KO mice (Fig. 4A and B). Surprisingly, male mice did not show major defects in cortical bone volume (Fig. 4A and B). Additionally, female Col1a1/Nfe2l2-KO mice did not show changes in either cortical or trabecular bone parameters (Fig. 4C and D). Expression of the NRF2 targets genes Nqo1 and Gclc were reduced in both male and female bones to a similar extent as that in Dmp1/Nfe2l2-KO mice. Col1a1/Nfe2l2-KO male and female mice also displayed reduced expression (about 50%) of the osteocyte genes Dmp1, Mepe, and Sost, but no major changes in other osteoblastic genes, such as Runx2 or Osx (Fig. 4E). As observed in Dmp1/Nfe2l2-KO mice, the number of osteocytes was unaltered and the number of osteoclasts was higher in male but not female Col1a1/Nfe2l2-KO mice (Suppl. Figure 4). In conclusion, our data demonstrate the sexually dimorphic relevant role of NRF2 in osteocyte gene expression and function in vivo that results in lower bone mass and an increased number of osteoclasts in male mice.
Fig. 4.
Bone phenotype of mice with Nfe2l2 conditional deletion in osteoblasts. (A–D) MicroCT analysis and representative images of femurs obtained from male (A and B) or female (C and D) Nfe2l2f/f;Col1a1-Cre and control (Nfe2l2f/f) mice (E) mRNA levels in calvaria obtained from NRF2 conditional knock-out (Nfe2l2f/f;Col1a1-Cre) and control (Nfe2l2f/f) mice. mRNA expression levels were measured by RT-qPCR and normalized to Tbp expression. Results are plotted as mean ± SEM of nine to eleven independent animals. *p < 0.05, **p < 0.01, and ***p < 0.001 using Student's t-test.
3.4. NRF2 activity regulates osteocyte-specific gene expression
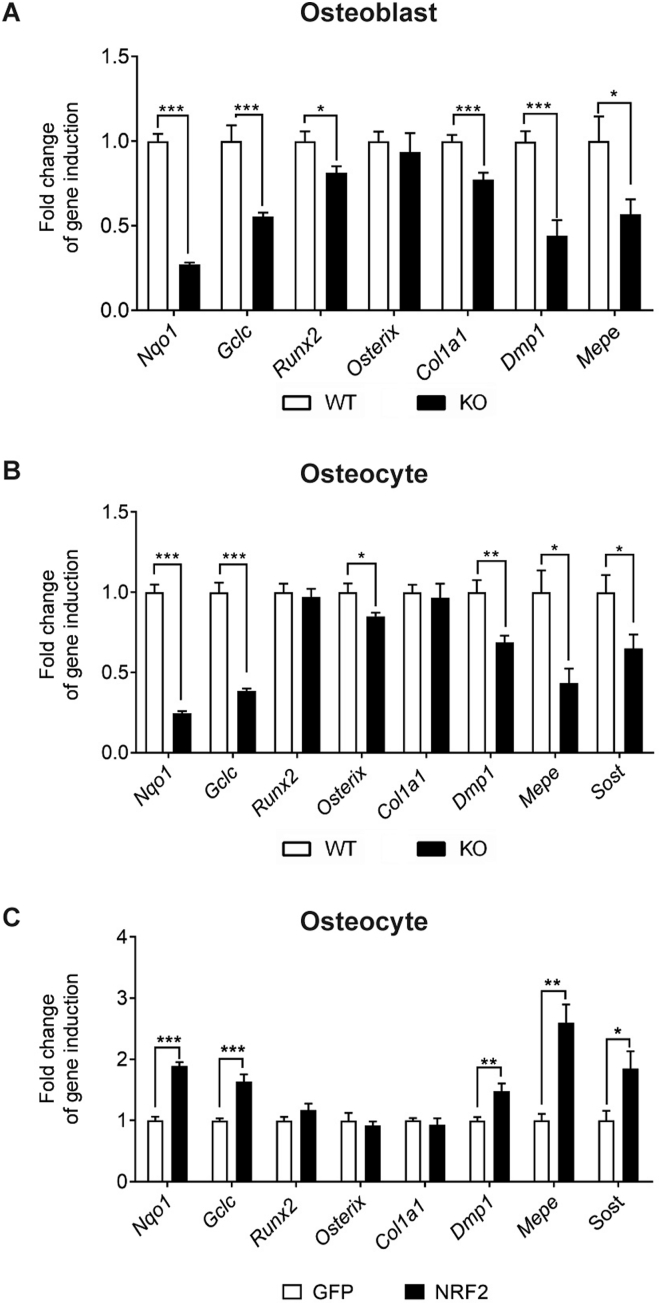
Our data suggest that some of the effects observed in vivo arise from the alteration of paracrine communication between distinct bone cell types. Therefore, to identify further cell-autonomous effects, we analyzed Nfe2l2 deletion in vitro on primary cultures of osteblasts and osteocytes after transducing them with Cre retroviruses. Expression of the NRF2 targets genes Nqo1 and Gclc, were strongly reduced. While major osteoblastic genes, such as Runx2, Osx, or Col1a1, were only slightly affected, osteoblasts and osteocytes displayed reduced expression of osteocytic genes, including Dmp1, Mepe, and Sost (Fig. 5A and B). Moreover, analysis of osteocytes overexpressing NRF2 showed inverse effects, including increased Nqo1 and Gclc mRNA levels and higher expression of the osteocytic genes Dmp1, Mepe, and Sost (Fig. 5C).
Fig. 5.
Activation of Nrf2 induce osteocyte-specific gene expression. (A and B) mRNA expression levels of osteoblastic and osteocytic genes in primary osteoblasts (A) (n = 9) and primary osteocytes (B) (n = 9) wild type and knockout for NRF2. (C) mRNA expression in primary osteocytes infected with NRF2 expression vectors (n = 6). mRNA expression levels were measured by RT-qPCR and normalized to Tbp expression. Results were plotted as expression relative to cells infected with GFP vector (mean ± SEM of six to eight independent experiments). *p < 0.05, **p < 0.01, and ***p < 0.001 using Student's t-test.
Mechanistically, increased expression of osteocytic genes upon activation of NRF2 could rely on the intrinsic transcriptional effects of NRF2 itself or alternatively, depending on decreased ROS upon activation of the anti-oxidative machinery. We tested this hypothesis by treatment of osteocytes deficient in Nfe2l2 with 1 mM N-acetyl-cysteine (NAC). Antioxidant treatment slightly reduced Nqo1 and Gclc expression levels in control osteocytes, whereas osteocytes deficient in Nfe2l2 were refractory to changes upon NAC addition (Suppl. Figure 5). Either treatment of NAC or deficiency of Nfe2l2 had no major effects on Runx2 or Col1a1 expression, and only slightly decreased Osx mRNA levels in Nfe2l2 deficient osteocytes (Suppl. Figure 5). As shown above, Nfe2l2 deficiency reduced the levels of Dmp1, Mepe, and Sost. More importantly, the antioxidant effects of NAC did not alter expression of Dmp1 in control cells, or after Nfe2l2 deletion, but reduced Mepe and Sost mRNA expression independent of the Nfe2l2 status (Suppl. Figure 5). NAC is not only an antioxidant but also a cysteine source that can change redox thiol signaling independently of ROS. Therefore, we also analyzed expression of osteocytic genes after addition of the non-thiol antioxidants Trolox and MitoQ. Trolox is an analog of vitamin E also used to reduce oxidative stress, whereas Mito Q is a mitochondria-targeted antioxidant designed to protect against oxidative damage. Treatment of osteocytes with this non-thiol antioxidants yielded similar results where osteocytes deficient in Nfe2l2 were refractory to changes upon antioxidant addition (Suppl. Figure 6).
Several drugs with the ability to activate NRF2 have been developed. Among them, dimethyl fumarate (DMF) was shown to activate NRF2 and an antioxidant response and it has been approved for the treatment of multiple sclerosis or psoriasis [33]. To further confirm whether the expression of osteocytic genes was modulated by NRF2, we treated osteocytes deficient for Nfe2l2 with 5 μM DMF. This dose is far below the maximum serum concentration of DMF when clinically administrated (www.accessdata.fda.gov/drugsatfda_docs/label/2013/204063lbl.pdf). DMF treatment increased Nqo1 and Gclc expression levels in control osteocytes, whereas in osteocytes deficient for Nfe2l2, these responses were blunted (Suppl. Figure 5). DMF had no major effects on Runx2, or Col1a1 expression but strongly induced expression of Dmp1, Mepe, and Sost mRNA expression in control cells. These responses were severely reduced in Nfe2l-deficient osteocytes (Suppl. Figure 5). Altogether, the data suggest that NRF2 activity led to transcriptional activation of these genes.
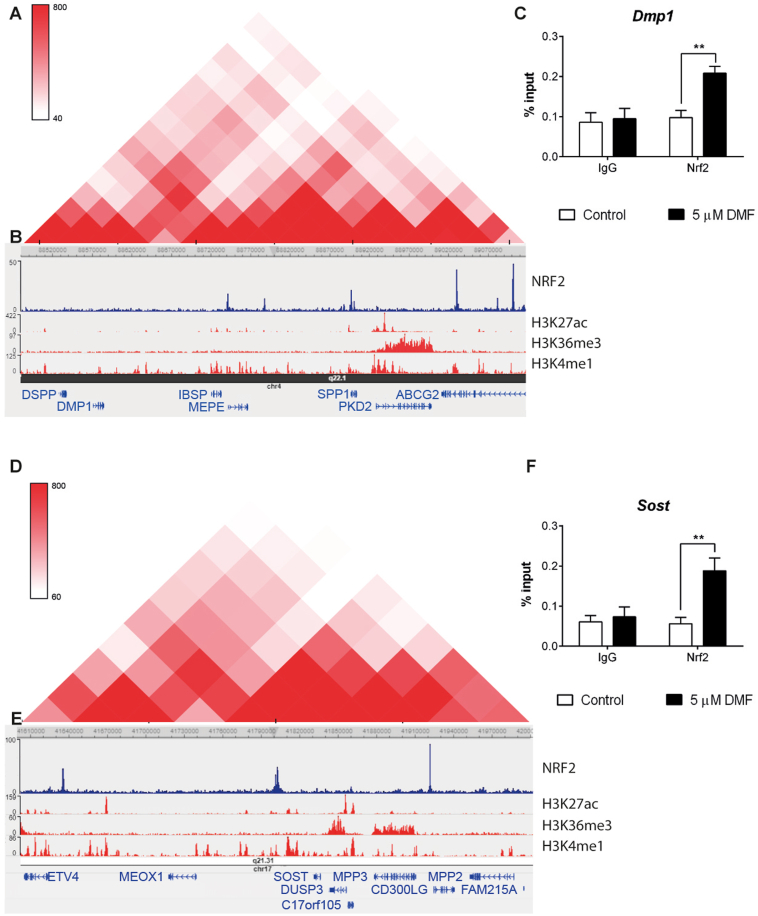
Genomic analysis indicated that Dmp1, Mepe, and additional SIBLING (Small Integrin-Binding LIgand, N-linked glycoproteins) family members, such as Dspp, Ibsp, or Spp1, share the same topologically associated domain (TAD) in human chromosome 4 (Fig. 6A). TADs play a critical role, favoring the contact of shared regulatory elements and preventing undesired interactions with neighboring TADs [34]. Bioinformatic analysis of whole-genome NRF2-ChipSeq data (GSE113497G) showed that this TAD is highly enriched for binding NRF2 (Fig. 6B) and correlates with the presence of the known NRF2 target Abcg2 in the close vicinity. We confirmed binding of NRF2 to the regulatory region of Dmp1 by ChIP assays in primary osteocytes after DMF activation (Fig. 6C). Similarly, whole-genome NRF2-ChipSeq data (GSE113497G) showed strong NRF2 binding in the distal enhancer located at 3′of the Sost gene (Fig. 6E). This distal enhancer has been shown to be sufficient for osteocyte specific expression of Sost and its deletion to be causative of Van Buchem disease [35,36]. We also confirmed binding of NRF2 to the regulatory region of Sost by ChIP assays in primary osteocytes after DMF activation (Fig. 6F). In conclusion, the data suggests that NRF2 activated expression of Dmp1, Mepe, and Sost through direct binding to their regulatory regions.
Fig. 6.
Osteocytic genes are organized in TADs regulated by NRF2 (A and D) Topological domain and looping structure indicated by Hi-C data from Dixon et al. (2015) in MSC. The schematic shows TAD (red-shaded triangle) containing the SIBLING family of proteins (A) and Sost (D). (B and E) ChIP-seq data showing NRF2 binding sites in A549 cells (data obtained from GSE113497), as well as active promoter/enhancer associated histone mark H3K27ac, gene body associated histone mark H3K36me3 and promoter associated histone mark H3K4me3 obtained from human osteoblast epigenome. (C and F). Chromatin immunoprecipitation from osteocytes cultured in the presence or absence of 5 μM DMF for 48 h. Results were normalized to input chromatin and plotted relative to untreated osteocytes (mean ± SEM; n = 5). *P < 0.05, **P < 0.01, and ***P < 0.001 using Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. DMF treatment preserves bone mass and osteocytic gene expression in ovariectomy-induced osteoporosis
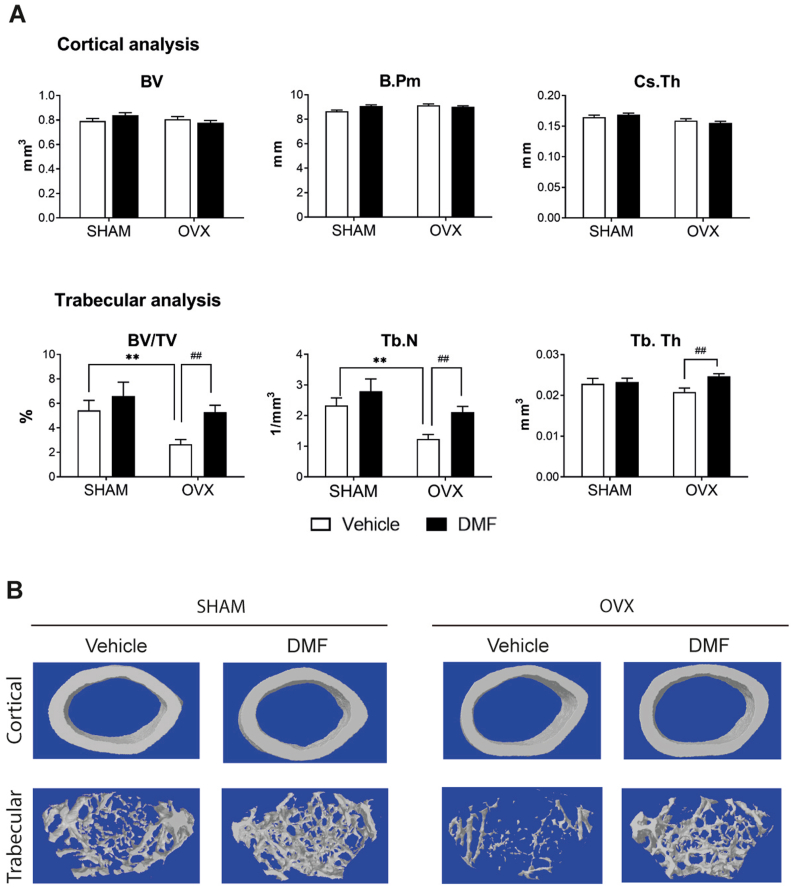
A number of pathological conditions are alleviated by NRF2 activation, including chronic kidney disease, multiple sclerosis, hepatotoxicity, or rheumatoid arthritis [16]. However, information about potential beneficial effects of pharmacological activation of NRF2 in bone pathologies is scarce [37,38]. We analyzed the effects of DMF in osteopenia induced by ovariectomy (OVX), an established model to induce osteoporotic bone loss. Ovariectomized mice were treated with 100 mg/kg of DMF for 8 weeks. MicroCT analysis indicated that cortical bone parameters were not significantly modified by either ovariectomy or DMF treatment (Fig. 7A and B). However, ovariectomized mice displayed decreased trabecular bone mass, mostly due to a reduced number of trabeculae. More importantly, treatment with DMF restored the trabecular bone mass and increased the number of trabeculae and their thickness in ovariectomized mice (Fig. 7A and B). Gene expression analysis indicated that ovariectomy led to decreased levels of Nfe2l2, whereas DMF treatment restored the levels of Nfe2l2 and increased the levels of its target Nqo1 (Suppl. Figure 7). Ovariectomized mice also had reduced expression of Dmp1, Osx, and Sost, whereas DMF treatment was able to rescue their gene expression in bone (Suppl Figure 7). Histomorphometric measurements showed that the number of osteoclasts was significantly higher in OVX mice, whereas this increase was reduced when these mice were treated with DMF (Suppl. Figure 7). These results demonstrate that DMF treatment restored the expression of the osteocytic genes Dmp1, Osx, and Sost and was effective in rescuing the osteopenic phenotype after ovariectomy.
Fig. 7.
Dimethylfumarate (DMF) reduces bone loss induced by ovariectomy. (A) MicroCT analysis of femurs obtained from ovariectomized (OVX) and sham operated (SHAM) mice treated with 100 mg/kg of dimethylfumarate (DMF) or vehicle. (B) Representative images of cortical and trabecular section of femurs obtained from OVX and SHAM mice treated with 100 mg/kg of DMF or vehicle. Results are plotted as mean ± SEM of seven to eleven independent animals. *p < 0.05, ** or ##p < 0.01, *** or ###p < 0.001 using Student's t-test. * refer to statistics performed against SHAM mice treated with vehicle. Similarly, # refers to significance between OVF mice treated with vehicle or DMF.
4. Discussion
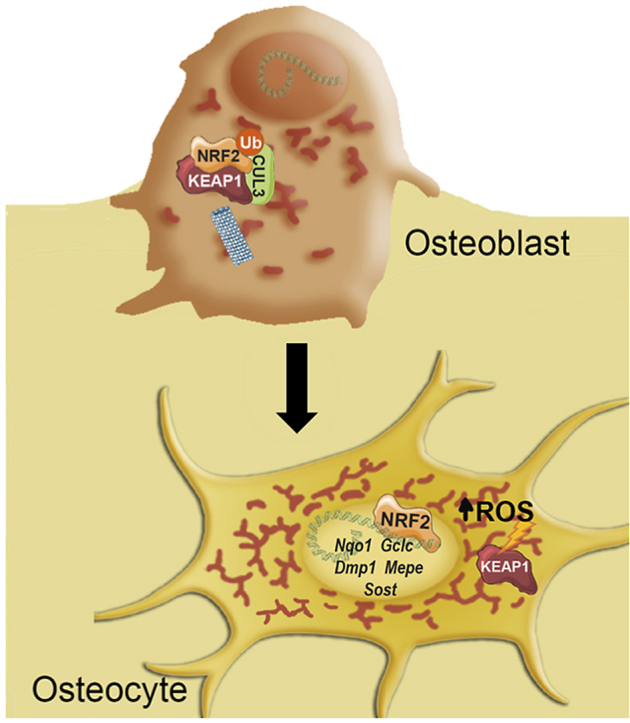
In this study, we identified that increased mitochondrial content led to increased levels of ROS during the transition from osteoblasts to osteocytes. As a major sensor of oxidative stress, the transcription factor NRF2 becomes activated during osteocytogenesis and directly transactivates a number of osteocyte-specific gens such as Dmp1, Mepe, and Sost. Furthermore, we found that mice deficient for NRF2 in osteocytes and mature osteoblasts were osteopenic with significant sexual dimorphism. Finally, treatment of ovariectomized mice with the NRF2 activator DMF restored their bone loss phenotype. Our comprehensive analysis in cells and genetic mouse models strongly support a relevant role for NRF2 in osteocytes for the maintenance of bone homeostasis.
In recent years, several studies have demonstrated that the metabolism of osteocytes optimize energy production to fulfill functional demands in a nutrient-restricted environment [39]. Our data showed a higher mitochondrial mass and function during the transition of osteoblasts into osteocytes. Accordingly, osteocytes in vivo contain numerous mitochondria to maintain their energy status [40,41]. As expected, since ROS mainly arise as byproducts of aerobic mitochondrial complexes I, and III, ROS levels increase during the transition from osteoblast to osteocyte. Unregulated accumulation of ROS has been shown to be a major contributor to aging and several diseases that include osteoporosis and bone diseases [12,14]. In contrast, we found that ROS at lower, non-toxic levels could actually contribute to osteocyte specification, as shown for other mesenchymal stem cell differentiation processes [13,42]. This link between ROS and cellular specification was already established for neurogenesis, astrocytogenesis, hepatic differentiation, and erythropoiesis [[43], [44], [45], [46]]. Consistent with its role as a regulator of oxidative stress, we found a correlation between NRF2 activity, mitochondrial content, and ROS levels during osteocytogenesis. Moreover, NRF2 activity increases have been associated with increased mitochondrial function as a feed-forward loop [47].
To address the unresolved issues of the role of NRF2 in specific bone cells in vivo, we found evidence that NRF2 activity has an indispensable role in the maintenance of bone homeostasis, both in the cortical and trabecular compartments. Previous studies with global deletion of NRF2 have shown sexual dimorphism in the skeletal phenotype [[17], [18], [19], [20],22]. Our data confirm this sexual dimorphism, with more profound effects of NRF2 deletion on bone mass in male mice. Interestingly, Dmp1/Nfe2l2-KO mice of both sexes display severe osteopenia, while Col1a1/Nfe2l2-KO mice only exhibit subtle osteopenia in males and non-significant changes in the female's skeleton. We observed that levels of CRE expression (Suppl. Figure 8), deletion of Nfe2l2, and expression of its target genes Nqo1 and Gclc were modified similarly in both models, arguing against different deleting efficiency. It is possible that deletion in the late embryonic and neonatal period (in Col1a1/Nfe2l2-KO), compared to deletion at postnatal P21-28 (in Dmp1/Nfe2l2-KO), could reprogram compensatory mechanisms in the former to keep bone formation and turnover. For instance, whereas Runx2 and Osx expression was significantly diminished in Dmp1/Nfe2l2-KO mice, it remains unaltered in Col1a1/Nfe2l2-KO of both sexes. Alternatively, it is possible that Nfe2l2 deletion in osteoblasts (Col1a1/Nfe2l2) or osteocytes (Dmp1/Nfe2l2) could somehow differentially modify their function and/or paracrine crosstalk to osteoclasts. More detailed studies are required to further explore these possibilities.
Our results also provide additional information about sexual dimorphism, observed previously after global Nfe2l2 deletion [[17], [18], [19], [20],22]. Although in our study deletion was performed only in the osteoblast/osteocyte lineages, dimorphic effects in bone parameters correlate with differential changes in osteoclast numbers. Interaction between NRF2 activation and signaling downstream of the sex steroid receptors could contribute to sex-specific differential regulation. For instance, deletion of estrogen receptors (ERα or β) differentially affects bone parameters in female and male mice and NRF2 displays transcriptional cooperation with ERs [48,49]. Similarly, androgen receptor (AR) function in osteocytes is required for proper bone homeostasis and NRF2 modifies AR-mediated transactivation [50,51]. Supporting this hypothesis, it has been shown that dissimilar expression of cytoprotective genes, Rankl/Opg ratio, or osteoclast numbers correlate with different effects of Nfe2l2 in bone maintenance, depending on sex [17,18]. NRF2 has been shown to negatively regulate the expression of cytokines that induce osteoclastogenesis, such as IL-1α, IL1β, IL6, TNFα, or RANKL [[52], [53], [54]]. Therefore, it is plausible to suggest that deletion of Nfe2l2 in osteocytes and mature osteoblasts would increase paracrine osteoclastogenic signaling.
Independent of the in vivo deletion model, sex, or in vitro genetic and pharmacological approaches, NRF2 activity induced transcriptional activation of a topologically associated domain (TAD), containing the SIBLING family of extracellular proteins. The SIBLING protein family is a structurally and phylogenetically homogeneous group that comprises Dspp, Dmp1, Mepe, Osteopontin Spp1, and Ibsp, located on human chromosome 4, which shows specific expression in bone cell types [55,56]. Sequencing data suggest that the SIBLING gene group appeared along with generation of the bony skeleton in the vertebrate lineage, highlighting their importance for bone tissue [[57], [58], [59], [60]]. SIBLING proteins are required for proper bone mineralization, as its defects led to osteoporosis and shared significant levels of functional redundancy [61,62]. Moreover, SIBLING proteins have been shown to inhibit osteoclast differentiation [61]. Whole-genome Chip-Seq assays and our Chip assays in osteocytes showed that this TAD, which also contains the canonical NRF2-activated gene Abcg2, bound NRF2, strongly suggesting that NRF2 activity has a key role in the regulation of SIBLING proteins in osteoblasts and osteocytes. Similarly, NRF2 bound to the 3′ region of the Sost gene in whole-genome Chip-Seq assays and our Chip assays. This region has been shown to be essential for osteocyte-specific expression and is responsible for Van Buchem disease [35]. Altogether, our results demonstrate the important role of NRF2 activity in the regulation of a number of relevant osteocyte-specific genes in vitro and in vivo.
Given these results we hypothesized that activation of NRF2 could become effective for bone pathologies. DMF (Tecfidera) was approved by the FDA and EMA after showing clinical effectiveness in patients with multiple sclerosis (www.accessdata.fda.gov/drugsatfda_docs/label/2014/204063s003s008s010lbl.pdf). DMF has a good tolerability and safety profile in humans and the most common adverse events were flushing and gastrointestinal events, which were of mild or moderate severity and appeared to be largely manageable [63]. Our data demonstrated that treatment with DMF prevented the deleterious effects of ovariectomy in the skeleton. Bone mass and architecture was mostly protected in the trabecular compartment. In addition, we found a decreased number of osteoclasts in DMF treated mice. Treatment with DMF also restored the expression of Dmp1, Osx, and Sost in ovariectomized mice, therefore improving osteocyte function and potential inhibitory effects on osteoclasts. However, we cannot exclude a direct effect of DMF on the reduction of recruitment and activation of osteoclasts through attenuation of ROS signaling as previously reported [37] or effects mediated by additional signaling pathways inhibited by DMF, such as NF-κB [64]. Therefore, further studies should be performed to optimize dosing and to fully understand the bone protecting mechanisms of DMF.
5. Conclusions
We report that increased mitochondrial biogenesis raised ROS levels and NRF2 activity during osteocytogenesis. NRF2 activity promoted osteocytic specification through transcriptional activation of osteocyte-specific genes. Furthermore, conditional ablation of Nfe2l2 in osteocytes or osteoblasts are evidence of its fundamental role in bone homeostasis. Finally, our results showed that treatment with the NRF2 activator dimethyl fumarate (DMF) might be beneficial for osteopenic bone pathologies. Altogether, we have uncovered the central role of NRF2 in bone homeostasis and the establishment of a transcriptional program leading to osteocyte specification.
Author contributions
Conceived and designed the experiments: CS-D, JLR, and FV. Performed the experiments: CS-D, LP, AM-M, CP-L, JAV, ND and FV. Analyzed the data: CS-D, LP, CP-L, JAV, PGR, JLR, and FV. Wrote the paper: CS-D and FV.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
We thank Dr. L. Bonewald for providing IDG-SW3 cells. We also thank B. Torrejón, E. Castaño, A. Aznar, and A. Gimeno for technical assistance. Cristina Sánchez de Diego, Arturo Martínez and Carolina Pimenta Lopes are recipients of an F.P.U. fellowship from the Spanish Ministry of Education. This research was supported by grants from the MEC and FEDER (BFU2017-82421-P).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101845.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- 1.Dallas S.L., Prideaux M., Bonewald L.F. The osteocyte: an endocrine cell . . . and more. Endocr. Rev. 2013;34:658–690. doi: 10.1210/er.2012-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karsenty G., Olson E.N. Bone and muscle endocrine functions: unexpected paradigms of inter-organ communication. Cell. 2016;164:1248–1256. doi: 10.1016/j.cell.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato M., Asada N., Kawano Y., Wakahashi K., Minagawa K., Kawano H., Sada A., Ikeda K., Matsui T., Katayama Y. Osteocytes regulate primary lymphoid organs and fat metabolism. Cell Metabol. 2013;18:749–758. doi: 10.1016/j.cmet.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Napoli N., Chandran M., Pierroz D.D., Abrahamsen B., Schwartz A.V., Ferrari S.L. Mechanisms of diabetes mellitus-induced bone fragility. Nat. Rev. Endocrinol. 2017;13:208–219. doi: 10.1038/nrendo.2016.153. [DOI] [PubMed] [Google Scholar]
- 5.Lai X., Price C., Modla S., Thompson W.R., Caplan J., Kirn-Safran C.B., Wang L. The dependences of osteocyte network on bone compartment, age, and disease. Bone Res. 2015;3 doi: 10.1038/boneres.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonewald L.F. The amazing osteocyte. J. Bone Miner. Res. 2011;26:229–238. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo S.M., Rosser J., Dusevich V., Kalajzic I., Bonewald L.F. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. J. Bone Miner. Res. 2011;26:2634–2646. doi: 10.1002/jbmr.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buenzli P.R., Sims N.A. Quantifying the osteocyte network in the human skeleton. Bone. 2015;75:144–150. doi: 10.1016/j.bone.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Piekarski K., Munro M. Transport mechanism operating between blood supply and osteocytes in long bones. Nature. 1977;269:80–82. doi: 10.1038/269080a0. [DOI] [PubMed] [Google Scholar]
- 10.Petrov N., Pollack S.R. Comparative analysis of diffusive and stress induced nutrient transport efficiency in the lacunar-canalicular system of osteons. Biorheology. 2003;40:347–353. [PubMed] [Google Scholar]
- 11.Wang L. Solute transport in the bone lacunar-canalicular system (LCS) Curr. Osteoporos. Rep. 2018;16:32–41. doi: 10.1007/s11914-018-0414-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sánchez-de-Diego C., Artigas N., Pimenta-Lopes C., Valer J.A., Torrejon B., Gama-Pérez P., Villena J.A., Garcia-Roves P.M., Rosa J.L., Ventura F. IScience; 2019. Glucose Restriction Promotes Osteocyte Specification by Activating a PGC-1α-dependent Transcriptional Program. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atashi F., Modarressi A., Pepper M.S. The role of reactive oxygen species in mesenchymal stem cell adipogenic and osteogenic differentiation: a review. Stem Cell. Dev. 2015 doi: 10.1089/scd.2014.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callaway D.A., Jiang J.X. Reactive oxygen species and oxidative stress in osteoclastogenesis, skeletal aging and bone diseases. J. Bone Miner. Metabol. 2015 doi: 10.1007/s00774-015-0656-4. [DOI] [PubMed] [Google Scholar]
- 15.Kanzaki H., Shinohara F., Kajiya M., Kodama T. The Keap1/Nrf2 protein axis plays a role in osteoclast differentiation by regulating intracellular reactive oxygen species signaling. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.478545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018 doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellegrini G.G., Cregor M., Mcandrews K., Morales C.C., Mccabe L.D., Mccabe G.P., Peacock M., Burr D., Weaver C., Bellido T. NRF2 regulates mass accrual and the antioxidant endogenous response in bone differently depending on the sex and age. PloS One. 2017 doi: 10.1371/journal.pone.0171161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin Y., Corry K.A., Loughran J.P., Li J. Moderate Nrf2 activation by genetic disruption of Keap1 has sex-specific effects on bone mass in mice. Sci. Rep. 2020 doi: 10.1038/s41598-019-57185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibáñez L., Ferrándiz M.L., Brines R., Guede D., Cuadrado A., Alcaraz M.J. Effects of Nrf2 deficiency on bone microarchitecture in an experimental model of osteoporosis. Oxid. Med. Cell. Longev. 2014 doi: 10.1155/2014/726590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park C.K., Lee Y., Kim K.H., Lee Z.H., Joo M., Kim H.H. Nrf2 is a novel regulator of bone acquisition. Bone. 2014 doi: 10.1016/j.bone.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Rana T., Schultz M.A., Freeman M.L., Biswas S. Loss of Nrf2 accelerates ionizing radiation-induced bone loss by upregulating RANKL. Free Radic. Biol. Med. 2012 doi: 10.1016/j.freeradbiomed.2012.10.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y.X., Li L., Corry K.A., Zhang P., Yang Y., Himes E., Mihuti C.L., Nelson C., Dai G., Li J. Deletion of Nrf2 reduces skeletal mechanical properties and decreases load-driven bone formation. Bone. 2015 doi: 10.1016/j.bone.2014.12.066. [DOI] [PubMed] [Google Scholar]
- 23.Nijweide P.J., van der Plas A., Alblas M.J., Klein-Nulend J. Osteocyte isolation and culture. Methods Mol. Med. 2003;80:41–50. doi: 10.1385/1-59259-366-6:41. [DOI] [PubMed] [Google Scholar]
- 24.Dacquin R., Starbuck M., Schinke T., Karsenty G. Mouse α1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev. Dynam. 2002;224:245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 25.Artigas N., Urena C., Rodriguez-Carballo E., Rosa J.L., Ventura F. Mitogen activated protein kinase (MAPK)-regulated interactions between osterix and Runx2 are critical for the transcriptional osteogenic program. J. Biol. Chem. 2014 doi: 10.1074/jbc.M114.576793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frezza C., Cipolat S., Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured filroblasts. Nat. Protoc. 2007 doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 27.Senft A.P., Dalton T.P., Shertzer H.G. Determining glutathione and glutathione disulfide using the fluorescence probe o-phthalaldehyde. Anal. Biochem. 2000;280:80–86. doi: 10.1006/abio.2000.4498. [DOI] [PubMed] [Google Scholar]
- 28.Gomes L.C., Di Benedetto G., Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell W.F., Barry K.J., Tulum I., Kobayashi T., Harris S.E., Bringhurst F.R., Pajevic P.D. Targeted ablation of the PTH/PTHrP receptor in osteocytes impairs bone structure and homeostatic calcemic responses. J. Endocrinol. 2011 doi: 10.1530/JOE-10-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W., Wang Z., Yang J., Wang Y., Li K., Huang B., Yan B., Wang T., Li M., Zou Z., Yang J., Xiao G., Cui Z.K., Liu A., Bai X. Osteocyte TSC1 promotes sclerostin secretion to restrain osteogenesis in mice. Open Biol. 2019 doi: 10.1098/rsob.180262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenzie J., Smith C., Karuppaiah K., Langberg J., Silva M.J., Ornitz D.M. Osteocyte death and bone overgrowth in mice lacking fibroblast growth factor receptors 1 and 2 in mature osteoblasts and osteocytes. J. Bone Miner. Res. 2019 doi: 10.1002/jbmr.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kedlaya R., Kang K.S., Hong J.M., Bettagere V., Lim K.E., Horan D., Divieti-Pajevic P., Robling A.G. Adult-onset deletion of β-Catenin in10kb dmp1-expressing cells prevents intermittent PTH-induced bone gain. Endocrinology. 2016 doi: 10.1210/en.2015-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linker R.A., Haghikia A. Dimethyl fumarate in multiple sclerosis: latest developments, evidence and place in therapy. Ther. Adv. Chronic Dis. 2016 doi: 10.1177/2040622316653307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acemel R.D., Maeso I., Gómez-Skarmeta J.L. Topologically associated domains: a successful scaffold for the evolution of gene regulation in animals, Wiley Interdiscip. Rev. Dev. Biol. 2017 doi: 10.1002/wdev.265. [DOI] [PubMed] [Google Scholar]
- 35.Sebastian A., Loots G.G. Transcriptional control of Sost in bone. Bone. 2017 doi: 10.1016/j.bone.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Collette N.M., Genetos D.C., Economides A.N., Xie L.Q., Shahnazari M., Yao W., Lane N.E., Harland R.M., Loots G.G. Targeted deletion of Sost distal enhancer increases bone formation and bone mass. Proc. Natl. Acad. Sci. U. S. A. 2012 doi: 10.1073/pnas.1207188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi Y., Kanzaki H., Katsumata Y., Itohiya K., Fukaya S., Miyamoto Y., Narimiya T., Wada S., Nakamura Y. Dimethyl fumarate inhibits osteoclasts via attenuation of reactive oxygen species signalling by augmented antioxidation. J. Cell Mol. Med. 2018 doi: 10.1111/jcmm.13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bollag A.E., Guo T., Ding K.-H., Choudhary V., Chen X., Zhong Q., Xu J., Yu K., Awad M.E., Elsalanty M., Johnson M.H., McGee-Lawrence M.E., Bollag W.B., Isales C.M. Monomethylfumarate protects against ovariectomy-related changes in body composition. J. Endocrinol. 2019 doi: 10.1530/joe-18-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riddle R.C., Clemens T.L. Bone cell bioenergetics and skeletal energy homeostasis. Physiol. Rev. 2017;97:667–698. doi: 10.1152/physrev.00022.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frikha-Benayed D., Basta-Pljakic J., Majeska R.J., Schaffler M.B. Regional differences in oxidative metabolism and mitochondrial activity among cortical bone osteocytes. Bone. 2016;90:15–22. doi: 10.1016/j.bone.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dallas S.L., Bonewald L.F. Dynamics of the transition from osteoblast to osteocyte. Ann. N. Y. Acad. Sci. 2010;1192:437–443. doi: 10.1111/j.1749-6632.2009.05246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higuchi M., Dusting G.J., Peshavariya H., Jiang F., Hsiao S.T.F., Chan E.C., Liu G.S. Differentiation of human adipose-derived stem cells into fat involves reactive oxygen species and forkhead box o1 mediated upregulation of antioxidant enzymes. Stem Cell. Dev. 2013 doi: 10.1089/scd.2012.0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agostini M., Romeo F., Inoue S., Niklison-Chirou M.V., Elia A.J., Dinsdale D., Morone N., Knight R.A., Mak T.W., Melino G. Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 2016;23:1502–1514. doi: 10.1038/cdd.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui S., Tanabe O., Lim K.-C., Xu H.E., Zhou X.E., Lin J.D., Shi L., Schmidt L., Campbell A., Shimizu R., Yamamoto M., Engel J.D. PGC-1 coactivator activity is required for murine erythropoiesis. Mol. Cell Biol. 2014;34:1956–1965. doi: 10.1128/MCB.00247-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wanet A., Caruso M., Domelevo Entfellner J.B., Najar M., Fattaccioli A., Demazy C., Evraerts J., El-Kehdy H., Pourcher G., Sokal E., Arnould T., Tiffin N., Najimi M., Renard P. The transcription factor 7-like 2–peroxisome proliferator-activated receptor gamma coactivator-1 alpha Axis connects mitochondrial biogenesis and metabolic shift with stem cell commitment to hepatic differentiation. Stem Cell. 2017;35:2184–2197. doi: 10.1002/stem.2688. [DOI] [PubMed] [Google Scholar]
- 46.Xing W., Singgih A., Kapoor A., Alarcon C.M., Baylink D.J., Mohan S. Nuclear factor-E2-related factor-1 mediates ascorbic acid induction of osterix expression via interaction with antioxidant-responsive element in bone cells. J. Biol. Chem. 2007;282:22052–22061. doi: 10.1074/jbc.M702614200. [DOI] [PubMed] [Google Scholar]
- 47.Holmström K.M., Baird L., Zhang Y., Hargreaves I., Chalasani A., Land J.M., Stanyer L., Yamamoto M., Dinkova-Kostova A.T., Abramov A.Y. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol. Open. 2013 doi: 10.1242/bio.20134853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J., Williams D., Walter G.A., Thompson W.E., Sidell N. Estrogen increases Nrf2 activity through activation of the PI3K pathway in MCF-7 breast cancer cells. Exp. Cell Res. 2014 doi: 10.1016/j.yexcr.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 49.Khalid A.B., Krum S.A. Estrogen receptors alpha and beta in bone. Bone. 2016 doi: 10.1016/j.bone.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinnesael M., Claessens F., Laurent M., Dubois V., Boonen S., Deboel L., Vanderschueren D. Androgen receptor (AR) in osteocytes is important for the maintenance of male skeletal integrity: evidence from targeted AR disruption in mouse osteocytes. J. Bone Miner. Res. 2012 doi: 10.1002/jbmr.1713. [DOI] [PubMed] [Google Scholar]
- 51.Schultz M.A., Hagan S.S., Datta A., Zhang Y., Freeman M.L., Sikka S.C., Abdel-Mageed A.B., Mondal D. Nrf1 and Nrf2 transcription factors regulate androgen receptor transactivation in prostate cancer cells. PloS One. 2014 doi: 10.1371/journal.pone.0087204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., Yamamoto M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016 doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amarasekara D.S., Yun H., Kim S., Lee N., Kim H., Rho J. Immune Netw; 2018. Regulation of Osteoclast Differentiation by Cytokine Networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narimiya T., Kanzaki H., Yamaguchi Y., Wada S., Katsumata Y., Tanaka K., Tomonari H. Nrf2 activation in osteoblasts suppresses osteoclastogenesis via inhibiting IL-6 expression. BoneKEy Rep. 2019 doi: 10.1016/j.bonr.2019.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouleftour W., Bouet G., Granito R.N., Thomas M., Linossier M.T., Vanden-Bossche A., Aubin J.E., hélène Lafage-Proust M., Vico L., Malaval L. Blocking the expression of both bone sialoprotein (BSP) and osteopontin (OPN) impairs the anabolic action of PTH in mouse calvaria bone. J. Cell. Physiol. 2015 doi: 10.1002/jcp.24772. [DOI] [PubMed] [Google Scholar]
- 56.Bouleftour W., Boudiffa M., Wade-Gueye N.M., Bouët G., Cardelli M., Laroche N., Vanden-Bossche A., Thomas M., Bonnelye E., Aubin J.E., Vico L., Lafage-Proust M.H., Malaval L. Skeletal development of mice lacking Bone Sialoprotein (BSP) - impairment of long bone growth and progressive establishment of high trabecular bone mass. PloS One. 2014 doi: 10.1371/journal.pone.0095144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venkatesh B., Lee A.P., Ravi V., Maurya A.K., Lian M.M., Swann J.B., Ohta Y., Flajnik M.F., Sutoh Y., Kasahara M., Hoon S., Gangu V., Roy S.W., Irimia M., Korzh V., Kondrychyn I., Lim Z.W., Tay B.H., Tohari S., Kong K.W., Ho S., Lorente-Galdos B., Quilez J., Marques-Bonet T., Raney B.J., Ingham P.W., Tay A., Hillier L.W., Minx P., Boehm T., Wilson R.K., Brenner S., Warren W.C. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014 doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staines K.A., MacRae V.E., Farquharson C. The importance of the SIBLING family of proteins on skeletal mineralisation and bone remodelling. J. Endocrinol. 2012 doi: 10.1530/JOE-12-0143. [DOI] [PubMed] [Google Scholar]
- 59.Fisher L.W., Fedarko N.S. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect. Tissue Res. 2003 doi: 10.1080/713713644. [DOI] [PubMed] [Google Scholar]
- 60.McKnight D.A., Fisher L.W. Molecular evolution of dentin phosphoprotein among toothed and toothless animals. BMC Evol. Biol. 2009 doi: 10.1186/1471-2148-9-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouleftour W., Juignet L., Bouet G., Granito R.N., Vanden-Bossche A., Laroche N., Aubin J.E., Lafage-Proust M.H., Vico L., Malaval L. The role of the SIBLING, Bone Sialoprotein in skeletal biology - contribution of mouse experimental genetics. Matrix Biol. 2016 doi: 10.1016/j.matbio.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 62.Jani P.H., Gibson M.P., Liu C., Zhang H., Wang X., Lu Y., Qin C. Transgenic expression of Dspp partially rescued the long bone defects of Dmp1-null mice. Matrix Biol. 2016 doi: 10.1016/j.matbio.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blair H.A. Drugs; 2019. Dimethyl Fumarate: A Review in Relapsing-Remitting MS. [DOI] [PubMed] [Google Scholar]
- 64.Meili-Butz S., Niermann T., Fasler-Kan E., Barbosa V., Butz N., John D., Brink M., Buser P.T., Zaugg C.E. Dimethyl fumarate, a small molecule drug for psoriasis, inhibits Nuclear Factor-κB and reduces myocardial infarct size in rats. Eur. J. Pharmacol. 2008 doi: 10.1016/j.ejphar.2008.02.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.